Abstract
The composition and diversity of animal microbiomes is shaped by a variety of factors, many of them interacting, such as host traits, the environment, and biogeography. Hybrid zones, in which the ranges of two host species meet and hybrids are found, provide natural experiments for determining the drivers of microbiome communities, but have not been well studied in marine environments. Here, we analysed the composition of the symbiont community in two deep-sea, Bathymodiolus mussel species along their known distribution range at hydrothermal vents on the Mid-Atlantic Ridge, with a focus on the hybrid zone where they interbreed. In-depth metagenomic analyses of the sulphur-oxidising symbionts of 30 mussels from the hybrid zone, at a resolution of single nucleotide polymorphism analyses of ~2500 orthologous genes, revealed that parental and hybrid mussels (F2–F4 generation) have genetically indistinguishable symbionts. While host genetics does not appear to affect symbiont composition in these mussels, redundancy analyses showed that geographic location of the mussels on the Mid-Atlantic Ridge explained most of the symbiont genetic variability compared to the other factors. We hypothesise that geographic structuring of the free-living symbiont population plays a major role in driving the composition of the microbiome in these deep-sea mussels.
Subject terms: Metagenomics, Population genetics, Symbiosis, Microbiome
Introduction
The community composition of an animal’s microbiome is the product of multiple interacting factors that include the environment, geography and host genetics [1–5]. To which extent host genetics affects microbiome composition is currently a topic of intense debate, in part as high-throughput sequencing is revealing the genetic makeup of host and symbiont populations in ever higher resolution [6–8]. Animal hybrids are useful for assessing the effects of host genotype on microbiomes [9]. Studies of lab-reared animal hybrids, such as wasps [10], fish [11–13], and mice [14, 15] found that these hosts had different gut microbiota compositions than their parental species, based on sequencing of the microbial 16S rRNA gene. These altered gut microbiomes of hybrids affected the fitness of some hosts, suggesting that microbiomes play an important role in determining species barriers [10]. Studies on lab-reared hosts cannot, however, fully reflect the environmental conditions animals experience in their natural habitat. Hybrid zones, in which parental species interbreed and produce hybrid offspring, are excellent natural experiments for teasing apart the impact of host genotype, environment and geographic distance on microbiome composition. Yet surprisingly few studies have investigated the microbiota of hybrids from the wild, and these have yielded mixed results. For example, in a hybrid zone of the European house mouse, the composition of gut microbiota of hybrids differed from that of their parental species [15]. In contrast, in African baboons, there were no significant differences between hybrids and their parental species, and gut community composition was best explained by the environment [16]. To date, all hybrid microbiome studies, whether on lab-reared animals or those from the wild, have been based on the sequencing of only a few microbial genes, with the vast majority of analyses based on the 16S rRNA gene, or a variable region of this gene. These studies were therefore limited to determining microbial community composition at the genus level or higher, and could not distinguish closely related species or strains.
Almost nothing is known about the microbial communities of hosts from marine hybrid zones, despite the pervasiveness of such zones in many regions of the oceans. Hydrothermal vents on the Mid-Atlantic Ridge (MAR), an underwater mountain range extending from the Arctic to the Southern Ocean, provide an ideal setting for investigating the microbiomes of hosts in natural hybrid zones. Many of the vents on the MAR are dominated by Bathymodiolus mussels that live in a nutritional symbiosis with chemosynthetic bacteria. Two mussel species colonise the northern MAR, Bathymodiolus azoricus, which is found at vents from 38° N to 36° N, and Bathymodiolus puteoserpentis, which inhabits vents further south from 23° N to 13° N. A hybrid zone between these two relatively young host species, with an estimated splitting time of 8.4 Mya [17], occurs at the Broken Spur vent field at 29° N on the MAR, where B. puteoserpentis co-occurs with hybrids between B. azoricus and B. puteoserpentis [18–20]. The sulphur-oxidising (SOX) symbionts of B. azoricus and B. puteoserpentis belong to a gammaproteobacterial clade within the Thioglobaceae, and co-occur in the mussel gills with methane-oxidising symbionts, which belong to a gammaproteobacterial clade within the Methylomonaceae [21]. The relative abundance of these two symbionts in these mussels is assumed to not depend on host genetics, but rather the availability of the energy sources these symbionts use in their environment [21, 22].
