Abstract
Missense mutations in leucine rich-repeat kinase 2 (LRRK2) cause forms of familial Parkinson’s disease and have been linked to ‘idiopathic’ Parkinson’s disease. Assessment of LRRK2 kinase activity has been very challenging due to its size, complex structure, and relatively low abundance. A standard in the field to assess LRRK2 kinase activity is to measure the level of substrate phosphorylation (pThr73-Rab10) or autophosphorylation of serine 1292 (i.e., phosphoserine 1292; pS1292). The levels of pS1292 have typically been assessed by western blotting, which limits cellular and anatomical resolution. Here, we describe the method for a novel proximity ligation assay (PLA) that can detect endogenous LRRK2 kinase activity (PLA LRRK2) in situ at cellular and subcellular resolutions. PLA is a fluorescence- or chromogen-based assay that can be used to either (1) detect protein-protein interactions or (2) detect and amplify post-translational modifications on proteins. We used PLA for in situ detection and amplification of LRRK2 autophosphorylation levels at serine 1292. Our findings demonstrate that PLA LRRK2 is a highly sensitive and specific assay that can be used for assessing kinase activity in cultured cells and postmortem tissues.
Keywords: Proximity ligation assay, Parkinson’s disease, LRRK2, Fluorescence
Background
Parkinson’s disease (PD) is the second most common neurodegenerative disease (Dorsey and Bloem, 2018), which is caused by both genetic and environmental factors. Approximately 10% of cases are caused by single gene mutations, while 90% are due to generally unknown causes and are termed idiopathic PD (iPD). Missense mutations in leucine rich-repeat kinase 2 (LRRK2) are the most frequent cause of autosomal dominant PD (Paisan- Ruiz et al., 2004 ; Zimprich et al., 2004 ). These mutations result in a toxic gain of function related to elevated LRRK2 kinase activity ( West et al., 2005 ; Greggio et al., 2006 ). The LRRK2 gene locus is also a risk factor for iPD (Simon- Sanchez et al., 2009 ), and increased kinase activity has been implicated in the pathogenesis of iPD ( Fraser et al., 2016 ; Di Maio et al., 2018 ).
Assessment of LRRK2 kinase activity has been challenging until recently. Phosphorylation of the LRRK2 substrate Rab10 has been used as an indicator of elevated kinase activity ( Steger et al., 2016 ). There has been growing consensus that phosphoserine 1292 (pS1292) is an effective surrogate for LRRK2 kinase activity, constituting a reliable readout of kinase activity ( Sheng et al., 2012 ). Several LRRK2 mutations (i.e., G2019S and R1441G) that result in elevated kinase activity also display an increase in pS1292 levels ( Sheng et al., 2012 ). Kinase activity is typically measured by immunoblotting for pS1292, which negates anatomical and cellular resolution. LRRK2 is believed to be a relatively low-abundance protein, and detection of pS1292 with traditional immunocytochemistry is difficult due to problems inherent in detecting a fraction of a sparse protein, such as the need for high antibody concentrations that result in nonspecific signals. Signal amplification, an important feature of the proximity ligation assay (PLA), provides higher sensitivity and allows the use of low concentrations of antibodies, which minimizes nonspecific signals. Additionally, the requirement for molecular proximity of the antibody binding sites to generate a signal effectively filters out off-target or nonspecific antibody binding.
We employed the PLA technique ( Weibrecht et al., 2010 ) to amplify the signal of pS1292 (PLA LRRK2) (Figure 1) which, in turn, allows us to visualize kinase active LRRK2 in a cellular context (Di Maio et al., 2018 ). This approach relies on two antibodies, one that binds to total LRRK2 protein and another antibody from a different species that binds to the pS1292 residue. Next, species-specific oligonucleotide-labeled secondary antibodies (PLA probes) bind to the respective primary antibody (i.e., anti-mouse PLA Probe Minus binds to mouse anti-LRRK2/N241A and anti-rabbit PLA Probe Plus binds to rabbit anti-LRRK2 pS1292). If both primary antibodies are bound to LRRK2 (in close proximity), the PLA probes will be connected and ligated together into circular DNA. This newly ligated circular DNA is used as template for a rolling circle amplification reaction that amplifies the signal several hundred-fold. Then, fluorescently labeled complementary detection oligonucleotides are hybridized to the amplified signal. The resulting fluorescent product can be visualized by confocal microscopy. We validated PLA LRRK2 in CRISPR/Cas9 genome-edited human embryonic kidney 293 (HEK293) cells LRRK2G2019S/G2019S (kinase active) and LRRK2-/- (kinase deficient). We also used PLA to look at LRRK2 kinase activity in rat brain and post-mortem human brain tissues (Di Maio et al., 2018 ). Importantly, independent research groups have used our PLA LRRK2 assay to measure LRRK2 kinase activity ( Fernandez et al., 2019 ; Obergasteiger et al., 2020 ; Bucher et al., 2020 ). Here, we describe in detail our method for how to investigate LRRK2 kinase activity in situ via PLA LRRK2.
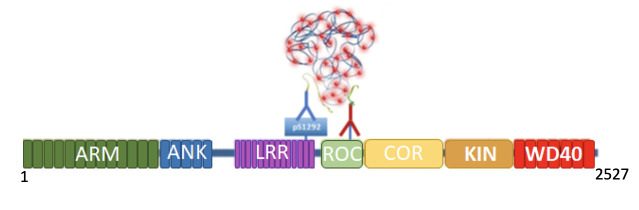
Figure 1. PLA LRRK2 Schematic.
