Abstract
Membrane-bound extracellular vesicles (EVs), secreted by cells from all three domains of life, transport various molecules and act as agents of intercellular communication in diverse environments. Here we demonstrate that EVs produced by a hyperthermophilic and acidophilic archaeon Sulfolobus islandicus carry not only a diverse proteome, enriched in membrane proteins, but also chromosomal and plasmid DNA, and can transfer this DNA to recipient cells. Furthermore, we show that EVs can support the heterotrophic growth of Sulfolobus in minimal medium, implicating EVs in carbon and nitrogen fluxes in extreme environments. Finally, our results indicate that, similar to eukaryotes, production of EVs in S. islandicus depends on the archaeal ESCRT machinery. We find that all components of the ESCRT apparatus are encapsidated into EVs. Using synchronized S. islandicus cultures, we show that EV production is linked to cell division and appears to be triggered by increased expression of ESCRT proteins during this cell cycle phase. Using a CRISPR-based knockdown system, we show that archaeal ESCRT-III and AAA+ ATPase Vps4 are required for EV production, whereas archaea-specific component CdvA appears to be dispensable. In particular, the active EV production appears to coincide with the expression patterns of ESCRT-III-1 and ESCRT-III-2, rather than ESCRT-III, suggesting a prime role of these proteins in EV budding. Collectively, our results suggest that ESCRT-mediated EV biogenesis has deep evolutionary roots, likely predating the divergence of eukaryotes and archaea, and that EVs play an important role in horizontal gene transfer and nutrient cycling in extreme environments.
Subject terms: Archaeal physiology, Archaea
Introduction
Extracellular vesicles (EVs) are membrane-bound particles of variable diameter secreted by the cells into extracellular milieu. Although known for several decades, EVs were broadly regarded as cellular waste products, debris or artifacts of lipid aggregation [1]. However, the growing body of data shows that EVs play multiple, biologically important roles in all three domains of life [1–7]. During the past decade, it was discovered that EVs are responsible for intercellular shuttling of diverse cargoes, including proteins, DNA, RNA, lipids and various signaling molecules [5, 7–10], and may promote certain human pathologies [11, 12], including cancer [13, 14]. Furthermore, EVs hold great promise as vehicles for drug targeting and delivery [15, 16]. Finally, EVs may play an important ecological role, especially, in aquatic ecosystems [5]. It has been shown that DNA-carrying EVs produced by diverse bacteria, including Prochlorococcus, a numerically dominant marine cyanobacterium, are abundant in coastal and open-ocean seawater samples [7]. Importantly, Prochlorococcus EVs could support the growth of heterotrophic bacterial cultures, which implicates EVs in marine carbon flux [7]. Archaea of the phyla Euryarchaeota and Crenarchaeota are also known to produce EVs under laboratory conditions. Thermococcus prieurii, but not other Thermococcus species, secrete EVs packed with elemental sulfur, presumably to prevent the accumulation of toxic levels of sulfur in the cytoplasm [17]. Furthermore, similar to bacteria, EVs produced by members of the phylum Euryarchaeota thriving in deep-sea hydrothermal vents (order Thermococcales) and saline lakes (order Halobacteriales) were shown to carry DNA [18–21]. Whether the same is true for EVs produced by crenarchaea of the order Sulfolobales, which represent major inhabitants of terrestrial acidic hot springs, remains unknown. It is also unclear whether archaeal EVs are secreted under natural growth conditions in the environment.
The mechanisms of EV biogenesis have been extensively studied in eukaryotes but remain poorly understood in bacteria and archaea [1, 3]. In eukaryotes, the most studied mechanism of EV formation relies on the endosomal sorting complex required for transport (ESCRT) machinery [22–24]. Many archaea also encode homologs of the ESCRT system but its involvement in EV biogenesis remains unclear. The ESCRT machinery is responsible for many key membrane remodeling processes in eukaryotic cells, including membrane abscission during cytokinesis, biogenesis of certain types of EVs and multivesicular bodies, and budding of enveloped viruses, such as HIV-1 and Ebola virus [22, 25, 26]. The ESCRT proteins assemble on the cytosolic face of the membrane and drive membrane bending and scission reaction [26]. The ESCRT machinery can be subdivided into several functionally distinct subcomplexes known as ESCRT-0, ESCRT-I, ESCRT-II, and ESCRT-III as well as AAA+ ATPase Vps4. Among these, ESCRT-III and Vps4 are universally involved in ESCRT-dependent membrane remodeling processes, whereas ESCRT-0, ESCRT-I, and ESCRT-II are compartment-specific and facilitate recruitment of ESCRT-III to diverse membranes in different cellular contexts [25, 27]. ESCRT-III proteins form a ring-like filament at the membrane, whereas the Vps4 ATPase binds directly to ESCRT-III and dynamically disassembles the ESCRT-III complex in an ATP-dependent manner, thereby driving membrane-remodeling [28, 29].
Similar to eukaryotes, most archaea of the TACK (for Thaumarchaeota, Aigarchaeota, Crenarchaeota and Korarchaeota) and Asgard superphyla encode an ESCRT machinery [30–33]. Interestingly, the ESCRT machinery encoded by Asgard archaea is phylogenetically more closely related to the eukaryotic homologs compared to those from other archaea and Asgard Vps4 could efficiently complement the vps4 null mutant of Saccharomyces cerevisiae [31, 34]. However, due to difficulties in cultivation and lack of genetic tools in Asgard archaea, the role of their ESCRT machinery in membrane remodeling remains to be investigated in this superphylum. The archaeal ESCRT system has been experimentally investigated in Sulfolobus and Nitrosopumilus species [33, 35–41], with Sulfolobus representing the model organism for elucidation of the role and functioning of the archaeal ESCRT machinery [30, 41, 42]. In hyperthermophilic archaea of the order Sulfolobales, the ESCRT machinery is the key component of cell division apparatus composed of AAA+ ATPase Vps4 (also known as CdvC), four ESCRT-III homologs (ESCRT-III [CdvB], ESCRT-III-1 [CdvB1], ESCRT-III-2 [CdvB2], ESCRT-III-3 [CdvB3]), and archaea-specific component CdvA. The latter protein binds to DNA [43, 44] and recruits ESCRT-III to the membrane [44]. CdvA is not homologous to the eukaryotic ESCRT-0, ESCRT-I, or ESCRT-II [45], and is missing in certain archaea, including some thaumarchaea [46] and aigarchaea [47]. Recently, it has been demonstrated that ESCRT machinery also mediates asymmetric cell division via budding in virus-infected S. islandicus cells [48]. It has been also proposed that the Sulfolobus ESCRT machinery is involved in viral assembly within the cytoplasm and in escape from the infected cell by using a unique lysis mechanism [49]. Whether the function of archaeal ESCRT machinery can be extended to other membrane remodeling processes, such as budding of enveloped viruses [50] and EVs [3], remains to be demonstrated. Notably, ESCRT-III-1, ESCRT-III-2, and Vps4 were identified among proteins present within EVs secreted by Sulfolobus acidocaldarius, S. solfataricus and S. tokodaii [51]. This finding suggested that ESCRT machinery is involved in EV biogenesis [51]. However, EVs are known to be produced by archaea which lack the functional ESCRT system and divide using the bacterial-like FtsZ-based cell division machinery, including halophilic archaea (class Halobacteria) and members of the order Thermococcales [18, 52].
Here we characterize the composition, role and biogenesis of EVs produced by a hyperthermophilic and acidophilic archaeon Sulfolobus islandicus. We demonstrate that besides proteins, Sulfolobus EVs carry chromosomal and plasmid DNA, and that EVs can transfer this DNA to recipient cells. We also investigate the role of the Sulfolobus ESCRT machinery in EV biogenesis and show that all four ESCRT-III homologs and Vps4 ATPase play an important role in this process, whereas CdvA appears to be dispensable. Using synchronized S. islandicus cultures, we demonstrate that EV production is linked to cell division and appears to follow the cell cycle-coordinated fluctuations in the expression of ESCRT proteins. Finally, we show that EVs similar to those produced by Sulfolobus cells under laboratory conditions can be also found in an environmental sample collected from a terrestrial hot spring. Collectively, our results suggest that the ESCRT-dependent mechanism of EV biogenesis is conserved in the archaeo-eukaryotic lineage and that EVs play an important role in gene transfer in extreme environments.
