Abstract
Background & Aims:
Tumor recurrence is frequent even in intrahepatic cholangiocarcinoma (ICC); and improved strategies are needed to identify patients at highest risk for such recurrence. We performed genome-wide expression profile analyses to discover and validate a gene signature associated with recurrence in patients with ICC.
Approach & Results:
For biomarker discovery, we analyzed genome-wide transcriptomic profiling in ICC tumors from two public datasets (TCGA, n=27 and GSE107943, n=28). We identified an 8-gene panel (BIRC5, CDC20, CDH2, CENPW, JPH1, MAD2L1, NEIL3, and POC1A), which robustly identified patients with recurrence in the discovery (area under the curve [AUC]=0.92) and in-silico validation cohorts (AUC=0.91). We next analyzed 241 specimens from patients with ICC (training cohort: n=64, validation cohort: n=177), followed by Cox proportional hazard regression analysis, to develop an integrated transcriptomic panel and establish a risk-stratification model for recurrence in ICC. We subsequently trained this transcriptomic panel in a clinical cohort (AUC=0.89, 95% confidence intervals [CI]=0.79–0.95), followed by evaluating its performance in an independent validation cohort (AUC=0.86, 95% CI=0.80–0.90). By combining our transcriptomic panel with various clinicopathologic features we established a risk-stratification model which was significantly superior for the identification of recurrence (AUC=0.89, Univariate: hazard ratio [HR]=6.08, 95% CI=3.55–10.41, P<0.01; Multivariate: HR=3.49, 95% CI=1.81–6.71, P<0.01). The risk-stratification model identified potential recurrence in 85% of high-risk patients and non-recurrence in 76% of low-risk patients, which is dramatically superior to currently used pathological features.
Conclusions:
We report a novel transcriptomic signature for risk-stratification and recurrence prediction that is superior to currently used clinicopathological features in patients with ICC.
Keywords: transcriptomic panel, detection biomarker, tissue-based identification, genome-wide gene profiling, risk-classification
INTRODUCTION
Intrahepatic cholangiocarcinoma (ICC) is the second most common primary liver cancer with increasing incidence and mortality worldwide (1–4). Surgical resection still remains the only potentially curative treatment option for patients with ICC according to the National Comprehensive Cancer Network (NCCN) guidelines (5). However, the 5-year survival rates following curative resection are only 20–30%, and the primary reason for such an unfavorable long-term outcome is the high incidence of tumor recurrence, which ranges from 50–70% (6–10). While cancer recurrence occurs frequently, the post-operative treatment options remain arbitrary. The NCCN guidelines recommend several treatment options including post-surgical surveillance alone, adjuvant chemo(radio)therapy in patients with a resectable disease, and enrollment of patients in various exploratory clinical trials. In addition, because of the rarity of this malignancy, presently there are no well-defined randomized or prospective studies comparing survival outcomes in patients subjected to surgery alone or together with adjuvant chemotherapy.
Some of the retrospective studies have demonstrated that postoperative adjuvant chemotherapy could result in improving the overall survival (OS) of patients with ICC (7, 11, 12). 5-Fluorouracil or gemcitabine-based chemotherapy is generally recommended and frequently used for treating high-risk patients who are lymph node metastasis-positive or have a positive surgical margin (13). Previous studies have also indicated the efficacy of adjuvant chemotherapy or chemoradiotherapy in biliary tract cancers following curative resection (14, 15). Finally, some studies have illustrated the therapeutic benefits of various surgical techniques, such as radical lymphadenectomy (16–18) and larger surgical resection margins (6, 19). Nonetheless, there remains a lack of consensus on the benefit of postoperative adjuvant chemotherapy should and which patients with ICC should be offered such treatments following resection of their primary tumor. In the context of surveillance alone, the use of computed tomography (CT) or magnetic resonance imaging (MRI) of abdomen and pelvis, and if needed, CT of chest every 6 months for 2 years is recommended. However, these criteria are inadequate as some patients require a more aggressive surveillance approach and potentially adjuvant chemotherapy as well, because ~40% of patients experience tumor recurrence within 1 year of their surgical treatment (20).
A few studies have investigated the significance of various clinical features (e.g. tumor size, tumor number, margin <10mm, and major hepatectomy) and tumor markers, which could help predict recurrence, following a surgical resection with a curative intent; however, the predictive accuracy of these biomarkers remains inadequate for their clinical use (21, 22). Accumulating evidence indicates that the risk of cancer recurrence generally associates with distinct biological factors indicative of tumor aggressiveness, as well as specific technical factors including a positive surgical margin (7–9). In addition, most of the recurrence events in patients with ICC following hepatic resection occurs in the remnant liver (23). The etiology of tumor recurrence if often attributed to either intrahepatic metastasis resulting from the initial tumor or the emergence of a de novo tumor (23, 24); highlighting the need for improved surveillance and the potential of timely detection of recurrence.
Emerging evidence indicates that the biological behavior of ICC varies among patients and the expression pattern of genes have the potential to reflect the physiological and pathological status in cancer patients (7–9, 25). In fact, several studies have identified that differential expression of specific genes is directly associated with ICC pathogenesis, as well as suggested their potential as disease biomarkers. These findings highlight the imperative need to develop molecular biomarkers that can predict recurrence, and potentially help characterize the treatment response and outcomes in patients with recurrent ICC (7, 17, 26–29). Furthermore, most of these studies were performed in small retrospective cohorts and based on single-institution experience, in which the resection status included a admixture of patients with various cholangiocarcinomas (26, 27, 30).
In the present study we have addressed these important gaps in knowledge, by performing a genome-wide, systematic and comprehensive biomarker discovery effort to identify a gene expression signature for the detection of recurrence following hepatectomy in patients with ICC. The signature was initially identified in multiple gene expression profiling datasets, followed by rigorous validation and performance evaluation in large independent clinical cohorts. Our final risk-stratification model is robust in identification of patients with high-risk ICC, which will improve overall management of patients by sparing the low-risk patients from the unnecessary toxicity and expense associated with adjuvant treatments.
MATERIALS AND METHODS
Biomarker discovery
To perform a comprehensive genome-wide biomarker discovery, we analyzed gene expression profiling results obtained from primary tumor tissues from two datasets (The Cancer Genome Atlas [TCGA] and GSE107943) in order to identify and establish a gene panel for the detection of recurrence in patients with ICC. TCGA gene expression profiling data was downloaded from the University of California–Santa Cruz Xena Browser (https://xenabrowser.net), while GSE107943 dataset (normalized gene profiling and clinical data) was downloaded from the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov). In total, gene expression profiling data was obtained from 55 patients, which included 27 patients from the TCGA cohort (14 with recurrence and 13 without recurrence) and 28 patients from the GSE107943 dataset (20 with recurrence and 8 without recurrence) as illustrated in Supplementary Fig. 1. To evaluate the diagnostic potential of the discovered gene signature, we first established a univariate Cox proportional hazard regression model using the selected biomarkers and subsequently determined the area under the curve (AUC) values for each of the receiver operator characteristic (ROC) plots (Supplementary Fig. 2) (31, 32).
Patient cohorts
For the clinical training and validation of the identified gene panel, we analyzed a total of 241 formalin-fixed paraffin-embedded (FFPE) specimens from three independent ICC patient cohorts, which included a training cohort (n = 64; 37 with recurrence and 27 non-recurrence) of patients enrolled at the Kyushu University, and a validation cohort (n = 177; 109 with recurrence and 68 non-recurrence) enrolled at the Tokushima University and Kumamoto University. All patients underwent radical surgeries between January 2000 and December 2018 in the training cohort, and between January 1994 and December 2018 in the validation cohort. All specimens were diagnosed as ICC by pathologists at each participating institution, according to the 7th edition of the American Joint Committee on Cancer TNM grading system. The follow-up protocol included a physical examination, evaluation of serum carcinoembryonic antigen (CEA) and carbohydrate antigen 19–9 (CA19–9) levels, as well as imaging modalities such as ultrasonography, CT and MRI. Only regions confirmed by imaging modalities were diagnosed as tumor recurrence sites. The study was conducted in accordance with the Declaration of Helsinki. A written informed consent was obtained from all patients, and the study was approved by the institutional review boards of all participating institutions.
