Abstract
Introduction
Although bacterial co- and superinfections are rarely present in patients with COVID-19, overall antibiotic prescribing in admitted patients is high. In order to counter antibiotic overprescribing, antibiotic stewardship teams need reliable data concerning antibiotic prescribing in admitted patients with COVID-19.
Methods
In this prospective observational cohort study, we performed a quantitative and qualitative evaluation of antibiotic prescriptions in patients admitted to the COVID-19 ward of a 721-bed Belgian university hospital between 1 May and 2 November 2020. Data on demographics, clinical and microbiological parameters and antibiotic consumption were collected. Defined daily doses (DDD) were calculated for antibiotics prescribed in the context of a (presumed) bacterial respiratory tract infection and converted into two indicators: DDD/admission and DDD/100 hospital bed days. A team of infectious disease specialists performed an appropriateness evaluation for every prescription. A driver analysis was performed to identify factors increasing the odds of an antibiotic prescription in patients with a confirmed COVID-19 diagnosis.
Results
Of 403 eligible participants with a suspected COVID-19 infection, 281 were included. In 13.8% of the 203 admissions with a COVID-19 confirmed diagnosis, antibiotics were initiated for a (presumed) bacterial respiratory tract co-/superinfection (0.86 DDD/admission; 8.92 DDD/100 bed days; 39.4% were scored as ‘appropriate’). Five drivers of antibiotic prescribing were identified: history of cerebrovascular disease, high neutrophil/lymphocyte ratio in male patients, age, elevated ferritin levels and the collection of respiratory samples for bacteriological analysis.
Conclusion
In the studied population, the antibiotic consumption for a (presumed) bacterial respiratory tract co-/superinfection was low. In particular, the small total number of DDDs in patients with confirmed COVID-19 diagnosis suggests thoughtful antibiotic use. However, antibiotic stewardship programmes remain crucial to counter unnecessary and inappropriate antibiotic use in hospitalized patients with COVID-19.
Trial Registration
The study is registered at ClinicalTrials.gov (NCT04544072).
Supplementary Information
The online version contains supplementary material available at 10.1007/s40121-021-00535-2.
Keywords: Antibiotics, Antimicrobial stewardship, Bacterial respiratory tract infection, Co-infection, COVID-19, Superinfection
Key Summary Points
| Coronavirus disease-19 (COVID-19) has caused great challenges for antibiotic stewardship teams (AST). |
| Drug use evaluation data combined with a driver analysis can help AST to target their interventions. |
| Assessing the quantity and quality of antibiotics prescribed for a (presumed) bacterial respiratory tract infection is an essential part of evaluating prescribing practice in an objective manner. |
| The diagnosis of bacterial respiratory tract infections in patients with COVID-19 is difficult. |
| In order to facilitate surveillance of antibiotic prescribing and decrease the risk of antimicrobial resistance, accurate diagnostic markers and infection control are complementary to prescribing data. |
Introduction
Since December 2019, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has spread worldwide, causing a major pandemic. The disease itself has been named coronavirus disease 2019 or COVID-19 [1]. While most attention focused on the management of COVID-19, the worldwide burden of antimicrobial resistance (AMR) pushes modern medicine silently back to a pre-antibiotic era. A recent report by the United Nations warned that without adequate response, AMR would lead to more than 10 million deaths per year by 2050. Death from AMR could become the primary cause of death worldwide, possibly leading to an economic crisis worse than that of 2008 [2, 3]. It is still unclear to what extent the COVID-19 pandemic will have a deleterious impact on AMR. However, different authors have highlighted the fact that routine antimicrobial stewardship efforts and infection prevention measures were downgraded because of a shift of activities, which was caused by COVID-19 [4].
Compared to other viral infections such as influenza, the reported rate of bacterial respiratory tract super/co-infections (bRTI) in COVID-19 is generally low (8.6–19% for COVID-19 vs. 11–35% for influenza) [5–7]. However, overall antibiotic prescribing has been high. In the beginning of the pandemic, Zhou et al. described that 95% of patients were treated with antibiotics [8]. In an international survey investigating antibiotic prescription patterns, 71% of the respondents reported to prescribe antibiotics for patients with COVID-19 admitted to a regular ward [9]. A recent systematic meta-analysis of 254 studies by Langford et al., reporting on 30,623 patients, found that almost three-quarters (74.6%) of patients with COVID-19 received antibiotic therapy [7]. This raises several issues relating to antibiotic prescribing in patients with COVID-19. First, overuse and misuse of empiric antibiotics can lead to an underestimation of real co-infections [10, 11]. Second, antibiotic overprescribing in general has been associated with adverse events and worse outcomes. Third, antibiotic overconsumption is problematic in light of global antimicrobial resistance (AMR) [3]. Reports on nosocomial infections in COVID-19 include clusters of vancomycin-resistant enterococci [12] and New Delhi metallo-beta-lactamase-producing Enterobacterales [13] or carbapenem-resistant Enterobacterales [14, 15].
Acute bRTIs should always be treated appropriately and in a timely manner. However, the diagnosis of such infections can be challenging, especially in patients with COVID-19. Although biomarkers for bacterial diseases and medical imaging can be useful diagnostic and prognostic tools, they lack specificity for diagnoses of bRTIs in patients with COVID-19 [16–19]. These are all reasons why the continuation of antimicrobial stewardship team (AST) programmes during the COVID-19 pandemic is highly recommended. AST programmes guide physicians in diagnosing infections, infection control and responsible antibiotic prescriptions [20, 21].
To promote judicious antibiotic use in presumed bRTIs in admitted patients with COVID-19, the antimicrobial stewards need to obtain good data to gain a better knowledge of the quantity and quality of antibiotics prescribed on their wards. Understanding the factors that drive individual physicians to prescribe antibiotics is a crucial element in their toolbox. In this prospective observational cohort study, we performed an evaluation of antibiotics used in patients admitted to the COVID-19 ward and studied factors associated with antibiotic prescribing, in order to help target future AST interventions.
Methods
The study was conducted in UZ Brussel, a Belgian university hospital with a 721-bed capacity. All adult patients admitted to the COVID-19 ward between 1 May 2020 and 2 November 2020 in the context of a (suspected) COVID-19 infection were eligible for inclusion. Included patients, or their legal representative, were required to sign an informed consent form (ICF). Patients eligible for inclusion had to be admitted for at least 24 h before inclusion. A COVID-19 confirmed infection was diagnosed either by clinical suspicion with positive SARS-CoV-2 polymerase chain reaction (PCR) on nasopharyngeal swab or by clinical suspicion together with strongly suggestive chest computed tomography (CT) scan. For the COVID-19 cases confirmed by PCR, the timing of COVID-19 confirmation was documented as ‘early’ (a positive PCR result in ambulatory setting or a positive rapid PCR test performed at the emergency department) or ‘late’ (PCR test result available after admission on a ward). For participants in whom the diagnosis of COVID-19 was eventually rejected by SARS-CoV-2 PCR on nasopharyngeal swab and/or on clinical grounds, the follow-up period continued until the moment of rejection. Data were extracted from the patients’ electronic medical file following a structured registration method. These data include baseline demographic data, comorbidities, information related to the patient’s hospital stay (e.g. length of stay and survival), symptoms, laboratory results, severity of COVID-19 (amount of oxygen needed, mechanical ventilation, intensive care unit (ICU) hospitalization), microbiological data and antibiotic use data. Radiological data were reassessed retrospectively by a thoracic radiology specialist. Definitions used for fever, Mycoplasma pneumonia infection and calculation of oxygen need can be found in the supplementary material. If participants were to be admitted to ICU, data collection stopped during the ICU admission. A graphical presentation of patient inclusion and follow-up is shown in Fig. 1.
