Abstract
Introduction
The absence of a diagnostic test for acute rheumatic fever (ARF) is a major impediment in managing this serious childhood condition. ARF is an autoimmune condition triggered by infection with group A Streptococcus. It is the precursor to rheumatic heart disease (RHD), a leading cause of health inequity and premature mortality for Indigenous peoples of Australia, New Zealand and internationally.
Methods and analysis
‘Searching for a Technology-Driven Acute Rheumatic Fever Test’ (START) is a biomarker discovery study that aims to detect and test a biomarker signature that distinguishes ARF cases from non-ARF, and use systems biology and serology to better understand ARF pathogenesis. Eligible participants with ARF diagnosed by an expert clinical panel according to the 2015 Revised Jones Criteria, aged 5–30 years, will be recruited from three hospitals in Australia and New Zealand. Age, sex and ethnicity-matched individuals who are healthy or have non-ARF acute diagnoses or RHD, will be recruited as controls. In the discovery cohort, blood samples collected at baseline, and during convalescence in a subset, will be interrogated by comprehensive profiling to generate possible diagnostic biomarker signatures. A biomarker validation cohort will subsequently be used to test promising combinations of biomarkers. By defining the first biomarker signatures able to discriminate between ARF and other clinical conditions, the START study has the potential to transform the approach to ARF diagnosis and RHD prevention.
Ethics and dissemination
The study has approval from the Northern Territory Department of Health and Menzies School of Health Research ethics committee and the New Zealand Health and Disability Ethics Committee. It will be conducted according to ethical standards for research involving Indigenous Australians and New Zealand Māori and Pacific Peoples. Indigenous investigators and governance groups will provide oversight of study processes and advise on cultural matters.
Keywords: paediatric rheumatology, valvular heart disease, immunology, diagnostic microbiology
Strengths and limitations of this study.
Searching for a Technology-Driven Acute Rheumatic Fever Test addresses a critically important health problem, acute rheumatic fever (ARF), which disproportionately affects global Indigenous populations and for which there is no diagnostic test nor effective treatment able to limit progression to rheumatic heart disease.
The large sample size recruited across international sites and the use of unbiased, comprehensive immune profiling will maximise the likelihood of being able to define a biomarker signature which can discriminate between ARF and other clinical conditions.
The most robust ‘gold-standard’ rheumatic fever diagnostic process currently available will be used, comprising the revised 2015 Jones Criteria applied by an expert clinical panel.
Culturally safe processes with Indigenous governance guide the development and conduct of the study.
An inherent limitation is heterogeneity among people with ARF with regard to demographic factors, ARF type (first or recurrent), diagnostic category (possible, probable, definite), clinical phenotype (carditis, chorea, arthritis, skin and soft-tissue) and timing in relation to ARF onset (variable time taken for healthcare to be reached after disease onset, consent gained and blood samples collected).
Introduction
The absence of a diagnostic test for acute rheumatic fever (ARF) is a major impediment to management and epidemiological understanding of this serious childhood condition. ARF is one of the best examples of human autoimmunity caused by an infection, with group A Streptococcus (GAS) the identified trigger. It is the precursor to rheumatic heart disease (RHD), a leading cause of health inequity and premature mortality for Indigenous peoples of Australia, New Zealand (NZ) and internationally.1 Prompt diagnosis of ARF allows timely commencement of secondary prophylaxis with long-acting penicillin to prevent repeated GAS infections that drive ARF recurrences. ARF recurrences are in turn the main mechanism responsible for development of RHD. Underdiagnosis of ARF appears to be a major contributor to the high rates of RHD in Aboriginal communities in Australia’s Northern Territory (NT), where 76% of patients with RHD lack a prior ARF diagnosis.2 Similarly a cohort study in NZ found the majority of RHD cases (65%) had never been previously hospitalised with ARF.3 Similar findings are reported in African countries.4
The Jones criteria (table 1) have been the diagnostic tool for ARF for nearly 80 years.5 This is a regularly reviewed clinical algorithm, now available in a user-friendly smart device application (‘app’),6 most recently modified to optimise sensitivity for use in high-risk settings and optimise specificity for low-risk populations.7 Despite these revisions, ARF diagnosis remains challenging and subjective. Diagnostic biomarkers for ARF that improve the performance of these criteria or provide a definitive diagnostic test would be a major advance in improving ARF detection, and thereby, RHD prevention.8 Accurate case ascertainment is also needed to monitor progress towards disease control targets.
Table 1.
Revised Jones criteria
| High-risk groups* | Low-risk groups | |
| Definite initial episode of ARF | 2 major manifestations+evidence of preceding GAS infection. 1 major +2 minor manifestations+evidence of preceding GAS infection.† |
|
| Definite recurrent‡ episode of ARF in a patient with a documented history of ARF or RHD | 2 major manifestations+evidence of preceding GAS infection. 1 major +2 minor manifestations+evidence of preceding GAS infection. 3 minor manifestations+evidence of a preceding GAS infection.† |
|
| Probable or possible ARF (first episode or recurrence‡) | A clinical presentation in which ARF is considered a likely diagnosis but falls short in meeting the criteria by either:
Such cases should be further categorised according to the level of confidence with which the diagnosis is made:
|
|
| Major manifestations | Carditis (including subclinical evidence of rheumatic valvulitis on echocardiogram) Polyarthritis§ or aseptic monarthritis or polyarthralgia Sydenham chorea¶ Erythema marginatum** Subcutaneous nodules |
Carditis (including subclinical evidence of rheumatic valvulitis on echocardiogram) Polyarthritis§ Sydenham chorea¶ Erythema marginatum** Subcutaneous nodules |
| Minor manifestations | Fever ††≥38°C Monoarthralgia ‡‡ ESR ≥30 mm/h or CRP ≥30 mg/L Prolonged P-R interval on ECG§§ |
Fever ≥38.5°C Polyarthralgia or aseptic monarthritis‡‡ ESR ≥60 mm/h or CRP ≥30 mg/L Prolonged P-R interval on ECG§§ |
Reproduced with permission from The 2020 Australian guideline for prevention, diagnosis and management of ARF and RHD (third edition).2
*High-risk groups are those living in communities with high rates of ARF (incidence >30/100 000 per year in 5–14 years old) or RHD (all-age prevalence >2/1000). Aboriginal and Torres Strait Islander peoples living in rural or remote settings are known to be at high risk. Data are not available for other populations but Aboriginal and Torres Strait Islander peoples living in urban settings, Māori and Pacific Islanders, and potentially immigrants from developing countries, may also be at high risk.