RNA extraction and RT-qPCR
Total RNA was isolated from 10 μm thick sections of FFPE surgical tissues by manual microdissection to enrich for cancer cells (>75% of tumor cells), and the RNA was extracted using the AllPrep DNA/RNA FFPE Kit (Qiagen, Hilden, Germany). Synthesis of complementary DNA (cDNA) was conducted with 500ng of total RNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA). Real-time quantitative reverse transcription polymerase chain reaction (RT-qPCR) analysis was performed using the SensiFAST™ Lo-ROX Kit (Bioline, London, UK) on the Quantstudio 7 Flex Real Time PCR System (Applied Biosystems, Foster City, CA), and expression levels were evaluated with Applied Biosystems QuantStudio 7 Flex Real Time PCR System Software. The relative abundance of target transcripts was evaluated and normalized to the expression of β-actin as an internal control using the 2−ΔDCt method. Normalized expression values were log10 transformed (33). The primer sequences for the target genes used in the present study are shown in Supplementary Table 1.
Statistical analysis
Clinicopathologic characteristics of the patient cohorts are shown in Table 1. The cutoff thresholds for continuous variables were divided by the median value in all participants. Several clinicopathologic characteristics were compared between recurrence and non-recurrence group, using the Chi-Square test or Mann-Whitney U test for categorical data. Cox’s proportional hazard regression analysis was used to train a classifier based on the expression of each gene. Of note, once the model was trained (in the training cohort), the same statistical model variables (weights and cutoff thresholds) were applied in the independent validation cohort, to better appreciate the translational potential of our risk-stratification model. The cutoff threshold for the recurrence risk scores was chosen as −0.47, which was determined by Youden’s index. For all cohorts, ROC curves and AUC values were used to evaluate the performance of the panel for recurrence detection in patients with ICC. The OS and relapse-free survival (RFS) times were calculated from the date of surgery to the date of death from any cause or recurrence, or last follow-up date. We estimated OS and RFS using the Kaplan-Meier method. A univariate and multivariate Cox proportional hazard regression model was established and a P value < 0.05 was considered statistically significant. Hazard ratios (HR) were presented with their 95% confidence intervals (CI). Statistical analyses were performed using JMP Genomics V 9.0 statistical software (SAS Institute Japan, Tokyo, Japan), Medcalc statistical software V.17.4.0 (Medcalc Software bvba, Ostend, Belgium), GraphPad Prism V8.0 (GraphPad Software, San Diego, CA, USA), and R (3.5.0, R Development Core Team, https://cran.r-project.org/).
Table 1:
Clinicopathological characteristics of clinical cohorts
| Characteristics | Training cohort (n=64), n (%) | Validation cohort (n=177), n (%) | P | |
|---|---|---|---|---|
| Age (years) | Median (range) | 67 (39–87) | 71 (37–96) | 0.11 |
| Gender | Male | 43 (67.2) | 107 (60.5) | |
| Female | 21 (32.8) | 70 (39.5) | 0.34 | |
| Recurrence status | Recurrence | 37 (57.8) | 109 (61.6) | |
| Non-recurrence | 27 (42.2) | 68 (38.4) | 0.60 | |
| Hepatitis virus infection | Positive | 13 (20.3) | 44 (24.9) | |
| Negative | 51 (79.7) | 133 (75.1) | 0.46 | |
| Liver cirrhosis | Cirrhosis | 22 (34.4) | 60 (33.9) | |
| Non-cirrhosis | 39 (60.9) | 111 (62.7) | ||
| Missing | 3 (4.7) | 6 (3.4) | 0.89 | |
| Tumor location | Perihilar | 17 (26.6) | 67 (37.8) | |
| Peripheral | 47 (73.4) | 110 (62.2) | 0.10 | |
| Tumor size (mm) | ≥35 | 39 (60.9) | 90 (50.9) | |
| <35 | 25 (39.1) | 87 (49.1) | 0.18 | |
| Bile duct type | Small | 49 (76.6) | 119 (67.2) | |
| Large | 15 (23.4) | 58 (32.8) | 0.16 | |
| CEA (ng/ml) | ≥5.0 | 29 (45.3) | 58 (32.8) | |
| <5.0 | 35 (54.7) | 119 (67.2) | 0.08 | |
| CA19–9 (U/ml) | ≥37 | 32 (50.0) | 87 (49.1) | |
| <37 | 32 (50.0) | 90 (50.9) | 0.91 | |
| Vascular invasion | Positive | 29 (45.3) | 90 (50.9) | |
| Negative | 35 (54.7) | 87 (49.1) | 0.42 | |
| Macro type | MF | 51 (79.7) | 120 (67.8) | |
| PI | 13 (14.0) | 57 (32.2) | 0.07 | |
| Adjuvant chemotherapy | Yes | 26 (40.6) | 51 (28.8) | |
| No | 38 (59.4) | 123 (69.5) | ||
| Missing | 0 (0) | 3 (1.7) | 0.10 | |
| Differentiation | Well | 32 (50.0) | 62 (35.0) | |
| Moderately | 28 (43.8) | 79 (44.6) | ||
| Poor | 4 (6.2) | 24 (13.6) | ||
| Others | 0 (0) | 12 (6.8) | 0.16 | |
CA19–9, Carbohydrate antigen 19–9; CEA, Carcinoembryonic antigen; MF, mass forming type; PI, periductal-infiltrative type
RESULTS
Genome-wide expression profiling resulted in the identification of an 8-gene panel for the detection of recurrence in patients with ICC
We performed a genome-wide, unbiased, comprehensive biomarker discovery analysis in two independent gene expression profiling datasets (TCGA and GSE107943) to identify a gene panel for the detection of recurrence in patients with ICC. We first compared the gene expression profiles in patients with and without recurrence in the GSE107943 cohort. This cohort included patients who had undergone curative surgery and identified a panel of differentially expressed candidates (P < 0.05, |log2FC| >1), with data availability in at least 50% of all cases and excluded highly correlated genes. Subsequent validation of these candidates in the TCGA dataset led us to finally identify a panel of 8 genes, that were consistent between both the cohorts. These genes included BIRC5, CDC20, CDH2, CENPW, JPH1, MAD2L1, NEIL3, and POC1A. This 8-gene panel in the GSE107943 resulted in an AUC of 0.92 (95% confidence interval [CI] = 0.75–0.99). (Supplementary Fig. 2A), which was successfully and robustly validated in the TCGA dataset (AUC = 0.91, 95% CI = 0.73–0.98); highlighting the diagnostic performance of this panel for the identification of recurrence in patients with ICC (Supplementary Fig. 2B). To evaluate the prognostic potential of our gene biomarkers, we performed survival analysis for RFS. The ICC patients within the high-risk group demonstrated a significantly worse survival in both cohorts (P < 0.01, Supplementary Fig. 2C, D); yet again, highlighting the clinical significance of our biomarker panel.
Training and validation efforts allowed us to establish a transcriptomic panel for the identification of recurrence in patients with ICC
Next, we wanted to validate this biomarker panel in the clinical cohorts. To this end, we first confirmed that the patients within our training and validation cohorts possessed similar clinicopathologic characteristics. The training cohort comprised of 64 patients, which included 37 patients with recurrence (58%); the median age of patients in this cohort was 67 years. The validation cohort consisted of 177 patients, which included 109 patients with recurrence (62%); the median age of patients in this cohort was 71 years. The detailed clinicopathological characteristics of patients within these cohorts are provided in Table 1. We observed no statistically significant differences in the prevalence of recurrence rates, nor any other clinicopathological characteristics between the two cohorts, eliminating the possibility of any inadvertent bias between the patient cohorts examined in our study.
