Abstract
Early prediction of COVID-19 in-hospital mortality relies usually on patients’ preexisting comorbidities and is rarely reproducible in independent cohorts. We wanted to compare the role of routinely measured biomarkers of immunity, inflammation, and cellular damage with preexisting comorbidities in eight different machine-learning models to predict mortality, and evaluate their performance in an independent population. We recruited and followed-up consecutive adult patients with SARS-Cov-2 infection in two different Italian hospitals. We predicted 60-day mortality in one cohort (development dataset, n = 299 patients, of which 80% was allocated to the development dataset and 20% to the training set) and retested the models in the second cohort (external validation dataset, n = 402).
Demographic, clinical, and laboratory features at admission, treatments and disease outcomes were significantly different between the two cohorts. Notably, significant differences were observed for %lymphocytes (p < 0.05), international-normalized-ratio (p < 0.01), platelets, alanine-aminotransferase, creatinine (all p < 0.001). The primary outcome (60-day mortality) was 29.10% (n = 87) in the development dataset, and 39.55% (n = 159) in the external validation dataset. The performance of the 8 tested models on the external validation dataset were similar to that of the holdout test dataset, indicating that the models capture the key predictors of mortality. The shap analysis in both datasets showed that age, immune features (%lymphocytes, platelets) and LDH substantially impacted on all models' predictions, while creatinine and CRP varied among the different models. The model with the better performance was model 8 (60-day mortality AUROC 0.83 ± 0.06 in holdout test set, 0.79 ± 0.02 in external validation dataset). The features that had the greatest impact on this model's prediction were age, LDH, platelets, and %lymphocytes, more than comorbidities or inflammation markers, and these findings were highly consistent in both datasets, likely reflecting the virus effect at the very beginning of the disease.
Keywords: COVID-19, SARS-CoV-2, Coronavirus, Lymphocytes, Platelets, CRP, LDH, In-hospital death
Graphical abstract
Highlights
-
•
Models for early prediction of COVID-19 mortality relies usually on the presence of preexisting comorbidities, and are rarely reproducible.
-
•
In two independent hospital populations, routinely measured biomarkers relative to the immune response and cellular damage contributed to COVID-19 prognosis prediction better than the preexisting comorbidities.
-
•
The features that had the greatest impact on the model's mortality prediction were age, LDH, platelets count, and % of lymphocytes, while C-reactive protein, kidney and liver biomarkers contributed less constantly.
1. Introduction
The outbreaks of the Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) first detected in Wuhan, China, in December 2019, evolved into a pandemic in the following weeks, raising concerns all over the world (Huang et al., 2020a). The infection can lead to coronavirus disease 2019 (COVID-19), which is characterized by a high rate of hospitalization, respiratory failure, and ultimately death (Guan et al., 2020; Onder et al., 2020; Zhou et al., 2020). To improve the recognition of the patients at higher risk of deterioration and death, efforts were undertaken to early predict the outcomes, ideally at the point of hospital admission.
Numerous articles on large cohorts of hospitalized patients affected by COVID-19 have been published so far (Geleris et al., 2020; Grasselli et al., 2020; Guan et al., 2020; Hamer et al., 2020; Huang et al., 2020b; Richardson et al., 2020; Wang et al., 2020; Zhou et al., 2020). Coexisting conditions, such as diabetes, hypertension, malignancy, chronic obstructive pulmonary disease (COPD), obesity and older age are risk factors for severe disease and poor outcome in hospitalized patients (Chow et al., 2020; Docherty et al., 2020; Du et al., 2020; Guan et al., 2020; Huang et al., 2020a; Petrilli et al., 2020; Simonnet et al., 2020; Zhang et al., 2020; Zhou et al., 2020). Along with these clinical predictors, several immune and inflammatory markers predicting worst outcomes have been identified. Patients with severe COVID-19 develop life-threatening hyperinflammatory response to the virus, which is characterized by a high circulating levels of C-reactive protein (CRP) and interleukin (IL)-1β, IL-6, IL-18, tumor-necrosis factor, granulocyte-macrophage colony stimulating factor and interferon-γ (Mehta et al., 2020) (Ruan et al., 2020). This response is detrimental and has been shown to anticipate intubation and mortality. On the other hand, more severe forms of COVID-19 were associated with peripheral lymphocyte subset alteration, and patients with higher lymphocyte counts were less likely to have cytokine storm syndrome and may experience more harm than benefit when receiving corticosteroids (Lu et al., 2021; Wang et al., 2020b). Among others, lactic dehydrogenase (LDH), lymphocyte and CRP have been shown to have a role in the stratification of COVID-19 hospitalized patient outcomes (Brinati et al., 2020; Yan et al., 2020).
With the attempt to offer incremental value for patient stratification to these univariable predictors, machine learning (ML) models were used to achieve a more accurate outcome prediction to support decision making when dealing with critically ill COVID-19 patients (Brinati et al., 2020; Yan et al., 2020). However, these ML models showed the challenges of the prediction of outcomes, since in most cases the reported performance was found to be overestimated in the tested population, when the model was validated in an external one (Gupta et al., 2020).
In this study, we aimed to compare the role of routinely measured biomarkers of immunity, inflammation, and organ damage at hospital admission with preexisting comorbidities in eight different machine learning models to predict 60-day mortality. Importantly, to assess the generalizability our findings, we aimed to evaluate the models’ performance in an unrelated, external population from a different hospital.
2. Material and methods
2.1. Setting and data sources
We conducted an observational retrospective study collecting 2 independent cohorts, one from Poliambulanza Hospital of Brescia, Italy, referred as the “Brescia cohort”, and one from Policlinico San Marco, Hospital of Zingonia, Bergamo, Italy, referred as the “Zingonia cohort”. Study participants were consecutive adult (≥18 years old) patients with documented COVID 19 infection (i.e., tested by reverse-transcriptase-polymerase-chain-reaction (RT-PCR) assay for SARS-CoV-2) at admission in the internal medicine units, from March 1st to April 1st, 2020. Follow-up continued until death or May 31st, 2020. The electronic medical records of the patients recruited were accessed by the respective providers and data were manually abstracted, allowing a detailed case ascertainment.
The study is reported in accordance with transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD) guidance for external validation studies (Collins et al., 2015). This study was conducted in compliance with the Good Clinical Practice protocol and the Declaration of Helsinki principles and was approved by the local institutional review board.
2.2. Case ascertainment and variable assessed
Laboratory exams and clinical data were withdrawn and collected at day 1 of patient admission (baseline). Treatment and outcome data were collected during the follow up, from day 1 forward. Severe patients that at admission which were deemed to be hospitalized directly in ICU were not included. Patients with clear evidence of bacterial pneumonia (i.e. clear imaging signs of bacterial pneumonia according to the radiological report) were also excluded.
