Abstract
Aim:
To investigate the role of the gut microbiome in regulating key insulin homeostasis traits (insulin sensitivity, insulin secretion and insulin clearance) whose dysfunction leads to type 2 diabetes (T2D).
Materials and Methods:
The Microbiome and Insulin Longitudinal Evaluation Study (MILES) focuses on African American and non-Hispanic white participants aged 40–80 years without diabetes. Three study visits are planned (at baseline, 15 and 30 months). Baseline measurements include assessment of the stool microbiome and administration of an oral glucose tolerance test, which will yield indexes of insulin sensitivity, insulin secretion and insulin clearance. The gut microbiome profile (composition and function) will be determined using whole metagenome shotgun sequencing along with analyses of plasma short chain fatty acids. Additional data collected include dietary history, sociodemographic factors, health habits, anthropometry, medical history, medications and family history. Most assessments are repeated 15 and 30 months following baseline.
Results:
After screening 875 individuals, 129 African American and 224 non-Hispanic white participants were enrolled. At baseline, African American participants have higher blood pressure, weight, body mass index, waist and hip circumferences but similar waist-hip ratio compared with the non-Hispanic white participants. On average, African American participants are less insulin-sensitive and have higher acute insulin secretion and lower insulin clearance.
Conclusions:
The longitudinal design and robust characterization of potential mediators will allow for the assessment of glucose and insulin homeostasis and gut microbiota as they change over time, improving our ability to discern causal relationships between the microbiome and the insulin homeostasis traits whose deterioration determines T2D, setting the stage for future microbiome-directed therapies to prevent and treat T2D.
Keywords: insulin resistance, insulin secretion, type 2 diabetes
1 |. INTRODUCTION
With an estimated prevalence of 10.5% of the population (all ages), the United States is experiencing an unprecedented epidemic of diabetes, the majority of which is type 2 diabetes (T2D).1 Evidence suggests that insulin resistance is the earliest deficit in the pathogenesis of T2D; yet the majority of individuals with insulin resistance do not develop T2D because they are able to increase insulin levels to overcome tissue insulin resistance and maintain normoglycaemia. Those who are not able to maintain this hyperinsulinaemic compensation develop T2D. Increased insulin secretion from pancreatic β-cells and reduction in insulin clearance (removal of insulin from the circulation) are responsible for hyperinsulinaemic compensation.2 We refer to insulin sensitivity, insulin secretion and insulin clearance as components of ‘insulin homeostasis’. While genetic factors play a critical role in the aetiology of T2D, the remarkable increase in T2D prevalence (>4% per year in US adults from 1990 to 20093) indicates that non-genetic factors are also contributing, including changes in lifestyle (diet, physical activity), demographic shifts in the population (race/ethnicity, socioeconomic status) and psychosocial trends (development of the ‘obesogenic’ environment). The Microbiome and Insulin Longitudinal Evaluation Study (MILES) is addressing a key non-genetic factor regulated by diet, the gut microbiota (microbiome), to define its role in regulating the key insulin homeostasis traits whose dysfunction leads to T2D.
Initially influenced by early life experiences, including mode of delivery, infant feeding and antibiotic exposure, the gut microbiome is established by age 2–3 years and remains comparatively stable during adulthood4; yet it can be altered by diet and medications.5,6 The human gastrointestinal tract contains a diverse community of 1013 to 1014 bacteria, archaea and eukaryotes.7 These organisms establish symbiotic relationships with their hosts, defining the mucosal immune system, maintaining epithelial barrier function, producing vitamins and fermenting food components (e.g. fibre) that humans cannot digest. The combined genomes of the gut microbiota represent more than 100-fold more genes than encoded in the human genome.8
Alterations in gut microbial populations have been associated with risk of obesity and T2D.9 The gut microbiota influences energy harvest from the diet and affects host metabolism and inflammation. The profile in individuals with T2D is different from healthy lean individuals as well as from obese, non-diabetic individuals who are not insulin-resistant.10 Obesity is induced in germ-free mice exposed to gut microbiota from obese mice or from humans with obesity, suggesting a causal relationship.11
Reduced intestinal bacterial diversity (bacterial species number or richness) is associated with increased inflammation and features of the metabolic syndrome.12 Bacterial metabolism may promote or ameliorate insulin resistance and insulin secretion, impacting T2D risk. Several studies have performed large-scale microbiome profiling in individuals with prediabetes or T2D versus controls, as recently reviewed.9 What emerged from most of these studies was a consistent finding of depletion in individuals with T2D of bacterial species that produce short chain fatty acids (SCFA), particularly butyrate (e.g. Faecalibacterium prausnitzii and Akkermansia muciniphilia). However, the current body of evidence consists of cross-sectional studies, precluding establishment of the temporality of the associations being observed. Thus, MILES has established a cohort free of diabetes at baseline (avoiding confounding effects of medication) and is prospectively assessing glucose and insulin homeostasis and gut microbiota as they change over time, improving our ability to discern causal relationships between the microbiome and the three insulin homeostasis traits whose deterioration determines T2D. Our overarching hypothesis is that the gut microbial composition at baseline and over time influences change (improvement or deterioration) in insulin homeostasis and that bacteria associated with deterioration in insulin homeostasis traits will, in part, be attributable to an unhealthy diet. We have a particular focus on bacteria that produce SCFA; for example, insulin sensitivity declining over time may be associated with reduced abundance of butyrate-producing bacteria. Our planned deep sequencing will also allow us to examine bacterial functions (in addition to species) that modulate insulin homeostasis changes. Given that the microbiome can be modified by diet, lifestyle or medication, MILES is poised to identify microbiome-based targets of future therapies to prevent and treat T2D.
