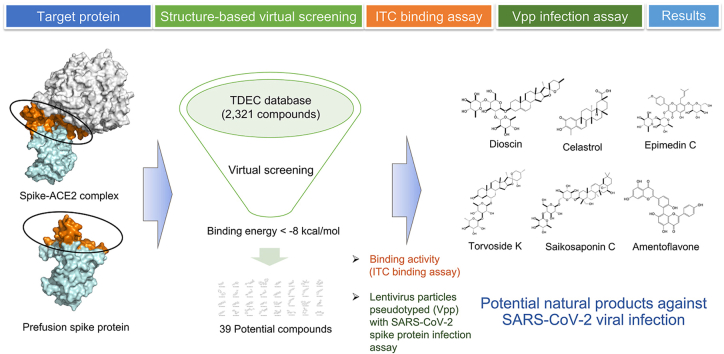
Abstract
Background and aim
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) enters cells through the binding of the viral spike protein with human angiotensin-converting enzyme 2 (ACE2), resulting in the development of coronavirus disease 2019 (COVID-19). To date, few antiviral drugs are available that can effectively block viral infection. This study aimed to identify potential natural products from Taiwan Database of Extracts and Compounds (TDEC) that may prevent the binding of viral spike proteins with human ACE2 proteins.
Methods
The structure-based virtual screening was performed using the AutoDock Vina program within PyRX software, the binding affinities of compounds were verified using isothermal titration calorimetry (ITC), the inhibitions of SARS-CoV-2 viral infection efficacy were examined by lentivirus particles pseudotyped (Vpp) infection assay, and the cell viability was tested by 293T cell in MTT assay.
Results and conclusion
We identified 39 natural products targeting the viral receptor-binding domain (RBD) of the SARS-CoV-2 spike protein in silico. In ITC binding assay, dioscin, celastrol, saikosaponin C, epimedin C, torvoside K, and amentoflavone showed dissociation constant (Kd) = 0.468 μM, 1.712 μM, 6.650 μM, 2.86 μM, 3.761 μM and 4.27 μM, respectively. In Vpp infection assay, the compounds have significantly and consistently inhibition with the 50–90% inhibition of viral infection efficacy. In cell viability, torvoside K, epimedin, amentoflavone, and saikosaponin C showed IC50 > 100 μM; dioscin and celastrol showed IC50 = 1.5625 μM and 0.9866 μM, respectively. These natural products may bind to the viral spike protein, preventing SARS-CoV-2 from entering cells.
Section: 1
Natural Products.
Taxonomy (classification by evise)
SARS-CoV-2, Structure-Based Virtual Screening, Isothermal Titration Calorimetry and Lentivirus Particles Pseudotyped (Vpp) Infection Assay, in silico and in vitro study.
Keywords: SARS-CoV-2, Spike protein, Virtual screening, Isothermal titration calorimetry, Lentivirus particles pseudotyped (Vpp) infection assay
Graphical abstract
List of abbreviations
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2
- ACE2
Angiotensin-converting enzyme 2
- COVID-19
Coronavirus disease 2019
- TDEC
Taiwan Database of Extracts and Compounds
- ITC
Isothermal titration calorimetry
- RBD
Receptor-binding domain
- Kd
Dissociation constant
- MERS-CoV
Middle East respiratory syndrome coronavirus
- CFR
Case fatality rate
- BatCoV
Bat coronavirus
- ORFs
Open reading frames
- WHO
World Health Organization
- TMPRSS2
Protease transmembrane protease, serine 2
- HR1
Heptad repeat 1
- HR2
Heptad repeat 2
- 6-HB
Six-helix bundle
- 3CLpro
3CL protease
- PLpro
Papain-like protease
- NTD
N-terminal domain
- ΔS
Entropy
- ΔH
Enthalpy
- ΔG
Gibbs free energy change
- FPLC
Fast protein liquid chromatography
- SDS-PAGE
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- CTD
C-terminal domain
- RMSD
Root-mean-square deviation
- MW
Molecular weight
- HBA
Hydrogen bond acceptor
- HBD
Hydrogen bond donor
- TPSA
Topological polar surface area
- AMR
molar refractivity
- nRB
Number rotation bond
- nAtom
Number atom
- RC
Number of Ring
- nRigidB
Number of rigid bond
- LR
Lipinski's rule
- GF
Ghose Filter
- CR
CMC-50 like rule
- VR
Veber rule
- MR
MDDR-like rule
1. Introduction
Recently, the World Health Organization (WHO, https://www.who.int/) reported the spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and the resultant outbreak of coronavirus disease 2019 (COVID-19), which has been confirmed in 188 million cases and has been associated with 4 million confirmed deaths across over 220 countries, areas, or territories. Most known coronaviruses cause only mild respiratory distress.1,2 However, SARS-CoV and SARS-CoV-2 had been identified as highly pathogenic, capable of being transmitted from animals to humans.3 The spread of SARS-CoV in 2003 was associated with a 10% case fatality rate (CFR).4, 5, 6 However, the CFR for the novel SARS-CoV-2 coronavirus remains unclear because the current pandemic is ongoing; however, the CFR has been estimated at 3.4%, and the infection transmission rate associated with SARS-CoV-2 appears to be higher than those associated with previous severe coronavirus outbreaks.7
SARS-CoV-2 primarily causes severe acute respiratory distress by attacking the lung cells and induces other complications in the heart, kidney, brain, and spleen, which can ultimately cause disastrous effects on coronavirus-afflicted patients.8 Currently, no available effective standard-of-care exists for the treatment of coronavirus-infected patients. Therefore, the identification of agents able to prevent and treat this disease represents an urgent need. Scientists globally are attempting to devise effective therapeutic agents. To design an effective antiviral drug, understanding the processes associated with viral infection, survival, and replication is essential. The SARS-CoV-2 genome showed a sequence similarity of 96.3% with the bat coronavirus (BatCoV) RaTG13 and 79% similarity with SARS-CoV.9,10 SARS-CoV-2 is an enveloped, single-stranded, positive-sense, RNA genome-containing virus. The RNA genome contains 29,891 nucleotides, which include 6–12 open reading frames (ORFs) that can be translated into approximately 28 identified non-structural and structural proteins (NCBI Reference Sequence: NC_045512.2).11
SARS-CoV-2 enters host cells that express angiotensin-converting enzyme 2 (ACE2), including cells found in the lungs, oral cavities, and nasal mucosa, through interactions between ACE2 and the viral glycosylated spike protein,12 facilitating SARS-CoV-2 entry into target cells.13 The viral glycosylated spike protein is composed of S1 and S2 subunits that allow the virus to access host cells. The receptor-binding domain (RBD) of the S1 subunit of the SARS-CoV-2 spike protein attaches to ACE2, resulting in the shedding of the S1 subunit and the subsequent cleavage of the S2 subunit by the host protease transmembrane protease, serine 2 (TMPRSS2), which can cleave the S1/S2 proteins at a protease cleavage site.13 Cleavage causes a conformational change, allowing the viral proteins heptad repeat 1 (HR1) and heptad repeat 2 (HR2) to form a six-helix bundle (6-HB), which facilitates membrane fusion and allows the virus to release its payload RNA into the cell cytoplasm. The first translation products typically produced by the RNA genome are the polyproteins pp1a and pp1ab. Subsequently, the viral 3CL protease (3CLpro) and papain-like protease (PLpro) cleave pp1a and pp1ab to generate the functional proteins required for genome amplification. The other viral structural proteins are produced, utilizing the host endoplasmic reticulum (ER)-Golgi system for assembly, facilitating the release of mature virus from infected cells.14,15 The current understanding of viral infection and propagation strongly supports targeting the surface proteins on the viral particle, which might prevent the viruses from entering the host cell.
The SARS-CoV-2 spike protein is trimeric. The RBD in each subunit can adopt two different conformations, referred to as either “up” or “down”. When the conformation is “up”, the viral spike protein can interact with the human ACE2 protein smoothly; otherwise, it is unable to interact well with ACE2 and enter the host cells once the conformation of all three subunits of the spike protein is “down”.16 The spike proteins expressed by SARS-CoV-2 and SARS-CoV feature similar amino acid sequences and functions.17 However, in the down conformation, the N-terminal region of the SARS-CoV spike protein RBD is packed tightly against the N terminal domain (NTD) of a neighboring protomer, whereas in SARS-CoV-2, the down conformation is angled closer to the central cavity of the trimer. Additionally, the binding affinity between the SARS-CoV-2 spike protein and ACE2 is 10–20-fold stronger than that for the SARS-CoV spike protein.18 These findings suggest that the structural conformation of the SARS-CoV-2 spike protein may be different from that of the SARS-CoV spike protein.
Drug discovery and development are time-consuming, costly, and complex processes, and typically years of effort are necessary to identify clinically successful interventions.19 However, the application of virtual screening methods to authentic databases and the use of advanced bioinformatics and cheminformatics analyses can reduce the time required to identify optimal drugs matched against selected target proteins This virtual screening process has become a gold standard method for the preliminary phases of drug design. After screening out potential compounds, the binding affinities of the identified compounds can be verified using bioassay studies, which represents another important step during drug development. Isothermic titration calorimetry (ITC) is a useful technique for estimating the binding affinities of compounds that target proteins.20,21 ITC measures changes in heat during reactions between compounds and target protein to further measure thermodynamic parameters, such as entropy (ΔS), enthalpy (ΔH), and Gibbs free energy change (ΔG). Based on ITC binding assay results, we can confirm the abilities of identified compounds to bind to the SARS-CoV-2 spike protein.
To identify compounds with the potential to act against SARS-CoV-2, we screened compounds that target the essential entry pathway used during the viral penetration of host cells. This study explored the compounds currently listed in the Taiwan Database of Extracts and Compounds (TDEC, https://tdec.kmu.edu.tw/) to identify potential compounds that are capable of targeting the SARS-CoV-2 spike protein. We focused on compounds identified as being able to interact with the SARS-CoV-2 spike protein and the spike-ACE2 complex, which are likely to prevent the delivery of the viral payload into the host cytoplasm. We have selected compounds that primarily target the RBD of spike proteins to prevent viral and host cell interactions. TDEC is an academic and scientific platform through which investigators in different fields can share their research findings. The TDEC includes information on the chemical structures, physicochemical properties, and biological activities of pure natural isolates, crude extracts, and synthesized extracts derived from plants, microbes, marine organisms, and Chinese herbal medicines. The present study attempted to virtually screen candidates from the TDEC to identify compounds capable of acting against the SARS-CoV-2 spike protein, including the application of high-throughput bioinformatics and cheminformatics analyses to obtain additional insight into the interactions between the spike protein and identified compounds.
In this study, several compounds in the TDEC were identified as being able to bind to the SARS-CoV-2 spike protein, based on structure-based docking calculations. The binding affinities of identified compounds were verified using ITC. The inhibitory activities of viral infection were examined by lentivirus particles pseudotyped (Vpp) with SARS-CoV-2 spike protein infection assay. The interactions between the identified compounds and ACE2-free spike protein were also explored using molecular simulation assays. Together, these analyses allowed the identification of potential natural products that may be able to target the SARS-CoV-2 spike protein for the prevention of viral infection.
2. Material and methods
2.1. Construction of expression vectors
The DNA sequence encoding the S1 domain of the SARS-CoV-2 spike protein was obtained from NCBI (accession number: NC_045512.2, Fig. S1). A DNA fragment of this sequence was purchased from AllBio Co., Ltd., which constructed it using gene synthesis. Subsequently, the DNA fragment was cloned into the pET-21 vector to form reconstructed plasmids, which were transformed into Escherichia coli BL21(DE3) competent cells. The obtained protein expression plasmids were able to continuously express the S1 domain of the SARS-CoV-2 viral spike protein in E. coli. The expressed S1 domain of the SARS-CoV-2 viral spike protein was extracted using a protein preparation technique.