In its kinase active form, S1292 undergoes autophosphorylation (pS1292). Therefore, both pS1292 (blue) and total LRRK2 (red) antibodies will bind to LRRK2. The species-specific PLA probes coupled with oligonucleotides recognize a primary antibody (pS1292 or total LRRK2). As both pS1292 and total LRRK2 antibodies are bound to the LRRK2 protein and are in close proximity, the PLA probes are connected and ligated together. This is amplified by a rolling circle amplification reaction, and fluorescent detection oligonucleotides (red dots) are hybridized to this amplified product. The fluorescent signal is visualized by confocal microscopy. ARM, armadillo; ANK, ankyrin; LRR, leucine-rich repeat; ROC, ras of complex; COR, c-terminal of ROC; KIN, kinase.
Materials and Reagents
Fisherbrand Microscope Cover Glass, 12 mm (Fisher Scientific, catalog number: 12-545-80)
Fisherbrand Microscope Cover Glass 22 × 60-1 (Fisher Scientific, catalog number: 205450J)
24-well cell culture plate tissue culture treated (Costar, catalog number: 3526)
Hydrophobic pen
Fisherbrand Superfrost Plus Microscope Slides (Fisher Scientific, catalog number: 22-037-246)
Chamber slides (Tek Chamber Slide w/ Cover glass slide sterile; Thermo Fisher, catalog number: 178599)
Poly-D-lysine hydrobromide (Sigma-Aldrich, catalog number: P6407)
Normal Donkey Serum (NDS) (Jackson Immuno, catalog number: 017-000-121)
Thermo Scientific Nalgene Laptop Cooler (-20°C) (Fisher Scientific, catalog number: 5115-0012)
Triton X-100 (Fisher Scientific, catalog number: BP151-100)
Rabbit anti-LRRK2 phospho-S1292 [MJFR-19-79] (Abcam, catalog number: Ab203181)
Mouse anti-LRRK2/Dardin Clone N241A/34 (UC Davis [Antibodies Inc.], catalog number: 75-253)
Anti-Mouse Minus (Sigma-Aldrich, catalog number: DUO92004-100RXN)
Anti-Rabbit Plus (Sigma-Aldrich, catalog number: DUO92002-100RXN)
Detection Reagents Orange (Sigma-Aldrich, catalog number: DUO92007-100RXN)
Duolink Wash Buffer A and Wash Buffer B (Sigma-Aldrich, catalog number: DUO82049-4L)
16% Paraformaldehyde (10 ml) (Fisher Scientific, catalog number: 50-980-487)
Polyvinyl alcohol (PVA) (Sigma-Aldrich, catalog number: P8136)
Glycerol (Sigma-Aldrich, catalog number: G9012)
Sodium Azide, crystalline (Fisher Scientific, catalog number: S277-100)
Sudan Black B (Sigma-Aldrich, catalog number: 199664)
Histoclear (manufactured by National Diagnostics) (Fisher Scientific, catalog number: HS200)
Sheep anti-tyrosine hydroxylase (Millipore, catalog number: Ab1542)
NDS (see Recipes)
1× PBS (see Recipes)
Wash Buffer A and Washer Buffer B (see Recipes)
Blocking Solution for cells (see Recipes)
Blocking Solution for rat brain tissue (see Recipes)
Blocking Solution for human brain tissue (see Recipes)
Gelvatol Mounting Media (see Recipes)
Equipment
Pipettes (ranging from 1 µl to 1,000 µl)
37°C, 5% CO2 cell culture incubator
Orbital shaker at room temperature (for fixation, permeabilization, and washes)
Orbital shaker at 4°C (for primary antibody incubation overnight)
-
Incu-shakerTM Mini with nonslip rubber mat (Benchmark Scientific, catalog number: H1001-M)
Note: You can use any other humified incubator at 37°C.
Olympus BX61 confocal microscope (or any confocal microscope)
Software
-
Olympus Fluoview 1000 Software was used in this analysis; however, any other software that is suitable for image analysis can also be used.
Nikon elements, ImageJ
GraphPad PRISM
Procedure
Part I: PLA LRRK2 in fixed cells
-
Cell culture and seeding
-
Seed 100,000 HEK293 cells in 1 ml of DMEM media supplemented with 10% FBS onto a 12 mm coverslip coated with poly-D-lysine in a 24-well plate. Place the plate in 37°C + 5% CO2 cell culture incubator overnight.
Note: At the time of fixation, cells should be around 70-80% confluent for best results. The assay can also be done in chamber slides (Tek Chamber Slide w/ Cover glass slide sterile, Thermo Fisher, catalog number: 178599). The volumes need to be adjusted accordingly as they are smaller than the coverslips (e.g., 100 µl of solution per well and 25 µl of PLA solution per reaction).
-
-
Cell Fixation, Permeabilization, and Primary Antibody Incubation
-
After 24 h, remove the media and fix cells in 4% paraformaldehyde for 20 min at room temperature with gentle agitation.
Note: Use 325 µl of solution per well.
Wash the fixed cells 3 × 10 min in 1× PBS (approximately 1 ml/well).
-
Permeabilize the cells with blocking solution for cells (0.02% Triton X-100 + 10% NDS made in 1× PBS) incubate for 1 h at room temperature with gentle agitation.
Note: Use 325 µl of solution per well.
Wash the fixed and permeabilized cells 3 × 10 min in 1× PBS (approximately 1 ml/well).
-
Dilute primary antibodies in 10% NDS in 1× PBS and incubate overnight at 4°C with gentle agitation.
Note: Use 325 µl of solution per well.
1:1,000 Rabbit anti-LRRK2 phospho-S1292.
