Abstract
A method for the detection and quantitation of oncogenic human papillomavirus (HPV) was developed by using the fluorescent 5′ exonuclease assay. The method is based on the amplification of a 180-bp fragment from the 3′ part of the E1 open reading frame in a single PCR with type-specific probes for HPV types 16, 18, 31, 33, and 35. The probes can be used separately or in combinations of up to three probes per assay. Quantitation over a range of 101 to 106 initial HPV copies was possible by using real-time detection of the accumulation of fluorescence with cycle number. Reconstitution experiments, performed to mimic mixed infections, showed that individual HPV types can be detected down to a ratio of about 1% in a mixture. The performance of the assay depends on DNA quality, the presence of PCR inhibitors, and the number of different probes used simultaneously. This homogeneous assay provides a fast and sensitive way of screening for oncogenic HPV types in biopsy specimens as well as cervical smear samples. The closed-tube nature of the assay and the inclusion of uracil N′-glycosylase reduces cross contamination of PCR products to a minimum. A similar assay for β-actin was used in parallel for quantitation of genomic DNA. After normalizing the samples for genomic DNA content, the mean number of HPV copies per cell could be calculated.
Infection with certain types of human papillomavirus (HPV) has been shown to be the single most important risk factor for the development of cervical cancer (15). More than 70 different types of HPV have been described, and about 30 of these infect the internal and external genitalia. Approximately 95% of biopsy specimens from patients with cervical cancer have been found to contain DNA of high-risk HPV types, most commonly HPV type 16 (HPV-16), followed by HPV types 18, 31, and 45 (3). Among the low-risk types, HPV-6 and HPV-11 are associated with condylomata acuminata but only rarely with carcinogenic progression (3, 18, 30, 34).
Given the importance of HPV infection as a risk factor for cervical cancer, considerable efforts have been made in the development of techniques for the detection of the virus as well as the associated cellular changes (11, 13, 17). Serological detection methods, in which conformational antibodies against self-assembled capsid proteins are used, have been employed to detect present or recent infection with HPV (5, 18). Since not every infected individual develops antibodies, this method has certain limitations (12). Other methods have been based on detection of viral nucleic acid by either in situ hybridization (6, 33), restriction fragment length polymorphism analysis and Southern blot analysis (4, 27, 31), hybrid capture (in which a DNA-RNA heteroduplex is recognized by monoclonal antibodies) (4, 22, 23), and finally, PCR. Many of the PCR systems for HPV detection involve an amplification step followed by any of a number of methods for distinguishing different HPV types (28, 29). The most common HPV DNA amplification systems are based either on the MY09-M411 primer pair (26) or the GP5+-GP6+ primer pair (8); both of these target the L1 reading frame. These primer pairs have been used in combination with microtiter plate hybridization (16) or with reverse dot blots (13a) for identification of viral subtype. A nested PCR has sometimes been used to increase the sensitivity of the assay when samples with limited DNA content, such as Formalin-fixed biopsy specimens and archival Papanicolaou (Pap)-stained smears (35), are analyzed.
To investigate aspects of the natural history of cervical cancer, such as the variation in the amount of HPV in low-grade lesions versus the amount in high-grade lesions, a highly sensitive detection system that allows the quantitation of viral copy numbers is required; none of the presently available methods appear to be suitable for such analyses. We have therefore converted our previously developed PCR assay for the E1 reading frame (35) to a detection system in which the 5′ exonuclease assay is used (14). The 5′ exonuclease assay is based on the ability of the 5′ to 3′ exonuclease activity of Taq polymerase to cleave a dually labeled, nonextendible hybridization probe during the extension phase of the PCR (14, 19, 21). The probe has one fluorescent dye attached as a reporter at the 5′ end, and in the undigested form, the emission from this reporter dye is quenched by a second fluorescent dye, which is attached to the 3′ end. Concomitant with the accumulation of PCR product, there is a release of the reporter dye. In this report we describe the characteristics of our HPV assay based on the 5′ exonuclease method. Recently, a fluorescent 5′ exonuclease assay for the detection of HPV with a series of type-specific PCRs was described (32). Our system differs from that of Swan et al. (32) in several ways. In particular, (i) we perform a single PCR and for detection use multiple HPV type-specific probes labeled with different fluorescent dyes instead of multiple PCRs and a single probe for each HPV type, and (ii) most importantly, we use real-time detection of the accumulation of fluorescence rather than a measure of the fluorescence at a single cycle number (endpoint reading) to quantitate the initial viral copy numbers.
MATERIALS AND METHODS
DNA extraction.
High-molecular-weight DNA was isolated from blood samples by a standard protocol that included proteinase K treatment, followed by phenol-chloroform extraction and ethanol precipitation (25). DNA extraction from archival formalin-fixed biopsy specimens was performed by using a deparaffination step with xylene, successive ethanol washes followed by proteinase K digestion, phenol-chloroform extraction, and finally, ethanol precipitation (17, 20). DNA from Pap-stained smears was purified by a modification (17) of the protocol described by Chua and Hjerpe (7). This protocol included xylene incubation, destaining, proteinase K treatment (60°C for a minimum of 1 h), and subsequently, a transfer of cells to sterile Eppendorf tubes. Saturated ammonium acetate was then added to precipitate the protein. The DNA supernatant was recovered with ethanol; and the pellet was washed with 70% ethanol, dried, and dissolved in 200 μl of TE-low (10 mM Tris-HCl [pH 7.4], 0.1 mM EDTA).
