Abstract
Antibiotic-resistant bacteria are considered one of the major global threats to human and animal health. The most harmful of the resistant bacteria are β-lactamases producing Gram-negative species (β-lactamases). β-lactamases constitute a paradigm shift in the evolution of antibiotic resistance. Therefore, it is imperative to present a comprehensive review of the mechanisms responsible for the development of antimicrobial resistance. Resistance due to β-lactamases develops through to a variety of mechanisms, and the number of resistant genes is involved that can be transferred between bacteria, mostly via plasmids. Over time, these new molecular-based resistance mechanisms have been progressively disclosed. The present review article provides information on the recent findings regarding the molecular mechanisms of resistance to β-lactams in Gram-negative bacteria, including CTX-M-type ESBLs with methylase activity, plasmids harbouring phages with β-lactam resistance genes, the co-presence of β-lactam resistant genes of unique combinations and the presence of β-lactam and non-β-lactam antibiotic-resistant genes in the same bacteria. By keeping in view, the molecular ways of resistance development, multifactorial and coordinated measures may be taken to counter the challenge of rapidly increasing β-lactam resistance.
Keywords: Antibiotics, resistance, Carbapenemas, microbial drug resistance, public health
Introduction:
Infections due to multidrug-resistant (MDR) bacteria are increasing worldwide, and the number of untreatable diseases is rapidly growing [1]. According to the Global Risk report of the World Economic Forum, antibiotic resistance is one of the worst universal risks to animal and human health [2]. Antibiotic resistance is not limited to human medicine, a similar trend has been found in the veterinary field because equivalent quantities of antibiotics are inadequately used in livestock to improve the health and production of animals, even resistance has been reported from organic farms [3].
According to Ambler’s classification, more than 1800 described variants are classified into four types of β-lactamases based on protein sequence: A (serine penicillinases), B (Metallo-β-lactamases), C (cephalosporinases), or D (oxacillinases), which show resistance to penicillin, most β-lactams, cephalosporins, and cloxacillin, respectively [4]. These variants can also be classified based on their phenotypic profiles Bush and Jacoby (2010); however, Ambler’s classification is mostly followed. Class A and Class D consist of ESBLs and are mainly composed of CTX-M, TEM, SHV and OXA enzymes, class B β-lactamases consist of Metallo-β-lactamases (NDM-1) while class C contains AmpC β-lactamases. Many β-lactamases producing genes with these mechanisms are also responsible for resistance against different classes of antibiotics, like aminoglycosides, β-lactams, and fluoroquinolones [6]. It is well noted that major microbial resistance is because of production of extended-spectrum β-lactamases (ESBLs), AmpC β-lactamases, and carbapenemases. The beta lactam ring in all of the first, second, third generation cephalosporins and aztreonam are hydrolyzed by β-lactamases while most of fourth generation cephalosporins are also affected by ESBL [5].
β-lactamase mediated resistance to β-lactam antibiotics developed in recent years as a significant clinical threat to these lifesaving drugs. Accordingly, two techniques were developed to preserve the utility of β-lactam antibiotics: (i) identify or develop β-lactam antibiotics that avoid bacterial enzymatic inactivation presented by β-lactamase enzymes, or (ii) repress β-lactamases so that the accomplice β-lactam can aptly target [7]. An understanding of bacterial resistance on a genetic basis and the activity range of antibiotics will allow researchers to develop new antibiotic combinations with a vast range of target species. Therefore, this review will provide an update on the most recent research on β-lactam resistance mechanisms and the possible preventive measures. Moreover, from a wider scope, the anti-microbial resistance has become a one-health issue affecting eco-health and sustainability at a faster pace. This review presents a comprehensive way forward to understand the molecular pathology and the ways to inhibit or reduce the rapid development of antibiotic resistance.
Development of β-lactams resistance
Primarily, there are four basic mechanisms of β-lactams resistance in bacteria. β-lactamase enzymes production is the most common resistance mechanism in Gram-negative bacteria, which is the focus of our review (fig 1). Secondly, changes in the dynamic site of penicillin-binding proteins (PBPs) that can reduce the attraction for β-lactam antibiotics and enhance resistance to these operators. Through characteristic change and recombination with DNA from different organisms, bacteria have procured extreme resistance and low attraction to PBPs. Similarly, horizontal gene transfer (HGT) plays an important role in β-lactam resistance [8]. Another mechanism of resistance is the low expression of external membrane proteins. With a specific goal of reaching the PBPs on the inner plasma membrane, β-lactams may either diffuse or cross porin diverts in the external membrane of the Gram-negative bacterial cell wall [9]. It is worth noting that the interruption porin proteins alone are not generally adequate to deliver the resistance phenotype, and this system is associated with β-lactamase expression [10]. Efflux pumps being another probable pathway to resistance development, are a major feature of either a procured or natural resistance phenotype, are equipped to send out an extensive variety of substrates from the periplasm to its surroundings [11]. These pumps are vital determinants of multidrug resistance in numerous Gram-negative pathogens, especially Pseudomonas aeruginosa and Acinetobacter spp. In Pseudomonas aeruginosa, the upregulation of the MexA-MexB-OprD framework, combined with the low permeability of the external membrane, can add to reduced acceptance of penicillins and cephalosporins, as well as tetracycline, chloramphenicol, and quinolones [12].
Figure 1:
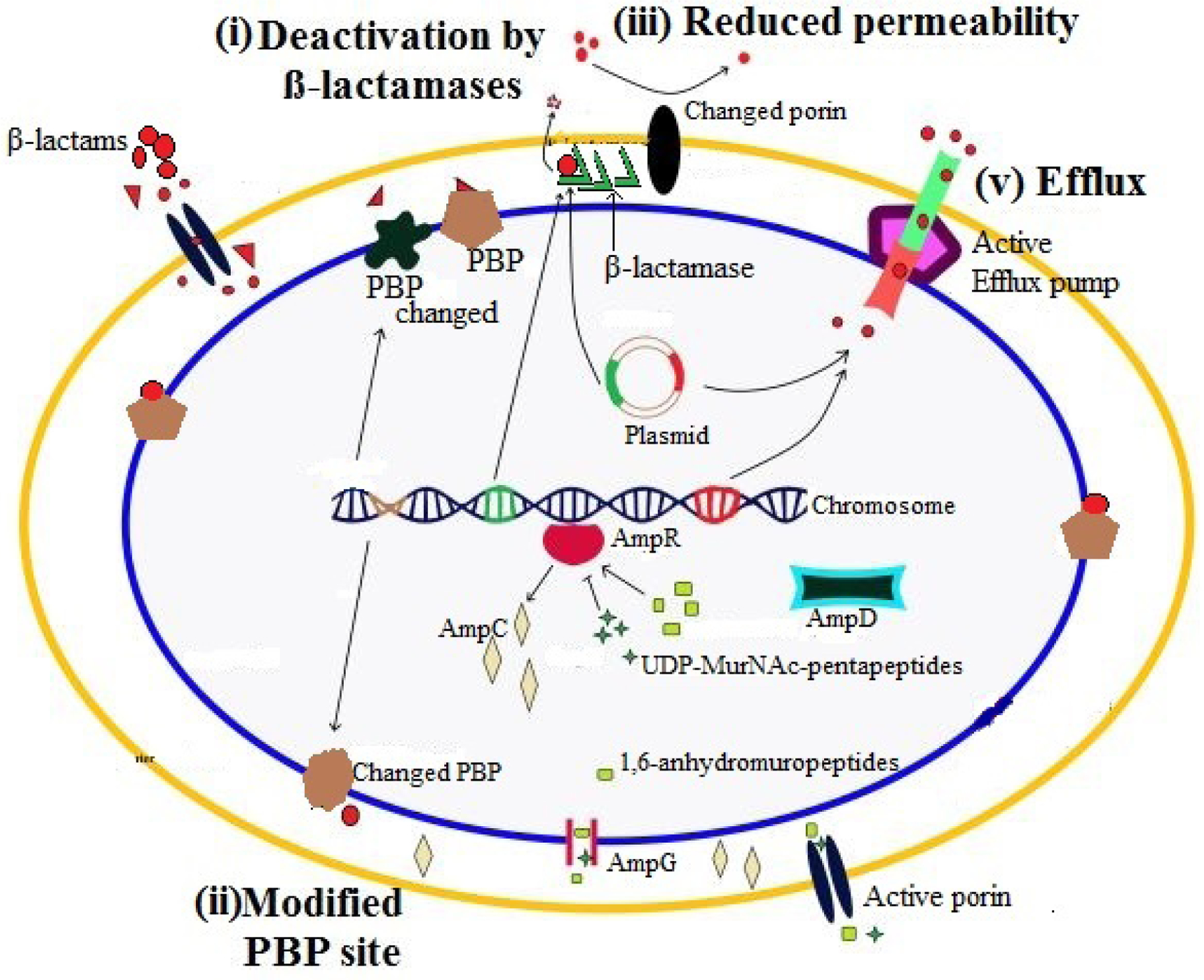
Basic mechanisms of β-lactam resistance in Gram-negative bacteria are illustrated. (i) deactivation of β-lactams by plasmid or chromosome encoded β-lactamases having hydrolytic activity, (ii) change in dynamic site of PBPs which hinder the binding of β-lactam molecules to PBPs, similarly, mecA gene encodes PBP2a in methicillin-resistant Staphylococcus spp. which has no affection for β-lactams, (iii) reduced permeability of outer membrane by porin modifications, change in the types of porins found in the outer membrane or complete loss of porin expression, (iv) efflux of β-lactams to outside of bacterium through activation of efflux pump (Nordmann et al., 2012).
Additionally, β-lactamases are either chromosomal or plasmid-related. The genes encoding β-lactamases may be located on chromosomes and expressed with or without the involvement or transported by the plasmids in single or various duplicates. In Gram-negative bacteria, the inducible expression of β-lactamases is usually found in chromosomal β-lactamases. However, plasmid-encoded β-lactamases are, for the most part, constitutively transferable to other bacteria [13]. Resistance was first appeared in Gram-negative bacteria against β-lactams such as cephalosporin and monobactams due to the mutation or overproduction of chromosomal AmpC β-lactamase. The plasmid-encoding class C of β-lactamases features a wider spectrum of resistance than chromosomal concern [14]. Moreover, chromosomal β-lactamases were found to move to plasmids or transposons; some plasmid-encoded β-lactamases can also be transferred to bacterial chromosome [15].
Most of AmpC β-lactamases are plasmid-mediated as well as carry the resistance genes for other antibiotic classes [16]. Furthermore, the plasmids harboring ESBL genes also develop resistance to trimethoprim/sulfamethoxazole and aminoglycosides which is mostly related to plasmid-evoked resistance to a cephalosporin [17]. β-lactamases are inhibited by β-lactamase inhibitors e.g., clavulanic acid. AmpC β-lactamases are also inhibited by β-lactamase inhibitors such as cloxacillin, aztreonam, and cefoxitin in addition to high concentrations of clavulanate [18].