Fig. 1.

Schematic overview of patient inclusion and follow-up
The primary objective of this study is to evaluate the use of antibiotics for a suspected or confirmed bacterial respiratory tract co-infection in patients with COVID-19, using indicators related to quantity and quality. Possible drivers of antibiotic prescription for a presumed or confirmed bRTI were investigated as secondary endpoints.
For all included patients admitted to the COVID-19 ward, the UZ Brussel electronic hospital information system was screened on a daily basis for every prescribed antibiotic of the Anatomical Therapeutic Chemical classification group J01 [22]. Following careful examination of the patient’s record by an infectious diseases (ID) specialist, only antibiotic prescriptions targeting (possible) respiratory tract infections were analysed. First, the daily defined dose (DDD) was calculated, as proposed by the World Health Organization (WHO) [22], both for single antibiotic formulations as well as for the total amount of administered antibiotics. The DDD is a commonly used technical measure to visualize and compare cross-national antibiotic consumption. This measure can also help surveillance organizations to assess antibiotic prescribing in a longitudinal way [23]. DDD per admission and per 100 days of admission are reported as complementary indicators. DDD per admission is indicative of the selection pressure and DDD per 100 bed days is indicative of the therapy duration [24]. Analyses of the quantitative endpoints were performed separately: on the one hand for the total group of patients (including the group with rejected COVID-19 diagnosis until the time of rejection); on the other hand for the group of patients with confirmed COVID-19 diagnosis. Second, the appropriateness of the antibiotic(s) was scored daily by an expert panel of ID specialists and every antibiotic categorized as ‘appropriate’, ‘inappropriate’, ‘suboptimal’ or ‘unnecessary’ by at least two independent ID specialists (see supplementary material) [25]. In case of disagreements between them, a third ID specialist was consulted in order to reach a consensus. Whenever indicated, written and oral antimicrobial stewardship advice was communicated to the treating physician as part of the standard AST activity.
The following variables of interest were included in the driver analysis: demographics, laboratory variables (C-reactive protein (CRP), neutrophilic count, neutrophil/lymphocyte ratio (NLR) [26], lymphopenia (below 1000 lymphocytes/mm3), D-dimers, troponin levels and ferritin level at admission), the presence of fever at admission, symptoms present before or at admission (dyspnoea, cough, chest pain, fever), comorbidities (pre-existing lung disease, congestive heart failure or ischemic heart disease, diabetes mellitus, solid or haematological neoplasia, dementia, cerebrovascular disease, chronic neurological disorder, Charlson comorbidity index (CCI)) [27], markers of severe disease (SpO2/FiO2 at admission, oxygen need at admission, quick Sequential Organ Failure Assessment (qSOFA) score [28], need for mechanical ventilation), the presence of (positive) respiratory microbiological samples, ICU stay during admission, length of stay and the timing of COVID-19 diagnosis confirmation. Only data of the (PCR- and clinically) confirmed patients with COVID-19 were used in the driver analysis to avoid selection bias.
Statistical analyses were performed using the statistical software RStudio version 1.1.463 running on R version 3.5.3 (R Foundation for Statistical Computing, Vienna, Austria). Descriptive statistics were applied to characterize the cohort, reporting percentages and medians with interquartile ranges (IQR) for the group of patients with definite or presumed COVID-19 diagnosis and the group of patients with solely definite COVID-19 diagnosis. The hospital pharmacist calculated the DDDs for each antibiotic in general and as a function of the appropriateness. In a final model to identify hidden drivers, a mixed effects logistic regression was fitted with a random intercept per patient admission to deal with clustering within patients. A combination of both forward and backward model selection based on the Akaike information criterion (AIC) was used. Only significant variables were retained in the final model. Finally, inclusion of possible interaction effects was also tested. Results with a p value less than 0.05 were considered statistically significant. Multicollinearity among variables was taken care of by verifying the variance inflation factors (VIF) of each driver (VIF ≤ 5). Statistical analyses were performed using the statistical software RStudio version 1.1.463 running on R version 3.5.3 (R Foundation for Statistical Computing, Vienna, Austria). The trial is registered at ClinicalTrials.gov (NCT04544072).
Compliance with Ethics Guidelines
This study was approved by the local ethics committee (Commissie Medische Ethiek, UZ Brussel) prior to data collection (B.U.N. 1432020000092) and was carried out in accordance with the ethical principles for medical research involving human subjects established by the Declaration of Helsinki, protecting the privacy of all participants, as well as the confidentiality of their personal information. All patients provided informed consent to participate in the study.
Results
Patients’ and Disease Characteristics
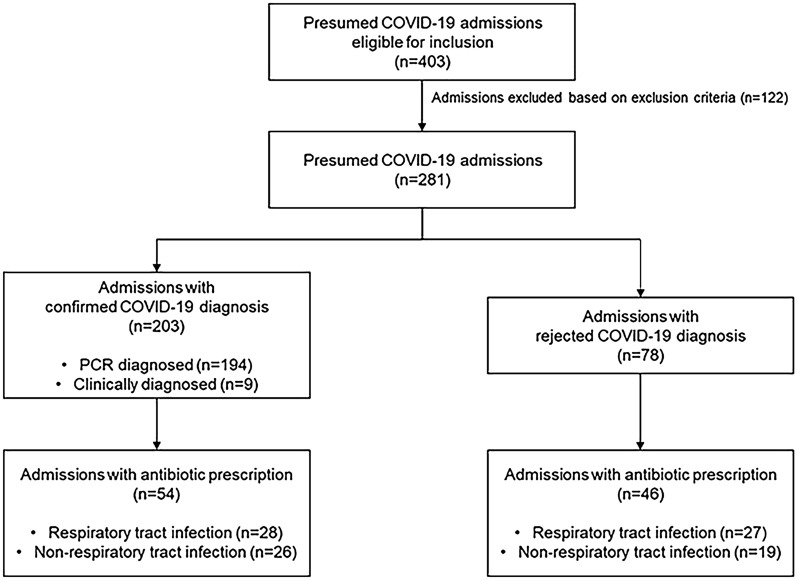
During the study period from 1 May 2020 until 2 November 2020, 403 patients were admitted with (suspected) COVID-19. After exclusion of admissions with a hospital stay of less than 24 h, and those unwilling or unable to sign the ICF, 281 participants were included in the prospective cohort. In 203/281 admissions (72%), COVID-19 diagnosis was confirmed clinically (n = 9) or by means of a positive PCR on nasopharyngeal swab (n = 194) (Fig. 2). Median age was 65 years with a male/female ratio of 1.38 and a median BMI of 27 kg/m2 (Table 1). In 123/194 (63%) of the participants with PCR-confirmed COVID-19 the diagnosis was classified as early. Only one participant was admitted more than once.