Results and discussion
EV production and purification
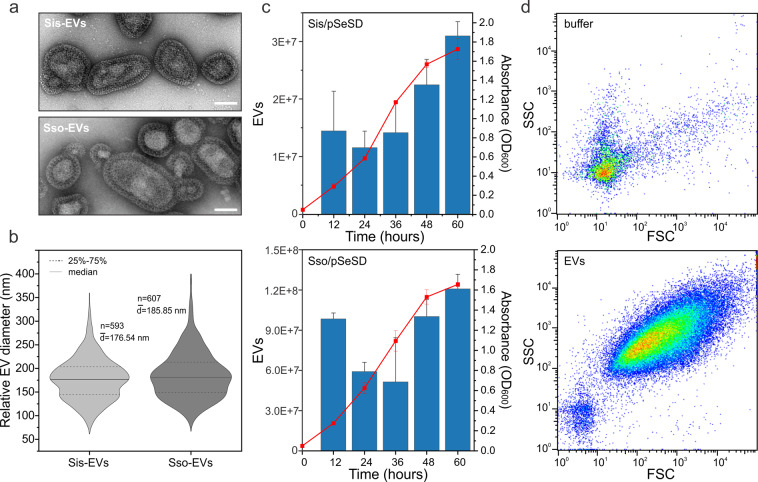
To study the composition and function of archaeal EVs, and to investigate the role of ESCRT in their biogenesis, we established a procedure for purification and quantification of EVs from Sulfolobus islandicus REY15A and Saccharolobus solfataricus PH1 cells (Sis-EVs and Sso-EVs, respectively), and compared the EV production at different growth stages in both strains. Consistent with the observations made for EVs isolated from other Sulfolobus species [51], Sis-EVs and Sso-EVs were visibly coated with the proteinaceous surface (S-)layer (Fig. 1a) typical of Sulfolobus cells [53, 54] and displayed considerable variation in shape and diameter. The median diameters of Sis-EVs and Sso-EVs were 176.54 nm and 185.85 nm, respectively, with the Sso-EVs being slightly more variable in size (Fig. 1b). The EVs were collected at different stages of cell growth (Fig. 1c) and could be reproducibly quantified by flow cytometry using tubes containing a calibrated number of fluorescent beads (Fig. 1d and Fig. S1). With a notable exception of the 12-h time point, EV production by S. islandicus and S. solfataricus followed a similar pattern: EV titer increased throughout the growth of the cells (Fig. 1c). Given the similarities in EV production patterns in S. islandicus and S. solfataricus, for all subsequent experiments, we focused on EVs from S. islandicus, for which more advanced genetic tools are available [55]. We next tested whether there is a link between Sis-EV production and increase in the fraction of the dead cells in the growing S. islandicus population by performing the live/dead staining at different time points (see “Materials and Methods”). From 12 to 60 h (early exponential to stationary phase) the ratio of dead cells remained at around 1% and only when the cells progressed into the “death” phase, the ratio of dead cells increased sharply, with around 30% of dead cells at 72 h and more than 90% of dead cells at 84 h (Fig. S2). These results suggest that Sis-EV production is not a consequence of cell death. To verify that the EV preparations were devoid of cellular contaminants, we performed the following procedures: (i) flow cytometry profiles of the EV samples were compared with those containing Sulfolobus cells (Fig. S1); (ii) purified EV preparations were subjected to semi-quantitative transmission electron microscopy analysis (Fig. S3a); (iii) EV preparations were also analyzed by fluorescence microscopy (Fig. S3b); and (iv) EV preparations were plated on solid medium supporting the growth of Sulfolobus cells (Fig. S4). None of these procedures revealed the presence of Sulfolobus cells, viable or otherwise, in the EV preparations.
Fig. 1. Characterization of the Sulfolobus islandicus EVs.
a Transmission electron micrographs of negatively stained Sis-EVs (top) and Sso-EVs (bottom). Scale bars, 100 nm. b Violin plots showing the size distributions of Sis-EVs (n = 593) and Sso-EVs (n = 607). The width of the distribution corresponds to the frequency of occurrence. c Growth curves of S. islandicus E233S and S. solfataricus PH1-16 harboring the vector pSeSD, and quantification of EVs released at indicated time points. Error bars represent standard deviation from three independent experiments. d Quantification of Sis-EVs by flow cytometry. Top panel shows the control with buffer only. SSC side scattering, FSC forward scattering.
Protein content of Sis-EVs
EVs are known to carry diverse cargo, including proteins [2–5]. Proteomics analysis of Sis-EVs and S. islandicus cells led to the identification of 413 and 1035 proteins, respectively (Supplementary Data 1). The number of proteins detected in Sis-EVs is considerably higher than that reported previously for EVs from S. acidocaldarius, S. solfataricus and S. tokodaii [51], possibly due to improved sensitivity of mass-spectrometry over the past decade. Notably, recent studies on the proteomics of EVs produced by diverse bacteria [56–59] as well as halophilic archaea [18] report the presence of hundreds of proteins in each type of EVs, consistent with our results. For instance, it has been shown that EVs produced by halophilic archaeon Halorubrum lacusprofundi contain 447 different proteins [18].
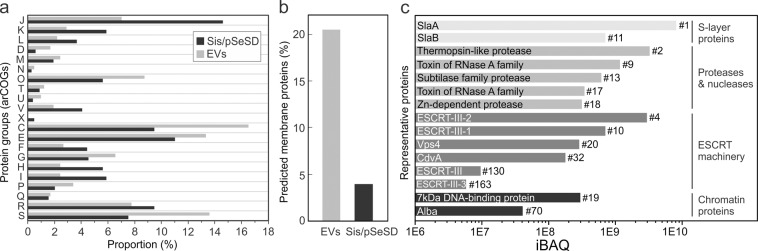
All but one functional protein categories found in S. islandicus proteome, as defined using the archaeal clusters of orthologous groups (arCOG; Table S1) [60], were represented in the Sis-EVs (Fig. 2a). Proteins of the arCOG category X (Mobilome: prophages, transposons) were not found in the Sis-EVs, likely due to the fact that only few proteins of this functional category are expressed in S. islandicus under normal growth conditions [61, 62]. The fractions of proteins of the categories J (Translation, ribosomal structure and biogenesis), K (Transcription), V (Defense mechanisms), H (Coenzyme transport and metabolism) and I (Lipid transport and metabolism) were more than twice smaller compared to their corresponding fractions in the total cellular proteome. By contrast, arCOG categories D (Cell cycle control, cell division, chromosome partitioning), N (Cell motility), O (Posttranslational modification, protein turnover, chaperones), U (Intracellular trafficking, secretion, and vesicular transport), C (Energy production and conversion), P (Inorganic ion transport and metabolism) and S (Function unknown) were enriched in Sis-EVs compared to the total cellular proteome (Fig. 2a). For instance, the D category proteins constitute only 0.6% of the total S. islandicus proteome, whereas in Sis-EVs, these proteins correspond to 1.7% of proteins (2.9-fold increase). There is also notable enrichment in Sis-EVs of proteins with predicted transmembrane domains compared to the cellular proteome (20% vs 4%; Fig. 2b). Of the top 100 most abundant proteins in the EVs, 65 have predicted transmembrane domains, whereas there are no such proteins among the top 100 most abundant cellular proteins. Thus, although Sis-EVs incorporate a considerable fraction of the total S. islandicus proteome, there is a strong enrichment for certain functional categories and, in particular, for membrane proteins. Presence in the Sis-EVs of proteins from nearly all functional categories suggests that many of these proteins are incorporated non-selectively, by entrapment of the cytosolic and membrane contents. It is possible, if not likely, that not all of the proteins are present within each Sis-EV, but are rather distributed across the EV population.
Fig. 2. Analysis of the Sis-EV protein content.
The EVs were collected during the exponential growth phase (24 h) of Sis/pSeSD, purified on sucrose gradient, treated with DNase I and subjected to mass spectrometry analysis. a Functional classification of proteins identified in highly purified Sis-EVs using archaeal clusters of orthologous groups (arCOGs). arCOG categories are indicated with capital Roman letters, with the annotation provided in Table S1. b Fraction of proteins with predicted transmembrane domains in Sis-EV and cellular (Sis/pSeSD) proteomes. c Label-free intensity-based absolute quantification (iBAQ) of selected Sis-EV proteins (the corresponding functional categories are indicated on the right). Numbers next to each bar indicate the abundance rank.
Sulfolobus EVs have been previously shown to carry toxins, dubbed sulfolobicins, active against closely related Sulfolobus species [63, 64]. However, homologs of these particular toxins are not encoded in the S. islandicus REY15A genome. Nevertheless, nearly one-third of proteins in the O category in Sis-EVs corresponded to diverse proteases and nucleases. Notably, we also detected two putative toxins (WP_014512538 and WP_014512541) of the RNase A family and several hydrolases of diverse specificities (Fig. 2c; Supplementary Data 1). This finding suggested that deployment of the Sis-EV payload could be toxic to recipient cells lacking necessary immunity. Incubation of Sis-EVs with Sulfolobus cells for 3 h indeed resulted in modest, albeit significant, decrease in colony forming units for S. solfataricus cells, but not for the more distantly related S. acidocaldarius or S. shibatae (Fig. S5a). However, the inhibitory effect of Sis-EVs on S. solfataricus was temporary and was lifted when the incubation was prolonged to 5 h (Fig. S5b). Thus, Sis-EVs do not appear to participate in intermicrobial conflicts, at least, not among the tested Sulfolobus species.
Sis-EVs contained all six components of the Sulfolobus ESCRT machinery (arCOG category D; Fig. 2c), consistent with the possibility that ESCRT machinery is involved in EV biogenesis [51]. Label-free intensity-based absolute quantification (iBAQ) [65] of protein abundances showed that two of the ESCRT components, ESCRT-III-2 and ESCRT-III-1, were among the top-10 most abundant proteins in Sis-EVs (Fig. 2c). Western blot analysis has confirmed that both proteins were present and strongly enriched in Sis-EVs (Fig. S6). As expected, both S-layer proteins were found in the EVs, with SlaA being the most abundant protein in Sis-EVs (Fig. 2c).
Sis-EVs carry plasmid and genomic DNA
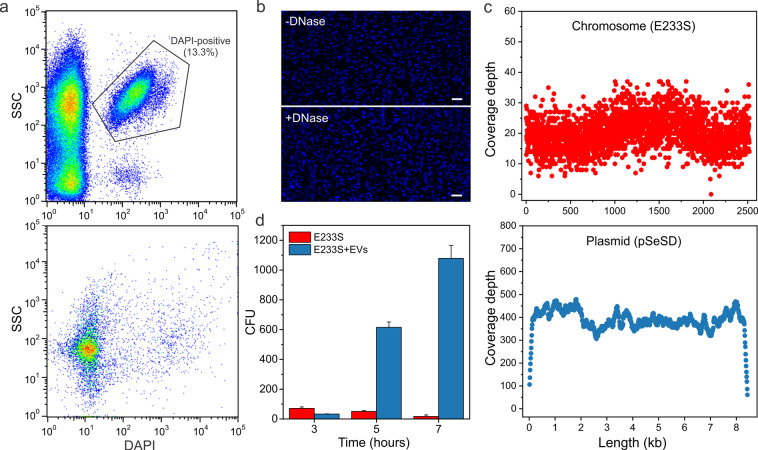
Sis-EVs carried the chromatin proteins Sac7d/Sso7d and Alba (Fig. 2c), responsible for compaction of the Sulfolobus chromosome [66, 67], suggesting that Sis-EVs contain DNA. Indeed, DNA has been previously observed in EVs from halophilic archaea and Thermococcus spp. (both in the phylum Euryarchaeota) [18, 19, 21, 68] but never reported in Sulfolobus EVs. To test if Sis-EVs carry DNA, purified EVs were treated with DNase I and stained with 4′,6-diamidino-2-phenylindole (DAPI). The DAPI-stained Sis-EVs could be detected by both flow cytometry (Fig. 3a) and fluorescence microscopy (Fig. 3b), consistent with the presence of DNA. Notably, only 13.3% of EVs detected by the flow cytometry were DAPI-positive, whereas the majority of EVs were DAPI-negative, indicating a heterogeneity of the EV content.
