This diagnostic study evaluates a deep learning model for whole-volume segmentation of 4 clinically important pathological features as well as clinical applicability.
Key Points
Question
Is deep learning–based segmentation of macular disease in optical coherence tomography (OCT) suitable for clinical use?
Findings
In this diagnostic study of OCT data from 173 patients with age-related macular degeneration or diabetic macular edema, model segmentations qualitatively ranked better or comparable for clinical applicability to 1 or more expert grader segmentations in 127 scans (73%) by a panel of 3 retinal specialists. Scans with high quantitative accuracy scores were not reliably associated with higher rankings.
Meaning
These findings suggest that qualitative evaluation adds to quantitative approaches when assessing clinical applicability of segmentation tools and clinician satisfaction in practice.
Abstract
Importance
Quantitative volumetric measures of retinal disease in optical coherence tomography (OCT) scans are infeasible to perform owing to the time required for manual grading. Expert-level deep learning systems for automatic OCT segmentation have recently been developed. However, the potential clinical applicability of these systems is largely unknown.
Objective
To evaluate a deep learning model for whole-volume segmentation of 4 clinically important pathological features and assess clinical applicability.
Design, Setting, Participants
This diagnostic study used OCT data from 173 patients with a total of 15 558 B-scans, treated at Moorfields Eye Hospital. The data set included 2 common OCT devices and 2 macular conditions: wet age-related macular degeneration (107 scans) and diabetic macular edema (66 scans), covering the full range of severity, and from 3 points during treatment. Two expert graders performed pixel-level segmentations of intraretinal fluid, subretinal fluid, subretinal hyperreflective material, and pigment epithelial detachment, including all B-scans in each OCT volume, taking as long as 50 hours per scan. Quantitative evaluation of whole-volume model segmentations was performed. Qualitative evaluation of clinical applicability by 3 retinal experts was also conducted. Data were collected from June 1, 2012, to January 31, 2017, for set 1 and from January 1 to December 31, 2017, for set 2; graded between November 2018 and January 2020; and analyzed from February 2020 to November 2020.
Main Outcomes and Measures
Rating and stack ranking for clinical applicability by retinal specialists, model-grader agreement for voxelwise segmentations, and total volume evaluated using Dice similarity coefficients, Bland-Altman plots, and intraclass correlation coefficients.
Results
Among the 173 patients included in the analysis (92 [53%] women), qualitative assessment found that automated whole-volume segmentation ranked better than or comparable to at least 1 expert grader in 127 scans (73%; 95% CI, 66%-79%). A neutral or positive rating was given to 135 model segmentations (78%; 95% CI, 71%-84%) and 309 expert gradings (2 per scan) (89%; 95% CI, 86%-92%). The model was rated neutrally or positively in 86% to 92% of diabetic macular edema scans and 53% to 87% of age-related macular degeneration scans. Intraclass correlations ranged from 0.33 (95% CI, 0.08-0.96) to 0.96 (95% CI, 0.90-0.99). Dice similarity coefficients ranged from 0.43 (95% CI, 0.29-0.66) to 0.78 (95% CI, 0.57-0.85).
Conclusions and Relevance
This deep learning–based segmentation tool provided clinically useful measures of retinal disease that would otherwise be infeasible to obtain. Qualitative evaluation was additionally important to reveal clinical applicability for both care management and research.
Introduction
Quantitative 3-dimensional (3-D) imaging tools that accurately measure various anatomical features have transformed multiple medical specialties,1 including radiotherapy,2 neuroimaging,3 and cardiology.4 In ophthalmology, 3-D optical coherence tomography (OCT) imaging has revolutionized retinal disease management5,6,7; however, few tools exist to accurately quantify abnormalities.8
Treatment decisions for common retinal conditions, including wet age-related macular degeneration (AMD) and diabetic macular edema (DME), rely on subjective assessment of retinal fluid. This assessment is prone to intergrader disagreement, particularly at lower volumes.9,10 Most commercial instruments include basic tools for 2-D measurements such as retinal thickness but are susceptible to errors in the presence of pathology.11,12,13 Such measures also do not elucidate underlying pathological features and thus correlate poorly with visual outcomes.14
Optical coherence tomography segmentation offers the potential to objectively quantify disease burden and standardize treatment decisions. Although theoretically possible through manual segmentation by specialists, OCT segmentation is infeasible in both clinical and research settings owing to the time-consuming task of delineating pathological features across hundreds of sections per scan. Deep learning–based methods offer a new approach, enabling quick and accurate segmentation of pathological features15,16 and providing more granular assessments of disease progression.17,18 De Fauw et al19 previously published a 2-stage deep learning system, involving segmentation as an intermediate representation, and assessed its accuracy when classifying macular diseases. Herein, we assessed the agreement of automated segmentations in deliberately challenging scans with AMD and DME against a reading center criterion standard that importantly involved manual human segmentation of entire OCT volumes, rather than selecting key sections as others have done. Given the challenges of OCT interpretation in complex macular disease where several clinical interpretations may be equally valid, we performed both quantitative and blinded qualitative evaluations with retinal specialists to assess potential clinical applicability.
Methods
This diagnostic study followed the Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis (TRIPOD) reporting guideline. This research received approval from the Cambridge East Research Ethics Committee. Deidentification was performed in line with the Anonymisation: Managing Data Protection Risk Code of Practice of the Information Commissioner’s Office20 and validated by Moorfields Eye Hospital information technology and information governance. Only anonymized retrospective data were used for research, without the active involvement of patients, and informed consent was not required. The study adhered to the tenets of the Declaration of Helsinki.21
Data Sets
Five data sets were used, split between an initial pilot study (set 1) and the main study (set 2). The pilot study was performed to evaluate feasibility of the task and for guideline iteration before the main study. For all data sets, 1 eye per patient was selected.
For set 1 (1 data set), 15 OCT scans from patients with new presentations of severe AMD to Moorfields Eye Hospital were randomly selected from the test set of De Fauw et al,19 acquired using the 3D OCT-2000 device from Topcon Corporation. For set 2 (4 data sets), a total of 164 OCT scans not previously used to train the models were randomly selected by Moorfields Ophthalmic Reading Centre from unique patients attending Moorfields Eye Hospital from January 1 to December 31, 2017, with either AMD or DME.19 Scans were acquired using the Topcon device or a Spectralis OCT device from Heidelberg Engineering GmbH, resulting in 4 subsets: (1) Topcon-AMD, (2) Heidelberg-AMD, (3) Topcon-DME, and (4) Heidelberg-DME. To ensure a representative data set, selection was enriched to fulfill different levels of disease severity (mild, moderate, and severe) (the definition of which is provided in the eMethods in the Supplement) and treatment status (at initial referral, at 3 months after intravitreal therapy, and at 12 months after initial presentation).
Each Topcon scan consisted of 128 B-scans. For Heidelberg, volumes with 25 or 49 B-scans were selected. All images were visually inspected for quality by a senior ophthalmologist (K.B.), consistent with previous work.19
Procedures
Grading Process
The full grading protocol is described in the eMethods in the Supplement. Segmentations were drawn using ImageJ (Fiji)22 and a drawing tablet (Wacom Co, Ltd). Gradings were performed between November 2018 and January 2020.
