ABSTRACT
Osteomyelitis can result from the direct inoculation of pathogens into bone during injury or surgery or from spread via the bloodstream, a condition called hematogenous osteomyelitis (HOM). HOM disproportionally affects children, and more than half of cases are caused by Staphylococcus aureus. Laboratory models of osteomyelitis mostly utilize direct injection of bacteria into the bone or implantation of foreign material and therefore do not directly interrogate the pathogenesis of pediatric hematogenous osteomyelitis. In this study, we inoculated mice intravenously and characterized the resultant musculoskeletal infections using two strains isolated from adults (USA300-LAC and NRS384) and five new methicillin-resistant S. aureus isolates from pediatric osteomyelitis patients. All strains were capable of creating stable infections over 5 weeks, although the incidence varied. Micro-computed tomography (microCT) analysis demonstrated decreases in the trabecular bone volume fraction but little effect on bone cortices. Histological assessment revealed differences in the precise focus of musculoskeletal infection, with various mixtures of bone-centered osteomyelitis and joint-centered septic arthritis. Whole-genome sequencing of three new isolates demonstrated distinct strains, two within the USA300 lineage and one USA100 isolate. Interestingly, this USA100 isolate showed a distinct predilection for septic arthritis compared to the other isolates tested, including NRS384 and LAC, which more frequently led to osteomyelitis or mixed bone and joint infections. Collectively, these data outline the feasibility of using pediatric osteomyelitis clinical isolates to study the pathogenesis of HOM in murine models and lay the groundwork for future studies investigating strain-dependent differences in musculoskeletal infection.
KEYWORDS: Staphylococcus aureus, clinical isolates, hematogenous osteomyelitis, pediatric infectious disease
INTRODUCTION
Infectious osteomyelitis (OM) is commonly bacterial in origin, with Staphylococcus aureus being the pathogen most frequently identified (1). Hematogenous osteomyelitis (HOM), initiated via the seeding of bacteria through the bloodstream, predominantly affects children, with 85% of cases occurring in children under 17 years of age (1–4). HOM in children is not rare, with a reported incidence between 1 in 1,000 and 1 in 20,000 (5–7), but it is notoriously difficult to treat, often requiring prolonged antibiotics and surgical debridement. The recurrence rate may be as high as 9% (7–10), and this problematic event can extend into adulthood (11). Typically, HOM involves the metaphysis of long bones, predominantly the distal femur or proximal tibia (12). A main concern with pediatric HOM is the development of chronic infection. This can cause pathological bone changes leading to fractures, dislocations, and devitalized bone segments known as sequestra (12). HOM infections can also spread to an adjacent joint, causing septic arthritis (SA), or systemically, leading to potentially lethal sepsis (12, 13). Additionally, antimicrobial-resistant strains, particularly methicillin-resistant S. aureus (MRSA), are highly prevalent in both health care and community settings (14).
Despite significant clinical challenges, studies of the specific mechanisms underlying HOM development and progression have been relatively limited. This is partly because most animal models recapitulate cases of bone infection related to orthopedic surgery and involve manipulations of the bone, including the creation of cortical defects or the placement of hardware into the bone (15–20). One study utilized the hematogenous route but relied upon the implantation of metal into the femur before the injection of bacteria (19). Although HOM has been modeled in mice without prior manipulation (21–24), characterization of bone changes and comparisons among strains have been limited.
In addition to two previously established community-acquired USA300 strains isolated from adults (16, 25, 26), we utilized several new clinical isolates of S. aureus from pediatric HOM patients. To mimic the bone microenvironment in children, the population in which HOM is most prevalent, we intravenously inoculated young (6-week-old), actively growing mice. The generation of stably bioluminescent bacteria allowed real-time, longitudinal assessment of infection localization and progression in vivo while preserving the bone for downstream analysis by micro-computed tomography (microCT) and histology. We compared measures of pathogenicity between these isolates, including the incidence of hindlimb infection, propensity toward developing bone-centered HOM or joint-centered SA, subsequent bone loss, cytotoxicity toward host cells, and intracellular proliferation. We also performed genomic sequence analysis of 3 of these isolates, discovering significant genetic differences among the clinical isolates. Taken together, these studies provide a robust platform for the analysis of host-pathogen interactions in HOM.
