Abstract
Metabolites control epigenetic mechanisms, and conversly, cell metabolism is regulated at the epigenetic level in response to changes in the cellular environment. In recent years, this metabolo-epigenetic control of gene expression has been implicated in the regulation of multiple stages of embryonic development. The developmental potency of stem cells and their embryonic counterparts is directly determined by metabolic rewiring. Here, we review the current knowledge on the interplay between epigenetics and metabolism in the specific context of early germ cell development. We explore the implications of metabolic rewiring in primordial germ cells in light of their epigenetic remodeling during cell fate determination. Finally, we discuss the relevance of concerted metabolic and epigenetic regulation of primordial germ cells in the context of mammalian transgenerational epigenetic inheritance.
Keywords: metabolism, epigenetics, primordial germ cells, alpha-ketoglutarate
The interplay between metabolism and epigenetics participates in the development of primordial germ cells and may play a role in transgenerational epigenetic inheritance.
Introduction
Primordial germ cells (PGCs) are the embryonic precursors of gametes in metazoans. As such, PGCs are at the origin of new organisms and ensure the faithful passage of genetic and epigenetic information across generations [1, 2]. During their development, mammalian PGCs are subjected to a unique and comprehensive epigenetic remodeling, coinciding with their transition toward totipotency [3–6]. Indeed, while PGCs are unipotent progenitor cells, they differentiate into gametes that possess the unique ability to reacquire totipotency upon fertilization, leading some authors to coin PGCs as dormant totipotent cells [7]. Similar to other stem cells [8], the dynamic changes in PGC epigenome are considered of crucial importance for their extended developmental potency and their capacity to differentiate into gametes.
An increasing body of literature points toward the interplay of metabolism and epigenetics in the acquisition and maintenance of potency programs in stem cells [9–12]. This has allowed a new paradigm to emerge where cellular metabolism, in addition to providing energy, also generates key metabolites for epigenetic modifications. Hence, the metabolic status of stem cells, rather than being merely a consequence of their lineage commitment, acts as a driver for cell fate decisions [9, 13]. As a consequence, harnessing metabolic reprogramming to manipulate stem cell fate could pave the way for the development of novel therapies [9, 14]. In addition to the metabolic control of epigenomes, metabolic pathways are conversely regulated at the epigenetic level in response to environmental cues [9]. While this research avenue is currently less explored in stem cells, studies in cancer cells highlight the importance of the bidirectional regulation between metabolism and epigenetics for the integration of changes in the cellular microenvironment [15].
The metabolo-epigenetic control of cell fate is important at several stages of mammalian embryonic development [16], including during zygotic genome activation [17]. In addition, recent studies have also started to address how cellular metabolism plays a role in PGC development and their reacquisition of totipotency [7, 18]. Here, we review these findings and further discuss what the interplay between metabolism and epigenetics might imply for the epigenetic remodeling of PGCs and for the phenomenon of transgenerational epigenetic inheritance.
Metabolic requirements during PGC development
Energetic metabolism of stem cells
Mammalian cells produce energy under the form of adenosine triphosphate (ATP) by varying the levels of glycolysis and oxidative phosphorylation (OXPHOS, Figure 1) [19–21]. Glycolysis is a metabolic pathway occurring in the cytoplasm during which glucose transported from the outer cellular environment is progressively reduced into pyruvate. In the presence of oxygen, pyruvate is carried to the mitochondria where it is irreversibly converted into acetyl coenzyme A (acetyl-CoA), which then enters the tricarboxylic acid (TCA) cycle (or Krebs cycle, Figure 1). Successive oxidation of acetyl-CoA in the TCA cycle generates only one molecule of ATP but provokes the accumulation of electron donors (NADH and FADH2), which feed the electron transport chain (ETC, Figure 1), ultimately driving ATP synthesis. The net energetic yield of glycolysis is two molecules of ATP per molecule of glucose, whereas OXPHOS provides up to 36 molecules of ATP per molecule of glucose. Purely in terms of energetic yield and in aerobic conditions, relying solely on glycolysis for energy production is thus inefficient. However, aerobic glycolysis is predominant in multiple cell types that share proliferating characteristics because growth and division require building materials in addition to energy. Indeed, glycolytic intermediates are essential for anabolic reactions and participate in the synthesis of fatty acids, amino acids, and nucleotides (reviewed in [19]).
Figure 1.
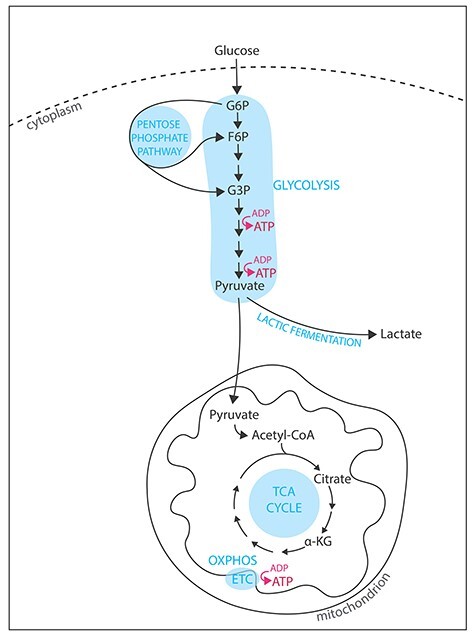
Energy pathways in mammalian cells. Metabolic pathways involved in energy production in mammalian cells. OXPHOS is the most efficient pathway for ATP production. However, glycolysis in aerobic conditions has been reported in cancer (where pyruvate is mainly fermented in lactate) and in ESCs (where the pentose phosphate pathway is more active).