For set 1, 2 specialist optometrist graders manually segmented all B-scans for each OCT for 3 pathological features: intraretinal fluid (IRF), subretinal fluid (SRF), and pigment epithelial detachment (PED) (eTable 1 in the Supplement). Segmentations were independently adjudicated by 2 senior ophthalmologists (P.A.K. and K.B.) with more than 10 years of experience and reading center certification for OCT segmentation.
For set 2, each subset was segmented by 2 certified graders (from a pool of 4 Moorfields Ophthalmic Reading Centre graders) and adjudicated by 1 senior ophthalmologist (K.B.). The DME scans were segmented for IRF and SRF only, and AMD scans were segmented for IRF, SRF, PED, and subretinal hyperreflective material (SHRM).
Adjudication involved an interactive process of feedback and revision until each grading was completed to reading center standards and approved by the senior ophthalmologist. The aim was to achieve a reasonable clinical interpretation without intent to converge toward a single ground truth. Grading and revisions for the pilot study were performed to the highest standard reasonably possible, without overt time constraints. All grading and adjudication were performed blinded to model predictions.
Model Segmentation
All OCTs underwent automated segmentation using a previously published deep learning network.19 Briefly, a 3-D U-Net architecture23 translates an OCT input to a map with 15 classes, including anatomical and pathological features. Five instances of the same network were trained with a different order of inputs and different random weight initializations, which were then ensembled to produce the final segmentation maps. Features of interest were IRF, SRF, SHRM, and PED. Model-predicted drusen, fibrovascular PED, and serous PED were combined to a singular PED feature.
Qualitative Evaluation
Owing to inherent variability in human grading, no single ground truth is available against which to compare the algorithm; 2 segmentations of the same OCT scan may vary volumetrically or geometrically owing to alternative interpretations, whereas clinical applicability of each may be equivalent. We therefore tasked 3 retinal specialists (S.V., H.K., and L.N.) (eTable 2 in the Supplement) to qualitatively assess 3 segmentations (2 experts and 1 model) for each OCT scan at the volume level. For each case, retinal specialists were presented with the original OCT, followed by 3 blinded segmentations in a randomly shuffled order (eFigure 1 in the Supplement).
To assess how representative segmentations were of the specialists’ impression of the scan, each specialist stack ranked the segmentations in the order that most closely reflected their interpretation and selected the magnitude of difference between each pair in the ranking as slightly better, moderately better, or considerably better. To assess clinical applicability, specialists were asked if they would be satisfied to use each segmentation within their clinical practice, using a 5-point Likert scale where 1 indicates strongly disagree; 2, disagree; 3, neither agree nor disagree; 4, agree; and 5 strongly agree.
Data Analysis
Qualitative Evaluation Analysis
For stack-ranking analysis, scans with slight or slight and moderate differences were considered separately as comparable to each other. Scans with moderate and considerable or only considerable differences were determined to be better or worse than one another. The number and percentage of scans for which the model was considered better or comparable to at least 1 of the expert gradings by most of the specialists was calculated. Likert ratings were analyzed by taking the median specialist rating per segmentation and determining the distribution of ratings for both model and expert gradings. We used the Krippendorff α to evaluate agreement between specialists for ordinal ranking and ratings. The 95% CIs were calculated using Wilson’s methods.24
Volumetric Analysis
Total feature volume (in cubic millimeters) was calculated by multiplying the number of labeled voxels for each pathological feature by the voxel volume. Bland-Altman plots with limits of agreement were used to visualize agreement between graders (intergrader) and between the model and each individual grader (model-grader) for total volume segmented for each feature. Because we expected that larger differences would be observed at larger segmented volumes, limits of agreement based on relative volume changes were calculated from log-transformed variables.25 The intraclass correlation coefficient (ICC) was calculated using the 2-way random-effects model for agreement, and 95% CIs were calculated using bootstrapping to ensure consistency between intergrader and model-grader metrics.
Geometric Analysis
The similarity of segmentations between model and expert gradings for each feature was evaluated using the Dice similarity coefficient (DSC)26 and was calculated only for scans in which the respective feature was present (defined as ≥1 voxel segmented in the whole scan) in all 3 gradings (both experts and model).
Statistical Analysis
Data were analyzed from February 2020 to November 2020. Statistical analysis was performed with Python, version 3.6.7 (Python Software Foundation). Volumetric and geometric analyses were stratified by disease, disease severity, treatment time point, and device type.
Results
A total of 173 scans were used for analysis after 6 scans were excluded (3 for poor quality; 3 owing to data extraction failures) (Table). This included 107 AMD scans and 66 DME scans, with 103 scans acquired using the Topcon device and 70 using the Heidelberg device. Expert grading took a mean of 50 hours per scan for set 1 and 7 hours per scan for set 2. The mean time taken by the model was less than 10 seconds per scan, running on graphics processing units (5 Tesla V100; Nvidia). Example segmentations are shown in Figure 1, the Video, and eFigures 2 to 12 in the Supplement.
Table. Data Set Characteristicsa.
| Characteristic | Set 1 | Set 2 | |||
|---|---|---|---|---|---|
| Topcon (3D OCT-2000) | Topcon (3D OCT-2000) | Heidelberg (Spectralis OCT) | |||
| Condition | Wet AMD | Wet AMD | DME | Wet AMD | DME |
| Features segmented | IRF, SRF, PEDb | IRF, SRF, SHRM, PED | IRF, SRF | IRF, SRF, SHRM, PED | IRF, SRF |
| Total No. of OCT volumes | 15 | 46 | 42 | 46 (19 with 49 B-scans, 27 with 25 B-scans) | 24 (7 with 49 B-scans, 17 with 25 B-scans) |
| Sex, % | |||||
| Female | 47 | 59 | 55 | 50 | 50 |
| Male | 53 | 41 | 45 | 50 | 50 |
| Age, mean (SD), y | 75 (10) | NC | NC | NC | NC |
| No. of OCT volumes | |||||
| First presentation (treatment-naive) | 15 | 16 | 14 | 20 | 10 |
| After first treatment | |||||
| 3 mo | 0 | 15 | 15 | 14 | 8 |
| 12 mo | 0 | 15 | 13 | 12 | 6 |
| Total No. of B-scans | |||||
| Requiring assessment, mean (SD) | 1920 (128) | 5888 (128) | 5376 (128) | 1606 (35) | 768 (32) |
| With features manually segmented, counting both graders (mean [SD] per grader volume) | 3002 (100 [24]) | 6062 (66 [23]) | 5350 (64 [29]) | 2133 (23 [13]) | 880 (18 [13]) |
| Mean time taken per grader to manually segment a volume, h | 50 | 7c | |||
Abbreviations: AMD, age-related macular degeneration; DME, diabetic macular edema; IRF, intraretinal fluid; NC, not collected; OCT, optical coherence tomography; PED, pigment epithelial detachment; SHRM, subretinal hyperreflective material; SRF, subretinal fluid.
Scans were obtained using the 3D OCT-2000 device from Topcon Corporation (Topcon) and the Spectralis device from Heidelberg Engineering GmbH (Heidelberg) for wet AMD and DME.
Graders were asked to segment PED in 13 of 15 volumes in this pilot set of scans.
Mean time for the entirety of set 2.
Figure 1. Examples of Segmentations of Optical Coherence Tomography (OCT).