Patients were treated for COVID-19 according medical judgment, following slightly different protocol in the two hospital. In the Poliambulanza hospital of Brescia, treatment option included hydroxychloroquine (HCQ) 200 mg/day; oral prednisolone or equivalents: 5–25 mg/day. Antiviral therapy (oral Lopinavir/ritonavir, 400 mg/100 mg 2 times/day) were available, and biologic therapy (subcutaneous tocilizumab, 162 mg single shot, eventually repeated after 12 h if no response were observed). In the Zingonia hospital, antiviral and biologic therapy were not available, while HCQ and Prednisone were variably used. Both structures used antibiotics, in the majority of cases azithromycin 500 mg/day or oral cefixime: 400 mg/day. In general, patients started with azithromycin with or without HCQ, and cefixime was added after 5 days if no improvement was seen, in case of macrolide allergy or in addition to previous treatments in patients with age ≥65 or ≥1 comorbidities. Prednisolone and HCQ were added according to clinical judgment. Low-flow O2 therapy were prescribed to patients with oxygen saturation <93% at resting in ambient air documented by pulse oximeter (<88% for patient affected by COPD) or heart rate >22 beats per minute. Data on patients’ demographic, baseline comorbidities, presenting symptoms, oxygen saturation in ambient air at presentation, historical and current medication list, low-flow O2 prescription by the general practitioners, inpatient hospitalization, invasive and non-invasive ventilator use data, and death were collected.
2.3. Variables of interest and outcome
Categorical and continuous variables already shown to have a prognostic value for COVID-19 patients were collected. Blood hypertension (HTN), smoking (current or former) ≥10 pack/year, chronic obstructive pulmonary disease (COPD), cardiovascular diseases (coronary artery disease, heart failure, atrial fibrillation), diabetes, and chronic kidney disease (CKD) ≥ grade III (eGFR<60 mL/min/1.73 m2), were identified and recorded as present or absent according to chart review. Age and sex were also included. The most recent patient weight and height, during the 12 months preceding the admission to the hospital were collected, and BMI was calculated; following the World Health Organization definitions (World Health Organization, n.d.), obesity was defined as having a BMI ≥30 kg/m2 (World Health Organization, n.d.). A routine panel of laboratory exams were performed at patients’ admission, including complete blood cell count, LDH, CRP, serum creatinine (sCr), aspartate aminotransferase (AST), alanine aminotransferase (ALT), and international normalized ratio (INR).
The eight numerical variables included in the “Numerical” models were: age at diagnosis, lymphocytes percentage, platelets count, CRP, LDH, ALT, sCr, INR. In the “Numerical and Categorical” models the following eight categorical variables were added to the previous numerical ones: sex, obesity, diabetes, HTN, COPD, CKD, cardiovascular disease, smoking.
The primary endpoint of this study was 60-day mortality. The time from index date (hospital admission) to death was also collected. Other outcomes collected were the need of O2 therapy, the need of non-invasive ventilation (NIV), the need of intubation in intensive care unit (ICU) during the observation period.
2.4. Data curation and statistical analysis
Categorical data were summarized as percentages, significant differences between the 2 independent cohorts or associations of outcomes with clinical features were analyzed using the X2 test or Fisher exact tests, where appropriate. Continuous variables were presented as mean ± standard deviation (SD) or median and interquartile range (IQR), depending on normality demonstrated by Kolmogorov–Smirnov test. Comparisons were performed with Student's t-test for independent samples (2-tailed). Kaplan-Meier survival plots were constructed and the survival curves for groups were compared using a log-rank test. Patients without a primary endpoint event had their data censored on May 31st, 2020.
All the analyses were performed using JMP Pro package (SAS Institute Inc., Cary, North Carolina) and SAS System for Windows, version 9.4 (SAS Institute), and scikit-learn (Pedregosa et al., 2011). A p-value of <0.05 was considered statistically significant for all the analysis.
All data processing was performed using scikit-learn (Pedregosa et al., 2011). In case of missing data, missing values were imputed using the Iterative Imputer functions, that models each feature with missing values as a function of other features in a round-robin fashion (Buuren and Oudshoorn, 2011). The Brescia cohort was randomly divided into a training and a test set: 80% of the Brescia cohort served as training set and 20% as test set. All the data from the Zingonia cohort served as the external validation dataset. After the train/test split, we normalized the numerical features of the training data using the Standard Scaler function, that standardize each feature by removing the mean and scaling to unit variance; for categorical variables, we performed one-hot encoding using the One Hot Encoder function. We applied the transformations learned on the training set on the two test sets (Brescia and Zingonia).
2.5. ML models: development, training, evaluation and interpretability
We evaluated four machine learning classifiers:
-
-
Decision tree (DT) (Safavian and Landgrebe, 1991)
-
-
Random forest (RF) (Ho, 1998)
-
-
Gradient boosting (GBOOST) (Friedman, 2001)
-
-
Support vector machine (SVM) (Williams, 2003)
All classifiers were developed in Python using the scikit-learn library (Pedregosa et al., 2011). We trained each of the four classifiers using only numerical features, or a combination of numerical and categorical features, for a total of 8 models.
Prior its training, each classifier required the definition of a set of parameters that will drive the training process (hyperparameters). To find the best combination of hyperparameters for each model, we performed a grid search analysis using a nested five-fold cross-validation on the training set, using the mean F1-score obtained in the five folds as the metric to select the best performing hyperparameters; we then used the selected parameters to re-train each model from scratch on the whole training set. Given the imbalance between the two classes being predicted, we also tested different combination of class weights to help the models focusing on the minority class.
After cross-validation, a total of 8 best-performing models (two for each classifier) were selected and used to perform predictions on both test sets (Supplementary Fig. 1). We evaluated the models using precision, recall, F1-score, and AUROC. These were defined as follow:
-
-
Precision = True Positive/(True Positive + False Positive)
-
-
Recall = True Positive/(True Positive + False Negative)
-
-
F1 Score = 2 * [ (Precision * Recall)/(Precision + Recall)]
We used the python package shap (Lundberg et al., 2018) to interpret the output of our models, and have a sense of the features that most influence the models' predictions. Briefly, SHAP (SHapley Additive exPlanations) uses classic Shapley values from game theory and their extension to connect optimal credit allocation with local explanation and assigns each feature an importance value for a particular prediction, allowing interpreting the predictions of complex models (Lanctot et al., 2017). We used the shap package to obtain the summary plots that show which features contributed the most to the model's predictions. We performed this detailed analysis on the model that showed the best performance on the external validation set.
3. Results
3.1. Demographic, clinical, and laboratory features at admission, treatments and outcomes were significantly different in the two datasets
A total of 302 and 411 patients were included from the Brescia and Zingonia cohorts, respectively. We excluded 3 and 9 patients respectively because they did not meet the inclusion criteria (i.e., evidence of bacterial pneumonia). A total of 299 and 402 patients, respectively, were therefore included in the analysis. For the model development, we allocated 80% of the patients (n = 239) of the Brescia cohort as training set and 20% of the patients (n = 60) as test set. The complete Zingonia cohort (402 patients) was used as the external validation dataset. The study design is summarized in Supplementary Fig. 1.
