Abstract
A portion of those infected with SARS-CoV-2 shed the virus and its genetic material in respiratory fluids, saliva, urine, and stool, thus giving the potential to monitor for infections via wastewater. Wastewater surveillance efforts to date have largely assumed that stool shedding has been the primary source of SARS-CoV-2 RNA signal; however, there are increasing questions about the possible contribution of other shedding routes, with implications for wastewater surveillance design and feasibility. In this study we used clinical SARS-CoV-2 RNA shedding data and a Monte Carlo framework to assess the relative contribution of various shedding routes on SARS-CoV-2 RNA loads in wastewater. Stool shedding dominated total SARS-CoV-2 RNA load for community-level surveillance, with mean contributions more than two orders of magnitude greater than other shedding routes. However, RNA loads were more nuanced when considering building-level monitoring efforts designed to identify a single infected individual, where any shedding route could plausibly contribute a detectable signal. The greatest source of model variability was viral load in excreta, suggesting that future modeling efforts may be improved by incorporating specific modeling scenarios with precise SARS-CoV-2 shedding data, and beyond that wastewater surveillance must continue to account for large variability during data analysis and reporting. Importantly, the findings imply that wastewater surveillance at finer spatial scales is not entirely dependent on shedding via feces for sensitive detection of infections thus enlarging the potential use cases of wastewater as a non-intrusive surveillance methodology.
Keywords: SARS-CoV-2 RNA, Shedding route, Wastewater, Wastewater surveillance, Wastewater-based epidemiology, COVID-19
Graphical abstract
1. Introduction
Individuals infected with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) shed the virus and its genetic material in their upper and lower respiratory fluids (Cevik et al., 2021), saliva (Wyllie et al., 2020), urine (Kashi et al., 2020), and stool (Cevik et al., 2021). Since these excreta are frequently shed into domestic sewage, wastewater surveillance of SARS-CoV-2 RNA has emerged as a non-intrusive approach to monitor and manage coronavirus disease 2019 (COVID-19) (Bivins et al., 2020; Fuschi et al., 2021; Lundy et al., 2021). Community-level COVID-19 dynamics have been assessed (Fernandez-Cassi et al., 2021) by measuring SARS-CoV-2 RNA in both raw sewage (Agrawal et al., 2021; Li et al., 2021) and settled solids (Wolfe et al., 2021) at wastewater treatment plants (WWTPs) of varying sizes (Rusiñol et al., 2021). Monitoring wastewater from buildings has also proven useful as an approach for managing COVID-19 at finer spatial scales on college campuses (Betancourt et al., 2021; Harris-Lovett et al., 2021), in schools (Crowe et al., 2021; Hassard et al., 2021), in skilled nursing facilities (Spurbeck et al., 2021), and even on aircraft and cruise ships (Ahmed et al., 2020b).
Normalization of SARS-CoV-2 RNA counts by various fecal markers such as crAssphage (Wilder et al., 2021) or pepper mild mottle virus (Graham et al., 2021; Wu et al., 2020) has been used to tune wastewater data for correlation with COVID-19 cases. These approaches reflect an assumption that SARS-CoV-2 RNA loading in domestic wastewater is primarily driven by fecal shedding of SARS-CoV-2 RNA (Ahmed et al., 2020a; Wannigama et al., 2021; Wolfe et al., 2021). However, a recent study reporting near-source monitoring of wastewater from primary and secondary schools raises the potential for other shedding routes to contribute to the SARS-CoV-2 RNA load in wastewater (Gutierrez et al., 2021). Herein, we use SARS-CoV-2 shedding datasets from the clinical literature along with a Monte Carlo framework to consider the likely contribution of various shedding routes to the daily SARS-CoV-2 RNA load produced by infected individuals and a collection of infected persons. The goal of our analysis is to consider the most probable source of SARS-CoV-2 RNA in wastewater at both the population and building level to further inform wastewater-based epidemiology.
2. Materials & methods
A Monte Carlo simulation was performed to assess the contribution of four SARS-CoV-2 shedding routes, stool, urine, sputum, and saliva, on the total viral load likely to be discharged to the sewer system. Monte Carlo modeling allows for propagation of the uncertainty surrounding the model inputs. Predictions therefore reflect the error associated with the collected data, creating a useful model despite high variability. Two models were created: a population model, which examines the contribution of the different excreta from a population of infected persons, and a single shedder model to examine the potential contributions of shedding route combinations that are likely for infected individuals.