2 |. MATERIALS AND METHODS
2.1 |. Study participants
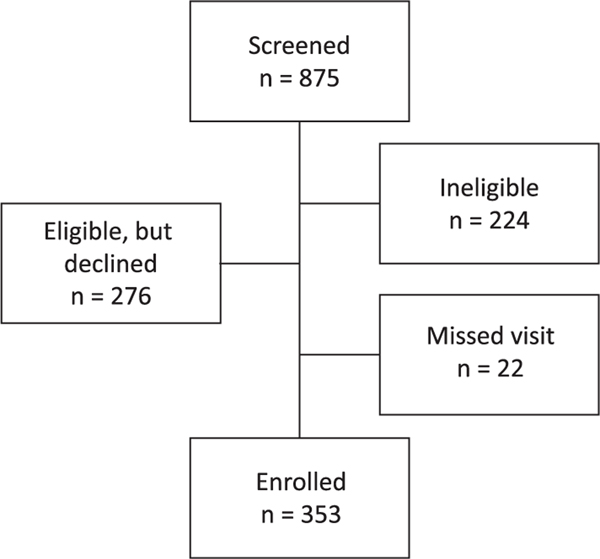
We screened 875 individuals and enrolled 353 non-diabetic individuals (129 African American and 224 non-Hispanic white) aged 40–80 years. Race and ethnicity were self-identified. A flowchart illustrating participant recruitment is presented in Figure 1. Participants were recruited in the Piedmont Triad area of North Carolina, including Forsyth, Davidson, Davie, Guilford, Stokes and Surry counties. We utilized several recruitment strategies, including targeted mailings and telephone calls, primarily based on lists of patients seen in the Wake Forest Baptist Health System. We also utilized advertisements on local media (newspaper, social media and radio) and conducted interviews with African American-oriented media outlets. Project team members also gave presentations at community settings (e.g. Young Women’s Christian Association (YWCA), public libraries and retirement communities) and attended community events at which recruitment materials were distributed. Flyers and advertising utilized one of two phrases: ‘Help us understand the relationship between diet and the risk of type 2 diabetes’ or ‘We have a gut feeling you can help us’. Most materials stated, ‘The Microbiome and Insulin Longitudinal Evaluation Study (MILES) will analyze the relationships among diet, gut bacteria, and insulin levels in order to better understand why some people develop type 2 diabetes’.
FIGURE 1.
Flowchart of participant recruitment. The flowchart provides the numbers of individuals screened, determined as ineligible, declined participation at the time of screening, and those that did not attend the first study visit scheduled, to arrive at the final number enrolled
Exclusion criteria at the time of enrolment included self-identified Hispanic ethnicity, severe illness (e.g. actively treated cancer) that might lead to failure to return for follow-up visits, antibiotic use in the prior month (or use of other medication with microbiome effects; i.e. metformin and proton pump inhibitors5,13), use of oral steroids, inflammatory bowel disease, surgery for weight loss, chronic constipation or diarrhoea requiring prescription therapy, pregnancy, end-stage renal disease and heavy alcohol use. Additionally, presence of diabetes at baseline (by history or point-of-care fasting glucose ≥126 mg/dL) was an exclusion criterion; however, diabetes developing during follow-up (second or third visit) will not lead to exclusion, allowing secondary analyses of microbiome determinants of incident T2D. Glucose tolerance (normal, prediabetes, T2D) is being defined using American Diabetes Association criteria.14
Eligible participants were invited to attend a clinic visit, which will be followed by two additional visits approximately 15 months apart, for a total of three visits. At this time, all participants have completed their baseline study visit, and no new participants are being recruited. Each visit includes anthropometric measurements (weight, height, waist and hip circumference), a full medication history, and documentation of any interim illness that may have occurred. Stool collection materials are mailed to every participant before each visit and participants complete comprehensive questionnaires to document health history, physical activity, health behaviours and other factors. The study was approved by institutional review boards at participating centres. All subjects gave written informed consent prior to participation.
2.2 |. Stool collection
Study participants collect a stool sample at home 1–2 days prior to each of the three clinic visits. Collection is aided by the use of a FecesCatcher (http://www.fecesvanger.nl/en_GB/) and stools are stored in the OMNIgene GUT collection kit. The OMNIgene GUT kit is an all-in-one system designed for self-collection and stabilization of microbial DNA from stools for gut microbiome profiling. After collection and homogenization, sample DNA is stable at an ambient temperature for 60 days without the requirement of a cold chain. Sampling with the OMNIgene GUT kit produces a microbial composition profile consistent with that of a direct stool sample.15 Participants are instructed to document stool consistency (Bristol score16) at the time of the collection. Whole metagenome shotgun sequencing methods are given in Appendix S1.
2.3 |. Diet and physical activity assessment
We are assessing both habitual diet over the past year and recent changes in overall diet pattern. At each of the three visits, habitual diet is assessed using a food frequency questionnaire. Participants are asked to record the frequency of consumption, and the usual portion size, of 124 food and drink items using the Diet History Questionnaire II, developed by the National Cancer Institute. Additional details concerning diet assessment are given in Appendix S1.