†Elevated or rising antistreptolysin O or other streptococcal antibody, or a positive throat culture or rapid antigen or nucleic acid test for GAS infection.
‡Recurrent definite, probable or possible ARF requires a time period of more than 90 days after the onset of symptoms from the previous episode of definite, probable or possible ARF.
§A definite history of arthritis is sufficient to satisfy this manifestation. Note that if polyarthritis is present as a major manifestation, polyarthralgia or aseptic monarthritis cannot be considered an additional minor manifestation in the same person.
¶Chorea does not require other manifestations or evidence of preceding GAS infection, provided other causes of chorea are excluded.
**Care should be taken not to label other rashes, particularly non-specific viral exanthems, as erythema marginatum.
††In high-risk groups, fever can be considered a minor manifestation based on a reliable history (in the absence of documented temperature) if anti-inflammatory medication has already been administered.
‡‡If polyarthritis is present as a major criterion, monarthritis or arthralgia cannot be considered an additional minor manifestation.
§§If carditis is present as a major manifestation, a prolonged P-R interval cannot be considered an additional minor manifestation.
ARF, acute rheumatic fever; CRP, C reactive protein; ESR, erythrocyte sedimentation rate; GAS, Group A Streptococcus; RHD, rheumatic heart disease.
A further barrier to ARF management is the absence of an identified effective immunomodulatory treatment to alter long-term cardiac outcomes. Non-steroidal anti-inflammatory drugs alleviate joint symptoms but do not alter the long-term disease trajectory. Corticosteroids appear ineffective in limiting development or severity of RHD,9 but in some settings are prescribed as a treatment of last resort for severe carditis.10 11 Hydroxychloroquine use has recently been reported in two cases of ARF with carditis, based on plausible in vitro data12 but efficacy is as yet unknown.13 Improved knowledge of ARF immune pathogenesis is needed to inform potential immune modulatory strategies targeted to the immune pathology. Compounding complexity, different pathological processes may occur, accounting for the heterogeneous clinical phenotypes of ARF (table 1, see major manifestations).
Most investigations into the immune basis of ARF have used biased (directed) approaches, with limited utility. For example, elevated tumour necrosis factor-α, interleukin 6 (IL-6) and IL-8 have been reported in patients with active ARF and RHD, but are unreliable disease markers.14–16 In difficult-to-diagnose diseases, the use of a multiplicity of biomarkers (‘multivariate’) to identify the presence or progression of disease has proven useful. Using microarrays, characteristic gene expression signatures have been reported in white blood cells from patients with Kawasaki disease (another inflammatory cardiac condition), lupus and tuberculosis demonstrating that multivariate expression profiling has potential to identify biomarker composite signatures as diagnostic tools.17–20 Multivariate approaches to analysing biomarkers in sera or plasma have also shown promise. In a bead-based multiplex assay, a number of Mycobacterium tuberculosis-reactive antibodies were successfully used to diagnose tuberculosis in non-human primates,21 and antibody glycosylation patterns can discriminate latent from active TB in humans.22 Most importantly, our recent studies show proof-of-principle for immunopathogenesis studies to understand ARF, identifying a dysregulated cytokine axis using multivariate approaches including RNAseq and flow cytometry12 as well as a linked IgG3-C4 response in early ARF with multiplex bead-based assays.23 Serumborne and plasmaborne biomarkers in addition to transcriptional profiling and circulating cellular components thus constitute a strong and accessible means of diagnosis once useful biomarkers have been identified.
Our hypothesis is that unbiased, multivariate analyses hold the greatest potential for identifying meaningful, translatable information on the immunopathogenesis of ARF. The aims of the ‘Searching for a Technology-Driven Acute Rheumatic Fever Test’ (START) study are to determine whether this approach can identify a biomarker signature that accurately classifies ARF diagnoses according to a defined ‘gold-standard’ diagnosis (the 2015 revised Jones criteria applied by a panel of clinical experts), and to better understand ARF immune pathogenesis (table 2). Specifically, we aim to develop, in a discovery cohort and validate in a second cohort, a profile of metabolic and immunological biomarkers that distinguishes ARF cases from a range of non-ARF conditions and healthy controls.
Table 2.
Control groups
| Group A (n=30): | Non-ARF streptococcal infections or toxin mediated condition or poststreptococcal condition Examples:
and
|
| Group B (n=30): | Other acute condition Examples:
and
|
| Group C (n=30): | RHD
|
| Group D (n=30): | Healthy
and
|
ARF, acute rheumatic fever; RHD, rheumatic heart disease.
Methods
Design
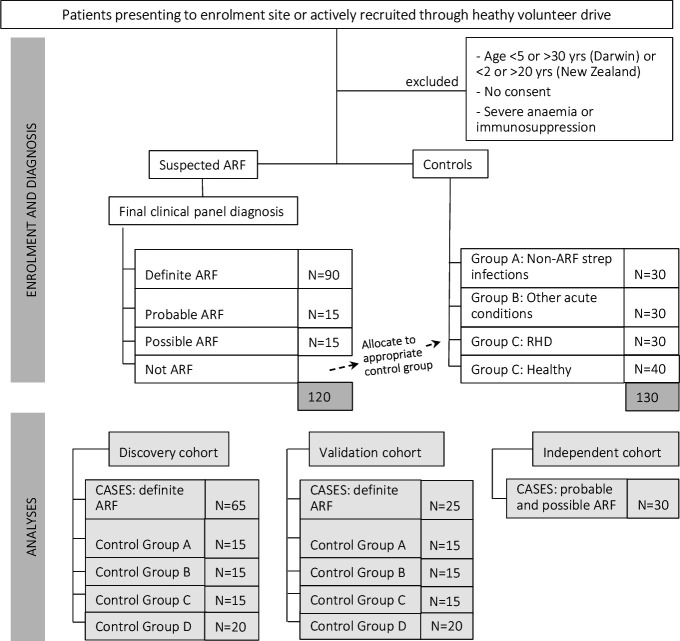
START is a cross-national, prospective, observational study with a quasi case–control design. We will prospectively recruit children and young adults with ARF, and people matched on age, sex and ethnicity who have eligible control diagnoses, in Australia and NZ. The study will comprise a discovery cohort (up to 65 definite cases and 65 controls) and a validation cohort (up to 25 definite cases and 65 controls). Probable and possible ARF diagnoses will then be tested independently (figure 1). Eligible control conditions are shown in table 2. Participants will have blood collected for comprehensive immune profiling.