2.2. Preparation of the S1 domain of SARS-CoV-2 spike protein
The S1 domain of SARS-CoV-2 spike protein was expressed in E. coli. BL21(DE3) competent cells, which were allowed to grow to an optical density at 600 nm (OD600) of 0.6 in LB medium at 37 °C. Protein expression was induced by the addition of 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG), followed by overnight incubation at 20 °C. For cell lysis, cell pellets obtained following centrifugation at 7,000 rpm for 20 min at 4 °C were resuspended in lysis buffer (25 mM Tris, pH 8.0, 500 mM NaCl, 10% glycerol, and 5 mM imidazole) and then lysed by sonication using a sonicator (Misonix Sonicator 3000). Subsequently, the sonicated cell lysates were centrifuged at 5,000 RPM for 40 at 4 °C, and the supernatants were purified through Ni2+ columns (General Electric Co., Ltd.). Next, the samples attached to the columns were washed with lysis buffer and eluted from the columns with elution buffer (25 mM Tris, pH 8.0, 500 mM NaCl, 10% glycerol, and 200 mM imidazole) using fast protein liquid chromatography (FPLC, ÄKTA prime plus, GE). The elution was collected in 36 total tubes, at 8mL/tube, and detection using an ultraviolet (UV) detector contained within the FPLC, at 280 nm, revealed the presence of protein in 19 tubes. Moreover, the purified S1 domain of the SARS-CoV-2 spike proteins was further confirmed to be of the correct size (75 kDa) by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie blue staining (Fig. S2). In this study, all solution buffers were filtered through 0.22-μm membrane filters (F-Millipore™) before use.
2.3. Estimation of binding affinities by ITC
In this study, ITC was performed by Nano ITC (TA Instruments, USA) to estimate the binding affinities between the compounds and the spike protein. Sample solutions containing the target protein (10 μM) and 8 natural product samples, including amentoflavone (200 μM), candidine (100 μM), cephalinone D (100 μM), dioscin (246 μM), epimedinc (200 μM), saikosaponin C (200 μM), torvoside K (200 μM), and celastrol (100 μM), were all dissolved in dimethyl sulfoxide (DMSO) and diluted with protein elution buffer. The detection of protein concentration was estimated by the Bradford method with BSA as the standard in this study. During the experiment, 50 μL of each compound was placed in a syringe and titrated into 300 μL of spike protein contained in the cell of the ITC instrument using 20 additions at 2.5 μL/time at 5 min intervals. The parameters for this instrument were set to a stirring rate of 300 rpm within the cell and a constant temperature of 25 °C. All solutions were degassed by a degassing station from TA Instruments (Lindon, UT, USA) and were filtered through a 0.22-μm membrane filter (F-Millipore™) before the titration experiments began. We titrated the DMSO diluted buffer into the same buffer to be the background signal. All samples are deducted background signal (The buffer into the buffer). To estimate the strength of the interaction between each compound and protein, the dissociation constant (Kd), reaction stoichiometry (n), c value (c), and the thermodynamic parameters, including entropy (ΔS), enthalpy (ΔH), and Gibbs free energy change (ΔG), were all measured by the NanoAnzalyze software (TA Instruments, USA). Thermodynamic parameters were calculated according to thermodynamics formulas22, 23, 24:
| ΔG = −RTlnKa = ΔH − TΔS |
(R: gas constant, T: absolute temperature, Ka: binding constant).
ITC data with c values between 1 and 1000 is an acceptable binding constants.24 The c values were calculated according to the equation:
| c = nKa[M]T = n[M]T/Kd |
(n: stoichiometry of interaction, Ka: binding constant, [M]T: total macromolecular concentration, Kd: dissociation constant).
2.4. Structural superimposition
To analyze changes in the SARS-CoV-2 spike protein conformation between the ACE2-free and ACE2-bound forms, structural superimpositions were performed using PyMOL (version 0.99rc6) software.25 Fig. S3 shows the homology modeling performed for the ACE2-free spike protein, which was obtained from the SWISS-MODEL website, and the ACE2-bound protein (PDB ID: 6M0J), which was obtained from the Protein Data Bank (PDB, https://www.rcsb.org/).18,26,27
2.5. Structure-based virtual screening
The structure-based virtual screenings were performed using the AutoDock Vina (version 1.1.2) program within PyRx (version 0.8) software to discover which potential compounds from the TDEC can bind to the viral spike protein.28,29 The 2,321 compounds downloaded (May 9, 2020) from TDEC were collected and saved in a .sdf format file by Discovery Studio 2019 visualizer software (DS 2019,30). Subsequently, energy minimization using the steepest descent algorithm was performed with the MMFF94 force field to appropriately add partial charges and polar hydrogens to the atoms in the compounds. Next, those compounds were individually saved as .pdbqt format files by the Open Babel program within PyRx software.31 The substrates, including ligands, metal ions, and water molecules, that are contained in both the ACE2-free and ACE2-bound spike proteins, as resolved by X-ray crystallography, were removed by DS 2019. Next, the atoms contained in the residues of the protein structures were modified by DS 2019 using the CHARMm force field to appropriately add polar hydrogens and partial charges. For docking calculations, the dimensions of docking search spaces were set to contain the residues of the ACE2 binding site of the spike protein RBD as follows. For the ACE2-bound spike protein structure, as resolved by X-ray crystallography, the coordinates of the docking search space were set to x = −38.6172, y = 28.4230, and z = 5.0328, and the size of the dimensions of the x-, y-, and z-axes (in angstroms) were respectively set to x = 30.2373, y = 57.9713, and z = 19.4614, at the connective interface of complex proteins. Similarly, for the ACE2-free spike protein, the coordinates of the docking search space were set to x = 226.271, y = 195.386, and z = 306.4952, and the sizes of the dimensions of the x-, y-, and z-axes (in angstroms) were respectively set to x = 47.2191, y = 38.0448, and z = 30.3288, at the protein. Additionally, the value of exhaustiveness used to search for molecular conformations was set to 4 for the docking calculation.
2.6. Molecular simulation
The best poses for each compound when docking with the ACE2-free spike protein or the spike-ACE2 complex protein, as established by the AutoDock Vina docking calculation, was displayed using PyMOL software. The interactions, including electrostatic hydrogen-bonding interactions, hydrophobic π-alkyl interactions, and hydrophobic alkyl-alkyl interactions, between the docked compounds and the proteins were identified by DS 2019 visualizer software.
2.7. Lentivirus particles pseudotyped (Vpp) with SARS-CoV-2 spike protein infection assay
Vpp contains SARS-CoV-2 Spike protein and luciferase reporter. SARS-CoV-2 was purchased from National RNAi Core Facility (NRC), Academia Sinica, Taipei, Taiwan. The Vpp was added to cells in a 96-well plate (MOI∼0.2) in the presence of polybrene (8 μg/mL). The plate was centrifuged at 1200 g for 30 min and then put back into the incubator. At 24 h postinfection (hpi), the culture supernatants were replaced with fresh media. At 24 hpi, the Cell Counting Kit-8 (CCK-8) assay (Abcam) was performed to measure cell viability. Each sample was mixed with an equal volume of ready-to-use luciferase substrate Bright-Glo Luciferase Assay System (Promega) afterward. The relative light unit (RLU) was measured immediately.32
2.8. Cell viability
Cell viability was assessed using the MTT assays. Torvoside K, epimedin C, amentoflavone, saikosaponin C, dioscin and celastrol were dissolved in DMSO. 293T cells were seeded on 96-well plates at 4 × 104 cells/well. After incubation for 24 h, cells were treated with 0.044828–100 μM natural compounds for 48 h. The culture medium was removed and cells were washed twice with phosphate-buffered saline (PBS). 100 μL MTT/medium solution (2.5 mg/mL) were added to each well and incubated with cells for 1 h. After incubation, the medium was removed and 100 μL aliquots of DMSO were added to each well to solubilize the formazan crystals. Absorbance was measured at 470 nm using a microplate reader (Bio-Tek, USA). The percentage of cell viability was expressed relative to the control.
2.9. Calculation of ADME/T and drug-likeness properties
The predictions of physiochemical properties and drug-likeness were estimated by Drug Likeness Tool (DruLiTo) program (http://www.niper.gov.in/pi_dev_tools/DruLiToWeb/DruLiTo_index.html). The chemical ADMET properties were predicted by the admetSAR website (version 2.0, http://lmmd.ecust.edu.cn/admetsar2). The data of the water solubility category were obtained from the annotations of the TDEC website (https://tdec.kmu.edu.tw/index_en.aspx).
The physicochemical properties included molecular weight (MW), hydrogen bond acceptor (HBA), hydrogen bond donor (HBD), topological polar surface area (TPSA), molar refractivity (AMR), number rotation bond (nRB), number atom (nAtom), number of ring (RC), and number of rigid bond (nRigidB) were calculated in this study. Additionally, the drug-likeness of compounds were estimated by five drug-likeness rules, including Lipinski's rule, Ghose Filter, CMC-50 like rule, Veber rule, and MDDR-like rule. The parameters of the five drug-likeness rules were set as default in the DruLiTo program.
3. Results
3.1. Structural superimposition
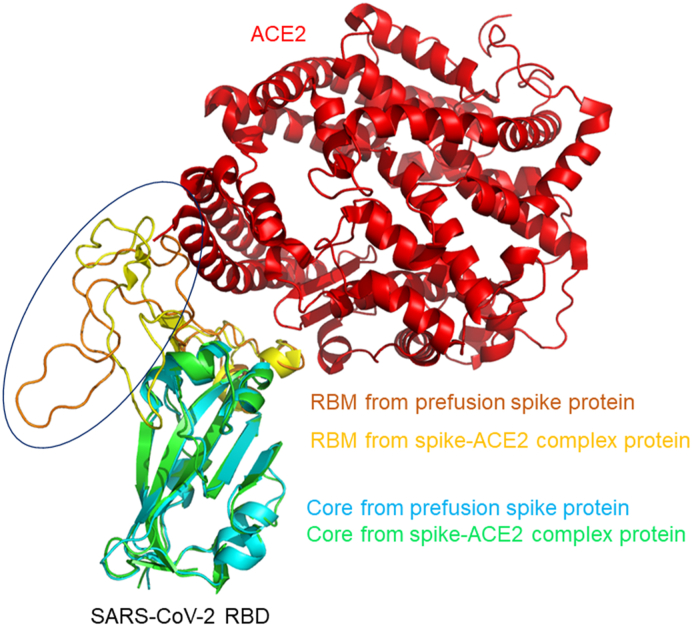
SARS-CoV-2 entry into host cells occurs through the binding of the spike protein on the viral surface with the human ACE2 protein.17 A previous study reported that SARS-CoV could alter the conformation of the C-terminal domain (CTD) of the spike protein to better fit the conformation of the ACE2 protein using various angles.10 To analyze the conformation of the SARS-Co-V-2 spike protein RBD and to determine whether conformational changes occurred between the ACE2-free state and the ACE2-bound state, structural superimposition was performed using PyMOL software. Fig. 1 shows the superimposition model that was constructed [root-mean-square deviation (RMSD) = 1.735 < 2 Å], showing the ACE2-free spike protein aligned with the ACE2-bound spike protein. We found that the conformation of the viral receptor-binding motif (RBM), which contains many of the residues that contact the human ACE2 protein,27 differed between the ACE2-free and ACE2-bound spike proteins. The RBM of the ACE2-bound spike protein was more proximal to the RBD core and the ACE2 protein than that of the ACE2-free spike protein (blue ring in Fig. 1). Compounds that bind to these regions have a high probability of causing a conformational change in the spike proteins, which would likely reduce the binding affinity between the spike protein and the ACE2 protein. To quickly find and identify potential compounds that target the spike proteins, structured-based virtual screening technology was applied.
Fig. 1.
The structural superimposition between the ACE2-free and ACE2-bound structures of the SARS-CoV-2 spike protein. The RBD cores of the SARS-CoV-2 spike protein in the ACE2-free structure and the ACE2-bound structure are respectively shown in cyan and green, and their RBMs are respectively shown in orange and yellow. The ACE2 receptor is also shown in red. The major difference between the RBD of the ACE2-free spike protein and the RBD of the spike-ACE2 complex protein is shown within the blue ring.