The physical activity survey employed in MILES is the well-validated instrument used in the Multi-Ethnic Study of Atherosclerosis (MESA). The MESA Typical Week Physical Activity Survey was based on the Cross-Cultural Activity Participation Study.17 How the MESA survey was implemented has been previously described.18 The survey yields estimates of different degrees of physical activity (light, moderate and vigorous) in metabolic equivalent (MET) minutes per week. Herein, we analysed total physical activity as the sum of light, moderate and vigorous physical activity.
2.4 |. Phenotyping insulin homeostasis
To achieve the best balance of quality phenotyping without undue burden on participants, the oral glucose tolerance test (OGTT) is being used to obtain measures of insulin homeostasis (Table 1). Following an overnight fast, venous blood samples are obtained for the measurement of plasma glucose, insulin and C-peptide before (fasting) and 30 and 120 minutes after the oral administration of a 75 g glucose load. While several OGTT-derived indices of insulin sensitivity/resistance have been developed, we will utilize the Matsuda insulin sensitivity index (ISI), which is highly correlated with directly quantified (by euglycaemic clamp) insulin sensitivity (r 0.7–0.8).19 Furthermore, the Matsuda ISI can be calculated using fewer than five OGTT time points, without reduction in correlation with directly measured insulin sensitivity.20 Our measure of insulin secretion is the area under the curve (AUC) for insulin from baseline to 30 minutes over the corresponding AUC for glucose (AUC-Ins30/AUC-Glu30). This measure was found to be highly correlated with first phase insulin secretion from the intravenous glucose tolerance test (r = 0.7).19 In addition, this AUC-based insulin secretion measure has been found to have a hyperbolic relationship with insulin sensitivity, consistent with the relationship found when insulin secretion and insulin sensitivity are measured with gold standard physiologic tests.21 This relationship allows calculation of the disposition index, the product of insulin secretion and insulin sensitivity (herein, DI30 = ISI x AUC-Ins30/AUC-Glu30), which represents an index of insulin secretion that accounts for its degree of compensation for insulin resistance. Insulin clearance is measured as the AUC of C-peptide over the AUC of insulin (AUC-Cpep/AUC-Ins), a commonly used index of hepatic insulin extraction given that the liver clears insulin but not C-peptide.22 We will also explore other emerging indices of insulin homeostasis traits.
TABLE 1.
Insulin homeostasis traits from the OGTT
| Trait | Definition |
|---|---|
| Insulin sensitivity index (ISI) | 10 000/square root of (Glu0 × Ins0 × Glumean × Insmean) |
| Insulin secretion | AUC-Ins30/AUC-Glu30 |
| Insulin clearance | AUC-Cpep/AUC-Ins |
Abbreviations: AUC, area under the curve; Cpep, C-peptide; Glu0, Ins0, fasting glucose, insulin; Glumean, Insmean, mean glucose and insulin concentrations during OGTT; G30, I30, 30 minute glucose, insulin.
The methodology for measurement of plasma SCFA is described in Appendix S1. We are also storing blood samples for future use, which may include assessment of additional metabolites that may mediate the relationship between the gut microbiome and metabolism, as well as DNA extraction for human genotyping.
2.5 |. Statistical analysis
T-tests (quantitative traits) or chi-square tests (sex, ethnicity) were used to compare baseline characteristics between African Americans and non-Hispanic whites and between women and men. Multivariable analyses were carried out using multiple regression wherein each insulin homeostasis trait was analysed as a dependent variable, and sex, ethnicity, age, body mass index (BMI) and physical activity were the independent variables. Regarding demographic variables (marital status, education, income, smoking, alcohol use, BMI categories, family history of prediabetes or diabetes), we conducted two-sided tests of difference of proportions (chi-square test) to test for differences in the distribution of proportions between racial groups. Microbiome analyses, power calculations and the handling of potential confounding factors are described in Appendix S1.
3 |. RESULTS
Baseline quantitative traits (Table 2) and demographic characteristics (Table 3) of the cohort are presented. Table 2 includes the insulin homeostasis traits (derived from the OGTT) that are the main endpoints of the study. African American participants are slightly younger, and have higher blood pressure, weight, BMI, waist and hip circumferences but a similar waist-hip ratio compared with the non-Hispanic white participants. In addition, African American participants generally are less insulin-sensitive but have higher acute insulin secretion and lower insulin clearance, compensatory responses that result in similar glucose levels, higher fasting insulin levels, and fairly similar disposition index. Physical activity did not differ between the two racial groups.
TABLE 2.