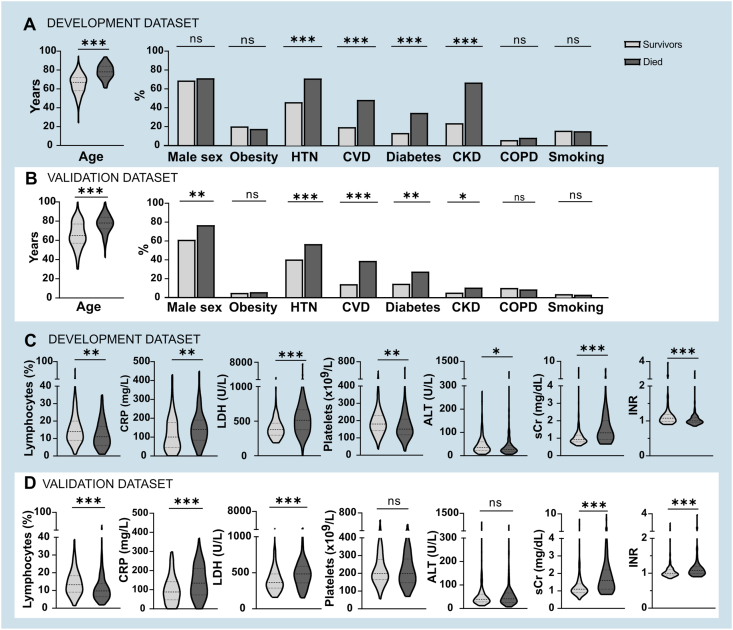
Baseline demographic and clinical features are described in Table 1. The frequency of obesity, smoking and the CKD ≥ III grade were higher in the development dataset (<0.0001). At admission, the proportions of patients with fever>37.5 °C and subjective dyspnea at resting were significantly lower in the development dataset, while the PaO2/FiO2 ratio were significantly higher in the validation dataset. Significant difference between the two datasets were observed for % of lymphocytes (p < 0.05), platelets count (p < 0.0001), alanine aminotransferase (p < 0.0001), international normalized ratio (p < 0.01), creatinine (p < 0.001), but not for white blood cell count, C-reactive protein, lactic dehydrogenase (LDH), aspartate amino transferase (p > 0.05). Treatment approach was significantly different as well: the frequency of antibiotics, HCQ and Prednisone was significantly lower in the validation cohort, and antiviral and biologic therapy was never used to treat these patients. As a consequence, the proportion of patients requiring NIV and the proportions of deaths were significantly higher in the validation cohort compared to the development one.
Table 1.
Baseline demographics, comorbidities, clinical features at presentation, treatments and outcomes of hospitalized patients with COVID-19 in the development dataset and external validation dataset. The variables used as input variables of the models are marked asb. Comparisons were performed with either X2 test or Fisher exact tests for categorical variables, and Student's t-test for continuous variables.
| Characteristics | Development dataset | External validation dataset | p value |
|---|---|---|---|
| N. | 299 | 402 | |
| Demographics | |||
| Age at diagnosis,bmean (±SD) | 68.79 (11.65) | 70.21 (13.17) | 0.1384 |
| Male sex,b% (number) | 69.57% (208) | 67.41% (271) | 0.5446 |
| Obesity,bBMI ≥ 30 kg/m2, % (number) | 19.40% (58) | 5.22% (21) | <.0001 |
| Ethnicity, white,% (number) | 99.33% (297) | 100% (402) | 0.1816 |
| Smoking,b( ≥ 10 pack/year), current or former, % (number) | 15.39% (46) | 3.48% (14) | <.0001 |
| Comorbidities | |||
| Diabetes,b% (number) | 19.39% (58) | 19.90% (80) | 0.8686 |
| HTN,b% (number) | 53.51% (160) | 46.77% (188) | 0.0773 |
| Cardiovascular Diseases,b% (number) | 28.09% (84) | 24.13% (97) | 0.2356 |
| CKD ≥ stage III,b % (number) | 36.12% (108) | 7.46% (30) | <.0001 |
| COPD,b % (number) | 6.35% (19) | 9.70 (39) | 0.1116 |
| Cancer (active or < 5 years), % (number) | 5.69% (17) | 6.22% (25) | 0.7686 |
| Previous stroke,% (number) | 3.34% (10) | 0.50% (2) | 0.0041 |
| Clinical presentation | |||
| Fever, temperature>37.5 °C,% (number) | 85.62% (256) | 98.01% (394) | <.0001 |
| Dry cough,% (number) | 51.51% (154) | NA | – |
| Dyspnea at resting,% (number) | 50.17% (150) | 96.52% (388) | <.0001 |
| Myalgias,% (number) | NA | 95.27% (383) | – |
| Gastrointestinal symptoms, % (number) | 6.02% (18) | 4.48% (18) | 0.3602 |
| Syncope/Presyncope, % (number) | 4.01% (12) | NA | – |
| Altered mental status, % (number) | 2.68% (8) | NA | – |
| Evidence of pneumonia at thoracic imaging,a % (number) | 96.66% (289) | 95.52% (384) | 0.4486 |
| PaO2/FiO2Ratio | 248.9 (73.6) | 355.6 (116.1) | <.0001 |
| Laboratory Characteristics | |||
| WBC, mean (±SD) | 7.89 (4.35) | 8.13 (4.32) | 0.4637 |
| Lymphocytes,b % of WBC, mean (±SD) | 14.75 (9.45) | 13.28 (7.73) | 0.0235 |
| PLT,b mean (±SD) | 187.000 (82.000) | 225.000 (98.000) | <.0001 |
| CRP,b mean (±SD) | 126.3 (88.58) | 122.8 (95.7) | 0.6260 |
| LDH,b median [25–75%IQR] | 395 [305.75–530] | 405 [304–524] | 0.9897 |
| AST, median [25–75%IQR] | 53 [38–75] | 50 [36–74.25] | 0.1225 |
| ALT,b median [25–75%IQR] | 32 [20–57] | 41 [27.75–62] | <.0001 |
| INR,b median [25–75%IQR] | 1.01 [0.96–1.12] | 1.04 [0.99–1.12] | 0.0018 |
| sCr,b (mg/dL), mean (±SD) | 1.26 (0.94) | 1.53 (1.13) | 0.0011 |
| Treatments | |||
| Antibiotics, % (number) | 83.28% (249) | 28.61% (115) | <.0001 |
| HCQ, % (number) | 22.75% (68) | 5.72% (23) | <.0001 |
| Lopinavir/ritonavir, % (number) | 21.07% (63) | 0% (0) | <.0001 |
| Prednisone, % (number) | 34.45% (103) | 0.75% (3) | <.0001 |
| Tocilizumab, % (number) | 4.01% (12) | 0% (0) | <.0001 |
| Outcomes | |||
| O2 therapy,b % (number) | 48.16% (144) | 35.57% (143) | 0.008 |
| NIV,c % (number) | 13.04% (39) | 19.65% (79) | 0.0207 |
| ICU with intubation,d % (number) | 10.03% (30) | 10.70% (43) | 0.7762 |
| Death, % (number) | 29.10% (87) | 39.55% (159) | 0.0041 |
Abbreviations: HTN: Blood hypertension, BMI: body mass index; Cardiovascular Disease: chronic heart failure, myocardial infarction, atrial fibrillation; CKD: chronic kidney disease, stage III correspond to estimated glomerular filtration rate <60 mL/min; COPD: Chronic obstructive pulmonary disease; WBC: White blood cells, PLT: platlets, CRP: C-reactive protein, LDH: lactic dehydrogenase, AST: aspartate aminotransferase; ALT: alanine aminotransferase; INR: international normalized ratio; sCr: serum Creatinine;; Antibiotics: oral Cefixime: 400 mg/day for ≥5 days; oral Azithromycin 500 mg/day for ≥5 days; oral Claritromicin 250 mg x 2/day for ≥5 days endovenoous Ceftriaxon 2 g/day for ≥5 days; endovenous piperacillina/tazobactam 4.5 mg x 3 or 4/day for ≥5 da; oral or endovenous Levofloxacin 500 mg/day for ≥5 days. HCQ: hydroxychloroquine, 200 mg 12 h apart for the first 2 doses, then 200 mg/day for ≥5 days; Oral Prednisolone or equivalents: range 5–25 mg/day for ≥5 days. NIV: Non-invasive ventilation; ICU: intesive care unit. SD = standard deviation.