RESULTS
Modeling hematogenous osteomyelitis using NRS384 and LAC.
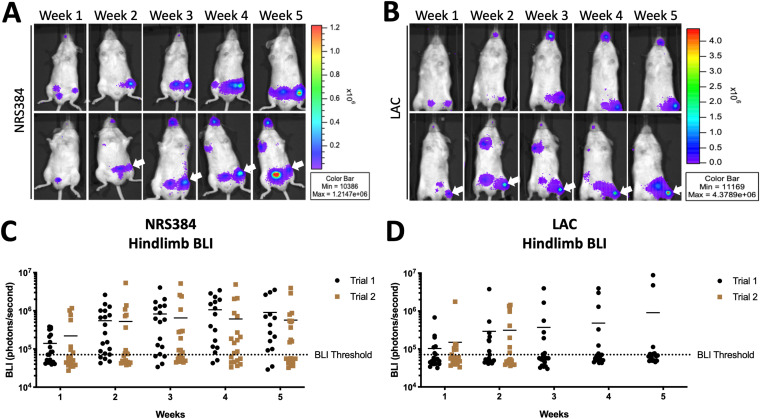
We first established HOM with two USA300 strains of S. aureus previously made spontaneously bioluminescent by us and another group, via the stable integration of a plasmid bearing the lux operon (27, 28), allowing the tracking of infection over time in vivo. We injected 2.5 × 106 CFU of NRS384 or LAC in phosphate-buffered saline (PBS) into the tail vein of wild-type C57BL/6-albino mice at 6 weeks of age, a period of rapid growth. With this inoculum, we did not observe any instances of lethal sepsis or other complications requiring euthanasia prior to 4 weeks after infection with either strain (see Fig. S1 in the supplemental material). Bioluminescence imaging (BLI) was performed weekly for 5 weeks. With both USA300 strains, distinct foci of signals were frequently seen in hindlimbs as well as bladder, likely indicative of bacteria being shed from the kidneys (Fig. 1A and B). Foci of signals were also observed in the rostrum and occasionally the forelimb. Based on this distribution of bone-centric signals, regions of interest were drawn over the hindlimbs, also referred to as legs (as in Fig. S2), allowing quantitation over time (Fig. 1C and D). We found no appreciable differences between signal intensities from either strain or the two cohorts of mice injected on different days (trial 1 and trial 2), and the signal values stabilized at around the 2-week time point, suggesting the establishment of chronic infection. Because of this BLI signal stabilization, the trial 2 LAC-injected mice were harvested at 2 weeks. Next, we used all the BLI values acquired to devise a BLI positivity threshold to stratify actively infected BLI-positive (BLI+) legs from BLI-negative (BLI−) legs for later analyses. This threshold was set at 70,000 photons/s because legs under this value consistently had no foci of BLI signals over the 5-week course of infection, excluding “bleed” from strong bladder signals (as in Fig. S2B, right leg), while the baseline signal for autoluminescence was around 35,000 photons/s. Furthermore, no legs that reached this threshold subsequently fell below it. Interestingly, we observed that the cohorts injected with NRS384 had a markedly higher percentage of mice that developed a hindlimb infection, as detected by BLI, than in LAC-injected cohorts (80% versus 35% cumulative incidence).
FIG 1.
Longitudinal tracking of NRS384 and LAC hematogenous osteomyelitis. (A and B) In vivo bioluminescence imaging (BLI) of two representative mice (top and bottom rows), performed weekly for 5 weeks, after injection with NRS384 (A) or LAC (B) strains of S. aureus. Arrows highlight BLI foci. (C and D) Quantification of NRS384 (C) and LAC (D) BLI signals measured in each hindlimb (2 data points per mouse) for two separate trials over the 5 weeks postinjection (n = 10 mice per trial) (the BLI threshold is set at 70,000 photons/second). LAC trial 2 purposely terminated at 2 weeks postinjection.