1:1,000 Mouse anti-LRRK2/Dardin Clone N241A/34.
-
-
PLA
Note:Allow PLA Wash Buffer A and B to reach room temperature prior to use as cold wash buffers lead to diffuse nonspecific background signal. Each coverslip represents one reaction, and 45 µl of solution is used per reaction. Enough solution is needed to completely cover the coverslip as the sample must not dry out. Adjust the volume if needed. The calculations below are only forone reaction.
Wash cells 3 × 10 min in 1× PBS with gentle agitation (approximately 1 ml/well).
-
Step 1: Probes
Note: Make probe solution and let sit at room temperature for 20 min prior to adding to cells.
Mix 9 µl PLA MINUS Probe (1:5 dilution), 9 µl PLA PLUS Probe (1:5 dilution), and 27 µl antibody diluent and add 45 µl of solution directly on top of the coverslip.
Add 1 drop of PLA Blocking Solution to the coverslip.
Incubate for 1 h in a 37°C humidified incubator.
Wash 2 × 5 min in Wash Buffer A (approximately 1 ml/well) with gentle agitation.
-
Step 2: Ligation
Note: Thaw 5×Ligation Buffer on ice; leave 40×Ligase in freezer block at -20°C and only remove when adding it to the 1×Ligation Solution.
Vortex 5× Ligation Buffer thoroughly.
Make 1× Ligation Solution by mixing 9 µl 5× Ligation Buffer (1:5 dilution) and 34.9 μl Milli-Q water.
After the second wash in Wash Buffer A, add 1.1 µl Ligase (1:40 dilution) to 1× Ligation Solution. Add 45 µl of ligation solution directly on top of the coverslip.
Incubate for 45 min in a 37°C humidified incubator.
Wash 2 × 2 min in Wash Buffer A (approximately 1 ml/well) with gentle agitation.
-
Step 3: Amplification
Note: Thaw 5×Amplification Buffer on ice; leave 80×Polymerase in freezer block at -20°C and only remove when adding it to the 1×Amplification Solution. The plate must be protected from light from this point forward.
Vortex 5× Amplification Buffer thoroughly.
Make 1× Amplification Solution by mixing 9 µl of 5× Amplification Orange Buffer (1:5 dilution) and 35.4 µl Milli-Q water.
After the second wash in Wash Buffer A, add 0.6 µl Polymerase (1:80 dilution) to 1× Amplification Solution. Add 45 µl of amplification solution directly on top of the coverslip.
Incubate for 100 min in a 37°C humidified incubator.
Wash 2 × 10 min in Wash Buffer B (approximately 1 ml/well) with gentle agitation.
-
Preparation for Imaging
Incubate cells in DAPI (1:5,000 dilution in 1× PBS) for 1 min at room temperature with gentle agitation.
Wash 3 × 10 min in 1× PBS.
-
Mount coverslips on Superfrost Plus Microscope Slides using 15 µl of gelvatol mounting medium per coverslip.
Note: See “Recipes” for how to make Gelvatol Mounting Medium.
Once coverslips are mounted, place in slide book, refrigerate, and let gelvatol mounting medium dry for 24-48 h.
-
Image within a few days after completing the assay.
Note: PLA fluorescent signal fades with time.
-
Imaging and Analysis (Figures 2A-2E)
-
Acquire images on 60× or 100× magnification using an Olympus BX61 confocal microscope or any confocal and/or fluorescence microscope available.
To ensure the signal detected is above background fluorescence, set up imaging parameters (laser power, exposure, and pinhole) on negative control (i.e., LRRK2 knockout cell line, cells treated with LRRK2 kinase inhibitor, LRRK2 knockdown cells, or primary delete). For quantitative comparisons, keep all imaging parameters constant across all groups.
Take multiple images per coverslip.
-
Analyze images by taking fluorescence measurements in regions of interest of each individual cell in a single field.
Note: Depending on the signal pattern, either fluorescence intensity, number of dots per cell, or nucleus may be appropriate for quantitation.
For each independent biological experiment, there are two technical replicates per experimental group. This helps control for assay reliability and variability. Use at least three to four independent biological replicates.
-
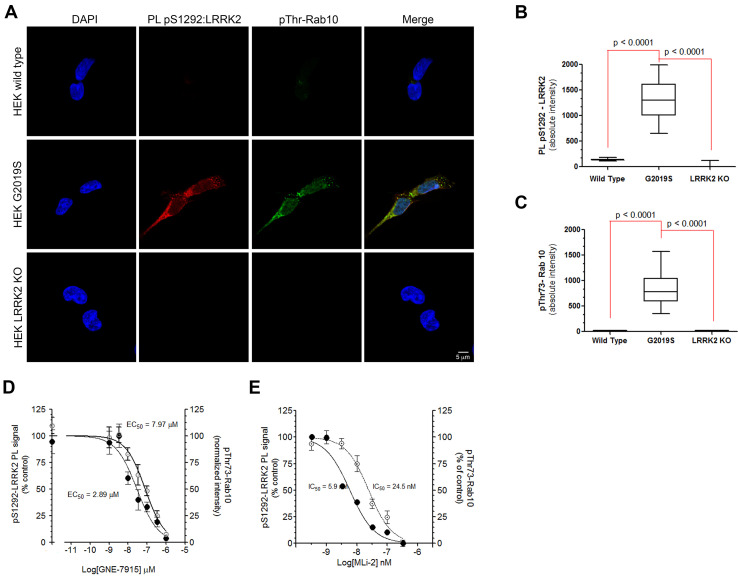
Figure 2. PLA LRRK2 assay validation in CRISPR/Cas9 genome-edited HEK293 cells. (A).