The symbionts of bathymodiolin mussels are transmitted horizontally from the environment to juvenile mussels, yet each mussel species harbours a highly specific symbiont community [23–25]. This specificity suggests that the genetics of bathymodiolin mussels plays an important role in determining symbiont composition. In this study, we took advantage of the natural hybrid zone of Bathymodiolus mussels at the Broken Spur vent field to investigate how host genotype, geographic distance, and the vent environment affect the composition of their SOX symbionts. The recent discovery of a high diversity of SOX symbiont strains in Bathymodiolus from the MAR, with as many as 16 strains co-occurring in single Bathymodiolus mussels [22, 26, 27], made it critical to resolve genetic differences at the strain level of the SOX symbiont community (strain is defined here as suggested by Van Rossum et al. [28], as subordinate to subspecies, in our study >99% average nucleotide identity). We achieved this resolution through multilocus phylogeny, genome-wide gene profiling, and single nucleotide polymorphism (SNP)-based population differentiation analyses of 30 Bathymodiolus hybrid and parental individuals collected in 1997 and 2001 at the Broken Spur vent field.
Materials and methods
A detailed description of samples (Supplementary Table S1) and methods is available in the Supplementary Information and an overview of the analyses of SOX symbionts used in this study is provided in Supplementary Table S2. Data files and scripts used for the analyses can be found in the GitHub repository (https://github.com/muecker/Symbionts_in_a_mussel_hybrid_zone).
Broken Spur parental mussels (13 B. puteoserpentis) and hybrids (17 F2–F4 generation hybrids, see Supplement) were identified as described previously [20, 29] (no parental B. azoricus were found at Broken Spur). Briefly, mussels were genotyped based on 18 species-diagnostic markers and identified as parental or hybrid mussels using bioinformatic analyses of population structure, admixture and introgression (Supplementary Table S3). After DNA extraction and sequencing, we assembled metagenomes per mussel individual from Illumina short-read sequences. Metagenome-assembled genomes (MAGs) of the SOX symbionts from each mussel individual were binned (for statistics of symbiont MAGs, see Supplementary Table S4), representing the consensus of all SOX symbiont strains in each host individual.
To evaluate genetic differences between symbionts from the northern MAR at the level of bacterial subspecies (sensu Van Rossum et al. [28], here above 97% average nucleotide identity), we used 171 single-copy, gammaproteobacterial marker genes for phylogenomic analysis of the SOX symbiont MAGs and their closest symbiotic relatives, e.g., symbionts of B. azoricus from vents north of Broken Spur and B. puteoserpentis mussels from vents south of Broken Spur, and free-living relatives (see Supplementary Table S5). To understand which factors affect symbiont composition on the strain level at the northern MAR, we assessed the influence of geographic distance, host species, vent type (basaltic versus ultramafic rock) and depth on SOX symbiont allele frequencies using redundancy analysis (RDA). We analysed Broken Spur symbiont MAGs at the genome-wide level by comparing their average nucleotide identities (ANI) to resolve differences on the subspecies level. To resolve strain-level differences between SOX symbionts from Broken Spur, we analysed pairwise FST values based on SNPs in 2496 orthologous genes from Broken Spur SOX symbiont MAGs. To identify genes that differed between hybrid and parental symbiont populations, we analysed the presence/absence and differential abundance of these orthologues, and further investigated pairwise FST values of all 2496 orthologous genes.
Results and discussion
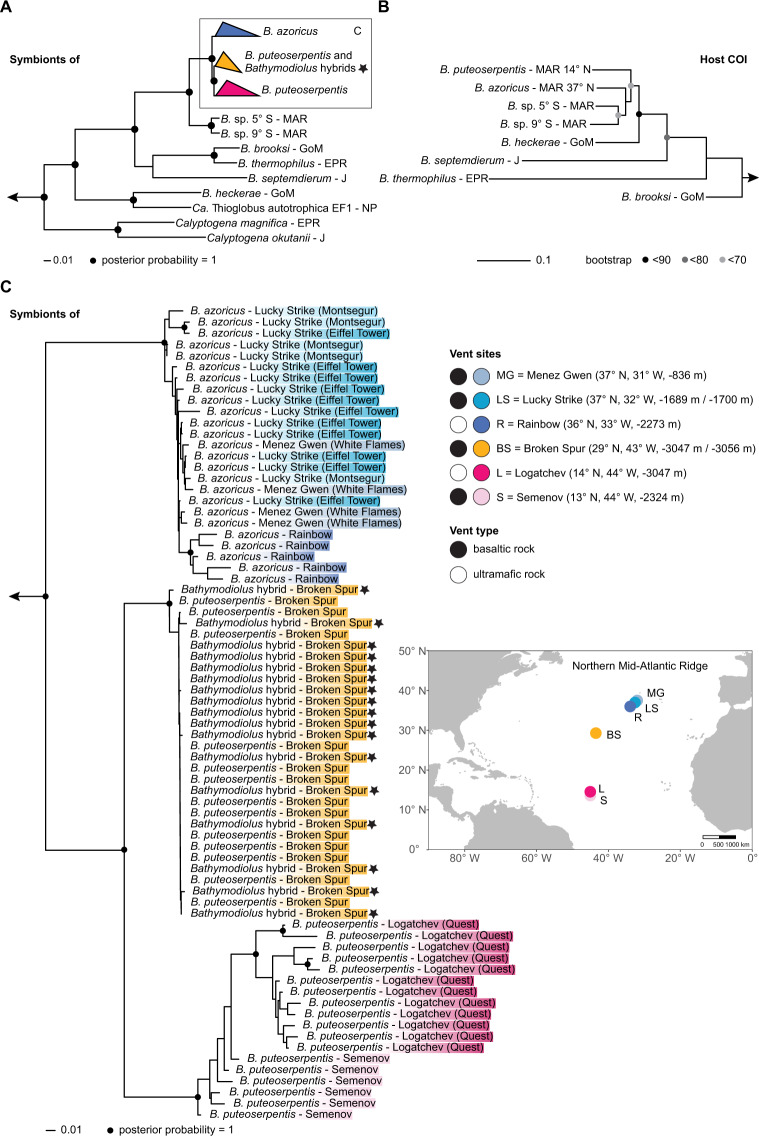
Phylogenomic analysis of 171 single-copy genes revealed the presence of two SOX symbiont subspecies, one specific to B. azoricus from the more northern vents Menez Gwen, Lucky Strike and Rainbow, and one specific to B. puteoserpentis from the vents further south, Logatchev and Semenov (Fig. 1A, C).