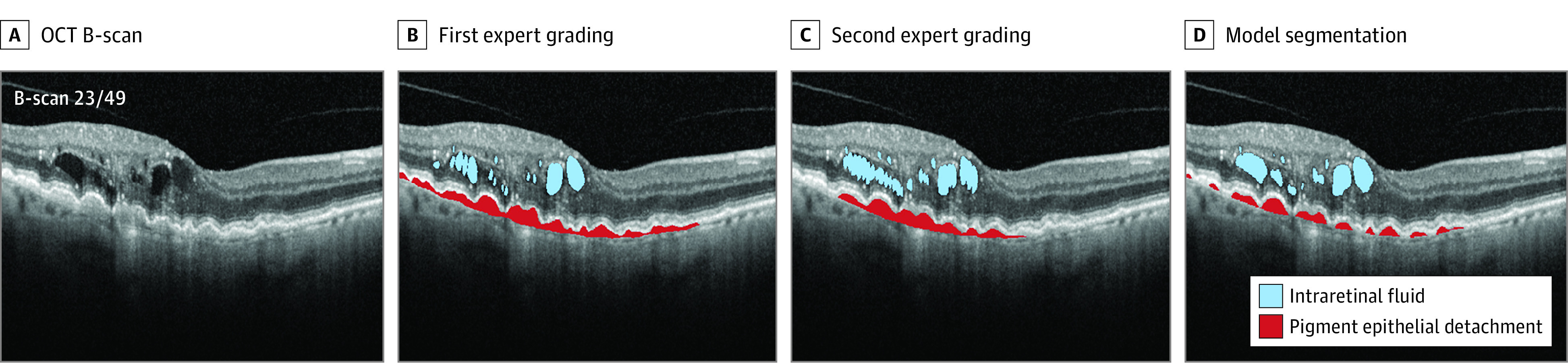
Scans are from set 2, using the Spectralis OCT device (Heidelberg Engineering GmbH) for wet age-related macular degeneration (AMD). For AMD scans, as many as 4 features were segmented: intraretinal fluid, subretinal fluid, subretinal hyperreflective material, and pigment epithelial detachment. The Video shows the whole volume segmentation.
Video. Whole-Volume Automated Segmentation of Retinal Optical Coherence Tomography (OCT) Images Using Deep Learning.
Examples of segmentations, model successes, model failures, and model-specialist disagreements when using deep learning to quantify volumes of clinically relevant pathology in OCT scans of patents with age-related macular degeneration and diabetic macular edema. See article text and eFigures 1-12 in the Supplement for full details.
Qualitative Evaluation by Retinal Specialists
When considering scans stack ranked with slight differences as comparable, the model was better or comparable to at least 1 expert in 127 of 173 (73%; 95% CI, 66%-79%) and worse in 46 of 173 (27%; 95% CI, 21%-34%) (eTable 3 in the Supplement). When both slight and moderate differences were considered comparable, the model was better or comparable to at least 1 expert in 149 of 173 scans (86%; 95% CI, 80%-90%) and worse in 24 of 173 (14%; 95% CI, 10%-20%). eFigure 13 in the Supplement visualizes the complete distribution. Where the model ranked highest, there was generally a slight difference from the next expert grade (range, 40%-75%) (eTable 4 in the Supplement).
Most of the specialists gave positive Likert ratings (4 or 5) to 85 of 173 model segmentations (49%; 95% CI, 42%-57%) and 225 of 346 expert segmentations (2 per scan) (65%; 95% CI, 60%-70%), which increased to 78% (95% CI, 71%-84%) and 89% (95% CI, 86%-92%), respectively, when also considering neutral ratings (Likert rating 3). Although specialist 3 generally rated all segmentations lower than specialists 1 and 2, the relative trend in ratings was similar among all 3 specialists (Figure 2). The full breakdown of ratings is provided in eTables 5 and 6 in the Supplement. The Krippendorff α for testing agreement between specialists was 0.57 (95% CI, 0.52-0.62) for ranking and 0.36 (95% CI, 0.30-0.41) for rating.
Figure 2. Diverging Stacked Bar Charts Showing Distribution of the Likert Ratings of the Segmentations Given to the Expert Gradings and to the Model.
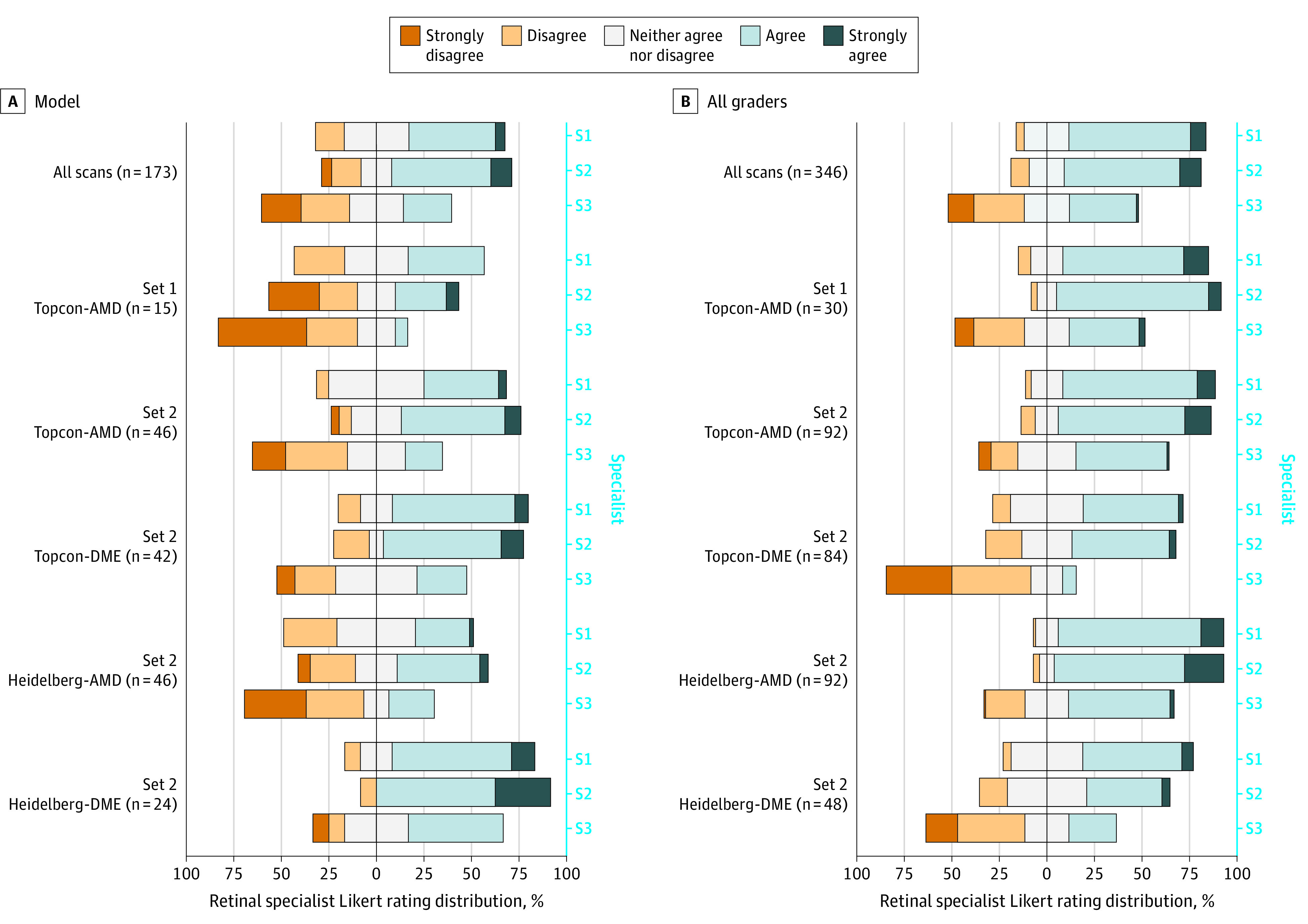
The distribution is shown for all scans and per subset for the statement “I would be satisfied to use this segmentation within my clinical practice.” Each bar represents ratings given by an individual specialist (S1, S2, and S3). Specialists selected a single point on the Likert scale. The bars are centered on the neutral rating, with negative ratings stacked to the left and positive ratings stacked to the right.