To confirm the diagnostic robustness of our discovered gene panel, we evaluated its performance in these clinical cohorts. First, we systematically interrogated the diagnostic accuracy of our transcriptomic panel for its ability to detect recurrence in ICC patients within the training cohort (37 with recurrence and 27 non-recurrence). Using the Cox’s proportional hazard regression analysis, we trained a model that allowed robust identification of recurrence in patients with ICC (AUC = 0.89, 95% CI = 0.79–0.95, Fig. 1A, B). We developed this risk-stratification panel based on the coefficients derived from individual markers by using the Cox’s proportional hazard regression analysis using the following model parameters: Logit (P) = (−0.4196*BIRC5) + (−0.6444*CDC20) + (0.06929*CDH2) + (0.06698*CENPW) + (−0.3512*JPH1) + (1.1956*MAD2L1) + (0.06698*NEIL3) + (0.3971*POC1A). Following the encouraging results of our panel for the detection of recurrence in the training cohort, we next evaluated its robustness and accuracy by applying the same statistical model (the coefficients and cutoff thresholds) in an independent validation cohort (with recurrence = 109, non-recurrence = 68). We were enthused to observe that the diagnostic performance of our transcriptomic risk-prediction model in this cohort was quite comparable to that observed in the training cohort (AUC = 0.86, 95% CI = 0.80–0.90; Fig. 1C, D). Taken together, our findings allowed us to successfully establish a novel transcriptomic panel for identification of recurrence in patients with ICC.
Figure 1. Training and validation phase of a transcriptomic panel for the identification of recurrence in patients with ICC.
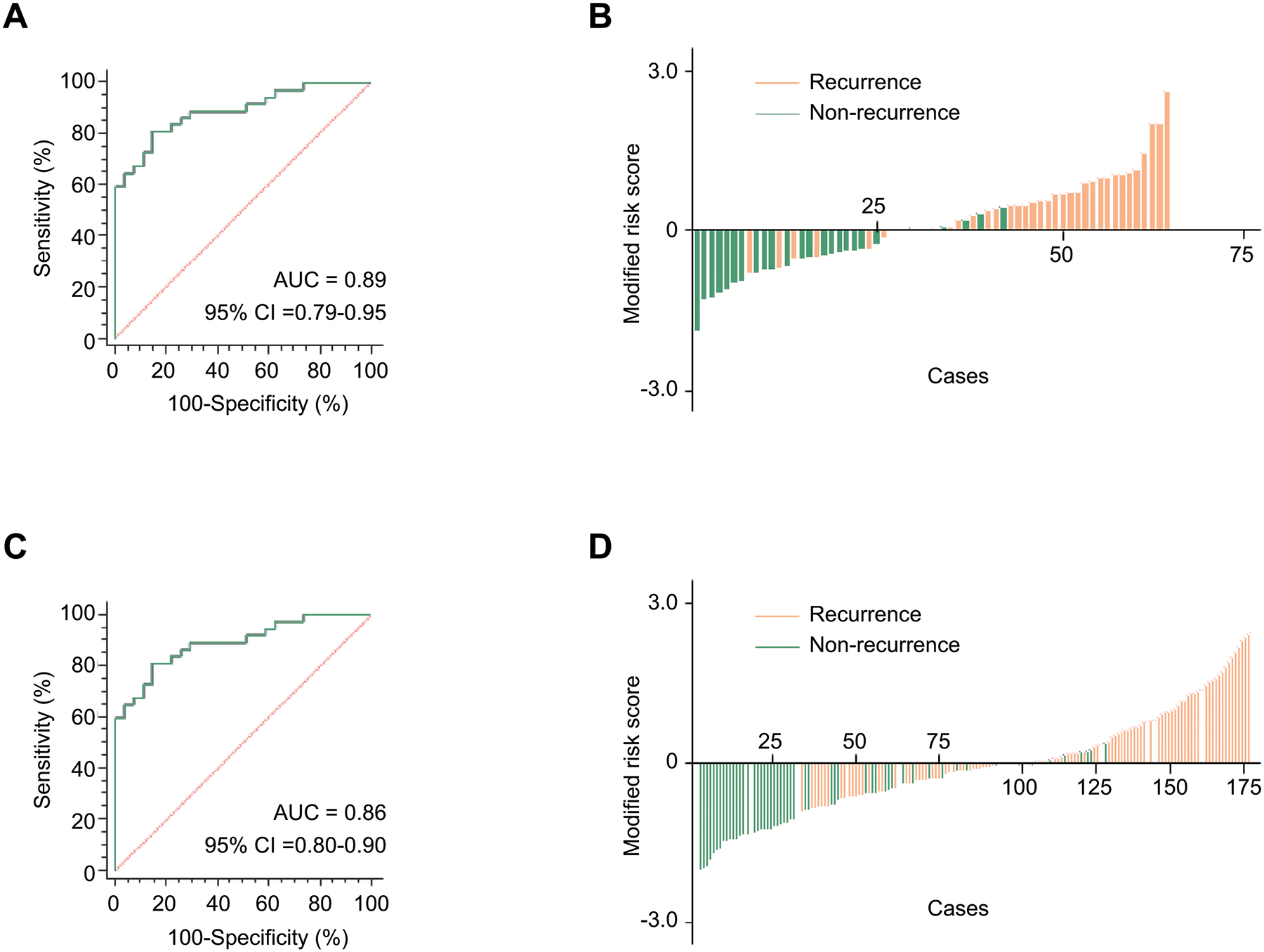
A) A ROC curve for a transcriptomic panel in tissue specimens from training cohort patients (with recurrence = 37, non-recurrence = 27, AUC = 0.89). B) Risk score distribution plot in training cohort patients. Modified risk scores were obtained from individual risk scores by using Youden’s index values from the risk model. C) A ROC curve for the transcriptomic panel in tissue specimens from validation cohort patients (with recurrence = 109, non-recurrence = 68, AUC = 0.86). D) Risk score distribution plot in validation cohort patients.
Establishment of a risk-nomogram for predicting survival outcomes in patients with ICC
Since cancer recurrence is often associated with poor survival in patients with ICC, we were curious to inquire the prognostic potential of our gene panel. In this regard, we performed survival analysis for RFS and OS in the independent patient cohorts. The median follow-up times were 26.1 months (95% CI = 24.7–54.4) in the training cohort and 26.3 months (95% CI = 23.2–44.9) in the validation cohort. More importantly, we observed that based upon our transcriptomic panel derived parameters, patients categorized within the high-risk group demonstrated a significantly worse prognosis in both the training cohort (RFS [P < 0.01]; OS [P < 0.01], Fig. 2A, B) and the validation cohort (RFS [P < 0.01]; OS [P < 0.01], Fig. 2C, D). In addition, for an easier translation of our transcriptomic panel into the clinic, we next evaluated its performance along with other clinicopathological risk features (i.e. CA19–9 or CEA, vascular invasion, tumor size), and established a nomogram. Although we incorporated various pathological and molecular risk features in the risk-nomogram, we noted that while a few pathological risk-stratification features added some weight to the model, our 8-gene panel carried the highest weight in this nomogram and emerged as an independent and the most significant predictor for recurrence in patients with ICC (Fig. 2E).
Figure 2. Prognostic potential of the transcriptomic panel in patients with ICC.