Plasmids containing HPV types 16, 18, 31, 33, and 35 were used as positive controls and to estimate the sensitivity of the assay (1, 9, 10, 24). The plasmids with integrated HPV were transformed into One Shot cells (INV α F′; Invitrogen TA Cloning kit), positive transformants were isolated and grown in 100 ml of TB (Terrific Broth) at 37°C overnight, and plasmid DNA was extracted with the Qiagen Maxiprep kit (Qiagen). A cell line containing integrated HPV-16 (SiHa) was grown on a solid phase and was harvested after trypsination in 3 ml of STV (137 mM sodium chloride, 5.4 mM potassium chloride, 5.6 mM glucose, 6.9 mM sodium carbonate, 0.5 mM EDTA, 5 mg of phenol red per ml) plus 1.5% trypsin for 2 min with incubation at 37°C. The cells were then washed, first in growing medium (Dulbecco’s modification of Eagle’s medium, 160 μM k-benzylpenicillin, 34.3 mM streptomycin-sulfate, 1.99 mM glutamine, 10% newborn calf serum) and then in 1× phosphate-buffered saline. The cell pellet was dissolved in 1 ml of phosphate-buffered saline, and the concentration of the cells was estimated.
To study the effect of the Pap staining procedure on assay sensitivity, microscope glass slides with SiHa cells were prepared, the cells were stained, and the DNA was extracted by a modification (17) of the protocol described by Chua and Hjerpe (7) as described above.
PCR.
The PCR amplification was performed in a 50-μl volume containing 1× Taqman buffer (50 mM KCl, 10 mM Tris-HCl [pH 8.4], 10 mM EDTA, 60 nM passive reference dye 6-carboxytetramethyl rhodamine [Rox]), 5 mM MgCl2, 0.5 mM HPV E1 5′ primer (see Table 1 and below), 0.5 mM HPV E1 3′ primer (see Table 1 and below), each HPV-specific dual-labeled probe at a concentration of 100 nM, dATP, dCTP, and dGTP each at a concentration of 200 μM, 400 μM dUTP, 0.5 U of uracil N′-glycosylase (AmpErase UNG; Perkin-Elmer), 1.25 U of DNA polymerase (Amplitaq Gold; Perkin-Elmer), 124 ng of bovine serum albumin (BSA) per μl, and 2 to 10 μl of DNA. The amount of DNA added to a PCR mixture represents 1 to 5% of the DNA obtained from a cervical smear. The uracil N′-glycosylase was included in order to cleave potential PCR products from previous PCRs.
TABLE 1.
Sequences of oligonucleotides used as primers in HPV assay
| Type and name | Direction | Specificity | Sequence |
|---|---|---|---|
| Degenerate oligonucleotide set | |||
| HPVE1 350La | 5′ | General | 5′-TRYRKGYYYTAAAACGAAAGT-3′ |
| HPVE1 547Rb | 3′ | General | 5′-TTCCACTTCAGWAYWGCCATA-3′ |
| Specific oligonucleotide set | |||
| HPVE116Lc | 5′ | HPV-16 | 5′-TACAGGTTCTAAAACGAAAGT-3′ |
| HPVE118Lc | 5′ | HPV-18 | 5′-TGCATGTTTTAAAACGAAAGT-3′ |
| HPVE116Rd | 3′ | HPV-16 | 5′-TTCCACTTCAGTATTGCCATA-3′ |
| HPVE118Rd | 3′ | HPV-18 | 5′-TTCCACTTCAGAACAGCCATA-3′ |
| HPVE1REd | 3′ | HPV types 30 to 50 | 5′-TRYRKGMNYTAAAACGAAAGT-3′ |
Used as the 5′ primer at a concentration of 250 nM in the PCR.
Used as the 3′ primer at a concentration of 500 nM in the PCR.
Used in a mixture in equimolar amounts as the 5′ primer.
Used in a mixture in equimolar amounts as the 3′ primer.
Individual HPV types differ substantially at the nucleotide level, complicating the development of an HPV typing system capable of detecting a range of genital types of HPV (2). We previously identified a region in the first part of the E1 gene that is highly conserved among genital HPV types and that is suitable as a PCR priming site. The product generated by using this site and an adjacent priming site is only 180 bp (35). In our previously described system (35) we used a pair of degenerate primers (Table 1). In the present system we have redesigned the primers to reduce the complexity of the primers added to the PCR mixture. In the new system the 5′ primer mix consisted of only two primers (HPVE116L and HPVE118L) and the 3′ primer mix consisted of three primers (HPVE116R, HPVE118R, and HPVE1RE) (Table 1). Each of the primer mixtures contained an equimolar mixture of the oligonucleotides included in the mixture.
Probes.
The sequences for the probes used in the fluorescence assay were modified from those used in our previous HPV detection system (35). The probes were 30 to 33 bp in length to ensure a higher melting temperature for the probes than for the primers. A fluorescent reporter dye was covalently linked to the 5′ end, and a quencher dye was linked to the 3′ end. As reporter dyes we used 6-carboxyfluorescein (FAM), tetrachloro-6-carboxyfluorescein (TET), or hexachloro-6-carboxyfluorescein (HEX). As a quencher we used 6-carboxytetramethylrhodamine (TAMRA; Applied Biosystems Division of Perkin-Elmer, Inc., Foster City, Calif.) (Table 2). The probes were made so that a guanosine was avoided at the most 5′ end and were purified by high-pressure liquid chromatography prior to use. The probes were found to be sensitive to the number of cycles of freezing and thawing. To ensure stability, fresh probe was aliquoted in an amount sufficient for about 1 day of experiments and frozen.
TABLE 2.
Sequence, fluorescent label, and specificity of the oligonucleotide probes used in the fluorescent 5′ exonuclease HPV assay
| Specificity | Sequence and labels |
|---|---|
| HPV-16 | FAM-5′-ATAATCTCCTTTTTGCAGCTCTACTTTGTTTTT-3′TAMRA |
| HPV-18 | TET-5′-CCGCCTTTTTGCCTTTTTCTGCCCACTATT-3′TAMRA |
| HPV-31 | FAM-5′-TCTTCGTTTTGCTGTTTTACTGTTATTTTCTAT-3′TAMRA |
| HPV-33 | TET-5′-TTTTCGTTTTCTGTATGTGCATTCTTTATTTTT-3′TAMRA |
| HPV-35 | HEX-5′-TCGTCGCTTTCGTGCTGTATTTTTATTTTCA-3′TAMRA |
Amplification and detection by the 5′ exonuclease method.