Fig. 2.
Flow chart of the included admissions with a presumed COVID-19 diagnosis admitted to the COVID-19 ward in UZ Brussel
Table 1.
Patients’ characteristics
| All admissions (n = 281) | Confirmed COVID-19 admissions (n = 203) | Rejected COVID-19 admissions (n = 78) | |
|---|---|---|---|
| Demographics | |||
| Age (years); (median, IQR) | 65 (26) | 62 (26) | 71 (29) |
| Gender (male); (n, %) | 163 (58) | 118 (58) | 45 (58) |
| BMI (kg/m2); (median, IQR) | 27 (7) | 27 (7) | 26 (7) |
| COVID-19 diagnosis (n (%)) | |||
| PCR | 194 (69) | 194 (69) | NA |
| Clinical diagnosis | 9 (3) | 9 (4) | NA |
| Rejected diagnosis | 78 (28) | 0 (0) | 78 (100) |
| Length of stay (median, IQR) | 8 (9) | 8 (9) | 5 (10) |
| Symptoms at admission (n, %) | |||
| Fatigue | 195 (69) | 142 (70) | 53 (68) |
| Cough | 160 (57) | 128 (63) | 32 (41) |
| Fever or history of fever | 166 (59) | 124 (61) | 42 (54) |
| Dyspnoea | 160 (57) | 115 (57) | 45 (58) |
| Muscle aches | 117 (42) | 98 (48) | 19 (24) |
| Gastrointestinal symptoms | 126 (45) | 96 (47) | 30 (38) |
| Headache | 114 (41) | 91 (45) | 23 (29) |
| Anosmia | 87 (31) | 76 (37) | 11 (14) |
| Thoracic pain | 87 (31) | 69 (34) | 18 (23) |
| Laboratory findings (median, IQR; except for lymphopenia) | |||
| White blood cell count (/mm3) | 6700 (5600) | 5600 (4200) | 9800 (7400) |
| Neutrophil count (/mm3) | 4442 (5220) | 3829 (3526) | 7636 (6703) |
| Lymphocyte count (/mm3) | 1070 (848) | 1051 (709) | 1122 (1338) |
| Neutrophil/lymphocyte count rate | 4.20 (5.43) | 3.73 (4.22) | 6.2 (10.8) |
| Lymphopenia (< 1000/mm3); (n, %) | 124 (44) | 90 (45) | 34 (44) |
| Ferritin (mcg/L) | 432 (618) | 480 (592) | 265 (456) |
| CRP (mg/dL) | 51 (93) | 47 (74) | 148 (159) |
| Ferritin (mcg/L) | 432 (618) | 480 (592) | 265 (456) |
| D-dimers (ng/mL) | 645 (1096) | 593 (974) | 1149 (2052) |
| Comorbidities | |||
| CCI (median, IQR) | 1 (3) | 1 (2) | 2 (2) |
| Diabetes mellitus (n, %) | 88 (31) | 67 (33) | 21 (27) |
| Pre-existing pulmonary disease (n, %) | 49 (17) | 37 (18) | 12 (15) |
| Moderate to severe renal diseasea(n, %) | 45 (16) | 29 (14) | 16 (8) |
| Ischaemic/congestive heart disease (n, %) | 35 (12) | 25 (12) | 10 (13) |
| Cerebrovascular disease (n, %) | 14 (5) | 12 (6) | 2 (3) |
| Dementia (n, %) | 16 (6) | 8 (4) | 8 (10) |
| Solid tumourb (n, %) | 27 (10) | 11 (5) | 16 (21) |
| Haematological neoplasia | 4 (1) | 2 (1) | 2 (3) |
| Prognostic factors | |||
| qSOFA score at admission (n, %) | |||
| 0 | 153 (55) | 105 (52) | 48 (62) |
| 1 | 112 (40) | 89 (44) | 23 (29) |
| 2 | 14 (5) | 8 (4) | 6 (8) |
| 3 | 0 (0) | 0 (0) | 0 (0) |
| ICU admission (n, %) | 41 (15) | 34 (17) | 7 (9) |
| Mechanical ventilation need (n, %) | 14 (5) | 14 (7) | 0 (0) |
| (SpO2/FiO2 × 100) at admission (median, IQR) | 428 (140) | 418 (139) | 442 (141) |
| (SpO2/FiO2 × 100) minc (median, IQR) | 313 (207) | 310 (201) | 365 (174) |
| Mortality (n, %) | 23 (8) | 20 (10) | 3 (4) |
IQR interquartile range, BMI body mass index, PCR polymerase chain reaction, NA not applicable, CRP C-reactive protein, CCI Charlson comorbidity index
aSerum creatinine > 3 mg/dL or kidney transplant or uraemia [25]
bActive solid neoplastic disease or solid neoplastic disease during the last 5 years
cSpO2/FiO2 × 100 min = the lowest value of the SpO2/FiO2 rate during the total stay on a COVID-19 ward or in the ICU
Most frequent symptoms at admission were fatigue (69%), cough (57%), fever (59%) and dyspnoea (57%). The occurrence of lymphopenia (below 1000/mm3) was similar for participants with confirmed and rejected COVID-19, respectively 45% and 44%. Mechanical ventilation was indicated in 7% of all participants with confirmed COVID-19 while none of the participants with rejected COVID-19 diagnosis were intubated. The overall mortality rate was 8% (respectively 10% and 4% for participants with confirmed and rejected COVID-19 diagnosis).
Pulmonary imaging (CT, X-ray or both) was performed in 243 (86%) of the 281 admissions, respectively 174/203 (86%) and 69/78 (88%) with confirmed and rejected COVID-19 diagnosis. The detailed radiological findings are presented in Table 2.
Table 2.
