Abstract
This paper describes the development and evaluation of a new nested reverse transcription (RT)-PCR for the detection of rhinovirus in clinical samples. The nucleotide sequences of the 5′ noncoding regions of 39 rhinoviruses were determined in order to map the most conserved subregions. We designed a set of rhinovirus-specific primers and probes directed to these subregions and developed a new nested RT-PCR. The new assay includes an optimal RNA extraction method and amplicon identification with probe hybridization to discriminate between rhinoviruses and the closely related enteroviruses. It proved to be highly sensitive and specific. When tested on a dilution series of cultured viruses, the new PCR protocol scored positive at 10- to 100-fold-higher dilutions than a previously used nested RT-PCR. When tested on a collection of clinical samples obtained from 1,070 acute respiratory disease patients who had consulted their general practitioners, the new assay demonstrated a rhinovirus in 24% of the specimens, including all culture-positive samples, whereas the previously used PCR assay or virus culture detected a rhinovirus in only 3.5 to 6% of the samples. This new assay should help determine the disease burden associated with rhinovirus infections.
Rhinovirus infections, the main cause of the common cold, are generally restricted to the upper airways and induce only mild symptoms. For elderly and very young individuals, however, it has been reported that rhinoviruses can also cause lower respiratory tract infections that may result in severe illness (23, 28, 29, 31). In addition, rhinovirus infections may exacerbate chronic bronchitis and asthma (10, 20, 33). The full scope of the disease burden associated with rhinovirus infections is only beginning to be recognized, since virus isolation is difficult (24, 25) and other methods for the detection of rhinovirus infections are hampered by extensive genetic variation. About 200 Picornaviridae serotypes have been identified, of which more than 100 belong to the genus Rhinovirus (36, 39). Sensitive and specific rhinovirus detection methods are needed to assess the full pathogenic potential of these viruses.
Numerous reports describing the development and application of PCRs for rhinovirus detection have been published. Most of these PCRs are based on primers that are exclusively directed towards sites with conserved nucleotide sequences in the 5′ noncoding region (NCR) (2, 12, 14, 16, 18, 21, 22, 38). Other rhinovirus PCRs also use primers directed towards the VP2 and VP4 capsid genes (11, 30, 34). Since most of these target sequences are shared with enteroviruses, which may also replicate in the respiratory tract, rhinovirus PCRs often include amplicon identification by probe hybridization (7, 11, 14, 16) or by restriction enzyme digestion (22). Other PCRs aim at rhinovirus specificity by amplifying a genome segment that differs in length between rhinoviruses and enteroviruses (4, 30, 34) or by selective secondary amplification (2, 18, 30). All rhinovirus PCRs developed to date are based on the same five rhinovirus prototype sequences presently available from the genetic databases and on the sequence homology of rhinoviruses and enteroviruses.
During a surveillance program on respiratory viral infections, we collected nose and throat swabs from individuals with acute respiratory disease who had consulted their general practitioners (GPs). These clinical samples were examined for the presence of rhinoviruses by using virus culture. The same samples were also tested with a rhinovirus PCR using primers described by Kämmerer et al. (22) in a locally developed protocol. A significant number of samples that were positive in virus culture were negative in this rhinovirus PCR. In order to improve rhinovirus detection, we determined the 5′ NCR nucleotide sequences of a panel of 39 rhinoviruses. On the basis of this sequence information, we developed a rhinovirus-specific nested reverse transcription (RT)-PCR (“new PCR”) that includes an optimal RNA extraction method and amplicon identification with probe hybridization to discriminate between rhinoviruses and enteroviruses. The sensitivity and specificity of the assay were evaluated with a dilution series of cultured viruses and with clinical samples.
MATERIALS AND METHODS
Clinical samples and patients.
Nose and throat swabs were obtained from patients with acute respiratory illness who had consulted their GPs. These GPs participate in the nationwide Continuous Morbidity Registration System of the Netherlands Institute of Primary Health Care (NIVEL, Utrecht, The Netherlands). Patients were included in the study when, in the prodromal stage for at most 4 days, at least one of a set of symptoms—cough, coryza, sore throat, frontal headache, retrosternal pain, and myalgia—was accompanied by a rise in body temperature. Clinical samples were taken from the August to May seasons of 1994 to 1995 (1994–1995) (n = 556) and 1996–1997 (n = 514). For both seasons, 9, 9, 53, 21, and 8% of the samples were taken from patients of ages 0 to 4, 5 to 14, 15 to 44, 45 to 64, and >64 years, respectively. The two sexes contributed almost equally to each age group except for the 0 to 4 group, which contained 61% boys. The nose/throat swabs were stored in 5 ml of transport medium (Hanks’ balanced salt solution containing gelatin, lactalbumin, yeast, and antibiotics) and sent by normal post to the laboratory at room temperature. Most samples were processed within 2 days (average of 1.6 days [± 1.2]). In the laboratory, transport medium was separated from the swabs and mixed, and a 1-ml aliquot was frozen (−70°C) for PCR analysis at a later stage. The remaining transport medium was centrifuged (10 min at 3,000 × g) and the supernatant was immediately used for virus isolation.
Virus culture.