Figure 1.
Study diagram showing target sample sizes. ARF, acute rheumatic fever; RHD, rheumatic heart disease.
Ethics and dissemination
The study has approval from the NT Department of Health and Menzies School of Health Research ethics committee (18/3126) and the NZ Health and Disability Ethics Committee (18/CEN/197). It will be conducted according to Good Clinical Practice, Good Laboratory Practice and the National Health and Medical Research Council and ethical standards for research involving Indigenous Australians and NZ Māori and Pacific Peoples.
Knowledge dissemination will be through academic channels and community discussion forums. The study team is well placed to foster translation of findings into practical tools, namely, an improved diagnostic and immunomodulatory therapy.
Study sites
Participating hospitals across the two international sites: Royal Darwin Hospital, Darwin, NT, Australia, and Starship and Middlemore Hospitals, Auckland, NZ. Clinical guidelines at all three study sites require that all people with suspected ARF are hospitalised for diagnostic workup and initiation of management.10 24 Royal Darwin Hospital is the major tertiary referral hospital in Australia’s NT. Thirty per cent of the NT population is Indigenous (Aboriginal or Torres Strait Islander).25 ARF occurs at rates >300/100 000 total Aboriginal population in highest-burden communities in the NT.26 Starship and Middlemore Hospitals serve a combined urban population of approximately 1.1 million people in the Auckland region. Māori and Pacific peoples make up approximately 27% of the population of greater Auckland region, where the incidence of ARF among Māori and Pacific children is approximately 33/100 000 and 93/100 000, respectively, for those 5–15 years old.24 27
Study participants
Inclusion criteria for ARF cases are: individuals with suspected or confirmed ARF (the vast majority of whom are expected to identify as being of Aboriginal, Torres Strait Islander, Māori or Pacific ethnicity) aged 5–30 years (Darwin) or 2–20 years (NZ). Written, informed consent will be obtained from guardians or the participant themself if aged ≥16 years in NZ or ≥18 years in Australia. Differences between sites are consistent with local epiodemiology and local ethical recommendations.
Exclusion criteria comprise individuals with severe anaemia in whom collection of the blood volume required for the study collection would be relatively contraindicated; profound immunosuppression other than corticosteroids for 2 months prior to study entry, or as appropriate depending on the half-life of the immunosuppressive agent and unstable social situation precluding discussion about consent for research.
Controls are recruited into one of four groups: (A) non-ARF Streptococcal infections; (B) other non-ARF acute inflammatory presentations; (C) established RHD without an intercurrent acute diagnosis and (D) healthy individuals (table 2). All must meet the above-stated age, ethnicity and consent inclusion criteria and none of the exclusion criteria. Controls will be matched at data analysis stage to cases by site on sex, age within 10 years, and ethnicity. Individuals in groups A, B and D will have an echocardiogram to exclude underlying RHD.
Cultural safety
Cultural safety28 is a core underpinning principle of the study. In NZ, the research is conducted in accordance with core Te Tiriti o Waitangi principles affording protection, participation and partnership for Māori participants. Research staff enrolling participants will engage appropriate family members (in NZ: whānau) and respect differences in decision-making processes. Interpreters and translated participant materials will be used where study participants and their families primarily speak another language. In Darwin, patient information has been recorded in six Aboriginal languages. In NZ, participant materials have been translated into Te Reo Māori, Samoan and Tongan.
Patient and public involvement
The Indigenous investigators and collaborators at each site form governance groups to provide oversight of study design, conduct, reporting and dissemination. Patient and public involvement, drawing on our established consumer networks, will support knowledge dissemination.
Enrolment
It will be made clear that participation is voluntary and any benefits of participation are to society through knowledge advancement, rather than to the individual. Participants will be advised that wherever possible, blood collection for the study will coincide with collection done for clinical purposes, to avoid extravenipuncture. If a participant or their parent/guardian withdraws consent, no further samples will be collected unless a new consent form is signed. Participants are asked whether their blood may be stored for future use.
People with suspected ARF will be enrolled as early as possible during admission, prior to a final diagnosis being assigned, to ensure that an acute sample is collected. A final diagnosis will be assigned once all required diagnostic information has become available. Participants in group C (RHD) may additionally be recruited through outpatient clinics. Group D (healthy) will be sought from among family members and friends of other enrolled participants, or healthy members of the community in Darwin.
Clinical data collection
After assigning a sequential study code, clinical details will be recorded on a paper case report form (CRF) then entered into an electronic database (Medrio Electronic Data Capture System), including: date of illness onset, date of blood sample collection, demographics, clinical presentation, Jones criteria, clinical laboratory results, medications received and diagnosis assigned by the clinical treating team.
Assignment of diagnosis
Patients presenting with suspected ARF may have a final diagnosis of definite ARF, probable ARF, possible ARF or a non-ARF condition. Probable ARF is defined in Australian guidelines as an acute presentation not fulfilling criteria, missing a major or minor criterion or lacking evidence of preceding streptococcal infection, but ARF is still considered the most likely diagnosis. Possible ARF applies to the same presentation type, but where ARF is considered uncertain but cannot be ruled out.10 For this study, participants ultimately diagnosed with a non-ARF condition will be allocated by the clinical panel as group A or B controls.