Radiological findings
| All admissions (n = 281) | Confirmed COVID-19 admissions (n = 203) | Rejected COVID-19 admissions (n = 78) | |
|---|---|---|---|
| Pulmonary CT performed or X-ray performed (n, %) | 243 (86) | 174 (86) | 69 (88) |
| Radiologist suggests a bacterial superinfection | 26 (11) | 11 (6) | 15 (19) |
| Presence of dense consolidation(s) | 52 (21) | 29 (17) | 23 (29) |
| > 3 pulmonary lobes involved | 152 (63) | 120 (69) | 32 (41) |
| Radiologist suggests COVID-19 infection | 117 (48) | 101 (58) | 16 (21) |
| Pulmonary CT performed (n, %) | 196 (70) | 135 (67) | 61 (78) |
| Ground glass infiltrates | 127 (65) | 109 (81) | 18 (23) |
| > 50% of the lung parenchyma affected | 57 (29) | 51 (38) | 6 (8) |
In 14% of the COVID-19 confirmed admissions, at least one respiratory sample was collected for bacteriological investigation. In 10 out of 28 admissions with at least one respiratory sample (5% of the 203 COVID-19 confirmed admissions), at least one respiratory sample with a significant result was identified, counting for a total of 16 significant microorganisms. Methicillin-sensitive Staphylococcus aureus (MSSA) (4/16; 25%) and Enterobacterales (7/16, 44%) were the most commonly isolated bacteria. One extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli was recorded. Further specification of the isolated respiratory pathogens is available in Table S1 in the electronic supplementary material. Bacteremia of respiratory origin (Streptococcus pneumonia) was identified in one patient with a confirmed COVID-19 diagnosis. No infections with Mycoplasma pneumoniae nor Legionella pneumophilia were diagnosed.
Antimicrobial Prescriptions and Associated Drivers
Antibiotics for respiratory or non-respiratory tract infections were initiated in 100/281 (36%) admissions. Overall, antibiotics were less frequently prescribed in COVID-19 confirmed admissions (54/203, 27%) than in admissions with a rejected COVID-19 diagnosis (46/78, 59%). Of all admissions, 19.6% (55/281) were prescribed at least one antibiotic for a (presumed) bRTI, counting for 289.2 DDDs (1.03 DDD/admission and 12.31 DDD/100 bed days), 244 days of antibiotic therapy and a total of 68 respiratory antibiotic prescriptions. In the COVID-19 confirmed group, the rate of prescriptions for a bRTI was lower (13.8%; 0.86 DDD/admission and 8.92 DDD/bed days) compared to the rejected diagnosis group (36.5%; 1.43 DDD/admission and 31.23 DDD/bed days). Beta-lactam antibiotics were administered most (83.8% of all DDDs). Further details on antibiotic consumption are shown in Table 3.
Table 3.
Quantitative data on antibiotic prescribing for (presumed) respiratory tract infection in participants admitted to the COVID-19 ward
| All admissions (n = 281) |
Confirmed COVID-19 admissions (n = 203) |
Rejected COVID-19 admissions (n = 78) |
|
|---|---|---|---|
| Total DDD (n, %) | 289.2 (100) | 177.7 (100) | 111.5 (100) |
| Beta-lactam antibiotics, penicillins | 28.3 (9.8) | 24 (13.5) | 4 (3.9) |
| Penicillins with beta-lactamase inhibitor | 182.1 (63.0) | 109.4 (61.6) | 72.7 (65.2) |
| Other beta-lactam antibiotics | 31.5 (10.9) | 26.0 (14.6) | 5.5 (4.9) |
| Sulfonamides and trimethoprim | 4 (1.4) | 4.0 (2.3) | NA |
| Macrolides, lincosamides and streptogramins | 23 (8.0) | 10.5 (5.9) | 12.5 (11.2) |
| Aminoglycosides | 1.8 (0.6) | 1.8 (1.0) | NA |
| Quinolones | 11 (3.8) | 2.0 (1.1) | 9 (8.1) |
| Other antibacterials | 7.5 (2.6) | NA | 7.5 (6.7) |
| DDD/admission | |||
| Total | 1.029 | 0.875 | 1.429 |
| Beta-lactam antibiotics, penicillins | 0.101 | 0.118 | 0.056 |
| Penicillins with beta-lactamase inhibitor | 0.648 | 0.539 | 0.932 |
| Other beta-lactam antibiotics | 0.112 | 0.128 | 0.071 |
| Sulfonamides and trimethoprim | 0.014 | 0.020 | NA |
| Macrolides, lincosamides and streptogramins | 0.082 | 0.052 | 0.160 |
| Aminoglycosides | 0.006 | 0.009 | NA |
| Quinolones | 0.039 | 0.010 | 0.115 |
| Other antibacterials | 0.027 | NA | 0.096 |
| DDD/100 bed days | |||
| Total | 12.317 | 8.921 | 31.232 |
| Beta-lactam antibiotics, penicillins | 1.206 | 1.205 | 1.214 |
| Penicillins with beta-lactamase inhibitor | 7.752 | 5.493 | 20.357 |
| Other beta-lactam antibiotics | 1.341 | 1.305 | 1.541 |
| Sulfonamides and trimethoprim | 0.170 | 0.201 | NA |
| Macrolides, lincosamides and streptogramins | 0.979 | 0.527 | 3.501 |
| Aminoglycosides | 0.077 | 0.090 | NA |
| Quinolones | 0.468 | 0.100 | 2.521 |
| Other antibacterials | 0.319 | NA | 2.101 |
DDD defined daily doses, NA not applicable
Among the respiratory antibiotic prescriptions (n = 68) in the 55 admissions who received at least one antibiotic for a (presumed) bRTI, prescription indications were a presumed COVID-19 bRTI (n = 24, 35%), treatment of pneumonia with low evidence of COVID-19 infection (n = 17, 25%), chronic obstructive pulmonary disease (COPD) exacerbation (n = 11, 16%), empiric coverage (n = 9, 13%) and other (n = 7, 10%; supplementary material). For these 55 admissions, the median lag time from the first day of respiratory symptoms until the start of the first respiratory antibiotic was 4 days (IQR 9). Time to treatment initiation was short with a median lag time of 0 (IQR 2) days between admission and antibiotic prescribing.
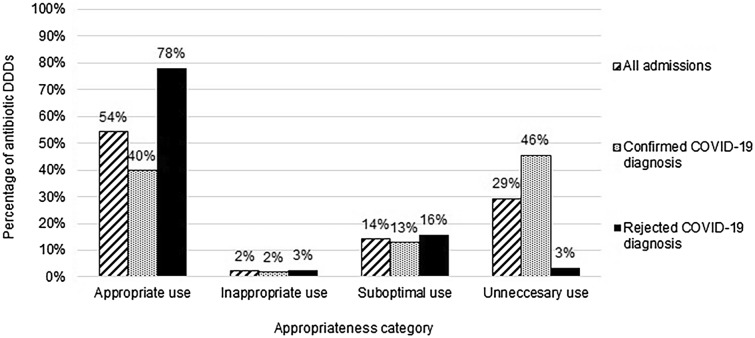
In the systematic evaluation of the antibiotic’s appropriateness, the two ID specialists disagreed in 7.2% (17/236) of all antibiotic days to be assessed. Overall, 54.3% of antibiotic consumption was evaluated as appropriate. The appropriateness of antibiotic prescriptions in COVID-19 confirmed participants was notably lower as only 39.4% of the prescriptions were considered appropriate, whereas 45.8% were considered unnecessary. In contrast, in the participants with a rejected COVID-19 diagnosis, the prescription was considered appropriate and unnecessary in, respectively, 77.9% and 3.4% of the participants. Suboptimal use was low (14.1%) (Fig. 3). The intravenous use of antibiotics throughout the inclusion period was high (rate of intravenous DDD to oral DDD = 2.9).