Virus isolation and identification were conducted according to standard protocols (19, 25). Clinical samples (200 μl of supernatant) were inoculated into four cell cultures in tubes including tertiary cynomolgus monkey kidney cells, human diploid lung fibroblasts, HEp-2 cells, and R-HeLa cells (a rhinovirus-sensitive subline of HeLa). Multiple rhinovirus-sensitive cell cultures were used because some rhinoviruses grow only in one of the cell cultures. The maintenance medium was slightly acidic, and the tubes were incubated in roller drums at 33°C to improve rhinovirus growth (19, 25). Rhinoviruses and enteroviruses were recognized by their (similar) cytopathic effects and host cell ranges and were distinguished in an acid lability assay (19, 25).
Sequence analysis of 5′ NCRs. (i) Selection of isolates.
The 5′ NCRs of a panel of 39 rhinovirus strains were sequenced. The panel comprised 30 Dutch isolates from the 1992–1993, 1993–1994, and 1994–1995 seasons (including 14 viruses from samples of the 1994–1995 season that were negative in the old PCR) and 9 prototype viruses (serotypes 1A, 7, 21, 29, 37, 58, 62, 72, and 87) displaying different sensitivities to antiviral compounds and using different cellular receptors (1, 40). The prototype viruses were obtained from the American Type Culture Collection (Manassas, Va.).
(ii) NCR amplification and sequence analysis.
Virion-associated RNA was released by heating 25 μl of culture supernatant for 30 min at 65°C in the presence of 5 units of RNase inhibitor (Amersham Life Science, Little Chalfont, United Kingdom). Five microliters of this preparation was subsequently used in a 25-μl single-tube RT-PCR, as described in “Nested RT-PCR protocol,” below. RT-PCR was performed with primers Pr7 and Pr13 (30) or with primers Rtheo1 (5′-TTAAAACTGGRTSTGGGTTGTTCCCAC-3′) and Pr13 (30). Three distinct Pr7 primers were used, each modified according to the nucleotide sequences of rhinovirus serotypes 2, 14, and 16. The secondary (seminested) amplification was performed with the same primers except for Pr13, which was replaced by either Pr12 (30) or primer 1 (35). The generated amplicons were purified with a Spin-X column (Costar, Corning, N.Y.) and directly sequenced with the Taq DyeDeoxy terminator cycle sequencing kit (Amersham Life Science) on a 373A DNA automated sequencer from Applied Biosystems (Foster City, Calif.). Sequence alignment and analysis were conducted by using the GeneWorks and DNASTAR software packages (Oxford Molecular Group, Oxford, United Kingdom, and Lasergene, Madison, Wis., respectively).
Primers.
Primers complementary to the most-conserved 5′ NCR subregions were designed by using the computer program Oligo (Oligo, Dortmund, Germany). Primer sequences are given in Fig. 1B. Oligonucleotides were purchased from Perkin-Elmer (Norwalk, Conn.).
FIG. 1.
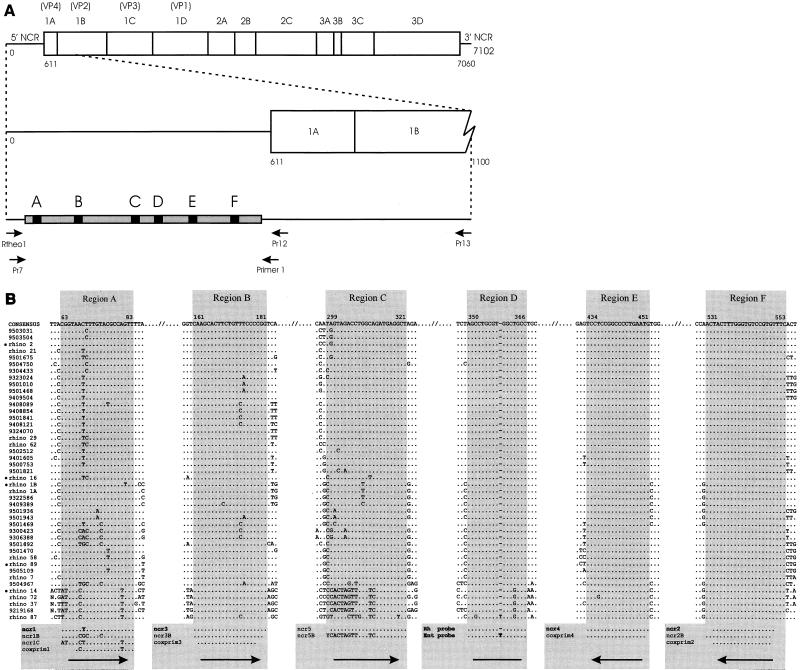
(A) Locations and orientations (arrows) of primers used to amplify the 5′ NCRs of 39 rhinovirus isolates and prototypes (see Materials and Methods). The gray bar represents the 5′ NCR from which the nucleotide sequence was determined. The black regions, labeled A to F, indicate the locations of the relatively long, well-conserved subregions. For reference, the complete rhinovirus genome is given at the top. (B) Nucleotide sequences (cDNA) of the six relatively long, well-conserved subregions of the rhinovirus 5′ NCR. At the top, the consensus sequence of each region is given together with its relative position based on the rhinovirus serotype 2 sequence. Identical residues are indicated with dots; a dash indicates that no nucleotide residue is present at that particular position. At the bottom, the sequences of the primers and probes used in the study are given. Arrows indicate primer orientations. The coxprim1 to coxprim4 primers were previously published by Kämmerer et al. (22). The codes of the rhinovirus field isolates and prototypes from which the sequences were derived are given on the left. Rhinovirus serotype x is abbreviated as rhino x. The seven-digit numbers represent field isolates; the first two digits indicate the year of isolation. The sequences of rhinoviruses marked with bullets were downloaded from the GenBank sequence database.