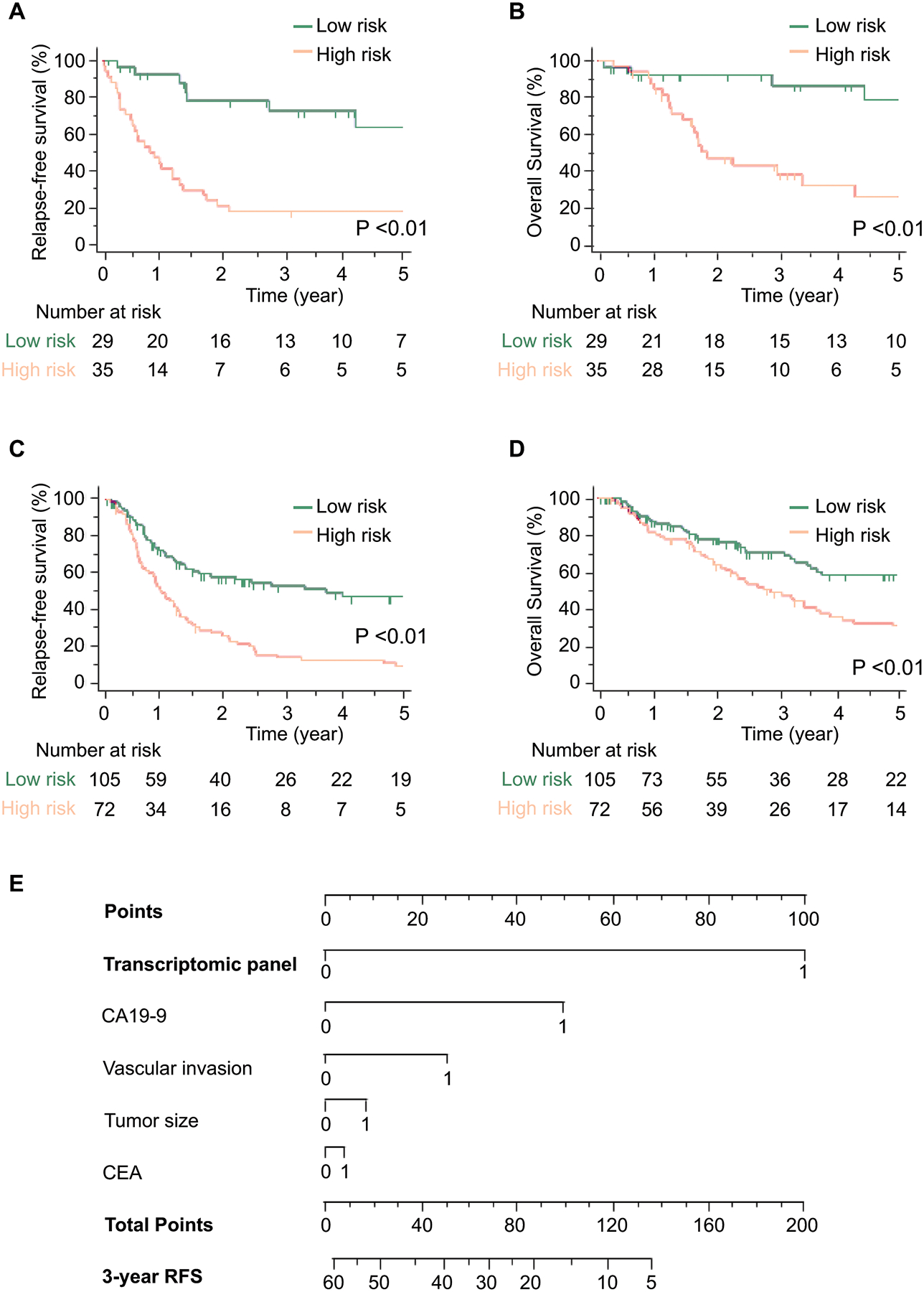
A–B) A comparison of (A) RFS and (B) OS between high and low-risk group estimated by the panel in the training cohort. C–D) A comparison of (C) RFS and (D) OS between high and low-risk group estimated by the panel in the validation cohort. E) A nomogram illustrating the probability of RFS. For clinical purposes, the scores of each covariate are added, and the total score is depicted on the total score point axis.
A risk-stratification model that combines transcriptomic biomarkers and clinicopathological features improves recurrence prediction in patients with ICC
Considering that some of the pathological risk factors currently used in the clinic offer some risk assessment potential in patients with ICC, we asked whether a risk-stratification model that includes some of these risk features along with our transcriptomic panel might further improve its accuracy in detecting recurrence. Interestingly, when we included certain classic clinicopathological features such as CA19–9, CEA, vascular invasion and tumor size into such a risk-stratification model, we observed that indeed this model exhibited a superior diagnostic accuracy compared to the transcriptomic panel and other individual risk features (AUC = 0.89, Fig. 3A).
Figure 3. Clinical validation of the risk-stratification model in patients with ICC.
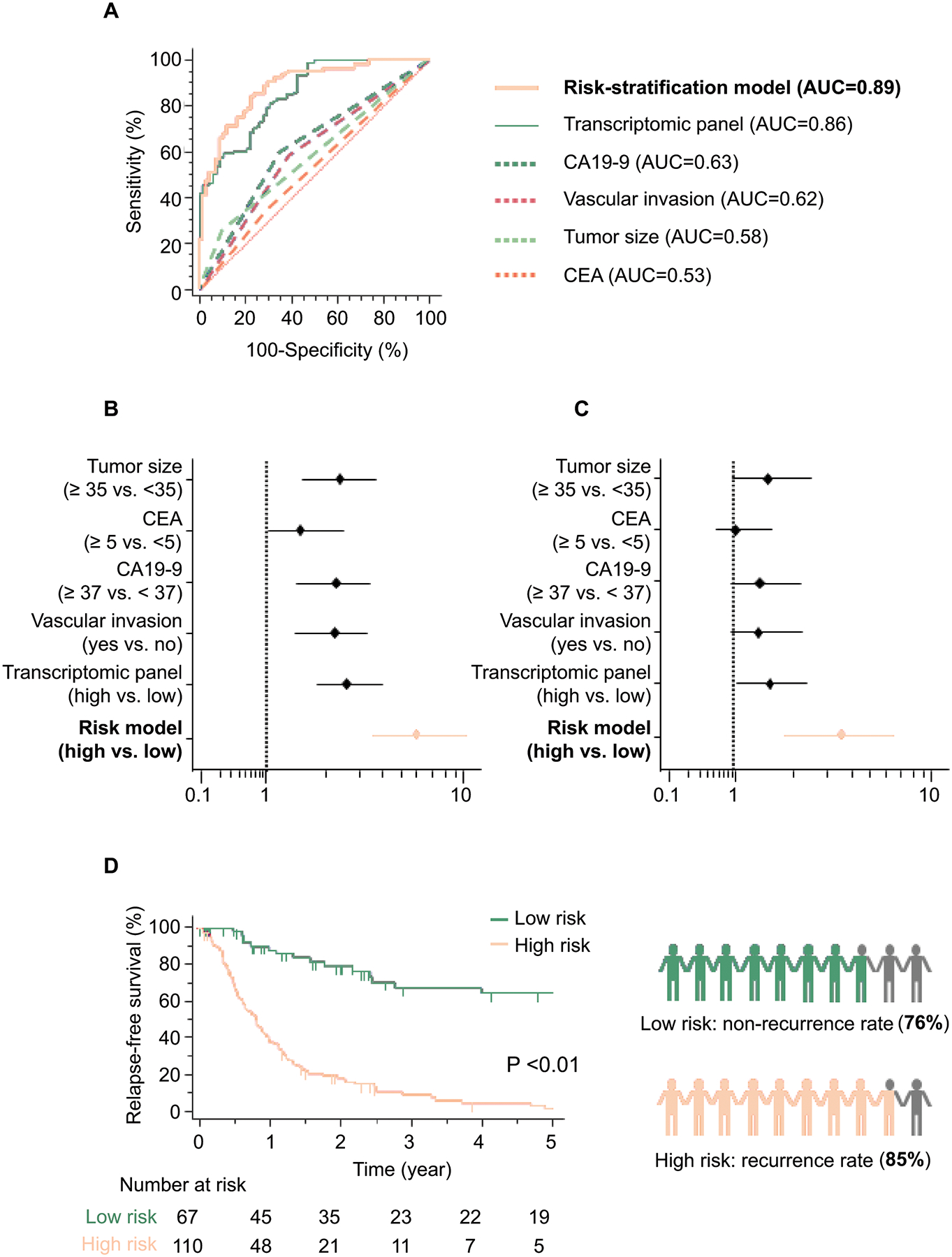
A) The risk-stratification model, which combines the transcriptomic panel and clinical risk factors, outperformed detection accuracy of the transcriptomic panel or risk factors alone in tissue specimens from validation cohort patients (AUC = 0.89). B–C) Forest plot with HRs of clinicopathological variables, transcriptomic panel, and risk-stratification model in univariate (B) and multivariate (C) Cox proportional hazard regression analysis in validation cohort patients. D) A comparison of RFS between high and low-risk group estimated by the risk-stratification model in the validation cohort (left panel). The risk-stratification model would have led to the 5-year recurrence rate of 24% patients with low-risk and 85% patients with high-risk ICC (right panel).