Fig. 1. Phylogenetic relationships of Bathymodiolus SOX symbionts and their mussel hosts.
A Overview tree based on 171 single-copy marker genes. The tree was reconstructed based on a 36,949 bp alignment using the LG + F + R6 amino acid model and 1000 samples for ultrafast bootstrap with IQ-TREE. The Bathymodiolus SOX symbionts from the northern Mid-Atlantic Ridge (blue, yellow and pink) form a clade within the gammaproteobacterial SUP05 clade. Thiomicrospira spp. and Ca. T. singularis PS1 were used as outgroups. MAG accessions are listed in Supplementary Table S5. B Host phylogeny based on published mitochondrial cytochrome oxidase subunit I (COI) sequences. “B.” childressi was used as an outgroup. Sequence accessions are listed in the Supplement (“1.3 Reconstruction of Bathymodiolus phylogeny”). C Zoom in of sequences shown in box in (A): Phylogeny of Bathymodiolus SOX symbionts from vents on the northern Mid-Atlantic Ridge. Black and white circles indicate the vent type (basaltic or ultramafic rock), colours correspond to vent sites shown in the map. Hybrid individuals from Broken Spur are marked with a black star. Bathymodiolus SOX symbionts from the vent sites Clueless (5° S) and Lilliput (9° S) were used as outgroups. B. Bathymodiolus, MAR Mid-Atlantic Ridge, GoM Gulf of Mexico, EPR East Pacific Rise, J Japan, NP North Pacific (colour figure online).
This substantiates previous analyses based on sequencing of the 16S rRNA gene and internal transcribed spacer that these two Bathymodiolus species harbour different SOX symbiont subspecies of the same bacterial species [21, 25, 30]. Our phylogenomic analyses revealed that all Bathymodiolus individuals from Broken Spur harboured a third SOX symbiont subspecies (Fig. 1A, C). This new subspecies is most closely related to the B. puteoserpentis SOX symbiont subspecies from mussels collected south of Broken Spur. These two symbiont subspecies form a sister group to the SOX symbiont subspecies of B. azoricus collected at vents north of Broken Spur.
