The use of self-collected specimens for the screening of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has gained interest as they may facilitate massive screening campaigns. Various authors have reported that mid-nasal swabs1 , 2 and saliva3, 4, 5, 6 are reliable specimens, alternative to nasopharyngeal swabs, to detect SARS-CoV-2 infections by RT-qPCR, irrespective of the age group tested.7 Despite drawing consistent conclusions, studies reported heterogeneous results regarding the performance of each type of sample, particularly sensitivity, which strongly depends on the viral load distribution of the investigated population and sample collection protocols. In the case of saliva, discrepancies regarding sensitivity might be even higher due to optimized protocols for RNA extraction adapted to the rheological properties of saliva.6 , 8 Therefore, there is a need for better characterizing-also from a quantitative perspective-the performance of self-collected specimens before using them as an alternative to nasopharyngeal swabs for SARS-CoV-2 screening.
In the context of a randomized clinical trial targeting mild COVID-19 patients (NCT04621123), we enrolled 130 adults in a sub-study to directly compare self-collected mid-nasal swabs and saliva specimens for SARS-CoV-2 detection by RT-qPCR, using nasopharyngeal swabs collected by the study nurses as a reference. Included patients had a mean age of 59 (SD 8.5) and a median of 4 days (95% IC 3–5) from symptoms onset; 43.2% were females. Patients received written instructions to self-collect a mid-nasal swab from both nostrils by introducing the swab 2,3 cm and rotating during 5 s, and 1 mL of saliva by spitting inside the funnel of a collection device (DANASALIVA™ sample collection kit). Participants were advised to avoid eating, drinking, smoking, and brushing their teeth within 30 min prior to sample collection. Self-collection was done in the presence of a study nurse, although they did not intervene during the collection process. Next, the study nurse collected a nasopharyngeal swab from both nostrils. Swab specimens were placed into sterile tubes containing viral transport media (DeltaSwab Virus). Saliva was mixed with 1 mL of saliva preservation solution in the collection device, following manufacturer's instructions. No additional pre-treatment step potentially increasing saliva sensitivity6 , 8 was used before RNA extraction. All three specimens were transported to the Microbiology laboratory of Hospital Germans Trias i Pujol and stored at 2–8 °C for up to 24 h before RT-qPCR. Paired samples were collected at baseline and 7 days after enrollment.
RNA was extracted using the STAR Mag reagent (Seegen) for the Microlab Starlet IV or Nimbus platforms (Hamilton life Science Robotics, USA), according to the manufacturer's instructions. PCR amplification was conducted according to the recommendations of the 2019-nCoV RT-qPCR Diagnostic Panel of the Centers for Disease Control and Prevention (CDC) using the Allplex™ 2019-nCoV assay (Seegene, South Korea) on the CFX96 (Bio-Rad, USA) according to manufacturer's instruction.
The three paired samples were successfully obtained in 129 of 130 patients, with 120 (92.3%) showing a positive nasopharyngeal swab at baseline. Compared to nasopharyngeal swabs, self-collected mid-nasal swabs and saliva samples showed a sensitivity of 99.2% (119/120) and 90.0% (108/120), respectively. This result is in line with a recently published head-to-head comparison, which included 38 positive COVID-19 cases.8 Of the nine participants with nasopharyngeal swabs testing negative, two had a positive saliva specimen and one a positive nasal swab. Given the successful internal PCR controls and the high cycle threshold (Ct) values (all above 30; days from symptom onset ranging from 4 to 6), we presume these patients were approaching the recovered state, with viral loads close to negativity.