Amplification and detection were performed with an ABI Prism 7700 Sequence Detection System (Perkin-Elmer, Inc.). The amplification ramp included two hold programs: (i) 2 min at 50°C to activate the uracil N′-glycosylase followed by (ii) 10 min at 95°C to inactivate the uracil N′-glycosylase and release the activity of the DNA polymerase. The two hold steps were followed by a two-step cycle consisting of 15 s at 95°C and 1 min at 50°C for a total of 50 cycles. Tubes that contained all PCR components but without template DNA (denoted no-template control [NTC] reactions) were used to ensure that the reagents were free of contamination.
Calculations.
The threshold cycle number (Ct) was calculated with Sequence Detection System software (Applied Biosystems Division, Perkin-Elmer, Inc.) and an automatic setting of the baseline (10 standard deviations [SDs] above the background in the first 3 to 15 cycles). Standard curves were generated by plotting the Ct values against the log of the copy numbers and the copy numbers for unknown samples inferred from the regression line.
RESULTS
Specificity of detection system.
The amplification efficiencies of the new primer system and that described previously (35) were similar. In the fluorescent 5′ exonuclease assay we have exclusively used the new primer set.
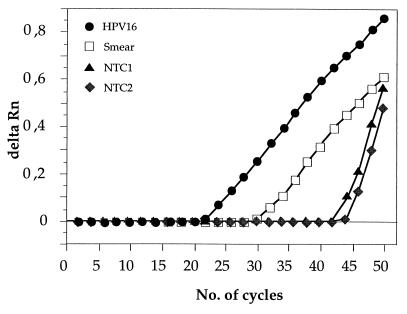
We first tested the ability of the probes for HPV types 16, 18, 31, 33, and 35 to discriminate between different HPV types. All probes showed an excellent ability to discriminate the correct from the incorrect HPV type, and no signal (i.e., Ct = 50) was seen with a mismatched combination of HPV DNA and probe (data not shown). When a large number of NTC reactions was performed, the probe for HPV-16 occasionally yielded a positive Ct value, despite the lack of added DNA template. This occurred in about 10% of the NTC reactions. When NTC reactions were run without the addition of uracil N′-glycosylase and the products of positive NTC reactions were examined electrophoretically, no product of the size expected for an HPV amplicon was found. Thus, the positive signal is likely to have been generated from an artifact product, such as a primer-dimer, rather than because of low levels of contamination with genomic DNA. Comparisons between primer and probe sequences did not reveal any regions with substantial homology, which could be responsible for the generation of such fluorescence. As can be seen from the amplification plot, HPV-16 artifact signals from NTC reactions are easily distinguished from the signal from HPV templates by the very steep amplification slope (Fig. 1). Also, the Ct values for specimens with positive NTC reactions were generally very high (Ct > 40) compared to those for smear or biopsy specimens. None of the other HPV probes tested gave any signal in the NTC reactions.
FIG. 1.
Comparison of the amplification plots for HPV-16 plasmid DNA (106 starting copies), a cervical smear DNA, and two NTCs yielding positive signals. All reactions were performed with the HPV-16 probe only. Delta Rn, reporter fluorescence of sample minus fluorescence of background (cycles 3 to 15).
Quantitation.
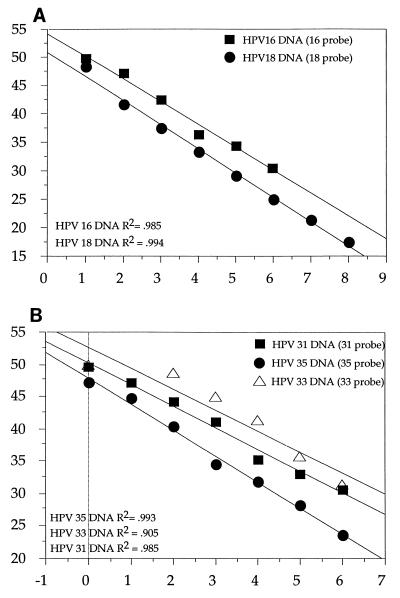
The sensitivity of the assay was studied with plasmid DNA with cloned HPV types 16, 18, 31, 33, and 35. Plasmid copy numbers were calculated from optical density (OD) measurements, and dilution series were made with 106 to 100 HPV copies (except for HPV-18, for which the dilution series was from 108 to 101 copies). The plasmids with cloned HPV were then mixed with a solution of 100 pg of high molecular weight human genomic DNA (lacking HPV inserts) per μl in order to mimic the complex nucleic acid environment present in an amplification from genomic DNA (biopsy specimens and cervical smears). To generate the standard curve for each HPV type, a minimum of five replications of each HPV copy number were performed. A highly significant linear relationship was found between the log of the input HPV copy number and threshold cycle (Ct) for all HPV types tested (Fig. 2). The slopes for the different HPV types were similar, but the intercepts differed, which is indicative of a difference in sensitivity. This could reflect either the efficiency of amplification of different HPV types or differences in efficiency of hybridization of the individual probes. Nevertheless, with the type-specific standard curves, quantitation of each HPV type was possible. The SD of the Ct values was found to be between 0.3 and 1.7 cycles, with the higher values seen for the lower copy numbers (Table 3). These SD values correspond to a variation in the estimated copy number of about 10 to 40%.
FIG. 2.
