Abstract
Spatial concentration gradients of antibiotics are prevalent in the natural environment. Yet, the microbial response in these heterogeneous systems remains poorly understood. We used a microfluidic reactor to create an artificial microscopic ecosystem that generates diffusive gradients of solutes across interconnected microenvironments. With this reactor, we showed that chemotaxis toward a soluble electron acceptor (nitrate) allowed Shewanella oneidensis MR-1 to inhabit and sustain metabolic activity in highly toxic regions of the antibiotic ciprofloxacin (>80× minimum inhibitory concentration, MIC). Acquired antibiotic resistance was not observed for cells extracted from the reactor, so we explored the role of transient adaptive resistance by probing multidrug resistance (MDR) efflux pumps, ancient elements that are important for bacterial physiology and virulence. Accordingly, we constructed an efflux pump deficient mutant (∆mexF) and used resistance-nodulation-division (RND) efflux pump inhibitors (EPIs). While batch results showed the importance of RND efflux pumps for microbial survival, microfluidic studies indicated that these pumps were not necessary for survival in antibiotic gradients. Our work contributes to an emerging body of knowledge deciphering the effects of antibiotic spatial heterogeneity on microorganisms and highlights differences of microbial response in these systems versus well-mixed batch conditions.
Subject terms: Water microbiology, Bacterial physiology, Biogeochemistry, Environmental sciences, Antibiotics
Introduction
Antibiotic pollution of terrestrial and aquatic environments can augment antibiotic-resistant genes that threaten the efficacy of antibiotic treatments and can impact the structure and function of nontarget microorganisms that modulate biogeochemical processes, such as nitrogen turnover [1–5]. As antibiotics are released to the environment, they exhibit dynamic spatial concentration heterogeneities (i.e., antibiotic gradients) due to solute transport phenomena and the cyclical nature of discharge from some sources [6–9]. While the impact of antibiotics on the nitrogen cycle has been investigated under various settings, the effect of antibiotic gradients is rarely considered. Antibiotic gradients exert complex patterns of selective pressures that allow microorganisms to survive and access nutrients in regions that would otherwise be considered inhospitable [10–12]. Most often microbial survival in the presence of antibiotic gradients is thought to be enabled by the development of genetically encoded, acquired resistance (i.e., mutational resistance); however, transient, non-inheritable resistance (i.e., adaptive resistance; sometimes defined as antibiotic tolerance) may facilitate such survival prior to or in lieu of mutational resistance [13, 14] (terminology refs: [15, 16]). Several studies have shown that migration, motility (swarming and swimming), mediated cell death, and threshold cell densities facilitate adaptive resistance in antibiotic gradients, but to date adaptive resistance remains poorly understood [17–19]. Herein, we focus on chemotaxis and multidrug resistance (MDR) elements, namely efflux pumps, as potential mechanisms of adaptive resistance that facilitate the survival of nontarget microorganisms and, in turn, affect nitrogen turnover in the presence of antibiotic gradients.
Microbial motility and migration enhance fitness in the presence of antibiotic gradients by providing access to resource-rich regions [20], a process that can be enabled by mutational or adaptive resistance mechanisms [17–19, 21–24]. For example, in a microfluidic study, swimming motility along a ciprofloxacin gradient allowed Escherichia coli to develop mutational resistance within 24 h that was beneficial for accessing resources in highly toxic compartments [22]. Another study, utilizing swim-agar plates with increasing concentrations of ciprofloxacin or trimethoprim, showed that E. coli depleted resources on one plate before gaining mutational resistance and proceeding to the next adjacent plate with higher antibiotic and resource concentrations [24]. Alternatively, other microfluidic studies showed that swimming motility and high cell densities allowed E. coli to colonize and remain active (i.e., not dormant persister cells) in the toxic region of a kanamycin gradient for over 24 h without the development of mutational resistance [18]. Moreover, we recently showed that Shewanella oneidensis MR-1, a nonpathogenic gram negative sediment-dwelling bacterium, was able to reduce nitrate to ammonium in toxic regions of a ciprofloxacin gradients for over 5 days in a microfluidic gradient chamber (MGC, Fig. 1) without observed mutational resistance [17]. The response was attributed to swimming motility and the migration of cells from nontoxic regions to toxic regions. Collectively, these studies identified the importance of motility and migration, and while chemotaxis was implied, none explicitly determined if chemotaxis or random motility allowed for habitability and metabolic processes in toxic regions. Because of this and the pronounced ecological role of chemotaxis [25, 26], we sought to determine if chemotaxis facilitated adaptive resistance along antibiotic gradients.
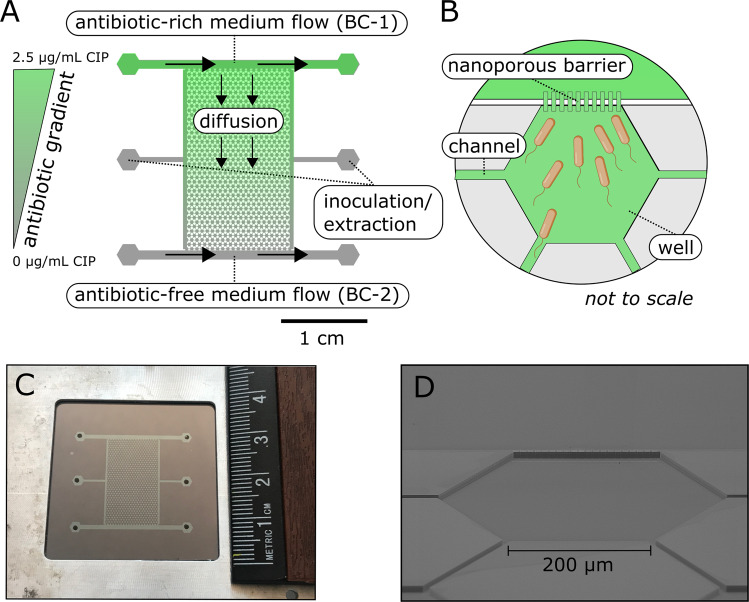
Fig. 1. The microfluidic gradient chamber (MGC).
A An illustration of the MGC with an antibiotic gradient of ciprofloxacin (CIP). The MGC consists of 850 interconnected hexagonal wells bound by two boundary channels (BC-1 and BC-2) on either side that allow for solute delivery and generation of gradients. The hexagonal well array is roughly 2 by 1.25 cm with a porosity of 0.3. Each well has a side length of 200 µm with a depth of 10 µm and support a total of 1 nL of fluid. The well array and the boundary channels are separated by a nanoporous barrier. The barriers have a depth of 0.2 µm to allow for diffusion, limit advection, and prevent cell washout. Inoculation and extraction ports allow for access into the well array. B A schematic demonstrating a single well, the nanoporous barrier that connects the boundary channels to the wells, and channels that allow bacteria to migrate between wells. C A picture of the actual reactor with a ruler for scale. D An SEM image depicting the rendered schematic of (B).
Efflux pumps can play important ecological role in microbial survival [27] and may contribute to adaptive resistance along antibiotic gradients by preventing the intracellular accumulation of antibiotics. The trimeric resistance-nodulation-cell-division family (RND) transporters are some of the more relevant efflux pumps in antibiotic resistance [28]. During mutational resistance, mutations in local or global regulatory genes can result in overexpression of these pumps [29]. This was evident in two aforementioned gradient studies [22, 24] where mutations associated with multiple antibiotic resistance (Mar) operons, the protein-coding genes known to regulate the expression of RND efflux systems [30, 31], contributed to antibiotic resistance. Alternatively, during adaptive resistance transient overexpression of efflux pumps can be induced as a response to environmental cues (e.g., chemical reagents [32–34], antibiotics themselves [35], and biofilm formation [36]). Even at the single-cell level, transient and stochastic expression of MarA can also induce adaptive resistance [37]. While these studies provide important insight into adaptive resistance via efflux pumps, the role of efflux pumps in facilitating adaptive resistance along antibiotic gradients has not been considered.