We surveyed available literature data of daily excreta production values, concentration of SARS-CoV-2 RNA in bodily excreta, and the prevalence of shedding SARS-CoV-2 RNA in bodily excreta. We used the extracted data (Table S1) to parameterize probability distributions for a Monte Carlo simulation. The resulting model parameter fittings can be found in Table S2. For daily excreta production values, the distributions were chosen from studies that included relevant populations (e.g., healthy populations for urine and saliva production and populations with respiratory illnesses for sputum production) and a wide range in values in order to be representative of the community. For urine, we used the gamma distribution reported by Rauch et al. (2003), for stool, the distribution reported by Rose et al. (2015), and distributions were assumed for sputum and saliva. For SARS-CoV-2 RNA concentrations in bodily excreta, we fit distributions to data reported in recent papers (Han et al., 2020; Jeong et al., 2020; Kashi et al., 2020; Lescure et al., 2020; Pan et al., 2020; Roshandel et al., 2020; Wölfel et al., 2020; Wyllie et al., 2020; Yoon et al., 2020; Zheng et al., 2020). For SARS-CoV-2 prevalence of shedding, distributions were fit to data reported in recent meta-analyses (Kashi et al., 2020; Khiabani and Amirzade-Iranaq, 2021; Roshandel et al., 2020). With the exception of the urine prevalence data, where the range of expected values was already reported in the paper, we used a weighted average to create a truncated normal distribution to describe the prevalence data. The fittings were conducted in RStudio Team (2020) using the ‘fitdistrplus’ package (Delignette-Muller and Dutang, 2015) using the Maximum Likelihood Estimation method. When data was sparse a uniform distribution was assigned.
The quantity of SARS-CoV-2 RNA shed to the sewer through urine, sputum, and stool was calculated by multiplying the concentration of SARS-CoV-2 RNA gene copies (GC per unit mass or volume) by the quantity (volume or mass) of excreta shed to the sewer per day. The quantity of SARS-CoV-2 GC shed to sewer through saliva was calculated by multiplying the concentration of SARS-CoV-2 GC in saliva by the number of tooth brushing events per day and by the amount of saliva shed to the sewer system per event. For the population model, an additional term is included to account for the likelihood of shedding SARS-CoV-2 GC associated with each route. This factor is the fraction of COVID-19 patients who have positive detection of SARS-CoV-2 GC in each excreta. For the single shedder model, four scenarios were considered. Case one examines a single person shedding in saliva and sputum alone, case two examines a person shedding in saliva, sputum, and urine, case three represents a person shedding in saliva, sputum, and stool, and case four represents a person shedding in saliva, sputum, stool, and urine. The total viral load in each case was calculated by adding the respective SARS-CoV-2 GC from each shedding route. Each model parameter was described as a distribution to propagate uncertainty and variation into the final estimates. A Monte Carlo simulation with 10,000 samplings drawn at random from the inputted parameters was used to perform the calculation.
3. Results and discussion
3.1. Population model
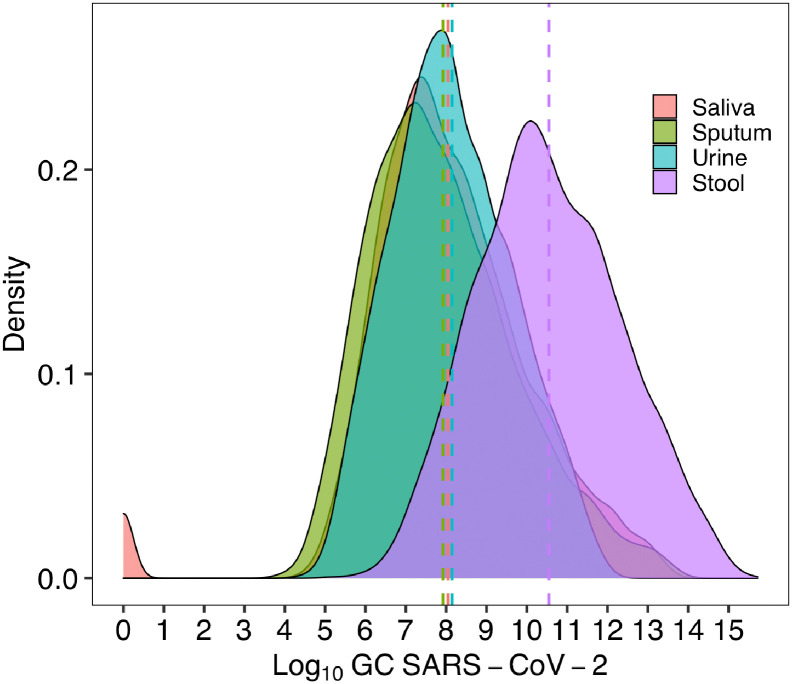
When considering wastewater-based epidemiology as a useful tool for monitoring COVID-19 in a community, the often-made assumption is that the primary contributor to the load of SARS-CoV-2 RNA in wastewater is stool. However, per current estimates, only about 40% of infected individuals excrete SARS-CoV-2 in stool (Roshandel et al., 2020). The likelihood of excreting in respiratory fluids is higher (~80%) (Khiabani and Amirzade-Iranaq, 2021), and while the likelihood of excreting in urine is lower (~1%) (Kashi et al., 2020), the volume of urine discharged to the sewer can be large compared to other body fluids. The goal of the population model is to examine the contribution of SARS-CoV-2 RNA to a common sewer system from an idealized population of 1000 infected individuals in order to determine which shedding route contributes the most RNA. Saliva contributes 8.05 log10 SARS-CoV-2 GC (25th-75th: 6.86-11.52), sputum contributes 7.92 log10 SARS-CoV-2 GC (25th-75th: 6.58-9.03), urine contributes 8.15 log10 SARS-CoV-2 GC (25th-75th: 7.07-9.18), and stool contributes 10.55 log10 SARS-CoV-2 GC (25th-75th: 9.27-11.79). The results of the population model are presented in Fig. 1 and the percentile values in Table S2. The similarity of the resulting cumulative distributions was assessed using the Kolmogorov-Smirnov test, and there was a significant difference between stool and all other excreta (Saliva: D = 0.42, p < 0.0001; Sputum: D = 0.46, p < 0.0001; Urine: D = 0.41, p < 0.0001).
