Abstract
This paper briefly outlines the aim, the objectives, the architecture and the main building blocks of the ongoing large scale integrating transatlantic research project CHIC (http://chic-vph.eu/).
I. Introduction
The impressive rate of generation of human biological data during the last decades has dictated the development of numerous statistical, computational and mathematical methods, in order to extract, analyze and exploit the hidden wealth of information. Unquestionably systems biology has been established as a key player in this arena. However, despite its maturation over the last decade a number of obstacles render it difficult for systems biology to be directly exploitable by clinical practice [1]. Recognizing that in most medical conditions crucial biological phenomena are manifested at several spatiotemporal scales, including scales lying far above the subcellular level - which is traditionally addressed by systems biology- researchers have proposed a number of ways to integrate super-cellular levels into systems biology approaches. Such initiatives have taken various forms and names such as systems physiology [2] systems medicine, multiscale modeling [3] and Virtual Physiological Human (VPH).
Cancer in the clinical context dictates the development of integrative hypermodels consisting of simpler and more manageable constituent component models which may already be available. Nevertheless, in order for models generally developed by different modellers or modelling groups to be reusable, there are a number of prerequisites that have to be satisfied. Models should be robust, reproduceable and interoperable. This implies that standardization of model description and operation is a sine qua non necessity if rational, coherent and comprehensive exploitation of the invaluable information hidden within human multiscale biological data is envisaged. Responding to this imperative in the context of both the broad (VPH) initiative and the paradigmal cancer domain, CHIC proposes the development of a suite of tools and services in a secure infrastructure that will support accessibility and reusability of VPH mathematical and computational hypermodels. The proposed objective is primarily centered around the development of a hypermodelling environment which, although will be applicable to the broad VPH space, it will be driven by and originally tested in the cancer domain. In order to ensure clinical relevance and foster clinical acceptance of hypermodelling in the future, the whole endeavour will in practice be driven by the clinical partners of the consortium. Cancer hypermodels to be collaboratively developed by the consortium cancer modellers will provide the framework and the testbed for the development of the CHIC technologies. Clinical adaptation and partial clinical validation [4–5] of hypermodels and hypermodel oncosimulators will be undertaken.
II. Aim
The CHIC proposal aims at developing cutting edge ICT tools, services and secure infrastructure to foster the development of elaborate and reusable integrative models (hypermodels) and larger repositories so as to demonstrate benefits of having both the multiscale data and the correponding models readily available. Although the broader VPH domain is the primary target of the hypermodelling infrastructure to be developed by CHIC, the primary application domain will be cancer and in silico oncology.
In the mid and long term CHIC aims to pave the way for reliable in silico clinical trials, lying at the heart of the vision of in silico medicine, and subsequently for patient individualized treatment optimization based on in silico experimentation [4–5].
III. Objectives
CHIC proposes the development of clinical trial driven tools, services and secure infrastructure that will support the creation of multiscale cancer hyper-models (integrative models). The latter are defined as choreographies of component models, each one describing a biological process at a characteristic spatiotemporal scale, and of relation models/metamodels defining the relations across scales. Integrative models can become component models for other integrative models. The development of a secure hypermodelling infrastructure consisting primarily of a hypermodelling editor and a hypermodelling execution environment is a central generic VPH geared objective of CHIC.
In order to render models developed by different modellers semantically interoperable, an infrastructure for semantic metadata management along with tools and services for ontology-based annotations will be developed. Existing approaches such as the one developed by the EC funded RICORDO project will be exploited and extended. Facilitated operations will range from automated dataset matching to model merging and managing complex simulation workflows. In this way standardization of cancer model and data annotation allowing multiscale hypermodelling will be fostered.
The following entities will also be developed: a hypermodel repository, a hypermodel-driven clinical data repository, a distributed metadata repository and an in silico trial repository for the storage of executed simulation scenarios, an image processing toolkit, a visualization toolkit and cloud and virtualization services.
In order to ensure that the entire project will be clinically driven and clinically oriented, three concrete clinical trials/studies will be adopted and addressed. They concern nephroblastoma treated by combined chemotherapy, glioblastoma treated by immunotherapy in combination with chemotherapy and radiotherapy and non-small cell lung cancer treated by a combination of chemotherapy and radiotherapy.
The multiscale data generated by these trials/studies will be exploited so as to both drive the development of a number of integrative multiscale cancer models (hypermodels) and hypermodel oncosimulators and clinically adapt and partly validate them.
The whole process will be supported by the technological tools, services and infrastructure to be developed and will serve as a paradigm of applicability and usability of the latter. Additional available multiscale data concerning colon and prostate cancer will be exploited in a similar way. The participation of five prominent multiscale cancer modelling groups from both EU and the US covering all spatiotemporal scales (from the molecular up to the organism and from nsecs up to years) and all the fundamental biological processes of cancer as well as some aspects of the treatment response of normal tissues will ensure a comprehensive coverage of the domain of cancer. The latter refers to both the process of annotating component models and hypermodels as well as pertinent multiscale data and the development of exemplary clinically driven and clinically validatable hypermodels.