Deactivation of β-lactams via hydrolysis
Initially, β-lactamases were resistant to first-generation β-lactams while later on extended spectrum β-lactamases (ESBLs) were discovered as resistant to oxyimoino-cephalosporins [19]. The presence of various carbapenemases and ESBLs, such as Klebsiella pneumoniae carbapenemase (KPC), IMP (imipenaemase), VIM (Verona integrin-encoded metallo β-lactamase), OXA (oxacillinose), and NDM enzymes in Gram-negative bacteria (e.g. E. coli, K. pneumoniae, A. baumannii and P. aeruginosa) has reinforced the bacteria to become resistant to all antibiotics containing β-lactam ring. This leads to dangerous conditions for the treatment of serious infections [20].
1. Extended-spectrum β-lactamases (ESBLs)
Single nucleotide transposition in genes that are encoding resistance to β-lactamases can result in resistance to extended-spectrum cephalosporins because these genes are related to SHV-1, TEM-1, and TEM-2 [21]. Many Gram-negative isolates show resistance against β-lactam antibiotics by hydrolysis because they contain ESBLs; therefore, the efficacy of those antibiotics for diseases developed by these bacteria is decreased. Furthermore, the plasmids harbouring ESBL genes often contain genes that develop resistance to trimethoprim/sulfamethoxazole and aminoglycosides, mostly due to plasmid-evoked resistance to cephalosporin. There is a clear relationship between ESBL production and quinolone resistance in the absence of a plasmid-mediated reduction in quinolone susceptibility [22]. Epidemiological studies found that Asia has the highest prevalence of ESBL enzymes in K. pneumonia and E. coli; China is at the top position in ESBL-producing E. coli [23].
1.1. CTX-M family
CTX is gene named because of ceftazidimase which were detected in 1986 from E. coli isolate of dog. Later these were identified in four-year-old child from Munich-Germany so named as CTX-M (Ceftazidimase Munich). These enzymes are the earliest ESBLs having no hydrolytic activity against cefotaxime. However, with type over other type of one or more cephalosporins, ESBL differed in substrate variability. These genes encode 291 amino acid and change in single amino acids leads to new CTX-M type. CTX-M enzymes are not developed as outcome of point mutations in parent enzymes rather the chromosomal bla genes of Kluyvera sps (Environmental bacteria isolated from water, sewage, soil, and animal-based food products) were mobilized into plasmids (Fig 2). Later to this mobilization of chromosome into plasmids were the mutations that give rise further diversification and providing opportunity for expansion of hydrolytic activity to ceftazidime. This is the reason that earlier CTX-M were only cefotaximases but recently more than 60% of CTX-M can hydrolyze cefotaxime as well as ceftazidime Cantón et al., 2012; Naseer et al., 2011)
Figure 2:
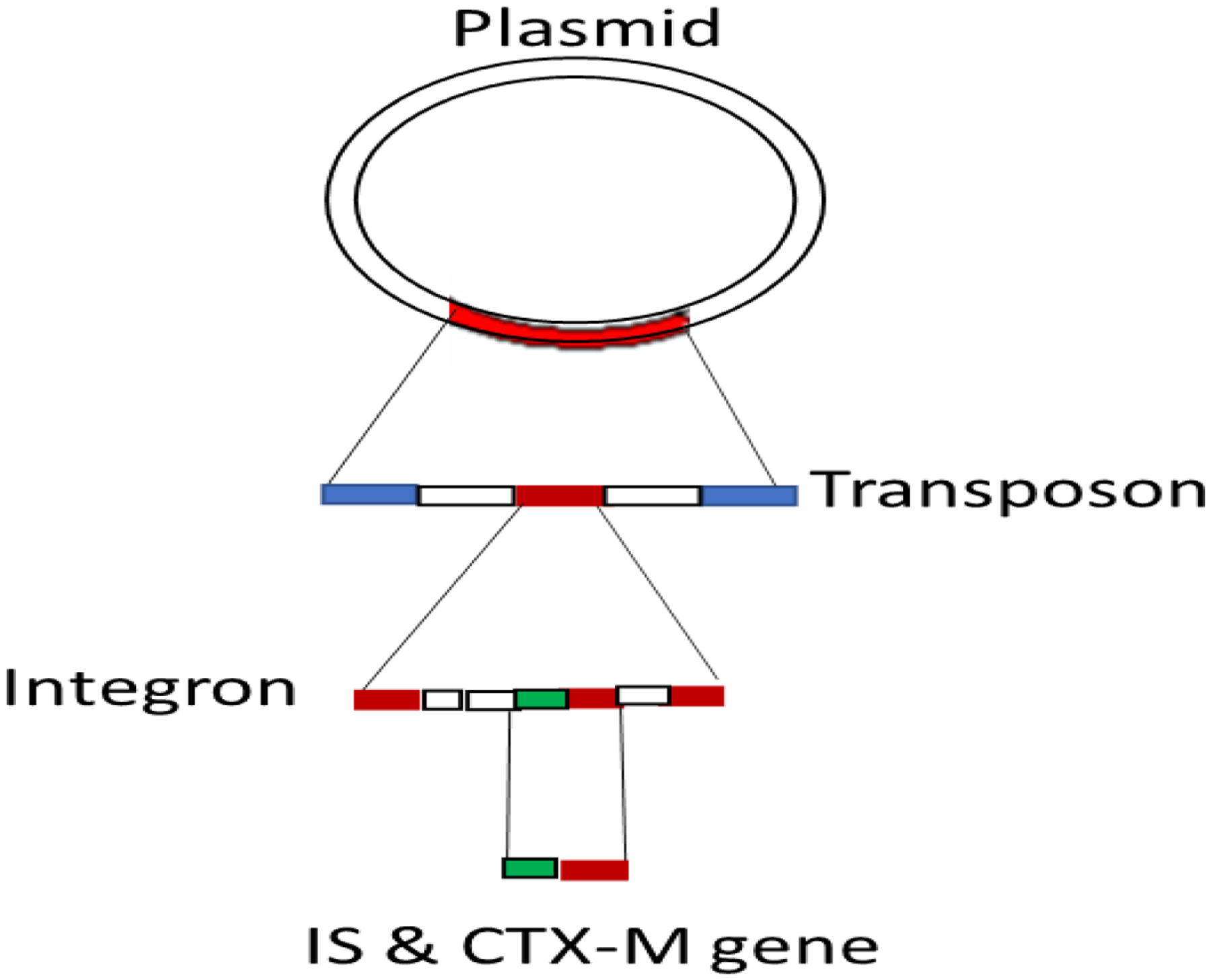
Insertion sequence (IS) and CTX-M gene upstream model.
Numerous forms of CTX-M genes encode ESBLs. CTX-M β-lactamases are likely the most widespread ESBLs, resulting in what is called the global “CTX-M pandemic” [24]. CTX-M genes originated from the chromosome of Kluyvera spp. and spread through mobile genetic elements. Their co-resistance with fluoroquinolones and aminoglycosides also facilitated the co-selection of these genes, along with other antibiotic resistance genes [25]. In some of cases, insertion sequences (IS) adjacent to blaCTX gene are identified in integron which are in turn part of transposon. Transposons integrate and make plasmids (Fig 2). These kinds of genes include ISEcp1, ISCR1, IS10 and IS26. ISEcp1 which were first identified adjacent to CTX-M-15 gene. CTX-M-15 gene is spotted 42–266 bp upstream of different blaCTX-M and provides a higher promoter expression for blaCTX-M. These genes are associated with all except CTX-M-8. Conjugative transposons and integrated conjugated plasmids are grouped as integrative and conjugate elements (ICE) are diverse mobile genetic elements which are found in integrated form in bacterial chromosome. These may excise themselves to get introduced into other chromosome and hence transfer bacteria to bacteria like plasmids. There are some ICE and IS that get fail to recognize terminal sequences of their own thus mobilize adjacent sequences through open-end transposition. Such cases may be seen with ISECP1and ISCR1 as well as Tn3 & Tn21 transposons (Cantón et al., 2012; Naseer et al., 2011).
Enterobacteriaceae isolates from animal and environmental origins contain the CTX-M-14 gene, frequently associated with IncK plasmid pCT [26]. Some of these isolates can hydrolyze ceftazidime more quickly than cefotaxime. These genes are divided into different subgroups, based on the escape of these genes from the chromosome. Enzymes CTX-M-2, CTX-M-3, and CTX-M-14 rank the highest ranking among ESBL isolation throughout the world, particularly in cephalosporin-resistant species of K. pneumonia and E. coli. Combinations of CTX-M 14 with OXA-1 and CTX-M-9 with SHV-12 have been studied on conjugative plasmids of Salmonella spp., which were resistant to a broad range of β-lactams. Similarly, combinations of CTX-M-15, SHV-12, and NDM-1 were detected in E. cloacae and demonstrated resistance towards many carbapenems. These resistant genes were present in IncHI2 plasmids with HI2 replicon as well [27]. A colistin-sensitive, ESBL-producing Klebsiella. pneumonia strain was treated with cefotaxime, which became resistant to cefotaxime due to the occurrence of the mgrB gene, which led to the cephalosporin selective pressure. However, the insertion of ISEcpI-blaCTX-M-15 locus into this gene also resulted in colistin resistance [28]. E. coli isolates with CTX-M-1, CTX-M-14, CTX-M-15, SHV-12, and TEM-167 have been identified as one of emerging mechanisms of resistance to 16S rRNA methylase. Therefore, ESBL-positive isolates paired with methylase activity are resistant to several antibiotics and susceptible to only colistin and carbapenems [29].