Fig. 1.
Daily viral load of SARS-CoV-2 produced to a sewer system by a population of 1000 SARS-CoV-2 infected individuals stratified by shedding route. The dotted vertical line represents the mean.
Despite lower shedding prevalence than respiratory fluids, the model output supports the assumption that stool dominates the RNA load in wastewater for community shedding. While the concentration of SARS-CoV-2 RNA in stool is comparable to respiratory fluids, the volume of stool produced to the sewer system per day is orders of magnitude higher than respiratory fluids, a contributing factor to the significant difference in viral load between saliva and sputum compared with stool. Additionally, while the volume of urine excreted to the sewer is high and comparable on an order of magnitude level with stool, the prevalence of shedding SARS-CoV-2 RNA in urine is much lower.
While stool dominates population-level SARS-CoV-2 RNA loading, non-stool sources still contribute meaningful SARS-CoV-2 RNA signal to wastewater. This result is robust even when the number of infected individuals is decreased (Fig. S1), suggesting that stool shedding dominates community-level SARS-CoV-2 wastewater monitoring efforts even as total infections decrease. However, besides community level surveillance, many efforts have focused on building level surveillance with the goal of detecting small numbers or single infected individuals. To evaluate this use case, we created a “single shedder” model to examine the contribution of shedding routes on a building-level scale, modeling individuals separately who excrete SARS-CoV-2 via a unique combination of routes.
3.2. Single shedder
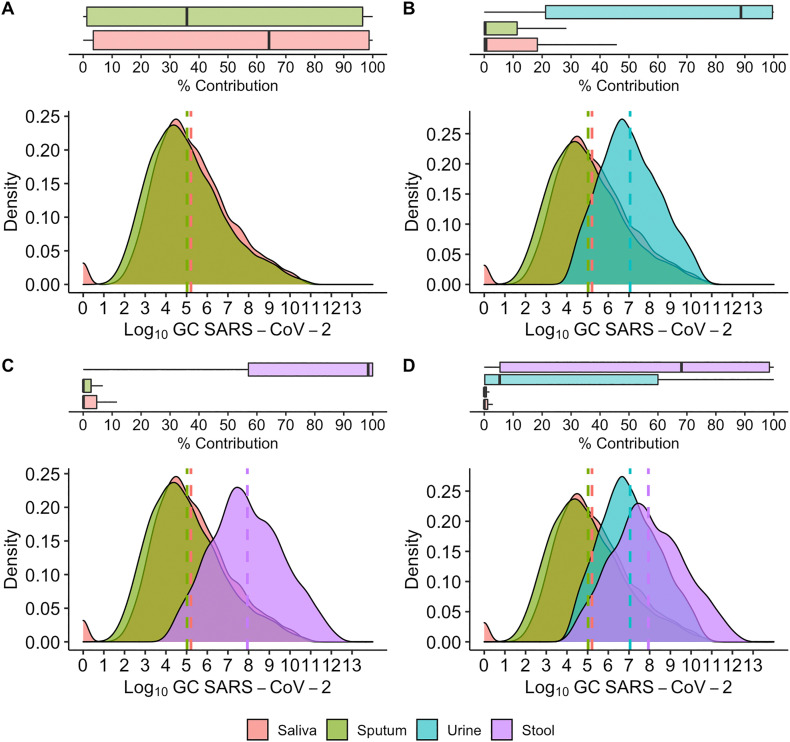
For the single shedder model, we focus on examining the probable contributions of saliva, sputum, stool, and urine in four different scenarios. The primary goal of this analysis is to assess the relative contribution of different shedding routes for detection of a single infected individual, e.g. for building-level surveillance. While there are a total of 16 possible shedding route combinations, we selected four as examples to interrogate the relative contribution of different shedding pathways. The results of all single shedder cases are presented in Fig. 2 and the percentile values in Table S3.