The objectives of this work were to determine the roles of chemotaxis and RND efflux pumps of S. oneidensis MR-1 in facilitating habitability and metabolic activity for nitrate reduction in toxic regions of a ciprofloxacin concentration gradient in an MGC. Ciprofloxacin was selected as a representative fluoroquinolone antibiotic. Ciprofloxacin and other fluoroquinolones are recalcitrant compounds in the environment [38, 39] and resistance to them can be acquired through efflux pumps [40]. A mexF in-frame deletion mutant (∆mexF) of S. oneidensis was constructed. This strain, the wild type strain (MR-1), and a motile but non-chemotactic strain (∆cheA) were individually placed into a diffusion-controlled well array within the MGC. The strains were subjected to nitrate-rich boundary conditions on either side of the well array, and antibiotic-rich boundary conditions on one side of the well array. In one case, MR-1 cells were exposed to an efflux pump inhibitor (EPI) at both boundaries. Microbial habitability and metabolic activity were monitored for 5 days in each experiment. Complementary minimum inhibitory concentration (MIC) assays, batch growth kinetic assays, and whole-genome sequencing of a ciprofloxacin-resistant mutant (SP-2) were performed to facilitate interpretation. This approach allowed us to test the hypotheses that chemotactic migration is required for habitability of antibiotic-rich regions and that RND efflux pumps facilitate survival in these regions to sustain metabolic activity for nitrate reduction.
Methods
Bacterial strains and growth conditions
The S. oneidensis strains used in this study can be found in Table 1. For all experiments, unless otherwise noted, cells were initially streaked on LB agar plates from a −80 °C clonal stock and incubated overnight at 30 °C. Single colonies were then selected and transferred to a piperazine-N,N′-bis(2-ethanesulfonicacid) (PIPES)-buffered anoxic media (PIPES medium), incubated overnight at 25 °C, and transferred for desired experiments. Chemicals and reagents, and the PIPES medium components and preparation are detailed in Note S1 and Note S2 of the Supporting Information.
Table 1.
S. oneidensis strains used in this study.
| S. oneidensis strains | Description | Reference |
|---|---|---|
| MR-1 | Wild type (WT), ATCC 700550 | |
| ∆cheA | WT with deletion in cheA-3; renders the strain motile but non-chemotactic | [68] |
| ∆mexF | WT with deletion in mexF | This study |
| SP-2 | A culture-based ciprofloxacin-resistant mutant derived from WT; generated through serial-passage experiments | [17] |
Three sets of batch experiments were developed to identify the minimum inhibitory concentrations (MIC), phenotype, and EPIs susceptibility for MR-1 and mutant cultures. In addition, five conditions in microfluidic reactors were tested and/or evaluated to investigate the microbial response to antibiotic gradients by MR-1 and the mutant cultures in the absence or presence of EPIs. Table S1 provides a list and summary of the experiments presented in the methods section.
Whole genome sequencing of a laboratory evolved ciprofloxacin-resistant mutant
The SP-2 antibiotic-resistant mutant was generated in a previous study via culture-based serial-passage experiments under nitrate-reducing condition [17]; a summary can be found in Note S3. The DNA from a ciprofloxacin-treated sample (SP-2) and the respective control sample from day 7 of the serial-passage experiment were submitted for whole genome sequencing. Antibiotic exposure on day 7 for the SP-2 culture was 130 ng/mL. An MIC assay indicated it was resistant up to 1.5 µg/mL of ciprofloxacin. The genomic DNA extraction was performed by using the Power Soil DNA Isolation Kit (Qiagen Inc., CA, US). The integrity of the extracted DNA was assessed using 0.8% agarose electrophoresis. The concentrations of the DNA were quantitated using a Qubit Fluorometric Quantification (Life Technologies, NY, US). The genomic DNA was submitted to the W. M. Keck Center at the University of Illinois at Urbana-Champaign for construction of genomic libraries and sequencing using an Illumina HiSeq 2500 platform. Our sequencing coverage was 200–250× for each of the genomic samples. The raw reads (mean, 4,541,900 reads/per sample) were quality trimmed using Trimmomatic v0.33 [41] under the command ‘TOPHRED33 SLIDINGWINDOW:4:20 LEADING:30 TRAILING:30 MINLEN:25’. Assembly was performed using SPAdes v3.9.0 (parameters: -t 24 -m 50 -careful). Annotations were retrieved using Prokka v1.11 (parameters: -addgenes -centre CBC -gram -cpus 24 -rfam). After being exposed to ciprofloxacin stress over several days, MR-1 was assumed to evolve into a suite of strains with increased resistance to ciprofloxacin. The pangenome of these strains were under mutation calling using breseq v0.32.1 (parameters: -j 6 -p -o B2 -n B2) [42] against the wild-type S. oneidensis MR-1 reference genome (NCBI Reference Sequence ID NZ_CP053946.1) to identify the mutations potentially associated with ciprofloxacin resistance. The DNA-sequencing data reported in this study were deposited in the Sequence Read Archive under accession SRR13626180 and SRR13644606.
Construction of the in-frame mexF deletion mutant strain
The ∆mexF mutant strain was constructed using suicide plasmid-mediated homologous recombination following previous protocols [43]. Primers used in this study to construct the suicide plasmid are detailed in Table S2. The plasmid was assembled by Golden Gate cloning [44] using the BsmBI restriction enzyme and T4 DNA ligase (New England Biolabs, MA, US). Genomic upstream, genomic downstream, and suicide vector DNA fragments were generated by PCR (Phusion High Fidelity DNA Polymerase, New England Biolabs). The Golden Gate assembly products were used to transform electrocompetent E. coli WM3064. The assembled plasmids were harvested and verified by Sanger sequencing (DNA Sequencing Facilities, University of Texas at Austin). Donor E. coli WM3064 maintaining the assembled suicide plasmid was conjugated with recipient wild-type S. oneidensis MR-1. A successful S. oneidensis transconjugant with the mexF deletion was then selected and stored. More details can be found in Note S4.
Minimum inhibitory concentration (MIC) assay
The MIC assays were conducted following EUCAST protocols under nitrate reducing conditions with PIPES medium in 96-well microtiter plates [17, 22, 45]. An inoculum of 106 cells/mL was used. The ciprofloxacin concentrations tested were 0, 10, 20, 30, 40, 50, 100, and 200 ng/mL. Ninety-six well plates were prepared and incubated in an anaerobic glove box containing a ~98% N2-2% H2 atmosphere. The optical density was measured over a 24 h period with a microtiter pate reader (Synergy-HT, Bio-Tek Instruments, VT, US) stationed within the anaerobic glove box at 29 °C (see Fig. S1 for growth curves). The MIC was defined as the concentration of ciprofloxacin that inhibited 100% visible growth within 24 h and was determined by fitting a modified Gompertz function to the inhibition profiles of ciprofloxacin against cell growth [46]. All data fitting was performed with Prism 8.4 (GraphPad Software). Five biological replicate experiments were conducted.
Three-day growth assay
Three-day growth assays were conducted under aerobic conditions following procedures from a previous study [27]. Overnight aerobic LB Miller grown cultures were used as the inoculate at a ratio of 1:50 in culture tubes containing 5 mL LB, and either 5 µg/mL chloramphenicol or 0.25 µg/mL ciprofloxacin. Cell densities were measured periodically with cuvettes (Nanodrop 2000c, Thermo, MA, US). Cells were incubated at 25 °C with shaking at 250 rpm for 72 h. Three biological replicate experiments were conducted.
Efflux pump inhibitor (EPI) susceptibility assay
The EPI assays were conducted under nitrate reducing conditions with PIPES medium following procedures from a previous study [47]. Phenylalanine-arginine-ß-naphthylamide dihydrochloride (PAßN) and 1-(1-naphthylmethyl)-piperazine (NMP) were the tested EPIs. The assays were conducted in a similar manner to an MIC assay but with the addition of 10, 25, 50, 100, 200 µg/mL of EPIs across the range of ciprofloxacin concentrations. The MIC was determined at 10 h due to fluctuations (early onset of death phase) induced by the EPIs after that period (see Fig. S2 for growth curves). Note that all nitrate is converted to ammonium by 10 h in batch conditions. Three biological replicate experiments were conducted.
Microfluidic gradient chamber (MGC) setup and operation
The design and fabrication of the MGC was presented in our previous work [17]. Briefly, the MGC was designed as an idealized representation of an interconnected porous network with solute heterogeneity. It was etched on a silicon substrate using standard photolithography and inductively coupled plasma reactive ion etching. Ports were ultrasonically drilled through the silicon substrate; a thin layer of silicon dioxide was grown on the surface by thermal oxidation, and it was sealed with a borosilicate wafer by anodic bonding. The reactor dimensions and assembly are presented in Figs. 1 and S3.