1.2. TEM family
TEM β-lactamases are the best-studied antibiotic resistance enzymes in that it is responsible for 90% of resistance in E. coli. Abbreviation of TEM comes from the name of Greek patient, Temoneira, who clinical samples yielded this gene. TEM variants were discovered during 1980, during which series of novel β-lactam antibiotics were discovered. At initial stages, these variants contained 1–3 amino acid substitutions whose extension later lead to one or more of new β-lactams. Hence, TEM alleles that are known today cover variants that present resistance to most of novel β-lactams introduced in the past three decades.Their mode of action lies in hydrolyzing β-lactam ring of penicillin, cephalosporins and related antibiotics. These resistant genes are isolated from hospitals and clinics at higher frequencies. Further looking into mode of action, it implies that amino acid substitution that is required for ESBL phenotype is clustered around active site of enzyme thus changing configuration and allowing access to oxyimino-beta-lacatm substrate. On the other hands, opening of active site may make it susceptible for beta-enzyme inhibitors e.g clavulanic acid that may destroy this enzyme. So use of penicillin in combination with clavulanic acid produces effective results against bacteria having TEM-1 resistant genes. It is evident from studies that single nucleotide mutations in amino acid of TEM beta lactamase give rise to substituted enzyme. Most of the substitution seen are at point 104 (Glu → Lys), point 164 (Arg → Ser or His), point 238 (Glu → Ser), point 240 (Glu → Lys) as per Ambler’s numbering scheme. TEM-harbouring microbes are equipped to resist the activity of β-lactams because they break down before attacking a specific target. These perceptions might be explained by the expansion of the protein’s stability [30]. ESBLs are most often encoded by large plasmids; however, single nucleotide mutations in genes producing TEM-1 β-lactamases lead to cephamycins and carbapenem resistance, attained during in vitro activities [31]. There are over 170 variants which are formed due to mutations in amino acids, mostly 1 to 5 substitutions, while some are those that emerge due to difference in resistance in phenotypes. TEM-1 only confer resistance to penicillin and early cephalosporin by hydrolyzing their βlactam rings. Mutations develop within the TEM structure and increase the plasticity of the active site due to the loss of hydrogen bonds, making the TEM more reactive to β-lactams and allowing for contact between oxyimino-β-lactams and the active site [32]. Thus, the exposure of the binding site to β-lactam substances as well as to the exposed TEM group members to β-lactamase blockers (e.g. clavulanic acid) leads to resistance, a dangerous condition in this case. A metagenomics study in nosocomial bacteriophages found that these phages confer resistant blaTEM genes. These results substantiate that bacteriophages act as a reservoir for these resistance genes and play an essential role in the development and spread of β-lactams resistance [33]. The transcription of TEM was affected by the exogenous addition of auto-inducer 2 (AI-2); the antibiotic’s breakpoints increased more than two-fold in a cow affected by mastitis. This suggests that exogenous AI-2 upregulates the TEM resistance. Furthermore, the transcription of lsrR increased, which encodes the AI-2 receptor. Moreover, TEM resistance is regulated by AI-2, which is itself reliant on LsrR. TEMs are becoming significant genes alone and in co with other genes isolated from various parts of the world. Five-year Swedish study revealed 63% of isoaltes positive for TEM alone, and 62% of clinical samples were identified for TEM and CTX-Ms. Global survey declared 73% of ESBL samples to be positive for TEM while expression of CTX-M was 65%. [34].
1.3. SHV family stands for sulfhydryl variable
Sulfhydryl-variable (SHV) are included in Ambler class A encoded as blaSHV that are divided as per their substrate and lactamase inhibitor on Bush-Jacoby-Medeiros (BJM) classification into class 2b type, class 2be type, and class 2br type. The type 2b hydrolyzes penicillin and 1st & 2nd generation cephalosporins, type 2be third generation cephalosporins, and type 2br becomes resistant to clavulanic acid and tazobactam. SHV-1 was isolated in 1970 from E. coli which was thought to be originated from chromosomal homology of K. Peneumoniae. Its plasmid named p453 was sequenced in 1988. The difficulty in identification with these genes as point mutation is differential aspect which are not commonly detected from routine PCR assays. SHV alleles express low clinical significance except their beta lactamase properties. It is another fact that process of transfer of blaSHV-1 from chromosome to plasmid has not been concluded because link with transposable element has not been confirmed so far. However, 189 SHV allelic variants has been reported as resistant to third generation cephalosporin, monobactams, and carbapenems. Only a portion of SHV has been genetically characterized so far which is the reason that half of the variants are yet to be classified into any group. Majority of SHVs are however found in E. coli and K. pneumoniae. Four clinical variants were found only in clinical E. coli isolates 1) blaSHV-15 Imported from India to United Kingdom; 2) blaSHV-24 found in Japan with 150kb highly resistant genes; 3) blaSHV-102 found from hospital in Spain, 4) blaSHV-129 identified from Italy. Three novel ESBLs variants were found in clinical E. cloacae:1) blaSHV-70, patient from china having been on ceftazidime treatment; 2) blaSHV-128, from Tunisia expressing IncFII conjugative plasmid, and expressing resistance to all β-lactams other than imipenem (Bourouis et al.,2015); 3) blaSHV-183, which is yet to be described. Whole genome sequencing, resistom of strain, and mobilome (mobile elements linked with antibiotic resistance diffusion) can make differentiation of SHV beta lactams. These information provide understanding of complex genome structure e.g. 1) 19 antibiotic resistance genes found in chromosome and plasmid, 2) beta lactam genes resistant against aminoglycosided, macrolides, sulphonamides, trimethoprim, and 3) Six various plasmids encoding for beta lactam and found on chromosome clusters
The prevalence of SHV-ESBL genes was associated with Klebsiella pneumonia outbreaks and to a lesser extent with E. coli [35]. The SHV-type ESBL enzymes evolved via point mutation from classical plasmid-encoded enzyme SHV-1, which has a limited hydrolytic capacity for extended-spectrum cephalosporins [36]. [37] [33,38]. The replacement of an amino acid found in an inhibitor-resistant SHV isolate (S130G in the SHV class-A β-lactamase, for instance) causes resistance to inhibition by avibactam, a unique non-β-lactam β-lactamase inhibitor. This resistance mechanism appears to be the S130G enzyme acylation by avibactam. The S130G replacement gives the impression of resistance to various classes of inhibitors and is mainly found in SHV-10. The absence of a hydroxyl group at location 130 in the S130G variant of SHV-1 substantially reduces the carbamylation of β-lactamase by avibactam in two ways. First, this absence eliminates a significant proton acceptor and donator for catalysis. Second, the quantity of hydrogen bonds is significantly reduced [39]. E. coli isolates (ST540 and ST58) harboring SHV-12 along with other non-β-lactam resistant genes (e.g. qnrS1) have been identified in China’s wastewater, posing a significant threat to the local community as well as its livestock [40].
1.4. Klebsiella pneumoniae carbapenemase (KPC)
KPC was identified in 1996 from isolates of Klebsiella pneumonia and thereafter found in several Enterobacteriaceae species [41]. Carbapenemases belong to Ambler molecular class A, which evolved from TEM-1. Nine KPC types have been identified, for which KPC-1 and KPC-2 enzymes are almost identical. The KPC carbapenemases, generally present on plasmids in Klebsiella pneumonia [42]. Recently, E. coli and Klebsiella pneumonia harbouring KPC-3 are resistant to colistin, where the primary mechanism of resistance was the inactivation of the mgrB gene [43]. There are numerous forms of kpc genes, which translate proteins, and they are differentiated on the basis of single amino acid. The kpc gene is present on a plasmid named pKP-Qil, or on its closely related variants [44]. Recently, E. coli and K. pneumonia harbouring KPC-3 have been found in an Indian hospital. These strains were resistant even to colistin, where the primary mechanism of resistance was the inactivation of the mgrB gene. The co-presence of KPC and NDM-1 has been studied in K. pneumoniae and E. coli on several plasmids that confers a significant threat of resistance development and treatment failure. KPC has also been found to be integrated into the prophage detected in K. pneumoniae strain ST258, which can be easily transmitted to other strains through transduction. In greece, carbapenem-resistant klebsiella pneumoniae (CRPK) was emerged in 2002 initiallly becausue of VIM carabpenemase production and later because of KPC, OXA-48 like carbapenemases, and NDM and apeared as endemic.The dynamicicity of CRPK strains is associated with antibiotic consumption and global travelling Porins defects inclusive of which are disruption of OmpK35 variants and have contributed to carbapenem resistance. It is considerable that CRKP would require stern preventive measures to control the spread of this pathogen. Molecular pattern would also be studied time to time to find suitable preventive measures (Karampatakis et al., 2016).
1.5. OXA β-lactamases
Plasmid encoded beta lactams in gram negative bacilli are of two types a) ubiquitous TEM enzyme, b) small group significantly hydrolyzing oxacillin. The latter was plasmid oriented having heterogenous substrate profile and encoded by narrow range plasmids showing properties like those found on R1818 that was later named as R46. OXA enzymes belong to class D carbapenemas of beta lactamases which are mostly isolated from Acinetobacter baumannii. These enzymes are effective against oxacillin and methicillin while narrow range of activity against penicillin and less inhibited by clavulanic acid. The resistance incurred by OXA β-lactams owes to use of flucloxacillin and methicillin. Initially three variants OXA-1, OXA-2 and OXA-3 were identified while the first two were inhibited by chloride ions. It is however also narrated in some other studies that not all the OXA beta lactams found on plasmids rather also found on chromosomes. It is noteworthy that plasmid linked resistance spread horizontally with greater speed.
Studies noted degree of conservation around serine active site at position number 71 (red text) and leucin at position number 179 (blue text)
| APDSTFKIALS | SLKISPEEQIQFLRK- | OXA-1 |
| SPASTFKIPHT | SLAISAQEQIAFLRK- | OXA-2 |
BlaOXA-1 was located in transposon Tn21 inserted between “aad gene” that encode aminoglycoside resistance and its promoter. Similarly, OXA-2 gene was also located in the same position which is indication presence of gene within bacteria, and it is carrying within transposon. This is the reason that OXA enzymes were given number according to their position on chromosome number which was later verified through amino acid sequences. It was however noteworthy that closely related enzymes meant for their position number not their similarities.
Most OXA group members are comparatively resistant to clavulanic acid-evoked suppression. Few show resistances to ceftazidime, but OXA-17 displays more resistance to cefixime and cefotaxime than ceftazidime [45]. ESBL-producing K. pneumonia and E. coli showed resistance to carbapenem due to the collective effect of β-lactamases with porin impenetrability and efflux pump’s function; in some microbes, this was traced to the presence of the carbapenemase-encoding gene OXA-48 and the more recently identified blaNDM-1. OXA-48 producers presented diverse stages of resistance to imipenem, ertapenem, and meropenem, perhaps as a result of the variations in the levels of gene appearance. The genome sequencing of Pseudomonas aeruginosa identified that, along with other resistance mechanisms for meropenem, the alteration of the β-lactam binding site of PBP3 and significant genomic deletion evolved. This provides bacteria with the ability to proliferate, even in the presence of an antibiotic. Acinetobacter spp., which were resistant to meropenem, contained enzyme oxacillinases, and their genetic evaluation showed the presence of OXA-23 and OXA-58-like genes associated with ISAba1 and ISAba3, respectively [46].
1.6. Other ESBLs
Rare ESBLs, for example, IBC-1, TLA-1, BES-1, and SFO-1, have only been identified in some species of Enterobacteriaceae family. Other ESBLs are mostly studied in the P. aeruginosa of different small territories [47]. Their multifactorial mechanism of resistance, (plasmid-mediated horizontal gene transfer, porin loss, carbapenem selective pressure, efflux pumps) and strains showed resistance to many antibiotics, including Piperacillin, cefotaxime, ceftriaxone, ceftazidime, aztreonam, and gentamicin. The co-presence of ESBL genes on the same plasmid constitutes a significant threat of transfer to other species [48].