Quantitative traits
| Trait | African American (n = 129) | Non-Hispanic white (n = 224) | P-value* | Women (n = 218) | Men (n = 135) | P-value† |
|---|---|---|---|---|---|---|
| Age (y) | 58 (13) | 61 (15) | <.01 | 58 (14) | 61 (16) | .29 |
| Female, n (%) | 85 (65.9) | 133 (59.4) | .22 | n/a | n/a | n/a |
| African American, n (%) | n/a | n/a | n/a | 85 (39.0) | 44 (32.6) | .22 |
| Weight (kg) | 85.3 (26.6) | 75.0 (26.5) | <.01 | 72.7 (22.8) | 89.6 (25.2) | <.01 |
| Height (cm) | 166.4 (14.2) | 167.9 (15.6) | .07 | 162.7 (8.1) | 177.9 (9.5) | <.01 |
| BMI (kg/m2) | 30.3 (9.5) | 26.2 (6.7) | <.01 | 27.8 (8.9) | 28.3 (6.1) | .81 |
| Waist circumference (cm) | 101.2 (23.5) | 95.2 (22.5) | <.01 | 94.8 (23.4) | 101.3 (19.5) | <.01 |
| Hip circumference (cm) | 108.9 (18.9) | 102.5 (14.4) | <.01 | 105.6 (18.2) | 103.1 (14.3) | .02 |
| Waist-hip ratio | 0.93 (0.11) | 0.92 (0.12) | .23 | 0.89 (0.095) | 0.99 (0.084) | <.01 |
| Systolic blood pressure (mmHg) | 128 (24.8) | 115.5 (21.4) | <.01 | 118.5 (25.5) | 121.0 (23.5) | .01 |
| Diastolic blood pressure (mmHg) | 76.5 (13.5) | 69.5 (11.4) | <.01 | 69.8 (14.5) | 75.0 (13.5) | <.01 |
| Physical activity (MET min/wk) | 8610 (9053) | 8295 (5796) | .47 | 8595 (7746) | 7631 (6553) | .33 |
| Fasting glucose (mg/dL) | 97 (14) | 96 (11.8) | .97 | 94 (12) | 100 (12) | <.01 |
| Fasting insulin (μU/mL) | 11.2 (9.7) | 8.7 (7.1) | <.01 | 8.6 (7.3) | 10.7 (11.2) | <.01 |
| Fasting C-peptide (ng/mL) | 2.2 (1.5) | 2.0 (1.2) | .76 | 2.0 (1.1) | 2.3 (1.6) | <.01 |
| ISI | 3.4 (2.9) | 4.4 (4.8) | <.01 | 4.3 (4.1) | 3.5 (3.7) | <.01 |
| AUC-Ins30/AUC-Glu30 | 0.49 (0.43) | 0.32 (0.23) | <.01 | 0.33 (0.26) | 0.38 (0.41) | .03 |
| DI30 | 1.6 (1.2) | 1.4 (1.0) | .04 | 1.6 (1.2) | 1.4 (1.0) | .03 |
| AUC-Cpep/AUC-Ins | 0.085 (0.042) | 0.12 (0.045) | <.01 | 0.11 (0.049) | 0.098 (0.055) | .05 |
Abbreviations: AUC-Ins30/AUC-Glu30, insulin secretion; BMI, body mass index; DI30, disposition index; AUC-Cpep/AUC-Ins, insulin clearance; ISI, insulin sensitivity index; MET, metabolic equivalent; n/a, not applicable.
Notes: Data are median (interquartile range) for quantitative traits and n (%) for female sex. Weight, waist, hip, BMI and all glucose/insulin homeostasis traits were log-transformed and physical activity was square root-transformed for the t-test P-values.
Comparing African American with non-Hispanic white.
Comparing women with men.
TABLE 3.
Participant demographic characteristics
| Total | Non-Hispanic white | African American | ||
|---|---|---|---|---|
| Characteristic | (n = 353) | (n = 224) | (n = 129) | P-value* |
| Marital status (current) – n (%) | <.01 | |||
| Married/co-habiting | 211 (60.3) | 155 (69.8) | 56 (43.4) | |
| Widowed | 16 (4.6) | 12 (5.4) | 4 (3.1) | |
| Never married | 38 (10.9) | 13 (5.9) | 25 (19.4) | |
| Divorced | 74 (21.1) | 38 (17.1) | 36 (27.9) | |
| Separated | 11 (3.1) | 4 (1.8) | 7 (5.4) | |
| Missing | 3 | 2 | 1 | |
| Education – n (%) | <.01 | |||
| <High school | 3 (0.9) | 1 (0.5) | 2 (1.6) | |
| High school diploma/GED | 30 (8.5) | 9 (4.0) | 21 (16.3) | |
| Some college | 102 (28.9) | 55 (24.6) | 47 (36.4) | |
| Bachelorʼs degree | 122 (34.6) | 91 (40.6) | 31 (24.0) | |
| Graduate or professional degree | 96 (27.2) | 68 (30.4) | 28 (21.7) | |
| Income (USD) – n (%) | <.01 | |||
| <$12 000 | 23 (7.3) | 8 (3.9) | 15 (13.6) | |
| $12 000–$24 999 | 32 (10.1) | 15 (7.3) | 17 (15.5) | |
| $25 000–$39 999 | 44 (13.9) | 25 (12.1) | 19 (17.3) | |
| $40 000–$49 999 | 25 (7.9) | 12 (5.8) | 13 (11.8) | |
| $50 000–$74 999 | 63 (19.9) | 45 (21.7) | 18 (16.4) | |
| $75 000–$99 999 | 42 (13.3) | 28 (13.5) | 14 (12.7) | |
| $100 000–$124 999 | 29 (9.2) | 22 (10.6) | 7 (6.4) | |
| $125 000–$149 999 | 18 (5.7) | 16 (7.7) | 2 (1.8) | |
| ≥$150 000 | 41 (12.9) | 36 (17.4) | 5 (4.6) | |
| Prefer not to report/missing | 36 | 17 | 19 | |
| Smoking status – n (%) | <.01 | |||
| Never | 207 (58.6) | 140 (62.5) | 67 (51.9) | |
| Former | 96 (27.2) | 70 (31.3) | 26 (20.2) | |
| Current | 50 (14.2) | 14 (6.3) | 36 (27.9) | |
| Alcohol use status – n (%) | <.01 | |||
| Never | 37 (10.5) | 20 (8.9) | 17 (13.2) | |
| Former | 54 (15.3) | 25 (11.2) | 29 (22.5) | |
| Current | 262 (74.2) | 179 (79.9) | 83 (64.3) | |
| BMI (kg/m2) – n (%) | <.01 | |||
| <18.5 | 6 (1.7) | 4 (1.8) | 2 (1.6) | |
| 18.5–24.9 | 102 (28.9) | 83 (37.1) | 19 (14.7) | |
| 25.0–29.9 | 123 (34.8) | 81 (36.2) | 42 (32.6) | |
| ≥30.0 | 122 (34.6) | 56 (25.0) | 66 (51.2) | |
| Family history of prediabetes or diabetes | (parents) – n (%) | .16 | ||
| Neither parent | 190 (55.1) | 129 (58.6) | 61 (48.8) | |
| One parent | 130 (37.7) | 78 (35.5) | 52 (41.6) | |
| Both parents | 25 (7.3) | 13 (5.9) | 12 (9.6) | |
| Missing | 8 | 4 | 4 | |
P-value for two-sided test of difference in proportions.