Standard curves generated for HPV types 16, 18, 31, 33, and 35. Dilution series were made with 106 to 100 HPV copies (except for HPV-18, for which the dilution series was from 108 to 101 copies). Linear regression was based on the titration series of HPV plasmid DNA in a background of 100 pg of genomic DNA per μl. Each curve is based on an average of 5 to 10 replicates. (A) Regression lines for HPV-16 DNA and HPV-18 DNA. Each reaction mixture contained only a single probe for either HPV-16 or HPV-18. (B) Regression lines for HPV-31 DNA, HPV-33 DNA, and HPV-35 DNA. Each reaction mixture contained only a single probe (for HPV-31, HPV-33, or HPV-35).
TABLE 3.
Mean and SD for Ct values for HPV titration series
| HPV type |
Ct value for the following no. of HPV copies
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 108 | 107 | 106 | 105 | 104 | 103 | 102 | 101 | 100 | |
| 16 | NDa | ND | 31 ± 1.0 | 34 ± 0.5 | 36 ± 0.3 | 42 ± 0.4 | 47 ± 1.6 | 50 ± 0.4 | 50 ± 0 |
| 18 | 17 ± 0.4 | 21 ± 0.3 | 25 ± 0.3 | 29 ± 0.4 | 33 ± 0.3 | 38 ± 0.8 | 42 ± 0.8 | 48 ± 2.7 | ND |
| 31 | ND | ND | 31 ± 3.6 | 33 ± 0.5 | 35 ± 0.5 | 41 ± 1.1 | 44 ± 0.9 | 47 ± 1.9 | 50 ± 0.8 |
| 33 | ND | ND | 31 ± 0.3 | 35 ± 0.7 | 41 ± 0.8 | 45 ± 1.2 | 48 ± 1.1 | 50 ± 0 | 50 ± 0.8 |
| 35 | ND | ND | 24 ± 1.5 | 28 ± 0.2 | 32 ± 0.4 | 34 ± 0.7 | 40 ± 0.8 | 45 ± 1.9 | 47 ± 2.5 |
ND, not determined.
Detection of infections with multiple HPV types.
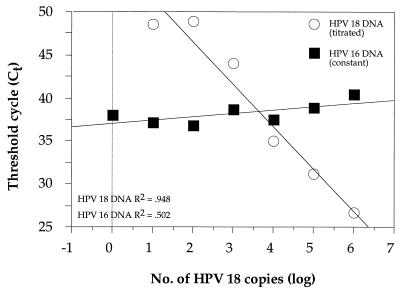
In a previous study (35) about 5% of the biopsy specimens from patients with cervical cancer were found to be infected with several HPV types. The ability of the fluorescent 5′ exonuclease assay to detect multiple infections was studied by mixing HPV plasmids in different combinations in a background of 100 pg of high-molecular-weight genomic DNA per μl. For instance, HPV-18 DNA was titrated from 106 to 100 copies in a constant background of 103 copies of HPV-16 DNA and was detected with a mixture of probes for HPV-16 and HPV-18 (Fig. 3). The results do not indicate a significant change in sensitivity for the detection of HPV-18 in the background of HPV-16 compared to the sensitivity from the standard curve (Fig. 2). HPV-18 could be identified down to a fraction of about 1% of the total number of HPV copies (Fig. 3). Similar results were obtained with several other combinations of HPV types (data not shown).
FIG. 3.
Analysis of a synthetic mixture of HPV types. Titration of HPV-18 plasmid DNA (106 to 100 copies) in a background of 103 copies of HPV-16 plasmid DNA. The reaction mixtures contained probes for both HPV-16 and HPV-18.
Effect of DNA quality on sensitivity.
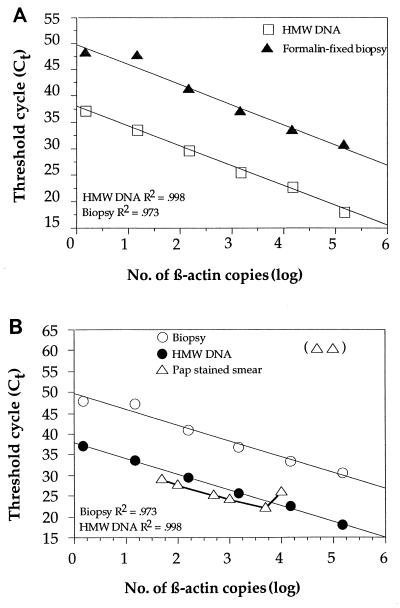
To study the effect of DNA quality on the performance of the assay, we compared standard curves for the amplification of β-actin using a fluorescent probe (Applied Biosystems Division, Perkin-Elmer, Inc.). The following DNA sources were used: (i) high-molecular-weight human genomic DNA purified from blood, (ii) DNA from an archival formalin-fixed, paraffin-embedded biopsy sample, and (iii) DNA from fresh cells prepared on microscope slides to mimic cervical smear samples and by fixation and staining methods identical to those used for cervical smear samples. DNA from the formalin-fixed, paraffin-embedded sample showed a very low amplification efficiency (high Ct value) compared to those for high-molecular-weight DNA samples with the same amount of DNA (on the basis of the OD measurement) (Fig. 4A). The Ct values for DNA from the formalin-fixed, paraffin-embedded sample were equivalent to a starting copy number of about 1/100 to 1/1,000 of that estimated from the OD measurement for the sample (Fig. 4A). Thus, the procedure for formalin fixation has a very strong effect on the amplification efficiency. To test the amplification efficiency with DNA from stained cervical smears, we used cells in cell culture and prepared series of smears containing different numbers of cells that were treated by the Pap staining procedure. Three to five replicates of each experiment were performed. No difference was found between the artificially made smears treated with the Pap stain and the high-molecular-weight genomic DNA samples, up to about 5 × 103 cells (Fig. 4B). With higher cell numbers, the Pap staining procedure had an adverse effect on the efficiency of the assay, and complete inhibition was seen for 5 × 104 cells (Fig. 4B). Since such inhibition is presumably caused by components of the Pap stain remaining in the DNA preparation and could not be removed by phenol-chloroform extraction, we investigated a number of additives that could remove the inhibitory effect. The addition of BSA at a concentration of 124 ng/μl to the PCR mixture entirely removed the inhibitory effect (data not shown). At much higher concentrations the BSA showed an inhibitory effect of its own. For cervical smears with a high Ct value (Ct > 40), the effect of the addition of BSA was pronounced (data not shown). Therefore, for PCR assays performed with cervical smears, as well as with several other types of clinical samples, we routinely add BSA to a final concentration of 124 ng/μl. This concentration of BSA does not reduce the efficiency of amplification for samples with no inhibitors, such as samples with high-molecular-weight genomic DNA.