This is expected to considerably advance the exploitation of both existing models and models to be developed in the future. An integrative platform dictated by the IT architecture of the project will provide access to all hypermodelling tools and services to be developed. Apart from the tools addressing semantic interoperability, a number of data pre-processing tools, services and resources will be developed and/or made available. These will include inter alia image segmentation, three-dimensional reconstruction, several forms of data and model prediction visualization and cloud computing.
The legal and ethical aspects of patients’ data handling will be addressed by a workpackage dealing with both the legal and the IT aspects of data anonymization and pseudonymization, patient’s consent etc. The same work package will also address the intellectual rights issues arising from the amalgamation of component models potentially developed by different modellers in order to construct integrative models.
The dissemination and exploitation of the CHIC proposal will target all stakeholders, namely clinicians, fundamental science researchers, IT specialists and engineers, industry and patients. Similarly, the project is expected to have a significant impact on all the corresponding domains. More precisely, CHIC aspires to make a breakthrough in multiscale cancer modelling through greatly facilitating multi-modeller cancer hypermodelling and its clinical adaptation and validation. Standardization of model description and model “fusion” will be two of the core means to achieve this goal. The creation of such elaborate and refined hypermodels is expected to sharply accelerate the clinical translation of multiscale cancer models and oncosimulators following their prospective clinical validation (in silico oncology). Addressing intellectual property issues in a multi modeller setting will foster the community spirit in the VPH domain.
IV. Architecture and MAIN Building Blocks of CHIC
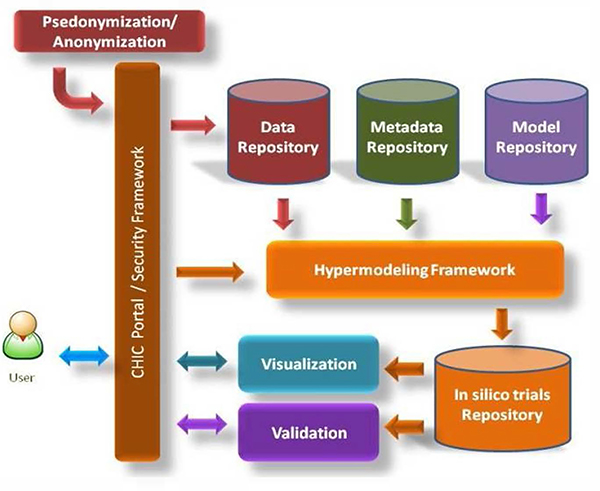
CHIC will develop a variety of tools and repositories that will assist the researcher in searching and retrieving models and data, composing and saving hypermodels, executing models and hypermodels and last but not least validating the outcome of the simulations (Fig. 1).
Figure 1.
Main technological components of CHIC directly related to the hypermodelling workflow
The core reference point for the users will be the CHIC portal. All the components of CHIC will reside under the “umbrella” of the security framework that will deal with the issues of secure and safe storing, acquisition and sharing of models and data.
Four individual repositories will be implemented in CHIC.
A model repository that will store the multiscale models, the complimentary tools and modules that will be needed in order to construct hypermodels and the hypermodels themselves. In the model repository will also reside the visualization and image processing tools that will be developed in CHIC.
A data repository that will store the heterogeneous multiscale data coming from clinical environment (clinical trials etc.). Especially for the storage of “sensitive” patient-specific data a special pseudo-anonymization/anonymization procedure will be followed in compliance with the legal and ethical framework. Due to legal limitations, the CHIC repositories, especially the ones that are dealing with patient data, will be implemented so as to be easily deployable in local or private cloud infrastructures of medical, educational and research institutions.
A metadata repository that will store the machine-readable documentation material that will semantically represent both models and data.
An in silico trial repository which will store the input and output of the in silico simulations along with the complete profile of each simulation, including the model/hypermodel used in the simulation and its version, the model/hypermodel configuration parameters etc.
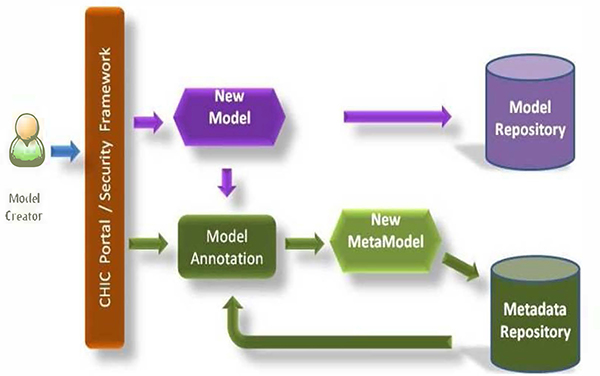
The users will upload their models and the complimentary tools in the model repository. In addition the user will use the model annotation framework to add semantic information to his/her models and data. This information will be used later on by the hypermodelling framework in order to construct and execute hypermodels (Fig.2).