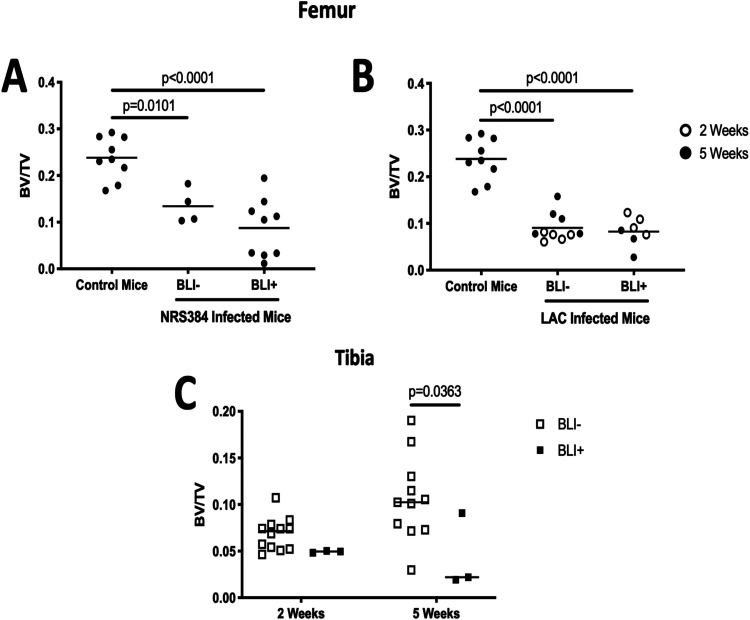
Since most BLI signals for NRS384-injected mice appeared to be proximal to the knee joint (Fig. 1A, arrows), femurs were assessed by microCT. Reconstructions indicated that the bone disruption is predominantly localized to the trabecular compartment (Fig. S3A). Quantification of the trabecular bone volume fraction (ratio of the bone volume to the total volume [BV/TV]) in the femur demonstrated significant decreases from both BLI− and BLI+ legs compared to those from noninjected control mice (Fig. 2A). While no statistical difference was found between BLI− and BLI+ legs of these NRS384-injected mice, extreme destruction of trabecular bone (BV/TV ratio below 0.1) was seen only in BLI+ legs. Interestingly, the BLI signal from LAC-injected mice appeared more distal than in NRS384-injected mice (Fig. 1B, arrows), so we measured the trabecular bone volumes from both femurs and tibias. The femoral BV/TV ratio in LAC-injected mice was lower than that in control mice, but there was no difference in BV/TV ratios between BLI− and BLI+ legs, nor was there a difference between femurs harvested at 2 and 5 weeks (Fig. 2B; Fig. S3B). However, the trabecular bone volumes in tibias of BLI+ legs were decreased compared to those in tibias from legs that were BLI− in LAC-injected mice, reaching statistical significance only at 5 weeks postinjection (Fig. 2C; Fig. S3B). Overall, we were able to induce consistent hindlimb HOM with both the NRS384 and LAC strains using our intravenous injection model.
FIG 2.
Trabecular bone volume fractions from NRS384- and LAC-injected mice. (A and B) Trabecular bone volume/total volume (BV/TV) ratio measured by micro-computed tomography (microCT) in femurs from noninjected control mice and NRS384 (A)- and LAC (B)-injected mice. The control group (age, sex, and strain matched) is the same in panels A and B. (C) Tibial trabecular BV/TV ratio from LAC-injected mice measured at 2 or 5 weeks postinjection. The BLI− or BLI+ designation is based on the threshold in the legend of Fig. 1. P values were calculated by one-way ANOVA with Tukey’s multiple-comparison post hoc test.
Characterizing HOM caused by S. aureus clinical isolates from children with OM.