RNA extraction methods.
We compared five RNA extraction methods. In extraction method A, a 25-μl sample was incubated for 30 min at 65°C in the presence of 5 units of RNase inhibitor (Amersham Life Science), and 5 μl was subsequently used in the PCR, as described in “Nested RT-PCR protocol,” below. Method B consisted of RNA extraction with Trizol reagent (Life Technologies, Rockville, Md.). Method C consisted of RNA extraction with the QIAamp viral RNA purification kit (Qiagen, Hilden, Germany). Method D consisted of RNA extraction with the High Pure RNA isolation kit (Boehringer, Mannheim, Germany). Method E was a silica-based RNA extraction method, according to Boom et al. (6). In methods B, C, D, and E, RNA was extracted from a 100-μl sample and the RNA was collected in either 50 μl (methods B, C, and D) or 25 μl (method E) of water, according to the instructions of the respective manufacturers; 5 μl of these RNA preparations were then used in the PCR (see below).
Nested RT-PCR protocol.
In the new PCR, viral RNA was extracted from 200 μl of transport medium by using the High Pure RNA isolation kit (Boehringer). Five microliters of the eluted RNA preparation was then used in a 25-μl single-tube RT-PCR. RT-PCR conditions were 50 mM Tris-HCl (pH 8.5), 50 mM NaCl, 6 mM MgCl2, 2 mM dithiothreitol, deoxynucleoside triphosphates (1 mM each), 2.5 units of RNase inhibitor (Amersham Life Science), 6 units of avian myeloblastosis virus reverse transcriptase (Boehringer), 1 unit of Taq polymerase (Perkin-Elmer), and 12.5 pmol of primers ncr2 and ncr1. Upon the RT reaction (0.5 h at 50°C), the 5′ NCR was amplified by 20 cycles at 95°C for 1 min, 50°C for 1 min, and 72°C for 2 min. Secondary amplification was performed on 1 μl of the RT-PCR mix in a final volume of 25 μl with primers ncr3 and ncr4 by 40 cycles at 95°C for 1 min, 64°C for 10 s, and 72°C for 2 min. Amplicon gel electrophoresis, blotting, hybridization with biotin-labeled rhinovirus- and enterovirus-specific probes, and subsequent detection were conducted according to standard protocols (37). Precautions for minimizing and checking for contamination included performing sample preparation, reaction mix preparation, and product analysis in separate rooms and the simultaneous processing of negative control samples, comprising 20% of the number of test samples. The old PCR protocol, initially used in the 1994–1995 season, was identical to the above protocol except that virion-associated RNA was released by incubating the clinical samples at 65°C (method A in “RNA extraction methods,” above) and that primers coxprim1 to coxprim4, as published by Kämmerer et al. (22), were used.
Evaluation of the newly developed rhinovirus PCR.
To evaluate the sensitivity of the new PCR, it was applied to serial dilutions of cell culture-grown rhinovirus preparations. Furthermore, the rhinovirus detection rate in clinical samples of the new PCR was compared with that of virus culture (1994–1995 and 1996–1997 seasons) and by our previously used old PCR assay (1994–1995 season only). The specificity of the new PCR was determined by sequence analysis and virus culture (combined with the acid lability test).
Nucleotide sequence accession numbers.
The nucleotide sequence data reported in this paper have been submitted to the GenBank database under accession no. AF108149 to AF108187.
RESULTS
NCR sequence analysis. (i) NCR amplification.
We sequenced the 5′ NCRs of 39 rhinoviruses, including recent isolates and prototypes, in order to design optimally matching primers for a rhinovirus-specific PCR. To obtain nucleotide sequences encompassing the most-conserved 5′ NCR subregions, amplification by seminested PCR and subsequent sequencing were done with primers directed towards less well conserved sites at the 5′ terminus of the 5′ NCR and in the open reading frames VP4 and VP2. First, primers (Pr7, Pr13, and Pr12) published by Mori and Clewley (30) were used (Fig. 1A). Using three versions of the 5′ primer Pr7, adapted to the nucleotide sequence of rhinovirus serotypes 2, 14, and 16, we were able to amplify the 5′ NCRs from eight of the nine selected prototype rhinoviruses and from 20 of the 30 selected field isolates. Subsequent attempts to amplify (parts of) the 5′ NCRs of the other selected rhinovirus isolates and prototype viruses with several combinations of primers indicated that PCR failure was caused by (additional) sequence variation in the Pr7 region. The 5′ NCRs of these isolates and prototype viruses, however, were amplified by using the primer Rtheol, which is directed towards the 5′ terminus of the rhinovirus genome (Fig. 1A). Finally, the 5′ NCR of rhinovirus serotype 87 was amplified by replacing primer Pr12 with primer 1, an enterovirus-specific primer (35).
(ii) NCR sequence variation.