Fig. 3. Sis-EVs promote gene transfer.
a Analysis of DAPI-stained Sis-EVs isolated from Sis/pSeSD (upper panel) by flow cytometry. All the events are shown, with the selected region indicating the DAPI-positive EVs harboring DNA. Note that most EVs are DAPI-negative. Bottom panel shows the control with the DAPI-containing buffer only. SSC, side scattering. b Fluorescence micrographs of DAPI-stained Sis-EVs prior (top) and after (bottom) DNase I treatment. Bar, 2 μm. c Sequencing depth across the chromosomal DNA (E233S) and plasmid (pSeSD). Each dot represents sequencing depth at the indicated position of the corresponding replicon. d Gene/plasmid transfer by Sis-EVs. Sis-EVs were treated with DNase I and then mixed with E233S cells, incubated for 3, 5, and 7 h and plated on selective plates. In the control experiment, E233S cells were mixed with the equal volume of PBS buffer. The number of obtained colony forming units (CFU) is plotted on the y-axis. Error bars represent standard deviation from three independent experiments.
High-throughput sequencing of the DNA extracted from Sis-EVs yielded reads aligning to both the S. islandicus chromosome and the resident extrachromosomal plasmid pSeSD. Both replicons were covered throughout their respective lengths (Fig. 3c), but the median sequencing depth of the plasmid was 19 times higher than that of the chromosome (386× versus 20× coverage, respectively). This difference cannot be explained by the higher copy number of the plasmid (3-5 copies per 1 chromosome copy) [69]. It is most probable that, as in the case of the Thermococcus [70] and bacterial [8] EVs, overlapping genomic fragments of variable sizes, rather than complete chromosome, are packed into the Sis-EVs.
To test the biological relevance of DNA incorporation into Sis-EVs, we investigated the ability of Sis-EVs to transfer the plasmid-borne pyrEF locus into a plasmid-free auxotrophic strain E233S of S. islandicus carrying a chromosomal deletion in the pyrEF operon responsible for uracil synthesis (Fig. S7) [69]. To this end, Sis-EVs produced by the pSeSD-carrying strain were purified, treated with DNase I, incubated with recipient E233S cells in liquid culture and plated on solid medium devoid of uracil. In the presence of EVs, strain E233S formed over 1000-fold more colonies than the control E233S cells (Fig. 3d), whereas plating of EVs alone did not yield colonies on either rich or uracil-deficient medium (Fig. S8a), further confirming that there were no cells contaminating EV preparation. Approximately half of the colonies obtained after incubation with Sis-EVs carried the pSeSD plasmid (Fig. S8b-e). To test if the plasmid-devoid strains carry the pyrEF cassette elsewhere on the chromosome, we performed PCR with the pyrF-specific primers (Fig. S7). However, pyrF gene was present exclusively in the pSeSD-carrying strains (Fig. S8f), indicating that there was no ectopic pyrF integration. Next, we verified if the ability of the plasmid-lacking strains to grow in the absence of uracil was inheritable. To this end, the plasmid-containing and plasmid-deficient cells from initial colonies were resuspended in the selective medium and spotted on the solid medium lacking uracil (Fig. S8g). Only plasmid-containing strains could grow, suggesting that initial growth of the colonies in the absence of uracil was supported by the nutrients provided by the EVs, whereas transfer of such colonies into fresh medium arrested the cell growth, unless the cells contained pSeSD. Collectively, these results demonstrate that Sis-EVs carry DNA and act as vehicles for gene transfer. The exact mechanism of EV-mediated gene transfer into recipient cells remains unclear but, presumably, it involves fusion between the EV and cell membranes.
Sis-EVs support heterotrophic growth of Sulfolobus
To further investigate if EVs can provide nutrients (other than uracil) to support heterotrophic cell growth, Sulfolobus cells were inoculated in media lacking nitrogen and/or carbon source. As expected, cells could not grow in a solution containing only mineral salts and a mix of vitamins (Fig. 4a), nor could they grow when only carbon source (sucrose) was added to this solution (Fig. 4b). Instead, slight decrease in optical density of the culture was observed, suggesting partial lysis. However, when either medium was supplemented with purified Sis-EVs, there was significant (two paired t test, p < 0.05), Sis-EV concentration-dependent increase in the optical density of S. islandicus culture, indicative of cell growth (Fig. 4). The same result was obtained with EVs isolated from Sis/pSeSD-CdvA (see below). These results strongly suggest that EVs can serve as a source of both carbon and nitrogen, and hence play an important role in nutrient cycling in extreme environments. Similarly, it has been shown that EVs produced by cyanobacteria can support the growth of heterotrophic bacteria by serving as a carbon source [7].
Fig. 4. Sis-EVs support heterotrophic growth of Sulfolobus cells.
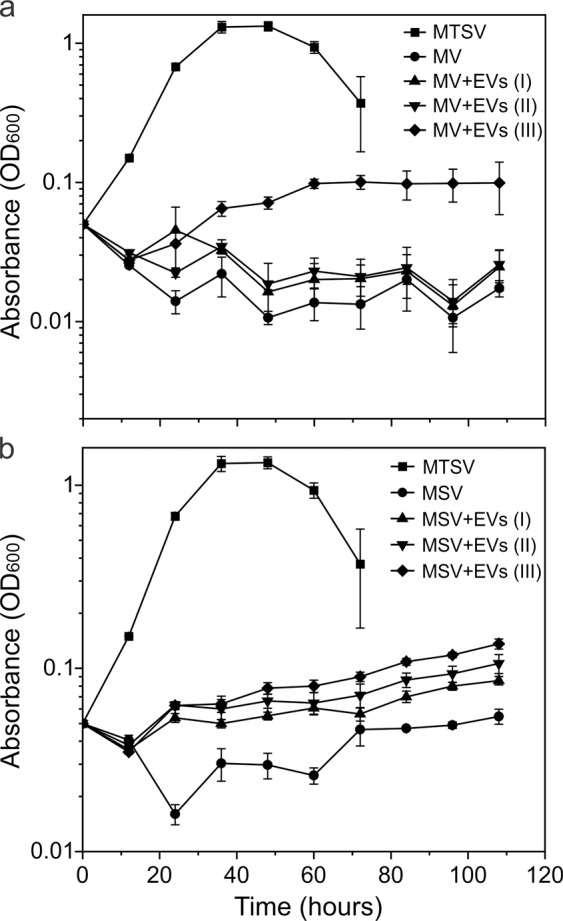
a Growth curves of S. islandicus REY15A in MV (M: mineral salts, V: vitamin mix) solution lacking organic carbon and nitrogen sources; MV solution supplemented with 1.5 ml preparations containing different amounts of EVs (MV + EVs): 24.8 μg for I, 49.6 μg for II, and 74.4 μg for III; and rich medium (MTSV), which in addition to MV solution contains 0.2% (wt/vol) sucrose (S) and 0.2% (wt/vol) tryptone (T). There was significant increase (two paired t test, p < 0.05) in optical density (OD600) of REY15A cells in the MV solution supplemented with EVs (III), as compared to the control culture lacking EVs. b Growth curves of S. islandicus REY15A in MSV (M: mineral salts, S: sucrose, V: vitamin mix) medium lacking organic nitrogen source; MSV medium supplemented with different concentrations of EVs (MSV + EVs), and rich MTSV medium. There was significant increase (two paired t test, p < 0.05) in optical density (OD600) of REY15A cells in the MSV medium supplemented with EVs, as compared to the control culture lacking EVs. Error bars represent standard deviations from three independent experiments.
To the best of our knowledge, production of EVs by different Sulfolobales strains or by any other archaeal strain has been reported only under laboratory cultivation conditions. To verify whether EVs are also produced in the environment, we analyzed a previously collected archaea-dominated hot spring sample [71] for the presence of EVs. The contents of the sample were directly concentrated by ultracentrifugation without prior cultivation in the laboratory and visualized by TEM. We observed multiple S-layer-coated EVs closely resembling those produced by Sulfolobales species (Fig. S9). The diameter of the observed EVs varied between 77 and 182 nm, which is considerably smaller than the size of the smallest known archaea, i.e., Nanoarchaea spp. with the diameter of ~400 nm [72, 73], confirming that these are subcellular structures. These results strongly suggest that Sis-EVs are not laboratory artifacts and are environmentally and biologically relevant.