Fig. 3.
Appropriateness of antibiotic consumption in bacterial respiratory tract infection
In the mixed effects logistic regression analysis of the antibiotic driver assessment, the following variables were associated with respiratory antibiotic prescription in the COVID-19 confirmed participants: a medical history of cerebrovascular disease (odds ratio (OR) 8.92; 95% CI 1.73–45.75) and the collection of at least one respiratory sample (OR 5.73; 95% CI 1.76–19.13). Per rise of the neutrophil/lymphocyte ratio by one unit in male participants, the probability of an antibiotic prescription for a potential bRTI increased by 31.7% (OR 1.32; 95% CI 1.11–1.59). Each augmentation of ferritin level by 100 μg/L increased the odds of antibiotic prescription by 4.6% (OR 1.05; 95% CI 1.00–1.09). Finally, per age rise of 1 year, the odds for antibiotic respiratory prescriptions rose by 5% on average (OR 1.05; 95% CI 1.01–1.09) (Table 4). No significant association was found between the timing of COVID-19 diagnosis (early versus late) and the initiation of respiratory antibiotic prescriptions.
Table 4.
Mixed effects logistic regression analysis of potential drivers associated with respiratory antibiotic prescribing
| Variable | OR (95% confidence interval) | p value |
|---|---|---|
| Confirmed COVID-19 admissions (n = 203) | ||
| Age (per increase of 1 years) | 1.05 (1.01–1.09) | 0.018* |
| History of cerebrovascular disease | ||
| No |
Ref 8.92 (1.73–45.75) |
0.007* |
| Yes | ||
| Neutrophil/lymphocyte ratio in male patients (per rise of 1 unit) | 1.32 (1.11–1.59) | 0.003* |
| Ferritin level (per rise of 100 μg/L) | 1.05 (1.00–1.09) | 0.022* |
| Respiratory sample present | 5.73 (1.76–19.13) | 0.004* |
*p < 0.05 is considered statistically significant
Discussion
It seems a paradox that a viral pandemic like COVID-19 has led to a rising threat of increased inappropriate antibiotic use in the hospital sector as well as in the community, undermining current and previous efforts to tackle AMR. In this prospective study, a thorough assessment of antibiotic use was performed in patients admitted to the COVID-19 ward, which indicates a decline in antibiotic prescription rates but also an important issue of inadequate antibiotic prescribing.
To date, prospective studies on antibiotic prescribing in patients with COVID-19 are scarce [7]. Reporting DDDs next to a qualitative evaluation can put antibiotic use in perspective, because the diagnosis of bacterial super- and co-infections is difficult. Most COVID-19 studies differentiate between bacterial co-infection and superinfection (or secondary infection), defining the identification of other respiratory pathogens at the time of COVID-19 diagnosis as co-infection and during care as superinfection [6]. Although the presence of a superinfection has been associated with a prolonged hospital stay, a higher mortality rate and different respiratory bacterial species, the pathophysiological mechanism of both entities has not been described separately, making it an artificial subdivision [11, 29, 30]. In patients with COVID-19, superinfections seem to be more frequent than co-infections, accounting for 70% of the infections in the ISARIC study [31]. Based on the timing of antibiotic prescribing, our study found an opposite trend, with 41/55 (75%) of the antibiotics started during the first 2 days of admission, assuming that antibiotics were initiated in the context of a presumed co-infection. However, our study did not include ICU admissions, where mechanical ventilation, a prolonged length of stay and nosocomial transmission play an important role in the pathogenesis of bacterial superinfection. Moreover, our study did not focus on the final diagnosis of super- or co-infection, because a certain diagnosis of bRTI is hard to make in the COVID-19 setting, as biomarkers are non-specific and respiratory tract colonization is difficult to distinguish from infection, especially in patients with previous underlying lung disease. Nevertheless, Stevens et al. found an association between the presence of positive respiratory samples and worse COVID-19 outcome, which highlights the importance of collecting microbiological samples [21]. Microbiological sampling was performed in 13.8% of the 203 COVID-19-positive participants, of which 4.9% had a significant positive result. This is in line with previous COVID-19 studies where significant respiratory samples ranged from 2% to 8.6% [7, 29, 31–34]. MSSA and Enterobacterales were the most commonly isolated pathogens, in analogy with the ISARIC study [31]. Unlike the high Mycoplasma spp. infection rate reported by Lansbury et al. (42%), none were identified. This is probably because we considered rising serological kinetics as a diagnostic test and did not solely rely on single positive immunoglobulin G titres [29]. Only one (0.5%) case of bacteremia of respiratory origin was identified, which indirectly supports current evidence on the low incidence rate of bRTIs in patients with COVID-19 [31].
Antibiotics for a presumed bRTI were initiated in only 14% of all COVID-19 confirmed admissions. This seems very low compared to other comparable studies, reporting rates of antibiotic prescription above 50%. A meta-analysis by Langford et al. (2021) reported an antibiotic prescription rate of 71% [7]. A comparable retrospective study observed a prescription rate of 59% for respiratory antibiotics in ward patients [35]. The reasons for lower antibiotic prescription rate observed in our study are multiple: the prospective design of the study, the successful functioning of the AST with close follow-up of antibiotic prescriptions and an inclusion period that succeeded the first COVID-19 wave. Langford et al. also report a decreasing antibiotic prescription rate in studies enrolling participants during the first weeks of the COVID-19 pandemic, compared to studies which ended patient enrolments by April 2020 (85.8% versus 62.6%) [7]. Similarly, the ISARIC study showed a decline in antibiotic prescriptions when comparing March–April 2020 to May 2020 [31]. A possible learning effect of the disease progression and the (mis)use of antibiotics in this setting could have meant physicians initiated an antibiotic in patients with (suspected) COVID-19 after the initial phase of the pandemic [7]. Additionally, we did not evaluate the patients during an ICU stay. It is known that patients in critical care are more prone to receive antibiotics [33]. Penicillins with a beta-lactamase inhibitor were prescribed most frequently (1.206 DDD/100 bed days). This is in line with previously reported results, which suggests a systematic preference for broad-spectrum antibiotics [7, 23, 33, 36]. Research suggests that the number of DDD may also be associated with a decreased antibiotic susceptibility [24]. In our study, the reported antibiotic consumption for COVID-19 confirmed admissions (0.87 DDD/admission and 8.92 DDD/100 hospital bed days) was lower than in a previously performed retrospective study by this research group (3.6 DDD/admission and 31.5 DDD/100 hospital bed days) [35]. Importantly, antibiotic consumption in ICU patients was taken into account in the retrospective study whereas this was not the case in the current prospective study. We were unable to compare our data with any other COVID-19 or historical influenza cohort, as other quantitative data regarding antibiotic DDDs in this context have not been published so far. Interestingly, the national average antibiotic consumption in Belgian acute care hospitals in 2018 was more than fourfold higher than the observed consumption in this study (respectively 3.28 DDD/admission and 46.2 DDD/100 hospital bed days versus 0.87 DDD/admission and 8.92 DDD/100 hospital bed days) [37]. However, this comparison is only indicative, as the national antibiotic consumption results do not distinguish types of infection. Moreover, in this study, the amount of DDD/admission only implied DDDs during admission on a COVID-19 ward. Because of those limitations, the case-by-case assessment by an ID specialist was a valuable tool to assess the antibiotic’s appropriateness. Although the total antibiotic consumption has decreased compared to a previous period, it appears that a high number of unnecessary, suboptimal or inappropriate use (45.7%) of antibiotics is still prevalent. In confirmed COVID-19 cases, almost half (45.8%) of antibiotic consumption was unnecessary. Therefore, AST interventions remain needed to reduce improper use of antibiotics, especially in patients with COVID-19, and to reduce the risk of antibiotic resistance development. Different studies implementing therapeutic guidelines combined with directed AST efforts on antibiotic use in the COVID-19 setting have proven efficacy with significant reduction of therapy duration and/or antibiotic quantity [38, 39]. Nevertheless, antibiotics can be indicated in selected cases, especially in severe disease or in immunocompromised patients. Certain guidelines also recommend a short course of antibiotics in case of radiological findings and inflammatory markers compatible with a bRTI, despite their low positive predictive value and the weak level of evidence [40, 41]. However, physicians can use a negative microbiological result of respiratory tract samples or blood cultures in their decision to interrupt antibiotic treatment, as well as the presence of low procalcitonin levels, which has a high negative predictive value for a bacterial pneumonia [16, 19, 42].