3.2. Structure-based virtual screening
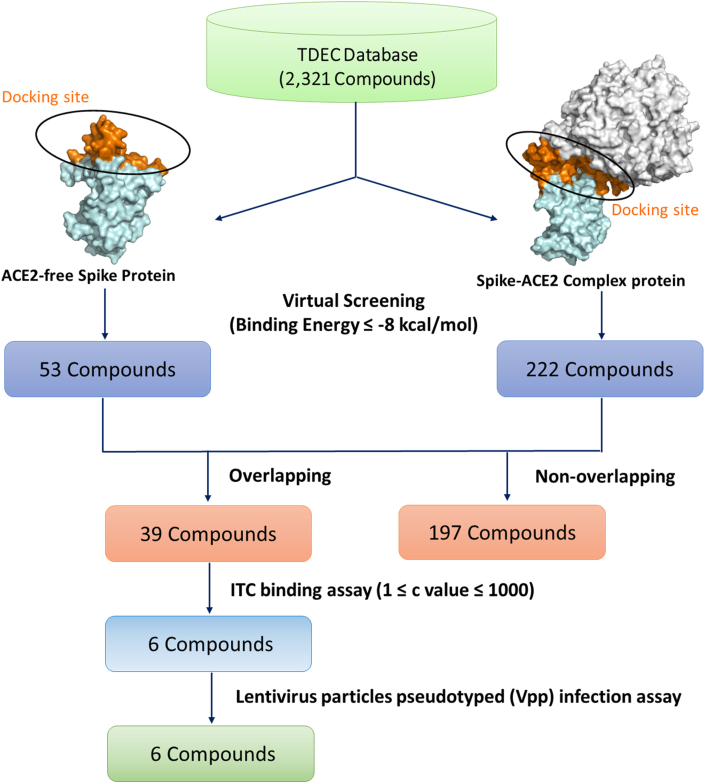
Structure-based virtual screening was performed using the AutoDock Vina program within PyRX software to discover compounds that not only bind to the RBD of the ACE2-free spike protein but also the interface between ACE2 and spike protein. Fig. 2 shows the flowchart followed during the virtual screening calculation performed in this study. The numbers of compounds that were able to bind to the ACE2-free spike protein and the spike-ACE2 complex protein were 53 and 222, respectively (binding energy ≤ −8 kcal/mol). Generally, if compounds with the value of the binding energy were higher than −6 kcal/mol, the binding ability was not expected.23 The data showed that the 39 potential compounds not only binding to the RBD of the ACE2-free spike protein but also binding to the interface between ACE2 and spike protein in silico (binding energy ≤ −8 kcal/mol, Table S1). The structures of the 39 compounds were also shown in Fig. S4.
Fig. 2.
The flowchart of the structure-based virtual screening process.
To further estimate the binding strength of the 39 compounds, the natural product glycyrrhizic acid was set as the positive control to do the comparison. Glycyrrhizic acid had been reported that it can bind to SARS-CoV-2 spike protein and has the inhibitory activity against the interaction between viral spike protein and ACE2 protein.33 The docking results showed that the binding energies of glycyrrhizic acid in ACE2-free spike protein and ACE2-spike complex were −7.8 kcal/mol and −8.2 kcal/mol, respectively (Fig. S5). The predicted binding strength of glycyrrhizic acid is similar to our set standard estimating the results of performing docking-based virtual screening from the database.
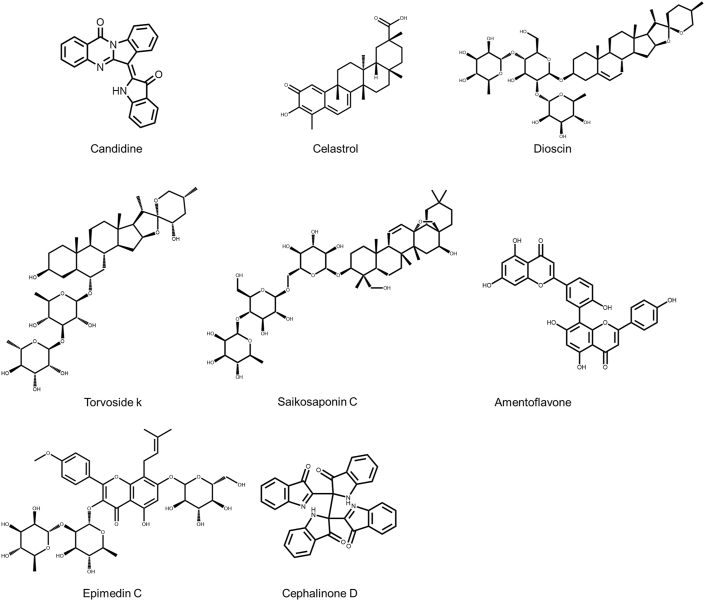
Among those 39 compounds, there were 8 natural products that our laboratory had easy access to. Fig. 3 shows the structures of the 8 natural products that were tested by our laboratory, including candidine, celastrol, dioscin, torvoside K, saikosaponin C, amentoflavone, epimedinc, and cephalinone D. The docking results of these 8 compounds are displayed in Table 1. Subsequently, the binding affinities of the obtained samples were further verified by bioassay.
Fig. 3.
The structures of 8 natural products that target the SARS-CoV-2 spike protein. The structures of candidine, celastrol, dioscin, torvoside K, saikosaponin C, amentoflavone, epimedinc, and cephalinone D are shown.
Table 1.
Molecular docking results and thermodynamic parameters of the natural products binding to the S1 domain of SARS-CoV-2 spike protein.
| Compound | The binding energy (kcal/mol) of Molecular docking calculation |
The thermodynamic parameters of ITC binding assaya |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| ACE2-free spike protein | spike-ACE2 complex protein |
ΔS (J/mol·K) | -TΔS (kJ/mol) | ΔH (kJ/mol) | ΔG (kJ/mol) | n | c | Kd (μM) | |
| Dioscin (TDEC2019CA001572) | −8.8 | −8.9 | 110.6 | −32.98 | −3.153 | −36.13 | 5.488 | 117.3 | 0.468 |
| Cephalinone D (TDEC2020CN000221) | −8.1 | −8.7 | 33.45 | −9.972 | −20.62 | −30.59 | 0.333 | 0.8 | 4.371 |
| Celastrol (TDEC2019CA001707) | −8.9 | −8.3 | 667.9 | −199.1 | 166.2 | −32.92 | 1.409 | 8.2 | 1.712 |
| Saikosaponin C (TDEC2019CA001664) | −8.6 | −9.1 | 476.6 | −142.1 | 112.5 | −29.55 | 2.802 | 4.2 | 6.65 |
| Epimedin C (TDEC2019CA001733) | −8.2 | −8.1 | 379.0 | −113.0 | 81.37 | −31.64 | 2.967 | 10.4 | 2.86 |
| Epimedin C (TDEC2019CA001733) | −8.2 | −8.1 | 383.30 | −114.30 | 82.65 | −31.62 | 2.965 | 10.2 | 2.908 |
| Candidine (TDEC2020CN000246) | −9.0 | −9.8 | −243.4 | 72.58 | −100 | −27.42 | 0.115 | 0.1 | 15.7 |
| Torvoside K (TDEC2019CN000617) | −8.8 | −8.3 | −130.8 | 39.01 | −69.97 | −30.96 | 2.823 | 7.5 | 3.761 |
| Amentoflavone (TDEC2019CA001644) | −8.5 | −9.1 | −172.7 | 51.48 | −82.13 | −30.65 | 2.731 | 6.4 | 4.27 |
1 kcal = 4.1868 kj.
3.3. ITC binding assay
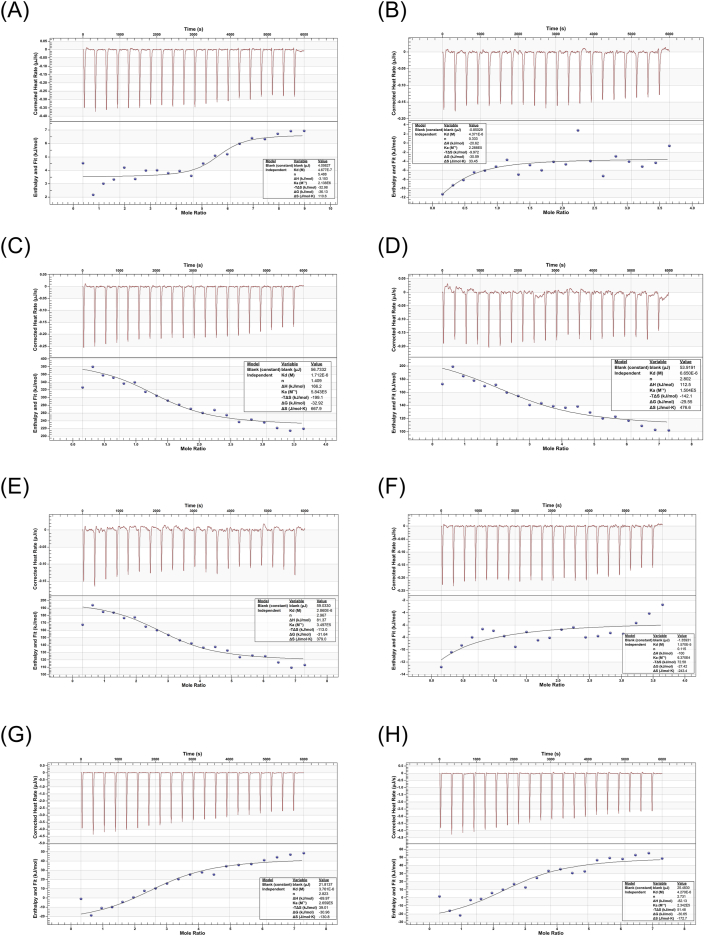
ITC has previously been reported as representing a reliable method for the estimation of binding affinities between compounds and proteins by measuring the heat change, as heat can be either absorbed or released during reaction processes.20,21 To verify the binding affinities of the 8 tested compounds, the ITC assay was performed. Fig. 4 shows the typical ITC profiles obtained for the 8 compounds during titration with the viral protein. According to the titration curve, all reaction equilibriums were achieved during the period between the 4000s and 5000s. The results of the thermodynamic parameters are documented in Table 1. Generally, the strength of the binding affinities between the compounds and the protein could be estimated by the Kd value.34 If the Kd value of the compound was smaller, the binding affinity was stronger. The data showed that the Kd of dioscin, celastrol, saikosaponin C, epimedin C, torvoside K, candidine, cephalinone D, and amentoflavone were 0.468 μM, 1.712 μM, 6.650 μM, 2.86 μM, 3.761 μM, 15.7 μM, 4.371 μM and 4.27 μM, respectively. However, the results of c value estimation reported that candidine and cephalinone D were illegal (c value < 1). Additionally, according to the thermodynamics formulas, the ΔG was primarily contributed by Kd. A negative ΔG indicates a spontaneous reaction process, and smaller ΔG values suggest stronger binding affinities. The results showed that the ΔG values of the 6 legal compounds ranged from −37 to −26 kJ/mol, and the binding between the S1 domain of the spike protein and these compounds occurred as the result of spontaneous reaction processes (ΔG < 0 kJ/mol). We discovered that dioscin, celastrol, epimedin C, amentoflavone, torvoside K, and Saikosaponin C might have the ability to bind to the S1 domain of the SARS-CoV-2 spike protein.
Fig. 4.
The ITC profiles of the potential natural products that might target the SARS-CoV-2 spike protein. (A) Dioscin, (B) Cephalinone D, (C) Celastrol, (D) Saikosaponin C, (E) Epimedin C, (F) Candidine, (G) Torvoside K, and (H) Amentoflavone.
3.4. Molecular simulation
To further analyze which amino acids of the ACE2-free and spike-ACE2 complex interacted with these identified compounds, the molecular simulation studies were performed using DS 2019 visualizer software. The results of the molecular simulation are documented in Table 2.