Analysis of the 39 obtained rhinovirus 5′ NCR sequences, together with the five rhinovirus prototype 5′ NCR sequences available from the genetic databases, revealed a particular pattern of sequence variation: short stretches of generally 1 to 6 relatively well conserved nucleotides separated by stretches (mostly similar in size) of highly variable residues (data not shown). This pattern resulted in a relatively high level of nucleotide sequence variation (up to 50%) between individual 5′ NCR sequences. Figure 2 shows a dendrogram based on an alignment of all 44 NCR sequences available (about 470 nucleotides bordered by regions A and F [see below]). It appears that these rhinovirus 5′ NCR sequences group into multiple clusters. The rhinovirus serotype 87 NCR sequence was the most divergent, which is in line with its particular phenotype. The cellular receptor used by rhinovirus serotype 87 is different from those used by all other rhinoviruses examined (13, 15, 27, 40). Comparison of the rhinovirus serotype 87 NCR sequence with the 5′ NCR sequences of enteroviruses revealed that this rhinovirus is genetically much more closely related to enteroviruses than to other rhinoviruses (data not shown).
FIG. 2.
Dendrogram based on the sequence alignment of a 530-nucleotide region (bordered by regions A and F) of the 5′ NCRs of 44 rhinoviruses. The codes of the rhinovirus field isolates (seven-digit numbers) and prototypes (“rhino x”) from which the sequences were derived are given on the right. ■, sequence obtained from the GenBank database.
(iii) Conserved 5′ NCR subregions.
Fortunately, a number of large and well-conserved subregions suitable for primer design are present in the variable rhinovirus 5′ NCR. Most of these regions, A to F (Fig. 1B), have already been used by others for rhinovirus detection by PCR. In our large set of 5′ NCR sequences, regions E and F appeared to be most conserved. Only one nucleotide substitution was observed for a single sequence in region E. Region D, which we used for probe hybridization to discriminate between rhinoviruses and enteroviruses, showed only very limited sequence variation (predominantly at nucleotide position 363). Discriminative probe hybridization is based on an extra thymidine residue present in enteroviruses at position 359 (16). In our data set, such a residue was absent from all but one rhinovirus 5′ NCR, namely that of rhinovirus serotype 87. Region C appeared as a new, previously unrecognized conserved region. It displayed a separate consensus sequence specific for a cluster of rhinoviruses, including the serotype 14 strain (Fig. 2). Similarly, region A showed sequence variation that is partly cluster specific. Finally, region B displayed little sequence variation, almost exclusively restricted to two central nucleotide positions.
Improvement of the rhinovirus PCR. (i) Primer selection and detection limits.
We first designed primers directed towards the conserved 5′ NCR subregions A to F (Fig. 1B). Since others have previously designed primers for the same regions, most of the primers used in this study are only marginally different in sequence. Six primer combinations were tested at various annealing temperatures in a nested RT-PCR format with three rhinoviruses, namely, rhinovirus serotype 2 and rhinoviruses 9219168 and 9504967. These rhinoviruses were selected to represent the most divergent 5′ NCR sequences (Fig. 1B). The assay sensitivities achieved by the various primer combinations were determined by testing the primers with a series of 10-fold dilutions of each of the three rhinoviruses (data not shown). Three of the primer sets that amplified these viruses most efficiently, together with a previously used primer set (set I [Table 1]), were subsequently tested with a larger panel of viruses consisting of 13 rhinoviruses and 7 enteroviruses (rhinoviruses 9219168, 9504967, serotype 2, serotype 87, serotype 14, 9503504, 9501469, 9306388, 9408089, 9501936, 9501943, 9409389, and serotype 72; enteroviruses coxsackievirus A9, coxsackievirus A21, coxsackievirus B3, coxsackievirus B4, echovirus 11, echovirus 12, and enterovirus 70). Primer set III proved to be superior to or performed as well as the three other primer sets for all rhinoviruses tested except for rhinovirus field strain 9409389 (data not shown). On average, the detection limit for rhinoviruses was 10 times lower with a nested RT-PCR based on primer set III (the new PCR) than with the previously used nested RT-PCR based on primer set I (the old PCR). Primer set III is not the best choice for the detection of enteroviruses. The nested RT-PCR based on primer set I appeared to be most sensitive for the detection of the—arbitrarily selected—enteroviruses. In the present study, we focused specifically on rhinovirus detection. In order to develop a PCR for the sensitive detection of both rhinoviruses and enteroviruses, additional primers that are more specific for enteroviruses (e.g., directed to region A) should be included.
TABLE 1.
Oligonucleotide primer setsa evaluated in a nested RT-PCR with dilution series of 13 rhinoviruses and 7 enteroviruses
| Primer pair | Primer set
|
|||
|---|---|---|---|---|
| I | II | III | IV | |
| Outer | coxprim1 + ncr2 | coxprim3 + ncr2 | ncr1 + ncr2 | coxprim3 + ncr2 |
| Inner | coxprim3 + coxprim4 | ncr5 + ncr4 | ncr3 + ncr4 | ncr3 + ncr4 |
See Fig. 1B for oligonucleotide sequences.
(ii) Selected primers and risk of mismatches.