2. Metallo-β-lactamases
Class B β-lactamases consist of metallo-β-lactamases (MBLs) with zinc in their active site [49], and consisted of imipenemase (IMP), Seoul imipenemase (SIM), Germany imipenemase (GIM), São Paulo metallo-β-lactamase (SPM), New Delhi metallo-β-lactamase (NDM) types, and Verona integron-encoded metallo-β-lactamase (VIM) as most prevalent types of acquired MBLs (Zavascki et al., 2011). MBLs may intrinsically be encoded by chromosomal genes or encoded by transferable genes. The later are reported extensively in Acinetobacter, Enterobacteriaceae, and Pseudomonas thus becoming as global broad-spectrum β-lactam resistance (Cornaglia et al., 2011). In a recent year 2020, prevalence of MBL genes in P. aeruginosa were noted to be 28.2, 18.8, 16.1, 9.4, 6.7, 6.0, 4.0, and 1.3% for blaIMP-1, blaVIM-2, blaVIM-1, blaNDM-1, blaIMP-2, blaSIM, blaSPM-1, and blaGIM genes, respectively. Combination of blaIMP-1 with blaIMP-2 was noted to be the most prevalent among other combinations. Increased prevalence of blaIMP-1 and blaVIM-2 alarms the increased resistance of carbapenem (Wang and Wang, 2020). Similar findings were also reported from Japan and Iran. New Delhi Metallo-β-lactamase (NDM) was first identified in India in 2009 and now is one of the most prevalent carbapenemases worldwide, isolated from Gram-negative bacteria, including E. coli, K. pneumonia, and A. baumannii [50,51]. NDM producing isolates are present in 55 countries. With the exception of aztreonam, NDM is resistant to all β-lactam antibiotics. The ndm genes are present in a variety of hosts and attach to plasmids belonging to numerous replicon types, including IncA, IncC, IncF, IncHI1, IncL-IncM, and other antibiotic-resistance genes [52,53]. The mobility of the blaNDM gene itself is responsible for NDM spread, facilitated by an ISAba125 component upstream of the ndm gene, rather than the extension of an endemic colon of isolates or plasmid, as occurs with kpc genes [54]. Many species of bacteria produce NDM enzymes, the genes of which are situated on the host chromosome, as well as the plasmid, and easily move from one to the other [55]. NDM-containing bacterial infections were initially epidemiologically limited to the Indo-Pak, Middle East, and Balkan states, but more recently have been reported in many other parts of the world [56]. Ancient-β-lactamases were initially identified in strains of P. aeruginosa; there is now a growing universal disaster with this class of β-lactamases, Enterobacteriaceae in particular. MDR E. coli and K. pneumoniae were more important in Enterobacteriaceae-containing, plasmid-harboring NDM-1, and were readily transferable through conjugation [57]. NDM-1 and KPC have similar ranges of antibiotic resistance and are spread by a wide variety of plasmids.
PCR mapping and DNA sequencing of Acinetobacter spp. revealed the presence of NDM-1 on a transposon between two copies of ISAba125. Most isolates were resistant to meropenem [58]. MDR E. coli contains not only NDM-1 but also other resistance genes, including qnrB, aac(6c)-ib-cr, and blaSHV-11. All of these strains were STs2 type with the exception of one strain that was ST594. Their MDR property was due to the presence of NDM-1 and the loss of the outer membrane. The conjugation experiment showed that drug resistance spread in the presence of IncN and STs2, which were horizontally transferred [59]. Previous research also found that the IncN plasmid-encoded very important resistance genes, such as blaNDM-1, blaCTX, and qnr in E. coli and K. pneumonia [60]. The genomic sequence of a nosocomial MDR E. cloacae showed the presence of NDM-1 plasmid on a novel class 1 integron, which is a mosaic between replicons, including IncA/C2 and IncR. This strain also carried blaKPC-3, gyrA, gyrB, parC, pare, and armA genes mutations and a simultaneous loss of the outer membrane porins, thus making it resistant to all β-lactams and many other antibiotics [57]. The metallo-β-lactamase fold (MBLf) in bacteria helped to hydrolyze many β-lactam drugs [61]. There is another gene identified and characterized in Pseudomonas aeruginosa in 2012 and named as blaFIM-1 gene. FIM-1 enzyme, belongs to subclass B1, expresses ca. 40% amino acid identity with NDM enzymes which is highest among other members of acquired MBLs. Kinetic parameters of FIM-1 revealed broad substrate specificity with penicillin and carbapenems. This activity can make it interesting model to further probe into structure cum functional relationships of metallo-β-lactamases (Polllini et al., 2013).
The blaSIM-1 gene was originally discovered from Acinetobacter pittii from hospitalized patient in Koreaa in year 2003 that later were also isolated from Acinetobacter bereziniae and Acinetobacter nosocomialis. It is also notable that blaSIM-1 is rare and limited to Acinetobacter from China and South Korea. Moreover, SIM-1 is rarely detected member of MBLs but displays more than 60% amino acid similarity with IMPs which are its nearest relatives. This gene is found embedded in class 1 integran with modular structure intI1-blaSIM-1-arr3- catB3-aadA1-qacED1.It is well noted that these genes are located on variable sized plasmids, but these are yet to be sequenced.
Recently, the presence of blaIMP-1, blaKPC-3, and the co-occurrence of blaNDM-1 and blaVIM-2 has been found in E. coli, and K. pneumonia isolates from India. The plasmid containing NDM-1 and VIM-2 belonged to IncL/M and IncA/C. Two predominant, transferable Metallo-β-lactamase genes have been found among the clinical bacteria (e.g. bla IMP and bla VIM) displayed on the gene cassette of the integrons and situated on the chromosome or plasmid. Verona integron-encoded MBLs (VIM-type enzymes) were first recognized in 1996 but holds 60 allelic variants among which VIM-2 is routinely reported MBL in the world. It has expressed itself in outbreaks of P. aeruginosa while rarely isolated in Enterobacteriaceae. Most VIM alleles are classified into VIM-1 and VIM-2. The latter is peculiar case which is VIM-1/VIM-2 hybrid giving rise to N-terminal sequence from VIM-1 (signal peptides are included) and C-terminus from VIM-2. The blaVIM-12 gene has been found in Enterobacteriaceae despite of occupation of VIM-2 in chromosomal cassette. This fact reveals that signal peptide is important than genetic environment of MBL gene in conferring host specificity. Hence, sequence speaks of host specificity and possible spread of MBL alleles (López et al., 2019). Another gene named as blaGIM-1 gene was identified in 2002 from Germany in clinical isolate of Pseudomonas aeruginosa consisting of 24-kb nontransferable plasmid. GIM-1 possesses three characteristics of B1 MBLs a) binuclear zinc site in an αβ/βα fold, b) mobile loops 1 & 2 adjacent to active site c) a lower-affinity Zn2 site) whereas some are unique properties with reference to Trp228 and Tyr233 producing narrower and more hydrophobic active-site groove. There are 9 independently pure models of GIM-1 that give rise to dynamic portfolio of active sites that to routinely provided crystallographic investigations. These reflect plasticity of active site groove which is feature of enzyme.
3. AmpC β-lactamases
In most bacterial genera, the AmpC gene was inducible via a mechanism involving the ampD, ampG, ampR and the intermediate molecules used in peptide glycan recycling [20]. AmpC β-lactamases were identified in many Gram-negative bacteria when mutations in significant genes caused increase in levels of expression and encouraged the development of cephalosporin resistance [48]. AmpC enzymes are either chromosomally encoded or plasmid-encoded. Most plasmid-encoded AmpC enzymes showed resistance to multiple antibiotics, e.g., tetracycline, aminoglycosides or sulfonamides as they conferred resistance to 7-α-methoxy-cephalosporins and were not affected by commercially available β-lactamase inhibitors. Among the plasmid-encoded AmpC enzymes, the most important subgroup is CMY, which features cephalosporins activity. CMY-2, DHA-1 and ACT-1 are chromosomally-located AmpC enzymes [7]. In Gram-negative bacteria, the production of chromosomal AmpC enzymes was lower compared to plasmid-encoded AmpC enzymes; however, this product can be enhanced by the induction of some β-lactams, such as cefoxitin. E. coli and K. pneumoniae strains harbouring AmpC β-lactamases have the ability to lose porin Omp-K35 from their outer membrane and show resistance against cephamycin because this porin permits water soluble β-lactams to passthrough and gain access into the cells [63]. Recent studies found that more than twenty plasmid-expressed AmpC β-lactamases exist. The regulatory pathway of ampC β-lactamases consists of 3 components: (1) AmpG, a permease present in the interior membrane; (2) AmpD, an amidase in the cytoplasm; and (3) AmpR, a transcription factor which is a member of the regulatory protein LysR family [64]. For the expression of AmpC β-lactamases in P. aeruginosa and Enterobacteriaceae Spp., these three components are necessary [65]. During the cell wall recycling process, 1,6-anhydromuropeptides are detached from the bacteria cell wall and relocated into the cytosol with the help of AmpG permease. AmpD protein cuts the 1,6-anhydromuropeptides and yields tripeptides, which are consequently changed into UDP-MurNAc-pentapeptides (UDP-MNPP). The UDP-MNPP combine with AmpR proteins in the intergenic region between ampC and ampR, and create a structure that monitors the stimulation of ampC. This results in decreased AmpC generation, low expression levels, and ultimately confines the β-lactamase to the periplasmic space. β-lactams penetrate the outer membrane of Gram-negative bacteria and bond with target PBPs after reaching such periplasmic spaces. 1,6-anhydromuropeptides increases and AmpD cannot competently handle such a large quantity of cell wall components. Anhydro-MurNAc-peptides (AMNP) substitute UDP-MNPP binding to AmpR, structurally altering the enzyme. The AmpR is changed into the role of a transcriptional promoter, a very high amount of AmpC is formed, and the quantity of AmpC increases in the periplasmic region. In cytoplasm, the quantity of AMNP decreases due to the fact that the concentration of β-lactam is lower than its “dangerous level”; as a result, the ability of AmpD to efficiently counter these peptides is reinstated. Even so, the mutation of nucleotides causes a shortage of AmpD and down-regulates the expression of AmpD damage in the process of cell wall fragment recycling, which produces a high concentration of AMNP in the cytosol. Consequently, the conjoining of AmpR with AMNP makes AmpR “protected” as a “transcriptional activator” of AmpC and yields a higher amount of AmpC β-lactamases (Table 1).
Table 1.