Abbreviations: GED, graduate equivalency degree.
Men had higher weight and height than women, but similar BMI. In having a larger waist circumference and lower hip circumference, men had a higher waist-hip ratio than women. Men also had higher blood pressure. Men were more insulin-resistant than women, with compensatory increases in insulin secretion and decreases in insulin clearance apparently insufficient, as their fasting glucose levels remained higher and disposition index lower, in contrast to the more effective compensation observed in African Americans versus non-Hispanic whites. Physical activity was similar between men and women.
Table 3 notes higher rates of being single, divorced or separated in African Americans. Educational attainment and income were higher in non-Hispanic whites. Rates of smoking were higher in African Americans while alcohol use was more common in non-Hispanic whites. The proportion of African Americans in the overweight and obese weight categories was higher. Family histories of prediabetes or diabetes were similar between the racial groups.
Multivariable analyses examining the correlates of age, sex, BMI, ethnicity and physical activity with insulin sensitivity, insulin secretion, disposition index and insulin clearance are presented in Table 4. BMI was highly correlated with all four traits, with the strongest effect on decreased insulin sensitivity. African American ancestry was independently associated with increased insulin secretion and disposition index and lower insulin clearance but did not correlate with insulin sensitivity. Female sex was associated with increased insulin sensitivity and insulin clearance. Older age was associated with decreased disposition index via decreased insulin sensitivity. In having an opposite direction of correlation with insulin sensitivity and insulin secretion, physical activity was not associated with disposition index.
TABLE 4.
Multivariable analyses
| Insulin sensitivity by ISI | Insulin secretion by AUC-Ins30/AUC-Glu30 | Disposition index by DI30 | Insulin clearance by AUC-Cpep/AUC-ins | |||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|||||
| β | P-value | β | P-value | β | P-value | β | P-value | |
| Female | .18 | <.01 | −.12 | .11 | .087 | .09 | .13 | <.01 |
| African American | −.63 | .17 | .21 | <.01 | .16 | <.01 | −.31 | <.01 |
| Age | −.18 | <.01 | −.028 | .58 | −.26 | <.01 | −.079 | .10 |
| BMI | −.52 | <.01 | .32 | <.01 | −.29 | <.01 | −.33 | <.01 |
| Physical activity | .12 | .01 | −.12 | .02 | .013 | .80 | .089 | .06 |
Abbreviations: AUC-Ins30/AUC-Glu30, insulin secretion; AUC-Cpep/AUC-Ins, insulin clearance; BMI, body mass index; DI30, disposition index; ISI, insulin sensitivity index; β, standardized beta coefficient. Each column represents a multiple regression analysis, wherein the insulin homeostasis trait was the dependent variable, and sex, ethnicity, age, BMI and physical activity were the independent variables.
4 |. DISCUSSION
This studyʼs deep sequencing is designed to find different bacterial profiles that associate with each insulin homeostasis trait change in the two racial groups. Whole metagenome shotgun sequencing also has the advantage of being able to reveal that the functions encoded by these bacteria may be similar between the different groups. This has been observed in the metagenomic sequencing studies in T2D; while different microbiome profiles were associated with T2D in Europeans compared with Chinese, these profiles encoded several similar functions.23,24 Identifying the fundamental microbial functions that influence insulin homeostasis changes may provide critical insight into how gut bacteria may predispose to T2D; interventions targeting these functions rather than specific bacteria may ultimately prove clinically useful.