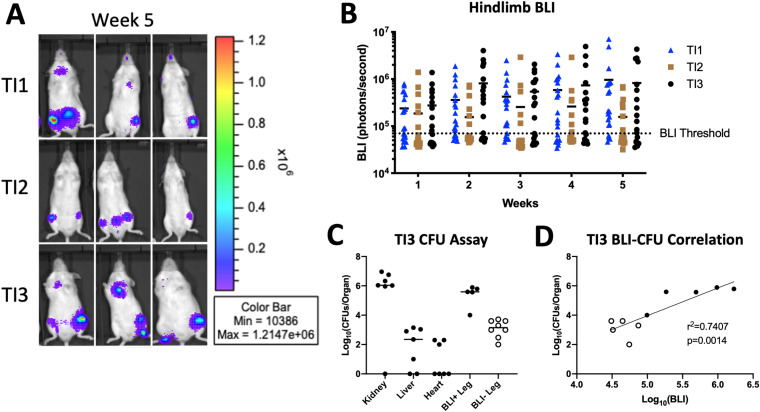
To expand the clinical significance of this model to pediatric HOM, we acquired five isolates of methicillin-resistant S. aureus (MRSA) from pediatric patients with HOM (Table 1). In order to employ these isolates for BLI during HOM in mice, we utilized the same stable chromosomal integration system for the lux operon used previously for NRS384 (27). To confirm that the bioluminescence of each isolate corresponds to an equivalent bacterial burden, we measured the BLI signal from in vitro serial dilutions of each isolate and correlated that with CFU enumerated from the same dilutions (Fig. S4). In liquid culture, the growth of TI2 and TI5 was minimally delayed (Fig. S5A). Growth curves of the lux isolates were slightly accelerated compared to their respective parental isolates, suggesting that the lux operon did not inhibit the growth kinetics of any of the isolates (Fig. S5B). Tail vein injection of 2.5 × 106 CFU of all 5 isolates led to reproducible infections with localization over hindlimbs (Fig. 3A; Fig. S6A and B) and limited lethality (Fig. S6C), comparable to those of NRS384 and LAC (Fig. S1). Interestingly, we discovered differences between the isolates in the rates at which they led to hindlimb infections in these mice, using our previously established BLI positivity threshold (Fig. 3B). Two of the isolates, TI1 and TI3, had a very high incidence of infection in at least one hindlimb, 72% and 89%, respectively, comparable to that of NRS384. However, TI2 had a much lower rate of infection, 25%, over the 5-week time course, similar to the incidence with LAC. The other two strains, TI4 and TI5, had infection rates between these extremes, 46% and 67%, respectively. For subsequent analyses, we focused primarily on TI3 and TI1, with the highest incidences of hindlimb infection, as well as TI2, with the lowest.
TABLE 1.
Source information for S. aureus clinical isolatesa
| Isolate | Osteomyelitis | Septic arthritis | Bacteremia | MRSA history | No. of surgical drainage procedures | No. of positive blood cultures | Sex | Age (yrs) |
|---|---|---|---|---|---|---|---|---|
| TI1 | Yes | No | Yes | Yes | 1 | 1 | Male | 6 |
| TI2 | Yes | No | No | No | 4 | Female | 8 | |
| TI3 | Yes | Yes | Yes | No | 2 | 3 | Male | 14 |
| TI4 | Yes | No | Yes | No | 2 | 1 | Female | 8 |
| TI5 | Yes | No | No | No | 1 | Male | 6 |
Each of the five clinical isolates used throughout these studies was obtained from white, non-Hispanic juvenile patients at Saint Louis Children’s Hospital, St. Louis, MO.
FIG 3.
Characterization of hematogenous osteomyelitis using clinical isolates of S. aureus. (A) In vivo bioluminescence imaging (BLI) of three representative mice for each isolate at 5 weeks postinjection. (B) Quantification of the BLI signal measured in the hindlimb of each injected mouse depicted over the 5 weeks postinjection (n = 20 per isolate) (the BLI threshold is 70,000 photons/s). (C) Enumerated bacterial burdens from the indicated organs, as measured by a CFU assay at 3 weeks postinjection. BLI+ and BLI− legs are the counts from femurs from legs with BLI signals higher and lower than 70,000 photons/s, respectively (n = 7 mice from a separate cohort from those in panels A and B injected with TI3) (each dot represents one leg). (D) Correlation of the bacterial burden (CFU) from panel C with the BLI signal of the corresponding leg. Open and closed dots correspond to the BLI+/− designations in panel C.
To further validate BLI as a proxy for bacterial load in vivo, we measured CFU obtained from homogenized femurs, along with the heart, liver, and kidneys, from mice injected with TI3 2 weeks previously (Fig. 3C). High levels (∼5 × 106 CFU/organ) of bacteria were found in the kidneys and hindlimb bones from BLI+ legs, as expected, and the correlation between CFU and the corresponding BLI signal in hindlimbs was strong (Fig. 3D).