PLA LRRK2 assay showing kinase activation by measuring phosphorylation of the autophosphorylation site, S1292 (red), and LRRK2 substrate phosphorylation Rab10 (green). In wild-type HEK293 cells (top row), there was very little pS1292:LRRK2 and pTh73-Rab10 signal indicating low kinase activities at baseline. In HEKG2019S/G2019S cells (middle row), there was a strong signal for both PL pS1292:LRRK2 and pThr73-Rab10. In LRRK2 knockout cells (bottom row), there was very little signal for both PL pS1292:LRRK2 and pThr73-Rab10. Scale bar 5 µM. (B). Quantification of PL pS1292:LRRK2 signal in wild-type, G2019S, and LRRK2 knockout cells. Results reflect three individual experiments. Statistical testing by ANOVA with post hoc Bonferroni correction. (C). Quantification of pTh73-Rab10 signal in wild-type, G2019S, and LRRK2 knockout cells. Results reflect three individual experiments. Statistical testing by ANOVA with post hoc Bonferroni correction. (D). Dose response curves for LRRK2 kinase inhibitor GNE-7915 assessed by PL pS1292:LRRK2 (filled circles) and pThr73-Rab10 (empty circles). HEKG2019S/G2019S cells were cultured in the presence of various concentrations of GNE-7915 for 24 h. Results are from three experiments. Symbols represent means ± SEM. IC50 values calculated with the GraphPad PRISM software. (E). Dose response curves for LRRK2 kinase inhibitor MLi-2 assessed by PL pS1292:LRRK2 (filled circles) and pThr73-Rab10 (empty circles) signal. HEKG2019S/G2019S cells were cultured in the presence of various concentrations of MLi-2 for 24 h. Results are from three experiments. Symbols represent means ± SEM. C50 values calculated with the GraphPad PRISM software.
Part II: LRRK2 PLA in rat brain tissue sections
Note: Steps A1-A7 are done in free floating, PFA-fixed brain sections (35 µm thick). It is important to avoid over-fixation. Each adult male Lewis rat is perfused with 250 ml of PBS followed by 150 ml of 4% PFA. Brains are extracted and submerged in 4% PFA for an additional 24 h at 4°C and then stored in 30% sucrose. Free-floating sections of 35 µm thickness are collected using a microtome and stored at -20°C in cryoprotectant until use. Sections are incubated with 1 ml of solution per well in a 12-well plate; 2-3 brain sections are used per animal.
-
Primary antibody incubation and secondary antibody incubation for neuronal marker
Wash sections 6 × 10 min in 1× PBS.
Permeabilize brain sections in blocking solution for rat brain tissue (10% NDS + 1% Triton X-100 in 1× PBS) for 1 h at room temperature with gentle agitation.
Wash brain sections 3 × 10 min in 1× PBS at room temperature with gentle agitation.
-
Incubate brain sections in neuronal marker primary antibody solution made up in 10% NDS in 1× PBS for 48 h at 4°C with gentle agitation.
Note: If assessing dopaminergic neurons, use Sheep anti-tyrosine hydroxylase at 1:2,000.
After neuronal marker primary antibody, wash sections in 1× PBS 3 × 10 min at room temperature with gentle agitation.
-
Incubate in secondary antibody at 1:500 for 1 h at room temperature with gentle agitation.
Note: From this point on, sections must be protected from light.
Wash brain sections in 3 × 10 min in 1× PBS.
-
PLA primary antibody incubation
-
Incubate brain sections in PLA primary antibody solution made up in 10% NDS in 1× PBS for 24 h at 4°C overnight with gentle agitation.
1:500 Rabbit anti-LRRK2 phospho-S1292.
1:500 Mouse anti-LRRK2/Dardin Clone N241A/34.
After PLA primary antibody incubation, wash brain sections 3× 10 min in 1× PBS.
Following washes, mount each brain section on Superfrost Plus Microscope Slides and let dry for 15-20 min; then, circle area around the sections with a hydrophobic pen (Video 1).
-
-
PLA
Note:Allow PLA Wash Buffer A and B to reach room temperature prior to use as cold wash buffers lead to diffuse nonspecific background signal.Each brain section circled with a hydrophobic pen is one reaction, and 40 µl of solution is used per reaction as this volume is sufficient to completely cover each section.Do not allow the section to dry out.If this volume is not enough to cover the section, increase the volume appropriately. The calculations below are only forone reaction.
-
Step 1: Probes
Note: Make probe solution and let sit at room temperature for 20 min prior to adding to brain section.
Mix 8 µl PLA MINUS Probe (1:5 dilution), 8 µl PLA PLUS Probe (1:5 dilution), and 24 µl antibody diluent and add 40 µl of probe solution directly on top of each brain section.
Add 1 drop of PLA Blocking Solution to brain section.
Incubate for 1 h in a 37°C humidified incubator.
Wash 2 × 5 min in Wash Buffer A (approximately 50 µl/section).
-
Step 2: Ligation
Note: Thaw 5× Ligation Buffer on ice; leave 40× Ligase in freezer block at -20°C and only remove it when adding it to the 1× Ligation Solution.
Vortex 5× Ligation Buffer thoroughly.
Make 1× Ligation Solution by mixing 8 µl of 5× Ligation Buffer (1:5 dilution) and 31 µl Milli-Q water.
After the second wash in Wash Buffer A, add 1 µl Ligase (1:40 dilution) to 1× Ligation Solution. Add 40 µl of ligation solution directly on top of each brain section (Video 1).