The main characteristics of pluripotent stem cells (PSCs) are their ability to differentiate into multiple cell types and to self-renew in vitro [22], which is reflected in their metabolic needs [9–12]. In the continuum of pluripotency, two stable pluripotent stem states have been derived from the mouse embryo: a naïve pluripotent stem state, such as embryonic stem cells (ESCs), and a primed pluripotent stem state, such as epiblast stem cells (EpiSCs) [23–25]. Mouse ESCs (mESCs) are derived from the inner cell mass of blastocysts and represent immortalization of the preimplantation embryo, whereas mouse EpiSCs (mEpiSCs) derived from the postimplantation epiblast [23]. Hence, mESCs have a more extensive developmental potency than mEpiSCs, as they can give rise to more cell types. In terms of metabolic requirements, mESCs are metabolic bivalent: they rely on both glycolysis and OXPHOS [26, 27]. Interestingly, the transition from naïve to primed pluripotency is marked by a metabolic switch toward aerobic glycolysis [26] and an increased activity of the pentose phosphate pathway (Figure 1), which stimulates nucleotide and lipid biosynthesis [12]. This metabolic switch sustaining anabolic pathways is reminiscent of the one observed in cancer cells, an effect described in the early 1920s by Otto Warburg [28]. However, during the so-called Warburg effect, metabolic rewiring of cancer cells and dependence on glycolysis generates an accumulation of lactate fermented from pyruvate [28]. Thus, metabolic rewiring in stem cells and cancer cells appears to differ in terms of the relative diversion of glycolytic metabolites to anabolic pathways, although it serves both cell populations to accumulate sufficient building materials for their proliferation.
PGCs development and in vitro modeling
In the mouse embryos, PGCs arise at the end of gastrulation, around embryonic day 7.5 (E7.5), from a founder population of about 30 to 40 cells (reviewed in [1, 29–31], Figure 2). PGC specification in the proximal epiblast is initiated by the signaling of WNT3 and bone morphogenetic proteins (BMP4, BMP2, and BMP8b) from the extraembryonic ectoderm and visceral endoderm. This signaling results in the induction of germline transcriptional programs via the transcription factors PRDM14, BLIMP1/PRDM1, and TFAP2C. As they migrate through the hindgut endoderm and start to colonize the genital ridge (from E9.5), PGCs proliferate and undergo a global epigenetic reprogramming (Figure 2) [32]. PGC transition toward totipotency is accompanied by a global loss of DNA methylation, to the lowest reported levels of 5-methylcytosines in mammalian epigenomes [33–39]. To preserve genomic stability, some loci, corresponding to transposable elements, such as the retrotransposon intracisternal A-particle, retain relatively higher levels of DNA methylation on their long-terminal repeats, thereby resisting the programmed global demethylation during PGC differentiation [5, 35]. Histone remodeling during PGCs transition to totipotency is also proposed to be functionally coordinated to safeguard the genome from DNA demethylation [4–6]. Indeed, there is an extensive reprogramming of histone post-translational modifications [40–42], including a progressive reduction in histone H3 lysine 9 dimethylation (H3K9me2) followed by an increase in H3 lysine 27 trimethylation (H3K27me3) and a decrease in symmetrical dimethylation of arginine 3 on H2A and H4 (H2A/H4R3me2s) [40–42].
Figure 2.
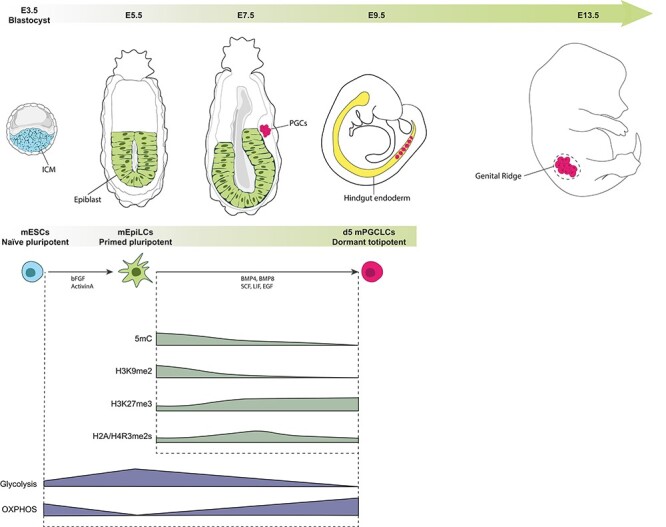
Mouse PGCs development and metabolo-epigenetic remodeling. Schematic representation of early embryonic development of PGCs in mouse, equivalent in vitro cell culture modeling, and main epigenetic remodeling events. mESCs and mouse PGCs (mPGCLCs) are both metabolically bivalent; however, distinct metabolic signatures are observed, where mESCs promote more glycolysis and mPGCLCs more OXPHOS. On the contrary, the epiblast-like cells intermediate (mEpiLCs) relies exclusively on glycolysis.
Because the in vivo scarcity of PGCs ordinarily precludes detailed molecular analyses, much work has focused on developing and improving in vitro models for PGCs. Direct differentiation of naïve mESCs is inefficient [43]. However, the groundbreaking work of Saitou and colleagues 10 years ago proved that by mimicking the in vivo development and introducing an epiblast-like cells (EpiLCs) intermediate, murine PGC-like cells (mPGCLCs) could be readily derived in vitro [43, 44]. In this two-step model, mESCs are first cultured with Activin A and FGF2 resulting in mEpiLCs that resemble epiblasts from the post-implantation embryo [43, 44]. The induction of mPGCLCs from mEpiLCs requires supplementing culture media with cytokines, mainly BMP4 and BMP8b (Figure 2). Importantly, mPGCLCs have been found to accurately recapitulate early developmental processes and epigenetic remodeling of mPGCs, especially with the recent development of extended PGCLC in vitro culture methods [39, 44, 45].