Thoracic X-ray as a screening test, followed by CT-scan in doubtful cases.
O2 therapy: administered when saturation were ≤92% at resting in ambient air; required nasal canula or Venturi mask; NIV: required non-inviasive ventilation.
NIV: patients non-responsive to high-flow O2-therapy, requiring.
ICU with intubation: required intensive care unit hospitalization with intubation.
3.2. Baseline clinical and laboratory features in survivors versus non-survivors were similarly distributed in the two datasets
The features included in the models are represented by outcomes for each dataset in Fig. 1. For clinical variables, age of the patients and the proportion of HTN, CVD, diabetes, and CKD were significantly higher in those that died during the 60-day observation period in both the datasets (Fig. 1A for the development dataset, Fig. 1B for the validation dataset). Similarly, lymphocyte percentage, CRP, LDH and sCr levels were higher in those patients that met the primary outcome (Fig. 1C for the development dataset, Fig. 1D for the validation dataset). Data imputation was very low (<0.01% in total, single variable ranging from 0 to 0.04%).
Fig. 1.
Clinical and laboratory features of the development dataset (A an C, in the blue panels) and of the validation dataset (B and D, in the white panels) by outcomes. * <0.05, ** <0.01, ***<0.001. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.3. Model training and evaluation in the development dataset and performance in the external validation set
The 8 models were developed and evaluated using F1-score and AUROC (Supplementary Table 1). When predicting the 60-days mortality after hospitalization in the test set, the performance was heterogeneous among the different models. Model 3 (GBOOST numerical) achieved the highest mean F1-score (weighted avg 0.83) followed by Model 7 (SVM, numerical; weighted avg 0.79), Model 8 (SVM numerical and categorical, weighted avg 0.78) and Model 5 (RF Numerical, weighted avg 0.78), Model 4 (GBOOST Numerical and Categorical, weighted avg 0.74), Model 2 (DT Numerical and Categorical, weighted avg 0.73) and Model 6 (RF Numerical and Categorical, weighted avg 0.73). Model 1 performed badly (DT Numerical, weighted avg 0.49). Compared to the test set, the training set showed overall lower F1 scores (Table 2).
Table 2.
Mean F1-score and AUROC obtained in the cross-validation on the training set (N = 239).
| F1-score (mean ± SD) | AUROC (mean ± SD) | |
|---|---|---|
| Model 1: Decision Tree Numerical | 0.60 ± 0.06 | 0.74 ± 0.11 |
| Model 2: Decision Tree Numerical and Categorical | 0.68 ± 0.07 | 0.83 ± 0.07 |
| Model 3: GBOOST Numerical | 0.66 ± 0.06 | 0.84 ± 0.05 |
| Model 4: GBOOST Numerical and Categorical | 0.69 ± 0.04 | 0.88 ± 0.04 |
| Model 5: Random Forest Numerical | 0.69 ± 0.15 | 0.86 ± 0.07 |
| Model 6: Random Forest Numerical and Categorical | 0.69 ± 0.05 | 0.87 ± 0.04 |
| Model 7: SVM Numerical | 0.72 ± 0.05 | 0.87 ± 0.04 |
| Model 8: SVM Numerical and Categorical | 0.68 ± 0.03 | 0.87 ± 0.03 |
Compared to the internal test set, the mean F1 scores on the external validation set were lower for all the models (Supplementary Table 2). Model 8 (SVM numerical and categorical) achieved the highest mean F1 score (weighted avg 0.72) followed by Model 6 (RF categorical, weighted avg 0.71) and Model 7 (SVM, numerical; weighted avg 0.70). All the other Models had a mean F1 score between 0.60 and 0.67, except Model 1, which performed worse (DT Numerical, weighted avg 0.49). Overall, although less accurate, the performance of the 8 tested models on the external validation dataset were similar to that of the holdout test dataset, indicating that the models capture the key predictors of patient mortality.
3.4. Immune and laboratory features at hospital admission impacted on mortality prediction more than concomitant clinical comorbidities or hyperinflammation
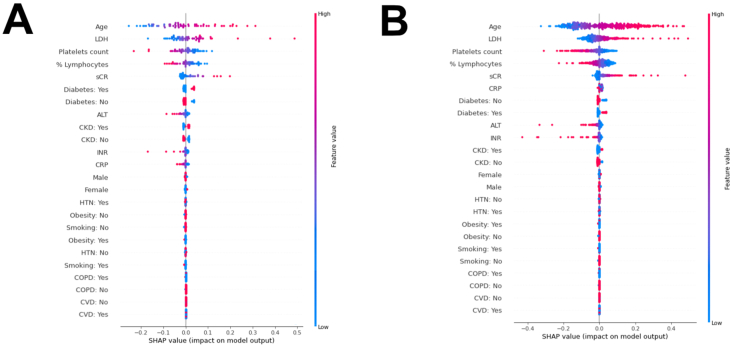
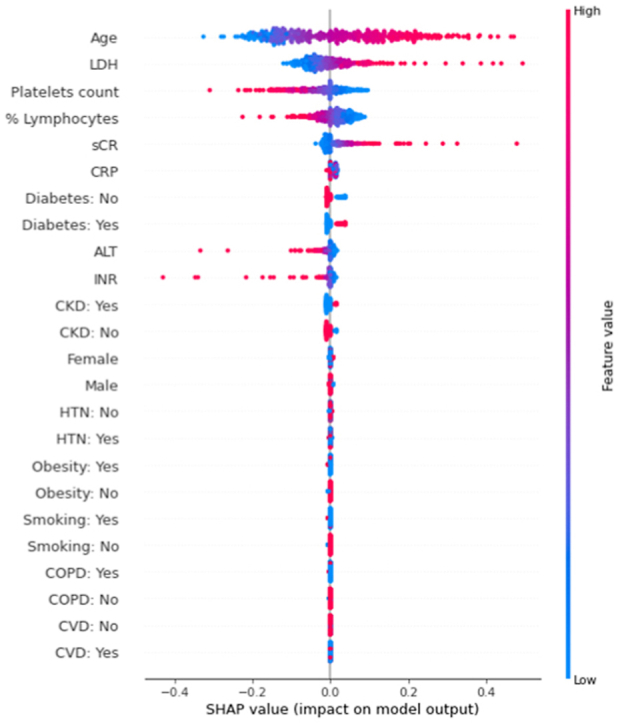
To make these ML models explainable in terms of the weight of each individual feature tested (i.e. age, sex, patient preexisting comorbidities, immune e laboratory parameters at hospital admission) for patient survival, we performed the shap analysis on all the 8 models in both the development test set and the external validation dataset (Supplementary Figs. 2 and 3). The shap analysis automatically orders the variables used based on the impact of each variable on the model output. In all models and of both the datasets, immune features (%lymphocytes, platelets), cellular damage (LDH in particular) substantially impacted on the models ranking constantly among the most influencing features. In all the models but one, age impacted significantly (first in the ranking), while the effect of sCr and CRP varied among the different models. Beside age, the weight of the preexisting comorbidities was substantially lower compared to laboratory features.