Sis-EVs biogenesis is ESCRT-dependent
Sis-EV biogenesis occurs through budding from the cytoplasmic membrane (Fig. 5a) and ESCRT system is a prime suspect implicated in membrane constriction and scission. To investigate the involvement of ESCRT machinery in EV biogenesis, we constructed a collection of knockdown strains in which expression of each of the six ESCRT machinery components was depleted by the endogenous type III-B CRISPR-Cas system of S. islandicus. The utility of this strategy for gene knockdown has been recently demonstrated in Sulfolobus [54, 74, 75]. Quantitative reverse transcription PCR (RT-qPCR) analysis has shown that whereas expression of escrt-III was decreased by ~30%, expression of all other genes was down by 60–70% (Fig. 5b). Western blot analysis of escrt-III-1 and escrt-III-2 knockdown strains has shown that the levels of the corresponding proteins have been decreased by 99% and 60%, respectively (Fig. S6b). It has been previously shown that escrt-III and vps4 are expressed from a bicistronic operon [44, 76] (Fig. S10a). Thus, we verified whether CRISPR targeting of the escrt-III gene has a polar effect on the expression of the vps4. There was no significant difference in the vps4 transcript levels between the escrt-III knockdown and control cells (Fig. S10b). This is consistent with the previous results showing that cleavage of a transcript by type III-B CRISPR system in S. islandicus REY15A occurs within 20 bp of the CRISPR spacer targeting [77]; that is, the fragment of the transcript encoding Vps4 is unaffected by the cleavage within the ESCRT-III-encoding region (Fig. S10a).
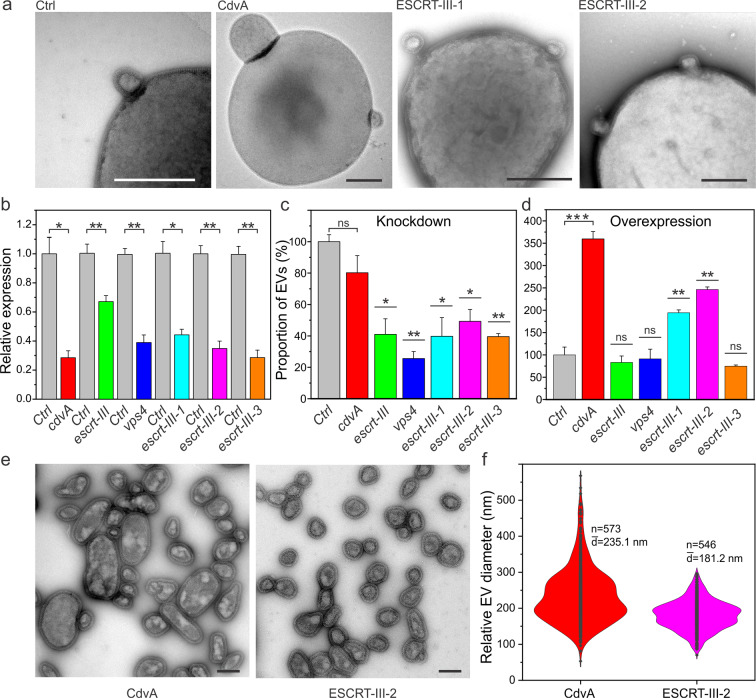
Fig. 5. ESCRT-dependent biogenesis of Sis-EVs.
a Representative transmission electron micrographs showing EV budding from S. islandicus strains overexpressing indicated proteins. Ctrl: control E233S cells carrying empty vector pSeSD. Bars, 400 nm. b RT-qPCR analysis of the RNA interference efficiency. Stars indicate the significance levels based on the paired two-tailed t test. The p values are 0.01512, 0.00514, 0.00737, 0.0146, 0.00883, 0.00733, respectively. Error bars represent standard deviation from three independent experiments. c Quantification of Sis-EVs released from strains in which different ESCRT machinery components were depleted by CRISPR targeting. Stars indicate the significance levels based on the paired two-tailed t test. The p values are 0.01047, 0.00316, 0.02337, 0.01763, and 0.00177. ns, non-significant. Error bars represent standard deviation from three independent experiments. d Quantification of Sis-EVs released from strains overexpressing indicated ESCRT machinery components. Stars indicated the significance levels based on the paired two-tailed t test. The p values are 0.001, 0.0094, and 0.00435. ns, non-significant. Error bars represent standard deviation from three independent experiments. e Representative transmission electron micrographs of negatively stained Sis-EVs isolated from cells overexpressing CdvA and ESCRT-III-2. Bars, 200 nm. f Violin plots showing the size distributions of Sis-EVs isolated from cells overexpressing CdvA (n = 573) and ESCRT-III-2 (n = 546). The width of the distribution corresponds to the frequency of occurrence.
Knockdown strains of cdvA, vps4, escrt-III, and escrt-III-1 displayed considerable cell growth defects, whereas those of escrt-III-2 and escrt-III-3 showed nearly normal cell growth (Fig. S11a). The depletion of cdvA, vps4, escrt-III and escrt-III-1 transcripts by CRISPR targeting resulted in obvious cell division defects (Fig. S11b, c), yielding cells 2–3 times larger than the control cells, and in slight increase (<7%) in the fraction of dead cells in the corresponding populations (Fig. S11d). Consistent with the growth dynamics (Fig. S11a), cell size of the escrt-III-2 and escrt-III-3 knockdown strains was similar to that of the control cells (Fig. S11b and S11c). The lack of growth retardation for the escrt-III-2 knockdown strain is somewhat unexpected, given that all previous attempts to delete this gene in S. islandicus were unsuccessful, whereas escrt-III-3 is known to be non-essential for normal growth [38, 78]. Notably, under the growth conditions used in this study, the expression of escrt-III-3 was much lower than that of all other cell division genes (Fig. S12). By contrast, the transcription level of escrt-III-2 was six and three times higher than those of escrt-III and escrt-III-1, respectively (Fig. S12). Thus, even with 60-70% decrease in transcript levels due to CRISPR targeting, the total level of escrt-III-2 transcripts would be comparable to that of other ESCRT-III homologs. Presumably, these levels are sufficient for normal growth of S. islandicus. Notably, escrt-III-2 is not essential in S. acidocaldarius [40]. Thus, we do not exclude the possibility that escrt-III-2 is also not strictly required for the growth of S. islandicus and that previous attempts to delete this gene were hindered by other factors.
Quantification of Sis-EVs produced by the knockdown strains has shown that whereas depletion of cdvA had no significant effect on Sis-EV titer, all other knockdown strains, including the non-essential escrt-III-3, produced significantly less EVs compared to the control strain (Fig. 5c). The vps4 knockdown strain displayed the strongest effect, with EV production being decreased by over 70%. Notably, it is possible that different ESCRT-III homologs can partially complement each other during EV biogenesis. Interestingly, overexpression of the ESCRT-III-1 and ESCRT-III-2 from a plasmid resulted in 200–250% increase in vesiculation (Fig. 5d) consistent with their role in Sis-EV budding. Unexpectedly, overexpression of CdvA resulted in hypervesiculation phenotype (Fig. 5d). However, the same effect was also observed when CdvA lacking the C-terminal domain (CdvAΔC) responsible for interaction with ESCRT-III [43, 44] was overexpressed (Fig. S13a), suggesting that excessive binding of CdvA to the membrane [44] precipitates the observed phenomenon. Overexpression of cdvA and cdvAΔC yielded cells with up to 2-5 fold larger diameters (Fig. S13b). By contrast, overexpression of ESCRT-III-1 and ESCRT-III-2 had no effect on cell size or cell viability (Fig. S13b and S13c). Taken together, the overexpression and knockdown results show that there is no apparent link between the cell size and EV biogenesis.
Budding of EVs from the control and overexpression strains was observed directly by TEM (Fig. 5a). Notably, EVs produced by the CdvA overexpression strain were considerably larger than those from the control (Fig. 1a) and ESCRT-III-2 overexpression (Fig. 5e) strains, with an average diameter of 235 nm versus 177 and 181 nm, respectively (Figs. 1b and 5f). To exclude the possibility that the large EVs produced by the CdvA overexpression strain represent small cells, the EV-containing supernatant was filtered through 0.45 μm filter and plated on the solid medium. No colonies were formed (Fig. S4b). Our results strongly suggest that EV budding in Sulfolobus is dependent on the ESCRT machinery, including Vps4 ATPase and the ESCRT-III ensemble, whereas CdvA appears to be dispensable for this process.
Sis-EVs biogenesis is linked to cell division
The expression of ESCRT-III homologs in S. acidocaldarius is linked to the cell cycle [37, 79]. To verify whether the same is true for S. islandicus and if EV biogenesis is linked to the cell cycle, we synchronized the S. islandicus culture by adapting a protocol previously used for S. acidcaldarius [79, 80]. The cells were arrested at the G2 phase using acetic acid and could progress into cell division phase following the removal of the acid (see Materials and Methods for details). Analysis of the DNA content by flow cytometry has shown that the cells started transitioning from G2 into the cell division phase at 90 min following the removal of the acetic acid (Fig. 6a). Western blot analysis of the synchronized cells has shown that ESCRT-III, ESCRT-III-1, and ESCRT-III-2 proteins were undetectable during the G2 phase and became detectable at 90 min after the removal of acetic acid (Fig. 6b). Notably, however, whereas ESCRT-III was abundantly expressed at this time point, ESCRT-III-1 and ESCRT-III-2 were barely detectable. Conversely, at 150 min time point, when the expression of ESCRT-III-1 and ESCRT-III-2 peaked, the expression of ESCRT-III started to decline (Fig. 6b). This dynamics is consistent with the recent suggestion that ESCRT-III is the first to form a ring in the mid-cell during cell division, which serves a platform for subsequent recruitment of ESCRT-III-1 and ESCRT-III-2 [79]. We next analyzed the production of Sis-EVs in synchronized cultures at 60 (G2 phase), 90 (beginning of cell division) and 135 (advanced cell division) min after removal of acetic acid (Fig. 6c). There was a dramatic increase in EV production at the 135 min time point which coincides with active cell division (Fig. 6c, Fig. S14). These results strongly suggest that Sis-EV production is linked to the cell division and might be triggered by the natural, cell cycle-linked changes in the expression of ESCRT-III homologs. In particular, the active EV production appears to coincide with the expression pattern of ESCRT-III-1 and ESCRT-III-2, rather than ESCRT-III, suggesting a prime role of these proteins in EV budding. This conclusion is fully consistent with the observation that the two proteins are strongly enriched in EVs as well as with the fact that overexpression of ESCRT-III-1 and ESCRT-III-2, but not ESCRT-III, dramatically increases EV biogenesis.