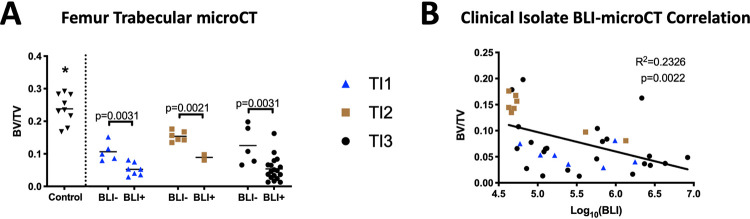
We next compared the trabecular BV/TV ratios from mice injected with these three clinical isolates, focusing on femurs because BLI signals were primarily proximal to the knee. As with the NRS384-injected mice, we found a significant decrease in all the bone volumes of femurs from mice injected with each clinical isolate compared to femurs of noninjected mice. However, with these mice, we also found a consistent decrease in the bone volume fractions of femurs from BLI+ legs compared to BLI− legs (Fig. 4A). Finally, we pooled the data from the BLI+ and BLI− legs of all three clinical isolates and found a weak but statistically significant correlation between the BLI signal and the BV/TV ratio from the corresponding femur (Fig. 4B). These data further validated our utilization of BLI to track infection incidence and location.
FIG 4.
Reduced bone volumes are associated with the local bacterial burden with HOM induced by clinical isolates. (A) Trabecular BV/TV ratio measured by microCT of femurs from noninjected control mice (same cohort as in Fig. 2) and mice injected with either TI1, TI2, or TI3. Classification as BLI− or BLI+ legs was determined by the threshold in the Fig. 3 legend. *, P ≤ 0.0014 for the control versus all other groups as calculated by one-way ANOVA with Tukey’s multiple-comparison post hoc test. P values between BLI− and BLI+ BV/TV ratios within each strain were calculated by Student’s t test. (B) Correlation of the BV/TV data from panel A with the BLI signal from the corresponding leg. Dot coding corresponds to the coding in panel A.
Osteomyelitis and septic arthritis result from hematogenous inoculation with S. aureus strains.
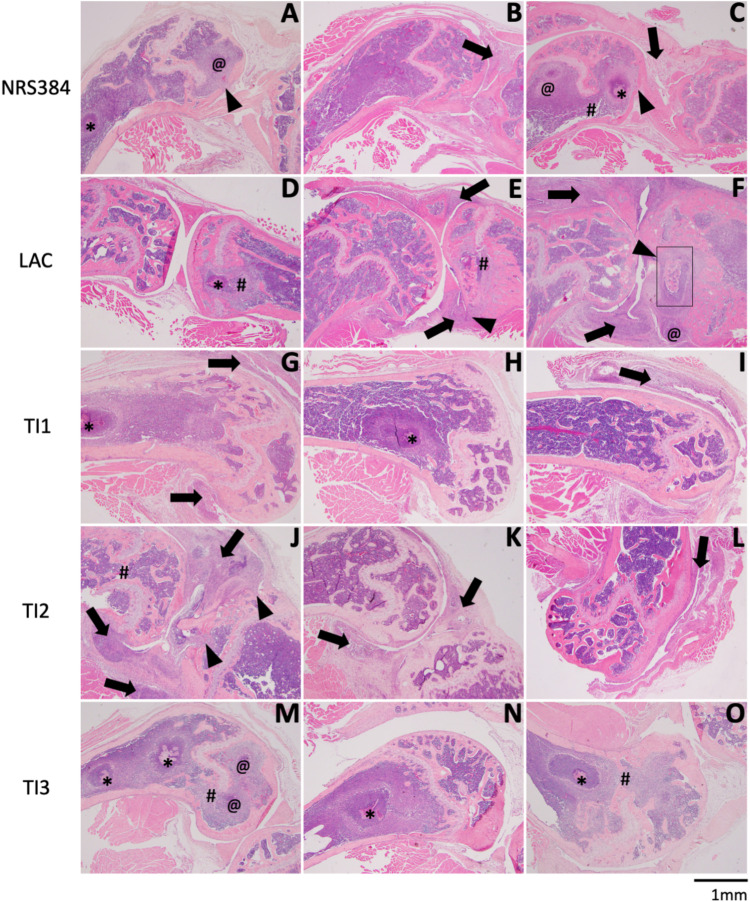
We next examined hindlimbs by standard histology 5 weeks after infection, finding various combinations of bone and joint involvement. Figure 5 depicts representative histological images of three separate samples of the distal femur and knee joint (one per column) from infections with NRS384 and LAC and three pediatric isolates (TI1, TI2, and TI3). Some samples (Fig. 5D, H, M, and N) appear to have infections localized entirely within the bone and medullary cavity, indicative of OM. Other samples (Fig. 5B, I, K, and L) show clear infection and inflammation within the joint space (black arrows), while the femur and tibia remain normal, demonstrating septic arthritis (SA). Finally, there are samples (Fig. 5A, C, E to G, and O) where the infection is present both in the bone and joint (OM+SA). However, in the latter samples, some infections seem to extend into the joint from severe bone involvement (Fig. 5A, C, and F, black arrowheads), while others seem to represent a joint-centered infection focally extending into the bone (Fig. 5J, black arrowheads). In all cases of OM infections, there is significant loss of trabecular bone, and in extreme