For both qualitative tasks, the model was ranked and rated highest in the Heidelberg-DME subgroup, followed by the Topcon-DME group. In these subgroups, expert gradings were rated least positively. The contrary was found for AMD subgroups: the model was ranked third most often in the Heidelberg-AMD set, and better or comparable in 24 of 46 scans (52%; 95% CI, 38%-66%) scans. A neutral or positive rating was given to 29 of 46 model segmentations (63%; 95% CI, 49%-75%). A greater proportion of model segmentations were ranked and rated higher in set 2 for Topcon-AMD (87%; 95% CI, 74%-94%) compared with set 1 (53%; 95% CI, 30%-75%), although the latter consisted of fewer scans. Expert gradings were rated neutral or positive in more than 90% of scans in each AMD subgroup.
eFigures 3 to 5 in the Supplement present examples of success cases where the model was rated 4 or 5 and/or ranked first compared with expert gradings. eFigures 7 to 10 in the Supplement are examples of model failure cases that were qualitatively rated poorly compared with expert gradings. Specialists were not always in consensus; eFigures 11 and 12 in the Supplement provide examples of substantial disagreements.
Quantifying Segmented Feature Volumes
Compared with expert graders, the model segmented greater volumes of IRF (mean difference, −0.16 mm3), comparable volumes of SRF and SHRM (mean differences, 0.01 and 0.002 mm3, respectively), and lower volumes of PED (mean difference, 0.14 mm3). For all segmented features, both linear and logarithmic limits of agreement were wider for model-grader agreement (Figure 3) compared with intergrader agreement (eFigure 14 in the Supplement).
Figure 3. Bland-Altman Plots Comparing Volumes of Individual Features Segmented Between the Model and Grader.
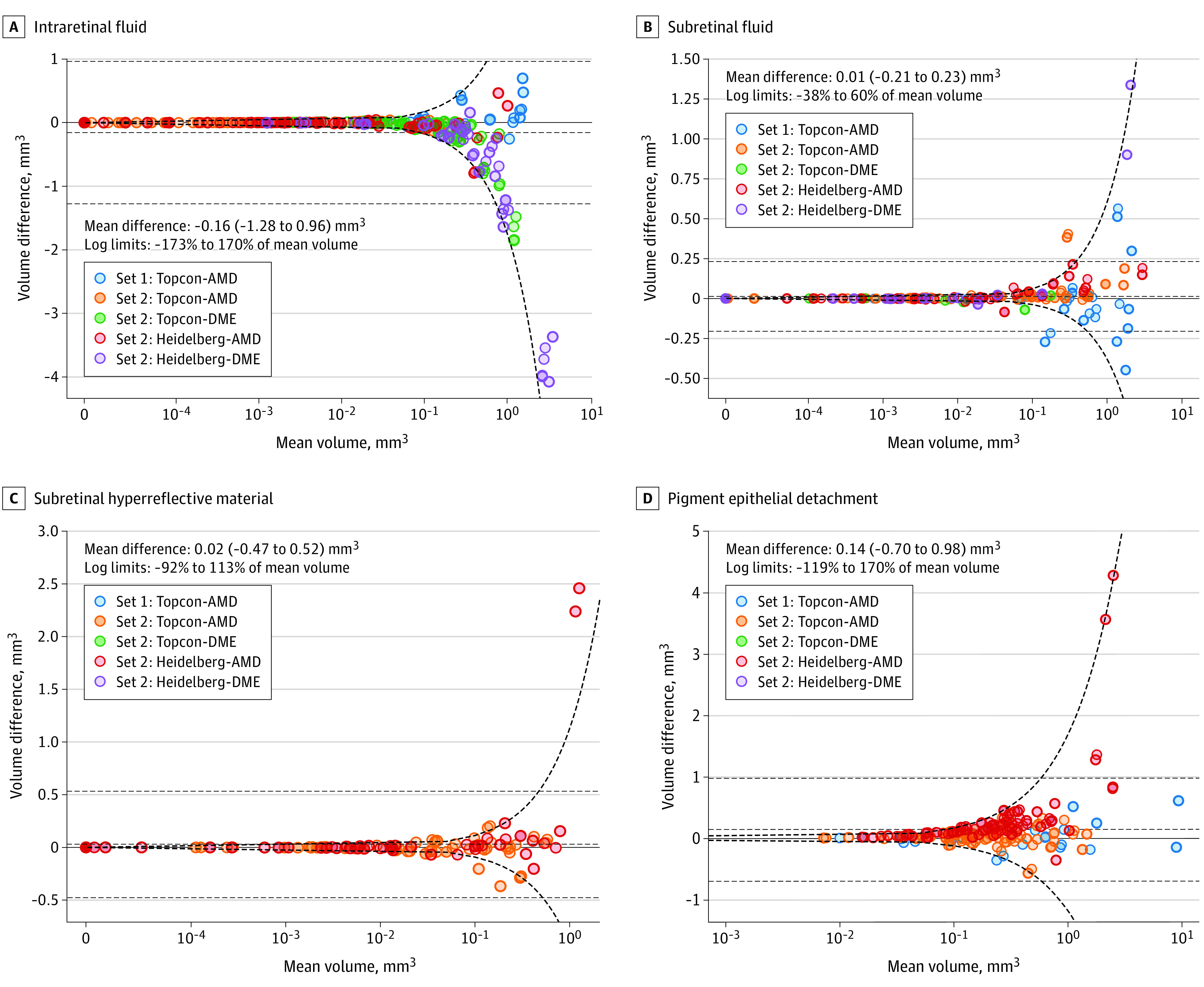
Negative differences on the plots on the right indicate that the model on average segmented greater volumes of the respective feature, whereas positive differences indicate the opposite. The mean value of the difference and the 95% limits of agreement (mean difference ±1.96 SD of the difference) are plotted with black dashed lines. Given that the differences are related to the magnitude of the mean volume, the limits of agreement are also calculated after log-transforming the data and are plotted in the linear space, as a ratio of the mean volume, with bold dashed lines. Optical coherence tomographic (OCT) scans were obtained using the 3D OCT-2000 device from Topcon Corporation (Topcon) and the Spectralis device from Heidelberg Engineering GmbH (Heidelberg) for wet age-related macular degeneration (AMD) and diabetic macular edema (DME).
Cases outside both conventional and log-transformed limits of agreement for IRF were almost all DME cases, particularly when the model segmented more fluid than the graders. Conversely, outliers for SHRM and PED were where the model had segmented less than the mean volume segmented by the expert graders. Similar numbers of outliers were seen on either side of the limits of agreement for SRF; however, the largest outliers were where the volume segmented by the model was greater than that segmented by the expert graders. Outliers were a mixture of model successes, model failures, and ambiguous cases (eFigures 3-10 in the Supplement).