Given its better performance on the external validation set, we focused our further evaluations on Model 8, a SVM classifier that uses both numerical and categorical variables; the hyperparameters for this model are listed in the Supplementary Table 3. This model had an AUROC for 60-day mortality of 0.83 ± 0.06 in the holdout test set, and an AUROC of 0.79 ± 0.02 in the external validation dataset (Supplementary Fig. 3). When considering the contribution of each of the features in this model, in both the development test set and the external validation dataset (Fig. 2A and B, respectively), age at admission had the greatest impact on the predictions, with older age driving the predictions towards deaths and younger age driving the predictions towards survival. This was followed by LDH (with higher levels driving prediction towards death), platelets count and %lymphocytes (with lower levels driving prediction towards death). The weight of these variables on the model predictions was highly consistent in both the datasets. Serum creatinine had also a significant weight in both dataset (with higher levels driving prediction towards death), while CRP did only the external validation dataset.
Fig. 2.
The impact of the input features on predictions. The shap analysis on the model with the best performance (Model 8), in the development test set (A) and the external validation dataset (B). The model includes both continuous and binary input features. Continuous features vary from low to high values, whereas binary features are either present or absent. Each dot represents the impact of a feature on the mortality prediction for one patient at entrance. The color indicates the level of contribution of each variable (with red indicating a higher impact on the prediction) and the direction the prediction towards death (right) or survival (left). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
Early prediction of COVID-19 in-hospital mortality relies usually on preexisting comorbidities and is rarely reproducible in independent cohorts of hospitalized patients. Our findings showed that immune and cellular damage markers at hospital admission impacted on mortality prediction substantially more than the presence of concomitant clinical comorbidities or systemic inflammation features (such as high CRP), and these results were reproducible in an independent population with different baseline features and outcomes. Numerous articles on hospitalized patients affected by COVID-19 showed that diabetes, hypertension, malignancy, COPD, obesity and older age are risk factors for severe disease and poor outcome in hospitalized patients (Chow et al., 2020; Docherty et al., 2020; Du et al., 2020; Guan et al., 2020; Huang et al., 2020a; Petrilli et al., 2020; Simonnet et al., 2020; Zhang et al., 2020; Zhou et al., 2020), while the role of immune and other laboratory parameters in mortality prediction were not reported so often. Patient with severe COVID-19 develop life-threatening hyperinflammatory response to the virus, characterized by a high circulating levels of CRP and IL-1β, IL-6, IL-18, tumor-necrosis factor, granulocyte-macrophage colony stimulating factor and interferon-γ. However, the attempt of blocking hyperinflammation with available agents inhibiting IL-6 (tocilizumab, sarilumab) and IL-1 (anakinra) has led to conflicting and ultimately marginal results in both clinical trials and real word settings (Campochiaro et al., 2020; Cavalli et al., 2020; Della-Torre et al., 2020; Guaraldi et al., 2020; Salvarani et al., 2021; Stone et al., 2017), suggesting that these agents may have a limited role in controlling the disease. On the other hand, more severe forms of COVID-19 were associated with peripheral lymphocyte subset alteration, and patients with higher lymphocyte counts were less likely to have cytokine storm syndrome and may experience more harm than benefit when receiving corticosteroids (Lu et al., 2021; Wang et al., 2020b). Consistently, CD8+ T cells tended to be an independent predictor for COVID-19 severity and treatment efficacy (Wang et al., 2020). In other studies, markers of cellular damage and in particular LDH has been shown to have a role in the stratification of COVID-19 hospitalized patient outcomes (Brinati et al., 2020; Yan et al., 2020). In our study, beside age, immune and laboratory features at hospital admission impacted on mortality prediction substantially more than the presence of concomitant clinical comorbidities or hyperinflammation. Taken altogether, we can speculate that this probably reflects the effect of the virus at the very beginning of disease onset, while the prediction of the risk may change dynamically during the disease and hospitalization course, i.e. as in ICU cohorts in which comorbidities may impact much more on patient survival or life-threatening hyperinflammatory response to the virus is usually reflected by higher circulating levels of CRP, IL-1β, IL-6, IL-18, and interferon-γ. Of course, other factors may be involved, as for example the genetic background of the patients or the virus genetic variant affecting patients.
In a rapidly evolving field like the COVID-19 research, discoveries accumulate rapidly. The strength of our approach is that it allows to interpret the clinical and laboratory variables imputed to perform a prediction, possibly favoring the selection of biomarker candidates for prospective trials. From this perspective, these models showed their potential as discovery tools rather than clinical tools, and their interpretable features makes them great candidates for this application. One thing to consider is the feasibility of incorporating the recent discoveries in a model like ours, that has built by imputing data from clinical routine. Recently, new potential immunologic biomarkers with prognostic value for COVID-19, such as mucosal-associated invariant T (MAIT) cells (Flament et al., 2021) or circulating NKT cells (Kreutmair et al., 2021), have been discovered. The methodology we used, i.e. the ML modelling, can be easily applied to these variables, contributing to reveal the immune dysregulation occurring during COVID-19 infection and with potential prediction of the outcome. The limitation of these ML modelling is that large numbers of patients are usually required to avoid overfitting.