The primers of set III were all elongated compared with the primers directed towards the same regions that were used in other PCRs. Only primer ncr1 had an altered nucleotide composition. The absence of sequence variation in region F allowed the crucial initiation step of cDNA synthesis to be primed with a perfectly matching primer (ncr2). We observed the most sequence variation in regions A and B. Therefore, primer mismatches are expected to predominantly affect the annealing of only one of the primers (the sense primer) of each primer pair. With this particular distribution of the observed sequence variation over the primer target sequences, the impact of variability in regions A and B on assay sensitivity is expected to be low, especially since both sense primers are elongated compared to those previously designed for these regions. Moreover, primer ncr1 no longer carries mismatched nucleotides to the consensus sequence of the majority of rhinoviruses in this region. Because of the elongation of the primer, the occasional mismatch expected in the target sequence of the sense primer for region B (ncr3) will now be located at a greater distance from the 3′ primer terminus (Fig. 1B) and therefore will have less influence on the efficiency of the priming process.
(iii) RNA extraction method.
Five RNA extraction methods were compared with respect to their efficiencies in recovering rhinovirus RNA from nose/throat swabs. Two positive clinical samples that were each serially diluted in pooled negative clinical samples were used. By methods C, D, and E, all based on the adherence of RNA to silica particles, the detection limit of the assay was about 10 times lower than that of methods A and B, which rely on RNA release by heating only and on ethanol precipitation after virus membrane solubilization, respectively (results not shown). The High Pure RNA isolation kit of Boehringer (method D) was selected for further experiments, since it was the least time consuming.
Evaluation of the newly developed rhinovirus PCR. (i) Sensitivity.
To determine whether the enhanced analytical sensitivity of the newly developed PCR (new PCR) did indeed result in an improved detection rate of rhinovirus genomic RNA in clinical samples, we tested a large number of nose/throat swabs from patients with acute respiratory disease who had consulted their GPs. We examined 556 swabs from the 1994–1995 season and 514 swabs from the 1996–1997 season by using virus isolation (both seasons), the old PCR (1994–1995 season), and the new PCR (both seasons). Table 2 shows the results obtained in both seasons. In the 1994–1995 season, the old PCR tested positive in about half of the culture-positive specimens and detected rhinovirus in another 3% of the culture-negative samples. In contrast, the new PCR scored positive in all culture-positive samples and identified rhinovirus in about 18% of the culture-negative specimens, raising the PCR detection rate for this season from 6.1 to 23.9%. No specimens from the 1996–1997 season were examined with the old PCR, but a very similar fraction of the samples in this season tested rhinovirus positive when assayed with the new PCR (23.5%). Virus culture detected only 3.5% positive samples in this season.
TABLE 2.
Rhinovirus detection in clinical samples
| Sample parameter | No. (%)
|
|
|---|---|---|
| 1994–1995 season | 1996–1997 seasona | |
| No. tested | 556 | 514 |
| Positive by virus culture | 35 (6.3) | 18 (3.5) |
| Positive by old PCR | 34 (6.1) | ND |
| Positive by virus culture and old PCR | 18 (3.2) | ND |
| Positive by new PCR | 133 (23.9) | 121 (23.5) |
ND, not done.
(ii) Specificity.
All rhinovirus culture-positive samples were also positive with the new PCR. Analysis of 63 rhinovirus PCR amplicon sequences from the 1996–1997 season revealed that all of these sequences were indeed rhinovirus sequences, indicating that the specificity of the new PCR is 100% (results not shown).
(iii) Clinical relevance.
To obtain information about the temporal association between a positive rhinovirus PCR test result and the clinical symptoms displayed by the patient, we resampled 26 patients on an average of 32 days after the first sample had tested positive with the new rhinovirus PCR. No rhinovirus was detected in any of the 19 follow-up samples taken from patients who had recovered from respiratory illness. In contrast, rhinovirus RNA was detected with the new PCR in 7 of 14 follow-up samples obtained from individuals (n = 14) still reporting respiratory illness.
DISCUSSION
To enhance the diagnostic sensitivity of the rhinovirus PCR, we analyzed a panel of 44 rhinovirus 5′ NCR sequences and designed optimally matching oligonucleotide primers. We developed a new nested RT-PCR with these primers that includes an optimal RNA extraction procedure and amplicon identification by probe hybridization to discriminate between rhinoviruses and enteroviruses. This assay proved to be highly sensitive and specific, as determined by titration of cultured rhinoviruses and by a comparative study with 1,070 clinical samples obtained during two seasons in The Netherlands.
The impulse for the present study was the observation that a number of virus culture-positive clinical specimens had tested negative with a previously used PCR. This underlines the importance of using both a virus isolation method and a PCR detection technique. These assays serve as mutual controls for sensitivity. The sequence variation displayed by the 5′ NCRs of 44 rhinoviruses was carefully mapped to assess whether primer mismatches due to sequence variation could explain the low sensitivity of the old rhinovirus PCR. Such mismatches are likely to occur, since the primers for all rhinovirus PCRs developed to date are based on the nucleotide sequences of only five rhinovirus prototypes isolated decades ago. Indeed, better matching primers could be designed for the same, but presently more accurately defined, conserved 5′ NCR subregions. When compared with the primers we initially used (coxprim1 to coxprim4), only three nucleotide substitutions were introduced into the new primers (all in the ncr1 primer [Fig. 1B]). However, the new sequence information also allowed us to elongate all four primers of the nested PCR. The increased lengths of the primers probably contributed the most to the increased sensitivity of the new PCR. One may conceive that, for example, the elongated primers ncr1 and ncr3 directed towards regions A and B still anneal efficiently even when a sequence variation that is similar to what we observed in our data set is encountered (Fig. 1B). In addition, the implemented efficient RNA extraction method also contributed significantly to the increased sensitivity of the new PCR. We have chosen to develop a nested PCR in order to obtain an assay with the highest possible sensitivity. In a pilot experiment, we obtained detection limits with a single PCR (no secondary amplification) that were 100-fold lower than we were able to achieve with a nested PCR. Only by using a nested PCR assay that incorporates an optimal RNA extraction method, optimally matching primers, and amplicon identification and detection by probe hybridization may we expect to develop a robust assay that detects most, if not all, rhinoviruses.