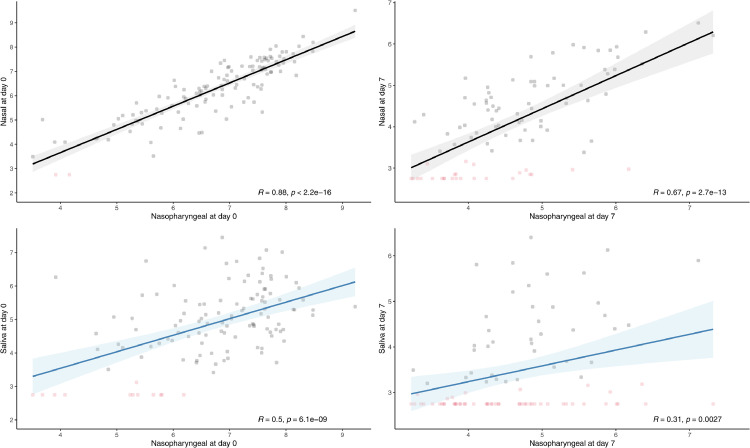
In addition to the qualitative and semi-quantitative analyses conducted in previous works, we estimated the viral load of each specimen and correlated the values observed in self-collected samples with those of nasopharyngeal swabs. The RT-qPCR Ct of positive specimens was, in mean (SD), 20.64 (3.43) for nasopharyngeal swabs, 22.90 (4.60) for mid-nasal swabs, and 29.56 (4.50) for saliva specimens (Fig. 1 A). The Pearson lineal correlation between viral loads obtained from nasopharyngeal swabs and self-collected specimens revealed a strong correlation for mid-nasal swabs (R = 0.88; p < 0.001) and a fair correlation for saliva specimens (R = 0.50; p < 0.001) (Fig. 2 A).
Fig. 1.
Distribution of RT-qPCR cycle threshold (Ct) of nasopharyngeal swabs, nasal swabs, and saliva specimens collected at baseline (A) and day 7 (B). Cts correspond to the viral RNA-dependent RNA polymerase (RdRP) gene.
Fig. 2.
Correlation of the viral load (VL) of nasopharyngeal swabs with that of nasal swabs and saliva specimens at baseline (A) and day 7 (B). Viral loads were estimated using a calibration line obtained with serial dilutions of SARS-CoV-2 control RNA, run in parallel to a set of samples covering all thermal cycles used in the analysis. Negative samples (in red) were assigned to a Ct of 40 (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
Of the 120 nasopharyngeal swabs obtained on day 7 (i.e., a mean of 11 days [range 10–12] from symptom onset), 28 tested negative and 92 positive. Mean Ct values were 31.85 (4.63), 33.95 (4.75), and 36.69 (4.42) for positive nasopharyngeal, mid-nasal and saliva specimens, respectively (Fig. 1B). As expected, the mean viral load was lower at day 7 than at baseline (p < 0.001). Compared to nasopharyngeal swabs, self-collected mid-nasal swabs and saliva specimens showed a sensitivity of 72.8% (67/92) and 42.4% (39/92), respectively, suggesting poorer performance at low viral loads. Of note, most negative self-collected samples with a positive paired nasopharyngeal swab yielded Ct values above 30 (92% and 52% of the nasal and saliva specimens, respectively). The viral load correlation with nasopharyngeal swab was poor for saliva specimens (R = 0.3; p = 0.003) and moderate for mid-nasal swabs (R = 0.67; p < 0.001) (Fig. 2B).
In summary, our findings show that self-collected mid-nasal swabs have better performance than saliva for detecting SARS-CoV-2 when compared with the gold-standard nasopharyngeal swabs. Of note, the sensitivity of saliva was remarkably high in samples with higher viral load, despite not using any of the RNA extraction protocols adapted to the rheological properties of this sample. Considering that respiratory specimens with Ct above 33,34 are unlikely to be contagious,9 our finding indicates that saliva would be sensitive enough to identify individuals at risk of transmission. Furthermore, the enhanced sensitivity achieved with adapted protocols for RNA extraction from saliva suggests that this might be the sample of choice for systematic screenings in settings in which a specific laboratory pathway can be implemented (e.g., school children).
Taken together, the existing literature and the results provided in our analysis encourage the use of self-collected specimens (mid-nasal when possible and saliva in vulnerable populations such as children) for massive screenings of SARS-CoV-2.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
We thank Gerard Carot-Sans for providing medical writing support with manuscript preparation and Roser Escrig for her support in the study design and medical writing assistance with the study documentation. We also thank Laia Bertran, Mireia Clua, Jordi Mitjà, Miquel Àngel Rodríguez, Claudia Laporte, Sergi Gavilan, Joan Mercado and Enric Nieto for the operational and financial management of the project. We thank the personnel from the Fight Aids and Infectious Diseases Foundation for their support in administration, human resources, and supply chain management. The authors declare no conflicts of interest.