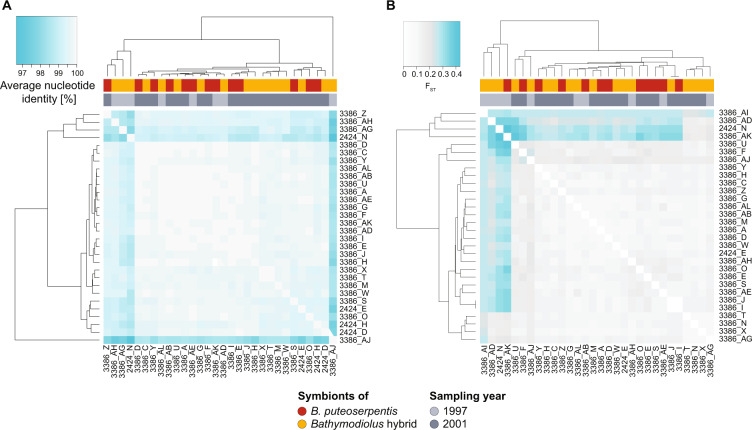
To evaluate if the SOX symbionts of Broken Spur parental and hybrid Bathymodiolus differed, we compared their ANI and estimated genomic differentiation (FST) based on ~2500 orthologous genes (for more information on the host, see Supplementary section 2 and Supplementary Table S3). Symbiont ANI values ranged from 96.7 to 99.9% with a median of 99.7%. We found no correlation between symbiont differentiation and the sampling year or genetic differentiation of the mussels (Mantel test of symbiont ANI and FST versus sampling year and host pairwise genetic distances based on 18 SNP markers, Fig. 2). Our analyses of SNPs per individual gene revealed that not even one of the ~2500 orthologous genes had significantly differing FST values (Mann-Whitney U test of FST per gene between versus within symbionts of hybrids and parental mussels). Similarly, there was also no significant difference between hybrids and parental individuals in the abundance of symbiont genes (based on a general linear model and Kruskal-Wallace test in ALDEx2 using Benjamini-Hochberg corrected p value < 0.05) or their presence/absence. These results indicate that the composition and gene repertoire of SOX symbionts in Broken Spur mussels was highly similar or identical in hybrids and parental B. puteoserpentis. A study of SOX symbionts in hybrids of Bathymodiolus thermophilus and Bathymodiolus antarcticus at 23° S in the eastern Pacific also found that these could not be distinguished from parental mussels, based on PCR analyses of seven bacterial marker genes in five parental and three hybrid individuals [31].
Fig. 2. Genome-wide differentiation of Bathymodiolus SOX symbionts at Broken Spur.
A Differentiation based on pairwise average nucleotide identity. B Differentiation based on pairwise average FST in 2496 orthologous genes. Colour bars represent host genotypes (red: B. puteoserpentis, yellow: hybrids) and the sampling year (light grey: 1997, dark grey: 2001). Turquoise indicates a higher differentiation or more dissimilar genomes. Based on a Mantel test, neither clustering based on ANI (A), nor FST (B) correlated with host pairwise genetic distances based on 18 SNP markers (A r = 0.054, p = 0.222, B r = 0.006, p = 0.435) or sampling year (A r = −0.191, p = 0.949, B r = 0.105, p = 0.150) (colour figure online).
Our results raise the question at what level of genetic divergence between two host species differences in their symbiont communities evolve. Bathymodiolus brooksi and Bathymodiolus heckerae, which regularly co-occur in the Gulf of Mexico, harbour different symbiont species that are only distantly related to each other (Fig. 1A, B). These two mussel species have an estimated splitting time of 15.4 Mya [17], and are not known to hybridise. More closely related hosts, such as B. thermophilus and B. antarcticus (estimated splitting time of 2.5–5.3 Mya [32]), and B. azoricus and B. puteoserpentis (estimated splitting time of 8.4 Mya [17]), produce fertile hybrids [19, 33], and have genetically indistinguishable symbionts in zones where they hybridise. This suggests that specificity at the symbiont species level in these horizontally transmitted symbioses evolves only after extended divergence times of tens of millions of years, during which these hosts become genetically dissimilar enough to evolve specific symbiont selection mechanisms.
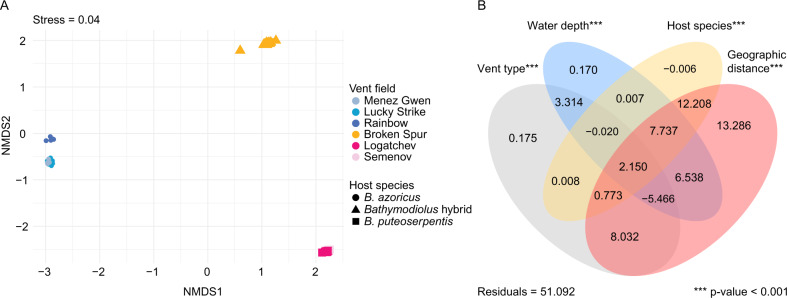
While Bathymodiolus mussels on the northern MAR host the same SOX symbiont species, our phylogenomic analyses revealed clear genetic differentiation in three SOX symbiont subspecies: B. azoricus, B. puteoserpentis and Broken Spur subspecies (Fig. 1). To better understand the factors that drive this symbiont differentiation, we tested which influence host species, geographic distance, vent type (basaltic versus ultramafic rock) and depth have on symbiont allele frequencies. All variables were highly collinear. For example, the water depth of the vents studied here increases with geographic distance, from 800 m at 37.8° N, to 3050 m at 14.7° N (only the southernmost vent at 13.5° N and 2320 m depth interrupted this pattern). When the four variables were considered individually, geographic distance explained 13% of symbiont differentiation, while the three other variables water depth, host species and vent type each explained 0.2%, 0.0%, and 0.2%, respectively. When interaction effects between the four variables were considered, geographic distance and interactions involving this variable explained 45% of symbiont differentiation, while the three other variables water depth, host species, vent type and the interactions with these explained 14%, 12%, and 9%, respectively (p value < 0.001, Fig. 3).