We designed PCR primers based on the sequence information obtained from a specific set of (tissue-cultured) rhinoviruses. We expect, however, that our new PCR will detect the vast majority of all circulating wild-type rhinoviruses since (i) the primers were designed on the basis of a large set (n = 44) of rhinoviruses, including viruses dependent on different cellular receptors and from both antiviral groups (A and B) (1, 40); (ii) we were able to obtain the 5′ NCR sequences of all rhinoviruses selected for analysis; (iii) presently circulating rhinoviruses and old prototype rhinoviruses, isolated decades ago, still appear to have the same conserved sequences; and (iv) the conserved primer regions are part of the internal ribosome entry site, whose function is essential for viral replication (5).
Since we chose to target a 5′ NCR region that is similar in length for both rhinoviruses and enteroviruses, we added a discriminative amplicon hybridization step to the rhinovirus PCR. Only nucleotide position 359, located in conserved subregion D, displayed rhinovirus- and enterovirus-associated variation that could be readily utilized. Our surveillance data indicate that the chance of rhinovirus misclassification is low since (i) we observed a 100% match between the results of probe hybridization and the results of acid lability testing for all rhinoviruses and enteroviruses cultured in the two illness seasons studied (n = 58), (ii) all 63 amplicon sequences that we have determined (rhinovirus probe positive) were true rhinovirus sequences, and (iii) only 1.6 and 3.8% of the samples were scored as enterovirus positive with the new PCR in the two monitored seasons (data not shown). Rhinovirus serotype 87 poses a special problem. It has the enterovirus-specific nucleotide at position 359 (Fig. 1B) and will therefore be falsely labeled as an enterovirus in our assay. This imperfection is difficult to avoid since we—quite unexpectedly—found that the entire 5′ NCR of rhinovirus serotype 87 is much more closely related to the 5′ NCR sequence of enteroviruses than to that of rhinoviruses (data not shown). Actually, the problem is that, irrespective of their genomic structures, rhinoviruses and enteroviruses are defined as acid-labile and acid-resistant picornaviruses, respectively.
With the new PCR, we detected a rhinovirus in about 24% of the clinical samples. This new PCR was able to detect a rhinovirus in at least four times as many specimens as could be detected by virus isolation (Table 2). This is even more significant since our culture techniques ensured optimal conditions for rhinovirus isolation, and significant inactivation of rhinovirus during transport is unlikely since the rhinovirus inactivation rate at room temperature is low (9). Still higher incidences (up to 53%) of rhinovirus infections have, however, been reported for several other PCR-based surveillance studies also targeting nonhospitalized individuals (3, 17, 18, 21, 26, 32). Several factors could explain this discrepancy. We sampled not only during the expected peak incidence of rhinovirus infection but nearly throughout the entire year, from August to May. Furthermore, most of the patients in our study suffered from relatively severe respiratory disease, which could be characterized as an influenza-like illness. Therefore, rhinovirus infections typically associated with milder respiratory diseases such as the common cold are less likely to be detected. Finally, our patients were not sampled at first symptoms but only when they had consulted their GPs, mostly because the respiratory disease had persisted. Considering these differences, our rhinovirus detection rate of 24% is surprisingly high.
There is experimental evidence that respiratory disease is positively correlated with the load of rhinoviruses in the respiratory tract (8). Introduction of an assay with a lower detection limit therefore has the inherent danger of a decreased clinical relevance of positive test results. The detection rate of rhinovirus infection in healthy individuals or, with the same line of reasoning, in patients with a respiratory disease from another cause may increase selectively relative to that in individuals actually suffering from respiratory disease caused by rhinoviruses. However, follow-up sample testing suggests a temporal association between a positive test result and the displayed respiratory disease, even when rhinoviruses were detected by the sensitive new PCR. We were unable to detect rhinoviruses in those patients who had recovered from disease, suggesting that rhinoviruses were not likely to be present as “commensal” organisms. In contrast, we detected a rhinovirus in 7 of the 14 follow-up samples when the initial respiratory symptoms persisted. In these cases, amplicon sequencing indicated viral persistence by the absence of sequence variation (data not shown). However, case control studies are required to provide further data on the causal relationship between rhinovirus infection and disease. Asymptomatic infections in 0 to 4% of adult control individuals and in as many as 12% of healthy children have been reported in other surveillance studies (18, 21).
In conclusion, we developed an improved, highly sensitive and specific nested RT-PCR for the detection of rhinoviruses in clinical specimens. Using the new PCR, we demonstrated that the acute respiratory tract disease of about 24% of the patients consulting their GPs was associated with a rhinovirus infection. The presently described assay should help determine the disease burden associated with rhinovirus infections in different categories of patients more accurately. In addition, evaluation of the clinical management of and the antiviral therapy for infections with rhinoviruses, as well as other respiratory pathogens inducing similar symptoms, can be improved with the presently described new rhinovirus PCR.