Metabolic remodeling of PGCs in the reacquisition of totipotency
Pioneer studies 50 years ago attempted to determine the importance of metabolism to several phases of embryonic development, including during PGC differentiation [46, 47]. Early biochemical characterization in murine germ cells showed that E15 germ cells oxidize 11 times more pyruvate than glucose, indicating that late PGCs mainly rely on OXPHOS [46]. The advent of in vitro mPGC modeling, enabling the acquisition of large numbers of cells, and the development of high-throughput metabolomic techniques have recently allowed the refinement of this observation over the span of PGC early development [48]. Using combined metabolomic and proteomic approaches, Hayashi et al. were able to examine the differences in metabolites and proteins in E13.5 mPGCs, gonadal somatic cells and mESCs [7]. These analyses revealed that both mESCs and E13.5 mPGCs are metabolic bivalent. However, each cell type has a distinct metabolomic and proteomic profile and, in particular, mESCs were found to promote more glycolysis than E13.5 mPGCs [7]. Furthermore, mPGCs accumulated more enzymes and metabolites involved in the early part of the TCA cycle, compared to gonadal somatic cells and mESCs [7], which further allows determining specific metabolic signatures for each cell type (Figure 2). The authors also directly assessed OXPHOS activity by measuring the oxygen consumption rate in mESCs, mPGCLCs (equivalent to E9.5 mPGCs), E11.5 mPGCs, and E13.5 mPGCs [7]. Higher consumption of oxygen, indicative of higher OXPHOS activity, was observed in the more differentiated PGCs (Figure 2) [7]. Interestingly, differences in OXPHOS rate in E13.5 male and female mPGCs were also observed [7], which indicate sexual dimorphism in the metabolism of early germ cells [49]. Building on these data, Tischler et al. investigated in further details how metabolic changes could facilitate cell state transitions in the mPGCLC model [18]. By analyzing transcriptional changes at the single-cell level during the differentiation of mESCs into mEpiLCs, they first confirmed that the naïve-to-primed transition was associated with an upregulation of glycolysis and a decreased TCA cycle entry (Figure 2) [18]. In particular, the level of the TCA cycle enzyme isocitrate dehydrogenase (Idh2) that produces alpha-ketoglutarate (α-KG) was particularly downregulated during mESCs to mEpiLC differentiation [18]. Alpha-KG has been shown to crucially control developmental potency transition in stem cells [50, 51]. Because mESCs and mPGCLCs share metabolic and transcriptional similarities (both being metabolic bivalent and both expressing naïve pluripotent genes) [7, 44], the authors next studied the impact of α-KG on mPGCLC fate. Remarkably, the addition of a cell-permeable and stable α-KG analogue led to a 50% increase in mPGCLC induction from mEpiLCs [18]. Beyond the consideration of cell culture and the potential to facilitate the in vitro differentiation of germ cells, these findings are important for a better understanding of the metabolo-epigenetic control of germ cell fate. Indeed, α-KG, a metabolite produced in the TCA cycle, is also an important cofactor for epigenetic dioxygenases such as the enzymes from the ten-eleven translocation (TET) family that participate in DNA demethylation and the Jumonji C (JmjC) domain-containing histone demethylases (JHDMs) [52, 53].
Interplay of metabolism and epigenetics
Metabolic control of epigenetic mechanisms
Epigenetic marks refer to the reversible and heritable modifications of gene function and/or chromatin states without changes into the primary nucleotide sequence [54, 55]. Epigenetic mechanisms include the positioning and remodeling of nucleosomes, histone post-translational modifications, chemical modifications of DNA itself, noncoding RNA-mediated modifications of the chromatin structure, and the three-dimensional organization of chromatin domains in the nucleus. Chemical modifications of the chromatin by epigenetic enzymes are metabolically regulated [56, 57]. Indeed, several metabolites act as cofactors for chromatin modifications (reviewed in [58, 59]). Here we briefly summarize current knowledge of the metabolic control in the best-studied cases of chromatin (DNA or histones) methylation and in the dynamics of histone acetylation. Of note, multiple new histone post-translational modifications have been described in recent years that are directly connected with metabolism. These include different forms of lysine acylation such as crotonylation [60, 61], malonylation, succinylation, and glutarylation [62].
Chromatin methylation refers to the chemical addition of methyl groups to DNA or to histone tails and is linked to one-carbon metabolism and the TCA cycle. DNA methylation in mammals occurs onto the C5 position of cytosine pyrimidine rings (5mC), mainly in the context of CpG dinucleotides [63, 64]. CpGs can be found in specific genomic regions termed CpG islands (CGIs) that are often associated with core promoters of housekeeping genes [65]. Methylation of promoter CGIs generally leads to transcriptional repression [66], and this epigenetic mechanism has been involved in a variety of biological processes [64, 67, 68]. Methylation of histone tails consists in the addition of one, two, or three methyl groups, on either lysine or arginine residues of histone tails [69]. Similar to DNA methylation, the outcome of histone methylation on transcription is locus-specific, depending on the methylated cis-regulatory region [69]. Importantly, methyltransferases responsible for DNA and histone methylation solely rely on S-adenosylmethionine (SAM) as a universal methyl group donor [64]. SAM production derives from the one-carbon metabolism, a series of interlinked pathways including the folate and methionine cycles (Figure 3) [70]. Hence, changes in the one-carbon metabolism will affect SAM levels, with epigenetic consequences on DNA and histone methylation [15, 70]. For instance, one study found that mESCs derived SAM mostly from threonine metabolism and that threonine restriction in culture medium decreased global levels of H3K4me3, leading to slower growth and loss of pluripotency [71]. Chromatin methylation is reversible, and the removal of methyl groups is catalyzed by demethylases. Active DNA demethylation involves TET enzymes, which iteratively oxidize 5mC into 5-hydroxymethylcytosine (5hmC), then into 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC), that can be efficiently removed by base-excision DNA repair machinery [72]. Histone demethylation can be achieved by two major types of chemical reactions that have led to categorize histone demethylases in two classes: the lysine-specific demethylases (LSD/KDMs) and the JHDMs [73]. As mentioned above, the TCA intermediate α-KG (that can also be produced by transamination of glutamate) serves as cofactor for TET and JHDMs (Figure 3) [52, 53]. Again, metabolic changes in α-KG have been found to affect epigenetic and transcriptional programs both during development and in tumorigenesis [9, 15]. For instance, mESCs actively use glucose and glutamate to maintain high levels of α-KG [50]. Exogenous supplementation of α-KG during mESC culture leads to the demethylation of repressive chromatin marks such as DNA methylation and H3K9me3, H3K27me3, and H4K20me3, in turn promoting the self-renewal of naïve pluripotency [50]. The example of mESCs combined use of SAM metabolism and α-KG metabolism to epigenetically promote pluripotency programs is particularly revealing of the complexity of the interplay between metabolism and epigenetics. Not only are epigenetic marks combinatorial but metabolic pathways are also interlinked. Hence, methylation and demethylation of certain loci can be coordinated in the control of cell fate and in response to environmental changes as dynamically sensed by metabolic pathways.
Figure 3.