Prior to each microfluidic experiment, a cleaning and disinfection procedure was conducted (Note S5). The MGC was inoculated with cells at 106 cells/mL in fresh PIPES medium containing a one-tenth strength concentration of lactate (0.4 mM) and nitrate (0.2 mM) to stimulate initial growth. The inoculum was passed through the well array via the inoculation and extraction ports resulting in an initial cell density of roughly 1 cell/well (equivalent to 106 cells/mL). The inoculation and extraction ports were then sealed to eliminate flow through the inner array. PIPES medium (containing 4 mM lactate and 2 mM nitrate) amended with 2.5 µg/mL ciprofloxacin (>80× MIC) and 10 µM fluorescein was continuously delivered through boundary channel 1 (BC-1) while PIPES medium only (also containing 4 mM lactate and 2 mM nitrate) was delivered through boundary channel 2 (BC-2). The fluorescein dye was added to confirm diffusive transport of solutes. Flow was delivered at a flowrate of 10 µL/h to each boundary channel and controlled with gas tight syringes (Hamilton, NV, US) and syringe pumps (Cole-Parmer, IL, US).
Images of four representative wells in every row perpendicular to the primary direction of diffusive transport were obtained daily (4 × 41 row; 164 images per day) to quantify the cell density and fluorescence intensity profiles. Image acquisition and processing details can be found in Note S6 and Fig. S4. Boundary channels effluent was collected daily for 5 days (240 µL per sample) in autoclaved HPLC vials and stored at −80 °C for subsequent nitrite quantification using ion chromatography (IC). Note S7 provides more details on the IC method. On day 5, cells were extracted and tested for increased ciprofloxacin resistance in like manner to an MIC assay (see Note S8). The MGC experiments were conducted for 5 days at 25 ± 1 °C with ∆cheA and ∆mexF cultures, and MR-1 cultures supplemented with 200 µg/mL PAßN (i.e., WT + PAßN). Two independent experiments were conducted for ∆cheA and ∆mexF and one independent experiment was conducted for WT + PAßN. Independent replicate results can be found in Figs. S5 and S6.
Statistical analyses
A summary of the statistical analyses performed in this study can be found in Note S9 and Tables S3, S4, and S5.
Results
Insight into mechanisms that may facilitate adaptive resistance in toxic regions of the MGC
Whole genome sequencing of the SP-2 mutant revealed three dominant (i.e., allele frequency >0.5) and distinct single-nucleotide polymorphism (SNP) mutations (Table 2). First, a missense A → G caused an Asp87 → Gly mutation in gyrA. Although not previously reported for S. oneidensis, it is not surprising to find a mutation in gyrA because ciprofloxacin targets the gyrase-DNA complex and inhibits its activity [48]. Known x-ray structures for a similar gyrase suggests that the mutated amino acid is near the active site that ciprofloxacin targets, suggesting that it is a functional SNP [22, 49]. Second, a missense G → A was identified in a region coding a MarR family transcriptional regulator. In E. coli, MarR is a global transcriptional repressor of the AcrAB-TolC RND efflux pump [50]. Mutations that relieve transcriptional repression by MarR consequently upregulate the expression of the efflux pump and increase antibiotic resistance [51, 52]. Therefore, this SNP may have altered the expression of efflux pumps in S. oneidensis for enhanced resistance. Third, a missense C → T was detected in the transcriptional repressor of the MexEF efflux pumps, MexR (SO3494). In P. aeruginosa, a MexR repressor modulates expression of the mexAB-oprM RND efflux operon and mutations in mexR have resulted in loss of repressor activity (i.e., higher expression levels of mexAB) [53, 54]. Also, in P. aeruginosa, hyperexpression of this operon results in antibiotic resistance to quinolones/fluoroquinolones and chloramphenicol [55–58]. The mexF gene in S. oneidensis shares 72% identity with that of P. aeruginosa, suggesting that hyperexpression by S. oneidensis may also result in antibiotic resistance [27]. In addition, Groh et al. (2007) showed that deletion of mexF in S. oneidensis increased susceptibility to chloramphenicol, lincomycin, nalidixic acid (a quinolone) and norfloxacin (a fluoroquinolone) [27]. Moreover, a chloramphenicol-adapted S. oneidensis mutant from the same study indicated that there were mutations in a TetR family regulatory protein (SO3494), which in fact is MexR from this study. Thus, being that MexR is the repressor for S. oneidensis MexEF efflux pumps, it is possible that the mutation in the mexR gene impeded S. oneidensis’s ability to repress MexEF efflux pumps, resulting in hyperexpression and increased ciprofloxacin resistance.
Table 2.
SNP mutations present in the ciprofloxacin resistant S. oneidensis mutant (SP-2).
| Allele Frequency | Mutation | Gene and Description |
|---|---|---|
| 0.70 | D87G (GAT → GGT) | gyrA, DNA gyrase subunit A (SO_2411) |
| 1.00 | V11I (GTT → ATT) | MarR family transcriptional regulator |
| 0.61 | A166V (GCC → GTC) | mexR, transcriptional repressor of MexEF efflux pump (SO_3494) |
Mutations with a frequency >0.5 are presented.
These results indicate that the resistance of the SP-2 mutant to batch concentrations of ciprofloxacin exceeding 50× MIC can be attributed to targeted site mutations (i.e., gyrA) and mutations that alter the expression of RND efflux pumps (i.e., MarR, MexR). While targeted site mutations are not candidates for transient adaptive resistance, resistance mechanisms related to altering expression levels are, because they do not require mutations to occur.
Deletion of the mexF gene inhibits growth of S. oneidensis at elevated ciprofloxacin concentrations in batch experiments
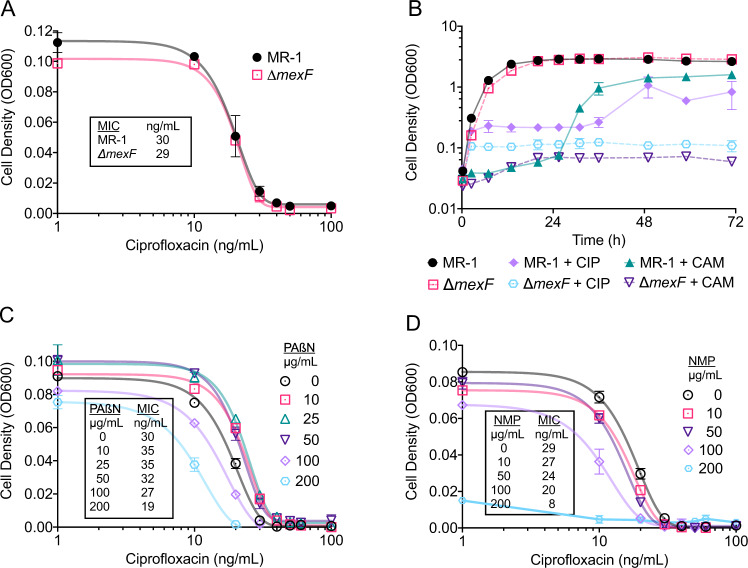
An MIC assay and a 3-day growth assay were conducted to verify a phenotypic response of an in-house constructed ∆mexF mutant. The MIC of our ∆mexF mutant culture indicated that there was not a significant difference in susceptibility to ciprofloxacin (P = 0.52, Fig. 2A). These results are consistent with previous work that compared the MICs of MR-1 and a ∆mexF mutant against chloramphenicol, norfloxacin, and nalidixic acid, and resulted in similar MIC values [27]. In contrast, results from the three-day growth assay showed that indeed deletion of the mexF gene increased ciprofloxacin susceptibility (Fig. 2B). Here, growth of MR-1 cultures was initially arrested for roughly 36 h during exposure to elevated ciprofloxacin concentrations, but thereafter recovered and achieved optical density values comparable to that of the non-treated cultures. In contrast, growth of ∆mexF mutant cultures was impaired, without recovery, throughout the experimental run. We also present results for a three-day growth assay with the antibiotic chloramphenicol (Fig. 2B). The results are identical to the phenotypic response presented for MR-1 and ∆mexF in Groh et al., 2007 [27].
Fig. 2. Measurements of antibiotic susceptibility for S. oneidensis strains.