Fig. 6. EV biogenesis is linked to cell division.
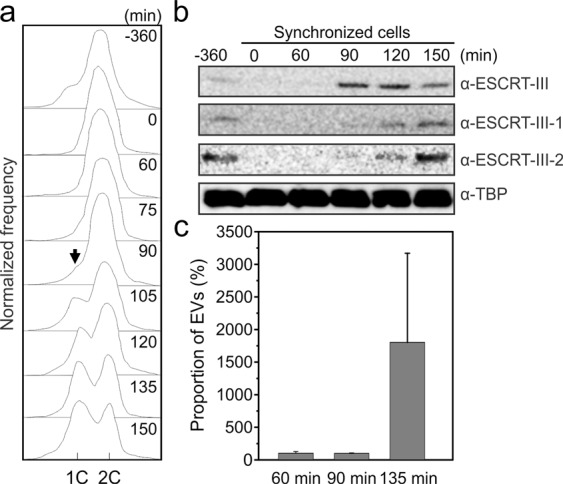
a Flow cytometry analysis of samples taken at the indicated time points during the progression of a synchronized culture of S. islandicus. The positions of peaks corresponding to one chromosome copy (1 C) and 2 C genome contents are indicated. Black arrow indicated the reappearance of the peak corresponding to the 1 C genome content, signifying cell division. b Western blot analysis of synchronized cells. Cells (~1×109) were collected at indicated time points and expression of ESCRT-III, ESCRT-III-1 and ESCRT-III-2 was analyzed using the corresponding antibodies. Tata-binding protein (TBP) was used as a loading control. c Flow cytometry analysis of Sis-EV production by synchronized Sis/pSeSD cells at different time points after removal of acetic acid: 60 min (prior to onset of cell division), 90 min (onset of cell division) and 135 min (active cell division). Error bars represent standard deviation from three independent experiments.
Concluding remarks
Here we have further characterized Sulfolobus EVs and showed that they carry DNA. Combined with the previous observation of DNA-containing EVs in euryarchaea (halobacteria and thermococci) [18, 19, 21, 52, 68, 81, 82], the finding that crenarchaeal EVs also contain DNA suggests that this property might be general across archaea. Horizontal gene transfer (HGT) is essential for the survival of microbial populations that otherwise deteriorate due to the Muller’s ratchet [83, 84]. Some bacteria and archaea are naturally competent and can uptake DNA from the environment [85, 86]. However, in low-density populations residing in high-temperature, acidic environments, as is the case for Sulfolobales, extracellular DNA might be neither stable nor readily available. In bacteria, conjugative plasmids, transducing bacteriophages and phage-derived gene transfer agents are considered the main drivers of the HGT. Although conjugative plasmids are known in Sulfolobus, their role in HGT has not been assessed [87]. By contrast, transducing viruses or dedicated gene transfer agents have not been described in Sulfolobales. Full-length genomic DNA could not be detected in the agarose gel, suggesting that only fragments of genomic DNA, which could represent byproducts of genome replication and repair, are incorporated into the Sis-EVs. Nevertheless, these DNA fragments collectively represented all genes present on the S. islandicus chromosome, as well as the resident plasmid. Importantly, Sis-EVs could successfully transfer the marker genes as well as the complete plasmid within the S. islandicus population. Furthermore, our data shows that S. islandicus can use EVs as carbon and nitrogen source, which is likely to be important in natural settings where nutrients are scarce. Collectively, these results indicate that EVs could play an important, yet overlooked role in gene transfer and nutrient flux in extreme environments. Indeed, we observed EVs resembling those produced by Sulfolobales directly in the environmental archaea-dominated sample, suggesting that properties of the EVs determined under laboratory conditions are biologically and environmentally relevant.
The mechanisms of EV biogenesis are poorly understood in prokaryotes [1]. Our results strongly suggest that Sulfolobus ESCRT machinery plays an important role in EVs formation. Importantly, EV budding appears to be specifically linked to cell division, when the expression of the ESCRT-III proteins is the highest. By contrast, CdvA appears to be dispensable for EV budding suggesting that there are mechanistic differences of the archaeal ESCRT functioning in different pathways of membrane remodeling. This would be similar to eukaryotes, where ESCRT-III complex is targeted to the membranes by different partner proteins [22]. We hypothesize that CdvA is substituted by a different targeting protein during EV budding. Notably, some archaea lack cdvA gene but encode ESCRT-III and Vps4 homologs [31, 45–47], suggesting that ESCRT-III targeting to the membrane in these organisms, similar to eukaryotes, is achieved by an unrelated protein or proteins. Alternatively, changes in membrane curvature at the EV budding sites might promote binding of ESCRT-III paralogs, without the necessary chaperoning of CdvA. Further in vitro experiments will be necessary to test this hypothesis. Regardless, our results show that the ESCRT-dependent mechanism of EV biogenesis is conserved in both archaea and eukaryotes, and likely represents one of the ancestral functions of the ESCRT system.
Materials and methods
Strains, growth conditions and transformation of Sulfolobus
Sulfolobus islandicus strains REY15A and E233S (REY15AΔpyrEFΔlacS) [69], and Sulfolobus solfataricus PH1-16 (PH1 pyrF mutant) [88], hereafter PH1-16, were grown aerobically with shaking (145 rpm) at 75 °C in 30 ml of STVU medium containing mineral salts (M), 0.2% (wt/vol) sucrose (S), 0.2% (wt/vol) tryptone (T), a mixed vitamin solution (V) and 0.01% (wt/vol) uracil (U); the pH was adjusted to 3.5 with sulfuric acid, as described previously [69]. SCV medium containing 0.2% (wt/vol) casamino acids (C) was used for selection of uracil prototrophic transformants. ATV medium containing 0.2% (wt/vol) arabinose (A) was used to induce protein overexpression and RNA interference. The plasmids and strains constructed and used in this study are listed in Tables S2 and S3, respectively. S. islandicus cells were synchronized using acetic acid (final concentration, 6 mM) as previously described for S. acidocaldarius [79, 80] (see Supplementary Methods for further details).
Isolation and purification of EVs
EVs were isolated from liquid cultures of S. islandicus E233S or S. solfataricus PH1-16 strains carrying shuttle vector pSeSD. The cells were grown at 75 °C in appropriate medium and EVs were harvested at the indicated times. Cells were removed by centrifugation at 7000 rpm at 4 °C for 20 min. The supernatant was filtered with 0.45 μm filter and EVs collected by ultracentrifugation at 40,000 rpm (Type 45 Ti rotor) at 4 °C for 2 h, followed by 100,000 rpm (TLA 100.2 rotor) at 4 °C for 1 h, and then re-suspended in 500 μl of PBS.
For mass spectrometry (see Supplementary Methods) and DNA content sequencing, the EVs were collected during the exponential growth phase (24 h) and further purified by ultracentrifugation in sucrose gradient (50%, 45%, 40%, 35%, 30%, and 25%) at 25,000 rpm (SW 60 rotor) at 4 °C for 10 min. The EVs formed an opalescent band in the region of the gradient corresponding to 30-40% sucrose (Fig. S15). The band was collected and EVs pelleted by ultracentrifugation at 100,000 rpm (TLA100.2 rotor) at 4 °C for 1 h. The resulting pellet was re-suspended in 500 μl of PBS.
Transmission electron microscopy
For TEM analysis, EVs or cell cultures were absorbed to glow-discharged copper grids with carbon-coated Formvar film and negatively stained with 2.0% (w/v) uranyl acetate. The samples were observed under FEI Tecnai Spirit BioTwin 120 microscope (FEI, Einthoven, The Netherlands) operated at 120 kV.
Flow cytometry and quantification of EVs
EVs were isolated from 50 ml cultures of E233S and PH1-16 cells carrying pSeSD vector at the given time points of cell growth, then 50 μl of the EV preparations were mixed with 250 μl PBS staining buffer containing 2.5 µg/ml DAPI (4’, 6-diamidino-2-phenylindole; Thermo Fisher Scientific, USA) and kept at 4 °C for 30 min. The EVs were analyzed and sorted on the MoFlo Astrios cell sorter (Beckman Coulter) equipped with an EQ module specifically developed to detect nanoparticles and with 488 nm and 561 nm lasers at 200 mW. The calibration of the machine was carried out using FITC-labeled Megamix-Plus SSC beads from BioCytex (Fig. S1). The sheath-liquid 0.9% NaCl (Revol, France) was filtered through a 0.04 μm filter. The analysis was performed using the side-scattered (SSC) light parameter of laser 561, with threshold set to 0.012% in order to have maximum of 300 events per second. An M2 mask was added in front of the forward-scattered (FSC) light.
To count the EVs, we used Trucount™ Tubes (BD Biosciences, San Diego, CA), which contain a defined number of fluorescently labeled beads and have been specifically designed for reproducible counting of various biological nanoparticles, including EVs [89, 90]. For quantification, we added the same volume (300 µl) of EV preparations into the tubes that contained the constant number of beads. The EV number was calculated using the following formula: EVtotal = (EV count/bead count) × total number of beads in the Trucount™ Tube. In each case, the samples were passed through the flow cytometer’s detector until 2000 beads were recorded. All quantifications by flow cytometry were done in triplicate (Fig. S14 and Fig. S16). Further details on cell cycle and cell size analysis by flow cytometry can be found in Supplementary Methods.