The early prediction of the prognosis of COVID-19 patients is of global interest. Much effort has been undertaken to understand which patients are at higher risk of deaths, in order to intensify treatment and care in these individuals. The growing body of literature offers many examples of studies aiming to stratify COVID-19 patients for early mortality prediction, by means of ML algorisms (Brinati et al., 2020; Gupta et al., 2020; Yan et al., 2020) or more conventional regression models (Chow et al., 2020; Docherty et al., 2020; Du et al., 2020; Guan et al., 2020; Huang et al., 2020a; Petrilli et al., 2020; Simonnet et al., 2020; Zhang et al., 2020; Zhou et al., 2020). Since none of the clinical or laboratory variables taken singularly was able to indisputably stratify the outcome of these patients at admission, several ML models were published. ML models have shown a great potential in predicting COVID-19 outcome and perform COVID-19 diagnosis (Chow et al., 2020; Docherty et al., 2020; Du et al., 2020; Geleris et al., 2020; Grasselli et al., 2020; Guan et al., 2020a, 2020b; Hamer et al., 2020; Huang et al., 2020a, 2020b; Petrilli et al., 2020; Richardson et al., 2020; Simonnet et al., 2020; Wang et al., 2020; Zhang et al., 2020; Zhou et al., 2020). A common limitation of ML models is that they might overfit to the population used to develop them, resulting in poorer performance when tested in different ones. The issue of overfitting has recently emerged also for COVID-19, since 22 published models, specifically developed for COVID-19 or routinely used in the clinical activity to assess the severity of pneumonia or general status (e.g. CURB65, NEWS2, etc.) performed sub-optimally when validated in an external cohorts (Gupta et al., 2020). It should also be noticed that most of these models were developed in a single center and not tested in an external population during the publication process, and that AUROC was used to assess their net benefit, both potentially leading to imprecision. Our work is unique since we had the opportunity work on 2 independent datasets, one used for development and one for external validation. This conferred robustness to our analysis. We developed and validated 8 models to predict 60-day mortality in two independent cohorts of hospitalized patients with COVID-19. We evaluated our models using the F1 score, a metric that considers both false positives and false negatives into account, and it is more accurate in the case of an uneven class distribution of the outcome, as in our case. Model 8 (SVM Numerical and Categorical) showed the best F-1 score on the external validation dataset, indicating the best performance, which corresponded to an AUROC of 0.79. To ensure comparability with previous ML models (Gupta et al., 2020), we calculated AUROC for Model 8 in the external validation population. The average of AUROCs were 0.60 of all the previous models when assessing mortality, with the highest being 0.76 for the models REMS and Xie (Gupta et al., 2020). Of note, the reason why nobody so far obtained a valid and reproducible prediction might be that the conventional parameters used for the modeling are not sufficient, and maybe more-disease specific features are needed to predict mortality, and this might be particularly true for patient preexisting comorbidities. Overall, even if the ultimate goal of ML modelling is the development of a risk prediction model at an individual patient level, collectively taken, most of these models failed the predictions in clinical practice. Although ML tools developed to assist in the management of COVID-19 have demonstrated high potential, the great majority of them (if not all) are not routinely used to support clinical decision making. The reasons might be many, i.e. the incapacity of the models to account for the changing nature of the predicted outcomes, or some of the input features do not have the anticipated impact on the predictions because rarer or less discriminating than expected. In this sense, we are aware of these limitations of ML, and to mitigate these potential issues we tested our models in a second, independent cohort of patients. Altogether, we believe that the best use of these ML models is probably to drive research questions, expand our knowledge of the disease, and to identify potential biomarkers by focusing on the variables that have shown to be the most important in the models’ predictions, to be tested in prospective studies. It is important to underline that this is possible only thanks to the complementary interpretability tools, that serves as agents that we can use to debug our models.
Finally, ML models tend to suffer whenever there is a change in either the input data or the population (i.e. population specific characteristics, like age and other demographics, comorbidities, etc.), but also changes in clinical practice, for example with the introduction of new drugs or therapeutic schemes. A possible application of our approach is that, given the interpretability of our models, we could test how they “react” to a change in clinical practice (e.g., will the same variables be important for prognosis?). In conclusion, while we wouldn't advise introducing these models in the clinical practice yet, they could be used experimentally to predict how patients respond to new therapies and, in general, to the improvement in the clinical management of these patients.
This study has some strengths and limitations. Compared to other previous papers, our work is characterized by a very low percentage of data imputation, a clear interpretability and an independent external validation dataset which increases the methodological rigor of our study and allows to test the reproducibility of the models. Most if not all the previous cohorts used for modeling were single-center, retrospective cohorts. Second, we used the nested cross validation and used mean F1 score instead of AUROC to select the models, contributing to the methodological rigor our analyses. A weakness of the current study is the observational retrospective design and the extraction of data from non-standardized medical records cannot completely exclude classification error. In addition, even if missing data were minimal (<5%), multiple imputation was performed. Laboratory data were collected only at baseline, and not longitudinal data were retrieved, likely reducing the performance of the tested models. However, most prognostic scores are intended to predict outcomes at the point of hospital admission.
In conclusion, beside age, in our ML models immune and laboratory features at hospital admission impacted on mortality prediction substantially more than the presence of concomitant clinical comorbidities or the presence of a systemic inflammatory status, and these findings were highly reproducible in independent populations. We can speculate that this probably reflects the effect of the virus at the very beginning of disease onset, while the prediction of the risk may change dynamically during the disease course. Future clinical and basic science studies are needed to have a better understanding of the immune and cellular perturbations that occurs during COVID-19, which may help to develop reliable and reproducible prognostic models for COVID-19.
Disclosure and author contributions
The authors have no financial or non-financial potential conflicts of interest to declare related to this project. Dr. Lombardi, Dr. Bigni, Dr. Roca, Dr. Ferrandina, Dr. Bertozzi, Dr. Franzin, Dr. Vivaldi, Dr. Del Poggio and Dr. D'Alessio acquired the data; Dr. Lombardi and conceived the study, while Dr. Conte and Dr. Berti designed the study. Dr. Lombardi and Dr. D'Alessio had full access to the data in the study and takes responsibility for the integrity of the data; Dr. Conte and Dr. Berti take responsibility for the accuracy of the data analysis and both drafted the article. All the authors were involved in the writing and editing of the manuscript, and approved the final version to be published.
Funding
The study was not supported by grants from any organization or institution.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We sincerely thank Dr. Renzo Rozzini, Chief of the Department of Geriatrics, COVID-19 Unit, Fondazione Poliambulanza Istituto Ospedaliero, Brescia, Italy.
This article is dedicated to all the Italian doctors and health personnel that sacrificed their own lives to save patient lives. We wish to honor their competence, braveness and generosity.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crimmu.2021.09.001.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Study overview and design.
Model 8 (SVM Numerical and Categorical). The area under the ROC curve (AUROC).
The shap analysis on all the 8 models in the development test set.
The shap analysis on all the 8 models in the external validation dataset.