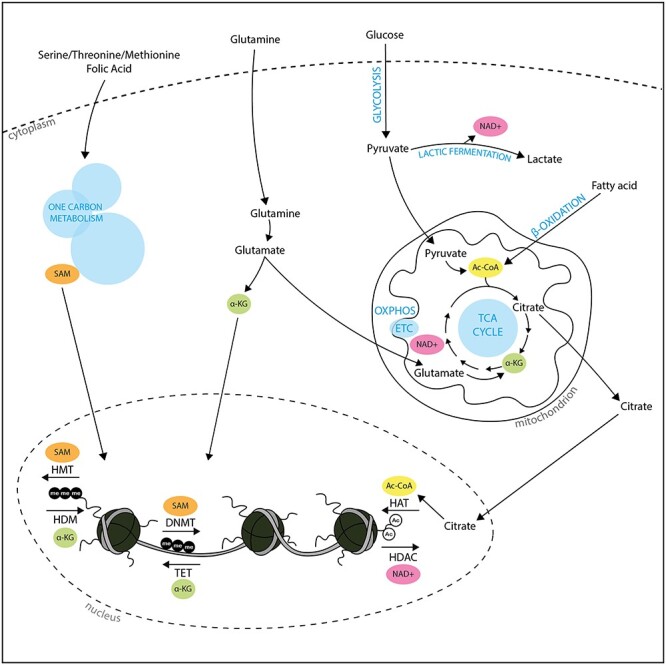
Metabolic control of epigenetic mechanisms. Four main metabolites are important in chromatin dynamics. Chromatin methylation depends on the nuclear level of SAM, deriving from the one-carbon metabolism, and nuclear levels of alpha-ketoglutarate (α-KG), deriving from glutamine or the TCA cycle. Histone acetylation, catalyzed by histone acetyltransferases (HATs) is dependent upon the levels of acetyl-coA (Ac-CoA), obtained by oxidation of pyruvate or fatty acids in the mitochondrion. One class of HDACs relies on NAD+ for its enzymatic activity.
The regulation of histone acetylation and deacetylation is tightly controlled by acetyl-CoA and NAD+ levels and represents another well-studied example of the interplay between metabolism and epigenetics in gene expression. Acetylation of histone tail lysines neutralizes their positive charge, weakening DNA:histones electrostatic interactions and resulting in an accessible chromatin structure generally favorable for transcription [74]. Acetylation is catalyzed by histone acetyltransferases that transfer an acetyl group from acetyl-CoA [74]. Acetyl-CoA can be synthesized in different cellular compartments from multiple sources, as a product from glycolysis through pyruvate, but also from translocation of mitochondrial citrate or from fatty acid through β-oxidation (Figure 3). However, glucose remains an important source for acetyl-CoA as a decrease of the glycolytic flux has been shown to significantly reduce intracellular levels of acetyl-CoA and acetylation of multiple histone lysines [75]. In fact, aerobic glycolysis in PSCs is crucial for maintaining acetyl-CoA levels and histone acetylation (particularly on H3K9 and H3K27), thereby promoting pluripotency programs [27]. Histone acetylation is reversible and histone deacetylases (HDACs) are a group of four classes of enzymes that erase the acetylation of lysines [76]. Of particular importance for the current topic, class III HDACs (or sirtuins) use NAD+ as a cofactor and can therefore directly integrate the cells energetic state into epigenetic remodeling [77] (Figure 3). During glycolysis, NAD+ is reduced to NADH, leading to lower NAD+/NADH ratios. Therefore, aerobic glycolysis is proposed to reduce sirtuin activity leading to histone hyperacetylation [15]. Indeed, studies showed that SIRT1 regulates pluripotency by promoting deacetylation of histones [78] and that during the naïve-to-primed transition SIRT1 activity is reduced [79]. However, the compartmentalization of both sirtuins and enzymes involved in NAD+ metabolism complexifies the role of sirtuins as sensors of cellular energetic states [59].
Epigenetic control of cell metabolism
The relationship between epigenetics and metabolism is bidirectional with examples of metabolic genes being regulated at the epigenetic level, particularly in several types of cancer [15]. For instance, the hexokinase isoform 2 (HK2), a key enzyme in glycolysis, is upregulated through DNA demethylation of their promoter-associated CGIs in liver cancer [80]. In addition, environment factors, such as stress, toxicant exposures, or nutrition, can modulate metabolism via epigenetic mechanisms [81–83]. For example, high-fat diet in rats results in increased DNA methylation and epigenetic silencing of hepatic enzymes promoters [84–86]. Ethanol exposure limits the availability of SAM, thereby altering DNA methylation profiles [87].
Few studies have addressed the environment-mediated epigenetic control of stem cell metabolism and how this affects potency programs. Indeed, studies on the impact of the surrounding environment on stem cell metabolism through epigenetic mechanisms have been motivated by in vitro culture systems, with metabolites increasing reprogramming efficiency of induced PSCs [14] or facilitating ESC culture [18, 50]. This is also true for the embryonic counterparts of stem cells, since studies on the environmental and epigenetic effects on the preimplantation embryo metabolism have largely been motivated by increased use of assisted reproductive technologies [88]. In this context, a fundamental understanding of how epigenetic mechanisms integrate environment changes in the metabolic control of cell fate is needed. For PGCs, this will provide insights both in their homeostatic development in utero and in their contribution to transgenerational inheritance of certain phenotypic traits.
Implications for PGCs development and transgenerational epigenetic inheritance
What causes metabolic switches during PGC reacquisition of totipotency in homeostatic conditions still needs to be answered. The metabolic switch occurring during naïve-to-primed transition has been proposed to be the result of the hypoxic environment of the endometrium where the blastocyst is implanted relative to the uterine cavity [26]. However, in this context, why OXPHOS progressively increases during PGC development is still unknown but appears important to actively promote germ cell epigenetic remodeling in a deterministic manner. Indeed, the reliance of TET and JHMDs on OXPHOS, and particularly on α-KG, suggests that they are involved in the chromatin demethylation observed during PGC reacquisition of totipotency (Figure 2). Both TET1 and TET2 participate in PGCs DNA demethylation [36] and α-KG elevation correlates with lower levels of the DNA methyltransferases DNMT3A/DNMT3B [18]. Thus, α-KG could stimulate active DNA demethylation while preventing de novo DNA methylation. Furthermore, α-KG elevation also correlates with both a global increase in H3K27me3 and a global decrease in H3K9me2 in EpiLCs [18], although the epigenetic enzymes involved have not been characterized. Interestingly, a recent study reported that JMJD1B (also known as KDM3B) possesses an arginine demethylase activity for H4R3me2s in addition to its lysine demethylase activity for H3K9me2 [89]. As JMJD1B demethylase activity is dependent on α-KG [89] and because both histone marks decrease along PGC development ([40–42], Figure 2), JMJD1B appears as an interesting candidate for the observed histone remodeling in PGCs. In addition to these global changes, local chromatin changes are expected to occur [15, 90], especially on certain cis-regulatory regions participating in the control of germline markers. Thus, current data, despite incomplete, support that actively promoting OXPHOS during PGC development facilitates their epigenetic remodeling.