Fig. 3. Differentiation of Bathymodiolus SOX symbionts at the northern Mid-Atlantic Ridge and the influence of geographic distance, host species and environmental parameters (vent type and water depth).
A NMDS plot of SOX symbiont allele frequencies show clear separation of the three symbiont subspecies at the northern MAR: B. azoricus, B. puteoserpentis, and Broken Spur symbiont subspecies. Symbionts from Broken Spur cluster together regardless of the species affiliation of their host (hybrid versus B. puteoserpentis). Colours correspond to vent fields, shapes to host species. B Variation partitioning of explanatory variables used in the RDA (Supplementary Fig. S3). Values are shown in percent. The variables vent type, water depth, host species, and geographic distance and their interaction effects explained 49% of the total variation, with 13% of the variation solely explained by geographic distance. Negative adjusted R values can occur for several reasons, e.g., negative eigenvalues in the model underlying the varpart function in R. P values are based on permutation tests with 1000 repetitions.
There are at least three explanations for why geographic distance has such a large effect on the SOX symbiont composition of Bathymodiolus mussels from the northern MAR. The first is that with increasing geographic distance, environmental differences between vents become larger and these environmental differences affect symbiont composition (genetic isolation by environment versus distance [34]). Bathymodiolus mussels acquire their symbionts horizontally from the environment, presumably when the larvae settle on the seafloor [25, 35], and it would be advantageous for the mussels if they selected symbionts that are best adapted to local environmental conditions. We tested the effect of vent type based on one of the key environmental variables at hydrothermal vents, basaltic and ultramafic rock. These two rock types have major effects on the biogeochemistry of vent fluids, including the relative concentrations of the symbiotic energy sources sulphide, methane and hydrogen [36]. However, vent type alone explained only 0.2% of symbiont genetic differentiation, similar to another environmental variable water depth, which also only explained 0.2% of symbiont differentiation. It is therefore unlikely that environmental differences between vents underlie the symbiont population structures we observed in this study.
The second explanation for why geographic distance has such a large effect on the SOX symbiont composition of Bathymodiolus mussels is that genetic differences between the hosts increased with geographic distance. However, population genetic analyses of B. azoricus and B. puteoserpentis from the same vents as in our study indicated no genetic structuring within each of these host species [29]. This indicates that host genetics did not play a major role in structuring the SOX symbiont composition. The third, and most likely explanation is that the free-living pool of SOX symbionts is geographically structured. At Broken Spur, hybrids and B. puteoserpentis host genetically indistinguishable symbionts, and these differed from the symbionts of B. azoricus and B. puteoserpentis from vents to the north and south of Broken Spur. This indicates that in these two closely related host species, geographic location but not host genetics drives the composition of their SOX symbiont communities. Furthermore, the environment, based on vent type, cannot explain why the mussels at Broken Spur had symbionts that are genetically distinct from other vent sites. Broken Spur is basalt-hosted, while the vents to the north are both basalt- (Menez Gwen and Lucky Strike) and ultramafic-hosted (Rainbow). Yet the symbionts from the vents to the north of Broken Spur are more closely related to each other than to the symbionts of Broken Spur mussels (Fig. 1C).
The validity of these three explanations could be tested in future studies by sampling the free-living microbial populations at hydrothermal vents on the MAR. This is, however, not as simple as it sounds because of multiple challenges including obtaining representative samples from the immediate environment of Bathymodiolus mussels, collecting environmental data at scales relevant to the microbial population, and characterising the free-living symbiont population at sufficiently high resolution.
Understanding the biogeography of the free-living stages of microbial symbionts and other as yet uncultured microorganisms is currently one of the biggest challenges in microbial ecology. While there is evidence that ‘everything is everywhere, but the environment selects’ [37, 38], there is also increasing data showing that dispersal limitation shapes the biogeography of marine microorganisms [39, 40]. Almost nothing is known about the biogeography of uncultivable marine microorganisms at the subspecies or strain level, as most species are rarely abundant enough to allow phylogenetic analyses at such high resolution. Advances in high-throughput short-read, and particularly long-read sequencing, coupled with bioinformatic methods for revealing genetic structuring of microbial populations, are now providing us with the tools for resolving the intraspecific diversity of environmental microorganisms. Our study highlights the importance of gaining a better understanding of the free-living community of microbial symbionts to disentangle the genetic, environmental, and geographic factors that contribute to the ecological and evolutionary success of animal–microbe associations in which the symbionts are acquired from the environment.