FIG. 4.
Effect of DNA quality on signal release in the β-actin assay. (A) Comparison of the linear regression lines for a titration of high-molecular-weight (HMW) DNA (1.5 × 105 to 1.5 × 100 copies) extracted from blood and titration series of DNA from archival formalin-fixed biopsy specimens (1.5 × 105 to 1.5 × 100 copies). The curves are based on three to five replicates. Only the β-actin probe was used for detection in each reaction mixture. (B) Comparison of titration series of DNA from formalin-fixed biopsy specimen (1.5 × 105 to 1.5 × 100 copies), high-molecular-weight (HMW) DNA isolated from blood (1.5 × 105 to 1.5 × 100 copies), and a titration series of Pap-stained DNA (1 × 105 to 5 × 101 copies) from a cell line applied on a microscope slide. The curves are based on three to five replicates. Only the β-actin probe was used for detection. The two open triangles at log copies of 4.5 and 5 have not been connected with a line, since no signal was obtained at a Ct of 60 (maximum number of cycles).
Mixed probe assay.
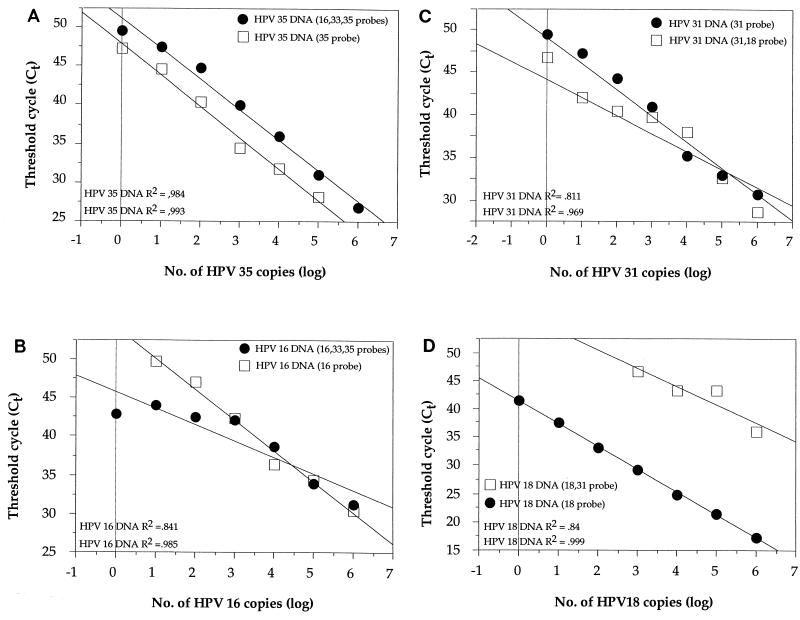
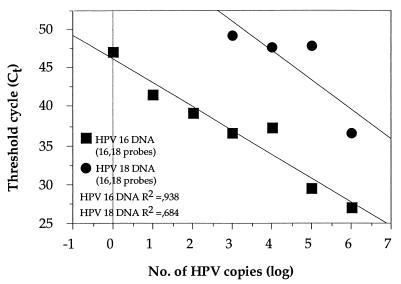
Since, in principle, up to three dyes can be detected in the same PCR, we investigated the effect of using multiple probes in each PCR on the sensitivity of detection of individual HPV types. Plasmids with HPV DNA were added in a titration series from 106 to 100 copies in a background of genomic DNA and with different combinations of probes. First, we compared the detection of HPV-35 DNA (106 to 100 copies) with a mixture of probes for HPV types 16, 33, and 35 compared to the detection of HPV-35 DNA (106 to 100 copies) with only the HPV-35 probe (Fig. 5A). A somewhat reduced sensitivity for the detection of HPV-35 was seen by the mixed probe assay. However, the assay was linear over the entire spectrum of copy numbers studied. Similarly, the detection of HPV-16 DNA (106 to 100 copies) was studied with a mixture of probes for HPV types 16, 33, and 35 and was compared to the detection of HPV-16 DNA with only the HPV-16 probe (Fig. 5B). The sensitivity of the three-probe assay was similar to that of the single-probe assay, in particular at low copy numbers, but the relationship between copy number and fluorescence was still linear. Similar results were obtained for the detection of HPV-31 DNA (106 to 100 copies) with a mixture of probes for HPV-18 and HPV-31 compared to the results obtained for the detection of HPV-31 DNA (106 to 100 copies) with only the HPV-31 probe (Fig. 5C). However, a substantial difference between the assay with a mixture of probes and the single-probe assay was found for the detection of HPV-18. The sensitivity for the detection of HPV-18 (106 to 100 copies) was dramatically reduced when a mixture of probes for HPV-18 and HPV-31 was used rather than when only the HPV-18 probe was used (Fig. 5D). Finally, we investigated the effect of using a mixture of HPV-16 and HPV-18 probes in the same reaction mixture. Therefore, we mixed both HPV-16 DNA and HPV-18 DNA in a parallel titration series (106 to 100 copies) with equal amounts of DNA of the two viral types and a mixture of probes for HPV-16 and HPV-18 (Fig. 6). For HPV-16 the slope was similar to that of the standard curve, while the sensitivity in detecting HPV-18 was reduced, and no HPV-18 signal was detected when HPV-18 was present at less than 103 copies.