In addition to these fundamental mechanistic questions, another unclear aspect is how the environment can affect PGC epigenetic remodeling through changes in the metabolism. The concept of environmental transgenerational epigenetic inheritance (TEI) proposes that environmental exposures can affect the phenotypes of successive generations in the absence of continued direct environmental influences and through germline epigenetic mechanisms (reviewed in [91]). While multiple studies report mammalian transgenerational inheritance in response to toxicants such as the agricultural fungicide vinclozolin, plastic-derived compound (such as bisphenol A), or hydrocarbons, the epigenetic mechanisms at play remain elusive [92–94]. So far, most studies have focused on later developmental stages of germ cells, such as mature gametes [95], with little attention to PGCs. However, PGCs epigenetic remodeling could constitute a sensitive developmental window for exposure memory. In addition, while most toxicants are known to impact metabolism, TEI mechanisms in PGCs have not been examined in light of metabolo-epigenetic control of gene expression. In this sense, the metabolism acts as an intermediate between environmental changes and epigenetic remodeling in the germline and its potential transmission to the next generation. Proving this hypothesis would, however, necessitate the identification of the epigenetic mechanisms of transgenerational inheritance through the early developmental stage of PGCs, whether they occur in specific loci in the endovirome [96] acting as hotspots for TEI [92, 97] and/or involving a concerted epigenetic and transcriptional control of gene expression [98].
Conclusion
An increasing number of studies points toward the importance of a metabolo-epigenetic control of developmental potency of stem cells. This is underlined by the conservation of the interplay between epigenetics and metabolism along development, and its relevancy at different embryonic stages and in different embryonic lineages, including now in germ cells. Crucial fundamental questions still need to be answered, including why is metabolism rewired and what are the consequences of these metabolic switches on PGC epigenetic and transcriptional programs. The metabolo-epigenetic control of germline fate also brings an additional layer of complexity in studies on environment exposures that alter cell metabolism and their potential transgenerational inheritance. Finally, harnessing metabolism in cell differentiation constitutes a formidable tool for in vitro differentiation of stem cells, which in the context of germ cells could provide therapeutic options in the treatment of infertility [47, 99].
Grant Support: The work in the Allard lab is supported by NIEHS R01 ES027487 and the John Templeton Foundation Grant 60742.
Contributor Information
Roxane Verdikt, Institute for Society and Genetics, University of California, Los Angeles, Los Angeles, CA, USA.
Patrick Allard, Institute for Society and Genetics, University of California, Los Angeles, Los Angeles, CA, USA; Molecular Biology Institute, University of California, Los Angeles, Los Angeles, CA, USA.
Data availability
The authors confirm that the data supporting this study are available within the article or within the cited references.
Conflict of interest
The authors declare no conflict of interest.
References
- [1].Saitou M, Yamaji M. Primordial germ cells in mice. Cold Spring Harb Perspect Biol 2012; 4:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Nikolic A, Volarevic V, Armstrong L, Lako M, Stojkovic M. Primordial germ cells: current knowledge and perspectives. Stem Cells Int 2016; 2016:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Mochizuki K, Matsui Y. Epigenetic profiles in primordial germ cells: global modulation and fine tuning of the epigenome for acquisition of totipotency. Develop Growth Differ 2010; 52:517–525. [DOI] [PubMed] [Google Scholar]
- [4].Leitch HG, Tang WWC, Surani MA. Primordial germ-cell development and epigenetic reprogramming in mammals. Curr Top Dev Biol 2013; 104:149–187. [DOI] [PubMed] [Google Scholar]
- [5].Matsui Y, Mochizuki K. A current view of the epigenome in mouse primordial germ cells. Mol Reprod Dev 2014; 81:160–170. [DOI] [PubMed] [Google Scholar]
- [6].Tang WWC, Kobayashi T, Irie N, Dietmann S, Surani MA. Specification and epigenetic programming of the human germ line. Nat Rev Genet 2016; 17:585–600. [DOI] [PubMed] [Google Scholar]
- [7].Hayashi Y, Otsuka K, Ebina M, Igarashi K, Takehara A, Matsumoto M, Kanai A, Igarashi K, Soga T, Matsui Y. Distinct requirements for energy metabolism in mouse primordial germ cells and their reprogramming to embryonic germ cells. Proc Natl Acad Sci U S A 2017; 114:8289–8294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Schlesinger S, Meshorer E. Open chromatin, epigenetic plasticity, and nuclear organization in pluripotency. Dev Cell 2019; 48:135–150. [DOI] [PubMed] [Google Scholar]
- [9].Ryall JG, Cliff T, Dalton S, Sartorelli V. Metabolic reprogramming of stem cell epigenetics. Cell Stem Cell 2015; 17:651–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Mathieu J, Ruohola-baker H. Metabolic remodeling during the loss and acquisition of pluripotency. Development 2017; 144:541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Zhang J, Zhao J, Dahan P, Lu V, Zhang C, Li H, Teitell MA. Metabolism in pluripotent stem cells and early mammalian development. Cell Metab 2018; 27:332–338. [DOI] [PubMed] [Google Scholar]