Evolutionary trends in β-lactamase resistance development.
| β-lactamases | Mechanisms | Resistance Strains | β-lactam Antibiotics | References |
|---|---|---|---|---|
| CTX-M family | ||||
| CTX-M-3, CTX-M-15, CTX-M-16, CTX-M-19, CTX-M-27 | Hydrolyze, mutation, ESBLs | E. coli | Ceftazidime | [73,74] |
| CTX-M-9, CTX-M-13, CTX-M-14. | Plasmid mediated, tnpA gene transposon, insert sequence ISEcp-1 on plasmid ESBLs |
Enterobacteriaceae, E. coli,
Klebsiella pneumoniae |
Cefotaxime, Ceftazidime | [75–77] |
| CTX-M-32 | Asp-240Gly substitution | E. coli | Cefotaxime, ceftazidime, and aztreonam | [78] |
| CTX-M-2, CTX-M-39 | Plasmid mediated ESBLs, substitution of arginine | E. coli | Cefotaxime | |
| CTX-M-32 | Asp-240Gly substitution of ESBLs | E. coli | Ceftazidime | [78] |
| CTX-M-14 CTX-M-15, CTX-M-57 along with qnrB and qnrS | Plasmid mediated substitution of ESBLs, Active efflux |
E. coli,
Klebsiella pneumoniae |
Ceftazidime and quinolones | [79] |
| CTX-M | Single nucleotide mutation | Salmonella spp. | Ceftriaxone | [42] |
| CTX-M-2, CTX-M-3, CTX-M-14 | Plasmid mediated substitution of ESBLs |
E. coli,
Klebsiella. pneumoniae |
Cephalosporin | [36,37] |
| CTX-M-14 | Plasmid mediated substitution of ESBLs |
E. coli,
Klebsiella. Pneumoniae |
Cefotaxime and ceftazidime | [35] |
| TEM Family | ||||
| TEM-30, TEM-34, TEM-40, TEM-51 | Single amino acid substitution | E. coli | Cefoxitin | |
| TEM-1, TEM-30, TEM-33, | Plasmid mediated ESBLs | E. coli | Cefotaxime | |
| TEM-3, TEM-4, TEM-133 | Mutations L21F, E104K, and R164S |
E. coli,
Klebsiella. pneumoniae |
Ceftazidime, Cefotaxime | [80] |
| TEM-1 along with qnrB and qnrS | Plasmid mediated ESBLs substitution, active efflux |
E. coli,
. Klebsiella. pneumoniae |
Ceftazidime and quinolones | [81] |
| TEM-34, TEM-40 | Single amino acid gene mutations |
E. coli,
. Klebsiella. pneumoniae |
Cefotaxime, Clavulanic acid | [44] |
| TEM-1 | Single nucleotide mutation ESBLs | Salmonella spp. | Nalidixic acid, ampicillin, ceftriaxone cotrimoxazole, chloramphenicol, and ciprofloxacin | [42] |
| SHV Family | ||||
| SHV-1 | Gene mutation, hydrolysis |
Enterobacteriaceae,
K. pneumoniae |
Penicillins, 1st generation cephalosporin |
[82] |
| SHV-5, SHV-12 | Plasmid mediated ESBLs, substitution |
E. coli,
K. pneumoniae |
Cefotaxime | |
| SHV-36 | Mutation | E. coli and E. cloacae | Cephalosporinss | [42] |
| SHV-2, SHV-5, SHV-12 along with qnrB and qnrS | Plasmid mediated ESBLs substitution, Active efflux |
E. coli,
K. pneumoniae |
Ceftazidime and quinolones | [81] |
| SHV-1, SHV-11, SHV-12, SHV-18, SHV-28 | Amino acid mutation |
E. coli,
K. pneumoniae |
Penicillin, amoxicillin, oxacillin, cefoxitin, Cefotaxime |
[38] |
| SHV-1, SHV-10 | Acylation of the S130G enzyme, Reduced carbamylation |
E. coli | Avibactam, Diazabicyclooctane |
[55] |
| KPC | ||||
| KPC-1 | Carbapenem-hydrolyzing β-lactamase, Alterations in porin expression |
K. pneumoniaee | Cephalosporins, aztreonam, imipenem, meropenem | [60] |
| KPC-1 | Carbapenem-hydrolyzing β-lactamase, | Enterobacteriaceae species | Cephalosporins, aztreonam, | [59] |
| KPC-2, KPC-3 | Carbapenem-hydrolyzing β-lactamase | K. pneumoniae ST258 | Cephalosporins, Penicillin, | [83] |
| KPC-2 | Plasmid mediated Carbapenem-hydrolyzing β-lactamase | K. pneumonia ST11 | Ceftazidime, cefoxitin, piperacillin/tazobactam, cefoperazone/sulbactam | [84] |
| KPC-kp | pkp28 plasmid mediated, hydrolysis | K. pneumoniae | Cephalosporins | [42] |
| Carbapenemase NDM | ||||
| NDM-1 | ESBLs, Porin impermeability |
K. pneumoniae,
E. coli |
Imipenem, ertapenem, meropenem | |
| NDM-1 | Plasmid mediated resistance |
K. pneumoniae,
E. coli |
Imipenem, ertapenem, Meropenem, aztreonam |
[85] |
| NDM-1 | Plasmid mediated blaNDM-1 |
K. pneumoniae ST11 K. pneumoniae ST14 |
Imipenem, ertapenem, meropenem | [86] |
| NDM-1 | Plasmid mediated blaNDM-1 | K. pneumoniae | All β-lactams except aztreonam | [87] |
| NDM+KPC | Plasmid mediated enzymatic degradation | Enterobacteriaceae | All carbapenem | |
| NDM-1 | bla NDM-1 | Salmonella Spp. | Ampicillin Amoxicillin-clavulanic acid Piperacillin-tazobactam Ceftriaxone Trimethoprim-sulfamethoxazole |
[88] |
| NDM-1 | IncN plasmid p46 with ISCR1 element |
K. pneumonia,
E. coli |
Carbapenems | [73,89] |
| NDM-1 | Plasmid mediated blaNDM-1 | Acinetobacter baumannii strains of ST1 | Carbapenems | [90] |
| NDM-1 | blaNDM-1 on transposon | Acinetobacter spp. | Meropenem | [58,66] |
| NDM-1 + qnrB + aac +SHV-11 | blaNDM-1 and outer membrane loss | E. coli | Multidrug resistance | [57] |
| NDM-1 + KPC-3, gyrA, gyrB, parC, pare and armA | Plasmid mediated blaNDM-1, and loss of porins | Enterobacter cloacae | Multidrug resistance | [91] |
| OXA Family | ||||
| OXA | Single amino acid mutation |
E. coli,
P. aeruginosa |
Cefoxitin | [92] |
| OXA-2 | Plasmid mediated ESBLs substitution | E. coli | Cefotaxime | |
| OXA-10, OXA17, OXA-74 | No OprD production | Pseudomonas aeruginosa | Cefipime and cefotaxime | [65] |
| OXA-48 | ESBLs, Porin impermeability |
K. pneumoniae,
E. coli |
Imipenem, ertapenem, meropenem | |
| OXA-23, OXA-58 | Hydrolysis by oxacillinase | Acinetobacter spp. | Meropenem | [58,66] |
| KPC | ||||
| KPC-1 | Carbapenem-hydrolyzing β-lactamase, Alterations in porin expression | K. pneumoniae | Cephalosporins, aztreonam, imipenem, meropenem | [60] |
| KPC-1 | Carbapenem-hydrolyzing β-lactamase, | Enterobacteriaceae species | Cephalosporins, aztreonam, | [59] |
| KPC-2, KPC-3 | Carbapenem-hydrolyzing β-lactamase | K. pneumoniae ST258 | Cephalosporins, Penicillin, | [83] |
| KPC-2 | Plasmid mediated Carbapenem-hydrolyzing β-lactamase | K. pneumonia ST11 | Ceftazidime, cefoxitin, piperacillin/tazobactam, cefoperazone/sulbactam | [93] |
| KPC-kp | pkp28 plasmid mediated, hydrolysis | K. pneumoniae | Cephalosporins | [61] |
| Carbapenemase NDM | ||||
| NDM-1 | ESBLs, Porin impermeability |
K. pneumoniae,
E. coli |
Imipenem, ertapenem, meropenem | [81] |
| NDM-1 | Plasmid mediated resistance |
K. pneumonia,
E. coli |
Imipenem, ertapenem, Meropenem, aztreonam |
Fisher and Mobashery, 2014; Bakour et al., 2015) |
| NDM-1 | Plasmid mediated blaNDM-1 |
K. pneumoniae ST11 K. pneumoniae ST14 |
Imipenem, ertapenem, meropenem | [50,51] |
| NDM-1 | Plasmid mediated blaNDM-1 | K. pneumoniae | All β-lactams except aztreonam | [69] |
| NDM+KPC | Plasmid mediated enzymatic degradation | Enterobacteriaceae | All carbapenem | [94] |
| NDM-1 | bla NDM-1 | Salmonella Spp. | Ampicillin Amoxicillin-clavulanic acid Piperacillin-tazobactam Ceftriaxone Trimethoprim-sulfamethoxazole |
|
| NDM-1 | IncN plasmid p46 with ISCR1 element |
K. pneumoniae,
E. coli |
Carbapenems | [95] |
| NDM-1 | Plasmid mediated blaNDM-1 | Acinetobacter baumannii strains of ST1 | Carbapenems | [84] |
| NDM-1 | blaNDM-1 on transposon | Acinetobacter spp. | Meropenem | [58,66] |
| NDM-1 + qnrB + aac +SHV-11 | blaNDM-1 and outer membrane loss | E. coli | Multidrug resistance | [73,74] |
| NDM-1 + KPC-3, gyrA, gyrB, parC, pare and armA | Plasmid mediated blaNDM-1, and loss of porins | Enterobacter cloacae | Multidrug resistance | [57] |
| AmpC β-lactamases | ||||
| AmpC | Mutations at positions −42 and −32 of the ampC promoter region | E. coli | Cefoxitin | [96,97] |
| DHA-1 | Plasmid mediated hydrolyzing β-lactamase | K. pneumoniae ST11 | Ceftazidime, cefoxitin, piperacillin/tazobactam, cefoperazone/sulbactam | [81] |
| AmpC | Overexpression of MexXY-OprM and MexAB-OprM | Pseudonomas aeruginosa | Cefipime and cefotaxime | [65] |
| AmpC | overexpression of the ampC gene and hydrolysis of oxyimino cephalosporins | Enterobacteriaceae | Cephalosporins | [23] |
| AmpC | Less generation and low activation of ampC leads to confine the β-lactams in periplasmic region | Enterobacteriaceae | Cephalosporin | [96,97] |
| AmpC + TEM | Mutation in genes | E. coli | Amoxicillin, cefixime, nalidixic acid, gentamicin, ceftriaxone, ceftazidime, TMP, difloxacin, cefotaxime, | |
| AmpC + CTX-M | Mutation in promoter at position −31 | E. coli | Cefpodoxime | [98] |
| AmpC | blaAmpC over production, Mutation in efflux parts, PBP3 mutation | Pseudomonas aeruginosa | Ceftazidime | [80] |
In summary, AmpC enzymes can be provoked for expression at resonant level through mutation which in turn lead to resistance to broad-spectrum cephalosporins inclusive of which are cefotaxime, ceftazidime, and ceftriaxone. Transmissible plasmids are holding acquired genes for AmpC that resultantly can emerge in bacteria such as E. coli, P. mirabilis, and K. pneumoniae which is lacking or poorly expressing chromosomal blaAmpC gene.