Management will be directed by treating clinicians. Clinical diagnoses will be assigned according to local Australian and NZ Diagnostic Criteria by a panel of ≥2 study clinicians expert in ARF. The decision on diagnostic category requires clinical judgement, after evaluating all aspects of the case. The Jones criteria app6 which provides a result of definite, probable or possible ARF or not ARF depending on information provided to the algorithm, will be used by the panel at their discretion. The diagnosis of carditis will be made if rheumatic valvulitis is evident on echocardiogram as per Jones criteria. The diagnosis of RHD will be made in accordance with features described by the World Heart Federation.29 The 2015 revised Jones Criteria7 30 will be applied as the gold standard diagnostic for definite ARF. This represents a departure from normal practice in NZ where local diagnostic criteria differ: polyarthralgia is not considered a major criterion in NZ.24 In addition to the Jones criteria, comprehensive clinical and laboratory information, such as results of synovial fluid testing, serology for alternative infectious aetiologies, autoantibodies, radiological findings, diagnosis assigned by the treating team, family history and local epidemiology, will be considered in adjudicating the diagnosis. ARF type will be further specified as initial episode or recurrence. Where the adjudication panel’s diagnosis differs from that of the treating team with implications for management, this will be communicated to the treating specialist. The clinical panel will also assign final diagnoses for controls.
Blood collection
Blood will be collected at baseline, and for patients with ARF, on follow-up occasions during the convalescent period as able, for example, during prolonged hospitalisation or at a later outpatient appointment (table 3). A safe maximum volume of 28.5 mL depending on age will be obtained in Australia,31 and 14.5 mL in NZ. Baseline samples will be collected as soon as possible during the acute presentation, timed to coincide with routine blood testing wherever possible. Convalescent samples will be used to determine persistence and decay of immunopathological signatures post ARF. After collection, samples will be transported immediately to research laboratories at Menzies School of Health Research (Australia) or the University of Auckland (NZ) for centrifugation, serum/plasma separation and viable peripheral blood mononuclear cell (PBMC) preparation. Timely freezing and storage of serum/plasma aliquots, PAXgene tubes at −80°C and PBMCs in gas-phase liquid nitrogen. Samples will be shipped periodically to relevant laboratories.
Table 3.
Blood collection
| Australian recruitment site | |||
| Participant age | Collection tube | ||
| Testing procedures | 5–9 years | ≥10 years | |
| Transcriptional profiling | 2.5 mL | 2.5 mL | PaxGene tube |
| Peripheral blood mononuclear cells for flow cytometry, metabolomics | 4.0 mL | 22.0 mL | Sodium heparin |
| Antibody studies | 4.0 mL | 4.0 mL | Serum |
| Total | 10.5 mL | 28.5 mL | |
| New Zealand recruitment sites | |||
| Participant age | Collection tube | ||
| Testing procedures | 2–10 years | ≥11 years | |
| Transcriptional profiling | 2.5 mL | 2.5 mL | PaxGene tube |
| Peripheral blood mononuclear cells for flow cytometry, metabolomics | 4.0 mL | 8.0 mL | Sodium heparin |
| Antibody studies | 4.0 mL | 4.0 mL | Serum |
| Total | 10.5 mL | 14.5 mL | |
Sample size
It is not possible to predict a priori the combined discriminatory ability of independently measured factors. This study has elected to enrol up to 120 ARF cases (comprising approximately 90 definite ARF, 15 probable, 15 possible) and 130 controls (30 each of control groups A, B and C, and 40 control group D; figure 1 and table 2). These numbers are feasible and should enable characterisation of different ARF clinical phenotypes (carditis, arthritis, chorea, skin/soft-tissue manifestations or a combination of these) and phases (first or recurrent episodes), acknowledging heterogeneity among participants (ethnicity, age, sex; on corticosteroids or not; at different stages of illness).
In the multivariate approach, it is expected that most, if not all, of the influential factors in the optimal model will be significant (p<0.05) in univariate testing. Assuming α of 5% and 95% power (β=0.05) the minimum sample size is 26 cases. These calculations are only a guideline for removing clear false positives, as the standard procedure for producing robust screening models is to have a separate blinded analysis of a test subset of samples. We will use discovery and validation cohorts, together with stratified bootstrap cross-validation to internally optimise the structural parameters in each model. We further used the standard inferential approach to sample size estimation in diagnostic test studies of biomedical informatics.32 An effective multivariate predictor of ARF with positive predictive value of 0.8 (clinically useful), and a 95% CI of±0.1, would require 65 ARF cases in the discovery cohort.
The discovery cohort will, therefore, comprise a target of 65 definite ARF cases and 65 controls. The validation cohort will comprise a target of 25 definite ARF cases and 65 controls. Probable and possible diagnoses (~15 in each group) will be tested independently (figure 1). Cases will be allocated to the discovery or validation cohorts using a computer-generated random selection of unique study identification numbers.
Laboratory methods
The overarching aim is to develop and validate a profile of host related biomarkers that distinguishes ARF cases from non-ARF conditions and healthy controls. This will be achieved through the following analyses on peripheral blood samples.
Immunophenotyping
PBMC will be stained with labelled antibody panels to identify specific cell populations. Flow cytometry raw data will be analysed manually using Flowjo software and via an automated gating platform as described.33 34 Such high throughput, automated analysis of big flow cytometry data offers several advantages over manual gating, including increased throughput while increasing quality control (such as preprocessing removal of anomalous events via the flowCut algorithm), and identification of specific cell populations with up to 50-dimensional datasets.34–36 Plasma samples will be analysed using multiplex cytokine assays to quantify cytokine and chemokine plasma concentrations, detailed elsewhere.33 37–39
Metabolome analyses
Untargeted metabolomic profiling (>1000 metabolites) will be performed on plasma samples using liquid chromatography coupled to high-resolution mass-spectrometry (LC-MS). Data will be acquired using three modes of operation: reverse-phase/ultraperformance liquid chromatography (UPLC)-MS/MS with positive ion mode electrospray ionisation (ESI), reverse-phase/UPLC-MS/MS with negative ion mode ESI, and Hydrophilic interaction (HILIC)/UPLC-MS/MS with negative ion mode ESI. All identified metabolites will be annotated using appropriate orthogonal analytical techniques applied to the metabolite of interest against a chemical reference standard.