Table 2.
The residues that interact with the 6 potential natural products within the ACE2-free spike and spike-ACE2 complex proteins.
| Compound | ACE2-free spike protein |
Spike-ACE2 complex protein |
||
|---|---|---|---|---|
| Electrostatic interactions (Å) | Hydrophobic interactions (Å) | Electrostatic interactions (Å) | Hydrophobic interactions (Å) | |
| Dioscin | Tyr351 (2.25 Å), Asn354 (2.45 Å) | Phe486 (5.05 Å), Arg346 (4.18 Å), Ala348 (3.46 Å), Ala352 (3.77 Å), Leu452 (4.34 Å, 5.36 Å), Leu492 (5.47 Å, 5.00 Å), Pro463 (3.91 Å, 5.23 Å) | Asp30 (2.51 Å), Lys417 (2.45 Å), Asp420 (2.59 Å), Asn460 (2.87 Å), Tyr421 (2.40 Å), Lys458 (2.35 Å) | His34 (5.02 Å, 5.47 Å), Tyr473 (5.41 Å), Lys417 (3.96 Å, 4.72 Å), Lys26 (4.45 Å) |
| Celastrol | Phe490 (2.05 Å, 2.48 Å), Glu465 (3.75 Å), | Ile468 (5.32 Å), Phe486 (5.41 Å), Leu492 (5.31 Å), Leu452 (3.95 Å, 4.67 Å, 4.69 Å), Lys462 (4.79 Å), Pro463 (3.51 Å), Leu492 (4.11 Å, 4.75 Å) | Arg393 (2.14 Å), Ala386 (2.56 Å) | Pro389 (4.66 Å), His34 (5.38 Å), Pro389 (4.79 Å, 5.10 Å, 5.27 Å), Leu29 (4.67 Å), Lys26 (4.73 Å) |
| Saikosaponin C | Glu471 (2.35 Å), Asn354 (2.83 Å), Asn450 (2.98 Å, 2.69 Å) | Phe486 (4.25 Å, 5.30 Å, 5.34 Å), Ile468 (4.20 Å, 5.25 Å), Leu492 (4.14 Å, 4.62 Å, 4.87 Å, 5.35 Å), Leu452 (3.96 Å, 4.68 Å, 4.85 Å, 5.48 Å) | Asp30 (2.61 Å), Asn460 (2.50 Å) | His34 (5.01 Å), Lys417 (4.34 Å) |
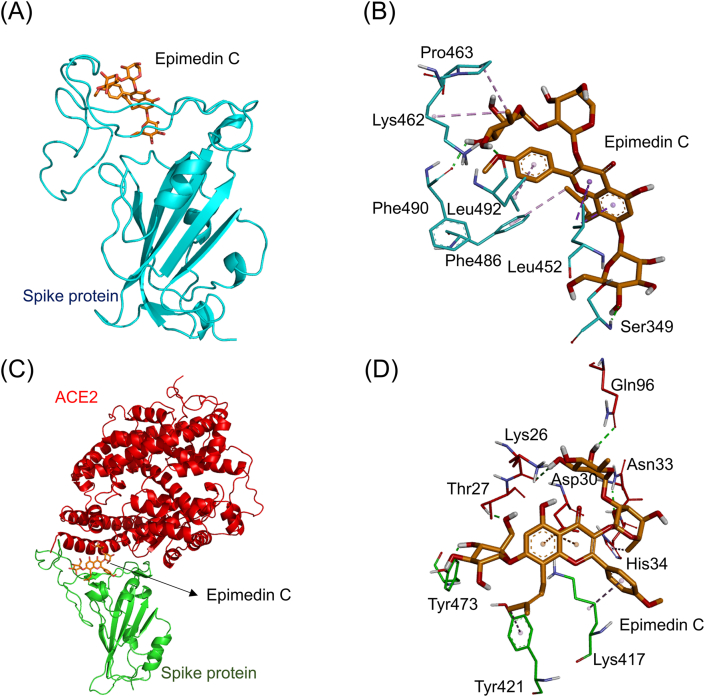
| Epimedin C | Ser349 (2.08 Å), Phe490 (2.16 Å), Leu492 (1.88 Å) | Leu492 (4.96 Å), Phe486 (4.92 Å), Lys462 (4.79 Å), Pro463 (4.62 Å), Leu452 (3.78 Å, 3.96 Å, 3.99 Å) | Lys26 (2.56 Å), Thr27 (1.89 Å), Asn33 (2.16 Å), Gln96 (2.49 Å), Tyr473 (2.36 Å), Asp30 (4.82 Å, 4.86 Å) | His34 (4.57 Å), Lys417 (5.19 Å), Tyr421 (5.24 Å) |
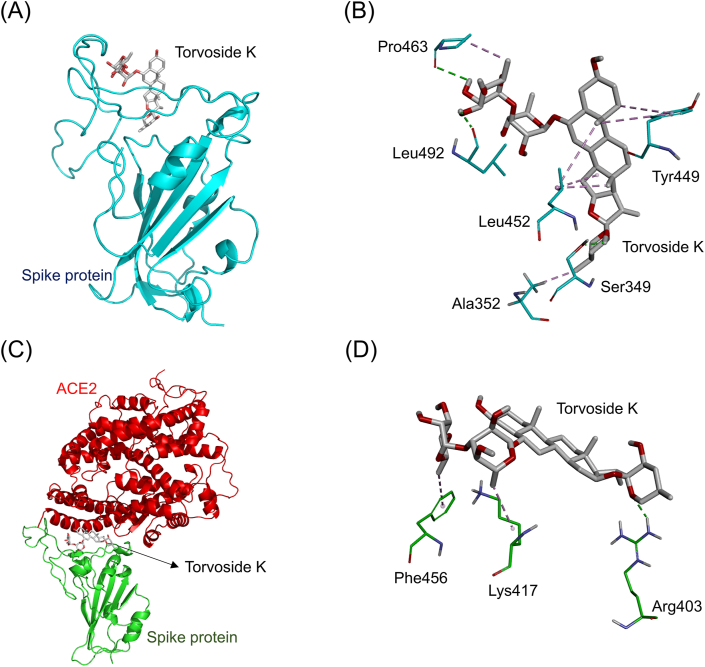
| Torvoside K | Ser349 (2.04 Å), Pro463 (3.02 Å), Leu492 (2.38 Å) | Tyr449 (4.77 Å, 5.44 Å), Leu452 (4.08 Å, 4.83 Å, 5.37 Å), Pro463 (3.94 Å), Ala352 (3.92 Å) | Arg403 (2.49 Å) | Phe456 (4.93 Å), Lys417 (4.09 Å) |
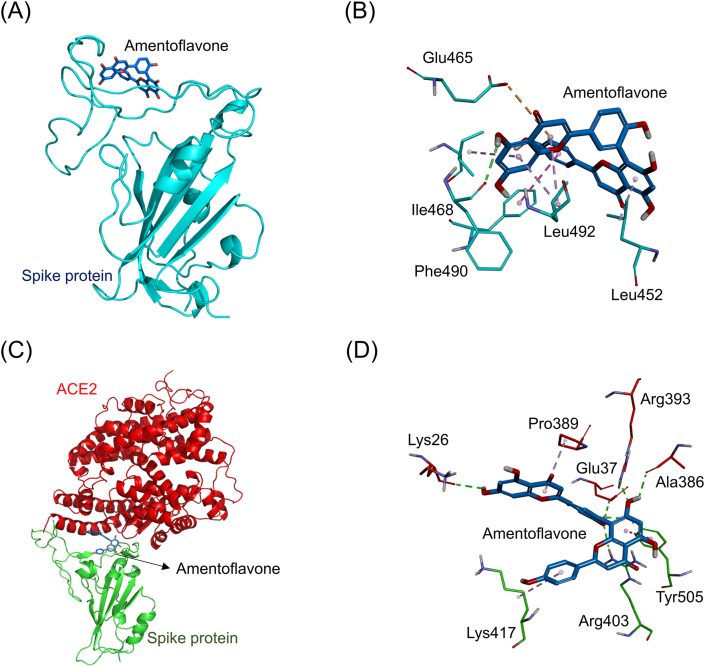
| Amentoflavone | Glu465 (4.44 Å), Phe490 (2.5 Å) | Leu452 (4.21 Å), Leu492 (4.39 Å, 4.6 Å), Ile468 (5.23 Å), Phe486 (4.81 Å) | Lys26 (2.56 Å), Glu37 (2.22 Å), Ala386 (2.25 Å), Arg393 (2.09 Å), Arg403 (2.23 Å), Tyr505 (2.51 Å) | Pro389 (4.47 Å), Lys417(5.04 Å), Tyr505 (5.94 Å) |
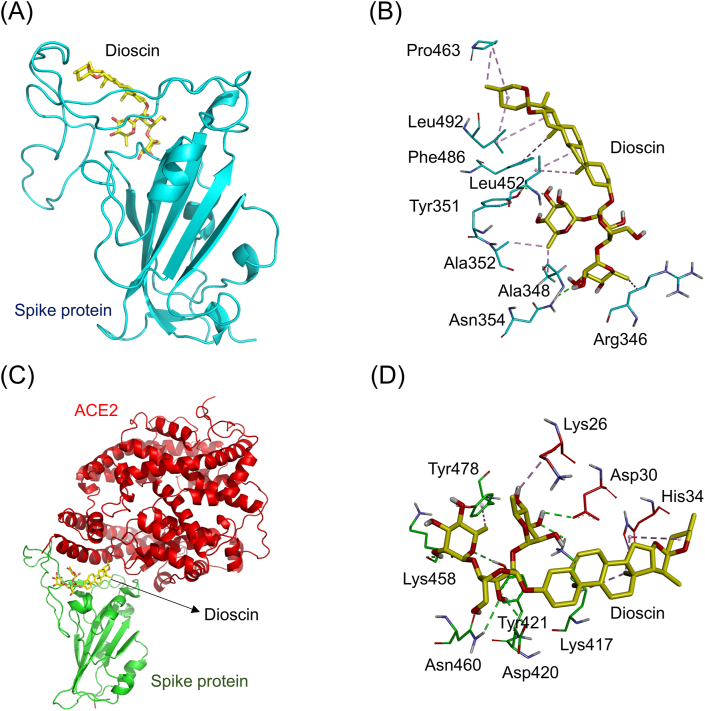
Fig. 5A shows the simulated docking between dioscin and the RBD of the ACE2-free spike protein (binding energy: −8.8 kcal/mol). Dioscin formed a hydrophobic π-alkyl interaction (light purple dashed line) with the amino acid Phe486 (5.05 Å). It also formed nine hydrophobic alkyl-alkyl interactions (dark purple dashed lines) with the amino acids Arg346 (4.18 Å), Ala348 (3.46 Å), Ala352 (3.77 Å), Leu452 (4.34 Å and 5.36 Å), Leu492 (5.47 Å and 5.00 Å), and Pro463 (3.91 Å and 5.23 Å). In addition, dioscin formed two electrostatic hydrogen-bonding interactions (green dashed lines) with the amino acids Tyr351 (2.25 Å) and Asn354 (2.45 Å, Fig. 5B). Fig. 5C shows the simulated docking between dioscin and the connective interface of the spike-ACE2 complex protein (binding energy: −8.9 kcal/mol). It formed three hydrophobic π-alkyl interactions (light purple dashed lines) with the amino acids His34 (5.02 Å and 5.47 Å) and Tyr473 (5.41 Å). It also formed three hydrophobic alkyl-alkyl interactions (dark purple dashed lines) with the amino acids Lys417 (3.96 Å and 4.72 Å) and Lys26 (4.45 Å). Moreover, it formed six electrostatic hydrogen-bonding interactions (green dashed lines) with the amino acids Asp30 (2.51 Å), Lys417 (2.45 Å), Asp420 (2.59 Å), Asn460 (2.87 Å), Tyr421 (2.40 Å), and Lys458 (2.35 Å, Fig. 5D).