References
- Brinati D., Campagner A., Ferrari D., Locatelli M., Banfi G., Cabitza F. Detection of COVID-19 infection from routine blood exams with machine learning: a feasibility study. J. Med. Syst. 2020;44 doi: 10.1007/s10916-020-01597-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buuren S. van, Oudshoorn K. MICE: multivariate imputation by chained equations in R. J. Stat. Softw. 2011;45 [Google Scholar]
- Campochiaro C., Della-Torre E., Cavalli G., De Luca G., Ripa M., Boffini N., Tomelleri A., Baldissera E., Rovere-Querini P., Ruggeri A., Monti G., De Cobelli F., Zangrillo A., Tresoldi M., Castagna A., Dagna L. Efficacy and safety of tocilizumab in severe COVID-19 patients: a single-centre retrospective cohort study. Eur. J. Intern. Med. 2020;76 doi: 10.1016/j.ejim.2020.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli G., De Luca G., Campochiaro C., Della-Torre E., Ripa M., Canetti D., Oltolini C., Castiglioni B., Tassan Din C., Boffini N., Tomelleri A., Farina N., Ruggeri A., Rovere-Querini P., Di Lucca G., Martinenghi S., Scotti R., Tresoldi M., Ciceri F., Landoni G., Zangrillo A., Scarpellini P., Dagna L. Interleukin-1 blockade with high-dose anakinra in patients with COVID-19, acute respiratory distress syndrome, and hyperinflammation: a retrospective cohort study. Lancet Rheumatol. 2020;2 doi: 10.1016/S2665-9913(20)30127-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow N., Fleming-Dutra K., Gierke R., Hall A., Hughes M., Pilishvili T., Ritchey M., Roguski K., Skoff T., Ussery E. Preliminary estimates of the prevalence of selected underlying health conditions among patients with coronavirus disease 2019 - United States, February 12-March 28, 2020. Morb. Mortal. Wkly. Rep. 2020 doi: 10.15585/MMWR.MM6913E2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins G.S., Reitsma J.B., Altman D.G., Moons K.G.M. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. BMJ. 2015;350 doi: 10.1136/bmj.g7594. [DOI] [PubMed] [Google Scholar]
- Della-Torre E., Campochiaro C., Cavalli G., De Luca G., Napolitano A., La Marca S., Boffini N., Da Prat V., di Terlizzi G., Lanzillotta M., Rovere Querini P., Ruggeri A., Landoni G., Tresoldi M., Ciceri F., Zangrillo A., De Cobelli F., Dagna L., Angelillo P., Assanelli A., Baldissera E., Bozzolo E.P., Calvisi S., Canetti D., Cariddi A., Castagna A., Cicalese M.P., Di Lucca G., Farina N., Fazio M., Mancuso G., Monti G., Moroni L., Oltolini C., Palumbo D., Ripa M., Rovere-Querini P., Sartorelli S., Scarpellini P., Spessot M., Tomelleri A. Interleukin-6 blockade with sarilumab in severe COVID-19 pneumonia with systemic hyperinflammation: an open-label cohort study. Ann. Rheum. Dis. 2020;79 doi: 10.1136/annrheumdis-2020-218122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Docherty A.B., Harrison E.M., Green C.A., Hardwick H.E., Pius R., Norman L., Holden K.A., Read J.M., Dondelinger F., Carson G., Merson L., Lee J., Plotkin D., Sigfrid L., Halpin S., Jackson C., Gamble C., Horby P.W., Nguyen-Van-Tam J.S., Dunning J., Openshaw P.J., Baillie J.K., Semple M.G. medRxiv; 2020. Features of 16,749 Hospitalised UK Patients with COVID-19 Using the ISARIC WHO Clinical Characterisation Protocol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du R.H., Liang L.R., Yang C.Q., Wang W., Cao T.Z., Li M., Guo G.Y., Du J., Zheng C.L., Zhu Q., Hu M., Li X.Y., Peng P., Shi H.Z. Predictors of mortality for patients with COVID-19 pneumonia caused by SARSCoV- 2: a prospective cohort study. Eur. Respir. J. 2020 doi: 10.1183/13993003.00524-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flament H., Rouland M., Beaudoin L., Toubal A., Bertrand L., Lebourgeois S., Rousseau C., Soulard P., Gouda Z., Cagninacci L., Monteiro A.C., Hurtado-Nedelec M., Luce S., Bailly K., Andrieu M., Saintpierre B., Letourneur F., Jouan Y., Si-Tahar M., Baranek T., Paget C., Boitard C., Vallet-Pichard A., Gautier J.F., Ajzenberg N., Terrier B., Pène F., Ghosn J., Lescure X., Yazdanpanah Y., Visseaux B., Descamps D., Timsit J.F., Monteiro R.C., Lehuen A. Outcome of SARS-CoV-2 infection is linked to MAIT cell activation and cytotoxicity. Nat. Immunol. 2021;22 doi: 10.1038/s41590-021-00870-z. [DOI] [PubMed] [Google Scholar]
- Friedman J.H. Greedy function approximation: a gradient boosting machine. Ann. Stat. 2001;29 doi: 10.1214/aos/1013203451. [DOI] [Google Scholar]
- Geleris J., Sun Y., Platt J., Zucker J., Baldwin M., Hripcsak G., Labella A., Manson D.K., Kubin C., Barr R.G., Sobieszczyk M.E., Schluger N.W. Observational study of hydroxychloroquine in hospitalized patients with covid-19. N. Engl. J. Med. 2020 doi: 10.1056/nejmoa2012410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasselli G., Zangrillo A., Zanella A., Antonelli M., Cabrini L., Castelli A., Cereda D., Coluccello A., Foti G., Fumagalli R., Iotti G., Latronico N., Lorini L., Merler S., Natalini G., Piatti A., Ranieri M.V., Scandroglio A.M., Storti E., Cecconi M., Pesenti A. Baseline characteristics and outcomes of 1591 patients infected with SARS-CoV-2 admitted to ICUs of the lombardy region. Italy. JAMA - J. Am. Med. Assoc. 2020 doi: 10.1001/jama.2020.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Wei-jie, Liang W., Zhao Y., Liang H., Chen Z., Li Y. Comorbidity and its impact on 1590 patients with covid-19 in China: a nationwide analysis. The European respiratory journal. 2020 doi: 10.1183/13993003.00547-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W., Ni Z., Hu Y., Liang W., Ou C., He J., Liu L., Shan H., Lei C., Hui D.S.C., Du B., Li L., Zeng G., Yuen K.Y., Chen R., Tang C., Wang T., Chen P., Xiang J., Li S., Wang J.L., Liang Z., Peng Y., Wei L., Liu Y., Hu Y.H., Peng P., Wang J.M., Liu J., Chen Z., Li G., Zheng Z., Qiu S., Luo J., Ye C., Zhu S., Zhong N. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guaraldi G., Meschiari M., Cozzi-Lepri A., Milic J., Tonelli R., Menozzi M., Franceschini E., Cuomo G., Orlando G., Borghi V., Santoro A., Di Gaetano M., Puzzolante C., Carli F., Bedini A., Corradi L., Fantini R., Castaniere I., Tabbì L., Girardis M., Tedeschi S., Giannella M., Bartoletti M., Pascale R., Dolci G., Brugioni L., Pietrangelo A., Cossarizza A., Pea F., Clini E., Salvarani C., Massari M., Viale P.L., Mussini C. Tocilizumab in patients with severe COVID-19: a retrospective cohort study. Lancet Rheumatol. 2020;2 doi: 10.1016/S2665-9913(20)30173-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R.K., Marks M., Samuels T.H.A., Luintel A., Rampling T., Chowdhury H., Quartagno M., Nair A., Lipman M., Abubakar I., van Smeden M., Wong W.K., Williams B., Noursadeghi M. Systematic evaluation and external validation of 22 prognostic models among hospitalised adults with COVID-19: an observational cohort study. Eur. Respir. J. 2020;56 doi: 10.1183/13993003.03498-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer M., Kivimäki M., Gale C.R., Batty G.D. Lifestyle risk factors, inflammatory mechanisms, and COVID-19 hospitalization: a community-based cohort study of 387,109 adults in UK. Brain. Behav. Immun. 2020;87 doi: 10.1016/j.bbi.2020.05.