- [12].Folmes CDL, Dzeja PP, Nelson TJ, Terzic A. Metabolic plasticity in stem cell homeostasis and differentiation. Cell Stem Cell 2012; 11:596–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Dahan P, Lu V, Nguyen RMT, Kennedy SAL, Teitell MA. Metabolism in pluripotency: both driver and passenger? J Biol Chem 2019; 294:5420–5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Spyrou J, Gardner DK, Harvey AJ. Metabolism is a key regulator of induced pluripotent stem cell reprogramming. Stem Cells Int 2019; 2019:7360121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Yu X, Ma R, Wu Y, Zhai Y, Li S. Reciprocal regulation of metabolic reprogramming and epigenetic modifications in cancer. Front Genet 2018; 9:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Miyazawa H, Aulehla A. Revisiting the role of metabolism during development. Development 2018; 145:1–11. [DOI] [PubMed] [Google Scholar]
- [17].Nagaraj R, Sharpley MS, Chi F, Braas D, Zhou Y, Kim R, Clark AT, Banerjee U. Nuclear localization of mitochondrial TCA cycle enzymes as a critical step in mammalian zygotic genome activation. Cell 2017; 168:210–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Tischler J, Gruhn WH, Reid J, Allgeyer E, Buettner F, Marr C, Theis F, Simons BD, Wernisch L, Surani MA. Metabolic regulation of pluripotency and germ cell fate through α-ketoglutarate. EMBO J 2019; 38:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lunt SY, Vander Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol 2011; 27:441–464. [DOI] [PubMed] [Google Scholar]
- [20].Locasale JW, Cantley LC. Metabolic flux and the regulation of mammalian cell growth. Cell Metab 2011; 14:443–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Bonora M, Patergnani S, Rimessi A, de Marchi E, Suski JM, Bononi A, Giorgi C, Marchi S, Missiroli S, Poletti F, Wieckowski MR, Pinton P. ATP synthesis and storage. Purinergic Signal 2012; 8:343–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Weinberger L, Ayyash M, Novershtern N, Hanna JH. Dynamic stem cell states: naive to primed pluripotency in rodents and humans. Nat Rev Mol Cell Biol 2016; 17:155–169. [DOI] [PubMed] [Google Scholar]
- [23].Nichols J, Smith A. Naive and primed pluripotent states. Cell Stem Cell 2009; 4:487–492. [DOI] [PubMed] [Google Scholar]
- [24].Hackett JA, Azim SM. Regulatory principles of pluripotency: from the ground state up. Cell Stem Cell 2014; 15:416–430. [DOI] [PubMed] [Google Scholar]
- [25].De Los AA, Ferrari F, Xi R, Fujiwara Y, Benvenisty N, Deng H, Hochedlinger K, Jaenisch R, Lee S, Leitch HG, Lensch MW, Lujan E et al. Hallmarks of pluripotency. Nature 2015; 525:469–478. [DOI] [PubMed] [Google Scholar]
- [26].Zhou W, Choi M, Margineantu D, Margaretha L, Hesson J, Cavanaugh C, Blau CA, Horwitz MS, Hockenbery D, Ware C, Ruohola-Baker H. HIF1α induced switch from bivalent to exclusively glycolytic metabolism during ESC-to-EpiSC/hESC transition. EMBO J 2012; 31:2103–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Moussaieff A, Rouleau M, Kitsberg D, Cohen M, Levy G, Barasch D, Nemirovski A, Shen-Orr S, Laevsky I, Amit M, Bomze D, Elena-Herrmann B et al. Glycolysis-mediated changes in acetyl-CoA and histone acetylation control the early differentiation of embryonic stem cells. Cell Metab 2015; 21:392–402. [DOI] [PubMed] [Google Scholar]
- [28].Liberti MV, Locasale JW. The Warburg effect: how does it benefit cancer cells? Trends Biochem Sci 2016; 41:211–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Magnúsdóttir E, Surani MA. How to make a primordial germ cell. Development 2014; 141:245–252. [DOI] [PubMed] [Google Scholar]
- [30].De Felici M. The formation and migration of primordial germ cells in mouse and man. Results Probl Cell Differ 2016; 58:23–46. [DOI] [PubMed] [Google Scholar]
- [31].Sybirna A, Wong FCK, Surani MA. Genetic basis for primordial germ cells specification in mouse and human: conserved and divergent roles of PRDM and SOX transcription factors. Curr Top Dev Biol 2019; 1351st ed. Elsevier Inc.:35–89. [DOI] [PubMed] [Google Scholar]
- [32].Kurimoto K, Saitou M. Epigenome regulation during germ cell specification and development from pluripotent stem cells. Curr Opin Genet Dev 2018; 52:57–64. [DOI] [PubMed] [Google Scholar]
- [33].Hajkova P, Erhardt S, Lane N, Haaf T, El-Maarri O, Reik W, Walter J, Surani MA. Epigenetic reprogramming in mouse primordial germ cells. Mech Dev 2002; 117:15–23. [DOI] [PubMed] [Google Scholar]
- [34].Guibert S, Forné T, Weber M. Global profiling of DNA methylation erasure in mouse primordial germ cells. Genome Res 2012; 22:633–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Seisenberger S, Andrews S, Krueger F, Arand J, Walter J, Santos F, Popp C, Thienpont B, Dean W, Reik W. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol Cell 2012; 48:849–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Hackett JA, Sengupta R, Zylicz JJ, Murakami K, Lee C, Down TA, Surani MA. Germline DNA demethylation dynamics and imprint erasure through 5-Hydroxymethylcytosine. Science (80- ) 2013; 339:448–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Gkountela S, Zhang KX, Shafiq TA, Liao WW, Hargan-Calvopiña J, Chen PY, Clark AT. DNA demethylation dynamics in the human prenatal germline. Cell 2015; 161:1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Shirane K, Kurimoto K, Yabuta Y, Yamaji M, Satoh J, Ito S, Watanabe A, Hayashi K, Saitou M, Sasaki H. Global landscape and regulatory principles of DNA methylation reprogramming for germ cell specification by mouse pluripotent stem cells. Dev Cell 2016; 39:87–103. [DOI] [PubMed] [Google Scholar]
- [39].von Meyenn F, Berrens RV, Andrews S, Santos F, Collier AJ, Krueger F, Osorno R, Dean W, Rugg-Gunn PJ, Reik W. Comparative principles of DNA methylation reprogramming during human and mouse in vitro primordial germ cell specification. Dev Cell 2016; 39:104–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Ancelin K, Lange UC, Hajkova P, Schneider R, Bannister AJ, Kouzarides T, Surani MA. Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat Cell Biol 2006; 8:623–630. [DOI] [PubMed] [Google Scholar]
- [41].Seki Y, Yamaji M, Yabuta Y, Sano M, Shigeta M, Matsui Y, Saga Y, Tachibana M, Shinkai Y, Saitou M. Cellular dynamics associated with the genome-wide epigenetic reprogramming in migrating primordial germ cells in mice. Development 2007; 134:2627–2638. [DOI] [PubMed] [Google Scholar]