Prevention and future prospectives
Several β-lactamase inhibitors have been used to minimize the development of β-lactamase inactivation and target the special features and characters of these enzymes. We must use the inhibitors with high affinity for the active site of the β-lactamase target so that the inhibitor can work efficiently; for example, phosphonates and methylidene penems attain an inhibition similar to classes D, C, and A [58,66]. By mimicking the available natural substrates, β-lactamase can therefore be inhibited. Like class C, β-lactamases show very high binding with those inhibitors that are structurally similar to natural cephalosporin substrates [67]. Similarly, β-lactamases can also be inhibited if the interaction between the β-lactamase inhibitor and the active site of β-lactamases is prolonged. Accordingly, the long-life, acyl-enzyme intermediate is crucial for a successful reaction by class An inhibitors. Another mechanism by which inhibitors cease the activity of β-lactamases is expedient diffusion in the cell membrane because β-lactamases are mostly found in the periplasm of Gram-negative bacteria. For example, cefipime, a β-lactam molecule, rapidly diffuses the bacterial cell membrane[68]. Therefore, drugs that easily diffuse the outer membrane of the cell through porins and rapidly filter the membrane back through efflux pumps should be formulated [7]. β-lactams should not only destroy the mechanism of β-lactamase during in vitro settings, but also inhibit the function of β-lactamases during in vivo conditions, which is our main goal. Similarly, different combinations of available drugs should be applied to control the activity of β-lactamases, including clavulanate with meropenem, cefpirome, or cefipime [50,51].
A combination of the fungal products (aspergillomarasmine) and meropenem are good inhibitors of NDM-1 in a murine infection model. Detection of ESBLs and plasmid-encoded AmpC lactamases is necessary for the effective surveillance and epidemiology, as well as to develop appropriate infection control strategies associated with resistance mechanism [47]. NDM-1 and KPC have the capacity to cross-genetic transfer through HGT and are distributed worldwide, so it is recommended to screen for their presence in nosocomial Enterobacteriaceae strains. Carbapenems should be widely used to limit the development of resistance. Avicaz, a combination of avibactam-ceftazidime, is a very effective β-lactamase inhibitor that was approved by the FDA in 2015 and is being widely used for systemic infections [69]. The blaNDM-1 genes have been recently found in wastewater treatment plants, and blaNDM-1 receiving rivers were located in a number of horizontally transferred plasmid isolates. This study tracked the pathway of NDM-1 genes as they spread into the ecosystem; accordingly, strict measures should be taken to combat the vectors that facilitate the dissemination of these genes. Non-therapeutic uses of antibiotics in agriculture and veterinary fields must be minimized; otherwise we will have no possible solutions to decrease antibiotic resistance in the future [70]. Worldwide actions are occurring in many forums. The World Health Assembly passed a resolution to develop a Global Action Plan on Antibiotic resistance published by WHO [57]. Likewise, a relevant proposal was passed by the G7 Ise-Shima Summit in Japan to take similar actions, aligned with the WHO and FAO resolutions. Available drugs should be used appropriately, and the unnecessary use of β-lactams with broad-spectrum activity (the main reason for resistance emergence) should be avoided [40]. Recently, in silico techniques have exposed the β-lactamases genetic evolution and the resistome in bacteria; therefore, these approaches will help to develop the specific molecular diagnostic tools for the detection and formulation of non-β-lactam β-lactamase inhibitors [71].
We are certain that controlling β-lactamase resistance is the greatest current challenge in the medical field. New β-lactams have been gradually integrated into clinical trials. This is a demonstration of the flexibility and multifaceted nature of β-lactamases; moreover, this review describes the unimaginable developmental capacity of the life forms harbouring β-lactamases. A thorough understanding of genetic pathways behind the development of anti-microbial resistance may help devise better solutions. For instance, an integrated approach to managing clinical infections that features the use of alternative ways to curb severe clinical symptoms. The use of anti-microbial adjuvants, nanomedicine and other easily biodegradable options for blocking resistance mechanisms may help counter the superbugs at initial stages [69,72].
Conclusion
In the last decade, some studies have focused β-lactam resistance; however, this field still warrants additional consideration of researchers in order to establish a paradigm for the development and spread of resistance. In Gram-negative bacteria, highly transferable plasmid-mediated β-lactamases that hydrolyze the most β-lactams pose the main challenge to the medical field. Every year new β-lactamase variants are identified. Moreover, an understanding of new mechanisms is necessary to combat this challenge. The co-presence of different classes of β-lactamase and combinations of β-lactamase genes with non-β-lactamase, antibiotic-resistant genes worsen the situation, as the presence of NDM- and KPC-type carbapenemases in Enterobacteriaceae enhance overall aminoglycoside resistance.
Recent investigation on the prevalence of ESBL- and AmpC-encoding Enterobacteriaceae in food products and river water has been reported in China. Accordingly, the priority should be to check for resistant bacteria in freshwater, food, animals, and food products. Through our knowledge of β-lactams resistance is not ample; we should consider the existing evidence in order to make policies and combat resistance. It is clear that the co-existence of β-lactamase genes and other non-β-lactam-resistant genes has been increasing due to the co-selection process, which ultimately will give rise to new opportunities for evolution. No single strategy will be entirely effective; therefore, numerous concurrent and synchronized measures should be taken on national and international levels. The most pressing task in the field is the best use of existing information, practical skills and scientific equipment to combat the influence of resistance along with the development of next-generation antibiotics. If we consider all of these aspects over the coming decades, resistance to β-lactams and all other antibiotics will become a controllable problem rather than a universal health disaster.
References:
- [1].Khaledi A, Weimann A, Schniederjans M, Asgari E, Kuo T-H, Oliver A, Cabot G, Kola A, Gastmeier P, Hogardt M, others, Predicting antimicrobial resistance in Pseudomonas aeruginosa with machine learning-enabled molecular diagnostics, EMBO Mol. Med 12 (2020) e10264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Holmes AH, Moore LSP, Sundsfjord A, Steinbakk M, Regmi S, Karkey A, Guerin PJ, V Piddock LJ, Understanding the mechanisms and drivers of antimicrobial resistance, Lancet. 387 (2016) 176–187. [DOI] [PubMed] [Google Scholar]
- [3].Usman Ishaq M, Rafique A, Cheema HMN, Umer Ashraf M, Rahman SU, Zahid Abbas R, Shahid Mahmood M, Role of cytosine-phosphate-guanosine-Oligodeoxynucleotides (CpG ODNs) as adjuvant in poultry vaccines, Worlds. Poult. Sci. J 74 (2018) 453–462. [Google Scholar]
- [4].Ambler RP, Coulson AFW, Frère J-M, Ghuysen J-M, Joris B, Forsman M, Levesque RC, Tiraby G, Waley SG, A standard numbering scheme for the class A $β$-lactamases, Biochem. J 276 (1991) 269–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Bush K, Jacoby GA, Updated functional classification of $β$-lactamases, Antimicrob. Agents Chemother 54 (2010) 969–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Doi Y, Wachino J, Arakawa Y, Aminoglycoside resistance: the emergence of acquired 16S ribosomal RNA methyltransferases, Infect. Dis. Clin 30 (2016) 523–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Drawz SM, Bonomo RA, Three decades of $β$-lactamase inhibitors, Clin. Microbiol. Rev 23 (2010) 160–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Drawz SM, Babic M, Bethel CR, Taracila M, Distler AM, Ori C, Caselli E, Prati F, Bonomo RA, Inhibition of the class C $β$-lactamase from Acinetobacter spp.: insights into effective inhibitor design, Biochemistry. 49 (2010) 329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Jacoby GA, Mills DM, Chow N, Role of $β$-lactamases and porins in resistance to ertapenem and other $β$-lactams in Klebsiella pneumoniae, Antimicrob. Agents Chemother. 48 (2004) 3203–3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Doumith M, Ellington MJ, Livermore DM, Woodford N, Molecular mechanisms disrupting porin expression in ertapenem-resistant Klebsiella and Enterobacter spp. clinical isolates from the UK, J. Antimicrob. Chemother 63 (2009) 659–667. [DOI] [PubMed] [Google Scholar]
- [11].Poole K, Efflux-mediated multiresistance in Gram-negative bacteria, Clin. Microbiol. Infect 10 (2004) 12–26. [DOI] [PubMed] [Google Scholar]
- [12].Li X-Z, Zhang L, Poole K, Interplay between the MexA-MexB-OprM multidrug efflux system and the outer membrane barrier in the multiple antibiotic resistance of Pseudomonas aeruginosa, J. Antimicrob. Chemother 45 (2000) 433–436. [DOI] [PubMed] [Google Scholar]
- [13].Cantón R, González-Alba JM, Galán JC, CTX-M enzymes: origin and diffusion, Front. Microbiol 3 (2012) 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Philippon A, Arlet G, Jacoby GA, Plasmid-determined AmpC-type $β$-lactamases, Antimicrob. Agents Chemother 46 (2002) 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Toleman MA, Bennett PM, Walsh TR, ISCR elements: novel gene-capturing systems of the 21st century?, Microbiol. Mol. Biol. Rev 70 (2006) 296–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Haenni M, Châtre P, Madec J-Y, Emergence of Escherichia coli producing extended-spectrum AmpC $β$-lactamases (ESAC) in animals, Front. Microbiol 5 (2014) 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Zykov IN, Sundsfjord A, Småbrekke L, Samuelsen Ø, The antimicrobial activity of mecillinam, nitrofurantoin, temocillin and fosfomycin and comparative analysis of resistance patterns in a nationwide collection of ESBL-producing Escherichia coli in Norway 2010--2011, Infect. Dis. (Auckl) 48 (2016) 99–107. [DOI] [PubMed] [Google Scholar]