Pairwise ICCs were highest for set 1, ranging from 0.90 (95% CI, 0.75-0.99) to 0.99 (95% CI, 0.90-1.00) for intergrader agreement and from 0.93 (95% CI, 0.85-0.99) to 1.00 (95% CI, 0.84-1.00) for model-grader agreement for the 3 segmented features (eTables 7 and 8 in the Supplement). The ICCs were lowest for IRF in the Topcon-DME (0.33; 95% CI, 0.22-0.63) and Heidelberg-DME (0.31; 95% CI, 0.17-0.48) subgroups. Overall, model-grader ICCs ranged from 0.33 (95% CI, 0.08-0.96) for SHRM to 0.96 (95% CI, 0.90-0.99) for SRF.
Similarity of Segmentations Between Experts and Model
Intraretinal fluid was present in both expert gradings and the model prediction in 121 of 173 scans (70%); SRF, in 67 of 173 scans (39%); SHRM, in 32 of 92 scans (35%); and PED, in 105 of 105 scans (100%) (eFigure 15 in the Supplement). There was no consensus on presence or absence of IRF in 42 of 173 scans (24%), SRF in 37 of 173 scans (21%), and SHRM in 37 of 92 scans (40%).
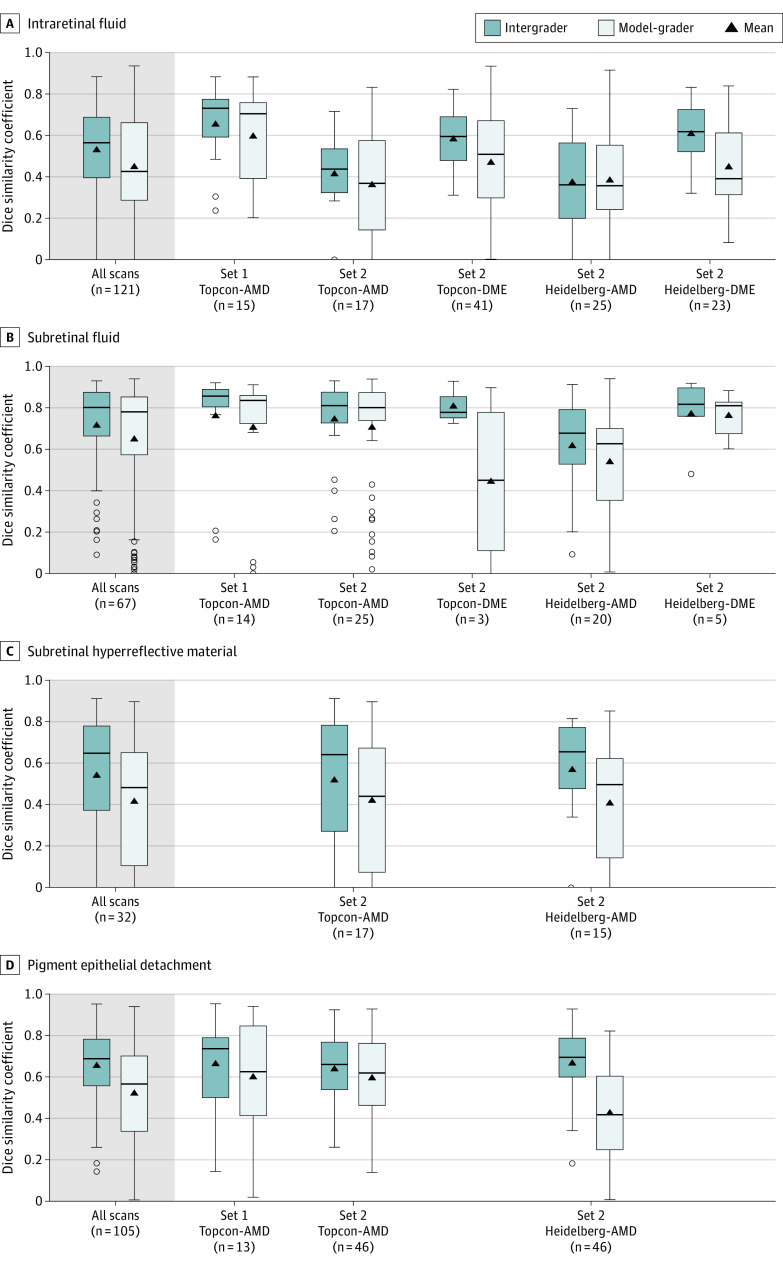
Across all the scans, SRF had the highest DSCs for both intergrader (0.80 [95% CI, 0.66-0.87]) and model-grader comparison (0.78 [95% CI, 0.57-0.85]), whereas IRF had the lowest DSCs (0.56 [95% CI, 0.40-0.69] and 0.43 [95% CI, 0.29-0.66], respectively). Similar patterns were found within each subset (Figure 4 and eTable 9 in the Supplement). There were too few DME scans with SRF present to make reliable conclusions for this feature. Of the subgroups, DSCs were consistently highest among segmented features for set 1, for both intergrader and model-grader comparisons. As intergrader DSC increased, model-grader DSC also increased (eFigure 16 in the Supplement). Mean manually segmented volumes were calculated and bucketed by quartiles. Median DSC increased as mean volume quartile increased for all 4 features (eFigure 17 in the Supplement). The DSCs stratified by severity and time point are presented in eTable 10 and eFigure 18 in the Supplement.
Figure 4. Distribution of Dice Similarity Coefficients (DSCs) for All Optical Coherence Tomographic (OCT) Scans and Stratified by Data Set Subgroup.
Each panel shows DSC distribution for intraretinal fluid, subretinal fluid, subretinal hyperreflective material, and pigment epithelial detachment. Boxes display the median and interquartile range. The whiskers extend up to 1.5 × interquartile range beyond the upper and lower quartiles; the isolated circles fall outside of this range. The black triangles represent the mean DSC. Scans were obtained using the 3D OCT-2000 device from Topcon Corporation (Topcon) and the Spectralis device from Heidelberg Engineering GmbH (Heidelberg) for wet age-related macular degeneration (AMD) and diabetic macular edema (DME).
Discussion
In this diagnostic study, we evaluated the clinical applicability of a deep learning model to segment clinically relevant pathology in a comprehensive external validation data set of real-world OCT scans, covering a wide range of different disease severities and treatment points. We compared model segmentations with carefully adjudicated expert gradings performed at a specialist reading center and rigorously evaluated quantitative agreement through both volumetric and spatial methods. Quantifying pathology through segmentation unlocks a wealth of novel information within OCT scans, but human segmentation is not feasible in practice owing to the vast time requirements to label even a single scan. Deep learning provides a route to consistent segmentation on a massive scale, potentially enabling new forms of disease management, clinical trial analysis, and scientific discovery.
We investigated the potential clinical applicability of the segmentations through an independent qualitative evaluation by 3 experienced retinal specialists and compared this with quantitative volumetric and 3-D geometric measures of agreement. We found that the model segmentations ranked at least as well as 1 expert grader’s segmentation in three-quarters of scans; however, 14% of model segmentations were considered worse than both expert gradings. Qualitatively, DME sets had the greatest proportion of first rankings and positive Likert scores for the model compared with the expert graders. However, from quantitative metrics alone, the model appears to have performed inferiorly to the experts in these sets. We observed numerous examples in which cases with lower quantitative agreement were still judged as clinically acceptable. We conclude that qualitative assessment of clinical applicability is essential and that quantitative evaluation alone is not sufficient.