We next determined specific diagnostic correlates for our biomarker panel in the validation cohort. Herein, we observed that its sensitivity, specificity, positive predictive value (PPV), and negative predictive values (NPV) were 79.8%, 70.6%, 81.3%, and 68.6%, respectively (Table 2). When we performed a similar analysis of the newly established risk-stratification model that also included pathological risk features, its performance yet again significantly superior with the sensitivity, specificity, PPV, and NPV values being 85.3%, 76.5%, 85.3%, and 76.5%, respectively. Collectively, this highlights the superiority of our newly developed risk-stratification model for identifying recurrence in patients with ICC.
Table 2:
Model performance in estimating the risk of recurrence
| Variable | Value (95% CI) | ||||
|---|---|---|---|---|---|
| GSE107943 | TCGA | Training cohort | Validation cohort (RNA panel) | Validation cohort (Risk model) | |
| Sensitivity, % | 85.0 (62.1–96.8) | 69.2 (38.6–90.9) | 81.1 (61.7–86.2) | 79.8 (71.1–86.9) | 85.3 (77.3–91.4) |
| Specificity, % | 87.5 (47.3–99.7) | 100.0 (76.8–100.0) | 85.2 (79.0–96.8) | 70.6 (58.3–81.0) | 76.5 (64.6–85.9) |
| AUC, % | 91.9 (75.2–98.8) | 90.7 (73.1–98.4) | 89.0 (78.7–95.4) | 85.5 (79.5–90.4) | 88.6 (82.9–92.9) |
| PPV, % | 94.4 (72.9–99.1) | 100.0 | 88.2 (75.0–94.9) | 81.3 (74.8–86.4) | 85.3 (79.0–90.0) |
| NPV, % | 70.0 (44.3–87.2) | 77.8 (58.7–88.2) | 76.7 (62.3–86.7) | 68.6 (59.3–76.6) | 76.5 (67.0–83.9) |
AUC, area under the curve; CI, confidence interval; NPV, negative predictive value; PPV, positive predictive value; TCGA, The Cancer Genome Atlas
We thereafter performed univariate Cox’s proportional hazard regression analysis, which revealed that our transcriptomic panel still was an independent predictor for recurrence in both clinical cohorts, compared to any singular clinical risk factors (training cohort: Hazard ratio (HR) = 3.22, 95% CI = 1.62–6.42, P < 0.01; validation cohort: HR = 2.73, 95% CI = 1.86–4.01, P < 0.01; Table 3). Furthermore, univariate and multivariate Cox’s proportional hazard regression analysis revealed that our novel risk-stratification model was superior to the transcriptomic panel and various clinicopathological features in terms of predicting recurrence (Univariate: HR = 6.08, 95% CI = 3.55–10.41, P < 0.01; Multivariate: HR = 3.49, 95% CI = 1.81–6.71, P < 0.01) in the patients within the validation cohort (Fig. 3B, C and Table 3). Collectively, these data highlight the potential clinical significance of our risk model in the identification of recurrence in ICC.
Table 3.
Univariate and multivariate Cox proportional hazard regression analysis for recurrence
| Univariate analysis | Multivariate analysis | |||||||
|---|---|---|---|---|---|---|---|---|
| Factors | HR | 95% CI | p | HR | 95% CI | p | ||
| Training cohort (n=64) | ||||||||
| Age | ||||||||
| (≥70 vs. <70) | 1.50 | 0.78–2.89 | 0.22 | |||||
| Gender | ||||||||
| (Male vs. Female) | 0.87 | 0.44–1.72 | 0.70 | |||||
| Hepatitis virus infection | ||||||||
| (Positive vs. Negative) | 0.68 | 0.28–1.64 | 0.39 | |||||
| Liver cirrhosis | ||||||||
| (Cirrhosis vs. Non-cirrhosis) | 1.70 | 0.87–3.34 | 0.12 | |||||
| Tumor location | ||||||||
| (Perihilar vs. Peripheral) | 1.74 | 0.88–3.42 | 0.11 | |||||
| Tumor size | ||||||||
| (≥35mm vs. 35mm) | 1.69 | 0.83–3.43 | 0.15 | |||||
| Bile duct type | ||||||||
| (Large vs. Small) | 2.03 | 1.01–4.06 | 0.05 | 1.53 | 0.74–3.15 | 0.25 | ||
| CEA | ||||||||
| (≥5.0ng/mL vs. <5.0ng/mL) | 1.83 | 0.96–3.49 | 0.07 | 1.80 | 0.93–3.49 | 0.08 | ||
| CA19–9 | ||||||||
| (≥37U/mL) vs. <37U/mL) | 1.79 | 0.93–3.45 | 0.08 | 1.72 | 0.87–3.41 | 0.12 | ||
| Vascular invasion | ||||||||
| (Positive vs. Negative) | 1.90 | 0.99–3.65 | 0.06 | 1.79 | 0.91–3.53 | 0.09 | ||
| Adjuvant chemotherapy | ||||||||
| (Positive vs. Negative) | 0.86 | 0.45–1.67 | 0.67 | |||||
| RNA panel | ||||||||
| (High risk vs. Low risk) | 3.22 | 1.62–6.42 | <0.01 | 2.87 | 1.42–5.81 | <0.01 | ||
| Validation cohort (n=177) | ||||||||
| Age | ||||||||
| (≥70 vs. <70) | 1.09 | 0.74–1.59 | 0.67 | |||||
| Gender | ||||||||
| (Male vs. Female) | 1.24 | 0.88–3.42 | 0.11 | |||||
| Hepatitis virus infection | ||||||||
| (Positive vs. Negative) | 0.70 | 0.45–1.10 | 0.12 | |||||
| Liver cirrhosis | ||||||||
| (Cirrhosis vs. Non-cirrhosis) | 1.28 | 0.86–1.90 | 0.23 | |||||
| Tumor location | ||||||||
| (Perihilar vs. Peripheral) | 1.14 | 0.74–1.76 | 0.54 | |||||
| Tumor size | ||||||||
| (≥35mm vs. 35mm) | 2.35 | 1.59–3.47 | <0.01 | 1.46 | 0.95–2.22 | 0.08 | ||
| Bile duct type | ||||||||
| (Large vs. Small) | 1.37 | 0.93–2.01 | 0.11 | |||||
| CEA | ||||||||
| (≥5.0ng/mL vs. <5.0ng/mL) | 1.57 | 1.03–2.41 | 0.04 | 1.04 | 0.68–1.57 | 0.87 | ||
| CA19–9 | ||||||||
| (≥37U/mL) vs. <37U/mL) | 2.17 | 1.48–3.20 | <0.01 | 1.35 | 0.88–2.08 | 0.17 | ||
| Vascular invasion | ||||||||
| (Positive vs. Negative) | 2.15 | 1.46–3.17 | <0.01 | 1.39 | 0.92–2.10 | 0.12 | ||
| Adjuvant chemotherapy | ||||||||
| (Yes vs. No) | 1.45 | 0.97–2.18 | 0.07 | |||||
| RNA panel | ||||||||
| (High risk vs. Low risk) | 2.73 | 1.86–4.01 | <0.01 | 1.58 | 1.02–2.46 | 0.04 | ||
| Risk model | ||||||||
| (High risk vs. Low risk) | 6.08 | 3.55–10.41 | <0.01 | 3.49 | 1.81–6.71 | <0.01 | ||
CA19–9, Carbohydrate antigen 19–9; CEA, Carcinoembryonic antigen; CI, confidence interval; HR, hazard ratio
Our risk-stratification model allows identification of high-risk ICC patients that have the highest likelihood of benefitting from post-surgical adjuvant chemotherapy
The ultimate translational goal of our study was to determine the clinical usefulness of our risk-stratification model in identifying patients who truly require post-operative therapy and spare the rest from unnecessary treatments. In this regard, we analyzed the patients within the validation cohort using our final risk-stratification classifier and dichotomized all patients into high and low risk groups. The Fig 3D illustrates that a comparison of RFS between high and low-risk groups estimated by the risk-stratification model in the validation cohort (P < 0.01; left panel) exhibited an even more accurate predictive efficacy compared to our transcriptomic panel alone, as it stratified 62% of patients into the high-risk group (110 of 177), and the remaining 38% (67 of 177) of patients were deemed as low risk. Of the 110 patients who were classified as high risk, 93 patients experienced tumor recurrence within 5 years (85%), indicating that these were indeed high-risk patients and will likely benefit from post-treatment adjuvant chemotherapy. Likewise, of the 67 patients who were classified as low risk, 51 patients didn’t experience recurrence within 5 years (76%), yet again emphasizing that these low-risk patients could potentially avoid unnecessary toxicity and expense from adjuvant therapy (Fig. 3D, right panel). These data highlight the potential for the clinical our risk-stratification model in patients with high- and low-risk ICC.