In the driver analysis, five different variables were identified that significantly increased the odds of at least one antibiotic prescription for a (suspected) bRTI: a history of cerebrovascular disease, a high neutrophil/lymphocyte ratio in male participants, increasing age, elevated ferritin level and the collection of at least one respiratory sample. Patients with a history of cerebrovascular disease are known to be at risk for aspiration pneumonia due to micro-aspiration, impaired airway clearance, insufficient tooth hygiene, relative immobilization and ineffective expectoration due to muscle weakness. In a study by He et al. patients with cardiovascular comorbidities had a higher probability of a bRTI [43]. However, evaluation of the medical history and physical examination in patients with a history of cerebrovascular disease often contribute poorly to the COVID-19 or bRTI diagnosis, which can also explain the lower threshold to start antibiotics [44–48]. Elevated inflammatory biomarkers such as C-reactive protein, procalcitonin, neutrophil count and NLR are associated with worse prognosis in COVID-19-infected patients. As these inflammatory markers rise in the context of the COVID-19-related cytokine storm, they show a low predictive value for bRTI [16, 19, 26, 49]. However, a high neutrophil count is generally seen as an important biomarker for the diagnosis of a bacterial infection. Alongside lymphopenia, one of the markers for COVID-19, it is comprehensible that a high neutrophil/lymphocyte ratio was identified as a driver of antibiotic administration [50–52]. The fact that this was only significant in male participants could be explained by the higher inclusion rate of male patients. Additionally, male gender is a risk factor of acquiring severe COVID-19 and hospital-acquired infection in patients with COVID-19 [34]. Both older age and high ferritin level showed a slight enhanced risk of antibiotic prescription. The two parameters are known to be associated with a higher risk of severe SARS-CoV-2 infection, which could explain the association with antibiotic prescribing [53–55].
Conclusions
Despite the undeniable evidence of low rates of bRTI in COVID-19 infection, reported antibiotic prescription rates are surprisingly high. In this study, antibiotic prescribing rates in patients with COVID-19 were low. In particular, the total number of DDDs in patients with a confirmed COVID-19 diagnosis suggests thoughtful antibiotic use. However, AST efforts are now more important than ever to promote the judicious use of antibiotics in COVID-19-infected patients and to halt unnecessary and inappropriate antibiotic use.
This study has some limitations that should be taken into account. First, patients were not evaluated during ICU stay, while severe illness is a known risk factor for bacterial superinfection and thus physicians tend to prescribe more antibiotics in these patients with COVID-19 [34]. Second, as a result of the low rate of antibiotic prescriptions for (presumed) bRTI in this study, potential significant drivers of antibiotic prescription might not have been detected. Third, the study was only observational which implied that respiratory samples for bacteriological examination were not collected systematically, which could explain the low rates of respiratory sampling. However, the rate of documented respiratory cultures was similar in the ISARIC study (13% versus 14% in this study) [31]. In order to be able to compare the reported results, prospective antibiotic evaluation studies are needed in similar settings and in the ICU. Future studies should also focus on the value of bRTI diagnostic markers. Robust predictors and/or excluders of bacterial co-/superinfection are urgently needed and should be incorporated in evidence-based guidelines.
To support and target AST interventions, the effectiveness of predictive inflammatory, microbiological and radiological tools to diagnose a bRTI should become more clear [6]. Alongside diagnostic parameters, knowledge about the drivers associated with antibiotic prescribing is crucial, as are quantitative and qualitative surveillance of prescribing practice, to increase the efficiency of an antibiotic stewardship programme.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
Funding
No funding or sponsorship was received for this study or publication of this article. The Rapid Service Fee was funded by the authors.
Medical Writing Assistance
We would like to thank Simon Planken for his assistance with the development of the electronic case report forms. We also thank Marco Moretti for the help provided during patient inclusions. We acknowledge Thomas Seyler for his advice concerning the study design. Finally, we thank all included participants for their participation.
Authorship
All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this article, take responsibility for the integrity of the work as a whole, and have given their approval for this version to be published.
Author Contributions
J. Van Laethem and S. Wuyts are both first authors and equally shared the workload: study concept, study design, data collection, data analysis and interpretation, writing and revision. S. Van Laere: data analysis and revision. S. Dirkx: study design, data collection, writing. B. Ilsen: data collection and revision. L. Seyler, R. Mertens, P. Lacor: revision. D. Pierard, S. Allard: contributed to the study concept, the study design, writing and revision.
Disclosures
J. Van Laethem, S. Wuyts, S. Dirkx, S. van Laere, L. Seyler, R. Mertens, B. Ilsen, P. Lacor, D. Pierard, and S.D. Allard have nothing to disclose.
Compliance with Ethics Guidelines
This study was approved by the local ethics committee (Commissie Medische Ethiek, UZ Brussel) prior to data collection (B.U.N. 1432020000092) and was carried out in accordance with the ethical principles for medical research involving human subjects established by the Declaration of Helsinki, protecting the privacy of all participants, as well as the confidentiality of their personal information. All patients provided informed consent to participate in the study.
Data Availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Johan Van Laethem and Stephanie Wuyts contributed equally.