A Inhibitory profiles of ciprofloxacin against MR-1 and ∆mexF. Symbols represent the mean ± s.d. of biological replicates (n = 5). Solid lines represent a fitted Gompertz function for MIC calculations. Text box shows the calculated MIC values. B Growth of MR-1 and ∆mexF in LB with 0.25 µg/mL ciprofloxacin (CIP) and 5 µg/mL chloramphenicol (CAM). Data represent the mean ± s.d. of biological replicates (n = 3). Inhibitory profiles of ciprofloxacin against MR-1 with the addition of (C) PAßN and (D) NMP. Symbols represent the mean ± s.d. of biological replicates (n = 3). Solid lines represent a fitted Gompertz function for MIC calculations. Text boxes show the calculated MIC values.
Our data suggest that in batch conditions deletion of the mexF gene inhibits cells from overcoming ciprofloxacin-mediated growth limitations. The results also strengthen the notion that the MexR repressor mutation in the SP-2 mutant increased ciprofloxacin resistance. Collectively, the results indicate that the MexEF operon of S. oneidensis plays an important role in susceptibility and resistance to elevated ciprofloxacin concentrations in batch settings.
Chemotaxis toward nitrate is required for habitation and enhanced nitrate reduction in toxic regions of the ciprofloxacin concentration gradient in the MGC
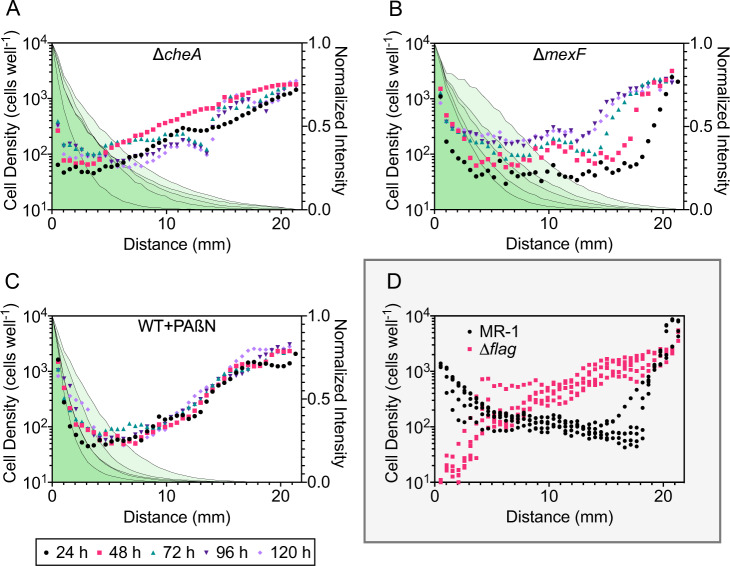
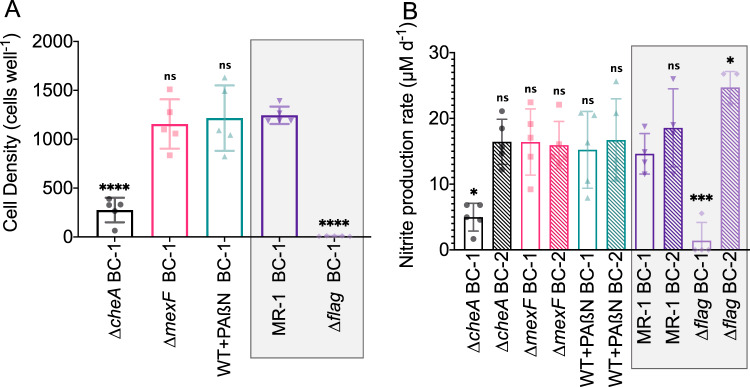
We directly tested the role of chemotaxis in facilitating habitation of toxic regions of the MGC by evaluating and comparing the cell density distributions of the ∆cheA mutant with our previous MGC study [17] that probed the MR-1 strain and a non-motile mutant strain (∆flag). The cell density profiles of the ∆cheA mutant along the ciprofloxacin gradient were inversely proportional to the antibiotic concentration throughout the duration of the experiment (Fig. 3A). These results were similar to the ∆flag mutant cell profiles, and contrasted MR-1 cell profiles that displayed high cell densities near the toxic boundary (Fig. 3D). The cell density at wells adjacent to the ciprofloxacin-rich boundary (BC-1) was significantly lower for ∆cheA mutant experiments in relation to MR-1 experiments (fourfold difference, P < 0.0001, Fig. 4A) and visually noticeable in the reactor (Fig. 5). Despite lower cell densities for ∆cheA adjacent to BC-1, cell densities for this mutant adjacent to BC-1 were still higher than for the ∆flag mutant (Fig. 4A). This may be due to effects of random motility and funneling aggregation overtime due to the geometric configuration of the MGC [59]. The MGC-extracted ∆cheA mutant cells did not exhibit increased ciprofloxacin resistance (Fig. S7A,B) indicating mutational resistance did not play a role in the observed response.
Fig. 3. Characterization of habitability in the MGC under various experimental conditions during exposure to a ciprofloxacin concentration gradient.
Cell density profiles along the ciprofloxacin concentration gradient in the MGC for (A) ∆cheA, (B) ∆mexF, and (C) MR-1 with the addition of 200 µg/mL PAßN (WT + PAßN). The origin of the horizontal axis represents the antibiotic-rich boundary channel (BC-1). Symbols represent the mean cell density of separate regions perpendicular to the gradient formation at 24 h intervals for 120 h (n = 4). Gray lines with shaded green fill represent the normalized mean fluorescence intensity of separate regions perpendicular to the gradient formation at 24 h intervals for 120 h (n = 4). Fluorescence intensity was normalized to the mean maximum and minimum intensity of either boundary channel at each time interval. Legend in bottom left corner is associated with (A, B, and C). D Cell density profiles along the ciprofloxacin concentration gradient in the MGC for MR-1 and ∆flag. Gray box indicates data reproduced from Alcalde et al. (2019). Data represent the mean cell density of separate regions perpendicular to the gradient formation at 24 h intervals for 120 h (n = 4). Time intervals are not resolved in (D). Independent replicate experiment data are in Fig. S5.
Fig. 4. Characterization of metabolic activity in the MGC under various experimental conditions during exposure to a ciprofloxacin concentration gradient.
A Cell density at wells adjacent to the antibiotic-rich boundary (BC-1) for experiments conducted with ∆cheA, ∆mexF, MR-1 with the addition of 200 µg/mL PAßN (WT + PAßN), MR-1, and ∆flag. B Nitrite production rates from either boundary channel for experiments conducted with ∆cheA, ∆mexF, MR-1 with the addition of 200 µg/mL PAßN (WT + PAßN), MR-1, and ∆flag. Antibiotic-rich boundary channel (BC-1) and antibiotic-free boundary channel (BC-2) are depicted by clear and fill bar patterns, respectively. Data for (A) and (B) represent the 5-day mean ± s.d. of daily cell counts and daily nitrite production, respectively (n = 5). Results of one-way ANOVA with Dunnett’s multiple comparisons test with MR-1 BC-1 as the control column are denoted as follows: ns, not significant; *P ≤ 0.05; ***P ≤ 0.001; ****P ≤ 0.0001. Gray boxes indicate data reproduced from Alcalde et al. (2019). Independent replicate experiment data are in Fig. S6.