Fig. 2.
Percent contribution and Log10GC SARS-CoV-2 contribution of shedding pathways to total SARS-CoV-2 viral load to sewer per day. A. represents an individual shedding in saliva and sputum alone, B. represents an individual shedding in saliva, sputum, and urine, C. represents an individual shedding in saliva, sputum, and stool, and D. represents an individual shedding in saliva, sputum, stool, and urine.
Case one assumes a single individual infected with COVID-19 shedding SARS-CoV-2 in saliva and sputum. This scenario was premised on the observation that there is a higher likelihood of observing SARS-CoV-2 RNA shedding in both sputum and saliva than either urine or stool (Table S1). As shown in Fig. 2A, for case one there is an almost equal proportion of SARS-CoV-2 RNA shed into the sewer system from saliva and sputum, as their total log10 SARS-CoV-2 GC averages converge at approximately 5 log10 and the distributions nearly overlap. This is due to the similar ranges of SARS-CoV-2 GC found in either excreta as well as the similar modeled volumes of either excreta shed into the sewer system each day.
For cases two and three, we assume that an individual sheds SARS-CoV-2 genome copies in either urine or stool, Figs. 2B and C respectively, in addition to both saliva and sputum. Our results suggest that both urine and stool yield a much larger percent contribution of the RNA shed into the sewer system than either sputum or saliva alone, specifically yielding total daily mean shedding of 7 and 8 log10 SARS-CoV-2 GC, respectively, when an individual with COVID-19 sheds either excreta in addition to saliva and sputum. This is explained by a larger daily production of urine and stool as shown in the distributions in Table S1 despite similar concentrations of GCs per quantity of excreta produced for the case of stool and a lower genome copies concentration for the case of urine.
Finally, case four assumes an individual shedding SARS-CoV-2 RNA in all four excreta (Fig. 2D). In case four, stool is the highest contributor to daily SARS-CoV-2 RNA load. However, urine also contributes a high amount of SARS-CoV-2 RNA. The urine probability density function overlaps the distribution observed for stool and can, at the distribution limits, result in a higher percentage contribution than stool. This is due to the fact that, despite the aforementioned lower concentration of the SARS-CoV-2 RNA per volume of urine, the volumetric amount of urine produced daily is likely to be greatest, by several orders of magnitude, compared to the other three excreta. For example, the general range of daily urine production shed to the sewer system is roughly 1 - 2 L (Rose et al., 2015) compared to sputum (0.01 - 1 mL) and saliva (1 - 3 mL).
The individual single shedder model indicates that in a scenario such as a building with only a few infected individuals, any of the four cases analyzed show potential for contributing detectable SARS-CoV-2 RNA to the sewer system. Thus, at the building level the primary contribution of SARS-CoV-2 GCs found in the sewer system may originate from any of the four shedding routes examined. However, when all four excreta contribute to SARS-CoV-2 GC shed into a sewer system (case four), stool is the most probable primary contributor by one order of magnitude on average. This is in contrast to the population model, where stool dominates the average total log10 SARS-CoV-2 GC contribution by nearly three orders of magnitude compared to the other shedding routes when the likelihood of excreting SARS-CoV-2-positive urine is less than 1% (Kashi et al., 2020).
3.3. Sensitivity analysis
A sensitivity analysis was performed using the Spearman's rank correlation coefficient. Coefficients and tornado plots can be seen in Fig. S2. Additionally, a CPS graph (Coefficient/p-value/sample size) (Lin et al., 2013) was created to determine the impact of the number of Monte Carlo samplings on significance testing (Figs. S3 and S4). The model was stable, and the test demonstrated statistical significance within 100 samplings. In both model assessments the primary contributors to variability were the SARS-CoV-2 GC concentration in stool, followed by concentrations in saliva, sputum, and urine. Shedding volumes did not have as strong an impact on the total RNA load. This is expected, as shedding volume variations are within an order of magnitude, while SARS-CoV-2 GC concentrations have distributions spanning many orders of magnitude. Future SARS-CoV-2 variants might produce varying concentrations in excreta, which could strongly impact model outcomes. For example, a two order of magnitude increase in volume of saliva shed during a toothbrushing event caused a 9% increase in the total RNA load in the population model, whereas the same increase in SARS-CoV-2 GC concentration in saliva causes a 4000% increase in the total SARS-CoV-2 RNA load. The same increase in sputum production resulted in a 5% increase in total concentration, and the similarity continues with the same increase in SARS-CoV-2 GC concentration in sputum causing about a 4500% increase in the final concentration. The sensitivity analysis results suggest that further model refinement should entail improved quantification of SARS-CoV-2 load and temporal variability in bodily fluids.