FIG. 5.
Detection by mixed-probe assays. (A) Titration series of HPV-35 DNA (106 to 100 copies) and a mixture of probes for HPV types 16, 33, and 35 compared to titration series of HPV-35 DNA (106 to 100 copies) and the HPV-35 probe. (B) Titration series for HPV-16 DNA (106 to 100 copies) detection and a mixture of probes for HPV type 16, 33, and 35 compared to titration series for HPV-16 DNA (106 to 100 copies) detection and the HPV-16 probe. (C) Titration series of HPV-31 DNA (106 to 100 copies) and a mixture of probes for HPV-18 and HPV-31 compared to titration series of HPV-31 DNA (106 to 100 copies) and the HPV-31 probe. (D) Titration series of HPV-18 DNA (106 to 100 copies) and a mixture of probes for HPV-18 and HPV-31 compared to titration series of HPV-18 DNA (106 to 100 copies) and the HPV-18 probe.
FIG. 6.
Detection of HPV-16 DNA and HPV-18 DNA in a titration series of 106 to 100 copies with equal amounts of DNA of the two types and a mixture of HPV-16 and HPV-18 probes.
Application to archival smears.
A series of 21 archival smears previously typed by the E1 dot blot system (35) were analyzed by the fluorescent 5′ exonuclease assay. In all these smears the typing result (with respect to HPV subtype) was identical to that obtained previously (data not shown).
DISCUSSION
We have evaluated the use of the real-time fluorescent 5′ exonuclease assay for HPV detection and quantitation. This assay has a number of potential advantages over existing PCR-based methods since it (i) requires no further analysis after the amplification step, because data collection occurs during amplification, (ii) prevents the contamination of PCR products due to the closed-tube system used and the inclusion of the carryover prevention enzyme uracil N′-glycosylase in each reaction mixture, (iii) allows the use of multiple detection probes in the same reaction mixture, and (iv) permits accurate quantitation over a very wide range of copy numbers. The fluorescent 5′ exonuclease assay was found to discriminate well between the HPV types investigated and can be used to quantify the HPV copy number over a wide range of HPV copy numbers. When a single type of HPV DNA and a single probe were used, all the HPV probes clearly discriminated among the HPV types (data not shown). Also, for all HPV types the standard curve obtained was linear over a spectrum of concentrations that greatly exceed the range normally found in actual clinical samples. Thus, under these circumstances the assay is quite suitable for use in the typing of HPV.
In certain synthetic mixtures of HPV types, made to mimic mixed infections, we observed a reduced sensitivity for the detection of individual HPV types, in particular, for HPV-18. In this case the reduced sensitivity is likely to be due to a competition between the two PCR products generated in the reaction or interference between probes. In synthetic mixtures the individual HPV types could be detected down to a fraction of about 1% of the total copy number, and in certain combinations it was even possible to detect the individual HPV types down to a fraction of 0.1% (data not shown). Thus, it should be possible to detect mixed infections, even when the different viral types occur in substantially different proportions. In analyzing a series of biopsy samples we have identified a number of mixed infections, attesting to the ability of the present assay to detect multiple HPV types in individual samples (data not shown). The present HPV assay can be performed either in a few reactions with multiple probes or in a large number of reactions each with a type-specific probe. It appears that the mixed probe assay is suitable solely for detection and has sufficient sensitivity. However, for accurate quantitation, the number of probes per reaction mixture may be reduced. In our system this appears to be required only for the quantitation of HPV-18, for which the strongest reduction in sensitivity was shown when a mixture of probes was used.
Our results point to the extensive variation in amplification efficiency between materials that have been subjected to different staining and fixation procedures. In particular, the DNA in formalin-fixed, paraffin-embedded tissue samples was shown to be amplified at an efficiency equivalent to about 1/100 to 1/1,000 of that of high-molecular-weight DNA. Thus, in analyzing formalin-fixed, paraffin-embedded samples, it is necessary to quantitate the extent of inhibition by using, for instance, the β-actin control assay before the pathogen titers can be accurately estimated. A similar problem may apply to cytological Pap-stained smears. For samples with low DNA concentrations, our results with samples mimicking Pap-stained smears showed that they had amplification efficiencies similar to that for high-molecular-weight DNA. However, with high DNA concentrations, Pap-stained smears completely inhibited the PCR. This indicates that reliable analysis of the HPV in archival smears will be possible only if the inhibitory effect can be removed. We found that the presence of a simple additive (BSA) at low concentrations can remove the inhibitory effect. Since this additive does not appear to have any effect on the efficiency of the reaction, it can be added to all reaction mixtures, including those used to generate the standard curve. This approach makes it possible to generate an appropriate standard curve that can be applied to cervical smear samples. However, BSA has no effect on the efficiency of amplification of DNA from formalin-fixed samples. For quantitation of viral copy numbers from tissue samples in which an additive, such as BSA, does not remove the inhibitory effect, the standard curves used must be derived from DNA samples handled in a manner identical to that in which the biological samples are handled.
The 5′ exonuclease assay for β-actin was used in parallel with the HPV assay to obtain an estimate of the amount of genomic DNA present in individual smear samples. By using the β-actin copy number, which was estimated from the standard curve, the smear samples can be normalized for the amount of genomic DNA. On the basis of such a normalization the variation in HPV copies per genomic DNA equivalent can be estimated (i.e., by dividing the HPV copy number by the β-actin copy number). Our preliminary results from the analysis of a large set of cervical smears indicate that the Ct values for β-actin can vary with more than six cycles between individual samples, indicating substantial differences in the amount of genomic DNA accessible to PCR (data not shown). An important aspect of any screening technique is the ability to identify samples with false-negative results resulting from either an insufficient amount of starting DNA or the presence of inhibitors. The β-actin assay was therefore used to determine whether the lack of an HPV signal (i.e., Ct = 50) could be due to the presence of insufficient amounts of genomic DNA or the presence of inhibitors rather than a lack of HPV infection. On the basis of an analysis of approximately 5,000 archival smears, about 90% of which gave a positive β-actin signal, it appears that the likelihood of finding an HPV-positive signal in a β-actin-negative sample is relatively low (data not shown).