Figure. 2.
Model and metamodel creation workflow
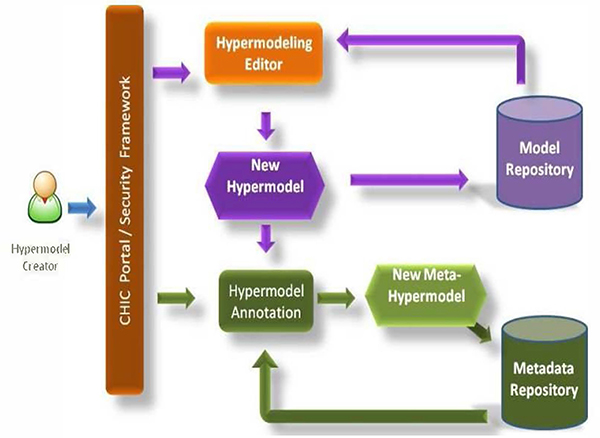
The composition and execution of hypermodels will be done by the Hypermodelling Framework. This will consist of the Hypermodelling Editor and the Hypermodel Executional Framework. The Hypermodelling Editor will communicate with the model and metadata repositories and will guide the user in easily and effectively constructing hypermodels (Fig.3) by
Figure 3.
Hypermodel and meta-hypermodel creation workflow
exposing information about existence and availability of models,
presenting interconnection possibilities,
indicating the model/modules that need to be developed in order to fill in the gaps,
visually constructing the hypermodels, provided that all needed components are available, either as implemented models/modules or as a “ to be implemented” dummy black boxes.
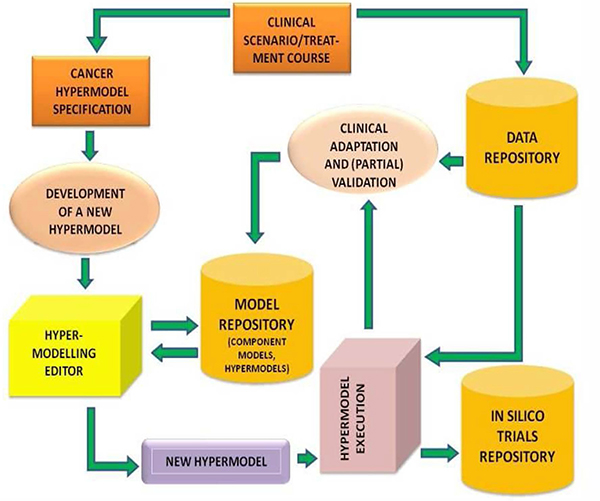
The Hypermodel Executional Framework will communicate with the model, the metadata and the data repository, in order to retrieve the relevant information to be used in the simulation (in silico trial). The outcome of the execution will be send to the in silico trial repository for persistent storage. The user will be able to retrieve the results of a simulation from the in silico trials repository (Fig.4).
Figure.4.
Clinical scenario driven hyper- model development
The CHIC image processing tools will be used in the preprocessing of imaging data in order to be prepared for usage in the simulations. The results of the simulations will use the CHIC visualization tools in order to be presented to the user.
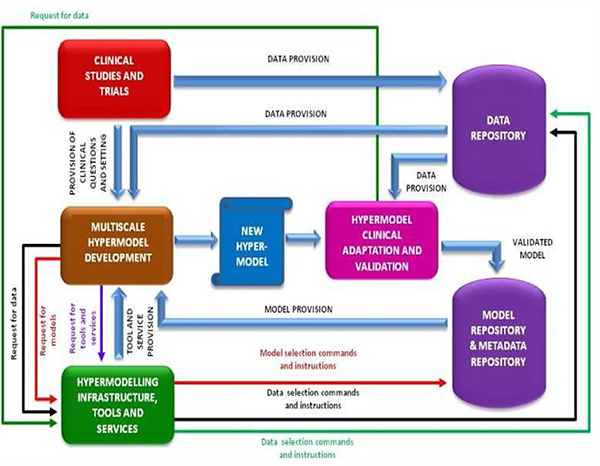
Fig.5 shows the gross overall CHIC architecture from a clinical study and trial centered perspective.
Figure 5.