- [18].Jacoby GA, AmpC $β$-lactamases, Clin. Microbiol. Rev 22 (2009) 161–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lynch III JP, Clark NM, Zhanel GG, Evolution of antimicrobial resistance among Enterobacteriaceae (focus on extended spectrum β-lactamases and carbapenemases), Expert Opin. Pharmacother 14 (2013) 199–210. [DOI] [PubMed] [Google Scholar]
- [20].Johnson AP, Woodford N, Global spread of antibiotic resistance: the example of New Delhi metallo-$β$-lactamase (NDM)-mediated carbapenem resistance, J. Med. Microbiol 62 (2013) 499–513. [DOI] [PubMed] [Google Scholar]
- [21].Sinha R, Kamath S, M Shenoy S, Association of risk factors, antimicrobial resistance trends and occurrence of blaTEM, bla SHV and blaCTX M in Escherichia coli causing bacteremia, Infect. Disord. Targets (Formerly Curr. Drug Targets-Infectious Disord 16 (2016) 95–100. [DOI] [PubMed] [Google Scholar]
- [22].Vala MH, Hallajzadeh M, Hashemi A, Goudarzi H, Tarhani M, Tabrizi MS, Bazmi F, Detection of Ambler class A, B and D ß-lactamases among Pseudomonas aeruginosa and Acinetobacter baumannii clinical isolates from burn patients, Ann. Burns Fire Disasters 27 (2014) 8. [PMC free article] [PubMed] [Google Scholar]
- [23].Jean S-S, Coombs G, Ling T, Balaji V, Rodrigues C, Mikamo H, Kim M-J, Rajasekaram DG, Mendoza M, Tan TY, others, Epidemiology and antimicrobial susceptibility profiles of pathogens causing urinary tract infections in the Asia-Pacific region: Results from the Study for Monitoring Antimicrobial Resistance Trends (SMART), 2010--2013, Int. J. Antimicrob. Agents 47 (2016) 328–334. [DOI] [PubMed] [Google Scholar]
- [24].Cantón R, Coque TM, The CTX-M $β$-lactamase pandemic, Curr. Opin. Microbiol 9 (2006) 466–475. [DOI] [PubMed] [Google Scholar]
- [25].Cantón R, Ruiz-Garbajosa P, Co-resistance: an opportunity for the bacteria and resistance genes, Curr. Opin. Pharmacol 11 (2011) 477–485. [DOI] [PubMed] [Google Scholar]
- [26].Dhanji H, Khan P, Cottell JL, V Piddock LJ, Zhang J, Livermore DM, Woodford N, Dissemination of pCT-like IncK plasmids harboring CTX-M-14 extended-spectrum $β$-lactamase among clinical Escherichia coli isolates in the United Kingdom, Antimicrob. Agents Chemother 56 (2012) 3376–3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Petrosillo N, Vranić-Ladavac M, Feudi C, Villa L, Fortini D, Barišić N, Bedenić B, Ladavac R, D’Arezzo S, Andrašević AT, others, Spread of Enterobacter cloacae carrying blaNDM-1, blaCTX-M-15, blaSHV-12 and plasmid-mediated quinolone resistance genes in a surgical intensive care unit in Croatia, J. Glob. Antimicrob. Resist 4 (2016) 44–48. [DOI] [PubMed] [Google Scholar]
- [28].Jayol A, Nordmann P, Desroches M, Decousser J-W, Poirel L, Acquisition of broad-spectrum cephalosporin resistance leading to colistin resistance in Klebsiella pneumoniae, Antimicrob. Agents Chemother 60 (2016) 3199–3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ayad A, Drissi M, de Curraize C, Dupont C, Hartmann A, Solanas S, Siebor E, Amoureux L, Neuwirth C, Occurence of ArmA and RmtB aminoglycoside resistance 16S rRNA methylases in extended-spectrum $β$-lactamases producing Escherichia coli in Algerian hospitals, Front. Microbiol 7 (2016) 1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Brown NG, Pennington JM, Huang W, Ayvaz T, Palzkill T, Multiple global suppressors of protein stability defects facilitate the evolution of extended-spectrum TEM $β$-lactamases, J. Mol. Biol 404 (2010) 832–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ahmed D, Ud-Din AIM, Wahid SUH, Mazumder R, Nahar K, Hossain A, others, Emergence of blaTEM type extended-spectrum $β$-lactamase producing Salmonella spp. in the urban area of Bangladesh, Int. Sch. Res. Not 2014 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Pimenta AC, Fernandes R, Moreira IS, Evolution of drug resistance: insight on TEM $β$-lactamases structure and activity and $β$-lactam antibiotics, Mini Rev. Med. Chem 14 (2014) 111–122. [DOI] [PubMed] [Google Scholar]
- [33].Subirats J, Sànchez-Melsió A, Borrego CM, Balcázar JL, Simonet P, Metagenomic analysis reveals that bacteriophages are reservoirs of antibiotic resistance genes, Int. J. Antimicrob. Agents 48 (2016) 163–167. [DOI] [PubMed] [Google Scholar]
- [34].Xue T, Yu L, Shang F, Li W, Zhang M, Ni J, Chen X, The role of autoinducer 2 (AI-2) on antibiotic resistance regulation in an Escherichia coli strain isolated from a dairy cow with mastitis, J. Dairy Sci 99 (2016) 4693–4698. [DOI] [PubMed] [Google Scholar]
- [35].Paterson DL, Bonomo RA, Extended-spectrum $β$-lactamases: a clinical update, Clin. Microbiol. Rev 18 (2005) 657–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ye Q, Wu Q, Zhang S, Zhang J, Yang G, Wang H, Huang J, Chen M, Xue L, Wang J, Antibiotic-resistant extended spectrum ss-lactamase-and plasmid-mediated AmpC-producing enterobacteriaceae isolated from retail food products and the pearl river in Guangzhou, China, Front. Microbiol 8 (2017) 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Thomson JM, Distler AM, Prati F, Bonomo RA, Probing Active Site Chemistry in SHV $β$-Lactamase Variants at Ambler Position 244 UNDERSTANDING UNIQUE PROPERTIES OF INHIBITOR RESISTANCE, J. Biol. Chem 281 (2006) 26734–26744. [DOI] [PubMed] [Google Scholar]
- [38].Li J, Ji X, Deng X, Zhou Y, Ni X, Liu X, Detection of the SHV genotype polymorphism of the extended-spectrum $β$-lactamase-producing Gram-negative bacterium, Biomed. Reports 3 (2015) 261–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Winkler ML, Papp-Wallace KM, Taracila MA, Bonomo RA, Avibactam and inhibitor-resistant SHV $β$-lactamases, Antimicrob. Agents Chemother 59 (2015) 3700–3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Röderova M, Halova D, Papousek I, Dolejska M, Masarikova M, Hanulik V, Pudova V, Broz P, Htoutou-Sedlakova M, Sauer P, others, Characteristics of quinolone resistance in Escherichia coli isolates from humans, animals, and the environment in the Czech Republic, Front. Microbiol 7 (2017) 2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Deshpande LM, Jones RN, Fritsche TR, Sader HS, Occurrence and characterization of carbapenemase-producing Enterobacteriaceae: report from the SENTRY Antimicrobial Surveillance Program (2000--2004), Microb. Drug Resist 12 (2006) 223–230. [DOI] [PubMed] [Google Scholar]
- [42].Marsh JW, Krauland MG, Nelson JS, Schlackman JL, Brooks AM, Pasculle AW, Shutt KA, Doi Y, Querry AM, Muto CA, Genomic Epidemiology of an Endoscope-Associated Outbreak of Klebsiella pneumoniae Carbapenemase (KPC)-Producing K. pneumoniae, PLoS One. 10 (2015) e0144310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Rahamathulla MP, Harish BN, Mataseje L, Mulvey MR, Carbapenem resistance mechanisms among blood isolates of Klebsiella pneumoniae and Escherichia coli, African J. Microbiol. Res 10 (2016) 45–53. [Google Scholar]
- [44].Leavitt A, Chmelnitsky I, Carmeli Y, Navon-Venezia S, Complete nucleotide sequence of KPC-3-encoding plasmid pKpQIL in the epidemic Klebsiella pneumoniae sequence type 258, Antimicrob. Agents Chemother 54 (2010) 4493–4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Vatcheva-Dobrevska R, Mulet X, Ivanov I, Zamorano L, Dobreva E, Velinov T, Kantardjiev T, Oliver A, Molecular epidemiology and multidrug resistance mechanisms of Pseudomonas aeruginosa isolates from Bulgarian hospitals, Microb. Drug Resist 19 (2013) 355–361. [DOI] [PubMed] [Google Scholar]
- [46].Chatterjee S, Datta S, Roy S, Ramanan L, Saha A, Viswanathan R, Som T, Basu S, Carbapenem resistance in Acinetobacter baumannii and other Acinetobacter spp. causing neonatal sepsis: focus on NDM-1 and its linkage to ISAba125, Front. Microbiol 7 (2016) 1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Arnaud I, Maugat S, Jarlier V, Astagneau P, others, Ongoing increasing temporal and geographical trends of the incidence of extended-spectrum beta-lactamase-producing Enterobacteriaceae infections in France, 2009 to 2013, Eurosurveillance. 20 (2015) 30014. [DOI] [PubMed] [Google Scholar]
- [48].Shayan S, Bokaeian M, Detection of ESBL-and AmpC-producing E. coli isolates from urinary tract infections, Adv. Biomed. Res 4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Nahid F, Khan AA, Rehman S, Zahra R, Prevalence of metallo-$β$-lactamase NDM-1-producing multi-drug resistant bacteria at two Pakistani hospitals and implications for public health, J. Infect. Public Health 6 (2013) 487–493. [DOI] [PubMed] [Google Scholar]
- [50].Hugonnet J-E, Tremblay LW, Boshoff HI, Barry CE, Blanchard JS, Meropenem-clavulanate is effective against extensively drug-resistant Mycobacterium tuberculosis, Science (80-.) 323 (2009) 1215–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Livermore DM, Defining an extended-spectrum $β$-lactamase, Clin. Microbiol. Infect 14 (2008) 3–10. [DOI] [PubMed] [Google Scholar]
- [52].Nordmann P, Poirel L, Walsh TR, Livermore DM, The emerging NDM carbapenemases, Trends Microbiol. 19 (2011) 588–595. [DOI] [PubMed] [Google Scholar]
- [53].Walsh TR, Weeks J, Livermore DM, Toleman MA, Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study, Lancet Infect. Dis 11 (2011) 355–362. [DOI] [PubMed] [Google Scholar]
- [54].Bush K, Bradford PA, Epidemiology of $β$-Lactamase-producing pathogens, Clin. Microbiol. Rev 33 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Tehrani KHME, Martin NI, $β$-lactam/$β$-lactamase inhibitor combinations: an update, Medchemcomm. 9 (2018) 1439–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Li S, Duan X, Peng Y, Rui Y, Molecular characteristics of carbapenem-resistant Acinetobacter spp. from clinical infection samples and fecal survey samples in Southern China, BMC Infect. Dis 19 (2019) 900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].W.H. Organization, others, Global action plan on antimicrobial resistance. Geneva: World Health Organization; 2015, Google Sch. (2019). [DOI] [PubMed] [Google Scholar]