DISCUSSION
The status of tumor recurrence remains one of the most important risk factors for deciding which subset of ICC patients should be offered post-operative adjuvant therapy, while spare the rest from its toxicity and expense. Our present study is a significant step forward in this regard as it helps alleviate some of the inadequacies of currently used clinicopathologic risk features for identifying recurrence in this malignancy. Our data demonstrate that analysis of a transcriptomic gene panel in the primary tissues from ICC patients can accurately estimate the risk probability for recurrence and has a tremendous clinical potential for more robust risk-stratification for the identification of recurrence. Identification of true high-risk patients and saving others from such unnecessary treatments will reduce patient complications, physician burden, and associated healthcare costs (34, 35). Previous data suggest that vascular invasion, CA19–9 levels, and tumor size were associated with advanced tumor characteristics and a higher risk of tumor recurrence (28, 36, 37). In this study, our newly developed model exhibited a notably superior diagnostic accuracy for recurrence (AUC = 0.89) vs. the clinical risk factors (including CA19–9, CEA, vascular invasion, and tumor size), in the training (AUC = 0.71) and validation cohorts (AUC = 0.69; Supplementary Fig 3). Our risk-stratification model identified that 85% of high-risk patients experienced tumor recurrence and 76% of low-risk patients did not experience recurrence, which is a significantly superior performance compared to currently used clinicopathological risk features in the clinic. Collectively, this highlights the potential significance of our findings for their clinical translation for recurrence identification and improving survival in patients with ICC.
Several previous reports have favored the importance of multidisciplinary treatments in ICC patients (38, 39). With the development of newer chemotherapeutic agents, there has been a significant improvement in the response rates of up to 30–50% (40–42). In addition, data from various clinical trials have suggested that combination of gemcitabine with platinum-based cytotoxic agents results in a better oncological control compared to gemcitabine alone in locally advanced biliary tract cancer (43). In meta-analysis, adjuvant chemotherapy was associated with improved OS and was suggested as a potential consideration in patients with ICC following curative resection (44, 45). The NCCN guidelines indicate that postoperative treatment for resectable ICC should be particularly considered in patients with high-risk features, and surveillance is recommended for the use of CT or MRI of abdomen and pelvis, and CT of chest every 6 months for 2 years. On the other hand, in clinical settings, adjuvant chemotherapy has been actively explored and is becoming a common treatment option regardless of resectability status in patients with ICC. Our ability to successfully validate our risk model underscores its clinical significance for improved treatment strategies such as more aggressive surveillance approach and potentially adjuvant chemotherapy in patients with ICC and often worst survival outcomes.
From a functional viewpoint, various genes in our biomarker panel have been shown to be bonafide candidates involved in cancer pathogenesis. For instance, BIRC5 is a member of the inhibitor of apoptosis gene family, which encodes negative regulatory proteins that prevent apoptotic cell death (46). Tumor growth was found to be reduced under the control of the BIRC5 promoter, a gene highly up-regulated in ICC (47). CDC20 is an important regulator that interacts with several other proteins at multiple cell cycle checkpoints, proliferation and apoptosis (48, 49). Likewise, CDC20 was involved in the modulation of the cell cycle progression from gap 2 to mitosis transition in ICC (50). Furthermore, CDH2, encodes a protein called N-cadherin, which is a member of cadherin family that regulates lots of biological processes (51). Similarly, the protein expression status of CDH2 might help subtyping and predicting clinical outcomes of ICC (52).
We would like to acknowledge a few potential limitations of our present study. First, our retrospective study design might result in a potential selection bias. Thus, a prospective clinical trial is required to further confirm the accuracy of our risk-stratification model. Second, our study used training and validation cohorts of patients from Japan, who showed similar clinicopathologic characteristics; such characteristics could potentially vary if one were to analyze patient populations from other countries. Therefore, it will be important to validate the selected biomarkers and our risk-stratification model in patient cohorts from other countries to further reinforce the generalizability of our findings. Third, although we used imaging modalities as reference standard to identify tumor recurrence sites, such an approach can potentially underestimate the actual extent of ICC. In future, we plan to perform prospective study in which we will focus on various morphological and biochemical parameters, as well as clinical follow up.
In conclusion, using a genome-wide gene expression profiling, we have established a novel risk-stratification model that was successfully validated in surgical tissue specimens for accurate identification of high-risk patients with ICC. Pending validation in future prospective studies, our findings highlight the potential clinical impact of our model for appropriate selection of patients with ICC, which may improve personalized management of patients suffering from this malignancy.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Drs. Tatsuhiko Kakisaka, Satoshi Nishiwada, Yasuyuki Okada, In-Seob Lee, Divya Sahu, Geeta Sharma, and Huanlin Wang for their thoughtful discussions and advice during the course of this project. We thank Drs. Masaaki Nishi, Shinichiro Yamada, Kazunori Tokuda, and Yumi Horikawa for collecting clinical specimens and information.
Funding:
This work was supported by CA72851, CA181572, CA184792, CA202797, and CA187956 grants from the National Cancer Institute, National Institutes of Health.
Abbreviations:
- AUC
Area under the curve
- CA19–9
Carbohydrate antigen 19–9
- CEA
Carcinoembryonic antigen
- CI
Confidence intervals
- CT
Computed tomography
- FFPE
Formalin-fixed paraffin-embedded
- HR
Hazard ratio
- ICC
Intrahepatic cholangiocarcinoma
- MRI
Magnetic resonance imaging
- NCCN
The National Comprehensive Cancer Network
- NPV
Negative predictive value
- OS
Overall survival
- PPV
Positive predictive value
- RFS
Relapse-free survival
- ROC
Receiver operator characteristic
- RT-qPCR
Real-time quantitative reverse transcription polymerase chain reaction
- TCGA
The Cancer Genome Atlas
Footnotes
Conflict of Interest: None of the authors has any potential conflicts to disclose.