- [42].Kurimoto K, Yabuta Y, Hayashi K, Ohta H, Kiyonari H, Mitani T, Moritoki Y, Kohri K, Kimura H, Yamamoto T, Katou Y, Shirahige K et al. Quantitative dynamics of chromatin remodeling during germ cell specification from mouse embryonic stem cells. Cell Stem Cell 2015; 16:517–532. [DOI] [PubMed] [Google Scholar]
- [43].Hayashi M, Kawaguchi T, Durcova-Hills G, Imai H. Generation of germ cells from pluripotent stem cells in mammals. Reprod Med Biol 2018; 17:107–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Hayashi K, Ohta H, Kurimoto K, Aramaki S, Saitou M. Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell 2011; 146:519–532. [DOI] [PubMed] [Google Scholar]
- [45].Ohta H, Kurimoto K, Okamoto I, Nakamura T, Yabuta Y, Miyauchi H, Yamamoto T, Okuno Y, Hagiwara M, Shirane K, Sasaki H, Saitou M. In vitro expansion of mouse primordial germ cell-like cells recapitulates an epigenetic blank slate. EMBO J 2017; 36:1888–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Brinster RL, Harstad H. Energy metabolism in primordial germ cells of the mouse. Exp Cell Res 1977; 109:111–117. [DOI] [PubMed] [Google Scholar]
- [47].Leese HJ. Metabolism of the preimplantation embryo: 40 years on. Reproduction 2012; 143:417–427. [DOI] [PubMed] [Google Scholar]
- [48].Hayashi Y, Matsui Y. Metabolomic and proteomic analyses of mouse primordial germ cells. In: Turksen K (ed.), Stem Cells and Aging, vol. 2045. 2nd ed. Springer Protocols, New York; 2019:259–269. [DOI] [PubMed] [Google Scholar]
- [49].Hayashi Y, Mori M, Igarashi K, Tanaka K, Takehara A, Ito-Matsuoka Y, Kanai A, Yaegashi N, Soga T, Matsui Y. Proteomic and metabolomic analyses uncover sex-specific regulatory pathways in mouse fetal germline differentiation. Biol Reprod 2020; 103:717–735. [DOI] [PubMed] [Google Scholar]
- [50].Carey BW, Finley LWS, Cross JR, Allis CD, Thompson CB. Intracellular α-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature 2015; 518:413–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].TeSlaa T, Chaikovsky AC, Lipchina I, Escobar SL, Hochedlinger K, Huang J, Graeber TG, Braas D, Teitell MA. α-Ketoglutarate accelerates the initial differentiation of primed human pluripotent stem cells. Cell Metab 2016; 24:485–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Accari SL, Fisher PR. Emerging roles of JmjC domain-containing proteins. Int Rev Cell Mol Biol 2015; 319:165–220. [DOI] [PubMed] [Google Scholar]
- [53].Islam S, Leissing TM, Chowdhury R, Hopkinson RJ, Schofield CJ. 2-Oxoglutarate-dependent oxygenases. Annu Rev Biochem 2018; 87:1–36. [DOI] [PubMed] [Google Scholar]
- [54].Bird A. Perceptions of epigenetics. Nature 2007; 447:396–398. [DOI] [PubMed] [Google Scholar]
- [55].Murr R. Interplay between different epigenetic modifications and mechanisms. Adv Genet 2010; 70:101–141. [DOI] [PubMed] [Google Scholar]
- [56].Meier JL. Metabolic mechanisms of epigenetic regulation. ACS Chem Biol 2013; 8:2607–2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Reid MA, Dai Z, Locasale JW. The impact of cellular metabolism on chromatin dynamics and epigenetics. Nat Cell Biol 2017; 19:1298–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Li X, Egervari G, Wang Y, Berger SL, Lu Z. Regulation of chromatin and gene expression by metabolic enzymes and metabolites. Nat Rev Mol Cell Biol 2018; 19:563–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Boon R, Silveira GG, Mostoslavsky R. Nuclear metabolism and the regulation of the epigenome. Nat Metab 2020; 2:1190–1203. [DOI] [PubMed] [Google Scholar]
- [60].Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, Buchou T, Cheng Z, Rousseaux S, Rajagopal N, Lu Z, Ye Z et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 2011; 146:1016–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Fang Y, Xu X, Ding J, Yang L, Doan MT, Karmaus PWF, Snyder NW, Zhao Y, Li JL, Li X. Histone crotonylation promotes mesoendodermal commitment of human embryonic stem cells. Cell Stem Cell 2021; 28:748–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Sabari BR, Zhang D, Allis CD, Zhao Y. Metabolic regulation of gene expression through histone acylations. Nat Rev Mol Cell Biol 2017; 18:90–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Bird A. DNA methylation patterns and epigenetic memory. Genes Dev 2002; 16:6–21. [DOI] [PubMed] [Google Scholar]
- [64].Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem 2005; 74:481–514. [DOI] [PubMed] [Google Scholar]
- [65].Haberle V, Stark A. Eukaryotic core promoters and the functional basis of transcription initiation. Nat Rev Mol Cell Biol 2018; 19:621–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Zhu H, Wang G, Qian J. Transcription factors as readers and effectors of DNA methylation. Nat Rev Genet 2016; 17:551–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Robertson KD, Wolffe AP. DNA methylation in health and disease. Nat Rev Genet 2000; 1:11–19. [DOI] [PubMed] [Google Scholar]
- [68].Klutstein M, Nejman D, Greenfield R, Cedar H. DNA methylation in cancer and aging. Cancer Res 2016; 76:3446–3450. [DOI] [PubMed] [Google Scholar]
- [69].Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet 2012; 13:343–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Clare CE, Brassington AH, Kwong WY, Sinclair KD. One-carbon metabolism: linking nutritional biochemistry to epigenetic programming of long-term development. Annu Rev Anim Biosci 2019; 7:263–287. [DOI] [PubMed] [Google Scholar]
- [71].Shyh-Chang N, Locasale JW, Lyssiotis CA, Zheng Y, Yi Teo R, Ratanasirintrawoot S, Zhang J, Onder T, Unternaehrer JJ, Zhu H, Asara JM, Daley GQ et al. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science (80- ) 2013; 339:222–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Wu X, Zhang Y. TET-mediated active DNA demethylation: mechanism, function and beyond. Nat Rev Genet 2017; 18:517–534. [DOI] [PubMed] [Google Scholar]