- [58].Majumdar S, Pratt RF, Intramolecular cooperativity in the reaction of diacyl phosphates with serine $β$-lactamases, Biochemistry. 48 (2009) 8293–8298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Du H, Chen L, Chavda KD, Pandey R, Zhang H, Xie X, Tang Y-W, Kreiswirth BN, Genomic characterization of Enterobacter cloacae isolates from China that coproduce KPC-3 and NDM-1 carbapenemases, Antimicrob. Agents Chemother 60 (2016) 2519–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Liu C, Qin S, Xu H, Xu L, Zhao D, Liu X, Lang S, Feng X, Liu H-M, New Delhi metallo-$β$-lactamase 1 (NDM-1), the dominant carbapenemase detected in carbapenem-resistant Enterobacter cloacae from Henan Province, China, PLoS One. 10 (2015) e0135044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Pettinati I, Brem J, Lee SY, McHugh PJ, Schofield CJ, The chemical biology of human metallo-$β$-lactamase fold proteins, Trends Biochem. Sci 41 (2016) 338–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Tsakris A, Poulou A, Kristo I, Pittaras T, Spanakis N, Pournaras S, Markou F, Large dissemination of VIM-2-metallo-$β$-lactamase-producing Pseudomonas aeruginosa strains causing health care-associated community-onset infections, J. Clin. Microbiol 47 (2009) 3524–3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Mishra M, Panda S, Barik S, Sarkar A, Singh DV, Mohapatra H, Antibiotic resistance profile, outer membrane proteins, virulence factors and genome sequence analysis reveal clinical isolates of Enterobacter are potential pathogens compared to environmental isolates, Front. Cell. Infect. Microbiol 10 (2020) 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Luan Y, Li G-L, Duo L-B, Wang W-P, Wang C-Y, Zhang H-G, He F, He X, Chen S-J, Luo D-T, DHA-1 plasmid-mediated AmpC $β$-lactamase expression and regulation of Klebsiella pnuemoniae isolates, Mol. Med. Rep 11 (2015) 3069–3077. [DOI] [PubMed] [Google Scholar]
- [65].Guérin F, Isnard C, Cattoir V, Giard JC, Complex regulation pathways of AmpC-mediated $β$-lactam resistance in Enterobacter cloacae complex, Antimicrob. Agents Chemother 59 (2015) 7753–7761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Bethel CR, Distler AM, Ruszczycky MW, Carey MP, Carey PR, Hujer AM, Taracila M, Helfand MS, Thomson JM, Kalp M, others, Inhibition of OXA-1 $β$-lactamase by penems, Antimicrob. Agents Chemother 52 (2008) 3135–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Morandi F, Caselli E, Morandi S, Focia PJ, Blázquez J, Shoichet BK, Prati F, Nanomolar inhibitors of AmpC $β$-lactamase, J. Am. Chem. Soc 125 (2003) 685–695. [DOI] [PubMed] [Google Scholar]
- [68].Chakraborty R, Storey E, van der Helm D, Molecular mechanism of ferricsiderophore passage through the outer membrane receptor proteins of Escherichia coli, Biometals. 20 (2007) 263–274. [DOI] [PubMed] [Google Scholar]
- [69].King DT, Sobhanifar S, Strynadka NCJ, One ring to rule them all: Current trends in combating bacterial resistance to the $β$-lactams, Protein Sci. 25 (2016) 787–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Graham DW, Knapp CW, Christensen BT, McCluskey S, Dolfing J, Appearance of $β$-lactam Resistance Genes in Agricultural Soils and Clinical Isolates over the 20 th Century, Sci. Rep 6 (2016) 21550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Brandt C, Braun SD, Stein C, Slickers P, Ehricht R, Pletz MW, Makarewicz O, In silico serine $β$-lactamases analysis reveals a huge potential resistome in environmental and pathogenic species, Sci. Rep 7 (2017) 43232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Zaheer Z, Rahman SU, Zaheer I, Younas T, Abbas G, others, Antimicrobial Adjuvants-An Innovative Strategy for Handling Antimicrobial Resistance Displayed by Microbes, J Bacteriol Mycol Open Access. 5 (2017) 144. [Google Scholar]
- [73].Bonnet R, Sampaio JLM, Labia R, De Champs C, Sirot D, Chanal C, Sirot J, A Novel CTX-M $β$-Lactamase (CTX-M-8) in Cefotaxime-ResistantEnterobacteriaceae Isolated in Brazil, Antimicrob. Agents Chemother 44 (2000) 1936–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Karim A, Poirel L, Nagarajan S, Nordmann P, Plasmid-mediated extended-spectrum $β$-lactamase (CTX-M-3 like) from India and gene association with insertion sequence IS Ecp1, FEMS Microbiol. Lett 201 (2001) 237–241. [DOI] [PubMed] [Google Scholar]
- [75].Bou G, Cartelle M, Tomas M, Canle D, Molina F, Moure R, Eiros JM, Guerrero A, Identification and broad dissemination of the CTX-M-14 $β$-lactamase in different Escherichia coli strains in the northwest area of Spain, J. Clin. Microbiol 40 (2002) 4030–4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Chanawong A, M’Zali FH, Heritage J, Xiong J-H, Hawkey PM, Three cefotaximases, CTX-M-9, CTX-M-13, and CTX-M-14, among Enterobacteriaceae in the People’s Republic of China, Antimicrob. Agents Chemother 46 (2002) 630–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Hernández JR, Mart L \’\inez-Mart\’\inez, Cantón R, Coque TM, Pascual A, others, Nationwide study of Escherichia coli and Klebsiella pneumoniae producing extended-spectrum $β$-lactamases in Spain, Antimicrob. Agents Chemother 49 (2005) 2122–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Cartelle M, del Mar Tomas M, Molina F, Moure R, Villanueva R, Bou G, High-level resistance to ceftazidime conferred by a novel enzyme, CTX-M-32, derived from CTX-M-1 through a single Asp240-Gly substitution, Antimicrob. Agents Chemother 48 (2004) 2308–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Kim MH, Lee HJ, Park KS, Suh JT, Molecular characteristics of extended spectrum $β$-lactamases in Escherichia coli and Klebsiella pneumoniae and the prevalence of qnr in extended spectrum $β$-lactamase isolates in a tertiary care hospital in Korea, Yonsei Med. J 51 (2010) 768–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Lewis J, The molecular epidemiology of ampC-mediated resistance in Escherichia coli: A study of clinical strains isolated from the South West region, 2020.
- [81].Qi Y, Wei Z, Ji S, Du X, Shen P, Yu Y, ST11, the dominant clone of KPC-producing Klebsiella pneumoniae in China, J. Antimicrob. Chemother 66 (2011) 307–312. [DOI] [PubMed] [Google Scholar]
- [82].Yousefipour M, Rasoulinejad M, Hadadi A, Esmailpour N, Abdollahi A, Jafari S, Khorsand A, Bacteria Producing Extended Spectrum $β$-lactamases (ESBLs) in Hospitalized Patients: Prevalence, Antimicrobial Resistance Pattern and its Main Determinants, Iran. J. Pathol 14 (2019) 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Woodford N, Zhang J, Warner M, Kaufmann ME, Matos J, MacDonald A, Brudney D, Sompolinsky D, Navon-Venezia S, Livermore DM, Arrival of Klebsiella pneumoniae producing KPC carbapenemase in the United Kingdom, J. Antimicrob. Chemother 62 (2008) 1261–1264. [DOI] [PubMed] [Google Scholar]
- [84].陳俊翰, others, A Common Flanking Region in Promiscuous Plasmids Encoding blaNDM-1 in Klebsiella pneumoniae Isolated in Singapore, Microb Drug Resist. (2015). [DOI] [PubMed] [Google Scholar]
- [85].Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, Chaudhary U, Doumith M, Giske CG, Irfan S, others, Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study, Lancet Infect. Dis 10 (2010) 597–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Giske CG, Fröding I, Hasan CM, Turlej-Rogacka A, Toleman M, Livermore D, Woodford N, Walsh TR, Diverse sequence types of Klebsiella pneumoniae contribute to the dissemination of blaNDM-1 in India, Sweden, and the United Kingdom, Antimicrob. Agents Chemother 56 (2012) 2735–2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Shakil S, Azhar EI, Tabrez S, Kamal MA, Jabir NR, Abuzenadah AM, Damanhouri GA, Alam Q, New Delhi Metallo-β-Lactamase (NDM-1): An Updates, J. Chemother 23 (2011) 263–265. [DOI] [PubMed] [Google Scholar]
- [88].Report C, Clinical Isolates of Salmonella enterica Serovar Agona Producing NDM-1 Metallo-  -Lactamase : First Report from Pakistan, 53 (2015) 346–348. 10.1128/JCM.02396-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Irfan S, Khan E, Jabeen K, Bhawan P, Hopkins KL, Day M, Nasir A, Meunier D, Woodford N, Clinical isolates of Salmonella enterica serovar Agona producing NDM-1 metallo-$β$-lactamase: first report from Pakistan, J. Clin. Microbiol 53 (2015) 346–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Chen C-J, Wu T-L, Lu P-L, Chen Y-T, Fung C-P, Chuang Y-C, Lin J-C, Siu LK, Closely related NDM-1-encoding plasmids from Escherichia coli and Klebsiella pneumoniae in Taiwan, PLoS One. 9 (2014) e104899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Bakour S, Touati A, Bachiri T, Sahli F, Tiouit D, Naim M, Azouaou M, Rolain J-M, First report of 16S rRNA methylase ArmA-producing Acinetobacter baumannii and rapid spread of metallo-$β$-lactamase NDM-1 in Algerian hospitals, J. Infect. Chemother 20 (2014) 696–701. [DOI] [PubMed] [Google Scholar]
- [92].Du J, Li B, Cao J, Wu Q, Chen H, Hou Y, Zhang E, Zhou T, Molecular characterization and epidemiologic study of NDM-1-producing extensively drug-resistant Escherichia coli, Microb. Drug Resist 23 (2017) 272–279. [DOI] [PubMed] [Google Scholar]
- [93].Briñas L, Zarazaga M, Sáenz Y, Ruiz-Larrea F, Torres C, $β$-Lactamases in ampicillin-resistant Escherichia coli isolates from foods, humans, and healthy animals, Antimicrob. Agents Chemother 46 (2002) 3156–3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Baroud áM, Dandache I, Araj GF, Wakim R, Kanj S, Kanafani Z, Khairallah M, Sabra A, Shehab M, Dbaibo G, Underlying mechanisms of carbapenem resistance in extended-spectrum β-lactamase-producing Klebsiella pneumoniae and Escherichia coli isolates at a tertiary care centre in Lebanon: role of OXA-48 and NDM-1 carbapenemases, Int. J. Antimicrob. Agents 41 (2013) 75–79. [DOI] [PubMed] [Google Scholar]
- [95].Pesesky MW, Hussain T, Wallace M, Wang B, Andleeb S, Burnham C-AD, Dantas G, KPC and NDM-1 genes in related Enterobacteriaceae strains and plasmids from Pakistan and the United States, Emerg. Infect. Dis 21 (2015) 1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Bakour S, Olaitan AO, Ammari H, Touati A, Saoudi S, Saoudi K, Rolain J-M, Emergence of colistin-and carbapenem-resistant Acinetobacter baumannii ST2 clinical isolate in Algeria: first case report, Microb. Drug Resist 21 (2015) 279–285. [DOI] [PubMed] [Google Scholar]
- [97].Fisher JF, Mobashery S, The sentinel role of peptidoglycan recycling in the $β$-lactam resistance of the Gram-negative Enterobacteriaceae and Pseudomonas aeruginosa, Bioorg. Chem 56 (2014) 41–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Lewis J, The molecular epidemiology of ampC-mediated resistance in Escherichia coli: A study of clinical strains isolated from the South West region, (2016).