Blood transcriptomics
RNA will be extracted from stabilised whole blood samples (PAXgene tubes) as described,40 41 libraries prepared (TruSeq Stranded Total RNA with Ribo-Zero Globin reduction, Illumina) and NextGen sequencing undertaken (Illumina HiSeq2500, 50 bp single-end reads). Read alignment and gene-level quantification (counts) will be performed using Hisat.42 Negative binomial models will be employed for differential expression analysis, with false discovery rate control for multiple testing.43 The analyses will be adjusted for batch effects and variations in cellular composition, which will be estimated employing Remove Unwanted Variation from RNA-Seq Data (RUVSeq) and CIBERSORT, respectively.44 45 Genes will be mapped to blood transcriptional modules to provide a systems-level view of the responses and reduce the dimensionality of the data.46
CD4 T cell transcriptomic responses to GAS
A two-phased approach will be used to examine CD4 T cell responses to GAS. First, PBMCs from a subset of definite ARF cases and healthy controls will be cultured under a variety of conditions to identify optimal conditions for the second phase. Cells will be harvested at multiple time points poststimulation (6 hours, 24 hours and 48 hours) with heat-killed and antibiotic-killed ARF-associated GAS strains12 47 48 and a selection of candidate ARF antigens. Total RNA from PBMCs cultured under each condition will be profiled by RNA-Seq (Illumina 100 bp paired-end reads, 20M reads), and culture supernatants will be examined by Luminex (Bio-Plex 48-plex Pro Human Cytokine Screening Panel) to identify optimal conditions for maximal discrimination of responses between cases and controls. In the second phase, an expanded set of ARF cases and controls for which PBMC samples are available will be examined following culture under the identified, optimal conditions. Gene expression patterns and cytokine production will be profiled by RNA-Seq and Luminex, as described above.
Antibody analysis
For unbiased investigation of autoantibodies, selected sera will be screened against planer protein microarrays comprised of over 42 000 protein fragments representing some 19 000 human proteins.49 Protein fragments that are more significantly bound by autoantibodies in ARF compared with controls will be identified using a p>0.05 and fold change of 2.0 as cut-off. A suspension bead array will then be designed50 composed of up to 380 potential autoantigens to assess autoantigen reactivity in all sera. The bead-antigens will be selected based on the planar array results, previously completed screens using high-content protein arrays,51 and targets from the literature. Finally, candidate ARF autoantigens identified in the suspension bead-array will be orthogonally validated as individual antigens in ELISA or Luminex bead-based assays to determine sensitivity and specificity.
Bioinformatics and statistical analyses
The data collected for each patient across the immune phenotyping, transcriptomic, proteomic and metabolomic technologies will be integrated computationally to identify patterns of covariance within and between designated clinical outcome groups. Convergence of signatures across diverse systems biology domains will provide independent functional validation. The use of ‘multiomic’ data integration techniques will also enable the possibility of deriving novel biological information that will not be revealed in a single dataset alone.
Data integration methods applying different computational strategies will be used to explore the complex covariance structure within and between the multiomic data blocks. Initially, analysis will focus on each data block in isolation employing a combination of classical generalised linear modelling, multivariate projection regression models (Principal Component Analysis (PCA), Partial Least Squares Discriminant Analysis (PLS-DA)) and unsupervised cluster analysis. The multiple blocks will then be integrated into a single computational model to enable multiomic functional mapping, which in turn will allow us to uncover the underlying biochemical mechanisms. Several methods for performing multiblock data integration will be investigated, from which a consensus model will be derived. These will include: protein−protein interaction networks using NetworkAnalyst,52 Multiblock component analysis,53 regularised canonical correlation analysis,54 the Data Integration Analysis for Biomarker discovery using Latent cOmponent framework55 and similarity network fusion.56
This systems biology analysis should result in both a domain-specific and domain-integrative summary of biological phenomena that are associated with clinical outcome. Likely outcomes are a signature biological pathway associated with ARF as well as individual candidate markers that may provide the basis for the development of novel diagnostic tests. Furthermore, the data generated through this effort will allow crisply defined power calculations for future narrowly targeted assessment of potential biomarkers in clinical trials.
Discussion
We anticipate that study findings will provide the most comprehensive knowledge of the immunopathogenesis of ARF to date, be used as the basis for development of a diagnostic test and provide a pathway towards development of targeted immunomodulatory treatment(s). START commenced recruitment in Australia in November 2018 and in NZ in May 2019. Anticipated completion of recruitment is end-2021. Laboratory analyses will be batched and run together at completion of recruitment.
Both underdiagnosisand overdiagnosis of ARF pose major challenges to individuals and health systems.10 Whether the use of the ‘probable’ and ‘possible’ ARF diagnostic categories is reducing underdiagnosis or contributing to overdiagnosis is currently unknown. Should findings from this study successfully identify a discriminatory biomarker profile differentiating definite ARF from non-ARF, this will provide a mechanism for accurate diagnosis to guide appropriate management. This will be a critically important advance in the diagnosis and management of ARF globally.