Supplementary information
Acknowledgements
We thank the captains, crews, and funding agencies of the sampling cruises AT-03 and AT-05, and the Monterey Bay Aquarium Research Institute and R.C. Vrijenhoek for providing samples. We are grateful to T.B.H. Reusch, J. Dierking, K. Trübenbach, and P. Weist for the opportunity to perform host genotyping at the GEOMAR and their assistance in the laboratory. We also thank J. Wippler for scientific input, T. Enders for support with troubleshooting, and A. Kupczok, L.G.E. Wilkens, and B. Geier for comments on the manuscript. This work was funded by the Max Planck Society, the MARUM Cluster of Excellence ‘The Ocean Floor’ (Deutsche Forschungsgemeinschaft (German Research Foundation) under Germany’s Excellence Strategy—EXC-2077–39074603), an European Research Council Advanced Grant (BathyBiome, 340535), and a National Science Foundation Grant (NSF OCE-1736932) to Roxanne Beinart (mentor of CB).
Author contributions
MÜ, RA, LS, and ND conceived the study. MÜ performed laboratory work and analyses of symbionts and hosts, prepared figures and tables, submitted data and code, and wrote the initial draft. YS, RA, and LS contributed to analyses of the symbionts. CB provided samples and contributed to analyses of the host. MÜ, RA, YS, and ND interpreted the results with advice from the other co-authors. MÜ, RA, YS, and ND revised the final manuscript with input from all co-authors.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Data availability
Sequence data (metagenomes and symbiont MAGs) are available in the European Nucleotide Archive (ENA) at EMBL-EBI under project accession number PRJEB36976. The data, together with their metadata, were deposited using the data brokerage service of the German Federation for Biological Data (GFBio [41]), with the standard information on sequence data provided as recommended [42].
Code availability
Additional data files and scripts used in this study are available in the GitHub repository (https://github.com/muecker/Symbionts_in_a_mussel_hybrid_zone).
Compliance with ethical standards
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-021-00927-9.
References
- 1.Benson AK, Kelly SA, Legge R, Ma F, Low SJ, Kim J, et al. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. PNAS. 2010;107:18933–8. doi: 10.1073/pnas.1007028107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davenport ER. Elucidating the role of the host genome in shaping microbiome composition. Gut Microbes. 2016;7:178–84. doi: 10.1080/19490976.2016.1155022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rothschild D, Weissbrod O, Barkan E, Kurilshikov A, Korem T, Zeevi D, et al. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555:210–5. doi: 10.1038/nature25973. [DOI] [PubMed] [Google Scholar]
- 4.Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol. 2011;9:279–90. doi: 10.1038/nrmicro2540. [DOI] [PubMed] [Google Scholar]
- 5.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–7. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Di Bella JM, Bao Y, Gloor GB, Burton JP, Reid G. High throughput sequencing methods and analysis for microbiome research. J Microbiol Methods. 2013;95:401–14. doi: 10.1016/j.mimet.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 7.Ellegren H. Genome sequencing and population genomics in non-model organisms. Trends Ecol Evol. 2014;29:51–63. doi: 10.1016/j.tree.2013.09.008. [DOI] [PubMed] [Google Scholar]
- 8.Luikart G, England PR, Tallmon D, Jordan S, Taberlet P. The power and promise of population genomics: from genotyping to genome typing. Nat Rev Genet. 2003;4:981–94. doi: 10.1038/nrg1226. [DOI] [PubMed] [Google Scholar]
- 9.Lim SJ, Bordenstein SR. An introduction to phylosymbiosis. Proc R Soc B. 2020;287:20192900. doi: 10.1098/rspb.2019.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brucker RM, Bordenstein SR. The hologenomic basis of speciation: gut bacteria cause hybrid lethality in the genus Nasonia. Science. 2013;341:667–9. doi: 10.1126/science.1240659. [DOI] [PubMed] [Google Scholar]