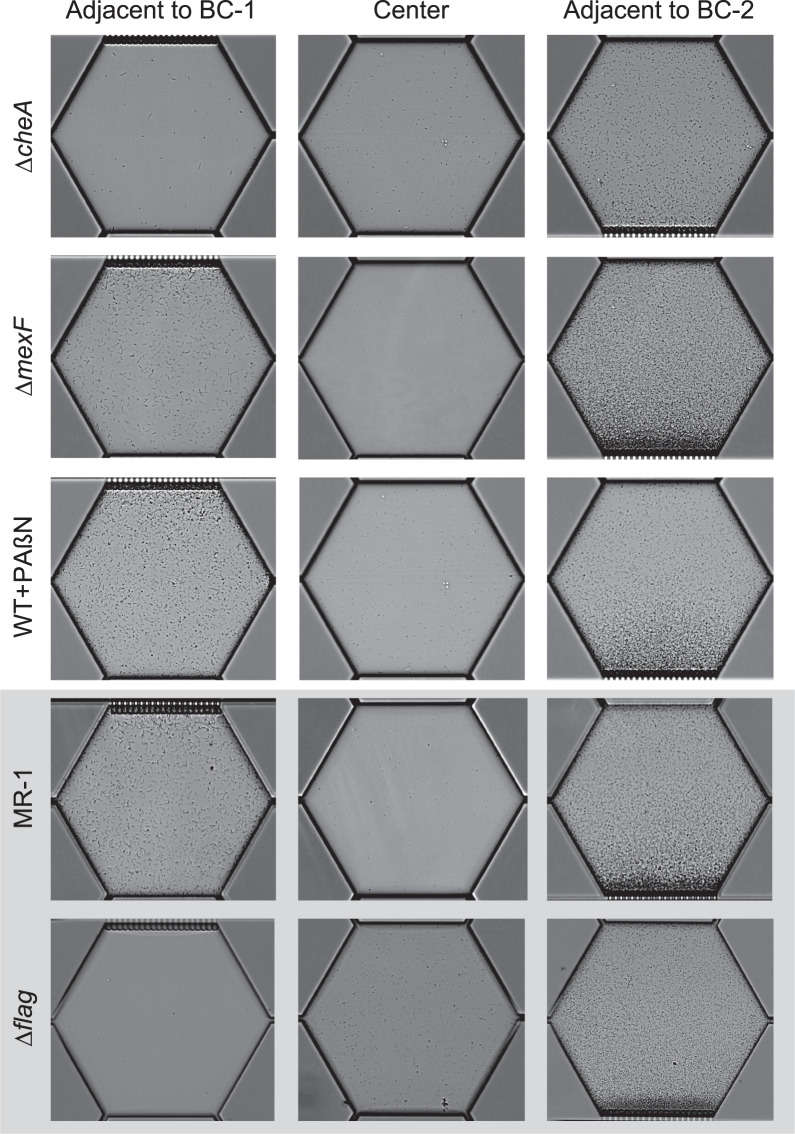
Fig. 5. Representative single well microscopy images of MGC experiments at 24 h.
Single well images of ∆cheA, ∆mexF, and WT + PAßN are presented. MR-1 and ∆flag images are reproduced from Alcalde et al., (2019). Note the lack of cell accumulation at BC-2 for ∆cheA due to the lack of chemotaxis. Some cells adjacent to BC-1 undergo cell elongation, a concept addressed in our previous work (Alcalde et al., (2019)) but not here. Overtime cells in wells adjacent to BC-2 accumulate to a point where single-cell counts are not possible. These data points are removed from Fig. 3 for clarity.
We next examined how respiratory nitrate reduction, an indicator of metabolic activity, differed between different S. oneidensis strains cultured within the MGC. Because the theoretical maximum conversion of the nitrate boundary concentration for a hydraulic-retention-time interval is 2.2% or ~44 µM (Note S10, Fig. S8), the most reliable measurement for quantifying metabolic activity is the production of nitrite, which has no initial background concentration, unlike nitrate (2 mM) and ammonium (0.5 mM). The 5-day mean nitrite production rates for ∆cheA mutant experiments are shown in Fig. 4B. Nitrate reduction rates in the antibiotic-free boundary (BC-2) for ∆cheA mutant cells were comparable to that of the MR-1 cells, while rates at the antibiotic-rich boundary (BC-1) were significantly slower (~threefold difference, P < 0.05, Fig. 4B).
Overall, cell density profiles, mean cell densities at wells adjacent to BC-1, and nitrate reduction rates are in agreement with each other for each of the tested strains (MR-1, ∆flag, and ∆cheA). The results demonstrate that indeed chemotaxis, not random motility, modulated habitation and, subsequently, enhanced nitrate reduction in toxic regions of the MGC.
MexEF efflux pumps are not required for habitation and enhanced nitrate reduction in toxic regions of the ciprofloxacin concentration gradient in the MGC
The ∆mexF mutant was evaluated in the MGC to test the role of MexEF pumps. Spatially, the ∆mexF cell density profiles were similar to those of MR-1 cells throughout the duration of the experiment (Fig. 3B, D). The mean cell density values of MR-1 and ∆mexF cultures in wells adjacent to the ciprofloxacin boundary support this observation, showing no significant difference between the strains (Figs. 4A and 5). Hence, like MR-1, the ∆mexF mutant was able to inhabit the toxic regions of the MGC. Notably, however, there was a temporal variability in the ∆mexF cell density profiles that was not observed in the MR-1 MGC experiments. The variability may be due to the deletion of the mexF gene, but this is difficult to interpret from our data resolution and not addressed further.
Nitrite concentrations from daily effluent samples collected from either boundary channel for ∆mexF mutant experiments are shown in Fig. 4B. The data show that deletion of mexF did not affect metabolic activity in toxic regions of the MGC. The mean nitrite production rates of ∆mexF mutant cells were not significantly different when comparing BC-1 to BC-2, nor were they significantly different to that of MR-1 cells at BC-1. The MIC for cells extracted at the conclusion of the ∆mexF MGC experiments did not differ (Fig. S7C, D), inferring that mutational resistance did not develop. Furthermore, PCR analysis of the mexF locus from extracted cells confirmed that the ∆mexF mutant strain was the only population present in the MGC (Fig. S9).
In summary, analysis of the MGC cell densities, boundary channel effluent, and cell extractions indicate that MR-1 MexEF efflux pumps are not required for successful habitation and sustained metabolic activity in toxic regions of the MGC. These results negate our initial hypothesis that suggested MR-1 was transiently expressing MexEF efflux pumps as a form of adaptive resistance to maintain cell viability and metabolic activity in the MGC.
Saturation of the MGC with the efflux pump inhibitor, PAßN, does not alter the ability of MR-1 to inhabit and reduce nitrate in toxic regions of the ciprofloxacin concentration gradient in the MGC
MDR efflux pumps can be redundant and unspecific in bacteria [60]. Therefore, it is possible that other homologous RND efflux pumps (Table S6) capable of substituting MexEF allowed for the apparent adaptive resistance in the MGC. In batch culture conditions, we exposed MR-1 to various concentrations of two well-characterized RND EPIs, PAßN and NMP [61, 62], to determine their efficacies in increasing ciprofloxacin susceptibility. An increase in susceptibility would suggest that the level of intrinsic resistance by constitutively expressed efflux pumps was decreased [61]. Both PAßN and NMP were able to decrease the MIC by 33%, when exposed to 200 µg/mL PAßN or 100 µg/mL NMP (Fig. 2C, D). PAßN was selected for the MGC experiments because NMP inhibited growth when ciprofloxacin was not present (Fig. S2).
We conducted MGC experiments with MR-1 cells and the addition of 200 µg/mL PAßN in both boundary channels (WT + PAßN). Cell density profiles for WT + PAßN were similar to both the MR-1 and ∆mexF MGC experiments (Fig. 3C, D). The mean cell density value at wells near BC-1 indicated that there was no significant difference between WT + PAßN, MR-1, and ∆mexF cultures (Figs. 4A and 5). Analysis of the boundary channel effluent showed that the combination of ciprofloxacin and PAßN did not have a significant effect on daily nitrite production rates (Fig. 4B). Similar to the ∆mexF MGC experiments, the cells extracted from the WT + PAßN MGC experiment had no increase in the MIC value (Fig. S7E).
These results show that the addition of 200 µg/mL PAßN does not affect the ability of MR-1 cells to successfully inhabit and sustain metabolic activity in the toxic regions of the MGC. They also support the notion that RND efflux pumps may not be a mechanism of adaptive resistance deployed by MR-1 cells for survival in the MGC.
Discussion
Artificial microfluidic microbial ecosystems show great potential in unraveling population dynamics in heterogeneous landscapes. These systems have begun to elucidate unique population dynamics during exposure to antibiotic gradients [17, 18, 22, 23, 63]. In this study, we used a microfluidic reactor (i.e., MGC) that generates antibiotic diffusive gradients to probe the role of chemotaxis and RND efflux pumps of S. oneidensis on the efficacy to inhabit and sustain metabolic activity in ciprofloxacin-rich regions of the MGC.