4.1 |. Focus on SCFA
SFCA (the most abundant [≥95%] of which are acetate, propionate and butyrate) are the most widely studied gut microbial metabolites. Intestinal microbes generate SCFA by fermenting dietary carbohydrates that humans cannot digest. SCFA have been extensively reported to improve glucose homeostasis and metabolism in adipose, muscle and liver.25 Examples include: (a) SCFA signal through the G-protein coupled receptors GPR41 and GPR43 (free fatty acid receptors FFAR3 and FFAR2, respectively) to promote the release of glucagon-like peptide-1 (GLP-1) and peptide YY (PYY) by intestinal L cells and leptin by adipose tissue, hormones that promote satiety and improve insulin secretion (GLP-1) and insulin sensitivity (PYY); (b) SCFA have insulin-sensitizing effects; and (c) SCFA may reduce mucosal and chronic systemic inflammation.25 While the balance of literature suggests that butyrate confers metabolic benefits, whether this is true for other SCFA is less certain.26
4.2 |. Detailed phenotyping of intermediate traits
Most large cohort studies have limited phenotypic data relevant to T2D, typically only fasting glucose and insulin. While convenient to measure in large epidemiological studies, these fasting measures imprecisely represent the components of insulin homeostasis (sensitivity, secretion and clearance). We found that fasting insulin was determined by insulin sensitivity and insulin clearance in almost equal proportions.27 We are using the OGTT to provide the three measures of insulin homeostasis. To date, several cross-sectional microbiome-wide studies have been performed comparing individuals with T2D with unaffected controls.9 The current study focuses on insulin homeostasis traits wherein derangement precedes and defines T2D. The associations observed between microbiome and change in components of insulin homeostasis, particularly those associated with reduced insulin sensitivity and reduced insulin secretion, may be enriched for organisms that may be causal for T2D.
4.3 |. Gut microbiota and insulin resistance
The role of the microbiome in mediating insulin resistance is emerging. A human study transplanted faecal material from lean healthy donors into the small intestines of individuals with the metabolic syndrome, resulting in improved peripheral muscle insulin sensitivity (by euglycaemic clamp) 6 weeks later that correlated with an increase in butyrate-producing bacteria (e.g. Roseburia intestinalis).28 In obese men, 1 week of vancomycin (which targets Gram-positive bacteria such as the butyrate producer F. prausnitzii) reduced gut bacterial diversity and peripheral insulin sensitivity, while amoxicillin (which targets Gram-negative bacteria) had no effect.6 A metagenomic sequencing study conducted with a focus on insulin sensitivity (notably, using the same Matsuda index we will use) found numerous species positively or negatively associated with insulin sensitivity,29 several of which were previously associated with T2D.23,24
4.4 |. Gut microbiota and insulin secretion
Insulin secretion is also influenced by the gut microbiota. Several studies suggest that particular bacterial taxa increase gut permeability, increasing the absorption of metabolites or toxins that affect pancreatic β-cells.30 SCFA, via signalling through G-protein coupled receptors on L cells, promote gut secretion of GLP-1, which potentiates insulin secretion.25 SCFA receptors are also found on β-cells, suggesting direct effects on insulin secretion.31 Bile acids, whose production is influenced by gut bacteria, may also affect GLP-1 secretion.6 Given the above-described prominent effects of SCFA on insulin sensitivity and insulin secretion, we hypothesize that levels of SCFA-producing bacteria, at baseline and as they change over time, will predict changes in these traits.
4.5 |. Gut microbiota and insulin clearance
We are not aware of any literature examining microbiota effects on insulin clearance; however, given the literature implicating gut microbiota alterations in non-alcoholic fatty liver disease,32 and the known effect of fatty liver to impair insulin clearance (the liver being the main organ that clears insulin),33 it is probable that we will observe microbial profiles associated with insulin clearance. The paucity of data for this insulin homeostasis trait is an opportunity for new discoveries in MILES.
4.6 |. Diet can modify the gut microbiota
Habitual (long-term) diet is a key determinant of the gut microbiota, as gut microbes appear to adapt to the host diet or co-evolve with the host.34 Given that bacteria have differing abilities to metabolize diet substrates, the diet can exert selective pressure on the composition of the microbiome. The microbiota of vegetarians differ from those of carnivores.35 Changes in diet composition can result in shifts in microbial populations, even as early as 1 day after the change in diet.36 Many of the effects of diet on glucose homeostasis may be mediated by SCFA produced by gut microbes. Consumption of resistant starch or fermentable fibres increases plasma and faecal SCFA levels and improves fasting glucose and insulin levels, whereas high-fat diets reduce SCFA levels.37 Short-term (1–3 days) intake of food rich in non-digestible carbohydrates improved glucose tolerance and peripheral insulin sensitivity, associated with an increase in circulating SCFA and GLP-1.38
4.7 |. Multi-racial study
Prior microbiome studies of T2D did not focus on high-risk minorities. Compared with non-Hispanic whites, diagnosed T2D is twice as common among African Americans.39 Many studies have established ethnic and racial differences in insulin sensitivity, insulin resistance and insulin clearance, which underlie differences in the prevalence of T2D. African Americans have increased insulin resistance, increased insulin secretion and lower hepatic insulin clearance compared with non-Hispanic whites.40 Increased insulin secretion and reduced insulin clearance in ethnic or racial groups with greater insulin resistance is consistent with our conceptual model, where T2D emerges when these compensatory mechanisms are insufficient. The Human Microbiome Project obtained samples from multiple body sites in 300 healthy individuals, and profiled the microbiome with 16S rDNA sequencing and shotgun metagenomic sequencing.7 Between individuals, race/ethnic background was the strongest determinant of compositional and functional differences. However, unlike our study, this research did not assess habitual diet, which differs between these groups. Inter-racial/ethnic microbiome variation may reflect differences in early and lifetime environmental exposures, social/cultural practices, diet and genetics. A key question addressed in our study is whether race-specific microbial profiles are associated with differences in T2D risk via effects on insulin homeostasis. Conversely, MILES provides a key opportunity to identify microbial associations with insulin homeostasis that are shared between racial groups.