The highest intergrader and model-grader DSCs and ICCs were achieved for set 1. These gradings were exhaustively segmented, taking a mean of 50 hours per volume. In related work, models developed and evaluated on single 2-D sections have achieved higher DSCs, where exhaustive grading can be performed more feasibly.27,28 Other reasons for higher DSCs in related work may be owing to a lack of independence of scans between the development and evaluation set,29 and the derivation of ground truth labels from the model being evaluated.30 Our study used a pool of 6 graders, all blinded to model predictions, with 2 graders segmenting each OCT volume. This provided multiple clinical interpretations resulting in the ability to benchmark the model against the natural, rich variation seen in clinical practice. Importantly, our data set did not overlap with the development set.
To be clinically applicable, it is important to test the robustness of the segmentation model when applied to images of different diseases, or from different device manufacturers. To do this, we used a representative data set of OCT images of 2 commonly treated retinal diseases of varying severities and points during treatment and from 2 widely used OCT scanners with fundamental differences in quality and resolution. Furthermore, the image quality inclusion criteria were broad: all scans were from real-world practice, excluding only 3 volumes where relevant features could not be delineated. The difference in performance between devices and diseases may be explained by the amount of data used to train the segmentation model, which included sparsely segmented B-scans of 877 Topcon OCTs and only 152 Heidelberg OCTs.19 Multiple groups have reported promising segmentation results using the RETOUCH (Retinal OCT Fluid Detection and Segmentation Benchmark and Challenge) test set comprising 14 volumes that are representative of multiple diseases and devices31; however, the set was too small to capture performance on different subsets.15,32 In our study, we observed substantial variability in performance among devices, diseases, and features.
Each retinal pathological feature has an individual role in functional and anatomical prognosis.33,34,35,36,37 Deep learning has facilitated automated analysis of these at scale for AMD38 and DME,17 and we show that each feature poses different challenges. Intraretinal fluid can be difficult to manually delineate owing to suboptimal resolution or contrast of the images, which reduces clarity of cystoid boundaries, especially in the presence of retinal thickening, common in DME. Subretinal fluid presented the strongest DSC and ICC and is simplest to delineate owing to high contrast between SRF and surrounding tissues. Subretinal hyperreflective material can be challenging owing to heterogeneous reflectivity and undefined margins and a distribution that is often mixed with other pathological features, including SRF and PED.
Limitations
This study has some limitations. A fundamental challenge for quantitative validation of deep learning–based segmentation tools is the imperfection of human-based reference standards. Two expert segmentations of the same OCT can vary considerably owing to alternative interpretations, whereas the clinical applicability of each may be equivalent. Severe pathology can be morphologically complex, making it difficult to be certain of underlying processes and features. For example, retinal specialists have low sensitivity for the binary detection task of fluid presence, more so for IRF than SRF.10 Therefore, simple binary detection models may help to ensure that abnormalities are not missed. In addition, volumetric estimation of fluid could help with objectively determining disease activity, especially in AMD, in which treatment decisions and follow-up intervals are guided by subjective interpretation. In DME, the presence of certain thresholds of fluid could be clinically useful to determine whether edema is substantial enough to warrant treatment.
Another limitation is the sheer scale of the OCT segmentation task. Each Topcon OCT scan consisted of nearly 60 million voxels. It is therefore unsurprising that DSC increased as the mean segmented volume increased for all pathological features, because the likelihood of overlap increases when more voxels are segmented. This presents a limitation of the DSC metric to compare segmentations owing to its sensitivity to small volumes and where a feature is segmented in one annotation but absent in another. This has been observed in other studies, including segmentation of brain lesions on computed tomography39 and magnetic resonance imaging.40
Conclusions
This diagnostic study evaluated the clinical applicability of a deep learning system to quantify volumes of clinically relevant pathology in OCT scans in AMD and DME that would otherwise be infeasible to obtain. Segmentations were acceptable to specialists in most cases, and qualitative evaluation provided valuable insights in addition to more traditional quantitative analysis. This tool has already advanced the understanding of the anatomical characteristics in AMD38,41 and in the future may provide novel quantitative end points for clinical trials to enable in-depth analysis. Automated segmentation systems offer the potential to transform clinical workflows; however, further research is needed to directly assess their utility for disease monitoring and management.
eMethods. Grading Protocol and Disease Severity Grading Definition
eFigure 1. Custom Viewer Used to Assess the 3 Segmentations for the Qualitative Evaluation
eFigure 2. Example Segmentations
eFigure 3. Example of a Scan Where the Model Was Qualitatively Rated Higher Than the 2 Manual Segmentations
eFigure 4. Example 1 of a Scan Where the Model Was Qualitatively Rated Higher Than the 2 Manual Segmentations
eFigure 5. Example 2 of a Scan Where the Model Was Ranked as Most Representative
eFigure 6. Example of a Scan Where All 3 Gradings Were Rated Highly
eFigure 7. Example of a Challenging Case Resulting in Large Disagreement in SRF Volume Between Model and Manual Gradings
eFigure 8. Example of Large Disagreement in SHRM and PED Volume Between Model and Manual Gradings
eFigure 9. Example of Model Failure and Largest Disagreement in PED Volume Between the Model and Expert Graders
eFigure 10. Example 1 of a Model Failure and Large Disagreement in IRF/SRF Volume
eFigure 11. Example 2 of a Disagreement Between Specialists for Qualitative Evaluation
eFigure 12. Example of Disagreement Between Specialists for Qualitative Evaluation
eFigure 13. Stacked Bar Chart Showing Distribution of the Stack Rank Positions for the Expert Gradings and the Model Segmentations
eFigure 14. Bland-Altman Plots Comparing Volumes of Individual Features Segmented Between Graders
eFigure 15. Matrix Showing the Number of Scans Segmented by the Model and Segmented by Neither, One or Both Human Expert Graders, per Feature
eFigure 16. Relationship Between Intergrader DSC and the Model-Grader DSC per Feature
eFigure 17. Distribution of Dice Similarity Coefficients (DSC) Stratified by 4 Ascending Mean Grader Volume Buckets