Incubate for 45 min in a 37°C humidified incubator.
Wash 2 × 2 min in Wash Buffer A (at least 50 µl/section).
-
Step 3: Amplification
Note: Thaw 5× Amplification Buffer on ice; leave 80× Polymerase in freezer block at -20°C and only remove when adding it to the 1× Amplification Solution.
Vortex 5× Amplification Buffer thoroughly.
Make 1× Amplification Solution by mixing 8 µl of 5× Amplification Orange Buffer (1:5 dilution) and 31.5 µl Milli-Q water.
After the second wash in Wash Buffer A, add 0.5 µl Polymerase (1:80 dilution) to 1× Amplification Solution. Add 40 µl of amplification solution directly on top of each brain section.
Incubate for 100 min in a 37°C humidified incubator.
Wash 2 × 10 min in Wash Buffer B (at least 50 µl/section).
Wash 1 × 10 min in 1× PBS (at least 50 µl/section).
-
-
Preparation for Imaging
-
Add 15 µl of gelvatol mounting media to each brain section and cover with Fisherbrand Microscope Cover Glass, 12 mm or any coverslip appropriate for sample.
Note: Use the amount of gelvatol mounting media that is needed for your sample size.
Once coverslips are mounted, place in slide book, refrigerate, and let gelvatol mounting media dry for 24-48 h.
-
Image within a few days after completing the assay.
Note: PL fluorescent signal fades with time.
-
-
Imaging and Analysis (Figures 3A-D)
-
Take images on 60× or 100× magnification using an Olympus BX61 confocal microscope or any confocal and/or fluorescence microscope available.
To ensure the signal detected is above background fluorescence, set up imaging parameters (laser power, exposure, and pinhole) on negative control (i.e., LRRK2 KO animals, animals treated with a LRRK2 kinase inhibitor, or primary delete). For quantitative comparisons, keep all imaging parameters constant across all groups.
Take multiple images per brain section.
Analyze images by taking fluorescence measurements in regions of interest of each individual cell in a single field (Depending on signal pattern, either fluorescence intensity or number of dots per cell or nucleus may be appropriate for quantitation).
-
Video 1. How to perform key steps of PLA LRRK2 in rat tissue sections.
This brief video provides a visual guide on how to handle rat tissue sections for PLA LRRK2. Part 1 shows how to draw the hydrophobic barrier surrounding the tissue section using a hydrophobic “PAP” pen. Part 2 demonstrates how to remove solutions from a tissue section without damaging it. Part 3 shows that the PLA solution should be added directly on top of the section to avoid drying out.
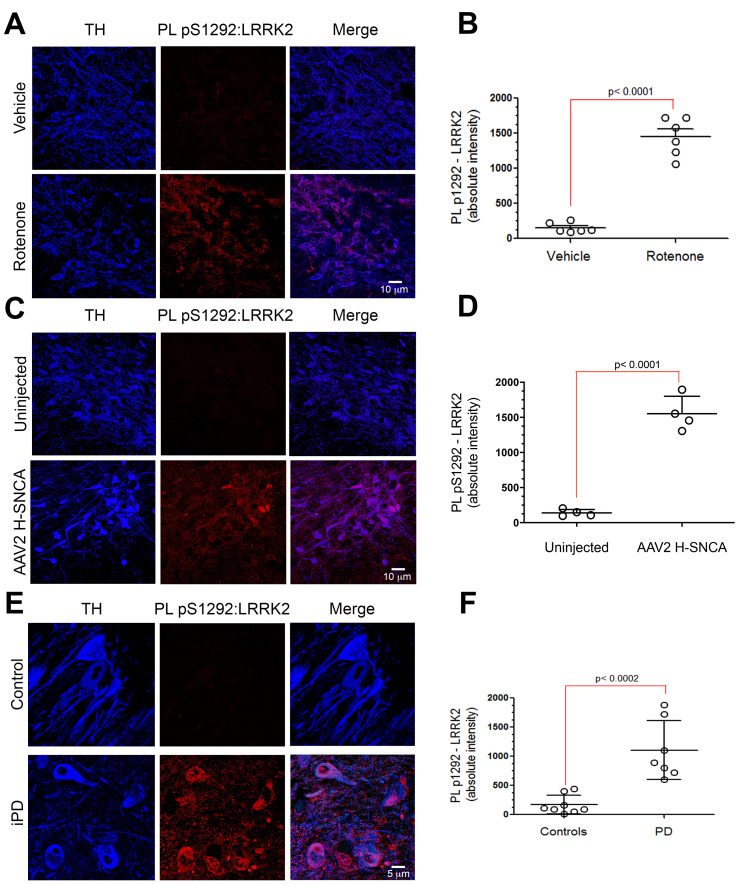
Figure 3. PLA LRRK2 assay in nigrostriatal neurons in two rat models of PD and human iPD brain tissue. (A).