5 |. CONCLUSION
In conclusion, several studies have documented compositional and functional differences in the gut microbiota in humans with T2D compared with controls9; however, these studies have been cross-sectional and results may be influenced by medication use (e.g. metformin in T2D).5 Therefore, whether the gut microbiome differences are a cause or a consequence of T2D remains unknown. In many instances, SCFA-producing bacteria were reduced in T2D; an extensive body of literature suggests that SCFA (particularly butyrate) produced by gut microbes may positively influence insulin sensitivity and insulin secretion, with effects that would prevent T2D. Given the public health implications, this evidence warrants further study to establish whether the gut microbiome contributes to T2D risk. Our study provides the much-needed prospective cohort study that will identify microbial compositional and functional changes that contribute to deterioration in insulin homeostasis and thus predispose to T2D, allowing determination of whether microbial shifts trigger host disease or vice versa, and the contributing role of diet and circulating SCFA. MILES has substantial translational potential, as it may identify eating patterns that warrant further investigation under more controlled dietary conditions for their ability to influence the trajectory of insulin homeostasis. It may also identify particular diabetogenic microbiomes independent of diet, setting the stage for prebiotic (foods that promote proliferation of beneficial species), probiotic or antibiotic trials to prevent and treat T2D.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported in part by National Institutes of Health grants from the National Institute of Diabetes and Digestive and Kidney Disease (R01-DK109588, P30-DK063491) and from the National Center for Advancing Translational Sciences (UL1TR001420, UL1TR001881). M.O.G. was supported by the Eris M. Field Chair in Diabetes Research. A.C.W. was supported, in part, by USDA/ARS cooperative agreement #58-3092-5-001. The contents of this publication do not necessarily reflect the views or policies of the U.S. Department of Agriculture, nor does mention of trade names, commercial products or organizations imply endorsement by the U.S. Government.
FUNDING INFORMATION
This study was supported in part by National Institutes of Health grants from the National Institute of Diabetes and Digestive and Kidney Disease (R01-DK109588, P30-DK063491) and from the National Center for Advancing Translational Sciences (UL1TR001420, UL1TR001881).
Funding information
National Center for Advancing Translational Sciences, Grant/Award Numbers: UL1TR001420, UL1TR001881; National Institute of Diabetes and Digestive and Kidney Diseases, Grant/Award Numbers: P30-DK063491, R01-DK109588; USDA/ARS, Grant/Award Number: 58-3092-5-001
Footnotes
CONFLICT OF INTEREST
J.F.P. is a consultant for Diversigen. E.T.J., A.G.B., O.L.C., K.L.H., A.C.W., Z.A., L.K.L., K.R., K.S., M.W., S.S.R., J.I.R., Y.I.C. and M.O.G. have no competing interests.
ENDNOTE
Notes: Data are median (interquartile range) for quantitative traits and n (%) for female sex. Weight, waist, hip, BMI and all glucose/insulin homeostasis traits were log-transformed and physical activity was square root-transformed for the t-test P-values.
PEER REVIEW
The peer review history for this article is available at https://publons.com/publon/10.1111/dom.14145.
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of this article.
REFERENCES
- 1.Centers for Disease Control and Prevention. National Diabetes Statistics Report. Estimates of Diabetes and its Burden in the United States, 2020. Atlanta, GA: US Department of Health and Human Services; 2020. [Google Scholar]
- 2.Mittelman SD, Van Citters GW, Kim SP, et al. Longitudinal compensation for fat-induced insulin resistance includes reduced insulin clearance and enhanced beta-cell response. Diabetes. 2000;49:2116–2125. [DOI] [PubMed] [Google Scholar]
- 3.Benoit SR, Hora I, Albright AL, Gregg EW. New directions in incidence and prevalence of diagnosed diabetes in the USA. BMJ Open Diabetes Res Care. 2019;7:e000657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Faith JJ, Guruge JL, Charbonneau M, et al. The long-term stability of the human gut microbiota. Science. 2013;341:1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forslund K, Hildebrand F, Nielsen T, et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. 2015;528:262–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vrieze A, Out C, Fuentes S, et al. Impact of oral vancomycin on gut microbiota, bile acid metabolism, and insulin sensitivity. J Hepatol. 2014;60:824–831. [DOI] [PubMed] [Google Scholar]
- 7.The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486: 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu T, Goodarzi MO. Metabolites linking the gut microbiome with risk for type 2 diabetes. Curr Nutr Rep. 2020;9:83–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Remely M, Aumueller E, Jahn D, Hippe B, Brath H, Haslberger AG. Microbiota and epigenetic regulation of inflammatory mediators in type 2 diabetes and obesity. Benef Microbes. 2014;5:33–43. [DOI] [PubMed] [Google Scholar]
- 11.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. [DOI] [PubMed] [Google Scholar]
- 12.Le Chatelier E, Nielsen T, Qin J, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500: 541–546. [DOI] [PubMed] [Google Scholar]