eFigure 18. Distribution of Dice Similarity Coefficients (DSC) for All Scans and Stratified by Scan Subgroup
eTable 1. Grader Experience and Set Assignment
eTable 2. Qualitative Evaluation Retinal Specialist Qualifications
eTable 3. Stack Rank Comparison Between the Model and Human Expert Graders Taking Into Account the Margins of Differences Within Each Stack
eTable 4. Magnitude of Difference Between Model and Next Expert Grader Where the Model Ranked Highest
eTable 5. Distribution of Specialist Likert Ratings for Model Segmentations
eTable 6. Distribution of Specialist Likert Ratings for Expert Gradings
eTable 7. Pairwise Intraclass Correlation Coefficient (ICC) With 95% CIs
eTable 8. Pairwise Intraclass Correlation Coefficient (ICC) and 95% CIs per Subgroup
eTable 9. Median and Interquartile Range of DSC for Scans Where the Feature Was Considered Present Across All 3 Segmentations, Stratified by Set
eTable 10. Median and Interquartile Range of DSC for Scans Where the Feature Was Considered Present Across All 3 Segmentations, Stratified by Device, Disease (Severity and Time Point), Eye, and Sex
References
- 1.Rosenkrantz AB, Mendiratta-Lala M, Bartholmai BJ, et al. Clinical utility of quantitative imaging. Acad Radiol. 2015;22(1):33-49. doi: 10.1016/j.acra.2014.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gurney-Champion OJ, Mahmood F, van Schie M, et al. Quantitative imaging for radiotherapy purposes. Radiother Oncol. 2020;146:66-75. doi: 10.1016/j.radonc.2020.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Man MY, Ong MS, Mohamad MS, et al. A review on the bioinformatics tools for neuroimaging. Malays J Med Sci. 2015;22(Spec Issue):9-19. [PMC free article] [PubMed] [Google Scholar]
- 4.Gomez J, Doukky R, Germano G, Slomka P. New trends in quantitative nuclear cardiology methods. Curr Cardiovasc Imaging Rep. 2018;11(1):1. doi: 10.1007/s12410-018-9443-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosenfeld PJ. Optical coherence tomography and the development of antiangiogenic therapies in neovascular age-related macular degeneration. Invest Ophthalmol Vis Sci. 2016;57(9):OCT14-OCT26. doi: 10.1167/iovs.16-19969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keane PA, Patel PJ, Liakopoulos S, Heussen FM, Sadda SR, Tufail A. Evaluation of age-related macular degeneration with optical coherence tomography. Surv Ophthalmol. 2012;57(5):389-414. doi: 10.1016/j.survophthal.2012.01.006 [DOI] [PubMed] [Google Scholar]
- 7.Costa RA, Skaf M, Melo LAS Jr, et al. Retinal assessment using optical coherence tomography. Prog Retin Eye Res. 2006;25(3):325-353. doi: 10.1016/j.preteyeres.2006.03.001 [DOI] [PubMed] [Google Scholar]
- 8.Wintergerst MWM, Schultz T, Birtel J, et al. Algorithms for the automated analysis of age-related macular degeneration biomarkers on optical coherence tomography: a systematic review. Transl Vis Sci Technol. 2017;6(4):10-10. doi: 10.1167/tvst.6.4.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toth CA, Decroos FC, Ying G-S, et al. Identification of fluid on optical coherence tomography by treating ophthalmologists versus a reading center in the comparison of age-related macular degeneration treatments trials. Retina. 2015;35(7):1303-1314. doi: 10.1097/IAE.0000000000000483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keenan TD, Clemons TE, Domalpally A, et al. Retinal specialist versus artificial intelligence detection of retinal fluid from OCT: Age-Related Eye Disease Study 2: 10-Year Follow-On Study. Ophthalmology. 2021;128(1):100-109. doi: 10.1016/j.ophtha.2020.06.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Waldstein SM, Gerendas BS, Montuoro A, Simader C, Schmidt-Erfurth U. Quantitative comparison of macular segmentation performance using identical retinal regions across multiple spectral-domain optical coherence tomography instruments. Br J Ophthalmol. 2015;99(6):794-800. doi: 10.1136/bjophthalmol-2014-305573 [DOI] [PubMed] [Google Scholar]
- 12.Sadda SR, Wu Z, Walsh AC, et al. Errors in retinal thickness measurements obtained by optical coherence tomography. Ophthalmology. 2006;113(2):285-293. doi: 10.1016/j.ophtha.2005.10.005 [DOI] [PubMed] [Google Scholar]
- 13.Giani A, Cigada M, Esmaili DD, et al. Artifacts in automatic retinal segmentation using different optical coherence tomography instruments. Retina. 2010;30(4):607-616. doi: 10.1097/IAE.0b013e3181c2e09d [DOI] [PubMed] [Google Scholar]
- 14.Ying G-S, Huang J, Maguire MG, et al. ; Comparison of Age-related Macular Degeneration Treatments Trials Research Group . Baseline predictors for one-year visual outcomes with ranibizumab or bevacizumab for neovascular age-related macular degeneration. Ophthalmology. 2013;120(1):122-129. doi: 10.1016/j.ophtha.2012.07.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu D, Heisler M, Lee S, et al. Deep-learning based multiclass retinal fluid segmentation and detection in optical coherence tomography images using a fully convolutional neural network. Med Image Anal. 2019;54:100-110. doi: 10.1016/j.media.2019.02.011 [DOI] [PubMed] [Google Scholar]
- 16.Schlegl T, Waldstein SM, Bogunovic H, et al. Fully automated detection and quantification of macular fluid in OCT using deep learning. Ophthalmology. 2018;125(4):549-558. doi: 10.1016/j.ophtha.2017.10.031 [DOI] [PubMed] [Google Scholar]
- 17.Roberts PK, Vogl W-D, Gerendas BS, et al. Quantification of fluid resolution and visual acuity gain in patients with diabetic macular edema using deep learning: a post hoc analysis of a randomized clinical trial. JAMA Ophthalmol. 2020;138(9):945-953. doi: 10.1001/jamaophthalmol.2020.2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmidt-Erfurth U, Vogl W-D, Jampol LM, Bogunović H. Application of automated quantification of fluid volumes to anti-VEGF therapy of neovascular age-related macular degeneration. Ophthalmology. 2020;127(9):1211-1219. doi: 10.1016/j.ophtha.2020.03.010 [DOI] [PubMed] [Google Scholar]
- 19.De Fauw J, Ledsam JR, Romera-Paredes B, et al. Clinically applicable deep learning for diagnosis and referral in retinal disease. Nat Med. 2018;24(9):1342-1350. doi: 10.1038/s41591-018-0107-6 [DOI] [PubMed] [Google Scholar]
- 20.Information Commissioner’s Office . Anonymisation: Managing Data Protection Risk Code of Practice. 2015. Accessed March 1, 2016. https://ico.org.uk/media/1061/anonymisation-code.pdf
- 21.World Medical Association . World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA. 2013;310(20):2191-2194. doi: 10.1001/jama.2013.281053 [DOI] [PubMed] [Google Scholar]
- 22.Schindelin J, Arganda-Carreras I, Frise E, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676-682. doi: 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation. In: Navab N, Hornegger J, Wells WM, Frangi AF, eds. Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015. Springer; 2015:234-241. doi: 10.1007/978-3-319-24574-4_28 [DOI] [Google Scholar]
- 24.Wilson EB. Probable inference, the law of succession, and statistical inference. J Am Stat Assoc. 1927;22(158):209-212. doi: 10.1080/01621459.1927.10502953 [DOI] [Google Scholar]