References
- 1.Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.UN. General Assembly (71st sess.: 2016–2017). President. Political declaration of the high-level meeting of the General Assembly on Antimicrobial Resistance: draft resolution. New York: UN; 2016. p. 6.
- 3.Interagency Coordination Group on Antimicrobial Resistance. No time to wait: securing the future from drug-resistant infections. Report to the Secretary-General of the United Nations. Geneva: World Health Organization; 2019 April 2019.
- 4.Rawson TM, Moore LSP, Castro-Sanchez E, et al. COVID-19 and the potential long-term impact on antimicrobial resistance. J Antimicrob Chemother. 2020;75(7):1681–1684. doi: 10.1093/jac/dkaa194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klein EY, Monteforte B, Gupta A, et al. The frequency of influenza and bacterial coinfection: a systematic review and meta-analysis. Influenza Other Respir Viruses. 2016;10(5):394–403. doi: 10.1111/irv.12398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Musuuza JS, Watson L, Parmasad V, et al. Prevalence and outcomes of co-infection and superinfection with SARS-CoV-2 and other pathogens: a systematic review and meta-analysis. PLoS One. 2021;16(5):e0251170. doi: 10.1371/journal.pone.0251170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Langford BJ, So M, Raybardhan S, et al. Antibiotic prescribing in patients with COVID-19: rapid review and meta-analysis. Clin Microbiol Infect. 2021;27(4):520–531. doi: 10.1016/j.cmi.2020.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beović B, Doušak M, Ferreira-Coimbra J, et al. Antibiotic use in patients with COVID-19: a 'snapshot' Infectious Diseases International Research Initiative (ID-IRI) survey. J Antimicrob Chemother. 2020;75(11):3386–3390. doi: 10.1093/jac/dkaa326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang CY, Chan KG. Underestimation of co-infections in COVID-19 due to non-discriminatory use of antibiotics. J Infect. 2020;81(3):e29–e30. doi: 10.1016/j.jinf.2020.06.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rawson TM, Wilson RC, Holmes A. Understanding the role of bacterial and fungal infection in COVID-19. Clin Microbiol Infect. 2021;27(1):9–11. doi: 10.1016/j.cmi.2020.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kampmeier S, Tönnies H, Correa-Martinez CL, Mellmann A, Schwierzeck V. A nosocomial cluster of vancomycin resistant enterococci among COVID-19 patients in an intensive care unit. Antimicrob Resist Infect Control. 2020;9(1):154. doi: 10.1186/s13756-020-00820-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nori P, Szymczak W, Puius Y, et al. Emerging co-pathogens: New Delhi metallo-beta-lactamase producing Enterobacterales infections in New York City COVID-19 patients. Int J Antimicrob Agents. 2020;56(6):106179. doi: 10.1016/j.ijantimicag.2020.106179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Porretta AD, Baggiani A, Arzilli G, et al. Increased risk of acquisition of New Delhi metallo-beta-lactamase-producing carbapenem-resistant Enterobacterales (NDM-CRE) among a cohort of COVID-19 patients in a teaching hospital in Tuscany, Italy. Pathogens. 2020;9(8):635. doi: 10.3390/pathogens9080635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tiri B, Sensi E, Marsiliani V, et al. Antimicrobial stewardship program, COVID-19, and infection control: spread of carbapenem-resistant Klebsiella pneumoniae colonization in ICU COVID-19 patients. What did not work? J Clin Med. 2020;9(9):2744. doi: 10.3390/jcm9092744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.May M, Chang M, Dietz D, et al. Limited utility of procalcitonin in identifying community-associated bacterial infections in patients presenting with coronavirus disease 2019. Antimicrob Agents Chemother. 2021;65(4):e02167–20. doi: 10.1128/AAC.02167-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwee TC, Kwee RM. Chest CT in COVID-19: what the radiologist needs to know. Radiographics. 2020;40(7):1848–1865. doi: 10.1148/rg.2020200159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang M, Hong KS, Chikontwe P, et al. Quantitative assessment of chest CT patterns in COVID-19 and bacterial pneumonia patients: a deep learning perspective. J Korean Med Sci. 2021;36(5):e46. doi: 10.3346/jkms.2021.36.e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vanhomwegen C, Veliziotis I, Malinverni S, et al. Procalcitonin accurately predicts mortality but not bacterial infection in COVID-19 patients admitted to intensive care unit. Ir J Med Sci. 2021:1–4. [DOI] [PMC free article] [PubMed]
- 20.Huttner BD, Catho G, Pano-Pardo JR, Pulcini C, Schouten J. COVID-19: don't neglect antimicrobial stewardship principles! Clin Microbiol Infect. 2020;26(7):808–810. doi: 10.1016/j.cmi.2020.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stevens MP, Patel PK, Nori P. Involving antimicrobial stewardship programs in COVID-19 response efforts: all hands on deck. Infect Control Hosp Epidemiol. 2020;41(6):744–745. doi: 10.1017/ice.2020.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.World Health Organization (WHO) Collaborating Centre for Drugs Statistics Methodology. DDD and ATC-classification Oslo: WHO Collaborating Centre for Drugs Statistics Methodology; 2020 [updated 2020 Dec 17; cited 2021 Aug 29]. https://www.whocc.no/atc_ddd_index/.
- 23.European Centre for Disease Prevention and Control. Antimicrobial resistance in the EU/EEA (EARS-Net) - Annual Epidemiological Report 2019. Stockholm; 2020.