- 13.Imhann F, Bonder MJ, Vich Vila A, et al. Proton pump inhibitors affect the gut microbiome. Gut. 2016;65:740–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.American Diabetes Association. Classification and diagnosis of diabetes: standards of medical care in diabetes-2020. Diabetes Care. 2020; 43:S14–S31. [DOI] [PubMed] [Google Scholar]
- 15.Choo JM, Leong LE, Rogers GB. Sample storage conditions significantly influence faecal microbiome profiles. Sci Rep. 2015;5:16350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewis SJ, Heaton KW. Stool form scale as a useful guide to intestinal transit time. Scand J Gastroenterol. 1997;32:920–924. [DOI] [PubMed] [Google Scholar]
- 17.Ainsworth BE, Irwin ML, Addy CL, Whitt MC, Stolarczyk LM. Moderate physical activity patterns of minority women: the cross-cultural activity participation study. J Womens Health Gend Based Med. 1999; 8:805–813. [DOI] [PubMed] [Google Scholar]
- 18.Bertoni AG, Whitt-Glover MC, Chung H, et al. The association between physical activity and subclinical atherosclerosis: the multi-ethnic study of atherosclerosis. Am J Epidemiol. 2009;169: 444–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stancakova A, Javorsky M, Kuulasmaa T, Haffner SM, Kuusisto J, Laakso M. Changes in insulin sensitivity and insulin release in relation to glycemia and glucose tolerance in 6,414 Finnish men. Diabetes. 2009;58:1212–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeFronzo RA, Matsuda M. Reduced time points to calculate the composite index. Diabetes Care. 2010;33:e93. [DOI] [PubMed] [Google Scholar]
- 21.Santos JL, Yevenes I, Cataldo LR, et al. Development and assessment of the disposition index based on the oral glucose tolerance test in subjects with different glycaemic status. J Physiol Biochem. 2016;72: 121–131. [DOI] [PubMed] [Google Scholar]
- 22.Semnani-Azad Z, Johnston LW, Lee C, et al. Determinants of longitudinal change in insulin clearance: the prospective metabolism and islet cell evaluation cohort. BMJ Open Diabetes Res Care. 2019;7: e000825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karlsson FH, Tremaroli V, Nookaew I, et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498:99–103. [DOI] [PubMed] [Google Scholar]
- 24.Qin J, Li Y, Cai Z, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. [DOI] [PubMed] [Google Scholar]
- 25.Canfora EE, Jocken JW, Blaak EE. Short-chain fatty acids in control of body weight and insulin sensitivity. Nat Rev Endocrinol. 2015;11: 577–591. [DOI] [PubMed] [Google Scholar]
- 26.Tirosh A, Calay ES, Tuncman G, et al. The short-chain fatty acid propionate increases glucagon and FABP4 production, impairing insulin action in mice and humans. Sci Transl Med. 2019;11:eaav0120. [DOI] [PubMed] [Google Scholar]
- 27.Goodarzi MO, Cui J, Chen YD, Hsueh WA, Guo X, Rotter JI. Fasting insulin reflects heterogeneous physiological processes: role of insulin clearance. Am J Physiol Endocrinol Metab. 2011;301:E402–E408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vrieze A, Van Nood E, Holleman F, et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology. 2012;143:913–916. [DOI] [PubMed] [Google Scholar]
- 29.Brahe LK, Le Chatelier E, Prifti E, et al. Specific gut microbiota features and metabolic markers in postmenopausal women with obesity. Nutr Diabetes. 2015;5:e159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.BosE, MoltenL, RadaellG, et al. Increased intestinal permeability precedes clinical onset of type 1 diabetes. Diabetologia. 2006;49: 2824–2827. [DOI] [PubMed] [Google Scholar]
- 31.Regard JB, Kataoka H, Cano DA, et al. Probing cell type-specific functions of Gi in vivo identifies GPCR regulators of insulin secretion. J Clin Invest. 2007;117:4034–4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Verdam FJ, Rensen SS, Driessen A, Greve JW, Buurman WA. Novel evidence for chronic exposure to endotoxin in human nonalcoholic steatohepatitis. J Clin Gastroenterol. 2011;45:149–152. [DOI] [PubMed] [Google Scholar]
- 33.Kotronen A, Vehkavaara S, Seppala-Lindroos A, Bergholm R, Yki-Jarvinen H. Effect of liver fat on insulin clearance. Am J Physiol Endocrinol Metab. 2007;293:E1709–E1715. [DOI] [PubMed] [Google Scholar]
- 34.De Filippo C, Cavalieri D, Di Paola M, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. 2010;107:14691–14696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zimmer J, Lange B, Frick JS, et al. A vegan or vegetarian diet substantially alters the human colonic faecal microbiota. Eur J Clin Nutr. 2012; 66:53–60. [DOI] [PubMed] [Google Scholar]
- 36.David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jakobsdottir G, Xu J, Molin G, Ahrne S, Nyman M. High-fat diet reduces the formation of butyrate, but increases succinate, inflammation, liver fat and cholesterol in rats, while dietary fibre counteracts these effects. PLoS One. 2013;8:e80476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nilsson AC, Johansson-Boll EV, Bjorck IM. Increased gut hormones and insulin sensitivity index following a 3-d intervention with a barley kernel-based product: a randomised cross-over study in healthy middle-aged subjects. Br J Nutr. 2015;114:899–907. [DOI] [PubMed] [Google Scholar]
- 39.Cowie CC, Rust KF, Byrd-Holt DD, et al. Prevalence of diabetes and impaired fasting glucose in adults in the U.S. population: National Health and Nutrition Examination Survey 1999–2002. Diabetes Care. 2006;29:1263–1268. [DOI] [PubMed] [Google Scholar]
- 40.Osei K, Schuster DP. Ethnic differences in secretion, sensitivity, and hepatic extraction of insulin in black and white Americans. Diabet Med. 1994;11:755–762. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.