- 25.Euser AM, Dekker FW, le Cessie S. A practical approach to Bland-Altman plots and variation coefficients for log transformed variables. J Clin Epidemiol. 2008;61(10):978-982. doi: 10.1016/j.jclinepi.2007.11.003 [DOI] [PubMed] [Google Scholar]
- 26.Dice LR. Measures of the amount of ecologic association between species. Ecology. 1945;26(3):297-302. doi: 10.2307/1932409 [DOI] [Google Scholar]
- 27.Lee CS, Tyring AJ, Deruyter NP, Wu Y, Rokem A, Lee AY. Deep-learning based, automated segmentation of macular edema in optical coherence tomography. Biomed Opt Express. 2017;8(7):3440-3448. doi: 10.1364/BOE.8.003440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mehta N, Lee CS, Mendonça LSM, et al. Model-to-data approach for deep learning in optical coherence tomography intraretinal fluid segmentation. JAMA Ophthalmol. 2020;138(10):1017-1024. doi: 10.1001/jamaophthalmol.2020.2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee H, Kang KE, Chung H, Kim HC. Automated segmentation of lesions including subretinal hyperreflective material in neovascular age-related macular degeneration. Am J Ophthalmol. 2018;191:64-75. doi: 10.1016/j.ajo.2018.04.007 [DOI] [PubMed] [Google Scholar]
- 30.Oakley JD, Sodhi SK, Russakoff DB, Choudhry N. Automated deep learning-based multi-class fluid segmentation in swept-source optical coherence tomography images. Preprint. Posted online September 2, 2020. bioRxiv 278259. doi: 10.1101/2020.09.01.278259 [DOI]
- 31.RETOUCH: Retinal OCT Fluid Challenge. September 14, 2017. Accessed September 8, 2020. https://retouch.grand-challenge.org/
- 32.Bogunovic H, Venhuizen F, Klimscha S, et al. RETOUCH: the Retinal OCT Fluid Detection and Segmentation Benchmark and Challenge. IEEE Trans Med Imaging. 2019;38(8):1858-1874. doi: 10.1109/TMI.2019.2901398 [DOI] [PubMed] [Google Scholar]
- 33.Pokroy R, Mimouni M, Barayev E, et al. Prognostic value of subretinal hyperreflective material in neovascular age-related macular degeneration treated with bevacizumab. Retina. 2018;38(8):1485-1491. doi: 10.1097/IAE.0000000000001748 [DOI] [PubMed] [Google Scholar]
- 34.Ritter M, Simader C, Bolz M, et al. Intraretinal cysts are the most relevant prognostic biomarker in neovascular age-related macular degeneration independent of the therapeutic strategy. Br J Ophthalmol. 2014;98(12):1629-1635. doi: 10.1136/bjophthalmol-2014-305186 [DOI] [PubMed] [Google Scholar]
- 35.Sophie R, Lu N, Campochiaro PA. Predictors of functional and anatomic outcomes in patients with diabetic macular edema treated with ranibizumab. Ophthalmology. 2015;122(7):1395-1401. doi: 10.1016/j.ophtha.2015.02.036 [DOI] [PubMed] [Google Scholar]
- 36.Gerendas BS, Prager S, Deak G, et al. Predictive imaging biomarkers relevant for functional and anatomical outcomes during ranibizumab therapy of diabetic macular oedema. Br J Ophthalmol. 2018;102(2):195-203. doi: 10.1136/bjophthalmol-2017-310483 [DOI] [PubMed] [Google Scholar]
- 37.Guymer RH, Markey CM, McAllister IL, Gillies MC, Hunyor AP, Arnold JJ; FLUID Investigators . Tolerating subretinal fluid in neovascular age-related macular degeneration treated with ranibizumab using a treat-and-extend regimen: FLUID Study 24-month results. Ophthalmology. 2019;126(5):723-734. doi: 10.1016/j.ophtha.2018.11.025 [DOI] [PubMed] [Google Scholar]
- 38.Moraes G, Fu DJ, Wilson M, et al. Quantitative analysis of optical coherence tomography for neovascular age-related macular degeneration using deep learning. Ophthalmology. 2021;128(5):693-705. doi: 10.1016/j.ophtha.2020.09.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Monteiro M, Newcombe VFJ, Mathieu F, et al. Multiclass semantic segmentation and quantification of traumatic brain injury lesions on head CT using deep learning: an algorithm development and multicentre validation study. Lancet Digit Health. 2020;2(6):e314-e322. doi: 10.1016/S2589-7500(20)30085-6 [DOI] [PubMed] [Google Scholar]
- 40.Anbeek P, Vincken KL, van Osch MJP, Bisschops RHC, van der Grond J. Automatic segmentation of different-sized white matter lesions by voxel probability estimation. Med Image Anal. 2004;8(3):205-215. doi: 10.1016/j.media.2004.06.019 [DOI] [PubMed] [Google Scholar]
- 41.Chopra R, Moraes G, Fu DJ, et al. Quantitative analysis of change in retinal tissues in neovascular age-related macular degeneration using artificial intelligence. Invest Ophthalmol Vis Sci. 2020;61(7):1152. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eMethods. Grading Protocol and Disease Severity Grading Definition
eFigure 1. Custom Viewer Used to Assess the 3 Segmentations for the Qualitative Evaluation
eFigure 2. Example Segmentations
eFigure 3. Example of a Scan Where the Model Was Qualitatively Rated Higher Than the 2 Manual Segmentations
eFigure 4. Example 1 of a Scan Where the Model Was Qualitatively Rated Higher Than the 2 Manual Segmentations
eFigure 5. Example 2 of a Scan Where the Model Was Ranked as Most Representative
eFigure 6. Example of a Scan Where All 3 Gradings Were Rated Highly
eFigure 7. Example of a Challenging Case Resulting in Large Disagreement in SRF Volume Between Model and Manual Gradings
eFigure 8. Example of Large Disagreement in SHRM and PED Volume Between Model and Manual Gradings
eFigure 9. Example of Model Failure and Largest Disagreement in PED Volume Between the Model and Expert Graders
eFigure 10. Example 1 of a Model Failure and Large Disagreement in IRF/SRF Volume
eFigure 11. Example 2 of a Disagreement Between Specialists for Qualitative Evaluation
eFigure 12. Example of Disagreement Between Specialists for Qualitative Evaluation
eFigure 13. Stacked Bar Chart Showing Distribution of the Stack Rank Positions for the Expert Gradings and the Model Segmentations
eFigure 14. Bland-Altman Plots Comparing Volumes of Individual Features Segmented Between Graders
eFigure 15. Matrix Showing the Number of Scans Segmented by the Model and Segmented by Neither, One or Both Human Expert Graders, per Feature
eFigure 16. Relationship Between Intergrader DSC and the Model-Grader DSC per Feature
eFigure 17. Distribution of Dice Similarity Coefficients (DSC) Stratified by 4 Ascending Mean Grader Volume Buckets
eFigure 18. Distribution of Dice Similarity Coefficients (DSC) for All Scans and Stratified by Scan Subgroup
eTable 1. Grader Experience and Set Assignment
eTable 2. Qualitative Evaluation Retinal Specialist Qualifications
eTable 3. Stack Rank Comparison Between the Model and Human Expert Graders Taking Into Account the Margins of Differences Within Each Stack
eTable 4. Magnitude of Difference Between Model and Next Expert Grader Where the Model Ranked Highest
eTable 5. Distribution of Specialist Likert Ratings for Model Segmentations
eTable 6. Distribution of Specialist Likert Ratings for Expert Gradings
eTable 7. Pairwise Intraclass Correlation Coefficient (ICC) With 95% CIs
eTable 8. Pairwise Intraclass Correlation Coefficient (ICC) and 95% CIs per Subgroup
eTable 9. Median and Interquartile Range of DSC for Scans Where the Feature Was Considered Present Across All 3 Segmentations, Stratified by Set
eTable 10. Median and Interquartile Range of DSC for Scans Where the Feature Was Considered Present Across All 3 Segmentations, Stratified by Device, Disease (Severity and Time Point), Eye, and Sex