REFERENCES
- 1.Aljiffry M, Abdulelah A, Walsh M, Peltekian K, Alwayn I, Molinari M. Evidence-based approach to cholangiocarcinoma: a systematic review of the current literature. J Am Coll Surg 2009;208:134–147. [DOI] [PubMed] [Google Scholar]
- 2.Bertuccio P, Malvezzi M, Carioli G, Hashim D, Boffetta P, El-Serag HB, La Vecchia C, et al. Global trends in mortality from intrahepatic and extrahepatic cholangiocarcinoma. J Hepatol 2019;71:104–114. [DOI] [PubMed] [Google Scholar]
- 3.Khan SA, Thomas HC, Davidson BR, Taylor-Robinson SD. Cholangiocarcinoma. Lancet 2005;366:1303–1314. [DOI] [PubMed] [Google Scholar]
- 4.Mavros MN, Economopoulos KP, Alexiou VG, Pawlik TM. Treatment and Prognosis for Patients With Intrahepatic Cholangiocarcinoma: Systematic Review and Meta-analysis. JAMA Surg 2014;149:565–574. [DOI] [PubMed] [Google Scholar]
- 5.Amini N, Ejaz A, Spolverato G, Kim Y, Herman JM, Pawlik TM. Temporal trends in liver-directed therapy of patients with intrahepatic cholangiocarcinoma in the United States: a population-based analysis. J Surg Oncol 2014;110:163–170. [DOI] [PubMed] [Google Scholar]
- 6.Farges O, Fuks D, Boleslawski E, Le Treut YP, Castaing D, Laurent A, Ducerf C, et al. Influence of surgical margins on outcome in patients with intrahepatic cholangiocarcinoma: a multicenter study by the AFC-IHCC-2009 study group. Ann Surg 2011;254:824–829; discussion 830. [DOI] [PubMed] [Google Scholar]
- 7.Hyder O, Hatzaras I, Sotiropoulos GC, Paul A, Alexandrescu S, Marques H, Pulitano C, et al. Recurrence after operative management of intrahepatic cholangiocarcinoma. Surgery 2013;153:811–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spolverato G, Kim Y, Alexandrescu S, Marques HP, Lamelas J, Aldrighetti L, Clark Gamblin T, et al. Management and Outcomes of Patients with Recurrent Intrahepatic Cholangiocarcinoma Following Previous Curative-Intent Surgical Resection. Ann Surg Oncol 2016;23:235–243. [DOI] [PubMed] [Google Scholar]
- 9.Sulpice L, Rayar M, Boucher E, Pracht M, Meunier B, Boudjema K. Treatment of recurrent intrahepatic cholangiocarcinoma. Br J Surg 2012;99:1711–1717. [DOI] [PubMed] [Google Scholar]
- 10.Zhang XF, Beal EW, Bagante F, Chakedis J, Weiss M, Popescu I, Marques HP, et al. Early versus late recurrence of intrahepatic cholangiocarcinoma after resection with curative intent. Br J Surg 2018;105:848–856. [DOI] [PubMed] [Google Scholar]
- 11.Hoehn RS, Wima K, Ertel AE, Meier A, Ahmad SA, Shah SA, Abbott DE. Adjuvant Chemotherapy and Radiation Therapy is Associated with Improved Survival for Patients with Extrahepatic Cholangiocarcinoma. Ann Surg Oncol 2015;22Suppl 3:S1133–1139. [DOI] [PubMed] [Google Scholar]
- 12.Wirasorn K, Ngamprasertchai T, Chindaprasirt J, Sookprasert A, Khantikaew N, Pakkhem A, Ungarereevittaya P. Prognostic factors in resectable cholangiocarcinoma patients: Carcinoembryonic antigen, lymph node, surgical margin and chemotherapy. World J Gastrointest Oncol 2013;5:81–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benson AB 3rd, D’Angelica MI, Abbott DE, Abrams TA, Alberts SR, Saenz DA, Are C, et al. NCCN Guidelines Insights: Hepatobiliary Cancers, Version 1.2017. J Natl Compr Canc Netw 2017;15:563–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin YK, Hsieh MC, Wang WW, Lin YC, Chang WW, Chang CL, Cheng YF, et al. Outcomes of adjuvant treatments for resectable intrahepatic cholangiocarcinoma: Chemotherapy alone, sequential chemoradiotherapy, or concurrent chemoradiotherapy. Radiother Oncol 2018;128:575–583. [DOI] [PubMed] [Google Scholar]
- 15.Shroff RT, Kennedy EB, Bachini M, Bekaii-Saab T, Crane C, Edeline J, El-Khoueiry A, et al. Adjuvant Therapy for Resected Biliary Tract Cancer: ASCO Clinical Practice Guideline. J Clin Oncol 2019;37:1015–1027. [DOI] [PubMed] [Google Scholar]
- 16.de Jong MC, Nathan H, Sotiropoulos GC, Paul A, Alexandrescu S, Marques H, Pulitano C, et al. Intrahepatic cholangiocarcinoma: an international multi-institutional analysis of prognostic factors and lymph node assessment. J Clin Oncol 2011;29:3140–3145. [DOI] [PubMed] [Google Scholar]
- 17.Endo I, Gonen M, Yopp AC, Dalal KM, Zhou Q, Klimstra D, D’Angelica M, et al. Intrahepatic cholangiocarcinoma: rising frequency, improved survival, and determinants of outcome after resection. Ann Surg 2008;248:84–96. [DOI] [PubMed] [Google Scholar]
- 18.Farges O, Fuks D, Le Treut YP, Azoulay D, Laurent A, Bachellier P, Nuzzo G, et al. AJCC 7th edition of TNM staging accurately discriminates outcomes of patients with resectable intrahepatic cholangiocarcinoma: By the AFC-IHCC-2009 study group. Cancer 2011;117:2170–2177. [DOI] [PubMed] [Google Scholar]
- 19.Ma KW, Cheung TT, She WH, Chok KS, Chan AC, Ng IO, Chan SC, et al. The effect of wide resection margin in patients with intrahepatic cholangiocarcinoma: A single-center experience. Medicine (Baltimore) 2016;95:e4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan KM, Tsai CY, Yeh CN, Yeh TS, Lee WC, Jan YY, Chen MF. Characterization of intrahepatic cholangiocarcinoma after curative resection: outcome, prognostic factor, and recurrence. BMC Gastroenterol 2018;18:180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He C, Zhang Y, Song Y, Wang J, Xing K, Lin X, Li S. Preoperative CEA levels are supplementary to CA19–9 levels in predicting prognosis in patients with resectable intrahepatic cholangiocarcinoma. J Cancer 2018;9:3117–3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu LS, Zhang XF, Weiss M, Popescu I, Marques HP, Aldrighetti L, Maithel SK, et al. Recurrence Patterns and Timing Courses Following Curative-Intent Resection for Intrahepatic Cholangiocarcinoma. Ann Surg Oncol 2019;26:2549–2557. [DOI] [PubMed] [Google Scholar]
- 23.Poon RT. Differentiating early and late recurrences after resection of HCC in cirrhotic patients: implications on surveillance, prevention, and treatment strategies. Ann Surg Oncol 2009;16:792–794. [DOI] [PubMed] [Google Scholar]
- 24.Poon RT, Fan ST, Lo CM, Liu CL, Ng IO, Wong J. Long-term prognosis after resection of hepatocellular carcinoma associated with hepatitis B-related cirrhosis. J Clin Oncol 2000;18:1094–1101. [DOI] [PubMed] [Google Scholar]