Fig. 5.
The docking simulation between dioscin and the SARS-CoV-2 spike protein. (A) Dioscin (yellow stick) docked with the RBD of the ACE2-free spike protein (cyan cartoon). (B) Dioscin formed several hydrophobic interactions (purple dashed lines) with amino acids (cyan sticks) in the RBD of the spike protein. (C) Dioscin docked with the site near the connective interface of the spike-ACE2 complex. The RBD of spike protein and ACE2 are respectively shown in green and red cartoons. (D) Dioscin formed several electrostatic interactions (green dashed lines) and hydrophobic interactions (purple dashed lines) with amino acids near the connective interface of spike-ACE2 complex protein.
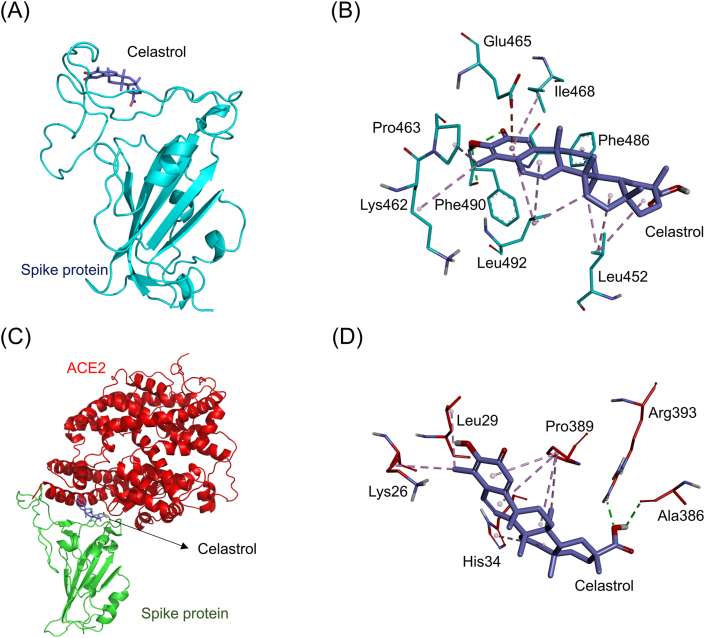
Fig. 6A shows the simulated docking between celastrol and the RBD of the ACE2-free spike protein (binding energy: −8.9 kcal/mol). Celestrol formed three hydrophobic π-alkyl interactions (light purple dashed lines) with the amino acids Ile468 (5.32 Å), Phe486 (5.41 Å), and Leu492 (5.31 Å). It also formed seven hydrophobic alkyl-alkyl interactions (dark purple dashed lines) with the amino acids Leu452 (3.95 Å, 4.67 Å, and 4.69 Å), Lys462 (4.79 Å), Pro463 (3.51 Å), and Leu492 (4.11 Å and 4.75 Å). Additionally, it formed two electrostatic hydrogen-bonding interactions (green dashed lines) with the amino acid Phe490 (2.05 Å and 2.48 Å). Moreover, it formed an electrostatic π-anion interaction (orange dashed line) with the amino acid Glu465 (3.75 Å, Fig. 6B). Fig. 6C shows the simulated docking between celestrol and the connective interface of the spike-ACE2 complex (binding affinity: −8.3 kcal/mol). It also formed two hydrophobic π-alkyl interactions (light purple dashed lines) with the amino acids Pro389 (4.66 Å) and His34 (5.38 Å). It also formed five hydrophobic alkyl-alkyl interactions (dark purple dashed lines) with the amino acids Pro389 (4.79 Å, 5.10 Å, and 5.27 Å), Leu29 (4.67 Å), and Lys26 (4.73 Å). In addition, it formed two electrostatic hydrogen-bonding interactions (green dashed lines) with the amino acids Arg393 (2.14 Å) and Ala386 (2.56 Å, Fig. 6D).
Fig. 6.
The docking simulation between celastrol and the SARS-CoV-2 spike protein. (A) Celastrol (purple stick) docked with the RBD of the ACE2-free spike protein (cyan cartoon). (B) Celastrol formed several electrostatic interactions (green dashed lines) and hydrophobic interactions (purple dashed lines) with amino acids (cyan sticks) in the RBD of the spike protein. (C) Celastrol docked with the site near the connective interface of the spike-ACE2 complex. The RBD of the spike protein and ACE2 are respectively shown in green and red cartoons. (D) Celastrol formed several electrostatic interactions (green dashed lines) and hydrophobic interactions (purple dashed lines) with amino acids near the connective interface of the spike-ACE2 complex protein.
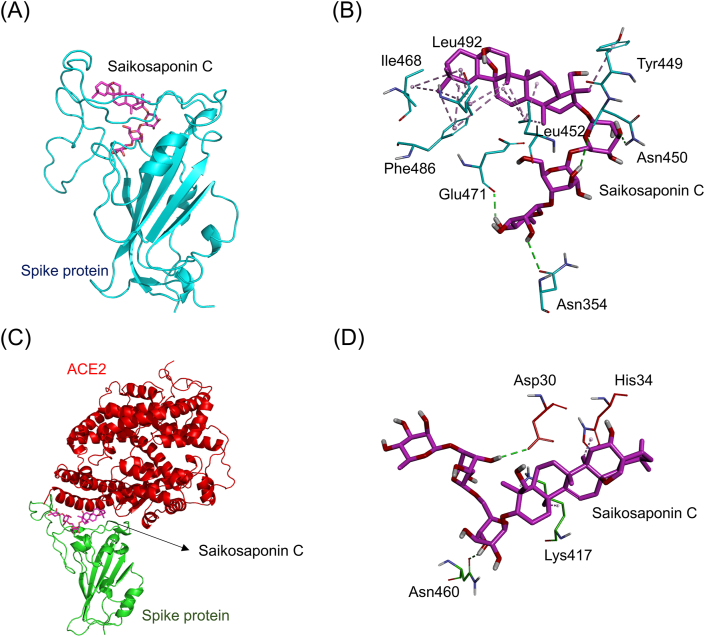
Fig. 7A showed the simulated docking between saikosaponin C and the RBD of the ACE2-free spike protein (binding energy: −8.6 kcal/mol). Saikosaponin C formed four hydrophobic π-alkyl interactions (light purple dashed lines) with the amino acids Phe486 (4.25 Å, 5.30 Å, and 5.34 Å) and Tyr449 (4.59 Å). It also formed ten hydrophobic alkyl-alkyl interactions (dark purple dashed lines) with the amino acids Ile468 (4.20 Å and 5.25 Å), Leu492 (4.14 Å, 4.62 Å, 4.87 Å, and 5.35 Å), and Leu452 (3.96 Å, 4.68 Å, 4.85 Å, and 5.48 Å). Additionally, it formed four electrostatic hydrogen-bonding interactions (green dashed lines) with the amino acids Glu471 (2.35 Å), Asn354 (2.83 Å), and Asn450 (2.98 Å and 2.69 Å, Fig. 7B). Fig. 7C showed the simulated docking between saikosaponin C and the connective interface of the spike-ACE2 complex (binding energy: 9.1 kcal/mol). It also formed a hydrophobic π-alkyl interaction (light purple dashed lines) with the amino acid His34 (5.01 Å). Besides, it also formed a hydrophobic alkyl-alkyl interaction (dark purple dashed lines) with the amino acid Lys417 (4.34 Å). In addition, it formed two electrostatic hydrogen-bonding interactions (green dashed lines) with the amino acids Asp30 (2.61 Å) and Asn460 (2.50 Å, Fig. 7D).
Fig. 7.
The docking simulation between saikosaponin C and the SARS-CoV-2 spike protein. (A) Saikosaponin C (magenta stick) docked with the RBD of the ACE2-free spike protein (cyan cartoon). (B) Saikosaponin C formed several electrostatic interactions (green dashed lines) and hydrophobic interactions (purple dashed lines) with amino acids (cyan sticks) in the RBD of the spike protein. (C) Saikosaponin C docked with the site near the connective interface of the spike-ACE2 complex. The RBD of the spike protein and ACE2 are respectively shown in green and red cartoons. (D) Saikosaponin C formed several electrostatic interactions (green dashed lines) and hydrophobic interactions (purple dashed lines) with amino acids near the connective interface of the spike-ACE2 complex protein.
Fig. 8A showed the simulated docking between epimedin C and the RBD of the ACE2-free spike protein (binding energy: −8.2 kcal/mol). Epimedin C formed two hydrophobic π-alkyl interactions (light purple dashed lines) with the amino acids Leu492 (4.96 Å) and Phe486 (4.92 Å). Besides, it also formed two hydrophobic alkyl-alkyl interactions (purple dashed lines) with the amino acids Lys462 (4.79 Å) and Pro463 (4.62 Å). Moreover, it also formed three hydrophobic π-sigma interactions (dark purple dashed line) with the amino acid Leu452 (3.78 Å, 3.96 Å, and 3.99 Å). Furthermore, it formed three electrostatic hydrogen-bonding interactions (green dashed lines) with the amino acids Ser349 (2.08 Å), Phe490 (2.16 Å), and Leu492 (1.88 Å, Fig. 8B). Fig. 8C showed the simulated docking between epimedin C and the connective interface of the spike-ACE2 complex (binding energy: 8.1 kcal/mol). It formed five electrostatic hydrogen-bonding interactions (green dashed line) with the amino acids Lys26 (2.56 Å), Thr27 (1.89 Å), Asn33 (2.16 Å), Gln96 (2.49 Å), and Tyr473 (2.36 Å). Besides, it also formed three hydrophobic π-alkyl interactions (light purple dashed lines) with the amino acids His34 (4.57 Å) and Lys417 (5.19 Å), Tyr421 (5.24 Å). In addition, it formed two electrostatic π-anion interactions (orange dashed line) with the amino acid Asp30 (4.82 Å and 4.86 Å, Fig. 8D).
Fig. 8.
The docking simulation between epimedin C and the SARS-CoV-2 spike protein. (A) Epimedin C (orange stick) docked with the RBD of the ACE2-free spike protein (cyan cartoon). (B) Epimedin C formed several electrostatic interactions (green dashed lines) and hydrophobic interactions (purple dashed lines) with amino acids (cyan sticks) in the RBD of the spike protein. (C) Epimedin C docked with the site near the connective interface of the spike-ACE2 complex. The RBD of the spike protein and ACE2 are respectively shown in green and red cartoons. (D) Epimedin C formed several electrostatic interactions (green dashed lines) and hydrophobic interactions (purple dashed lines) with amino acids near the connective interface of the spike-ACE2 complex protein.
Fig. 9A showed the simulated docking between torvoside K and the RBD of the ACE2-free spike protein (binding energy: 8.8 kcal/mol). Torvoside K formed two hydrophobic π-alkyl interactions (light purple dashed lines) with the amino acid Tyr449 (4.77 Å and 5.44 Å). It also formed five hydrophobic alkyl-alkyl interactions (dark purple dashed lines) with the amino acids Leu452 (4.08 Å, 4.83 Å, and 5.37 Å), Pro463 (3.94 Å), and Ala352 (3.92 Å). In addition, it formed three electrostatic hydrogen-bonding interactions (green dashed lines) with the amino acids Ser349 (2.04 Å), Pro463 (3.02 Å), and Leu492 (2.38 Å, Fig. 9B). Fig. 9C showed the simulated docking between torvoside K and the connective interface of the spike-ACE2 complex (binding energy: 8.3 kcal/mol). It formed an electrostatic hydrogen-bonding interaction (green dashed line) with the amino acid Arg403 (2.49 Å). Besides, it also formed a hydrophobic π-alkyl interaction (light purple dashed lines) with the amino acid Phe456 (4.93 Å). Likewise, it also formed a hydrophobic alkyl-alkyl interaction (dark purple dashed lines) with the amino acid Lys417 (4.09 Å, Fig. 9D).
Fig. 9.