PL pS1292:LRRK2 (red) signal in the substantia nigra from vehicle and rotenone treated rats. In rats treated with rotenone (2.8 mg/kg, i.p.), there was an increase in signal (bottom) compared to vehicle (top). Tyrosine hydroxylase (TH; blue) is a marker for dopaminergic neurons. Scale bar: 10 µm. (B). Quantification of pS1292:LRRK2 PL signal in nigrostriatal dopaminergic neurons from vehicle and rotenone treated rats. Symbols represent individual animals. Statistical analysis by unpaired two tailed t-test. (C). PL pS1292:LRRK2 signal (red) in the substantia nigra from rats that received unilateral injection of AAV2-hSNCA. In the hemisphere overexpressing α-synuclein, there was an increase in signal (bottom) compared to the control injected hemisphere (top). TH (blue) is a marker for dopaminergic neurons. Scale bar: 10 µm. (D). Quantification of pS1292:LRRK2 PL signal in nigrostriatal dopaminergic neurons from control and AAV2-hSNCA injected hemispheres. Symbols represent mean values from each hemisphere. Statistical analysis by paired two tailed t-test. (E). The PL pS1292:LRRK2 signal (red) is shown in sections of the substantia nigra from a healthy aged-matched control human brain (top) and a brain from an individual with idiopathic Parkinson’s disease (iPD) (bottom). In control brains, there was very little PL pS1292:LRRK2 signal in the surviving dopaminergic neurons compared to the iPD brains. TH (blue) is a marker for dopaminergic neurons. Scale bar: 5 µm. (F). Quantification for PL pS1292:LRRK2 signal in eight control brains and seven iPD brains. Statistical analysis by unpaired two-tailed t-test.
Part III: PLA LRRK2 in human brain tissue sections
Note: We receive tissue from the Brain Bank at the University of Pittsburgh. These sections are paraffin embedded on microscope slides.
-
Deparaffinization and Sudan Black
Place slides containing human tissue sections into a plastic slide holder or a coplin jar.
-
Deparaffinize the human brain tissue by following these steps.
-
Incubate tissue at 60°C for 30 min.
Note: If using a glass coplin jar, you must gradually heat jar up prior to using so the glass jar does not shatter.
Incubate tissue in Histoclear 3 × 4 min.
Incubate tissue in 100% ethanol 2 × 4 min.
Incubate tissue in 95% ethanol 2 × 4 min.
Incubate tissue in 70% ethanol 2 × 4 min.
Incubate tissue in Milli-Q water for 5 min.
-
To quench autofluorescence in human tissue sections, incubate tissue in sudan black for 5 min at room temperature.
Wash the slides in Milli-Q water for 3 min.
Circle area around the tissue section on the slide with a hydrophobic pen.
-
Primary antibody incubation and secondary antibody incubation for neuronal marker
Permeabilize and block brain tissue sections blocking solution for human brain tissue (10% NDS + 0.3% Triton X-100 in 1× PBS) for 1 h.
Wash brain sections 3 × 10 min in 1× PBS at room temperature.
-
Incubate brain sections in neuronal marker primary antibody solution made up in 10% NDS in 1× PBS for 48 h at 4°C.
Note: If assessing dopaminergic neurons, use Sheep anti-tyrosine hydroxylase at 1:2,000.
After neuronal marker primary antibody, wash sections in 1× PBS for 3 × 10 min at room temperature with gentle agitation.
-
Incubate in secondary antibody at 1:500 for 1 h at room temperature with gentle agitation.
Note: From this point on, sections must be protected from light.
Wash brain sections 3 × 10 min in 1× PBS.
-
PLA primary antibody incubation
-
Incubate brain sections in PLA primary antibody solution made up in 10% NDS in 1× PBS for 48 h at 4°C.
1:500 Rabbit anti-LRRK2 phospho-S1292.
1:500 Mouse anti-LRRK2/Dardin Clone N241A/34.
-
-
PLA
Note:Allow PLA Wash Buffer A and B to reach room temperature prior to use as cold wash buffers lead to diffuse nonspecific background signal. Each brain section circled with a hydrophobic pen is one reaction, and 60 µl of solution is used per reaction. As this volume is sufficient to completely cover each section. If this volume is not enough to cover the section, increase the volume appropriately. The sample must not dry out. The calculations below are only forone reaction.
After PLA primary antibody incubation, wash brain sections 3 × 10 min in 1× PBS.
-
Step 1: Probes
Note: Make probe solution and let sit at room temperature for 20 min prior to adding to brain sections.
Mix 12 µl PLA MINUS Probe (1:5 dilution), 12 µl PLA PLUS Probe (1:5 dilution), and 36 µl antibody diluent and add 60 µl of probe solution directly on top of each brain section.
Add 1 drop of PLA Blocking Solution to brain section.
Incubate for 1 h in a 37°C humidified incubator.
Wash 2 × 5 min in Wash Buffer A (approximately 100 µl/section).
-
Step 2: Ligation
Note: Thaw 5× Ligation Buffer on ice; leave 40× Ligase in freezer block at -20°C and only remove when adding it to the 1× Ligation Solution.
Vortex 5× Ligation Buffer thoroughly.
Make 1× Ligation Solution by mixing 12 µl of 5× Ligation Buffer (1:5 dilution) and 46.5 µl Milli-Q water.
After the second wash in Wash Buffer A, add 1.5 µl Ligase (1:40 dilution) to 1× Ligation Solution. Add 60 µl of ligation solution directly on top of each brain section.
Incubate for 45 min in a 37°C humidified incubator.
Wash 2 × 2 min in Wash Buffer A (approximately 100 µl/section).
-
Step 3: Amplification
Note: Thaw 5× Amplification Buffer on ice; leave 80× Polymerase in freezer block at -20°C and only remove when adding it to the 1× Amplification Solution.
Vortex 5× Amplification Buffer thoroughly.
Make 1× Amplification Solution by mixing 12 µl of 5× Amplification Orange Buffer (1:5 dilution) and 47.3 µl Milli-Q water.
After the second wash in Wash Buffer A, add 0.8 µl Polymerase (1:80 dilution) to 1× Amplification Solution. Add 60 µl of amplification solution directly on top of each brain section.
Incubate for 100 min in a 37°C humidified incubator.