DNA isolation from EVs and sequencing
To remove the traces of extravesicular nucleic acids, prior to DNA extraction, EVs were incubated with DNase I (15 U/ml) and RNase (100 μg/ml), in the presence of MgCl2 (10 mM), at 37 °C for 30 min, followed by addition of EDTA (20 mM). EVs were disrupted by proteinase K (100 μg/ml) and SDS (0.5%) treatment at 55 °C for 30 min. The DNA was extracted by standard phenol/chloroform procedure, precipitated with 0.3 M sodium acetate (pH 5.3) and isopropanol. The resultant pellet was resuspended in DNase/RNase-free water and used for sequencing. Sequencing libraries were prepared from 100 ng of DNA with the TruSeq DNA PCR-Free library Prep Kit from Illumina and sequenced on Illumina MiSeq platform with 150-bp paired-end read lengths (Institut Pasteur, France). Raw sequence reads were processed with Trimmomatic v.0.3.6 and mapped to the reference genomes of REY15A and pSeSD plasmid using Bowtie2 [91] with default parameters and analyzed with Sequana [92].
EV-mediated gene transfer
EVs isolated and purified from 6 L of exponentially growing Sis/pSeSD culture (24 h) were used for gene transfer experiments as described previously [93], with some modification (see Supplementary Methods).
Live/Dead staining and fluorescence microscopy analysis
Live/Dead staining was carried out using the LIVE/DEAD BacLightTM Bacterial Viability Kit (Invitrogen, US) [94, 95] according to the supplier’s protocols. See Supplementary Methods for the detailed protocol.
Overexpression of ESCRT proteins
Plasmids expressing different ESCRT machinery components and their mutants were described previously [38] (Table S2). Briefly, cells harboring the plasmids were first inoculated into 30 ml of the MTSV medium and when the OD600 reached ~0.6–0.8, they were transferred into the ATV medium containing 0.2% (wt/vol) arabinose with an initial OD600 of 0.05 to induce protein expression. All plasmids are listed in Table S3.
Construction of the CRISPR type III-B-based RNA interference plasmids and RNA interference
The CRISPR type III-B-based RNA interference plasmids were constructed according to the methods described previously [74, 77]. The spacers selected and used in this study are listed in Table S4, whereas all other oligonucleotides are listed in Table S5. See Supplementary Methods for further details.
Western blot
ESCRT proteins were detected using antibodies against ESCRT-III, ESCRT-III-1 and ESCRT-III-2 (HuaAn Biotechnology Co., Hangzhou, Zhejiang, China), as described previously [38]. See Supplementary Methods for further details.
Supplementary information
Acknowledgements
This work was supported by l’Agence Nationale de la Recherche (#ANR-17-CE15-0005-01) and Ville de Paris Emergence(s) program (project MEMREMA) grants to MK, and European Research Council grant from the European Union’s Seventh Framework Program (FP/2007-2013)/Project EVOMOBIL-ERC Grant Agreement 340440 to PF. JL was partly supported through the PRESTIGE post-doctoral program from European Union’s Seventh Framework Programme, the National Key Research and Development Program of China (No. 2020YFA0906800), and the post-doctoral fellowship from the State Key Laboratory of Microbial Technology, Shandong University. We would like to thank Thibault Chaze and Mariette Matondo (Proteomics Platform, Institut Pasteur) as well as Thomas Cokelaer and Marc Monot (Biomics Platform, Institut Pasteur) for help with the proteomics and sequencing analyses, respectively. We are also grateful to the Ultrastructural BioImaging (UTechS UBI) unit of Institut Pasteur for access to electron microscopes. We gratefully acknowledge the UtechS Photonic BioImaging (Imagopole), C2RT, Institut Pasteur, supported by the French National Research Agency (France BioImaging; ANR-10–INSB–04; Investments for the Future).
Compliance with ethical standards
Conflict of interest
The authors declare no competing interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-021-00984-0.
References
- 1.Coelho C, Casadevall A. Answers to naysayers regarding microbial extracellular vesicles. Biochem Soc Trans. 2019;47:1005–12. doi: 10.1042/BST20180252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Woith E, Fuhrmann G, Melzig MF. Extracellular vesicles-connecting kingdoms. Int J Mol Sci. 2019;20:E5695. doi: 10.3390/ijms20225695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gill S, Catchpole R, Forterre P. Extracellular membrane vesicles in the three domains of life and beyond. FEMS Microbiol Rev. 2019;43:273–303. doi: 10.1093/femsre/fuy042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown L, Wolf JM, Prados-Rosales R, Casadevall A. Through the wall: extracellular vesicles in Gram-positive bacteria, mycobacteria and fungi. Nat Rev Microbiol. 2015;13:620–30. doi: 10.1038/nrmicro3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schatz D, Vardi A. Extracellular vesicles—new players in cell-cell communication in aquatic environments. Curr Opin Microbiol. 2018;43:148–54. doi: 10.1016/j.mib.2018.01.014. [DOI] [PubMed] [Google Scholar]
- 6.Soler N, Krupovic M, Marguet E, Forterre P. Membrane vesicles in natural environments: a major challenge in viral ecology. ISME J. 2015;9:793–6. doi: 10.1038/ismej.2014.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biller SJ, Schubotz F, Roggensack SE, Thompson AW, Summons RE, Chisholm SW. Bacterial vesicles in marine ecosystems. Science. 2014;343:183–6. doi: 10.1126/science.1243457. [DOI] [PubMed] [Google Scholar]
- 8.Biller SJ, McDaniel LD, Breitbart M, Rogers E, Paul JH, Chisholm SW. Membrane vesicles in sea water: heterogeneous DNA content and implications for viral abundance estimates. ISME J. 2017;11:394–404. doi: 10.1038/ismej.2016.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denham J, Spencer SJ Emerging roles of extracellular vesicles in the intercellular communication for exercise-induced adaptations. Am J Physiol Endocrinol Metab. 2020;319:E320-E329. [DOI] [PubMed]
- 10.Abramowicz A, Story MD The long and short of it: The emerging roles of non-coding RNA in small extracellular vesicles. Cancers. 2020;12:1445. [DOI] [PMC free article] [PubMed]
- 11.Gho YS, Lee J. Special issue on the role of extracellular vesicles in human diseases. Exp Mol Med. 2019;51:34. doi: 10.1038/s12276-019-0208-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malloci M, Perdomo L, Veerasamy M, Andriantsitohaina R, Simard G, Martinez MC. Extracellular vesicles: Mechanisms in human health and disease. Antioxid Redox Signal. 2019;30:813–56. doi: 10.1089/ars.2017.7265. [DOI] [PubMed] [Google Scholar]
- 13.Linxweiler J, Junker K. Extracellular vesicles in urological malignancies: an update. Nat Rev Urol. 2020;17:11–27. doi: 10.1038/s41585-019-0261-8. [DOI] [PubMed] [Google Scholar]
- 14.Xu R, Rai A, Chen M, Suwakulsiri W, Greening DW, Simpson RJ. Extracellular vesicles in cancer - implications for future improvements in cancer care. Nat Rev Clin Oncol. 2018;15:617–38. doi: 10.1038/s41571-018-0036-9. [DOI] [PubMed] [Google Scholar]
- 15.Ramasubramanian L, Kumar P, Wang A Engineering extracellular vesicles as nanotherapeutics for regenerative medicine. Biomolecules. 2019;10:48. [DOI] [PMC free article] [PubMed]
- 16.Villa F, Quarto R, Tasso R Extracellular vesicles as natural, safe and efficient drug delivery systems. Pharmaceutics. 2019;11:557. [DOI] [PMC free article] [PubMed]
- 17.Gorlas A, Marguet E, Gill S, Geslin C, Guigner JM, Guyot F, et al. Sulfur vesicles from Thermococcales: A possible role in sulfur detoxifying mechanisms. Biochimie. 2015;118:356–64. doi: 10.1016/j.biochi.2015.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Erdmann S, Tschitschko B, Zhong L, Raftery MJ, Cavicchioli R. A plasmid from an Antarctic haloarchaeon uses specialized membrane vesicles to disseminate and infect plasmid-free cells. Nat Microbiol. 2017;2:1446–55. doi: 10.1038/s41564-017-0009-2. [DOI] [PubMed] [Google Scholar]
- 19.Gaudin M, Krupovic M, Marguet E, Gauliard E, Cvirkaite-Krupovic V, Le Cam E, et al. Extracellular membrane vesicles harbouring viral genomes. Environ Microbiol. 2014;16:1167–75. doi: 10.1111/1462-2920.12235. [DOI] [PubMed] [Google Scholar]
- 20.Soler N, Gaudin M, Marguet E, Forterre P. Plasmids, viruses and virus-like membrane vesicles from Thermococcales. Biochem Soc Trans. 2011;39:36–44. doi: 10.1042/BST0390036. [DOI] [PubMed] [Google Scholar]
- 21.Choi DH, Kwon YM, Chiura HX, Yang EC, Bae SS, Kang SG, et al. Extracellular vesicles of the hyperthermophilic archaeon Thermococcus onnurineus NA1T. Appl Environ Microbiol. 2015;81:4591–9. doi: 10.1128/AEM.00428-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vietri M, Radulovic M, Stenmark H. The many functions of ESCRTs. Nat Rev Mol Cell Biol. 2020;21:25–42. doi: 10.1038/s41580-019-0177-4. [DOI] [PubMed] [Google Scholar]
- 23.Juan T, Furthauer M. Biogenesis and function of ESCRT-dependent extracellular vesicles. Semin Cell Dev Biol. 2018;74:66–77. doi: 10.1016/j.semcdb.2017.08.022. [DOI] [PubMed] [Google Scholar]