4. Limitations and implications
The purpose of this study was to examine the contribution of SARS-CoV-2 shedding pathways to RNA loads in the sewer system for wastewater surveillance. The model results should not be interpreted as absolute quantitative values for the SARS-CoV-2 RNA concentrations found in each of the excreta studied. There is uncertainty on how the data is reported by all studies included here. Additionally, these studies neglected to include SARS-CoV-2 variants, and were conducted on different populations, age, and genders. All of these have an impact on the quality of the reported data. However, the purpose of this short paper is not to provide a comprehensive systematic review of current SARS-CoV-2 RNA shedding data. The data included were considered to be relevant representatives of the heterogeneity of the current available data. There is also significant uncertainty and variation in the model input parameters, especially the distribution of SARS-CoV-2 RNA concentrations, the likelihood of shedding RNA via each pathway, and the volume of excreta produced to the sewer system. The scope of this paper is limited towards available data, and future models can build on the framework here with updated inputs that include more specific and representative data. However, this uncertainty does not preclude the models’ usefulness to broadly describe the contributions of shedding pathways. Additionally, as infections decrease to single digits within a community, any shedding route could constitute a major contributor to the RNA load in a sewer system, due to high variability within and between individuals (Fig. S1).
Ultimately, for building level surveillance where the interest is in detecting one or a few shedding individuals, the SARS-CoV-2 RNA load in the sewer system could originate from any single shedding route. Importantly, this broadens the human behavior relevant to wastewater surveillance at the building level from defecation to include urination, mouth rinsing, spitting, or any other pathway that SARS-CoV-2 RNA could enter the sewer system. A recent publication suggested that SARS-CoV-2 RNA detection in wastewater from primary and secondary schools may indicate that students’ bathroom behavior at school could include defecation (Gutierrez et al., 2021). But the results of the current analysis might also explain the frequent detection of RNA in wastewater from schools. The results also suggest that even in settings where defecation is less likely, such as airplanes (Ahmed et al., 2020b), other behaviors could lead to the detection of SARS-CoV-2 RNA via wastewater surveillance. This could greatly expand the application of wastewater surveillance to detect SARS-CoV-2 infections at the building level.
At the community level, when the emphasis is on surveilling an aggregation of shedding individuals, the SARS-CoV-2 RNA load is most likely dominated by stool shedding. This result supports the use of various fecal-associated markers to normalize SARS-CoV-2 RNA concentrations in municipal wastewater and solids (Graham et al., 2021; Wilder et al., 2021; Wu et al., 2020). However, when the number of infections within a community decreases, wastewater surveillance at the community level may become more comparable to building level surveillance where non-defecation behaviors such as teeth brushing, mouth rinsing, and tissue disposal could become large contributors to daily RNA loading.
CRediT authorship contribution statement
Conceptualization – AB, KB
Investigation – KC, WC, SL
Writing - Original Draft – KC, WC, AB
Writing - Review & Editing – KC, WC, AB, KB
Visualization – KC
Supervision – KB
Funding Acquisition - KB
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This study was funded by United States National Science Foundation under grant 2027752.
Editor: Kevin V Thomas
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2021.150376.
Appendix A. Supplementary data
Supplementary material
References