- 11.Li W, Liu J, Tan H, Yang C, Ren L, Liu Q, et al. Genetic effects on the gut microbiota assemblages of hybrid fish from parents with different feeding habits. Front. Microbiol. 2018;9:2972. doi: 10.3389/fmicb.2018.02972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rennison DJ, Rudman SM, Schluter D. Parallel changes in gut microbiome composition and function during colonization, local adaptation and ecological speciation. Proc R Soc B. 2019;286:20191911. doi: 10.1098/rspb.2019.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sevellec M, Laporte M, Bernatchez A, Derome N, Bernatchez L. Evidence for host effect on the intestinal microbiota of whitefish (Coregonus sp.) species pairs and their hybrids. Ecol Evol. 2019;9:11762–74. doi: 10.1002/ece3.5676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Korach-Rechtman H, Freilich S, Gerassy-Vainberg S, Buhnik-Rosenblau K, Danin-Poleg Y, Bar H, et al. Murine genetic background has a stronger impact on the composition of the gut microbiota than maternal inoculation or exposure to unlike exogenous microbiota. Appl Environ Microbiol. 2019;85:e00826–19. doi: 10.1128/AEM.00826-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J, Kalyan S, Steck N, Turner LM, Harr B, Künzel S, et al. Analysis of intestinal microbiota in hybrid house mice reveals evolutionary divergence in a vertebrate hologenome. Nat Commun. 2015;6:1–10. doi: 10.1038/ncomms7440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grieneisen LE, Charpentier MJE, Alberts SC, Blekhman R, Bradburd G, Tung J, et al. Genes, geology and germs: gut microbiota across a primate hybrid zone are explained by site soil properties, not host species. Proc R Soc B. 2019;286:20190431. doi: 10.1098/rspb.2019.0431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faure B, Schaeffer SW, Fisher CR. Species distribution and population connectivity of deep-sea mussels at hydrocarbon seeps in the Gulf of Mexico. PLoS ONE. 2015;10:e0118460. doi: 10.1371/journal.pone.0118460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Won Y, Hallam SJ, O’Mullan GD, Vrijenhoek RC. Cytonuclear disequilibrium in a hybrid zone involving deep-sea hydrothermal vent mussels of the genus Bathymodiolus. Mol Ecol. 2003;12:3185–90. doi: 10.1046/j.1365-294X.2003.01974.x. [DOI] [PubMed] [Google Scholar]
- 19.O’Mullan GD, Maas PA, Lutz RA, Vrijenhoek RC. A hybrid zone between hydrothermal vent mussels (Bivalvia: Mytilidae) from the Mid-Atlantic Ridge. Mol Ecol. 2001;10:2819–31. doi: 10.1046/j.0962-1083.2001.01401.x. [DOI] [PubMed] [Google Scholar]
- 20.Breusing C, Vrijenhoek RC, Reusch TBH. Widespread introgression in deep-sea hydrothermal vent mussels. BMC Evol Biol. 2017;17:13. doi: 10.1186/s12862-016-0862-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duperron S, Bergin C, Zielinski F, Blazejak A, Pernthaler A, McKiness ZP, et al. A dual symbiosis shared by two mussel species, Bathymodiolus azoricus and Bathymodiolus puteoserpentis (Bivalvia: Mytilidae), from hydrothermal vents along the northern Mid-Atlantic Ridge. Environ Microbiol. 2006;8:1441–7. doi: 10.1111/j.1462-2920.2006.01038.x. [DOI] [PubMed] [Google Scholar]
- 22.Ansorge R, Romano S, Sayavedra L, González Porras MÁ, Kupczok A, Tegetmeyer HE, et al. Functional diversity enables multiple symbiont strains to coexist in deep-sea mussels. Nat Microbiol. 2019;4:2487–97. doi: 10.1038/s41564-019-0572-9. [DOI] [PubMed] [Google Scholar]
- 23.Dubilier N, Bergin C, Lott C. Symbiotic diversity in marine animals: the art of harnessing chemosynthesis. Nat Rev Microbiol. 2008;6:725–40. doi: 10.1038/nrmicro1992. [DOI] [PubMed] [Google Scholar]
- 24.Van Dover CL, German CR, Speer KG, Parson LM, Vrijenhoek RC. Evolution and biogeography of deep-sea vent and seep invertebrates. Science. 2002;295:1253–7. doi: 10.1126/science.1067361. [DOI] [PubMed] [Google Scholar]
- 25.Won Y-J, Hallam SJ, O’Mullan GD, Pan IL, Buck KR, Vrijenhoek RC. Environmental acquisition of thiotrophic endosymbionts by deep-sea mussels of the genus Bathymodiolus. Appl Environ Microbiol. 2003;69:6785–92. doi: 10.1128/AEM.69.11.6785-6792.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ikuta T, Takaki Y, Nagai Y, Shimamura S, Tsuda M, Kawagucci S, et al. Heterogeneous composition of key metabolic gene clusters in a vent mussel symbiont population. ISME J. 2016;10:990–1001. doi: 10.1038/ismej.2015.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Picazo DR, Dagan T, Ansorge R, Petersen JM, Dubilier N, Kupczok A. Horizontally transmitted symbiont populations in deep-sea mussels are genetically isolated. ISME J. 2019;13:2954–68. doi: 10.1038/s41396-019-0475-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Rossum T, Ferretti P, Maistrenko OM, Bork P. Diversity within species: interpreting strains in microbiomes. Nat Rev Microbiol. 2020;18:491–506. doi: 10.1038/s41579-020-0368-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Breusing C, Biastoch A, Drews A, Metaxas A, Jollivet D, Vrijenhoek RC, et al. Biophysical and population genetic models predict the presence of “phantom” stepping stones connecting Mid-Atlantic Ridge vent ecosystems. Curr Biol. 2016;26:2257–67. doi: 10.1016/j.cub.2016.06.062. [DOI] [PubMed] [Google Scholar]