A minority of any population are at risk of ARF after exposure to group A streptococci. The estimated lifetime cumulative incidence of ARF was previously calculated at 5.7% in Australia’s NT.57 This was considered likely to be an underestimate, and indeed more recent data highlight the burden of unrecognised ARF in Aboriginal communities in the NT where repeated infection with GAS is ubiquitous from early childhood,2 58 suggesting that the lifetime risk of ARF in these populations is higher than 6%. In US military camps, 2%–3% of recruits developed ARF after a single bout of streptococcal pharyngitis.59
Previous efforts to develop ARF diagnostic tests and elucidate the immunopathogenesis have laid foundations for the START study design and analysis. Kim et al12 analysed responses to GAS challenge of PBMC from ARF patients at Royal Darwin Hospital using multiplex cytokine array, flow cytometric analysis and global gene expression analysis by RNA sequencing. They identified a dysregulated IL-1β-granulocyte-macrophage colony-stimulating factor cytokine axis in PBMCs from ARF patients, and the potential to suppress this response by hydroxychloroquine. The authors proposed that hydroxychloroquine, an immunomodulatory agent, could be repurposed to reduce the risk or severity of RHD after ARF.12 Clinical use of hydroxychloroquine, now reported as safe in two cases,13 requires further investigation. A study using sera from patients with ARF in NZ combined multiplex bead-based assays and systems immunology data analysis to identify a linked IgG3-C4 response that may have utility as a clinical biomarker in early ARF.23 However, small sample sizes have been a limiting factor. Key strengths of the current study include the larger sample size, multicentre enrolment across different countries to maximise relevance and use of the latest laboratory approaches.
Limitations include that capture of eligible participants early in disease is challenging, due to the variable time taken for participants to be hospitalised after disease onset, to identify and gain consent from participants, and obtain blood samples. At the Australian site, the majority of eligible participants will be Aboriginal children living in remote communities, often hundreds of kilometres distant from the enrolment site. Although Auckland is a large urban city inequitable healthcare access is experienced by many Māori and Pacific people living in socioeconomic deprivation and this can lead to delayed hospitalisation for many children presenting with ARF. Given these inherent delays, a proportion of baseline blood samples will be collected after the peak inflammatory phase has passed. Another limitation is heterogeneity among people with ARF with regards to ARF type (first or recurrent), diagnostic category (possible, probable, definite), clinical phenotype (carditis, chorea, arthritis, etc) and demographic variations; however, we limited enrolment to people below 30 years and only of Aboriginal, Torres Strait Islander, Māori or Pacific ethnicity to minimise heterogeneity. Although a distinguishing biomarker signature may be identified, the ability to readily translate study findings into a feasible diagnostic able to be used in health service laboratories outside a research environment, is uncertain.
Successful completion of this study will considerably improve knowledge of ARF immunology. Innovative strategies to improve the clinical management of ARF are core components of the overall suite of activities required to achieve the goal of RHD elimination.60
Supplementary Material
Acknowledgments
We are very grateful to Caroline Motteram for project management, Yolanda Hernandez, Laura Francis and Vincent Mithen for recruiting participants in Darwin and Shirley Lawrence, Rhonda Holloway, Renee Clark, Alicia Stanley and Roxanne Buchanan for recruiting participants in New Zealand. We thank Alex Kaethner and NT Cardiac for facilitating echocardiography. Vanessa Rigas, Erin Gargan, Gabriella Minigo and Michael Serralha are gratefully acknowledged for laboratory support; Kyran Kuzio, Tasha Cole, Alana Whitcombe, Lauren Carlton and Timothy Ho for laboratory assistance; Amy Baker and Rebecca Trowman for program and financial management.
Footnotes
Twitter: @annapralph
Contributors: JC, AR, RW, NJM, RM, AB, DB, TCB, MM, GP and TK conceived and designed the study. NJM, RM, RM, AB, DB, TL, TCB, RB, JB and TK will perform the assays and analyse the data. GP and MM provide project governance. AR, RW, JY, BR, NW and JC provided clinical care, patient recruitment, assessment and clinical review panel participation. AR created the first draft and managed the edits to the manuscript. All authors reviewed and approved the study protocol.
Funding: The study is funded by the Australian National Health and Medical Research Council (NHMRC) grant #1147531. AR is supported by NHMRC fellowship #1142011. TCB is supported by the Western Australia Department of Health and a Career Development Fellowship from Improving Health Outcomes in the Tropical North (NHMRC #1131932).
Competing interests: None declared.
Patient and public involvement: Patients and/or the public were involved in the design, or conduct, or reporting, or dissemination plans of this research. Refer to the Methods section for further details.
Provenance and peer review: Not commissioned; peer reviewed for ethical and funding approval prior to submission.
Ethics statements
Patient consent for publication
Not required.
References
- 1.Ralph AP, Read C, Johnston V, et al. Improving delivery of secondary prophylaxis for rheumatic heart disease in remote Indigenous communities: study protocol for a stepped-wedge randomised trial. Trials 2016;17:51. 10.1186/s13063-016-1166-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hardie K, Ralph AP, de Dassel JL. Rhd elimination: action needed beyond secondary prophylaxis. Aust N Z J Public Health 2020;44:427. 10.1111/1753-6405.13002 [DOI] [PubMed] [Google Scholar]
- 3.Oliver J, Robertson O, Zhang J, et al. Progression of acute rheumatic fever to recurrence, rheumatic heart disease, and death in New Zealand children and youth: a cohort study. Heart, Lung and Circulation 2019;28:S4. 10.1016/j.hlc.2019.05.013 [DOI] [Google Scholar]