- 24.Kuster SP, Ruef C, Ledergerber B, et al. Quantitative antibiotic use in hospitals: comparison of measurements, literature review, and recommendations for a standard of reporting. Infection. 2008;36(6):549–559. doi: 10.1007/s15010-008-7462-z. [DOI] [PubMed] [Google Scholar]
- 25.Spivak ES, Cosgrove SE, Srinivasan A. Measuring appropriate antimicrobial use: attempts at opening the black box. Clin Infect Dis. 2016;63(12):1639–1644. doi: 10.1093/cid/ciw658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tjendra Y, Al Mana AF, Espejo AP, et al. Predicting disease severity and outcome in COVID-19 patients: a review of multiple biomarkers. Arch Pathol Lab Med. 2020;144(12):1465–1474. doi: 10.5858/arpa.2020-0471-SA. [DOI] [PubMed] [Google Scholar]
- 27.Charlson ME, Pompei P, Ales KL, MacKenzie CR. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis. 1987;40(5):373–383. doi: 10.1016/0021-9681(87)90171-8. [DOI] [PubMed] [Google Scholar]
- 28.Singer M, Deutschman CS, Seymour CW, et al. The third international consensus definitions for sepsis and septic shock (Sepsis-3) JAMA. 2016;315(8):801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lansbury L, Lim B, Baskaran V, Lim WS. Co-infections in people with COVID-19: a systematic review and meta-analysis. J Infect. 2020;81(2):266–275. doi: 10.1016/j.jinf.2020.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garcia-Vidal C, Sanjuan G, Moreno-García E, et al. Incidence of co-infections and superinfections in hospitalized patients with COVID-19: a retrospective cohort study. Clin Microbiol Infect. 2020;27(1):83–88. doi: 10.1016/j.cmi.2020.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Russell CD, Fairfield CJ, Drake TM, et al. Co-infections, secondary infections, and antimicrobial use in patients hospitalised with COVID-19 during the first pandemic wave from the ISARIC WHO CCP-UK study: a multicentre, prospective cohort study. Lancet Microbe. 2021;2(8):e354–e365. doi: 10.1016/S2666-5247(21)00090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vaughn VM, Gandhi T, Petty LA, et al. Empiric antibacterial therapy and community-onset bacterial co-infection in patients hospitalized with COVID-19: a multi-hospital cohort study. Clin Infect Dis. 2020;72(10):e533–e541. doi: 10.1093/cid/ciaa1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Townsend L, Hughes G, Kerr C, et al. Bacterial pneumonia coinfection and antimicrobial therapy duration in SARS-CoV-2 (COVID-19) infection. JAC Antimicrob Resist. 2020;2(3):dlaa71. doi: 10.1093/jacamr/dlaa071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kubin CJ, McConville TH, Dietz D, et al. Characterization of bacterial and fungal infections in hospitalized patients with coronavirus disease 2019 and factors associated with health care-associated infections. Open Forum Infect Dis. 2021;8(6):ofab201. doi: 10.1093/ofid/ofab201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Laethem J, Wuyts S, Van Laere S, et al. Antibiotic prescriptions in the context of suspected bacterial respiratory tract superinfections in the COVID-19 era: a retrospective quantitative analysis of antibiotic consumption and identification of antibiotic prescription drivers. Intern Emerg Med. 2021:1–11. 10.1007/s11739-021-02790-0. [DOI] [PMC free article] [PubMed]
- 36.Wang L, Amin AK, Khanna P, et al. An observational cohort study of bacterial co-infection and implications for empirical antibiotic therapy in patients presenting with COVID-19 to hospitals in North West London. J Antimicrob Chemother. 2021;76(3):796–803. doi: 10.1093/jac/dkaa475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vandael E, Latour K, Catry B. Antimicrobial consumption in Belgium: 10-year evolution (2010–2019) in community, nursing homes and hospitals. Brussels, Belgium: Sciensano; 2021. Report No.: D/2021/14.440/10.
- 38.Pettit NN, Nguyen CT, Lew AK, et al. Reducing the use of empiric antibiotic therapy in COVID-19 on hospital admission. BMC Infect Dis. 2021;21(1):516. doi: 10.1186/s12879-021-06219-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Staub MB, Beaulieu RM, Graves J, Nelson GE. Changes in antimicrobial utilization during the coronavirus disease 2019 (COVID-19) pandemic after implementation of a multispecialty clinical guidance team. Infect Control Hosp Epidemiol. 2021;42(7):810–816. doi: 10.1017/ice.2020.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sieswerda E, de Boer MGJ, Bonten MMJ, et al. Recommendations for antibacterial therapy in adults with COVID-19 - an evidence based guideline. Clin Microbiol Infect. 2021;27(1):61–66. doi: 10.1016/j.cmi.2020.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.World Health Organization. COVID-19 clinical management: living guidance. 2021 January 25 2021. Report No.: WHO/2019-nCoV/clinical/2021.1. Geneva: WHO.
- 42.Hariyanto TI, Japar KV, Kwenandar F, et al. Inflammatory and hematologic markers as predictors of severe outcomes in COVID-19 infection: a systematic review and meta-analysis. Am J Emerg Med. 2021;41:110–119. doi: 10.1016/j.ajem.2020.12.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He S, Liu W, Jiang M, et al. Clinical characteristics of COVID-19 patients with clinically diagnosed bacterial co-infection: a multi-center study. PloS one. 2021;16(4):e0249668. doi: 10.1371/journal.pone.0249668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Terpenning MS, Taylor GW, Lopatin DE, et al. Aspiration pneumonia: dental and oral risk factors in an older veteran population. J Am Geriatr Soc. 2001;49(5):557–563. doi: 10.1046/j.1532-5415.2001.49113.x. [DOI] [PubMed] [Google Scholar]
- 45.Muder RR. Pneumonia in residents of long-term care facilities: epidemiology, etiology, management, and prevention. Am J Med. 1998;105(4):319–330. doi: 10.1016/s0002-9343(98)00262-9. [DOI] [PubMed] [Google Scholar]
- 46.Ebihara S, Sekiya H, Miyagi M, Ebihara T, Okazaki T. Dysphagia, dystussia, and aspiration pneumonia in elderly people. J Thorac Dis. 2016;8(3):632–639. doi: 10.21037/jtd.2016.02.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marik PE. Aspiration pneumonitis and aspiration pneumonia. N Engl J Med. 2001;344(9):665–671. doi: 10.1056/NEJM200103013440908. [DOI] [PubMed] [Google Scholar]
- 48.Calcagno P, Ruoppolo G, Grasso MG, De Vincentiis M, Paolucci S. Dysphagia in multiple sclerosis - prevalence and prognostic factors. Acta Neurol Scand. 2002;105(1):40–43. doi: 10.1034/j.1600-0404.2002.10062.x. [DOI] [PubMed] [Google Scholar]
- 49.Verroken A, Scohy A, Gérard L, et al. Co-infections in COVID-19 critically ill and antibiotic management: a prospective cohort analysis. Crit Care. 2020;24(1):410. doi: 10.1186/s13054-020-03135-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tanrıverdi H, Örnek T, Erboy F, et al. Comparison of diagnostic values of procalcitonin, C-reactive protein and blood neutrophil/lymphocyte ratio levels in predicting bacterial infection in hospitalized patients with acute exacerbations of COPD. Wien Klin Wochenschr. 2015;127(19–20):756–763. doi: 10.1007/s00508-014-0690-6. [DOI] [PubMed] [Google Scholar]
- 51.Bhuiyan MU, Blyth CC, West R, et al. Combination of clinical symptoms and blood biomarkers can improve discrimination between bacterial or viral community-acquired pneumonia in children. BMC Pulm Med. 2019;19(1):71. doi: 10.1186/s12890-019-0835-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li S, Huang X, Chen Z, et al. Neutrophil CD64 expression as a biomarker in the early diagnosis of bacterial infection: a meta-analysis. Int J Infect Dis. 2013;17(1):e12–23. doi: 10.1016/j.ijid.2012.07.017. [DOI] [PubMed] [Google Scholar]
- 53.Del Valle DM, Kim-Schulze S, Huang HH, et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat Med. 2020;26(10):1636–1643. doi: 10.1038/s41591-020-1051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese center for disease control and prevention. JAMA. 2020;323(13):1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 55.Richardson S, Hirsch JS, Narasimhan M, et al. Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the New York City area. JAMA. 2020;323(20):2052–2059. doi: 10.1001/jama.2020.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.