The docking simulation between torvoside K and the SARS-CoV-2 spike protein. (A) Torvoside K (gray stick) docked with the RBD of the ACE2-free spike protein (cyan cartoon). (B) Torvoside K formed several electrostatic interactions (green dashed lines) and hydrophobic interactions (purple dashed lines) with amino acids (cyan sticks) in the RBD of the spike protein. (C) Torvoside K docked with the site near the connective interface of the spike-ACE2 complex. The RBD of the spike protein and ACE2 are respectively shown in green and red cartoons. (D) Torvoside K formed several electrostatic interactions (green dashed lines) and hydrophobic interactions (purple dashed lines) with amino acids near the connective interface of the spike-ACE2 complex protein.
Fig. 10A showed the simulated docking between amentoflavone and the RBD of the ACE2-free spike protein (binding energy: 8.5 kcal/mol). Amentoflavone formed four hydrophobic π-alkyl interactions (light purple dashed lines) with the amino acids Leu452 (4.21 Å), Leu492 (4.39 Å and 4.6 Å), and Ile468 (5.23 Å). Besides, it also formed a hydrophobic pi-pi T-shaped interaction (pinking purple dashed line) with the amino acid Phe486 (4.81 Å). Moreover, it formed an electrostatic π-anion interaction (orange dashed line) with the amino acid Glu465 (4.44 Å). Furthermore, it formed an electrostatic hydrogen-bonding interaction (green dashed lines) with the amino acid Phe490 (2.5 Å, Fig. 10B). Fig. 10C showed the simulated docking between amentoflavone and the connective interface of the spike-ACE2 complex (binding energy: 9.1 kcal/mol). It formed six electrostatic hydrogen-bonding interactions (green dashed lines) with the amino acids Lys26 (2.56 Å), Glu37 (2.22 Å), Ala386 (2.25 Å), Arg393 (2.09 Å), Arg403 (2.23 Å), and Tyr505 (2.51 Å). In addition, it also formed two hydrophobic π-alkyl interactions (light purple dashed lines) with the amino acids Pro389 (4.47 Å) and Lys417(5.04 Å). Likewise, it also formed a hydrophobic pi-pi T-shaped interaction (pinking purple dashed line) with the amino acid Tyr505 (5.94 Å, Fig. 10D).
Fig. 10.
The docking simulation between amentoflavone and the SARS-CoV-2 spike protein. (A) Amentoflavone (marine stick) docked with the RBD of the ACE2-free spike protein (cyan cartoon). (B) Amentoflavone formed several electrostatic interactions (green dashed lines) and hydrophobic interactions (purple dashed lines) with amino acids (cyan sticks) in the RBD of the spike protein. (C) Amentoflavone docked with the site near the connective interface of the spike-ACE2 complex. The RBD of the spike protein and ACE2 are respectively shown in green and red cartoons. (D) Amentoflavone formed several electrostatic interactions (green dashed lines) and hydrophobic interactions (purple dashed lines) with amino acids near the connective interface of the spike-ACE2 complex protein.
3.5. Lentivirus particles pseudotyped (Vpp) with SARS-CoV-2 spike protein infection assay
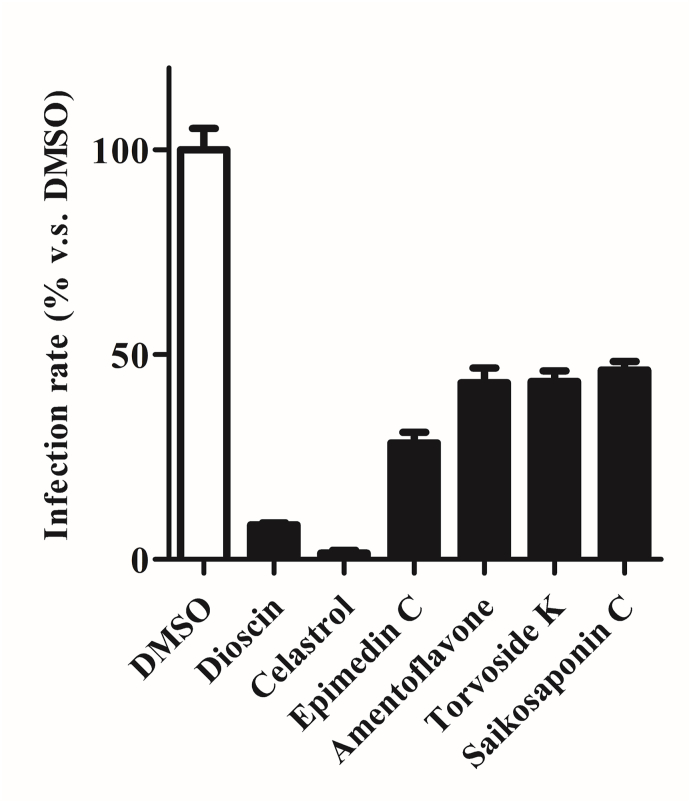
We selected the dioscin, celastrol, epimedin C, amentoflavone, torvoside K, and Saikosaponin C to treat 293T/hACE2 cells to examine their inhibitory activity. We discovered that cells with dioscin, celastrol, epimedin C, amentoflavone, torvoside K, and Saikosaponin C treatments significantly and consistently inhibited the 50–90% of SARS-CoV-2 viral infection efficacy (Fig. 11).
Fig. 11.
ACE2 overexpressing 293T cells were pre-treated with DMSO only, dioscin (10 μM), celastrol (10 μM), epimedin C (10 μM), amentoflavone (10 μM), torvoside K (10 μM), and Saikosaponin C (10 μM) for 2 h, respectively and then inoculated with lentivirus particles pseudotyped (Vpp) with SARS-CoV-2 Spike protein for 24 h.
3.6. Cell viability
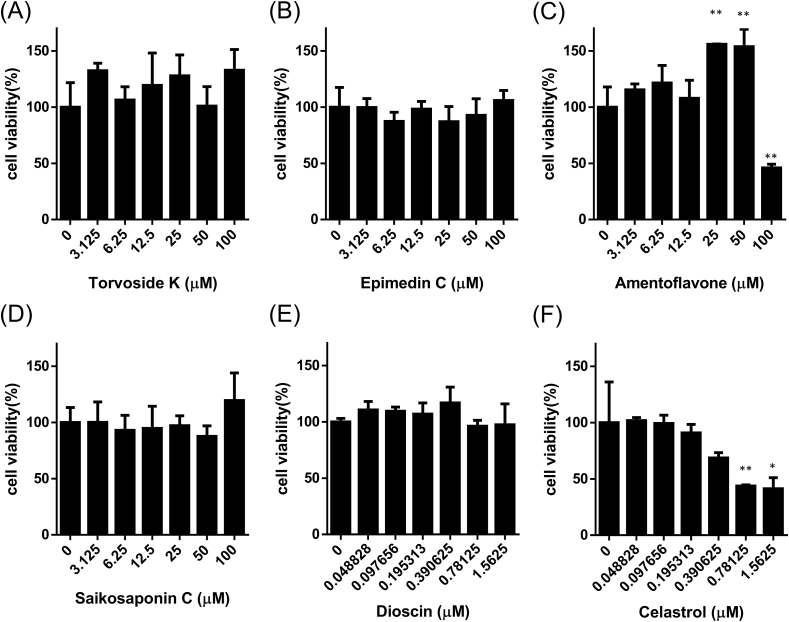
To observe the toxicity of these antiviral potential compounds in normal cells, cytotoxicity assessment of these compounds was carried out by 293T cells in MTT assay. We found that the IC50 value of torvoside K, epimedin, amentoflavone, and saikosaponin C were greater than 100 μM; the IC50 value of dioscin and celastrol were 1.5625 μM and 0.9866 μM, respectively (Fig. 12).
Fig. 12.
Effects of natural compounds on cell viability 293T cells were treated with different concentrations of (A) Torvoside K, (B) Epimedin C, (C) Amentoflavone, (D) Saikosaponin C, (E) Dioscin, and (F) Celastrol for 48 h in MTT assay. Data are the mean ± S.D. of three independent experiments. ∗p < 0.05 and ∗∗p < 0.01 versus control.
3.7. Estimation of ADME/T and drug-likeness properties
The predictions of the molecular physicochemical properties and drug-likeness are useful for drug development. The web tools, such as the admetSAR (version 2.0) website, Drug Likeness Tool (DruLiTo) website, and Chemicalize website were useful for the prediction of molecular physicochemical properties and estimation of drug-likeness. In Table 3, the data reported that candidine, dioscin, and saikosaponin C had higher possibilities to become successful drugs than others through the estimations of five drug-likeness rules (Lipinski's rule, Ghose Filter, CMC-50 like rule, Veber rule, MDDR-like rule). Table 4 showed the molecular physicochemical properties of the 6 potential natural products. The results predicted that celastrol, saikosaponin C, epimedin C, and amentoflavone had the positive (+) ability in human intestinal absorption; saikosaponin C, epimedin C, and torvoside K had higher water solubility than others; The 6 compounds were safe in carcinogenicity.
Table 3.
The physiochemical properties and drug-likeness in 6 potential natural products.
| Compound | Physiochemical properties |
Druglikeness rules |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MWa | logp | Alogp | HBAa | HBDa | TPSAa | AMRa | nRBa | nAtoma | RCa | nRigidBa | LRa | GFa | CRa | VRa | MRa | |
| Dioscin | 795.92 | 2.836 | −3.964 | 16 | 0 | 73.84 | 208.67 | 7 | 61 | 9 | 62 | 0 | 0 | 0 | 1 | 1 |
| Celastrol | 412.99 | 7.98 | 2.483 | 4 | 0 | 34.14 | 134.33 | 1 | 34 | 5 | 36 | 0 | 0 | 0 | 1 | 0 |
| Saikosaponin C | 863.91 | 2.417 | −4.103 | 18 | 0 | 64.61 | 230.27 | 9 | 66 | 9 | 65 | 0 | 0 | 0 | 1 | 1 |
| Epimedin C | 771.9 | 0.258 | −3.863 | 19 | 0 | 90.91 | 205.56 | 11 | 58 | 6 | 52 | 0 | 0 | 0 | 0 | 1 |
| Torvoside K | 675.93 | 2.734 | −3.142 | 13 | 0 | 55.38 | 178.15 | 4 | 52 | 8 | 55 | 0 | 0 | 0 | 1 | 0 |
| Amentoflavone | 519.95 | 2.03 | −0.839 | 10 | 0 | 52.6 | 157.86 | 3 | 40 | 6 | 42 | 0 | 0 | 0 | 1 | 0 |
MW: Molecular weight; HBA: Hydrogen bond acceptor; HBD: Hydrogen bond donor; TPSA: Topological polar surface area; AMR: molar refractivity; nRB: Number rotation bond; nAtom: Number atom; RC: Number of Ring; nRigidB: Number of rigid bond; LR: Lipinski's rule; GF: Ghose Filter; CR: CMC-50 like rule; VR: Veber rule; MR: MDDR-like rule.
Table 4.