- 4.Zühlke L, Engel ME, Karthikeyan G, et al. Characteristics, complications, and gaps in evidence-based interventions in rheumatic heart disease: the global rheumatic heart disease registry (the remedy study). Eur Heart J 2015;36:1115–22. 10.1093/eurheartj/ehu449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones TD. The diagnosis of rheumatic fever. J Am Med Assoc 1944;126:481–4. 10.1001/jama.1944.02850430015005 [DOI] [Google Scholar]
- 6.RHDAustralia . iPhone and android Apps, 2020. Available: http://www.rhdaustralia.org.au/apps
- 7.Gewitz MH, Baltimore RS, Tani LY, et al. Revision of the Jones criteria for the diagnosis of acute rheumatic fever in the era of Doppler echocardiography: a scientific statement from the American heart association. Circulation 2015;131:1806–18. 10.1161/CIR.0000000000000205 [DOI] [PubMed] [Google Scholar]
- 8.McMillan DJ, Rafeek RAM, Norton RE, et al. In search of the Holy Grail: a specific diagnostic test for rheumatic fever. Front Cardiovasc Med 2021;8:674805. 10.3389/fcvm.2021.674805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cilliers A, Adler AJ, Saloojee H, et al. Anti-Inflammatory treatment for carditis in acute rheumatic fever. Cochrane Database of Systematic Reviews 2015;262:CD003176. 10.1002/14651858.CD003176.pub3 [DOI] [PubMed] [Google Scholar]
- 10.RHDAustralia (ARF/RHD writing group) . The 2020 Australian guideline for prevention, diagnosis and management of acute rheumatic fever and rheumatic heart disease (3rd edition. Darwin Northern Territory, Australia: Menzies School of Health Research, 2020. https://www.rhdaustralia.org.au/arf-rhd-guideline [Google Scholar]
- 11.Dean GL, Edwards SG, Ives NJ, et al. Treatment of tuberculosis in HIV-infected persons in the era of highly active antiretroviral therapy. AIDS 2002;16:75–83. 10.1097/00002030-200201040-00010 [DOI] [PubMed] [Google Scholar]
- 12.Kim ML, Martin WJ, Minigo G, et al. Dysregulated IL-1β-GM-CSF axis in acute rheumatic fever that is limited by hydroxychloroquine. Circulation 2018;138:2648–61. 10.1161/CIRCULATIONAHA.118.033891 [DOI] [PubMed] [Google Scholar]
- 13.Wilson NJ, Concannon A, Malcolm J, et al. The treatment of acute rheumatic fever: novel use of hydroxychloroquine. Pediatr Infect Dis J 2020;39:e120–2. 10.1097/INF.0000000000002647 [DOI] [PubMed] [Google Scholar]
- 14.Hafez M, Morsy ZEL-, Shennawy FEL-, et al. Susceptibility to over production of cytokines in acute rheumatic carditis and their role in the pathogenesis. Journal of Medical Sciences 2002;2:65–73. 10.3923/jms.2002.65.73 [DOI] [Google Scholar]
- 15.Yeğin O, Coşkun M, Ertuğ H. Cytokines in acute rheumatic fever. Eur J Pediatr 1997;156:25–9. 10.1007/s004310050545 [DOI] [PubMed] [Google Scholar]
- 16.Narin N, Kütükçüler N, Narin F, et al. Anticardiolipin antibodies in acute rheumatic fever and chronic rheumatic heart disease: is there a significant association? Clin Exp Rheumatol 1996;14:567–9. [PubMed] [Google Scholar]
- 17.Wright VJ, Herberg JA, Kaforou M, et al. Diagnosis of Kawasaki disease using a minimal whole-blood gene expression signature. JAMA Pediatr 2018;172:e182293. 10.1001/jamapediatrics.2018.2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berry MPR, Graham CM, McNab FW, et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature 2010;466:973–7. 10.1038/nature09247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bennett L, Palucka AK, Arce E, et al. Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. J Exp Med 2003;197:711–23. 10.1084/jem.20021553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anderson ST, Kaforou M, Brent AJ, et al. Diagnosis of childhood tuberculosis and host RNA expression in Africa. N Engl J Med 2014;370:1712–23. 10.1056/NEJMoa1303657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khan IH, Ravindran R, Yee J, et al. Profiling antibodies to Mycobacterium tuberculosis by multiplex microbead suspension arrays for serodiagnosis of tuberculosis. Clin Vaccine Immunol 2008;15:433–8. 10.1128/CVI.00354-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu LL, Chung AW, Rosebrock TR, et al. A functional role for antibodies in tuberculosis. Cell 2016;167:433–43. 10.1016/j.cell.2016.08.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chung AW, Ho TK, Hanson-Manful P, et al. Systems immunology reveals a linked IgG3-C4 response in patients with acute rheumatic fever. Immunol Cell Biol 2020;98:12–21. 10.1111/imcb.12298 [DOI] [PubMed] [Google Scholar]
- 24.Heart Foundation of New Zealand . New Zealand guidelines for rheumatic fever: diagnosis, management and secondary prevention of acute rheumatic fever and rheumatic heart disease: 2014 update, 2014. Available: https://www.heartfoundation.org.nz/resources/acute-rheumatic-fever-and-rheumatic-heart-disease-guideline
- 25.Australian Bureau of Statistics . Estimates of aboriginal and torres strait islander Australians, 2016. Available: https://www.abs.gov.au/ausstats/abs@.nsf/mf/3238.0.55.001
- 26.Francis J R, Gargan C, Remenyi B. A cluster of acute rheumatic fever cases among Aboriginal Australians in a remote community with high baseline incidence. Aust N Z J Public Health 2019;4:288–93. [DOI] [PubMed] [Google Scholar]
- 27.Institute of Environmental Science and Research Limited . Rheumatic fever report, January-December 2018. EpiSurv science for communities, 2018. Available: https://surv.esr.cri.nz/PDF_surveillance/RheumaticFever/Rheumaticfeverbi-annualreportJan-Dec2018.pdf
- 28.Curtis E, Jones R, Tipene-Leach D, et al. Why cultural safety rather than cultural competency is required to achieve health equity: a literature review and recommended definition. Int J Equity Health 2019;18:174. 10.1186/s12939-019-1082-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reményi B, Wilson N, Steer A, et al. World Heart Federation criteria for echocardiographic diagnosis of rheumatic heart disease--an evidence-based guideline. Nat Rev Cardiol 2012;9:297–309. 10.1038/nrcardio.2012.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beaton A, Carapetis J. The 2015 revision of the Jones criteria for the diagnosis of acute rheumatic fever: implications for practice in low-income and middle-income countries. Heart Asia 2015;7:7–11. 10.1136/heartasia-2015-010648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seattle Children’s Hospital . Maximum allowable blood draw from infants. Mayo foundation for medical education and research, 2020. Available: https://seattlechildrenslab.testcatalog.org/show/1000721-1
- 32.Hajian-Tilaki K. Sample size estimation in diagnostic test studies of biomedical informatics. J Biomed Inform 2014;48:193–204. 10.1016/j.jbi.2014.02.013 [DOI] [PubMed] [Google Scholar]