ACKNOWLEDGMENTS
We thank Klaas Bijlsma and Cees Verweij for excellent technical support. Clinical samples were generously supplied by GPs participating in the Continuous Morbidity Registration System of NIVEL, Utrecht, The Netherlands. We are especially grateful to Aad Bartelds, the coordinator of this system.
REFERENCES
- 1.Andries K, Dewindt B, Snoeks J, Wouters L, Moereels H, Lewi P J, Janssen P A. Two groups of rhinoviruses revealed by a panel of antiviral compounds present sequence divergence and differential pathogenicity. J Virol. 1990;64:1117–1123. doi: 10.1128/jvi.64.3.1117-1123.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arruda E, Hayden F G. Detection of human rhinovirus RNA in nasal washings by PCR. Mol Cell Probes. 1993;7:373–379. doi: 10.1006/mcpr.1993.1055. [DOI] [PubMed] [Google Scholar]
- 3.Arruda E, Pitkäranta A, Witek T J, Jr, Doyle C A, Hayden F. Frequency and natural history of rhinovirus infections in adults during autumn. J Clin Microbiol. 1997;35:2864–2868. doi: 10.1128/jcm.35.11.2864-2868.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atmar R L, Georghiou P R. Classification of respiratory tract picornavirus isolates as enteroviruses or rhinoviruses by using reverse transcription-polymerase chain reaction. J Clin Microbiol. 1993;31:2544–2546. doi: 10.1128/jcm.31.9.2544-2546.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belsham G J, Sonenberg N. RNA-protein interactions in regulation of picornavirus RNA translation. Microbiol Rev. 1996;60:499–511. doi: 10.1128/mr.60.3.499-511.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boom R, Sol C J, Salimans M M, Jansen C L, Wertheim van Dillen P M, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruce C B, Gama R E, Hughes P J, Stanway G. A novel method of typing rhinoviruses using the product of a polymerase chain reaction. Arch Virol. 1990;113:83–87. doi: 10.1007/BF01318355. [DOI] [PubMed] [Google Scholar]
- 8.Couch R B. Rhinoviruses. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 713–734. [Google Scholar]
- 9.Dimmock N J. Differences between the thermal inactivation of picornaviruses at “high” and “low” temperatures. Virology. 1967;31:338–353. doi: 10.1016/0042-6822(67)90179-1. [DOI] [PubMed] [Google Scholar]
- 10.Fraenkel D J, Bardin P G, Sanderson G, Lampe F, Johnston S L, Holgate S T. Lower airways inflammation during rhinovirus colds in normal and in asthmatic subjects. Am J Respir Crit Care Med. 1995;151:879–886. doi: 10.1164/ajrccm/151.3_Pt_1.879. [DOI] [PubMed] [Google Scholar]
- 11.Freymuth F, Vabret A, Galateau Salle F, Ferey J, Eugene G, Petitjean J, Gennetay E, Brouard J, Jokik M, Duhamel J F, Guillois B. Detection of respiratory syncytial virus, parainfluenzavirus 3, adenovirus and rhinovirus sequences in respiratory tract of infants by polymerase chain reaction and hybridization. Clin Diagn Virol. 1997;8:31–40. doi: 10.1016/s0928-0197(97)00060-3. [DOI] [PubMed] [Google Scholar]
- 12.Gama R E, Horsnell P R, Hughes P J, North C, Bruce C B, Al Nakib W, Stanway G. Amplification of rhinovirus specific nucleic acids from clinical samples using the polymerase chain reaction. J Med Virol. 1989;28:73–77. doi: 10.1002/jmv.1890280204. [DOI] [PubMed] [Google Scholar]
- 13.Greve J M, Davis G, Meyer A M, Forte C P, Yost S C, Marlor C W, Kamarck M E, McClelland A. The major human rhinovirus receptor is ICAM-1. Cell. 1989;56:839–847. doi: 10.1016/0092-8674(89)90688-0. [DOI] [PubMed] [Google Scholar]
- 14.Halonen P, Rocha E, Hierholzer J, Holloway B, Hyypiä T, Hurskainen P, Pallansch M. Detection of enteroviruses and rhinoviruses in clinical specimens by PCR and liquid-phase hybridization. J Clin Microbiol. 1995;33:648–653. doi: 10.1128/jcm.33.3.648-653.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hofer F, Gruenberger M, Kowalski H, Machat H, Huettinger M, Kuechler E, Blass D. Members of the low density lipoprotein receptor family mediate cell entry of a minor-group common cold virus. Proc Natl Acad Sci USA. 1994;91:1839–1842. doi: 10.1073/pnas.91.5.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hyypiä T, Auvinen P, Maaronen M. Polymerase chain reaction for human picornaviruses. J Gen Virol. 1989;70:3261–3268. doi: 10.1099/0022-1317-70-12-3261. [DOI] [PubMed] [Google Scholar]
- 17.Hyypiä T, Puhakka T, Ruuskanen O, Mäkelä M, Arola A, Arstila P. Molecular diagnosis of human rhinovirus infections: comparison with virus isolation. J Clin Microbiol. 1998;36:2081–2083. doi: 10.1128/jcm.36.7.2081-2083.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ireland D C, Kent J, Nicholson K G. Improved detection of rhinoviruses in nasal and throat swabs by seminested RT-PCR. J Med Virol. 1993;40:96–101. doi: 10.1002/jmv.1890400204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Isenberg H D, editor. Clinical microbiology procedures handbook. Washington, D.C: American Society for Microbiology; 1992. [Google Scholar]