Our previous work, along with the work of others, has shown the importance of swimming motility, swarming motility, cell migration, and threshold cell densities on the ability to occupy antibiotic-rich microenvironments [17–19, 22–24]. We expanded on this literature by confirming, with a chemotactic deficient mutant (∆cheA) that chemotaxis toward chemoattractants (e.g., nitrate) is required for successful habitation of and metabolic activity in antibiotic-rich zones along antibiotic concentration gradients. Moreover, due to a lack of observed mutational resistance, we attribute chemotaxis as an element that enables adaptive resistance. Although mutational resistance was not observed, others have suggested that adaptive resistance can favor the acquisition of mutational resistance [14, 18, 29]. If so, chemotaxis may also play a role in the development of mutational antibiotic resistance, but that was not observed in our 5-day MGC experiments. Alternatively, continuous chemotactic migration of nonresistant cells may be the reason for the lack of observed mutational resistance. That is, mutant-cell development might be outcompeted because these cells must channel more energy toward cell maintenance (e.g., from overexpression of MDR elements). To this end, mutational resistance has previously developed in the MGC when migration of a nonresistant population was restricted or limited in the MGC (e.g., ∆flag cells or nutrient limitations) as indicated by our previous work [17]. In addition, experiments by Deng et al. (2019), comparing the development of mutational resistance by E. coli in temporal (batch) versus spatial (microfluidic) platforms support this theory [63].
Recognizing that RND efflux pumps are particularly involved in the rise of both adaptive and mutational antibiotic resistance [29, 31, 60], we explored their role on the habitation of toxic regions of the MGC. Interestingly, deletion of mexF and the use EPIs were not inhibiting to cells in the MGC, unlike batch experiments. These results align with previous work that found that removal of various RND efflux pumps did not affect swarming populations of P. aeruginosa [21]. We attribute our response to potential metabolic burdens and fitness costs associated with the expression of RND efflux pumps [64] that are outcompeted by other mechanisms in a configuration like ours. However, it is possible that transcriptional regulators associated with Mar-like proteins in MR-1, which we did not study in the MGC, play an important role and may compensate for the mexF deletion by regulating expression of other MDR elements. For example, MarR (repressor) and MarA (activator) of the marRAB operon in E. coli regulate over 40 downstream genes that not only include efflux pumps but also outer membrane porins [65]. Also, PAßN was used to inhibit homologous RND efflux pumps but there have been no other studies on the efficacy of PAßN to suppress MR-1 RND pumps, only our MIC assays. Confirming EPI suppression of relevant RND efflux pump was beyond the scope of this work. Ideally, we would query a suite of MDR elements in the MGC via gene targeting (e.g., gene deletion, anaerobic reporters) [66]. Therefore, although our data clearly indicate that RND pumps are less critical for cell viability in antibiotic gradients relative to batch conditions, it would be inappropriate to rule them out completely.
Our results challenge current computational population dynamic models that address antibiotic spatial gradients because these models typically assume that migration up gradient requires the development of mutational antibiotic resistance [11, 20]. Although this development has been previously observed in large-scale swimming agar plate and microfluidic reactors [22, 24], we show that it is not required. We believe these computational models necessitate an intimate interplay between rates of migration and rates of mutations that consider fitness costs and adaptive resistance. In the context of terrestrial and aquatic environments, our results suggest that in water-saturated porous networks where antibiotic gradients arise, chemotaxis can provide a competitive advantage for habitability and metabolic activity, while efflux pumps might not. Similarly, this competitive advantage could occur in soil communities where antibiotic gradients are locally generated from antibiotic producing bacteria (e.g., the rhizosphere). This may also help interpret or predict shifts in community structure based on adaptive traits [67]. Our results suggest that in regions of co-contamination with nitrate and antibiotics (e.g., agricultural settings), chemotactic bacteria may be able to reduce nitrate without the development of mutational resistance. It is worth noting that the boundary channel concentration of ciprofloxacin we used in the MGC (2.5 µg/mL) is greater than the bulk concentrations reported in most environmental matrices. However, because this boundary represents a source of antibiotic, and sources such as activated sludge have reported concentrations that exceed 5 µg/mL, it is not unrealistic [38]. The use of subinhibitory antibiotic concentrations at the boundaries, along with the use of microbial microcosms, is an area of future work that could be compared to this data.
In closing, we presented a specific scenario where nitrate and antibiotics are present in a heterogeneous landscape. We determined that chemotaxis, but not RND efflux pumps, facilitated adaptive antibiotic resistance of S. oneidensis MR-1 and, subsequently, enhanced nitrate reduction. Although our focus was narrow, adaptive resistance to chemical stressor gradients is diverse and we suspect that these mechanisms extend beyond antibiotics, for example, in response to gradients of organic and inorganic contaminants (chlorophenols, mercuric chloride, chloroacetophenone) or oxygen to obligate anaerobes (sulfate reducing bacteria). Future work could focus on addressing other underlying mechanisms that allow for adaptive resistance under gradient conditions, such as the role of quorum sensing molecules and porins, or the interplay between adaptive and mutational resistance as it relates to rates of cell migration, population densities, and nutrient availability. Lastly, we recognize this is a reductionist approach for addressing the complex interactions that occur at the pore scale, but notwithstanding, these interactions can inevitably drive large-scale dynamics. With this in mind, we challenge future microfluidic work to draw connections between scales (e.g., micro, meso, macro) for comprehensive interpretation and engineered applicability.
Supplementary information
Acknowledgements
This work was supported by the National Aeronautics and Space Administration (NASA) through the NASA Astrobiology Institute under Cooperative Agreement No. NNA13AA91A issued through the Science Mission Directorate. This work was also supported by the National Science Foundation Graduate Research Fellowship Program (NSF GRFP) under Grant No. DGE-1610403 to REA. Any opinion, findings, and conclusions or recommendations expressed in this work are those of the authors and do not necessarily reflect the views of the NASA Astrobiology Institute and the National Science Foundation. We acknowledge the W. M. Keck Center at UIUC and the ICMB Core Facilities at UT Austin for performing DNA sequencing. We thank Dr. Christopher Fields for bioinformatics support. Lastly, we thank Dr. Mandy J. Ward for generously gifting the ∆cheA-3 mutant.
Compliance with ethical standards
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-021-00975-1.