- 25.Miyata T, Yamashita YI, Yoshizumi T, Shiraishi M, Ohta M, Eguchi S, Aishima S, et al. CXCL12 expression in intrahepatic cholangiocarcinoma is associated with metastasis and poor prognosis. Cancer Sci 2019;110:3197–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen LP, Li C, Wang C, Wen TF, Yan LN, Li B. Predictive factors of recurrence for patients with intrahepatic cholangiocarcinoma after hepatectomy. Hepatogastroenterology 2012;59:1765–1768. [DOI] [PubMed] [Google Scholar]
- 27.Hanazaki K, Kajikawa S, Shimozawa N, Shimada K, Hiraguri M, Koide N, Adachi W, et al. Prognostic factors of intrahepatic cholangiocarcinoma after hepatic resection: univariate and multivariate analysis. Hepatogastroenterology 2002;49:311–316. [PubMed] [Google Scholar]
- 28.Miwa S, Miyagawa S, Kobayashi A, Akahane Y, Nakata T, Mihara M, Kusama K, et al. Predictive factors for intrahepatic cholangiocarcinoma recurrence in the liver following surgery. J Gastroenterol 2006;41:893–900. [DOI] [PubMed] [Google Scholar]
- 29.Nuzzo G, Giuliante F, Ardito F, De Rose AM, Vellone M, Clemente G, Chiarla C, et al. Intrahepatic cholangiocarcinoma: prognostic factors after liver resection. Updates Surg 2010;62:11–19. [DOI] [PubMed] [Google Scholar]
- 30.Kneuertz PJ, Cosgrove DP, Cameron AM, Kamel IR, Geschwind JF, Herman JM, Pawlik TM. Multidisciplinary management of recurrent hepatocellular carcinoma following liver transplantation. J Gastrointest Surg 2012;16:874–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ozawa T, Kandimalla R, Gao F, Nozawa H, Hata K, Nagata H, Okada S, et al. A MicroRNA Signature Associated With Metastasis of T1 Colorectal Cancers to Lymph Nodes. Gastroenterology 2018;154:844–848.e847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sonohara F, Gao F, Iwata N, Kanda M, Koike M, Takahashi N, Yamada Y, et al. Genome-wide Discovery of a Novel Gene-expression Signature for the Identification of Lymph Node Metastasis in Esophageal Squamous Cell Carcinoma. Ann Surg 2019;269:879–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402–408. [DOI] [PubMed] [Google Scholar]
- 34.Eamer GJ, Clement F, Pederson JL, Churchill TA, Khadaroo RG. Analysis of postdischarge costs following emergent general surgery in elderly patients. Can J Surg 2018;61:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Studdert DM, Mello MM, Sage WM, DesRoches CM, Peugh J, Zapert K, Brennan TA. Defensive medicine among high-risk specialist physicians in a volatile malpractice environment. Jama 2005;293:2609–2617. [DOI] [PubMed] [Google Scholar]
- 36.Hu LS, Weiss M, Popescu I, Marques HP, Aldrighetti L, Maithel SK, Pulitano C, et al. Impact of microvascular invasion on clinical outcomes after curative-intent resection for intrahepatic cholangiocarcinoma. J Surg Oncol 2019;119:21–29. [DOI] [PubMed] [Google Scholar]
- 37.Kim BH, Kim E, Kim K, Jang JY, Kim SW, Oh DY, Chie EK. The impact of perioperative CA19–9 change on the survival and recurrence patterns after adjuvant chemoradiotherapy in resectable extrahepatic cholangiocarcinoma. J Surg Oncol 2018;117:380–388. [DOI] [PubMed] [Google Scholar]
- 38.Le Roy B, Gelli M, Pittau G, Allard MA, Pereira B, Serji B, Vibert E, et al. Neoadjuvant chemotherapy for initially unresectable intrahepatic cholangiocarcinoma. Br J Surg 2018;105:839–847. [DOI] [PubMed] [Google Scholar]
- 39.Yadav S, Xie H, Bin-Riaz I, Sharma P, Durani U, Goyal G, Borah B, et al. Neoadjuvant vs. adjuvant chemotherapy for cholangiocarcinoma: A propensity score matched analysis. Eur J Surg Oncol 2019;45:1432–1438. [DOI] [PubMed] [Google Scholar]
- 40.Eckel F, Schmid RM. Chemotherapy in advanced biliary tract carcinoma: a pooled analysis of clinical trials. Br J Cancer 2007;96:896–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hezel AF, Zhu AX. Systemic therapy for biliary tract cancers. Oncologist 2008;13:415–423. [DOI] [PubMed] [Google Scholar]
- 42.Verderame F, Russo A, Di Leo R, Badalamenti G, Santangelo D, Cicero G, Valerio MR, et al. Gemcitabine and oxaliplatin combination chemotherapy in advanced biliary tract cancers. Ann Oncol 2006;17Suppl 7:vii68–72. [DOI] [PubMed] [Google Scholar]
- 43.Valle J, Wasan H, Palmer DH, Cunningham D, Anthoney A, Maraveyas A, Madhusudan S, et al. Cisplatin plus gemcitabine versus gemcitabine for biliary tract cancer. N Engl J Med 2010;362:1273–1281. [DOI] [PubMed] [Google Scholar]
- 44.Ma KW, Cheung TT, Leung B, She BWH, Chok KSH, Chan ACY, Dai WC, et al. Adjuvant chemotherapy improves oncological outcomes of resectable intrahepatic cholangiocarcinoma: A meta-analysis. Medicine (Baltimore) 2019;98:e14013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang ML, Ke ZY, Yin S, Liu CH, Huang Q. The effect of adjuvant chemotherapy in resectable cholangiocarcinoma: A meta-analysis and systematic review. Hepatobiliary Pancreat Dis Int 2019;18:110–116. [DOI] [PubMed] [Google Scholar]
- 46.Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med 1997;3:917–921. [DOI] [PubMed] [Google Scholar]
- 47.Lozano E, Macias RIR, Monte MJ, Asensio M, Del Carmen S, Sanchez-Vicente L, Alonso-Peña M, et al. Causes of hOCT1-Dependent Cholangiocarcinoma Resistance to Sorafenib and Sensitization by Tumor-Selective Gene Therapy. Hepatology 2019;70:1246–1261. [DOI] [PubMed] [Google Scholar]
- 48.Chen Z, Yu Y, Fu D, Li Z, Niu X, Liao M, Lu S. Functional roles of PC-PLC and Cdc20 in the cell cycle, proliferation, and apoptosis. Cell Biochem Funct 2010;28:249–257. [DOI] [PubMed] [Google Scholar]
- 49.Hartwell LH, Mortimer RK, Culotti J, Culotti M. Genetic Control of the Cell Division Cycle in Yeast: V. Genetic Analysis of cdc Mutants. Genetics 1973;74:267–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamanaka S, Campbell NR, An F, Kuo SC, Potter JJ, Mezey E, Maitra A, et al. Coordinated effects of microRNA-494 induce G₂/M arrest in human cholangiocarcinoma. Cell Cycle 2012;11:2729–2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ma T, Zhao Y, Wei K, Yao G, Pan C, Liu B, Xia Y, et al. MicroRNA-124 Functions as a Tumor Suppressor by Regulating CDH2 and Epithelial-Mesenchymal Transition in Non-Small Cell Lung Cancer. Cell Physiol Biochem 2016;38:1563–1574. [DOI] [PubMed] [Google Scholar]
- 52.Rhee H, Ko JE, Chung T, Jee BA, Kwon SM, Nahm JH, Seok JY, et al. Transcriptomic and histopathological analysis of cholangiolocellular differentiation trait in intrahepatic cholangiocarcinoma. Liver Int 2018;38:113–124. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