- Agrawal S., Orschler L., Lackner S. Long-term monitoring of SARS-CoV-2 RNA in wastewater of the Frankfurt metropolitan area in Southern Germany. Sci. Rep. 2021;11:5372. doi: 10.1038/s41598-021-84914-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P.M., Angel N., Bibby K., Bivins A., Dierens L., Edson J., Ehret J., Gyawali P., Hamilton K.A., Hosegood I., Hugenholtz P., Jiang G., Kitajima M., Sichani H.T., Shi J., Shimko K.M., Simpson S.L., Smith W.J.M., Symonds E.M., Thomas K.V., Verhagen R., Zaugg J., Mueller J.F. Detection of SARS-CoV-2 RNA in commercial passenger aircraft and cruise ship wastewater: a surveillance tool for assessing the presence of COVID-19 infected travellers. J. Travel Med. 2020;27 doi: 10.1093/jtm/taaa116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt W.Q., Schmitz B.W., Innes G.K., Prasek S.M., Pogreba Brown K.M., Stark E.R., Foster A.R., Sprissler R.S., Harris D.T., Sherchan S.P., Gerba C.P., Pepper I.L. COVID-19 containment on a college campus via wastewater-based epidemiology, targeted clinical testing and an intervention. Sci. Total Environ. 2021;779 doi: 10.1016/j.scitotenv.2021.146408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivins A., North D., Ahmad A., Ahmed W., Alm E., Been F., Bhattacharya P., Bijlsma L., Boehm A.B., Brown J., Buttiglieri G., Calabro V., Carducci A., Castiglioni S., Cetecioglu Gurol Z., Chakraborty S., Costa F., Curcio S., de los Reyes F.L., Delgado Vela J., Farkas K., Fernandez-Casi X., Gerba C., Gerrity D., Girones R., Gonzalez R., Haramoto E., Harris A., Holden P.A., Jones D.L., Kasprzyk-Hordern B., Kitajima M., Kotlarz N., Kumar M., Kuroda K., La Rosa G., Malpei F., Mautus M., McLellan S.L., Medema G., Meschke J.S., Mueller J., Newton R.J., Nilsson D., Noble R.T., van Nuijs A., Peccia J., Perkins T.A., Pickering A.J., Rose J., Sanchez G., Smith A., Stadler L., Stauber C., Thomas K., van der Voorn T., Wigginton K., Zhu K., Bibby K., Islam Md.T. Wastewater-based epidemiology: global collaborative to maximize contributions in the fight against COVID-19. Environ. Sci. Technol. 2020;54:7754–7757. doi: 10.1021/acs.est.0c02388. [DOI] [PubMed] [Google Scholar]
- Cevik M., Tate M., Lloyd O., Maraolo A.E., Schafers J., Ho A. SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: a systematic review and meta-analysis. Lancet Microbe. 2021;2:e13–e22. doi: 10.1016/S2666-5247(20)30172-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe J., Schnaubelt A.T., Schmidt-Bonne S., Angell K., Bai J., Eske T., Nicklin M., Pratt C., White B., Crotts-Hannibal B., Staffend N., Herrera V., Cobb J., Conner J., Carstens J., Tempero J., Bouda L., Ray M., Lawler J.V., Campbell W.S., Lowe J.-M., Santarpia J., Bartelt-Hunt S., Wiley M., Brett-Major D., Logan C., Broadhurst M.J. medRxiv; 2021. Pilot Program for Test-based SARS-CoV-2 Screening and Environmental Monitoring in an Urban Public School District. 2021.04.14.21255036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delignette-Muller M., Dutang C. Fitdistrplus: an R package for fitting distributions. J. Stat. Softw. 2015;64:1–34. [Google Scholar]
- Fernandez-Cassi X., Scheidegger A., Bänziger C., Cariti F., Tuñas Corzon A., Ganesanandamoorthy P., Lemaitre J.C., Ort C., Julian T.R., Kohn T. Wastewater monitoring outperforms case numbers as a tool to track COVID-19 incidence dynamics when test positivity rates are high. Water Res. 2021;200 doi: 10.1016/j.watres.2021.117252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuschi C., Pu H., Negri M., Colwell R., Chen J. Wastewater-based epidemiology for managing the COVID-19 pandemic. ACS EST Water. 2021 doi: 10.1021/acsestwater.1c00050. [DOI] [PubMed] [Google Scholar]
- Graham K.E., Loeb S.K., Wolfe M.K., Catoe D., Sinnott-Armstrong N., Kim S., Yamahara K.M., Sassoubre L.M., Mendoza Grijalva L.M., Roldan-Hernandez L., Langenfeld K., Wigginton K.R., Boehm A.B. SARS-CoV-2 RNA in wastewater settled solids is associated with COVID-19 cases in a large urban sewershed. Environ. Sci. Technol. 2021;55:488–498. doi: 10.1021/acs.est.0c06191. [DOI] [PubMed] [Google Scholar]