- 20.Johnston S L, Pattemore P K, Sanderson G, Smith S, Lampe F, Josephs L, Symington P, O’Toole S, Myint S H, Tyrrell D A, et al. Community study of role of viral infections in exacerbations of asthma in 9–11 year old children. Br Med J. 1995;310:1225–1229. doi: 10.1136/bmj.310.6989.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnston S L, Sanderson G, Pattemore P K, Smith S, Bardin P G, Bruce C B, Lambden P R, Tyrrell D A, Holgate S T. Use of polymerase chain reaction for diagnosis of picornavirus infection in subjects with and without respiratory symptoms. J Clin Microbiol. 1993;31:111–117. doi: 10.1128/jcm.31.1.111-117.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kämmerer U, Kunkel B, Korn K. Nested PCR for specific detection and rapid identification of human picornaviruses. J Clin Microbiol. 1994;32:285–291. doi: 10.1128/jcm.32.2.285-291.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krilov L, Pierik L, Keller E, Mahan K, Watson D, Hirsch M, Hamparian V, McIntosh K. The association of rhinoviruses with lower respiratory tract disease in hospitalized patients. J Med Virol. 1986;19:345–352. doi: 10.1002/jmv.1890190407. [DOI] [PubMed] [Google Scholar]
- 24.Larson H E, Reed S E, Tyrrell D A. Isolation of rhinoviruses and coronaviruses from 38 colds in adults. J Med Virol. 1980;5:221–229. doi: 10.1002/jmv.1890050306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lennette E H, editor. Laboratory diagnosis of viral infections. New York, N.Y: Marcel Dekker; 1992. [Google Scholar]
- 26.Mäkelä M J, Puhakka T, Ruuskanen O, Leinonen M, Saikku P, Kimpimäki M, Blomqvist S, Hyypiä T, Arstila P. Viruses and bacteria in the etiology of the common cold. J Clin Microbiol. 1998;36:539–542. doi: 10.1128/jcm.36.2.539-542.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin S, Martin A, Staunton D E, Springer T A. Functional studies of truncated soluble intercellular adhesion molecule 1 expressed in Escherichia coli. Antimicrob Agents Chemother. 1993;37:1278–1284. doi: 10.1128/aac.37.6.1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McMillan J A, Weiner L B, Higgins A M, Macknight K. Rhinovirus infection associated with serious illness among pediatric patients. Pediatr Infect Dis J. 1993;12:321–325. doi: 10.1097/00006454-199304000-00011. [DOI] [PubMed] [Google Scholar]
- 29.Monto A S, Bryan E R, Ohmit S. Rhinovirus infections in Tecumseh, Michigan: frequency of illness and number of serotypes. J Infect Dis. 1987;156:43–49. doi: 10.1093/infdis/156.1.43. [DOI] [PubMed] [Google Scholar]
- 30.Mori J, Clewley J P. Polymerase chain reaction and sequencing for typing rhinovirus RNA. J Med Virol. 1994;44:323–329. doi: 10.1002/jmv.1890440403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nicholson K G, Kent J, Hammersley V, Cancio E. Risk factors for lower respiratory complications of rhinovirus infections in elderly people living in the community: prospective cohort study. Br Med J. 1996;313:1119–1123. doi: 10.1136/bmj.313.7065.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nicholson K G, Kent J, Hammersley V, Cancio E. Acute viral infections of upper respiratory tract in elderly people living in the community: comparative, prospective, population based study of disease burden. Br Med J. 1997;315:1060–1064. doi: 10.1136/bmj.315.7115.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nicholson K G, Kent J, Ireland D C. Respiratory viruses and exacerbations of asthma in adults. Br Med J. 1993;307:982–986. doi: 10.1136/bmj.307.6910.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olive D M, Al Mufti S, Al Mulla W, Khan M A, Pasca A, Stanway G, Al Nakib W. Detection and differentiation of picornaviruses in clinical samples following genomic amplification. J Gen Virol. 1990;71:2141–2147. doi: 10.1099/0022-1317-71-9-2141. [DOI] [PubMed] [Google Scholar]
- 35.Rotbart H A. Enzymatic RNA amplification of the enteroviruses. J Clin Microbiol. 1990;28:438–442. doi: 10.1128/jcm.28.3.438-442.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rueckert R R. Picornaviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 609–654. [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 38.Santti J, Hyypiä T, Halonen P. Comparison of PCR primer pairs in the detection of human rhinoviruses in nasopharyngeal aspirates. J Virol Methods. 1997;66:139–147. doi: 10.1016/s0166-0934(97)00049-9. [DOI] [PubMed] [Google Scholar]
- 39.Stanway G. Structure, function and evolution of picornaviruses. J Gen Virol. 1990;71:2483–2501. doi: 10.1099/0022-1317-71-11-2483. [DOI] [PubMed] [Google Scholar]
- 40.Uncapher C R, DeWitt C M, Colonno R J. The major and minor group receptor families contain all but one human rhinovirus serotype. Virology. 1991;180:814–817. doi: 10.1016/0042-6822(91)90098-v. [DOI] [PubMed] [Google Scholar]