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho T.K. The random subspace method for constructing decision forests. IEEE Trans. Pattern Anal. Mach. Intell. 1998;20 doi: 10.1109/34.709601. [DOI] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020 doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreutmair S., Unger S., Núñez N.G., Ingelfinger F., Alberti C., De Feo D., Krishnarajah S., Kauffmann M., Friebel E., Babaei S., Gaborit B., Lutz M., Jurado N.P., Malek N.P., Goepel S., Rosenberger P., Häberle H.A., Ayoub I., Al-Hajj S., Nilsson J., Claassen M., Liblau R., Martin-Blondel G., Bitzer M., Roquilly A., Becher B. Distinct immunological signatures discriminate severe COVID-19 from non-SARS-CoV-2-driven critical pneumonia. Immunity. 2021;54 doi: 10.1016/j.immuni.2021.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanctot M., Zambaldi V., Gruslys A., Lazaridou A., Tuyls K., Pérolat J., Silver D., Graepel T. A unified game-theoretic approach to multiagent reinforcement learning. Advances in Neural Information Processing Systems. 2017 [Google Scholar]
- Lu C., Liu Y., Chen B., Yang H., Hu H., Liu Y., Zhao Y. Prognostic value of lymphocyte count in severe COVID-19 patients with corticosteroid treatment. Signal Transduct. Target. Ther. 2021 doi: 10.1038/s41392-021-00517-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg S.M., Nair B., Vavilala M.S., Horibe M., Eisses M.J., Adams T., Liston D.E., Low D.K.W., Newman S.F., Kim J., Lee S.I. Explainable machine-learning predictions for the prevention of hypoxaemia during surgery. Nat. Biomed. Eng. 2018;2 doi: 10.1038/s41551-018-0304-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020 doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onder G., Rezza G., Brusaferro S. Case-fatality rate and characteristics of patients dying in relation to COVID-19 in Italy. JAMA - J. Am. Med. Assoc. 2020 doi: 10.1001/jama.2020.4683. [DOI] [PubMed] [Google Scholar]
- Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V., Vanderplas J., Passos A., Cournapeau D., Brucher M., Perrot M., Duchesnay É. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 2011;12 [Google Scholar]
- Petrilli C.M., Jones S.A., Yang J., Rajagopalan H., O'Donnell L.F., Chernyak Y., Tobin K., Cerfolio R.J., Francois F., Horwitz L.I. medRxiv; 2020. Factors Associated with Hospitalization and Critical Illness Among 4,103 Patients with COVID-19 Disease in New York City. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson S., Hirsch J.S., Narasimhan M., Crawford J.M., McGinn T., Davidson K.W., Barnaby D.P., Barnaby D.P., Becker L.B., Chelico J.D., Cohen S.L., Cookingham J., Coppa K., Diefenbach M.A., Dominello A.J., Duer-Hefele J., Falzon L., Gitlin J., Hajizadeh N., Harvin T.G., Hirschwerk D.A., Kim E.J., Kozel Z.M., Marrast L.M., Mogavero J.N., Osorio G.A., Qiu M., Zanos T.P. Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the New York city area. JAMA - J. Am. Med. Assoc. 2020 doi: 10.1001/jama.2020.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Q., Yang K., Wang W., Jiang L., Song J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020 doi: 10.1007/s00134-020-05991-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safavian S.R., Landgrebe D. A survey of decision tree classifier methodology. IEEE Trans. Syst. Man Cybern. 1991;21 doi: 10.1109/21.97458. [DOI] [Google Scholar]
- Salvarani C., Dolci G., Massari M., Merlo D.F., Cavuto S., Savoldi L., Bruzzi P., Boni F., Braglia L., Turrà C., Ballerini P.F., Sciascia R., Zammarchi L., Para O., Scotton P.G., Inojosa W.O., Ravagnani V., Salerno N.D., Sainaghi P.P., Brignone A., Codeluppi M., Teopompi E., Milesi M., Bertomoro P., Claudio N., Salio M., Falcone M., Cenderello G., Donghi L., Del Bono V., Colombelli P.L., Angheben A., Passaro A., Secondo G., Pascale R., Piazza I., Facciolongo N., Costantini M. Effect of tocilizumab vs standard care on clinical Worsening in patients hospitalized with COVID-19 pneumonia: a randomized clinical trial. JAMA Intern. Med. 2021;181 doi: 10.1001/jamainternmed.2020.6615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonnet A., Chetboun M., Poissy J., Raverdy V., Noulette J., Duhamel A., Labreuche J., Mathieu D., Pattou F., Jourdain M., Caizzo R., Caplan M., Cousin N., Duburcq T., Durand A., El kalioubie A., Favory R., Garcia B., Girardie P., Goutay J., Houard M., Jaillette E., Kostuj N., Ledoux G., Mathieu D., Sophie Moreau A., Niles C., Nseir S., Onimus T., Parmentier E., Préau S., Robriquet L., Rouze A., Six S., Verkindt H. High prevalence of obesity in severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) requiring invasive mechanical ventilation. Obesity. 2020 doi: 10.1002/oby.22831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone J.H., Tuckwell K., Dimonaco S., Klearman M., Aringer M., Blockmans D., Brouwer E., Cid M.C., Dasgupta B., Rech J., Salvarani C., Schett G., Schulze-Koops H., Spiera R., Unizony S.H., Collinson N. Trial of tocilizumab in giant-cell arteritis. N Engl J Med. 2017;377:317–328. doi: 10.1056/NEJMoa1613849. [DOI] [PubMed] [Google Scholar]
- Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., Zhao Y., Li Y., Wang X., Peng Z. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA - J. Am. Med. Assoc. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Nie J., Wang H., Zhao Q., Xiong Y., Deng L., Song S., Ma Z., Mo P., Zhang Y. Characteristics of peripheral lymphocyte subset alteration in covid-19 pneumonia. J. Infect. Dis. 2020;221 doi: 10.1093/INFDIS/JIAA150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C.K.I. Learning with kernels: support vector machines, regularization, optimization, and beyond. J. Am. Stat. Assoc. 2003;98 doi: 10.1198/jasa.2003.s269. [DOI] [Google Scholar]
- World Health Organization, n.d. No Title [WWW Document]. Obes. Overweight. https//www.who.int/news-room/fact-sheets/detail/obesity-and-overweight.
- Yan L., Zhang H.-T., Goncalves J., Xiao Yang, Wang M., Guo Y., Sun C., Tang X., Jing L., Zhang M., Huang X., Xiao Ying, Cao H., Chen Y., Ren T., Wang F., Xiao Yaru, Huang S., Tan X., Huang N., Jiao B., Cheng C., Zhang Y., Luo A., Mombaerts L., Jin J., Cao Z., Li S., Xu H., Yuan Y. An interpretable mortality prediction model for COVID-19 patients. Nat. Mach. Intell. 2020;2 doi: 10.1038/s42256-020-0180-7. [DOI] [Google Scholar]
- Zhang J. jin, Dong X., Cao Y., yuan Yuan, dong Y., Yang, bin Y., Yan Y., qin, Akdis C.A., Gao Y. dong. Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan, China. Allergy Eur. J. Allergy Clin. Immunol. 2020 doi: 10.1111/all.14238. [DOI] [PubMed] [Google Scholar]
- Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., Guan L., Wei Y., Li H., Wu X., Xu J., Tu S., Zhang Y., Chen H., Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020 doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Study overview and design.
Model 8 (SVM Numerical and Categorical). The area under the ROC curve (AUROC).
The shap analysis on all the 8 models in the development test set.
The shap analysis on all the 8 models in the external validation dataset.