- Gutierrez V.C., Hassard F., Vu M., Leitao R., Burczynska B., Wildeboer D., Stanton I., Rahimzadeh S., Baio G., Garelick H., Hofman J., Kasprzyk-Hordern B., Kwiatkowska R., Majeed A., Priest S., Grimsley J., Lundy L., Singer A.C., Cesare M.D. medRxiv; 2021. Monitoring Occurrence of SARS-CoV-2 in School Populations: A Wastewater-based Approach. 2021.03.25.21254231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M.S., Seong M.-W., Kim N., Shin S., Cho S.I., Park H., Kim T.S., Park S.S., Choi E.H. Viral RNA load in mildly symptomatic and asymptomatic children with COVID-19, Seoul, South Korea. Emerg. Infect. Dis. 2020;26:2497–2499. doi: 10.3201/eid2610.202449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris-Lovett S., Nelson K.L., Beamer P., Bischel H.N., Bivins A., Bruder A., Butler C., Camenisch T.D., De Long S.K., Karthikeyan S., Larsen D.A., Meierdiercks K., Mouser P.J., Pagsuyoin S., Prasek S.M., Radniecki T.S., Ram J.L., Roper D.K., Safford H., Sherchan S.P., Shuster W., Stalder T., Wheeler R.T., Korfmacher K.S. Wastewater surveillance for SARS-CoV-2 on college campuses: initial efforts, lessons learned, and research needs. Int. J. Environ. Res. Public Health. 2021;18:4455. doi: 10.3390/ijerph18094455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassard F., Lundy L., Singer A.C., Grimsley J., Di Cesare M. Innovation in wastewater near-source tracking for rapid identification of COVID-19 in schools. Lancet Microbe. 2021;2:e4–e5. doi: 10.1016/S2666-5247(20)30193-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H.W., Kim S.-M., Kim H.-S., Kim Y.-I., Kim J.H., Cho J.Y., Kim S., Kang H., Kim S.-G., Park S.-J., Kim E.-H., Choi Y.K. Viable SARS-CoV-2 in various specimens from COVID-19 patients. Clin. Microbiol. Infect. 2020;26:1520–1524. doi: 10.1016/j.cmi.2020.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashi A.H., de la Rosette J.J.M.C.H., Amini E., Abdi H., Fallah-Karkan M., Vaezjalali M. 2020. Urinary Viral Shedding of COVID-19 and Its Clinical Associations: A Systematic Review and Meta-analysis of Observational Studies. [DOI] [PubMed] [Google Scholar]
- Khiabani K., Amirzade-Iranaq M.H. Are saliva and deep throat sputum as reliable as common respiratory specimens for SARS-CoV-2 detection? A systematic review and meta-analysis. Am. J. Infect. Control. 2021 doi: 10.1016/j.ajic.2021.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescure F.-X., Bouadma L., Nguyen D., Parisey M., Wicky P.-H., Behillil S., Gaymard A., Bouscambert-Duchamp M., Donati F., Le Hingrat Q., Enouf V., Houhou-Fidouh N., Valette M., Mailles A., Lucet J.-C., Mentre F., Duval X., Descamps D., Malvy D., Timsit J.-F., Lina B., Yazdanpanah Y., van-der-Werf S. Clinical and virological data of the first cases of COVID-19 in Europe: a case series. Lancet Infect. Dis. 2020;20:697–706. doi: 10.1016/S1473-3099(20)30200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Di D.Y.W., Saingam P., Jeon M.K., Yan T. Fine-scale temporal dynamics of SARS-CoV-2 RNA abundance in wastewater during a COVID-19 lockdown. Water Res. 2021;197 doi: 10.1016/j.watres.2021.117093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M., Lucas H.C., Shmueli G. Research commentary—too big to fail: large samples and the p-value problem. Inf. Syst. Res. 2013;24:906–917. doi: 10.1287/isre.2013.0480. [DOI] [Google Scholar]
- Lundy L., Fatta-Kassinos D., Slobodnik J., Karaolia P., Cirka L., Kreuzinger N., Castiglioni S., Bijlsma L., Dulio V., Deviller G., Lai F.Y., Alygizakis N., Barneo M., Baz-Lomba J.A., Béen F., Cíchová M., Conde-Pérez K., Covaci A., Donner E., Ficek A., Hassard F., Hedström A., Hernandez F., Janská V., Jellison K., Hofman J., Hill K., Hong P.-Y., Kasprzyk-Hordern B., Kolarević S., Krahulec J., Lambropoulou D., de Llanos R., Mackuľak T., Martinez-García L., Martínez F., Medema G., Micsinai A., Myrmel M., Nasser M., Niederstätter H., Nozal L., Oberacher H., Očenášková V., Ogorzaly L., Papadopoulos D., Peinado B., Pitkänen T., Poza M., Rumbo-Feal S., Sánchez M.B., Székely A.J., Soltysova A., Thomaidis N.S., Vallejo J., van Nuijs A., Ware V., Viklander M. Making waves: collaboration in the time of SARS-CoV-2 - rapid development of an international co-operation and wastewater surveillance database to support public health decision-making. Water Res. 2021;199 doi: 10.1016/j.watres.2021.117167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Zhang D., Yang P., Poon L.L.M., Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect. Dis. 2020;20:411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch W., Brockmann D., Peters I., Larsen T.A., Gujer W. Combining urine separation with waste design: an analysis using a stochastic model for urine production. Water Res. 2003;37:681–689. doi: 10.1016/S0043-1354(02)00364-0. [DOI] [PubMed] [Google Scholar]