Wash 2 ×10 min in Wash Buffer B (approximately 100 µl/section)
Wash 1 × 10 min in 1× PBS (approximately 100 µl/section).
-
Preparation for Imaging
-
Add 30 µl of gelvatol mounting media to middle of each brain section and around the edges of the slide and cover with Fisherbrand Microscope Cover Glass 22 × 60 or any coverslip appropriate for size sample.
Note: Use the amount of gelvatol mounting media that is needed for your sample size.
Once coverslips are mounted, place in slide book, refrigerate, and let gelvatol mounting media dry for 24-48 h.
-
Image within a few days after completing the assay.
Note: PL fluorescent signal fades with time.
-
-
Imaging and Analysis (Figures 3E-3F)
-
Take images on 60× or 100× magnification using an Olympus BX61 confocal microscope or any confocal microscope available.
To ensure the signal detected is above background fluorescence, set up imaging parameters (laser power, exposure, and pinhole) on negative control (i.e., primary delete). For quantitative comparisons, keep all imaging parameters constant across all groups.
Take multiple images per brain section.
Analyze images by taking fluorescence measurements in regions of interest of each individual cell in a single field (Depending on signal pattern, either fluorescence intensity, number of dots per cell, or nucleus may be appropriate for quantitation).
-
Data analysis
Statistical analysis
The statistical analysis description has been adapted from our publication (Di Maio et al., 2018 ). Each result presented here was derived from at least three independent experiments for in vitro studies. For in vivo rotenone rat studies, six independent biological replicates were analyzed per group. For in vivo AAV-hSNCA rat studies, independent biological replicates were analyzed per group. Seven iPD brain samples and eight aged-matched control brain samples were analyzed in human tissue stains. For simple comparisons of two experimental conditions, two-tailed, unpaired t tests were used. Where variances were not equal, Welch’s correction was used. When AAV vector was injected into one hemisphere of the rat brain and the other hemisphere was used as a control, two-tailed paired t tests were used. For comparisons of multiple experimental conditions, one-way or two-way ANOVA was used, and if significant, overall post hoc corrections (with Bonferroni test) for multiple pairwise comparisons were made. P values less than 0.05 were considered significant.
Notes and Conclusions
As demonstrated, PLA LRRK2 is a highly specific and sensitive assay that can be used to quantitate LRRK2 kinase activity. Here, we describe in detail how to perform the assay in multiple different types of specimens. We used the PLA approach to amplify a post-translational modification on LRRK2, the autophosphorylation site pS1292. We hope that the PLA LRRK2 becomes a widely used method to assess kinase activity in various specimens to help uncover the role LRRK2 plays in health and disease. Furthermore, the idea of utilizing PLA to study post-translational modifications on proteins may be extremely beneficial for defining the roles of other proteins.
Recipes
NDS: make up following manufacturers’ instructions
1× PBS
Wash Buffer A and Washer Buffer B: make up following manufacturers’ instructions
-
Blocking Solution for cells
10% NDS + 0.02% Triton X-100 in 1× PBS
-
Blocking Solution for rat brain tissue
10% NDS + 1% Trition X-100 in 1× PBS
-
Blocking Solution for human brain tissue
10% NDS + 0.3% Triton X-100 in 1× PBS
-
Gelvatol mounting media (recipe courtesy of the Center for Biologic Imaging at The University of Pittsburgh)
Add 21 g PVA to 42 ml glycerol
Add 52 ml Milli-Q water
Add a few crystals of sodium azide
Add 106 ml Tris (0.2 M, pH = 8.5)
Stir with low heat for a few hours or until reagents dissolve
Clarify the mixture by centrifugation at 5,000 × g for 15 min
Aliquot and store at 4°C
Note: This protocol is an approximation. You will have to add PVA in Step D until the solution is clear and is slightly less viscous than molasses. Refrigerate the beaker of gelvatol mounting media overnight at 4°C, after Step E and before Step F, and check it the next day to be sure that the viscosity is now that of molasses. If it is, continue on to Step F. If it is too viscous, add a little more glycerol to bring the viscosity down and then go on to Step F. If it is not viscous enough, add more PVA with heat and refrigerate for a few more hours to check the viscosity before going on to Step F.
Acknowledgments
This work was supported by research grants from the NIH (NS100744, R21ES027470, and NS095387) (J.T.G.). Support was also provided by the American Parkinson Disease Association Center for Advanced Research at the University of Pittsburgh and The Michael J. Fox Foundation PATH to PD. We would like to thank Christopher R. Bodle, PhD for filming the video. The protocol and figures are adapted from our publication LRRK2 Activation in idiopathic Parkinson’s disease (Di Maio et al., 2018 ).
Competing interests
The authors declare they have no competing interests.