The assay for HPV described here can be extended in a number of ways. Additional HPV types can be identified by increasing the number of probes. Presently, only up to three probes can be used simultaneously in the same reaction mixture. Also, our results indicate that for accurate quantitation, the number of probes per assay should be kept down. In the present application of the 5′ exonuclease assay, only a single measurement from the amplification curve, the Ct value, is being used. However, the remaining parts of the curve also contain valuable information, e.g., information that can be used to calculate the overall efficiency of the amplification. A more extensive analysis of the entire amplification curve would yield valuable information, e.g., for evaluation of the degree of inhibition of the individual PCRs and its effect on the Ct value.
In summary, the fluorescent 5′ exonuclease assay described here has a number of advantages over the existing methodologies for HPV detection, in particular, with respect to the very powerful quantitation ability. This method may be applied to studies of a number of issues related to the natural history of cervical cancer, such as the amounts of HPV in high- and low-grade lesions.
ACKNOWLEDGMENTS
This study was supported by the Swedish Cancer Foundation, the Beijer Foundation, and the National Institutes of Health (grant 1 RO: CA61197-01A3).
We are grateful to Catharina Hemström-Nilsson for the SiHa cell line, Margit Gustavsson for helping with the Pap staining, Marita Möllenberg for organizational help, and Patrik Magnusson for comments on this paper.
REFERENCES
- 1.Beaudenon S, Kremsdorf D, Croissant O, Jablonska S, Wain-Hobson S, Orth G. A novel type of human papillomavirus associated with genital neoplasias. Nature. 1986;321:246–249. doi: 10.1038/321246a0. [DOI] [PubMed] [Google Scholar]
- 2.Bonnez W. Taxonomy of the human papillomaviruses. Clin Infect Dis. 1993;17:1080. doi: 10.1093/clinids/17.6.1080. [DOI] [PubMed] [Google Scholar]
- 3.Bosch F X, Manos M M, Munoz N, Sherman M, Jansen A M, Peto J, Schiffman M H, Moreno V, Kurman R, Shah K V. Prevalence of human papillomavirus in cervical cancer: a worldwide perspective. International Biological Study on Cervical Cancer (IBSCC) Study Group. J Natl Cancer Inst. 1995;87:796–802. doi: 10.1093/jnci/87.11.796. [DOI] [PubMed] [Google Scholar]
- 4.Brown D R, Bryan J T, Cramer H, Fife K H. Analysis of human papillomavirus types in exophytic condylomata acuminata by hybrid capture and Southern blot techniques. J Clin Microbiol. 1993;31:2667–2673. doi: 10.1128/jcm.31.10.2667-2673.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carter J J, Koutsky L A, Wipf G C, Christensen N D, Lee S-K, Kuypers J, Kiviat N, Galloway D A. The natural history of human papillomavirus type 16 capsid antibodies among a cohort of university students. J Infect Dis. 1996;174:927–936. doi: 10.1093/infdis/174.5.927. [DOI] [PubMed] [Google Scholar]
- 6.Chardonnet Y, Bejui-Thivolet F, Guerin-Reverchon I. Human papillomavirus detection in cervical cells by in situ hybridization with biotinylated probes. Cytopathology. 1992;3:341–350. doi: 10.1111/j.1365-2303.1992.tb00059.x. [DOI] [PubMed] [Google Scholar]
- 7.Chua K L, Hjerpe A. Polymerase chain reaction analysis of human papillomavirus in archival cervical cytologic smears. Anal Quant Cytol Histol. 1995;17:221–229. [PubMed] [Google Scholar]
- 8.De Roda Husman A M, Walboomers J M, van den Brule A J, Meijer C J, Snijders P J. The use of general primers GP5 and GP6 elongated at their 3′ ends with adjacent highly conserved sequences improves human papillomavirus detection by PCR. J Gen Virol. 1995;76:1057–1062. doi: 10.1099/0022-1317-76-4-1057. [DOI] [PubMed] [Google Scholar]
- 9.de Villiers E M. Heterogeneity of the human papillomavirus group. J Virol. 1989;63:4898–4903. doi: 10.1128/jvi.63.11.4898-4903.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Villiers E M. Human pathogenic papillomavirus types: an update. Curr Top Microbiol Immunol. 1994;186:1–12. doi: 10.1007/978-3-642-78487-3_1. [DOI] [PubMed] [Google Scholar]
- 11.de Villiers E M. Hybridization methods other than PCR: an update. IARC Sci Publ. 1992;119:111–19. [PubMed] [Google Scholar]
- 12.Galloway D A. Papillomavirus capsids: a new approach to identify serological markers of H PV infection. J Natl Cancer Inst. 1994;86:474–475. doi: 10.1093/jnci/86.7.474. [DOI] [PubMed] [Google Scholar]
- 13.Gravitt P E, Manos M M. Polymerase chain reaction-based methods for the detection of human papillomavirus DNA. IARC Sci Publ. 1992;119:121–133. [PubMed] [Google Scholar]
- 13a.Gravitt P E, Peyton C L, Apple R J, Wheeler C M. Genotyping of 27 human papillomavirus types by using L1 consensus PCR products by a single-hybridization, reverse line blot detection method. J Clin Microbiol. 1998;36:3020–3027. doi: 10.1128/jcm.36.10.3020-3027.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holland P M, Abramson R D, Watson R, Gelfand D H. Detection of specific polymerase chain reaction product by utilizing the 5′–3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci USA. 1991;88:7276–7280. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.International Agency for Research on Cancer. IARC monographs on the evolution of carcinogenic risks to humans. Vol. 64. Lyon, France: International Agency for Research on Cancer; 1995. Human papillomaviruses. [Google Scholar]