The pharmacokinetics, water solubility, and toxicity prediction in 6 potential natural products.
| Compound | Pharmacokinetics |
Water solubility |
Toxicity prediction |
||||
|---|---|---|---|---|---|---|---|
| Human intestinal absorption | logS | Solubility category | Human either-a-go-go inhibition | Ames mutagenesis | Acute oral toxicity (c) | Carcinogenicity (trinary) | |
| Dioscin | – | −4.1278 | Moderate | + | – | III | Non-required |
| Celastrol | + | −3.8627 | low | + | – | III | Non-required |
| Saikosaponin C | + | −3.7177 | High | + | – | I | Non-required |
| Epimedin C | + | −3.4068 | High | + | – | III | Non-required |
| Torvoside K | – | −3.9448 | High | + | – | I | Non-required |
| Amentoflavone | + | −3.3648 | Low | – | – | II | Non-required |
4. Discussion
Notably, we identified the 6 natural products, including dioscin, celastrol, saikosaponin C, epimedin C, torvoside K, and amentoflavone, which have been respectively isolated from Dioscorea nipponica Makino,35 Tripterygium wilfordii Hook.f. [Celastraceae],36 Bupleurum chinense DC. [Apiaceae],37 Epimedium brevicornu Maxim. [Berberidaceae],38 Solanum torvum Sw. [Solanaceae],39 and Cunninghamia lanceolata (Lamb.) Hook.,40 These 6 compounds passed through all of the filters applied, with good binding energies (−10.0 kcal/mol ≤ binding energy ≤ −8.0 kcal/mol), strong ΔG values (−37 kJ/mol ≤ ΔG ≤ −26 kJ/mol), strong binding affinities, based on the Kd values (0.4 μM < Kd < 16 μM), and the good inhibitory activity of viral infection (50 ∼ 90% inhibitory rate). The other compounds listed in Table S1 have reasonable binding energies of approximately −8 kcal/mol, which may be considered in future phenotypic assays. These compounds may be able to reduce the binding between the viral spike protein and the human ACE2 protein. Among the 39 identified compounds, we also found that dioscin, actinomycin D, and saikosaponin C have previously been reported to exert antiviral activity against other viruses (Table S2), which combined with our binding affinity results and druglikeness predictions, suggest that dioscin might be the most promising candidate for further development into a potential drug against SARS-CoV-2 activity. The 6 natural products that we discovered from the database have the potential ability to bind to the viral spike protein to affect the binding affinity between the viral spike protein and the human ACE2 protein in vitro. However, the efficacy of the compounds required additional testing using in vivo assays.
Natural products have been found to be beneficial and have long been used to develop effective drugs against several diseases.41 Therefore, we examined our endemic TDEC, which is rich and diverse in natural product resources derived from traditional medicine, domestic microbes, and marine organisms. The results reported here describe the performance of a structure-based virtual screening combined with ITC binding assay and Vpp infection assay to identify natural products from among an existing database to identify compounds with the potential to prevent the interaction between the SARS-CoV-2 spike protein RBD and the ACE2 receptor of the host cell. Although, the bioactivity of the 6 compounds was verified to have good binding affinities with the S1 domain of the spike protein through the ITC binding assay and the ability of inhibitory SARS-CoV-2 virus infection was verified by in vitro lentivirus particles pseudotyped (Vpp) infection assay, the in vivo investigations are remaining necessary to confirm the abilities of those potential compounds for against SARS-CoV-2 infections.
Until now, many approaches have been devoted to finding compounds binding to ACE2-free SARS-CoV-2 spike protein against viral infection.42,43 Besides, several kinds of research using protein-protein interaction analysis to find compounds, such as zanamivir, for the inhibition of viral spike protein binding to human ACE2 protein through spike-ACE2 complex analysis.42,44 For example, hesperidin, a natural product, had been reported its disrupt the binding interface between the spike protein and ACE2.42 The above approaches are valuable and useful strategies to design antiviral activity compounds for preventing binding between viral spike protein and human ACE2 protein. In this study, the major aim of our approach is to discover potential compounds binding to viral spike protein to prevent viral infection. Our strategy is coupled with the above two strategies that were based on the ACE2-free spike protein and ACE2-complex proteins analysis. The benefit of our virtual screening strategy is not only finding the potential compounds binding to prefusion viral spike protein but also finding compounds binding to the connective interface of the spike-ACE2 complex. Besides, we confirmed the bioactivity of the 6 potential natural products by ITC binding assay and Vpp infection assay. We expected that the 6 compounds not only inhibit the binding between viral spike protein and ACE2 protein but also interfere with the complex process of the viral entry mechanism, such as the coordination between receptor binding and spike protein (S1/S2), at the ACE2-spike complex states.42 Moreover, the 6 compounds are natural products, we also expected those compounds are safe and beneficial for drug developments.
ACE2 plays an important role in Renin-Angiotensin System which regulates blood volume and systemic vascular resistance. SARS-CoV-2 spike protein inhibitors inhibit viral infection by blocking the interaction between spike protein and ACE2 protein, and these inhibitors may interfere with ACE2 physiological function to abolish blood pressure regulation. Therefore, the side effect of spike protein inhibitors is necessary to be considered. ACE2 inhibitors have been reported that inhibit spike protein-mediated SARS virus infection, and decrease ACE2 enzymatic activity in a dose-dependent manner. In contrast, the ACE-2-interacting Domain of SARS-CoV-2 (AIDS) peptide can decrease SARS-CoV-2 infection by blocking ACE2 and SARS-CoV-2 interaction without drug-related side effects. Therefore, the binding site of spike protein inhibitors may modulate Renin-Angiotensin System-associated side effects.45, 46, 47
We found that dioscin, actinomycin D and saikosaponin C had been reported that they had anti-virus activity in previous studies (Table S2). Dioscin can inhibit the initial stage of adenovirus infection in 293 cells48; in our Vpp infection assay, dioscin prevented more than 80% of the virus to infect cells in 8.69 mg/L (10 μM), which is lower than the solubility of the compound (0.2 g/L). Actinomycin D can inhibit measles virus replication and RNA synthesis in Vero cells.49 Saikosaponin C has anti-HBV replication activity.50 Among that, dioscin and saikosaponin C had been identified that they can inhibit the SARS-CoV-2 viral infection in vitro. Besides, the estimation of ADME/T and drug-likeness also indicated that dioscin and saikosaponin C had passed the Veber rule and MDDR-like rule estimation (Tables 3 and 4). Dioscin and saikosaponin C may be considered as potential compounds for developing anti-SARS-CoV-2 drugs.
The quantification of protein is one of the important processes in carrying out ITC binding assay. There are many common and useful methods had been applied to the biochemical analysis of protein, such as Lowry, Bradford, bicinchoninic acid (BCA), UV spectroscopic, and 3-(4-carboxybenzoyl)quinoline-2-carboxaldehyde (CBQCA) assays.51 Among them, the Bradford assay is a popular, simple, rapid, inexpensive, and sensitive assay. It is based on the direct binding of Coomassie brilliant blue G-250 (CBBG) dye to the proteins.51 In our study, we employ the Bradford assay to analyze the quantification of SARS-CoV-2 spike protein. It is based on the following reasons. Firstly, the Bradford assay has widely been used in biochemical analysis of protein quantitation in ITC binding assay. Secondly, it is faster, cheaper, and easier for us than other protein analyses. Therefore, the Bradford assay is adopted to carry out quantification of protein analysis in this study. Although some of the methods for the quantification of protein is a high accuracy, it is less used to carry out protein analysis for ITC binding assay. For example, amino acid analysis (AAA) is one of the accurate assays for protein quantification.52 However, it is much less to be adopted to implement quantification of protein by AAA for ITC binding assay. Until now, we do not find the literature that quantification of protein was carried out by AAA for ITC binding assay. Moreover, we lack the necessary types of equipment and skills for carrying out AAA. Therefore, we employ the Bradford assay for protein analysis in this study. It is one of the limitations of this study.
The SARS-CoV-2 virus enters human cells through viral spike protein binding to human ACE2 protein. Considering the SARS-CoV-2 virus is a biosafety-level-3 virus, a simplified assay using pseudotyped biosafety-level-2 viral particles with viral spike protein is necessary.53 In this study, we employ lentivirus particles pseudotyped (Vpp) with SARS-CoV-2 spike protein infection assay to further verify the antiviral activity of these potential natural products. In this assay system, the 293T cells are transfected with an HIV-based lentiviral system that can produce SARS-CoV-2 Spike-pseudotyped lentiviral particles. And then, to make viral particles infect 293T cells expressing human ACE2.53 Finally, to measure the Luciferase expression to estimate the inhibitory of viral infection of test compounds.53 Our results showed that dioscin, celastrol, saikosaponin C, epimedin C, torvoside K, and amentoflavone display good inhibitory activity decreasing the binding between SARS-CoV-2 spike and ACE2 protein. Therefore, these potential natural products may have the ability against SARS-CoV-2 viral infection.
5. Conclusion
This study was performed to identify potential drugs for COVID-19 therapy. We virtually screened more than 2,000 drugs against the RBD of the SARS-CoV-2 spike protein. After data mining and filtering out unfitted compounds, we identified 39 compounds with high estimated binding affinities with the targeted spike protein. Among these identified compounds, the 6 natural products, including dioscin, celastrol, saikosaponin C, epimedin C, torvoside K, and amentoflavone, were further analyzed to verify the binding affinity with the target protein using ITC. Besides, we also discovered that cells with dioscin, celastrol, epimedin C, amentoflavone, torvoside K, and Saikosaponin C treatments significantly and consistently inhibited the 50–90% of SARS-CoV-2 viral infection efficacy. Our results suggested that these 6 natural products binding to the viral spike protein with strong affinities and antiviral efficacy. We believe that these compounds could represent potential drugs for the treatment and prevention of SARS-CoV-2 infection. These identified compounds require further validation in animal-based tests to determine their potential to be developed into anti-SARS-CoV-2 therapies.
Disclosure statement
Author Shu-Ting Chan was employed by the company TCI CO., Ltd., Taiwan. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Author's contribution
G.Y. Chen designed, performed the simulation experiments, analyzed the data and did the manuscript writing. Y.C. Pan assisted in the literature survey and ITC assay. T.Y. Wu assisted the compounds' substance for assay. T.Y. Yao assisted in planning the research, literature survey and manuscript writing. W.J. Wang and W.J. Shen assisted the lentivirus particles pseudotyped (Vpp) with SARS-CoV-2 spike protein infection assay. S.T. Chan supported partial plant materials for studying. A. Ahmed, C.H. Tang, W.C. Huang assisted with the manuscript proofreading. M.C. Hung guided the Vpp with SARS-CoV-2 spike protein infection assay. J.C. Yang and Y.C. Wu supervised the study and approved the final manuscript.
Highlights of the findings and novelties
The 8 natural products, including dioscin, cephalinone D, celastrol, saikosaponin C, epimedin C, candidine, torvoside K, and amentoflavone have the potential activity binding to SARS-CoV-2 spike protein. In addition, dioscin, celastrol, saikosaponin C, epimedin C, torvoside K, and amentoflavone have the potential with anti-virus infection activity to prevent SARS-CoV-2 to bind with ACE2 protein in vitro.
Declaration of competing interest
Author Shu-Ting Chan was employed by the company TCI CO., Ltd., Taiwan. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported by grants from Ministry of Science and Technology, and China Medical University, Taiwan (Grant No. MOST-108-2320-B-039-062, MOST-107-2320-B-037-001, MOST-107-2321-B-037-004, MOST-106-2320-B-037-007, MOST-106-2321-B-037-004, CMU-104-S-14-01, and CMU-108-MF-22). We thank the Taiwan Database of Extracts and Compounds website for the assistance in offering chemical and biological information of extracts and compounds. Besides, we are grateful to the National Center for High-Performance Computing for computer time and facilities. We also thank the Center for Resources, Research, and Development of Kaohsiung Medical University for the ChemBio3D Ultra 11.0 technical support. The plant materials in this study were partly supported by TCI CO., Ltd., Taiwan.
Footnotes
Peer review under responsibility of The Center for Food and Biomolecules, National Taiwan University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jtcme.2021.09.002.
Contributor Information
Guan-Yu Chen, Email: markchen19822001@gmail.com.
Yi-Cheng Pan, Email: alce2943@gmail.com.
Tung-Ying Wu, Email: kuma0401@gmail.com.
Tsung-You Yao, Email: saviorano1@gmail.com.
Wei-Jan Wang, Email: cvcsky@cmu.edu.tw.
Wan-Jou Shen, Email: wshen@cmu.edu.tw.
Azaj Ahmed, Email: azajahmed92@gmail.com.
Shu-Ting Chan, Email: Rebecca.Chan@tci-bio.com.
Chih-Hsin Tang, Email: chtang@mail.cmu.edu.tw.
Wei-Chien Huang, Email: whuang@mail.cmu.edu.tw.
Mien-Chie Hung, Email: mhung@cmu.edu.tw.