Ethics
All animal procedures were performed in accordance with National Institute of Health guidelines and were approved by the Institutional Animal Care and Use Committee (IACUC) at University of Pittsburgh.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1.Bucher M. L., Barrett C. W., Moon C. J., Mortimer A. D., Burton E. A., Greenamyre J. T. and Hastings T. G.(2020). Acquired dysregulation of dopamine homeostasis reproduces features of Parkinson’s disease. NPJ Parkinson's Disease 6(1): 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Di Maio R., Hoffman E. K., Rocha E. M., Keeney M. T., Sanders L. H., De Miranda B. R., Zharikov A., Van Laar A., Stepan A. F., Lanz T. A., Kofler J. K., Burton E. A., Alessi D. R., Hastings T. G. and Greenamyre J. T.(2018). LRRK2 activation in idiopathic Parkinson's disease. Sci Transl Med 10(451). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dorsey E. R. and Bloem B. R.(2018). The Parkinson Pandemic-A Call to Action. JAMA Neurol 75(1): 9-10. [DOI] [PubMed] [Google Scholar]
- 4.Fernandez B., Lara Ordonez A. J., Fdez E., Mutez E., Comptdaer T., Leghay C., Kreisler A., Simonin C., Vandewynckel L., Defebvre L., Destee A., Bleuse S., Taymans J. M., Chartier-Harlin M. C. and Hilfiker S.(2019). Centrosomal cohesion deficits as cellular biomarker in lymphoblastoid cell lines from LRRK2 Parkinson's disease patients. Biochem J 476(19): 2797-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fraser K. B., Rawlins A. B., Clark R. G., Alcalay R. N., Standaert D. G., Liu N., C. Parkinson's Disease Biomarker Program and West A. B.(2016). Ser(P)-1292 LRRK2 in urinary exosomes is elevated in idiopathic Parkinson's disease. Mov Disord 31(10): 1543-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greggio E., Jain S., Kingsbury A., Bandopadhyay R., Lewis P., Kaganovich A., van der Brug M. P., Beilina A., Blackinton J., Thomas K. J., Ahmad R., Miller D. W., Kesavapany S., Singleton A., Lees A., Harvey R. J., Harvey K. and Cookson M. R.(2006). Kinase activity is required for the toxic effects of mutant LRRK2/dardarin. Neurobiol Dis 23(2): 329-341. [DOI] [PubMed] [Google Scholar]
- 7.Obergasteiger J., Castonguay A.-M., Frapporti G., Lobbestael E., Baekelandt V., Hicks A. A., Pramstaller P. P., Gravel C., Corti C., Lévesque M. and Volta M.(2020). RIT2 reduces LRRK2 kinase activity and protects against alpha-synuclein neuropathology. bioRxiv: 2020.2010.2021.348144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paisan-Ruiz C., Jain S., Evans E. W., Gilks W. P., Simon J., van der Brug M., Lopez de Munain A., Aparicio S., Gil A. M., Khan N., Johnson J., Martinez J. R., Nicholl D., Carrera I. M., Pena A. S., de Silva R., Lees A., Marti-Masso J. F., Perez-Tur J., Wood N. W. and Singleton A. B.(2004). Cloning of the gene containing mutations that cause PARK8-linked Parkinson's disease. Neuron 44(4): 595-600. [DOI] [PubMed] [Google Scholar]
- 9.Sheng Z., Zhang S., Bustos D., Kleinheinz T., Le Pichon C. E., Dominguez S. L., Solanoy H. O., Drummond J., Zhang X., Ding X., Cai F., Song Q., Li X., Yue Z., van der Brug M. P., Burdick D. J., Gunzner-Toste J., Chen H., Liu X., Estrada A. A., Sweeney Z. K., Scearce-Levie K., Moffat J. G., Kirkpatrick D. S. and Zhu H.(2012). Ser1292 autophosphorylation is an indicator of LRRK2 kinase activity and contributes to the cellular effects of PD mutations. Sci Transl Med 4(164): 164ra161. [DOI] [PubMed] [Google Scholar]
- 10.Simon-Sanchez J., Schulte C., Bras J. M., Sharma M., Gibbs J. R., Berg D., Paisan-Ruiz C., Lichtner P., Scholz S. W., Hernandez D. G., Kruger R., Federoff M., Klein C., Goate A., Perlmutter J., Bonin M., Nalls M. A., Illig T., Gieger C., Houlden H., Steffens M., Okun M. S., Racette B. A., Cookson M. R., Foote K. D., Fernandez H. H., Traynor B. J., Schreiber S., Arepalli S., Zonozi R., Gwinn K., van der Brug M., Lopez G., Chanock S. J., Schatzkin A., Park Y., Hollenbeck A., Gao J., Huang X., Wood N. W., Lorenz D., Deuschl G., Chen H., Riess O., Hardy J. A., Singleton A. B. and Gasser T.(2009). Genome-wide association study reveals genetic risk underlying Parkinson's disease. Nat Genet 41(12): 1308-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steger M., Tonelli F., Ito G., Davies P., Trost M., Vetter M., Wachter S., Lorentzen E., Duddy G., Wilson S., Baptista M. A., Fiske B. K., Fell M. J., Morrow J. A., Reith A. D., Alessi D. R. and Mann M.(2016). Phosphoproteomics reveals that Parkinson's disease kinase LRRK2 regulates a subset of Rab GTPases. Elife 5: e12813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weibrecht I., Leuchowius K. J., Clausson C. M., Conze T., Jarvius M., Howell W. M., Kamali-Moghaddam M. and Soderberg O.(2010). Proximity ligation assays: a recent addition to the proteomics toolbox. Expert Rev Proteomics 7(3): 401-409. [DOI] [PubMed] [Google Scholar]
- 13.West A. B., Moore D. J., Biskup S., Bugayenko A., Smith W. W., Ross C. A., Dawson V. L. and Dawson T. M.(2005). Parkinson's disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proc Natl Acad Sci U S A 102(46): 16842-16847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zimprich A., Biskup S., Leitner P., Lichtner P., Farrer M., Lincoln S., Kachergus J., Hulihan M., Uitti R. J., Calne D. B., Stoessl A. J., Pfeiffer R. F., Patenge N., Carbajal I. C., Vieregge P., Asmus F., Muller-Myhsok B., Dickson D. W., Meitinger T., Strom T. M., Wszolek Z. K. and Gasser T.(2004). Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron 44(4): 601-607. [DOI] [PubMed] [Google Scholar]