- 24.van Niel G, D’Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018;19:213–28. doi: 10.1038/nrm.2017.125. [DOI] [PubMed] [Google Scholar]
- 25.Christ L, Raiborg C, Wenzel EM, Campsteijn C, Stenmark H. Cellular functions and molecular mechanisms of the ESCRT membrane-scission machinery. Trends Biochem Sci. 2017;42:42–56. doi: 10.1016/j.tibs.2016.08.016. [DOI] [PubMed] [Google Scholar]
- 26.Schöneberg J, Lee IH, Iwasa JH, Hurley JH. Reverse-topology membrane scission by the ESCRT proteins. Nat Rev Mol Cell Biol. 2017;18:5–17. doi: 10.1038/nrm.2016.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hurley JH. ESCRTs are everywhere. Embo J. 2015;34:2398–407. doi: 10.15252/embj.201592484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mierzwa BE, Chiaruttini N, Redondo-Morata L, von Filseck JM, Konig J, Larios J, et al. Dynamic subunit turnover in ESCRT-III assemblies is regulated by Vps4 to mediate membrane remodelling during cytokinesis. Nat Cell Biol. 2017;19:787–98. doi: 10.1038/ncb3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adell MA, Vogel GF, Pakdel M, Muller M, Lindner H, Hess MW, et al. Coordinated binding of Vps4 to ESCRT-III drives membrane neck constriction during MVB vesicle formation. J Cell Biol. 2014;205:33–49. doi: 10.1083/jcb.201310114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Samson RY, Dobro MJ, Jensen GJ, Bell SD. The structure, function and roles of the archaeal ESCRT apparatus. Subcell Biochem. 2017;84:357–77. doi: 10.1007/978-3-319-53047-5_12. [DOI] [PubMed] [Google Scholar]
- 31.Zaremba-Niedzwiedzka K, Caceres EF, Saw JH, Backstrom D, Juzokaite L, Vancaester E, et al. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature. 2017;541:353–8. doi: 10.1038/nature21031. [DOI] [PubMed] [Google Scholar]
- 32.Makarova KS, Yutin N, Bell SD, Koonin EV. Evolution of diverse cell division and vesicle formation systems in Archaea. Nat Rev Microbiol. 2010;8:731–41. doi: 10.1038/nrmicro2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lindås AC, Karlsson EA, Lindgren MT, Ettema TJ, Bernander R. A unique cell division machinery in the Archaea. Proc Natl Acad Sci USA. 2008;105:18942–6. doi: 10.1073/pnas.0809467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu Z, Fu T, Li T, Liu Y, Zhang S, Li J, et al. Coevolution of eukaryote-like Vps4 and ESCRT-III subunits in the Asgard archaea. mBio. 2020;11:e00417–20. doi: 10.1128/mBio.00417-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ng KH, Srinivas V, Srinivasan R, Balasubramanian M. The Nitrosopumilus maritimus CdvB, but not FtsZ, assembles into polymers. Archaea. 2013;2013:104147. doi: 10.1155/2013/104147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pelve EA, Lindas AC, Martens-Habbena W, de la Torre JR, Stahl DA, Bernander R. Cdv-based cell division and cell cycle organization in the thaumarchaeon Nitrosopumilus maritimus. Mol Microbiol. 2011;82:555–66. doi: 10.1111/j.1365-2958.2011.07834.x. [DOI] [PubMed] [Google Scholar]
- 37.Samson RY, Obita T, Freund SM, Williams RL, Bell SD. A role for the ESCRT system in cell division in archaea. Science. 2008;322:1710–3. doi: 10.1126/science.1165322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu J, Gao R, Li C, Ni J, Yang Z, Zhang Q, et al. Functional assignment of multiple ESCRT-III homologs in cell division and budding in Sulfolobus islandicus. Mol Microbiol. 2017;105:540–53. doi: 10.1111/mmi.13716. [DOI] [PubMed] [Google Scholar]
- 39.Yang N, Driessen AJ. Deletion of cdvB paralogous genes of Sulfolobus acidocaldarius impairs cell division. Extremophiles. 2014;18:331–9. doi: 10.1007/s00792-013-0618-5. [DOI] [PubMed] [Google Scholar]
- 40.Pulschen AA, Mutavchiev DR, Culley S, Sebastian KN, Roubinet J, Roubinet M et al. Live imaging of a hyperthermophilic archaeon reveals distinct roles for two ESCRT-III homologs in ensuring a robust and symmetric division. Curr Biol. 2020;30:2852–2859.e4. [DOI] [PMC free article] [PubMed]
- 41.Dobro MJ, Samson RY, Yu Z, McCullough J, Ding HJ, Chong PL, et al. Electron cryotomography of ESCRT assemblies and dividing Sulfolobus cells suggests that spiraling filaments are involved in membrane scission. Mol Biol Cell. 2013;24:2319–27. doi: 10.1091/mbc.e12-11-0785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Samson RY, Duggin IG, Bell SD. Analysis of the archaeal ESCRT apparatus. Methods Mol Biol. 2019;1998:1–11. doi: 10.1007/978-1-4939-9492-2_1. [DOI] [PubMed] [Google Scholar]
- 43.Moriscot C, Gribaldo S, Jault JM, Krupovic M, Arnaud J, Jamin M, et al. Crenarchaeal CdvA forms double-helical filaments containing DNA and interacts with ESCRT-III-like CdvB. PLoS One. 2011;6:e21921. doi: 10.1371/journal.pone.0021921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Samson RY, Obita T, Hodgson B, Shaw MK, Chong PL, Williams RL, et al. Molecular and structural basis of ESCRT-III recruitment to membranes during archaeal cell division. Mol Cell. 2011;41:186–96. doi: 10.1016/j.molcel.2010.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Caspi Y, Dekker C. Dividing the archaeal way: the ancient Cdv cell-division machinery. Front Microbiol. 2018;9:174. doi: 10.3389/fmicb.2018.00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abby SS, Melcher M, Kerou M, Krupovic M, Stieglmeier M, Rossel C, et al. Candidatus Nitrosocaldus cavascurensis, an ammonia oxidizing, extremely thermophilic archaeon with a highly mobile genome. Front Microbiol. 2018;9:28. doi: 10.3389/fmicb.2018.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nunoura T, Takaki Y, Kakuta J, Nishi S, Sugahara J, Kazama H, et al. Insights into the evolution of Archaea and eukaryotic protein modifier systems revealed by the genome of a novel archaeal group. Nucleic Acids Res. 2011;39:3204–23. doi: 10.1093/nar/gkq1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu J, Cvirkaite-Krupovic V, Baquero DP, Yang Y, Zhang Q, Shen Y, et al. Virus-induced cell gigantism and asymmetric cell division in archaea. Proc Natl Acad Sci USA. 2021;118:e2022578118. doi: 10.1073/pnas.2022578118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Snyder JC, Samson RY, Brumfield SK, Bell SD, Young MJ. Functional interplay between a virus and the ESCRT machinery in archaea. Proc Natl Acad Sci USA. 2013;110:10783–7. doi: 10.1073/pnas.1301605110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Quemin ER, Chlanda P, Sachse M, Forterre P, Prangishvili D, Krupovic M. Eukaryotic-like virus budding in Archaea. mBio. 2016;7:e01439–16. doi: 10.1128/mBio.01439-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ellen AF, Albers SV, Huibers W, Pitcher A, Hobel CF, Schwarz H, et al. Proteomic analysis of secreted membrane vesicles of archaeal Sulfolobus species reveals the presence of endosome sorting complex components. Extremophiles. 2009;13:67–79. doi: 10.1007/s00792-008-0199-x. [DOI] [PubMed] [Google Scholar]
- 52.Marguet E, Gaudin M, Gauliard E, Fourquaux I, le Blond du Plouy S, Matsui I, et al. Membrane vesicles, nanopods and/or nanotubes produced by hyperthermophilic archaea of the genus Thermococcus. Biochem Soc Trans. 2013;41:436–42. doi: 10.1042/BST20120293. [DOI] [PubMed] [Google Scholar]
- 53.Zhang C, Wipfler RL, Li Y, Wang Z, Hallett EN, Whitaker RJ. Cell structure changes in the hyperthermophilic crenarchaeon Sulfolobus islandicus lacking the S-layer. mBio. 2019;10:e01589–19. doi: 10.1128/mBio.01589-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zink IA, Pfeifer K, Wimmer E, Sleytr UB, Schuster B, Schleper C. CRISPR-mediated gene silencing reveals involvement of the archaeal S-layer in cell division and virus infection. Nat Commun. 2019;10:4797. doi: 10.1038/s41467-019-12745-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peng N, Han W, Li Y, Liang Y, She Q. Genetic technologies for extremely thermophilic microorganisms of Sulfolobus, the only genetically tractable genus of crenarchaea. Sci China Life Sci. 2017;60:370–85. doi: 10.1007/s11427-016-0355-8. [DOI] [PubMed] [Google Scholar]
- 56.Morales-Aparicio JC, Lara Vasquez P, Mishra S, Barran-Berdon AL, Kamat M, Basso KB, et al. The impacts of sortase A and the 4’-phosphopantetheinyl transferase homolog Sfp on Streptococcus mutans extracellular membrane vesicle biogenesis. Front Microbiol. 2020;11:570219. doi: 10.3389/fmicb.2020.570219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rodovalho VR, da Luz BSR, Rabah H, do Carmo FLR, Folador EL, Nicolas A, et al. Extracellular vesicles produced by the probiotic Propionibacterium freudenreichii CIRM-BIA 129 mitigate inflammation by modulating the NF-kappaB pathway. Front Microbiol. 2020;11:1544. doi: 10.3389/fmicb.2020.01544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tartaglia NR, Nicolas A, Rodovalho VR, Luz B, Briard-Bion V, Krupova Z, et al. Extracellular vesicles produced by human and animal Staphylococcus aureus strains share a highly conserved core proteome. Sci Rep. 2020;10:8467. doi: 10.1038/s41598-020-64952-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zwarycz AS, Livingstone PG, Whitworth DE. Within-species variation in OMV cargo proteins: the Myxococcus xanthus OMV pan-proteome. Mol Omics. 2020;16:387–97. doi: 10.1039/D0MO00027B. [DOI] [PubMed] [Google Scholar]