- 16.Jacobs M V, van den Brule A J, Snijders P J, Helmerhorst T J, Meijer C J, Walboomers J M. A non-radioactive PCR enzyme-immunoassay enables a rapid identification of HPV 16 and 18 in cervical scrapes after GP5+/6+ PCR. J Med Virol. 1996;49:223–229. doi: 10.1002/(SICI)1096-9071(199607)49:3<223::AID-JMV11>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 17.Josefsson A, Magnusson P K E, Gyllensten U. Human papillomavirus. In: Peeling R S, Sparling P F, editors. Methods in molecular medicine: molecular protocols for sexually transmitted diseases. Vol. 11. 1998. pp. 171–193. [Google Scholar]
- 18.Kirnbauer R, Hubbert N L, Wheeler C M, Becker T M, Lowy D R, Schiller J T. A virus-like particle enzyme-linked immunosorbent assay detects serum antibodies in a majority of women infected with human papillomavirus type 16. J Natl Cancer Inst. 1994;86:494–499. doi: 10.1093/jnci/86.7.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee L G, Connell C R, Bloch W. Allelic discrimination by nick-translation PCR with fluorogenic probes. Nucleic Acids Res. 1993;21:3761–3766. doi: 10.1093/nar/21.16.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindqvist A K, Magnusson P K, Balciuniene J, Wadelius C, Lindholm E, Alarcon-Riquelme M E, Gyllensten U B. Chromosome-specific panels of tri- and tetranucleotide microsatellite markers for multiplex fluorescent detection and automated genotyping: evaluation of their utility in pathology and forensics. Genome Res. 1996;6:1170–1176. doi: 10.1101/gr.6.12.1170. [DOI] [PubMed] [Google Scholar]
- 21.Livak K J, Marmaro J, Todd J A. Towards fully automated genome-wide polymorphism screening. Nat Genet. 1995;9:341–342. doi: 10.1038/ng0495-341. [DOI] [PubMed] [Google Scholar]
- 22.Lorincz A. Hybrid capture. Clin Chem. 1998;44(6 Pt 1):1363. . (Letter and comment.) [PubMed] [Google Scholar]
- 23.Lorincz A T. Molecular methods for the detection of human papillomavirus infection. Obstet Gynecol Clin N Am. 1996;23:707–730. [PubMed] [Google Scholar]
- 24.Lorincz A T, Quinn A P, Lancaster W D, Temple G F. A new type of papillomavirus associated with cancer of the uterine cervix. Virology. 1987;159:187–190. doi: 10.1016/0042-6822(87)90366-7. [DOI] [PubMed] [Google Scholar]
- 25.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. 2nd ed, Appendix E3. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 26.Manos M M, Ting Y, Wright D K, Lewis A J, Broker T R, Wolinsky S M. The use of polymerase chain reaction amplification for the detection of genital human papillomavirus. Cancer Cells. 1989;7:209–214. [Google Scholar]
- 27.Meyer T, Arndt R, Stockfleth E, Flammann H T, Wolf H, Reischl U. Strategy for typing human papillomaviruses by RFLP analysis of PCR products and subsequent hybridization with a generic probe. BioTechniques. 1995;19:632–639. [PubMed] [Google Scholar]
- 28.Pizzighella S, Pisoni G, Bevilacqua F, Vaona A, Palu G. Simultaneous polymerase chain reaction detection and restriction typing for the diagnosis of human genital papillomavirus infection. J Virol Methods. 1995;55:245–256. doi: 10.1016/0166-0934(95)00063-z. [DOI] [PubMed] [Google Scholar]
- 29.Pizzighella S, Rassu M, Piacentini I, Maschera B, Palu G. Polymerase chain reaction amplification and restriction enzyme typing as an accurate and simple way to detect and identify human papillomaviruses. J Med Microbiol. 1993;39:33–38. doi: 10.1099/00222615-39-1-33. [DOI] [PubMed] [Google Scholar]
- 30.Roden R B S, Greenstone H L, Kirnbauer R, Booy F P, Jessie J, Lowy D R, Schiller J T. In vitro generation and type-specific neutralization of a human papillomavirus type 16 virion pseudotype. J Virol. 1996;70:5875–5883. doi: 10.1128/jvi.70.9.5875-5883.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 32.Swan D C, Tucker R A, Holloway B P, Icenogle J P. A sensitive, type-specific, fluorogenic probe assay for detection of human papillomavirus DNA. J Clin Microbiol. 1997;35:886–891. doi: 10.1128/jcm.35.4.886-891.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Unger E R, Vernon S D, Thoms W W, Nisenbaum R, Spann C O, Horowitz I R, Icenogle J P, Reeves W C. Human papillomavirus and disease-free survival in FIGO stage Ib cervical cancer. J Infect Dis. 1995;172:1184–1190. doi: 10.1093/infdis/172.5.1184. [DOI] [PubMed] [Google Scholar]
- 34.Van Den Brule A J, Walboomers J M, Du Maine M, Kenemans P, Meijer C J. Difference in prevalence of human papillomavirus genotypes in cytomorphologically normal cervical smears is associated with a history of cervical intraepithelial neoplasia. Int J Cancer. 1991;48:404–408. doi: 10.1002/ijc.2910480317. [DOI] [PubMed] [Google Scholar]
- 35.Ylitalo N, Bergstrom T, Gyllensten U. Detection of genital human papillomavirus by single-tube nested PCR and type-specific oligonucleotide hybridization. J Clin Microbiol. 1995;33:1822–1828. doi: 10.1128/jcm.33.7.1822-1828.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]