- 30.DeChaine EG, Bates AE, Shank TM, Cavanaugh CM. Off-axis symbiosis found: characterization and biogeography of bacterial symbionts of Bathymodiolus mussels from Lost City hydrothermal vents. Environ Microbiol. 2006;8:1902–12. doi: 10.1111/j.1462-2920.2005.01113.x. [DOI] [PubMed] [Google Scholar]
- 31.Ho P-T, Park E, Hong SG, Kim E-H, Kim K, Jang S-J, et al. Geographical structure of endosymbiotic bacteria hosted by Bathymodiolus mussels at eastern Pacific hydrothermal vents. BMC Evol Biol. 2017;17:121. doi: 10.1186/s12862-017-0966-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Won Y-J, Young CR, Lutz RA, Vrijenhoek RC. Dispersal barriers and isolation among deep-sea mussel populations (Mytilidae: Bathymodiolus) from eastern Pacific hydrothermal vents. Mol Ecol. 2003;12:169–84. doi: 10.1046/j.1365-294X.2003.01726.x. [DOI] [PubMed] [Google Scholar]
- 33.Johnson SB, Won Y-J, Harvey JB, Vrijenhoek RC. A hybrid zone between Bathymodiolus mussel lineages from eastern Pacific hydrothermal vents. BMC Evol Biol. 2013;13:21. doi: 10.1186/1471-2148-13-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sexton JP, Hangartner SB, Hoffmann AA. Genetic isolation by environment or distance: which pattern of gene flow is most common? Evolution. 2014;68:1–15. doi: 10.1111/evo.12258. [DOI] [PubMed] [Google Scholar]
- 35.Laming SR, Duperron S, Cunha MR, Gaudron SM. Settled, symbiotic, then sexually mature: adaptive developmental anatomy in the deep-sea, chemosymbiotic mussel Idas modiolaeformis. Mar Biol. 2014;161:1319–33. doi: 10.1007/s00227-014-2421-y. [DOI] [Google Scholar]
- 36.Petersen JM, Zielinski FU, Pape T, Seifert R, Moraru C, Amann R, et al. Hydrogen is an energy source for hydrothermal vent symbioses. Nature. 2011;476:176–80. doi: 10.1038/nature10325. [DOI] [PubMed] [Google Scholar]
- 37.Baas Becking LGM. Geobiologie of inleiding tot de milieukunde. Den Haag: W.P. Van Stockum & Zoon; 1934. [Google Scholar]
- 38.Wit RD, Bouvier T. ‘Everything is everywhere, but, the environment selects’; what did Baas Becking and Beijerinck really say? Environ Microbiol. 2006;8:755–8. doi: 10.1111/j.1462-2920.2006.01017.x. [DOI] [PubMed] [Google Scholar]
- 39.Dick GJ. The microbiomes of deep-sea hydrothermal vents: distributed globally, shaped locally. Nat Rev Microbiol. 2019;17:271–83. doi: 10.1038/s41579-019-0160-2. [DOI] [PubMed] [Google Scholar]
- 40.Martiny JBH, Bohannan BJM, Brown JH, Colwell RK, Fuhrman JA, Green JL, et al. Microbial biogeography: putting microorganisms on the map. Nat Rev Microbiol. 2006;4:102–12. doi: 10.1038/nrmicro1341. [DOI] [PubMed] [Google Scholar]
- 41.Diepenbroek M, Glöckner FO, Grobe P, Güntsch A, Huber R, König-Ries B, et al. Towards an integrated biodiversity and ecological research data management and archiving platform: the German federation for the curation of biological data (GFBio). In: Plödereder E, Grunske L, Schneider E, Ull D, editors. Informatik 2014. Bonn: Gesellschaft für Informatik e.V.; 2014, p. 1711–24.
- 42.Yilmaz P, Kottmann R, Field D, Knight R, Cole JR, Amaral-Zettler L, et al. Minimum information about a marker gene sequence (MIMARKS) and minimum information about any (x) sequence (MIxS) specifications. Nat Biotechnol. 2011;29:415–20. doi: 10.1038/nbt.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data (metagenomes and symbiont MAGs) are available in the European Nucleotide Archive (ENA) at EMBL-EBI under project accession number PRJEB36976. The data, together with their metadata, were deposited using the data brokerage service of the German Federation for Biological Data (GFBio [41]), with the standard information on sequence data provided as recommended [42].
Additional data files and scripts used in this study are available in the GitHub repository (https://github.com/muecker/Symbionts_in_a_mussel_hybrid_zone).