Funding
The trial was sponsored by the Fight AIDS and Infectious Diseases Foundation with funding from the pharmaceutical company Grifols S.A and the crowdfunding campaign YoMeCorono (www.yomecorono.com). The study received support of the Hospital Universitari Germans Trias i Pujol, and Banc de Sang i Teixits de Catalunya (BST). ISGlobal receives support from the Spanish Ministry of Science and Innovation through the “Centro de Excelencia Severo Ochoa 2019–2023″ Program (CEX2018–000806-S), and support from the Generalitat de Catalunya through the CERCA Program. CISM is supported by the Government of Mozambique and the Spanish Agency for International Development (AECID). BB was supported by a Beatriu de Pinós postdoctoral fellow granted by the Government of Catalonia's Secretariat for Universities and Research, and by Marie Sklodowska-Curie Actions COFUND Program (BP3, 801370). OM was supported by the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation program.
Ethics
The study was conducted according to the Helsinki Declaration of the World Medical Association. The study protocol was approved by the Ethics Committee at Hospital Germans Trias i Pujol (number PI 20–313) and the institutional review boards of participating centers. All patients provided written informed consent before enrolling the study, which was supervised by an independent data and safety monitoring board.
References
- 1.McCulloch Denise J., Kim Ashley E., Wilcox Naomi C., Logue Jennifer K., Greninger Alex L., Englund Janet A., et al. Comparison of unsupervised home self-collected midnasal swabs with clinician-collected nasopharyngeal swabs for detection of SARS-CoV-2 infection. JAMA Netw Open. 2020;3(7):22–25. doi: 10.1001/jamanetworkopen.2020.16382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kojima N., Turner F., Slepnev V., Bacelar A., Deming L., Kodeboyina S., et al. Self-collected oral fluid and nasal swabs demonstrate comparable sensitivity to clinician collected nasopharyngeal swabs for coronavirus disease 2019 detection. Clin Infect Dis. 2020:1–4. doi: 10.1093/cid/ciaa1589. Xx Xxxx. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lorenzo A., Giulio C., Francesco G., Paolo G., Daniela D., Angelo G., et al. Since January 2020 elsevier has created a COVID-19 resource center with free information in English and Mandarin on the novel coronavirus COVID- 19 . The COVID-19 resource centre is hosted on elsevier connect, the company ’ s public news and information. J Infect. 2020;81:e45–e50. January. [Google Scholar]
- 4.Jialou Z., Jiubiao G., Yuzhong Xu, Xinchun C. Viral dynamics of SARS-CoV-2 in saliva from infected patients. J Infect. 2020;81(3) doi: 10.1016/j.jinf.2020.06.059. e48–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monique T., Katherine C., Mariah C., Maureen R., Riley W.C., Lily K., et al. Comparison of saliva and nasopharyngeal swab SARS-CoV-2 RT-qPCR testing in a community setting. J Infect. 2021;82(4):84–123. doi: 10.1016/j.jinf.2020.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wyllie Anne L., John F., Arnau C.M., Melissa C., Maria T., Pavithra V., et al. Saliva or nasopharyngeal swab specimens for detection of SARS-CoV-2. N Engl J Med. 2020;383(13):1283–1286. doi: 10.1056/NEJMc2016359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pedro B., Amaresh P.A., Cristian L., Francesc T., Pilar S.M., Jesica S., et al. Validation and implementation of a direct RT-qPCR method for rapid screening of SARS-CoV-2 infection by using non-invasive saliva samples. Int J Infect Dis. 2021;110:363–370. doi: 10.1016/j.ijid.2021.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aimee B.C., Seanne B., Emily F.C., Paul J.J., Matthew B.J., Katelyn R., et al. Evaluation of self-collected mid-turbinate nasal swabs and saliva for detection of SARS-CoV-2 RNA. J Clin Microbiol. 2021 doi: 10.1128/jcm.00848-21. June. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernard L.S., Marion L.B., Julien A., Thuan H.V., Clio G., Philippe C., et al. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur J Clin Microbiol Infect Dis. 2020;39(6):1059–1061. doi: 10.1007/s10096-020-03913-9. 0.1007/s10096-020-03913-9. [DOI] [PMC free article] [PubMed] [Google Scholar]