- 33.Ben-Othman R, Cai B, Liu AC, et al. Systems biology methods applied to blood and tissue for a comprehensive analysis of immune response to hepatitis B vaccine in adults. Front Immunol 2020;11:580373. 10.3389/fimmu.2020.580373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rahim A, Meskas J, Drissler S, et al. High throughput automated analysis of big flow cytometry data. Methods 2018;134-135:164–76. 10.1016/j.ymeth.2017.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malek M, Taghiyar MJ, Chong L, et al. flowDensity: reproducing manual gating of flow cytometry data by automated density-based cell population identification. Bioinformatics 2015;31:606–7. 10.1093/bioinformatics/btu677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ge Y, Sealfon SC. flowPeaks: a fast unsupervised clustering for flow cytometry data via k-means and density peak finding. Bioinformatics 2012;28:2052–8. 10.1093/bioinformatics/bts300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amenyogbe N, Dimitriu P, Cho P, et al. Innate immune responses and gut Microbiomes distinguish HIV-exposed from HIV-Unexposed children in a population-specific manner. J Immunol 2020;205:2618–28. 10.4049/jimmunol.2000040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Amenyogbe N, Dimitriu P, Smolen KK, et al. Biogeography of the relationship between the child gut microbiome and innate immune system. mBio 2021;12. 10.1128/mBio.03079-20. [Epub ahead of print: 12 01 2021]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moncunill G, Dobaño C, González R, et al. Association of maternal factors and HIV infection with innate cytokine responses of delivering mothers and newborns in Mozambique. Front Microbiol 2020;11:1452. 10.3389/fmicb.2020.01452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee AH, Shannon CP, Amenyogbe N, et al. Dynamic molecular changes during the first week of human life follow a robust developmental trajectory. Nat Commun 2019;10:1092. 10.1038/s41467-019-08794-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Idoko OT, Smolen KK, Wariri O, et al. Clinical protocol for a longitudinal cohort study employing systems biology to identify markers of vaccine immunogenicity in newborn infants in the Gambia and Papua New Guinea. Front Pediatr 2020;8:197. 10.3389/fped.2020.00197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pertea M, Kim D, Pertea GM, et al. Transcript-level expression analysis of RNA-Seq experiments with HISAT, StringTie and Ballgown. Nat Protoc 2016;11:1650–67. 10.1038/nprot.2016.095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anders S, McCarthy DJ, Chen Y, et al. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat Protoc 2013;8:1765–86. 10.1038/nprot.2013.099 [DOI] [PubMed] [Google Scholar]
- 44.Risso D, Ngai J, Speed TP, et al. Normalization of RNA-Seq data using factor analysis of control genes or samples. Nat Biotechnol 2014;32:896–902. 10.1038/nbt.2931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453–7. 10.1038/nmeth.3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li S, Rouphael N, Duraisingham S, et al. Molecular signatures of antibody responses derived from a systems biology study of five human vaccines. Nat Immunol 2014;15:195–204. 10.1038/ni.2789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smoot JC, Barbian KD, Van Gompel JJ, et al. Genome sequence and comparative microarray analysis of serotype M18 group A Streptococcus strains associated with acute rheumatic fever outbreaks. Proc Natl Acad Sci U S A 2002;99:4668–73. 10.1073/pnas.062526099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bryant PA, Smyth GK, Gooding T, et al. Susceptibility to acute rheumatic fever based on differential expression of genes involved in cytotoxicity, chemotaxis, and apoptosis. Infect Immun 2014;82:753–61. 10.1128/IAI.01152-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sjöberg R, Mattsson C, Andersson E, et al. Exploration of high-density protein microarrays for antibody validation and autoimmunity profiling. N Biotechnol 2016;33:582–92. 10.1016/j.nbt.2015.09.002 [DOI] [PubMed] [Google Scholar]
- 50.Just D, Månberg A, Mitsios N, et al. Exploring autoantibody signatures in brain tissue from patients with severe mental illness. Transl Psychiatry 2020;10:401. 10.1038/s41398-020-01079-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McGregor R, Tay ML, Carlton LH, et al. Mapping autoantibodies in children with acute rheumatic fever. Front Immunol 2021;12:702877. 10.3389/fimmu.2021.702877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xia J, Gill EE, Hancock RE, et al. Visual and network-based meta-analysis of gene expression data. Nat Protoc 2015;10:823–44. [DOI] [PubMed] [Google Scholar]
- 53.De Roover K, Ceulemans E, Timmerman ME. How to perform multiblock component analysis in practice. Behav Res Methods 2012;44:41–56. 10.3758/s13428-011-0129-1 [DOI] [PubMed] [Google Scholar]
- 54.Wilms I, Croux C. Robust sparse canonical correlation analysis. BMC Syst Biol 2016;10:72. 10.1186/s12918-016-0317-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Singh A, Shannon CP, Gautier B, et al. DIABLO: an integrative approach for identifying key molecular drivers from multi-omics assays. Bioinformatics 2019;35:3055–62. 10.1093/bioinformatics/bty1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang B, Mezlini AM, Demir F, et al. Similarity network fusion for aggregating data types on a genomic scale. Nat Methods 2014;11:333–7. 10.1038/nmeth.2810 [DOI] [PubMed] [Google Scholar]
- 57.Carapetis JR, Currie BJ, Mathews JD. Cumulative incidence of rheumatic fever in an endemic region: a guide to the susceptibility of the population? Epidemiol Infect 2000;124:239–44. 10.1017/S0950268800003514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Francis JR, Fairhurst H, Hardefeldt H, et al. Hyperendemic rheumatic heart disease in a remote Australian town identified by echocardiographic screening. Med J Aust 2020;213:118–23. 10.5694/mja2.50682 [DOI] [PubMed] [Google Scholar]
- 59.Rammelkamp CH, Wannamaker LW, Denny FW. The epidemiology and prevention of rheumatic fever. Bull N Y Acad Med 1952;28:321–34. [PMC free article] [PubMed] [Google Scholar]
- 60.Wyber R, Noonan K, Halkon C, et al. Ending rheumatic heart disease in Australia: the evidence for a new approach. Med J Aust 2020;213 Suppl 10:S3–31. 10.5694/mja2.50853 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.