References
- 1.DeVries SL, Zhang P. Antibiotics and the Terrestrial Nitrogen Cycle: a review. Curr Pollut Rep. 2016;2:51–67. doi: 10.1007/s40726-016-0027-3. [DOI] [Google Scholar]
- 2.Kümmerer K. Antibiotics in the aquatic environment – A review – Part I. Chemosphere. 2009;75:417–34. doi: 10.1016/j.chemosphere.2008.11.086. [DOI] [PubMed] [Google Scholar]
- 3.Franklin AM, Aga DS, Cytryn E, Durso LM, McLain JE, Pruden A, et al. Antibiotics in Agroecosystems: introduction to the Special Section. J Environ Qual. 2016;45:377–93. doi: 10.2134/jeq2016.01.0023. [DOI] [PubMed] [Google Scholar]
- 4.Roose-Amsaleg C, Laverman AM. Do antibiotics have environmental side-effects? Impact of synthetic antibiotics on biogeochemical processes. Environ Sci Pollut Res. 2016;23:4000–12. doi: 10.1007/s11356-015-4943-3. [DOI] [PubMed] [Google Scholar]
- 5.Grenni P, Ancona V, Barra, Caracciolo A. Ecological effects of antibiotics on natural ecosystems: a review. Microchemical J. 2018;136:25–39. doi: 10.1016/j.microc.2017.02.006. [DOI] [Google Scholar]
- 6.Chee-Sanford JC, Mackie RI, Koike S, Krapac IG, Lin Y-F, Yannarell AC, et al. Fate and Transport of Antibiotic Residues and Antibiotic Resistance Genes following Land Application of Manure Waste. J Environ Qual. 2009;38:1086. doi: 10.2134/jeq2008.0128. [DOI] [PubMed] [Google Scholar]
- 7.Mehrtens A, Licha T, Broers HP, Burke V. Tracing veterinary antibiotics in the subsurface – A long-term field experiment with spiked manure. Environ Pollut. 2020;265:114930. doi: 10.1016/j.envpol.2020.114930. [DOI] [PubMed] [Google Scholar]
- 8.Kivits T, Broers HP, Beeltje H, van Vliet M, Griffioen J. Presence and fate of veterinary antibiotics in age-dated groundwater in areas with intensive livestock farming. Environ Pollut. 2018;241:988–98. doi: 10.1016/j.envpol.2018.05.085. [DOI] [PubMed] [Google Scholar]
- 9.Gros M, Mas-Pla J, Boy-Roura M, Geli I, Domingo F, Petrović M. Veterinary pharmaceuticals and antibiotics in manure and slurry and their fate in amended agricultural soils: Findings from an experimental field site (Baix Empordà, NE Catalonia) Sci Total Environ. 2019;654:1337–49. doi: 10.1016/j.scitotenv.2018.11.061. [DOI] [PubMed] [Google Scholar]
- 10.Baquero F, Negri M-C. Challenges: selective compartments for resistant microorganisms in antibiotic gradients. BioEssays. 1997;19:731–6. doi: 10.1002/bies.950190814. [DOI] [PubMed] [Google Scholar]
- 11.Hermsen R, Deris JB, Hwa T. On the rapidity of antibiotic resistance evolution facilitated by a concentration gradient. Proc Natl Acad Sci. 2012;109:10775–80. doi: 10.1073/pnas.1117716109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andersson DI, Hughes D. Microbiological effects of sublethal levels of antibiotics. Nat Rev Microbiol. 2014;12:465–78. doi: 10.1038/nrmicro3270. [DOI] [PubMed] [Google Scholar]
- 13.Levin-Reisman I, Ronin I, Gefen O, Braniss I, Shoresh N, Balaban NQ. Antibiotic tolerance facilitates the evolution of resistance. Science. 2017;355:826–30. doi: 10.1126/science.aaj2191. [DOI] [PubMed] [Google Scholar]
- 14.Cohen NR, Lobritz MA, Collins JJ. Microbial Persistence and the Road to Drug Resistance. Cell Host Microbe. 2013;13:632–42. doi: 10.1016/j.chom.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hughes D, Andersson DI. Environmental and genetic modulation of the phenotypic expression of antibiotic resistance. FEMS Microbiol Rev. 2017;41:374–91. doi: 10.1093/femsre/fux004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Venter H, Arzanlou M, Chai WC, Venter H. Intrinsic, adaptive and acquired antimicrobial resistance in Gram-negative bacteria. Essays Biochem. 2017;61:49–59. doi: 10.1042/EBC20160063. [DOI] [PubMed] [Google Scholar]
- 17.Alcalde RE, Michelson K, Zhou L, Schmitz EV, Deng J, Sanford RA, et al. Motility of Shewanella oneidensis MR-1 Allows for Nitrate Reduction in the Toxic Region of a Ciprofloxacin Concentration Gradient in a Microfluidic Reactor. Environ Sci Technol. 2019;53:2778–87. doi: 10.1021/acs.est.8b04838. [DOI] [PubMed] [Google Scholar]
- 18.Hol FJH, Hubert B, Dekker C, Keymer JE. Density-dependent adaptive resistance allows swimming bacteria to colonize an antibiotic gradient. ISME J. 2016;10:30–38. doi: 10.1038/ismej.2015.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Butler MT, Wang Q, Harshey RM. Cell density and mobility protect swarming bacteria against antibiotics. Proc Natl Acad Sci. 2010;107:3776–81. doi: 10.1073/pnas.0910934107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steel H, Papachristodoulou A. The effect of spatiotemporal antibiotic inhomogeneities on the evolution of resistance. J Theor Biol. 2020;486:110077. doi: 10.1016/j.jtbi.2019.110077. [DOI] [PubMed] [Google Scholar]
- 21.Lai S, Tremblay J, Déziel E. Swarming motility: a multicellular behaviour conferring antimicrobial resistance. Environ Microbiol. 2009;11:126–36. doi: 10.1111/j.1462-2920.2008.01747.x. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Q, Lambert G, Liao D, Kim H, Robin K, Tung C-k, et al. Acceleration of Emergence of Bacterial Antibiotic Resistance in Connected Microenvironments. Science. 2011;333:1764–7. doi: 10.1126/science.1208747. [DOI] [PubMed] [Google Scholar]
- 23.Wu A, Loutherback K, Lambert G, Estevez-Salmeron L, Tlsty TD, Austin RH, et al. Cell motility and drug gradients in the emergence of resistance to chemotherapy. Proc Natl Acad Sci. 2013;110:16103–8. doi: 10.1073/pnas.1314385110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baym M, Lieberman TD, Kelsic ED, Chait R, Gross R, Yelin I, et al. Spatiotemporal microbial evolution on antibiotic landscapes. Science. 2016;353:1147–51. doi: 10.1126/science.aag0822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alexandre G, Greer-Phillips S, Zhulin IB. Ecological role of energy taxis in microorganisms. FEMS Microbiol Rev. 2004;28:113–26. doi: 10.1016/j.femsre.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 26.Fenchel T. Microbial Behavior in a Heterogeneous World. Science. 2002;296:1068–71. doi: 10.1126/science.1070118. [DOI] [PubMed] [Google Scholar]
- 27.Groh JL, Luo Q, Ballard JD, Krumholz LR. Genes That Enhance the Ecological Fitness of Shewanella oneidensis MR-1 in Sediments Reveal the Value of Antibiotic Resistance. Appl Environ Microbiol. 2007;73:492–8. doi: 10.1128/AEM.01086-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blair JM, Piddock LJ. Structure, function and inhibition of RND efflux pumps in Gram-negative bacteria: an update. Curr Opin Microbiol. 2009;12:512–9. doi: 10.1016/j.mib.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 29.Fernández L, Hancock REW. Adaptive and mutational resistance: role of porins and efflux pumps in drug resistance. Clin Microbiol Rev. 2012;25:661–81. doi: 10.1128/CMR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alvarez-Ortega C, Olivares J, Martinez JL. RND multidrug efflux pumps: what are they good for? Front Microbiol. 2013;4:7. doi: 10.3389/fmicb.2013.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anes J, McCusker MP, Fanning S, Martins M. The ins and outs of RND efflux pumps in Escherichia coli. Front Microbiol. 2015;6:587. doi: 10.3389/fmicb.2015.00587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma D, Alberti M, Lynch C, Nikaido H, Hearst JE. The local repressor AcrR plays a modulating role in the regulation of acrAB genes of Escherichia coli by global stress signals. Mol Microbiol. 1996;19:101–12. doi: 10.1046/j.1365-2958.1996.357881.x. [DOI] [PubMed] [Google Scholar]
- 33.Nies DH. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol Rev. 2003;27:313–39. doi: 10.1016/S0168-6445(03)00048-2. [DOI] [PubMed] [Google Scholar]
- 34.Fraud S, Poole K. Oxidative Stress Induction of the MexXY Multidrug Efflux Genes and Promotion of Aminoglycoside Resistance Development in Pseudomonas aeruginosa. Antimicrobial Agents Chemother. 2011;55:1068–74. doi: 10.1128/AAC.01495-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.El Garch F, Lismond A, Piddock LJV, Courvalin P, Tulkens PM, Van Bambeke F. Fluoroquinolones induce the expression of patA and patB, which encode ABC efflux pumps in Streptococcus pneumoniae. J Antimicrob Chemother. 2010;65:2076–82. doi: 10.1093/jac/dkq287. [DOI] [PubMed] [Google Scholar]