The overall CHIC architecture from a clinical study and trial centered perspective
The Hypermodel Oncosimulator is an extension of the notion and the system of the original Oncosimulator [4–5] so as to make use of cancer and normal tissue hypermodels. The (hypermodel) Oncosimulator is at the same time a concept of multilevel integrative cancer biology, a complex algorithmic construct, a biomedical engineering system and eventually in the future a clinical tool which primarily aims at supporting the clinician in the process of optimizing cancer treatment in the patient individualized context through conducting experiments in silico i.e. on the computer. Additionally it is a platform for simulating, investigating, better understanding and exploring the natural phenomenon of cancer, supporting the design and interpretation of clinicogenomic trials and finally training doctors, researchers and interested patients alike. A synoptic outline of the clinical utilization of a specific version of the Oncosimulator, as envisaged to take place following an eventually successful completion of its clinical adaptation, optimization and validation process is provided in the form of steps (Fig.6).
Figure 6.
A synoptic diagram of the hypermodel based Oncosimulator
In this paper a short description of the main technical architecture and components of the CHIC project has been provided. Initial successful results (http://chic-vph.eu/) have demonstrated that the design of the project is realistic and possesses great potential for the semi-automatic development of cancer hypermodels. Since the technologies developed are quite generic, an extension to domains beyond cancer will be an obvious additional outcome of the project.
Acknowledgements
The scientific support of E.Kolokotroni, E.Ouzounoglou, E.Georgiadi all from the In Silico Oncology Group, ICCS, NTUA as well as the contributions of other consortium person-members including C.Hahn, Eurice during the preparation of the CHIC Proposal are duly acknowledged.
This work has been supported by the European Commission under the project Computational Horizons In Cancer (CHIC): Developing Meta- and Hyper-Multiscale Models and Repositories for In Silico Oncology (FP7-ICT-2011–9, Grant agreement no: 600841).
Contributor Information
G. Stamatakos, In Silico Oncology Group, Institute of Communication and Computer Systems, National Technical University of Athens, Greece.
Dimitra Dionysiou, In Silico Oncology Group, Institute of Communication and Computer Systems, National Technical University of Athens, Greece.
Fay Misichroni, In Silico Oncology Group, Institute of Communication and Computer Systems, National Technical University of Athens, Greece.
Norbert Graf, University of Saarland, Pediatric Oncology and Hematology Clinic, Germany.
Stefaan van Gool, Catholic University of Leuven, Pediatric Oncology Clinic, Belgium.
Rainar Bohle, University of Saarland, Dept. of Pathology, Germany.
Feng Dong, University of Bedfordshire, UK.
Marco Viceconti, University of Sheffield, UK.
Kostas Marias, Foundation for Research and Technology, Hellas, Greece.
Vangelis Sakkalis, Foundation for Research and Technology, Hellas, Greece.
Nikolaus Forgo, G.W.Leibnitz University of Hannover, Germany.
Ravi Radhakrishnan, University of Pennsylvania, USA.
Helen Byrne, University of Oxford, UK.
Caterina Guiot, University of Torino.
Philippe Buechler, University of Bern, Switzerland.
Elias Neri, Custodix NV, Belgium.
Anca Bucur, Philips Electronics Nederland B.V., The Netherlands.
Bernard de Bono, University College London, UK.
Debora Testi, Consorzio Universitario CINECA, Italy.
Manolis Tsiknakis, Technological Educational Institute of Crete, Greece.
References
- [1].Clermont G, Auffray C, Moreau Y, Rocke DM, Dalevi D, Dubhashi D, Marshall DR, Raasch P, Dehne F, Provero P, Tegner J, Aronow BJ, Langston MA, and Benson M, “Bridging the gap between systems biology and medicine,” Genome Medicine vol 1, p. 88, Sep. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Kitano H, “Grand challenges in systems physiology,” Frontiers in Physiology, vol 1, pp. 1–3, May 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3]. http://ecancer.org/tv/pubdate/105.
- [4].Stamatakos G, “In Silico Oncology Part I: Clinically Oriented Cancer Multilevel Modeling Based on Discrete Event Simulation,” in Multiscale Cancer Modeling, Deisboeck T and Stamatakos G, Eds. 407–436 2011–01-01 CRC Press, Print ISBN: 978–1-4398–1440-6 eBook ISBN: 978–1-4398–1442-0 DOI: 10.1201/b10407-19 Boca Raton, Florida, USA, 2011 [DOI] [Google Scholar]
- [5].Stamatakos G, Dionysiou D, Lunzer A, Belleman R, Kolokotroni E, Georgiadi E, Erdt M, Pukacki J, Rueping S, Giatili S, d’Onofrio A, Sfakianakis S, Marias K, Desmedt C, Tsiknakis M, and Grat N; “The Technologically Integrated Oncosimulator: Combining Multiscale Cancer Modeling with Information Technology in the In Silico Oncology Context,” IEEE J Biomedical and Health Informatics, vol. 18, no. 3, pp. 840–854, May 2014. DOI: 10.1109/JBHI.2013.2284276 [DOI] [PubMed] [Google Scholar]