- 60.Makarova KS, Wolf YI, Koonin EV. Archaeal Clusters of Orthologous Genes (arCOGs): An update and application for analysis of shared features between Thermococcales, Methanococcales, and Methanobacteriales. Life. 2015;5:818–40. doi: 10.3390/life5010818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vorontsov EA, Rensen E, Prangishvili D, Krupovic M, Chamot-Rooke J. Abundant lysine methylation and N-terminal acetylation in Sulfolobus islandicus revealed by bottom-up and top-down proteomics. Mol Cell Proteom. 2016;15:3388–404. doi: 10.1074/mcp.M116.058073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chu Y, Zhu Y, Chen Y, Li W, Zhang Z, Liu D, et al. aKMT catalyzes extensive protein lysine methylation in the hyperthermophilic archaeon Sulfolobus islandicus but is dispensable for the growth of the organism. Mol Cell Proteom. 2016;15:2908–23. doi: 10.1074/mcp.M115.057778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ellen AF, Rohulya OV, Fusetti F, Wagner M, Albers SV, Driessen AJ. The sulfolobicin genes of Sulfolobus acidocaldarius encode novel antimicrobial proteins. J Bacteriol. 2011;193:4380–7. doi: 10.1128/JB.05028-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Prangishvili D, Holz I, Stieger E, Nickell S, Kristjansson JK, Zillig W. Sulfolobicins, specific proteinaceous toxins produced by strains of the extremely thermophilic archaeal genus Sulfolobus. J Bacteriol. 2000;182:2985–8. doi: 10.1128/JB.182.10.2985-2988.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–42. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 66.Zhang Z, Guo L, Huang L. Archaeal chromatin proteins. Sci China Life Sci. 2012;55:377–85. doi: 10.1007/s11427-012-4322-y. [DOI] [PubMed] [Google Scholar]
- 67.White MF, Bell SD. Holding it together: chromatin in the Archaea. Trends Genet. 2002;18:621–6. doi: 10.1016/S0168-9525(02)02808-1. [DOI] [PubMed] [Google Scholar]
- 68.Forterre P, Soler N, Krupovic M, Marguet E, Ackermann HW. Fake virus particles generated by fluorescence microscopy. Trends Microbiol. 2013;21:1–5. doi: 10.1016/j.tim.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 69.Deng L, Zhu H, Chen Z, Liang YX, She Q. Unmarked gene deletion and host-vector system for the hyperthermophilic crenarchaeon Sulfolobus islandicus. Extremophiles. 2009;13:735–46. doi: 10.1007/s00792-009-0254-2. [DOI] [PubMed] [Google Scholar]
- 70.Gaudin M, Gauliard E, Schouten S, Houel-Renault L, Lenormand P, Marguet E, et al. Hyperthermophilic archaea produce membrane vesicles that can transfer DNA. Environ Microbiol Rep. 2013;5:109–16. doi: 10.1111/j.1758-2229.2012.00348.x. [DOI] [PubMed] [Google Scholar]
- 71.Baquero DP, Contursi P, Piochi M, Bartolucci S, Liu Y, Cvirkaite-Krupovic V, et al. New virus isolates from Italian hydrothermal environments underscore the biogeographic pattern in archaeal virus communities. ISME J. 2020;14:1821–33. doi: 10.1038/s41396-020-0653-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hamm JN, Erdmann S, Eloe-Fadrosh EA, Angeloni A, Zhong L, Brownlee C, et al. Unexpected host dependency of Antarctic Nanohaloarchaeota. Proc Natl Acad Sci USA. 2019;116:14661–70. doi: 10.1073/pnas.1905179116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Huber H, Hohn MJ, Rachel R, Fuchs T, Wimmer VC, Stetter KO. A new phylum of Archaea represented by a nanosized hyperthermophilic symbiont. Nature. 2002;417:63–7. doi: 10.1038/417063a. [DOI] [PubMed] [Google Scholar]
- 74.Li Y, Pan S, Zhang Y, Ren M, Feng M, Peng N, et al. Harnessing Type I and Type III CRISPR-Cas systems for genome editing. Nucleic Acids Res. 2016;44:e34. doi: 10.1093/nar/gkv1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zebec Z, Manica A, Zhang J, White MF, Schleper C. CRISPR-mediated targeted mRNA degradation in the archaeon Sulfolobus solfataricus. Nucleic Acids Res. 2014;42:5280–8. doi: 10.1093/nar/gku161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wurtzel O, Sapra R, Chen F, Zhu Y, Simmons BA, Sorek R. A single-base resolution map of an archaeal transcriptome. Genome Res. 2010;20:133–41. doi: 10.1101/gr.100396.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Peng W, Feng M, Feng X, Liang YX, She Q. An archaeal CRISPR type III-B system exhibiting distinctive RNA targeting features and mediating dual RNA and DNA interference. Nucleic Acids Res. 2015;43:406–17. doi: 10.1093/nar/gku1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang C, Phillips APR, Wipfler RL, Olsen GJ, Whitaker RJ. The essential genome of the crenarchaeal model Sulfolobus islandicus. Nat Commun. 2018;9:4908. doi: 10.1038/s41467-018-07379-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tarrason Risa G, Hurtig F, Bray S, Hafner AE, Harker-Kirschneck L, Faull P et al. The proteasome controls ESCRT-III-mediated cell division in an archaeon. Science. 2020; 369:eaaz2532. [DOI] [PMC free article] [PubMed]
- 80.Lundgren M, Andersson A, Chen L, Nilsson P, Bernander R. Three replication origins in Sulfolobus species: synchronous initiation of chromosome replication and asynchronous termination. Proc Natl Acad Sci USA. 2004;101:7046–51. doi: 10.1073/pnas.0400656101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Soler N, Forterre P. Vesiduction: the fourth way of HGT. Environ Microbiol. 2020;22:2457–60. doi: 10.1111/1462-2920.15056. [DOI] [PubMed] [Google Scholar]
- 82.Soler N, Marguet E, Verbavatz JM, Forterre P. Virus-like vesicles and extracellular DNA produced by hyperthermophilic archaea of the order Thermococcales. Res Microbiol. 2008;159:390–9. doi: 10.1016/j.resmic.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 83.Koonin EV Horizontal gene transfer: essentiality and evolvability in prokaryotes, and roles in evolutionary transitions. F1000Res. 2016; 5:F1000 Faculty Rev-1805. [DOI] [PMC free article] [PubMed]
- 84.Takeuchi N, Kaneko K, Koonin EV. Horizontal gene transfer can rescue prokaryotes from Muller’s ratchet: benefit of DNA from dead cells and population subdivision. G3 (Bethesda) 2014;4:325–39. doi: 10.1534/g3.113.009845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Blokesch M. Natural competence for transformation. Curr Biol. 2016;26:R1126–R1130. doi: 10.1016/j.cub.2016.08.058. [DOI] [PubMed] [Google Scholar]
- 86.Hileman TH, Santangelo TJ. Genetics techniques for Thermococcus kodakarensis. Front Microbiol. 2012;3:195. doi: 10.3389/fmicb.2012.00195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stedman KM, She Q, Phan H, Holz I, Singh H, Prangishvili D, et al. pING family of conjugative plasmids from the extremely thermophilic archaeon Sulfolobus islandicus: insights into recombination and conjugation in Crenarchaeota. J Bacteriol. 2000;182:7014–20. doi: 10.1128/JB.182.24.7014-7020.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Martusewitsch E, Sensen CW, Schleper C. High spontaneous mutation rate in the hyperthermophilic archaeon Sulfolobus solfataricus is mediated by transposable elements. J Bacteriol. 2000;182:2574–81. doi: 10.1128/JB.182.9.2574-2581.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Alkhatatbeh MJ, Enjeti AK, Baqar S, Ekinci EI, Liu D, Thorne RF, et al. Strategies for enumeration of circulating microvesicles on a conventional flow cytometer: Counting beads and scatter parameters. J Circ Biomark. 2018;7:1849454418766966. doi: 10.1177/1849454418766966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Inglis HC, Danesh A, Shah A, Lacroix J, Spinella PC, Norris PJ. Techniques to improve detection and analysis of extracellular vesicles using flow cytometry. Cytom A. 2015;87:1052–63. doi: 10.1002/cyto.a.22649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Desvillechabrol D, Bouchier C, Kennedy S, Cokelaer T Sequana coverage: detection and characterization of genomic variations using running median and mixture models. Gigascience. 2018;7:giy110. [DOI] [PMC free article] [PubMed]
- 93.Tran F, Boedicker JQ. Plasmid characteristics modulate the propensity of gene exchange in bacterial vesicles. J Bacteriol. 2019;201:e00430–18. doi: 10.1128/JB.00430-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Robertson J, McGoverin C, Vanholsbeeck F, Swift S. Optimisation of the protocol for the LIVE/DEAD((R)) BacLight(TM) bacterial viability kit for rapid determination of bacterial load. Front Microbiol. 2019;10:801. doi: 10.3389/fmicb.2019.00801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Leuko S, Legat A, Fendrihan S, Stan-Lotter H. Evaluation of the LIVE/DEAD BacLight kit for detection of extremophilic archaea and visualization of microorganisms in environmental hypersaline samples. Appl Environ Microbiol. 2004;70:6884–6. doi: 10.1128/AEM.70.11.6884-6886.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.