Juan-Cheng Yang, Email: T21087@mail.cmuh.org.tw.
Yang-Chang Wu, Email: yachwu@mail.cmu.edu.tw.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Song Z.Q., Xu Y.F., Bao L.L., et al. From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses-Basel. 2019;11(1):59. doi: 10.3390/v11010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Corman V.M., Lienau J., Witzenrath M. Coronaviruses as the cause of respiratory infections. Internist. 2019;60(11):1136–1145. doi: 10.1007/s00108-019-00671-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rabaan A.A., Al-Ahmed S.H., Haque S., et al. SARS-CoV-2, SARS-CoV, and MERSCOV: a comparative overview. Inf Med. 2020;28:174–184. [PubMed] [Google Scholar]
- 4.Tai W.B., He L., Zhang X.J., et al. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell Mol Immunol. 2020:1–8. doi: 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Du L.Y., He Y.X., Zhou Y.S., Liu S.W., Zheng B.J., Jiang S.B. The spike protein of SARS-CoV - a target for vaccine and therapeutic development. Nat Rev Microbiol. 2009;7(3):226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367(19):1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 7.Organization W.H. 2020. WHO Director-General's Opening Remarks at the Media Briefing on COVID-19-11 March 2020. Geneva, Switzerland. [Google Scholar]
- 8.Madjid M., Safavi-Naeini P., Solomon S.D., Vardeny O. Potential effects of coronaviruses on the cardiovascular system: a review. JAMA Cardiol. 2020 doi: 10.1001/jamacardio.2020.1286. [DOI] [PubMed] [Google Scholar]
- 9.Paraskevis D., Kostaki E.G., Magiorkinis G., Panayiotakopoulos G., Sourvinos G., Tsiodras S. Full-genome evolutionary analysis of the novel corona virus (2019-nCoV) rejects the hypothesis of emergence as a result of a recent recombination event. Infect Genet Evol. 2020;79:104212. doi: 10.1016/j.meegid.2020.104212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu R., Zhao X., Li J., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luk H.K.H., Li X., Fung J., Lau S.K.P., Woo P.C.Y. Molecular epidemiology, evolution and phylogeny of SARS coronavirus. Infect Genet Evol. 2019;71:21–30. doi: 10.1016/j.meegid.2019.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamming I., Timens W., Bulthuis M.L., Lely A.T., Navis G., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203(2):631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoffmann M., Kleine-Weber H., Schroeder S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–+. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xia S., Liu M.Q., Wang C., et al. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30(4):343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ziebuhr J., Siddell S.G. Processing of the human coronavirus 229E replicase polyproteins by the virus-encoded 3C-like proteinase: identification of proteolytic products and cleavage sites common to pp1a and pp1ab. J Virol. 1999;73(1):177–185. doi: 10.1128/jvi.73.1.177-185.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuan M., Wu N.C., Zhu X., et al. A highly conserved cryptic epitope in the receptor-binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020 doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lan J., Ge J., Yu J., et al. 2020. Crystal Structure of the 2019-nCoV Spike Receptor-Binding Domain Bound with the ACE2 Receptor. bioRxiv. [Google Scholar]
- 18.Wrapp D., Wang N., Corbett K.S., et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strømgaard K., Krogsgaard-Larsen P., Madsen U. CRC press; 2017. Textbook of Drug Design and Discovery. [Google Scholar]
- 20.Demarse N.A., Quinn C.F., Eggett D.L., Russell D.J., Hansen L.D. Calibration of nanowatt isothermal titration calorimeters with overflow reaction vessels. Anal Biochem. 2011;417(2):247–255. doi: 10.1016/j.ab.2011.06.014. [DOI] [PubMed] [Google Scholar]
- 21.Pierce M.M., Raman C.S., Nall B.T. Isothermal titration calorimetry of protein-protein interactions. Methods. 1999;19(2):213–221. doi: 10.1006/meth.1999.0852. [DOI] [PubMed] [Google Scholar]
- 22.Nunes N.M., Coelho Y.L., Castro J.S., et al. Naringenin-lactoferrin binding: impact on naringenin bitterness and thermodynamic characterization of the complex. Food Chem. 2020;331:127337. doi: 10.1016/j.foodchem.2020.127337. [DOI] [PubMed] [Google Scholar]
- 23.Li F.J., Liu Y., Yuan Y., Yang B., Liu Z.M., Huang L.Q. Molecular interaction studies of acetylcholinesterase with potential acetylcholinesterase inhibitors from the root of Rhodiola crenulata using molecular docking and isothermal titration calorimetry methods. Int J Biol Macromol. 2017;104(Pt A):527–532. doi: 10.1016/j.ijbiomac.2017.06.066. [DOI] [PubMed] [Google Scholar]
- 24.Velázquez-Campoy A., Ohtaka H., Nezami A., Muzammil S., Freire E. Isothermal titration calorimetry. Curr. Protoc. Cell Biol. 2004;23(1) doi: 10.1002/0471143030.cb1708s23. 17.18. 11-17.18. 24. [DOI] [PubMed] [Google Scholar]
- 25.DeLano W., DeLano Scientific L. DeLano Scientific; San Carlos, CA: 2002. PyMOL Version 0.99. [Google Scholar]
- 26.Waterhouse A., Bertoni M., Bienert S., et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46(W1):W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Backer J.A., Klinkenberg D., Wallinga J. Incubation period of 2019 novel coronavirus (2019-nCoV) infections among travellers from Wuhan, China, 20-28 January 2020. Euro Surveill. 2020;25(5):10–15. doi: 10.2807/1560-7917.ES.2020.25.5.2000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dallakyan S., Olson A.J. Springer; Chemical biology: 2015. Small-molecule Library Screening by Docking with PyRx; pp. 243–250. [DOI] [PubMed] [Google Scholar]
- 30.Visualizer DS. San Diego: Dassault Systèmes BIOVIA; 2019.
- 31.O'Boyle N., Banck M., James C., Morley C., Vandermeersch T., Hutchison G. Open babel: an open chemical toolbox. J Cheminf. 2011;3(1):33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang S.C., Chen Y., Wang Y.C., et al. Tannic acid suppresses SARS-CoV-2 as a dual inhibitor of the viral main protease and the cellular TMPRSS2 protease. Am J Cancer Res. 2020;10(12):4538–4546. [PMC free article] [PubMed] [Google Scholar]
- 33.Yu S., Zhu Y., Xu J., et al. Glycyrrhizic acid exerts inhibitory activity against the spike protein of SARS-CoV-2. Phytomedicine. 2021;85:153364. doi: 10.1016/j.phymed.2020.153364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krainer G., Broecker J., Vargas C., Fanghanel J., Keller S. Quantifying high-affinity binding of hydrophobic ligands by isothermal titration calorimetry. Anal Chem. 2012;84(24):10715–10722. doi: 10.1021/ac3025575. [DOI] [PubMed] [Google Scholar]
- 35.Cho J., Choi H., Lee J., Kim M.S., Sohn H.Y., Lee D.G. The antifungal activity and membrane-disruptive action of dioscin extracted from Dioscorea nipponica. Biochim Biophys Acta. 2013;1828(3):1153–1158. doi: 10.1016/j.bbamem.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 36.Calixto J.B., Campos M.M., Otuki M.F., Santos A.R. Anti-inflammatory compounds of plant origin. Part II. modulation of pro-inflammatory cytokines, chemokines and adhesion molecules. Planta Med. 2004;70(2):93–103. doi: 10.1055/s-2004-815483. [DOI] [PubMed] [Google Scholar]
- 37.Yuan B.C., Yang R., Ma Y.S., Zhou S., Zhang X.D., Liu Y. A systematic review of the active saikosaponins and extracts isolated from Radix Bupleuri and their applications. Pharmaceut Biol. 2017;55(1):620–635. doi: 10.1080/13880209.2016.1262433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H.B., Chen F. Separation and purification of epimedin A, B, C, and icariin from the medicinal herb Epimedium brevicornum maxim by dual-mode HSCCC. J Chromatogr Sci. 2009;47(5):337–340. doi: 10.1093/chromsci/47.5.337. [DOI] [PubMed] [Google Scholar]
- 39.Abhishek R.U., Thippeswamy S., Manjunath K., Mohana D.C. Antifungal and antimycotoxigenic potency of Solanum torvum Swartz. leaf extract: isolation and identification of compound active against mycotoxigenic strains of Aspergillus flavus and Fusarium verticillioides. J Appl Microbiol. 2015;119(6):1624–1636. doi: 10.1111/jam.12956. [DOI] [PubMed] [Google Scholar]
- 40.Zhang M., Liu J., Liu P., et al. Study on chemical constituents of the branches and leaves of Cunninghamia lanceolata. J Shanghai Jiaotong Univ Agric Sci. 2011;29(5):67–71. [Google Scholar]
- 41.Beutler J.A. Natural products as a foundation for drug discovery. Curr Protoc Pharmacol. 2019;86(1):e67. doi: 10.1002/cpph.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Papageorgiou A.C., Mohsin I. The SARS-CoV-2 spike glycoprotein as a drug and vaccine target: structural insights into its complexes with ACE2 and antibodies. Cells. 2020;9(11):2343. doi: 10.3390/cells9112343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheng F.J., Huynh T.K., Yang C.S., et al. Hesperidin is a potential inhibitor against SARS-CoV-2 infection. Nutrients. 2021;13(8) doi: 10.3390/nu13082800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Panda P.K., Arul M.N., Patel P., et al. Structure-based drug designing and immunoinformatics approach for SARS-CoV-2. Sci Adv. 2020;6(28) doi: 10.1126/sciadv.abb8097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paidi R.K., Jana M., Mishra R.K., Dutta D., Raha S., Pahan K. ACE-2-interacting domain of SARS-CoV-2 (AIDS) peptide suppresses inflammation to reduce fever and protect lungs and heart in mice: implications for COVID-19 therapy. J Neuroimmune Pharmacol. 2021;16(1):59–70. doi: 10.1007/s11481-020-09979-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shagufta Ahmad I. The race to treat COVID-19: potential therapeutic agents for the prevention and treatment of SARS-CoV-2. Eur J Med Chem. 2021;213:113157. doi: 10.1016/j.ejmech.2021.113157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tikellis C., Thomas M.C. Angiotensin-converting enzyme 2 (ACE2) is a key modulator of the Renin angiotensin system in Health and disease. Int J Pept. 2012;2012:256294. doi: 10.1155/2012/256294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu C., Wang Y., Wu C., et al. Dioscin's antiviral effect in vitro. Virus Res. 2013;172(1-2):9–14. doi: 10.1016/j.virusres.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 49.Schluederberg A., Williams C.A., Black F.L. Inhibition of measles virus replication and RNA synthesis by actinomycin D. Biochem Biophys Res Commun. 1972;48(3):657–661. doi: 10.1016/0006-291x(72)90398-1. [DOI] [PubMed] [Google Scholar]
- 50.Pan Y., Ke Z., Ye H., et al. Saikosaponin C exerts anti-HBV effects by attenuating HNF1alpha and HNF4alpha expression to suppress HBV pgRNA synthesis. Inflamm Res. 2019;68(12):1025–1034. doi: 10.1007/s00011-019-01284-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Olson B.J., Markwell J. Assays for determination of protein concentration. Curr Protoc Protein Sci. 2007;48(1) doi: 10.1002/0471140864.ps0304s48. 3.4. 1-43.4. 29. [DOI] [PubMed] [Google Scholar]
- 52.Kambhampati S., Li J., Evans B.S., Allen D.K. Accurate and efficient amino acid analysis for protein quantification using hydrophilic interaction chromatography coupled tandem mass spectrometry. Plant Methods. 2019;15:46. doi: 10.1186/s13007-019-0430-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Crawford K.H.D., Eguia R., Dingens A.S., et al. Protocol and reagents for pseudotyping lentiviral particles with SARS-CoV-2 spike protein for neutralization assays. Viruses. 2020;12(5) doi: 10.3390/v12050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.