- [73].Kooistra SM, Helin K. Molecular mechanisms and potential functions of histone demethylases. Nat Rev Mol Cell Biol 2012; 13:297–311. [DOI] [PubMed] [Google Scholar]
- [74].Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res 2011; 21:381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Cluntun AA, Huang H, Dai L, Liu X, Zhao Y, Locasale JW. The rate of glycolysis quantitatively mediates specific histone acetylation sites. Cancer Metab 2015; 3:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Gregoretti IV, Lee YM, Goodson HV. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J Mol Biol 2004; 338:17–31. [DOI] [PubMed] [Google Scholar]
- [77].Anderson KA, Madsen AS, Olsen CA, Hirschey MD. Metabolic control by sirtuins and other enzymes that sense NAD+, NADH, or their ratio. Biochim Biophys Acta Bioenerg 1858; 2017:991–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Calvanese V, Lara E, Suárez-Álvarez B, Dawud RA, Vázquez-Chantada M, Martínez-Chantar ML, Embade N, López-Nieva P, Horrillo A, Hmadcha A, Soria B, Piazzolla D et al. Sirtuin 1 regulation of developmental genes during differentiation of stem cells. Proc Natl Acad Sci U S A 2010; 107:13736–13741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Fang Y, Tang S, Li X. Sirtuins in metabolic and epigenetic regulation of stem cells. Trends Endocrinol Metab 2019; 30:177–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Johnson C, Warmoes MO, Shen X, Locasale JW. Epigenetics and cancer metabolism. Cancer Lett 2015; 356:309–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Remely M, Stefanska B, Lovrecic L, Magnet U, Haslberger AG. Nutriepigenomics: the role of nutrition in epigenetic control of human diseases. Curr Opin Clin Nutr Metab Care 2015; 18:328–333. [DOI] [PubMed] [Google Scholar]
- [82].Landecker H. Food as exposure: nutritional epigenetics and the new metabolism. Biosocieties 2011; 6:167–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Shaughnessy DT, McAllister K, Worth L, Haugen AC, Meyer JN, Domann FE, Van Houten B, Mostoslavsky R, Bultman SJ, Baccarelli AA, Begley TJ, Sobol RW et al. Mitochondria, energetics, epigenetics, and cellular responses to stress. Environ Health Perspect 2014; 122:1271–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Jiang M, Zhang Y, Liu M, Lan MS, Fei J, Fan W, Gao X, Lu D. Hypermethylation of hepatic glucokinase and L-type pyruvate kinase promoters in high-fat diet-induced obese rats. Endocrinology 2011; 152:1284–1289. [DOI] [PubMed] [Google Scholar]
- [85].Schwenk RW, Jonas W, Ernst SB, Kammel A, Jähnert M, Schürmann A. Diet-dependent alterations of hepatic scd1 expression are accompanied by differences in promoter methylation. Horm Metab Res 2013; 45:786–794. [DOI] [PubMed] [Google Scholar]
- [86].Zhang Y, Wang H, Zhou D, Moody L, Lezmi S, Chen H, Pan YX. High-fat diet caused widespread epigenomic differences on hepatic methylome in rat. Physiol Genomics 2015; 47:514–523. [DOI] [PubMed] [Google Scholar]
- [87].Vadigepalli R, Hoek JB. Alcohol and epigenetic regulation: do the products of alcohol metabolism drive epigenetic control of gene expression in alcohol-related disorders? Alcohol Clin Exp Res 2018; 42:845–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Chason RJ, Csokmay J, Segars JH, DeCherney AH, Armant DR. Environmental and epigenetic effects upon preimplantation embryo metabolism and development. Trends Endocrinol Metab 2011; 22:412–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Li S, Ali S, Duan X, Liu S, Du J, Liu C, Dai H, Zhou M, Zhou L, Yang L, Chu P, Li L et al. JMJD1B demethylates H4R3me2s and H3K9me2 to facilitate gene expression for development of hematopoietic stem and progenitor cells. Cell Rep 2018; 23:389–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Zylicz JJ, Dietmann S, Günesdogan U, Hackett JA, Cougot D, Lee C, Surani MA. Chromatin dynamics and the role of G9a in gene regulation and enhancer silencing during early mouse development. elife 2015; 4:1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Nilsson EE, Sadler-Riggleman I, Skinner MK. Environmentally induced epigenetic transgenerational inheritance of disease. Environ Epigenetics 2018; 4:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Blake GET, Watson ED. Unravelling the complex mechanisms of transgenerational epigenetic inheritance. Curr Opin Chem Biol 2016; 33:101–107. [DOI] [PubMed] [Google Scholar]
- [93].Boskovic A, Rando OJ. Transgenerational epigenetic inheritance. Annu Rev Genet 2018; 52:21–41. [DOI] [PubMed] [Google Scholar]
- [94].Liberman N, Wang SY, Greer EL. Transgenerational epigenetic inheritance: from phenomena to molecular mechanisms. Curr Opin Neurobiol 2019; 59:189–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Daxinger L, Whitelaw E. Understanding transgenerational epigenetic inheritance via the gametes in mammals. Nat Rev Genet 2012; 13:153–162. [DOI] [PubMed] [Google Scholar]
- [96].Friedli M, Trono D. The developmental control of transposable elements and the evolution of higher species. Annu Rev Cell Dev Biol 2015; 31:429–451. [DOI] [PubMed] [Google Scholar]
- [97].van Otterdijk SD, Michels KB. Transgenerational epigenetic inheritance in mammals: how good is the evidence? FASEB J 2016; 30:2457–2465. [DOI] [PubMed] [Google Scholar]
- [98].Kremsky I, Corces VG. Protection from DNA re-methylation by transcription factors in primordial germ cells and pre-implantation embryos can explain trans-generational epigenetic inheritance. Genome Biol 2020; 21:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Hikabe O, Hamazaki N, Nagamatsu G, Obata Y, Hirao Y, Hamada N, Shimamoto S, Imamura T, Nakashima K, Saitou M, Hayashi K. Reconstitution in vitro of the entire cycle of the mouse female germ line. Nature 2016; 539:299–303. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that the data supporting this study are available within the article or within the cited references.