- Rose C., Parker A., Jefferson B., Cartmell E. The characterization of feces and urine: a review of the literature to inform advanced treatment technology. Crit. Rev. Environ. Sci. Technol. 2015;45:1827–1879. doi: 10.1080/10643389.2014.1000761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roshandel M.R., Nateqi M., Lak R., Aavani P., Sari Motlagh R., F Shariat S., Aghaei Badr T., Sfakianos J., Kaplan S.A., Tewari A.K. Diagnostic and methodological evaluation of studies on the urinary shedding of SARS-CoV-2, compared to stool and serum: a systematic review and meta-analysis. Cell. Mol. Biol. Noisy--Gd. Fr. 2020;66:148–156. [PubMed] [Google Scholar]
- RStudio Team . PBC; Boston, MA: 2020. RStudio: Integrated Development Environment for R. [Google Scholar]
- Rusiñol M., Zammit I., Itarte M., Forés E., Martínez-Puchol S., Girones R., Borrego C., Corominas Ll, Bofill-Mas S. Monitoring waves of the COVID-19 pandemic: inferences from WWTPs of different sizes. Sci. Total Environ. 2021;787 doi: 10.1016/j.scitotenv.2021.147463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurbeck R.R., Minard-Smith A., Catlin L. Feasibility of neighborhood and building scale wastewater-based genomic epidemiology for pathogen surveillance. Sci. Total Environ. 2021;789 doi: 10.1016/j.scitotenv.2021.147829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wannigama D.L., Amarasiri M., Hurst C., Phattharapornjaroen P., Abe S., Hongsing P., Rad S.M.A.H., Pearson L., Saethang T., Luk-in S., Kueakulpattana N., Storer R.J., Ounjai P., Jacquet A., Leelahavanichkul A., Chatsuwan T. Tracking COVID-19 with wastewater to understand asymptomatic transmission. Int. J. Infect. Dis. 2021 doi: 10.1016/j.ijid.2021.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilder M.L., Middleton F., Larsen D.A., Du Q., Fenty A., Zeng T., Insaf T., Kilaru P., Collins M., Kmush B., Green H.C. Co-quantification of crAssphage increases confidence in wastewater-based epidemiology for SARS-CoV-2 in low prevalence areas. Water Res. X. 2021;11 doi: 10.1016/j.wroa.2021.100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe M.K., Archana A., Catoe D., Coffman M.M., Dorevich S., Graham K.E., Kim S., Grijalva L.M., Roldan-Hernandez L., Silverman A.I., Sinnott-Armstrong N., Vugia D.J., Yu A.T., Zambrana W., Wigginton K.R., Boehm A.B. Scaling of SARS-CoV-2 RNA in settled solids from multiple wastewater treatment plants to compare incidence rates of laboratory-confirmed COVID-19 in their sewersheds. Environ. Sci. Technol. Lett. 2021;8:398–404. doi: 10.1021/acs.estlett.1c00184. [DOI] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wu F., Zhang J., Xiao A., Gu X., Lee W.L., Armas F., Kauffman K., Hanage W., Matus M., Ghaeli N., Endo N., Duvallet C., Poyet M., Moniz K., Washburne A.D., Erickson T.B., Chai P.R., Thompson J., Alm E.J. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. mSystems. 2020;5 doi: 10.1128/mSystems.00614-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyllie A.L., Fournier J., Casanovas-Massana A., Campbell M., Tokuyama M., Vijayakumar P., Warren J.L., Geng B., Muenker M.C., Moore A.J., Vogels C.B.F., Petrone M.E., Ott I.M., Lu P., Venkataraman A., Lu-Culligan A., Klein J., Earnest R., Simonov M., Datta R., Handoko R., Naushad N., Sewanan L.R., Valdez J., White E.B., Lapidus S., Kalinich C.C., Jiang X., Kim D.J., Kudo E., Linehan M., Mao T., Moriyama M., Oh J.E., Park A., Silva J., Song E., Takahashi T., Taura M., Weizman O.-E., Wong P., Yang Y., Bermejo S., Odio C.D., Omer S.B., Dela Cruz C.S., Farhadian S., Martinello R.A., Iwasaki A., Grubaugh N.D., Ko A.I. Saliva or nasopharyngeal swab specimens for detection of SARS-CoV-2. N. Engl. J. Med. 2020;383:1283–1286. doi: 10.1056/NEJMc2016359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J.G., Yoon J., Song J.Y., Yoon S.-Y., Lim C.S., Seong H., Noh J.Y., Cheong H.J., Kim W.J. Clinical significance of a high SARS-CoV-2 viral load in the saliva. J. Korean Med. Sci. 2020;35 doi: 10.3346/jkms.2020.35.e195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S., Fan J., Yu F., Feng B., Lou B., Zou Q., Xie G., Lin S., Wang R., Yang X., Chen W., Wang Q., Zhang D., Liu Y., Gong R., Ma Z., Lu S., Xiao Y., Gu Y., Zhang J., Yao H., Xu K., Lu X., Wei G., Zhou J., Fang Q., Cai H., Qiu Y., Sheng J., Chen Y., Liang T. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: retrospective cohort study. BMJ. 2020;369 doi: 10.1136/bmj.m1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material