- 36.Zhang L, Mah T-F. Involvement of a Novel Efflux System in Biofilm-Specific Resistance to Antibiotics. J Bacteriol. 2008;190:4447–52. doi: 10.1128/JB.01655-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.El Meouche I, Siu Y, Dunlop MJ. Stochastic expression of a multiple antibiotic resistance activator confers transient resistance in single cells. Scientific Rep. 2016;6:1–9. doi: 10.1038/s41598-016-0001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frade VMF, Dias M, Teixeira ACSC, Palma MSA, Frade VMF, Dias M, et al. Environmental contamination by fluoroquinolones. Braz J Pharm Sci. 2014;50:41–54. doi: 10.1590/S1984-82502011000100004. [DOI] [Google Scholar]
- 39.Riaz L, Mahmood T, Yang Q, Coyne MS, D’Angelo E. Bacteria-assisted removal of fluoroquinolones from wheat rhizospheres in an agricultural soil. Chemosphere. 2019;226:8–16. doi: 10.1016/j.chemosphere.2019.03.081. [DOI] [PubMed] [Google Scholar]
- 40.Llanes C, Köhler T, Patry I, Dehecq B, Delden C, van, Plésiat P. Role of the MexEF-OprN Efflux System in Low-Level Resistance of Pseudomonas aeruginosa to Ciprofloxacin. Antimicrobial Agents Chemother. 2011;55:5676–84. doi: 10.1128/AAC.00101-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deatherage DE, Barrick JE. Identification of mutations in laboratory evolved microbes from next-generation sequencing data using breseq. Methods Mol Biol. 2014;1151:165–88. doi: 10.1007/978-1-4939-0554-6_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saltikov CW, Newman DK. Genetic identification of a respiratory arsenate reductase. PNAS. 2003;100:10983–8. doi: 10.1073/pnas.1834303100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Engler C, Kandzia R, Marillonnet S. A One Pot, One Step, Precision Cloning Method with High Throughput Capability. PLOS ONE. 2008;3:e3647. doi: 10.1371/journal.pone.0003647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.European Committee for Antimicrobial Susceptibility Testing (EUCAST) of the European Society of Clinical Microbiology and Infectious Diseases (ESCMID) Determination of minimum inhibitory concentrations (MICs) of antibacterial agents by broth dilution. Clin Microbiol Infect. 2003;9:ix–xv. doi: 10.1046/j.1469-0691.2003.00790.x. [DOI] [PubMed] [Google Scholar]
- 46.Lambert RJ, Pearson J. Susceptibility testing: accurate and reproducible minimum inhibitory concentration (MIC) and non-inhibitory concentration (NIC) values. J Appl Microbiol. 2000;88:784–90. doi: 10.1046/j.1365-2672.2000.01017.x. [DOI] [PubMed] [Google Scholar]
- 47.Sonnet P, Izard D, Mullié C. Prevalence of efflux-mediated ciprofloxacin and levofloxacin resistance in recent clinical isolates of Pseudomonas aeruginosa and its reversal by the efflux pump inhibitors 1-(1-naphthylmethyl)-piperazine and phenylalanine-arginine-β-naphthylamide. Int J Antimicrobial Agents. 2012;39:77–80. doi: 10.1016/j.ijantimicag.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 48.Lindgren PK, Karlsson Å, Hughes D. Mutation Rate and Evolution of Fluoroquinolone Resistance in Escherichia coli Isolates from Patients with Urinary Tract Infections. Antimicrobial Agents Chemother. 2003;47:3222–32. doi: 10.1128/AAC.47.10.3222-3232.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klaus W, Ross A, Gsell B, Senn H. Backbone resonance assignment of the N-terminal 24 kDa fragment of the gyrase B subunit from S. aureus complexed with novobiocin. J Biomol NMR. 2000;16:357–8. doi: 10.1023/A:1008383508219. [DOI] [PubMed] [Google Scholar]
- 50.Müller RT, Pos KM. The assembly and disassembly of the AcrAB-TolC three-component multidrug efflux pump. Biol Chem. 2015;396:1083–9. doi: 10.1515/hsz-2015-0150. [DOI] [PubMed] [Google Scholar]
- 51.Oethinger M, Podglajen I, Kern WV, Levy SB. Overexpression of the marA or soxS Regulatory Gene in Clinical Topoisomerase Mutants of Escherichia coli. Antimicrob Agents Chemother. 1998;42:2089–94. doi: 10.1128/AAC.42.8.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Praski Alzrigat L, Huseby DL, Brandis G, Hughes D. Fitness cost constrains the spectrum of marR mutations in ciprofloxacin-resistant Escherichia coli. J Antimicrob Chemother. 2017;72:3016–24. doi: 10.1093/jac/dkx270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Srikumar R, Paul CJ, Poole K. Influence of Mutations in the mexR Repressor Gene on Expression of the MexA-MexB-OprM Multidrug Efflux System of Pseudomonas aeruginosa. J Bacteriol. 2000;182:1410–4. doi: 10.1128/JB.182.5.1410-1414.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sánchez P, Rojo F, Martínez JL. Transcriptional regulation of mexR, the repressor of Pseudomonas aeruginosa mexAB-oprM multidrug efflux pump. FEMS Microbiol Lett. 2002;207:63–68. doi: 10.1111/j.1574-6968.2002.tb11029.x. [DOI] [PubMed] [Google Scholar]
- 55.Fukuda H, Hosaka M, Hirai K, Iyobe S. New norfloxacin resistance gene in Pseudomonas aeruginosa PAO. Antimicrobial Agents Chemother. 1990;34:1757–61. doi: 10.1128/AAC.34.9.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fukuda H, Hosaka M, Iyobe S, Gotoh N, Nishino T, Hirai K. nfxC-type quinolone resistance in a clinical isolate of Pseudomonas aeruginosa. Antimicrobial Agents Chemother. 1995;39:790–2. doi: 10.1128/AAC.39.3.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fetar H, Gilmour C, Klinoski R, Daigle DM, Dean CR, Poole K. mexEF-oprN Multidrug Efflux Operon of Pseudomonas aeruginosa: Regulation by the MexT Activator in Response to Nitrosative Stress and Chloramphenicol. Antimicrobial Agents Chemother. 2011;55:508–14. doi: 10.1128/AAC.00830-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Köhler T, Michea-Hamzehpour M, Plesiat P, Kahr AL, Pechere JC. Differential selection of multidrug efflux systems by quinolones in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1997;41:2540–3. doi: 10.1128/AAC.41.11.2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Galajda P, Keymer J, Dalland J, Park S, Kou S, Austin R. Funnel ratchets in biology at low Reynolds number: choanotaxis. J Mod Opt. 2008;55:3413–22. doi: 10.1080/09500340802495826. [DOI] [Google Scholar]
- 60.Alcalde-Rico M, Hernando-Amado S, Blanco P, Martínez JL. Multidrug Efflux Pumps at the Crossroad between Antibiotic Resistance and Bacterial Virulence. Front Microbiol. 2016;7:1483. doi: 10.3389/fmicb.2016.01483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lomovskaya O, Warren MS, Lee A, Galazzo J, Fronko R, Lee M, et al. Identification and Characterization of Inhibitors of Multidrug Resistance Efflux Pumps in Pseudomonas aeruginosa: novel Agents for Combination Therapy. Antimicrobial Agents Chemother. 2001;45:105–16. doi: 10.1128/AAC.45.1.105-116.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pannek S, Higgins PG, Steinke P, Jonas D, Akova M, Bohnert JA, et al. Multidrug efflux inhibition in Acinetobacter baumannii: comparison between 1-(1-naphthylmethyl)-piperazine and phenyl-arginine-beta-naphthylamide. J Antimicrob Chemother. 2006;57:970–4. doi: 10.1093/jac/dkl081. [DOI] [PubMed] [Google Scholar]
- 63.Deng J, Zhou L, Sanford RA, Shechtman LA, Dong Y, Alcalde RE, et al. Adaptive Evolution of Escherichia coli to Ciprofloxacin in Controlled Stress Environments: Contrasting Patterns of Resistance in Spatially Varying versus Uniformly Mixed Concentration Conditions. Environ Sci Technol. 2019;53:7996–8005. doi: 10.1021/acs.est.9b00881. [DOI] [PubMed] [Google Scholar]
- 64.Olivares J, Álvarez-Ortega C, Martinez JL. Metabolic Compensation of Fitness Costs Associated with Overexpression of the Multidrug Efflux Pump MexEF-OprN in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2014;58:3904–13. doi: 10.1128/AAC.00121-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Barbosa TM, Levy SB. The impact of antibiotic use on resistance development and persistence. Drug Resist Updates. 2000;3:303–11. doi: 10.1054/drup.2000.0167. [DOI] [PubMed] [Google Scholar]
- 66.Chia HE, Marsh ENG, Biteen JS. Extending fluorescence microscopy into anaerobic environments. Curr Opin Chem Biol. 2019;51:98–104. doi: 10.1016/j.cbpa.2019.05.008. [DOI] [PubMed] [Google Scholar]
- 67.Laverman AM, Cazier T, Yan C, Roose-Amsaleg C, Petit F, Garnier J, et al. Exposure to vancomycin causes a shift in the microbial community structure without affecting nitrate reduction rates in river sediments. Environ Sci Pollut Res. 2015;22:13702–9. doi: 10.1007/s11356-015-4159-6. [DOI] [PubMed] [Google Scholar]
- 68.Li J, Romine MF, Ward MJ. Identification and analysis of a highly conserved chemotaxis gene cluster in Shewanella species. FEMS Microbiol Lett. 2007;273:180–6. doi: 10.1111/j.1574-6968.2007.00810.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.