cases, there is disruption of the growth plate as well (Fig. 5C to E, J, M, and O, #). We did not observe extension through the cortical bone to the periosteal surface in affected femurs. The various patterns of bone and joint involvement for each strain were then enumerated (Table 2). NRS384 and TI1 produced mixtures of bone-centered OM and joint-centered SA infections. However, LAC and TI3 predominantly produced OM, with no case of SA that was not coincidental with OM. TI2 predominantly produced SA, with only one case of OM that was not associated with SA. Interestingly, LAC produced OM lesions in the tibias but not the femurs of 6 out of 7 mice, as opposed to all other strains, which consistently produced OM in the femurs. In order to further confirm the utility of BLI as a measure of OM, 15 BLI− hindlimbs from across all strains were examined histologically, and all appeared normal, except for only one that had evidence of minor inflammation in the joint space. We also examined 3 hindlimbs that were BLI+ at low levels for only 2 to 3 weeks out of the 5 weeks of imaging. These also all exhibited only low levels of inflammation in the bone or joint but no bacterial colonies or well-formed abscesses.
FIG 5.
Histological changes to hindlimbs with HOM. Each row depicts representative histology of the distal femur and knee joint, with or without the proximal tibia, from three separate samples of mice injected with NRS384 (A to C), LAC (D to F), TI1 (G to I), TI2 (J to L), or TI3 (M to O). In panels with both bones, the femur is on the left, and the tibia is on the right. Symbols denote specific histological characteristics: *, abscess with visible bacterial colonies; @, abscess without visible bacterial colonies; #, growth plate disruption; black arrows, inflammation within the joint space and/or synovium; black arrowheads, infection focally extending from the joint into the bone. The box in panel F depicts an area of higher magnification in Fig. 6C.
TABLE 2.
Infection localization in BLI+ limbs as assessed histologicallya
| Strain | No. of samples with infection localization in BLI+ limbs/total no. of samples |
|||
|---|---|---|---|---|
| OM | SA | OM+SA | No changes | |
| NRS384 | 5/9 | 1/9 | 1/9 | 2/9 |
| LAC | 3/7 | 0/7 | 4/7 | 0/7 |
| TI1 | 4/12 | 2/12 | 3/12 | 3/12 |
| TI2 | 1/8 | 3/8 | 3/8 | 1/8 |
| TI3 | 12/18 | 0/18 | 5/18 | 1/18 |
Fractions are listed as (number of samples that exhibit the indicated histological characteristic)/(total number of samples examined). Characteristics were determined as follows: osteomyelitis (OM) if abscesses and/or inflammation was present within the bone tissue or medullary cavity, septic arthritis (SA) if abscesses and/or inflammation was present in the joint space, OM+SA if both OM and SA were present, and no changes if no overt histological changes were seen in either the bone or the joint, on at least 2 to 3 sections per sample.
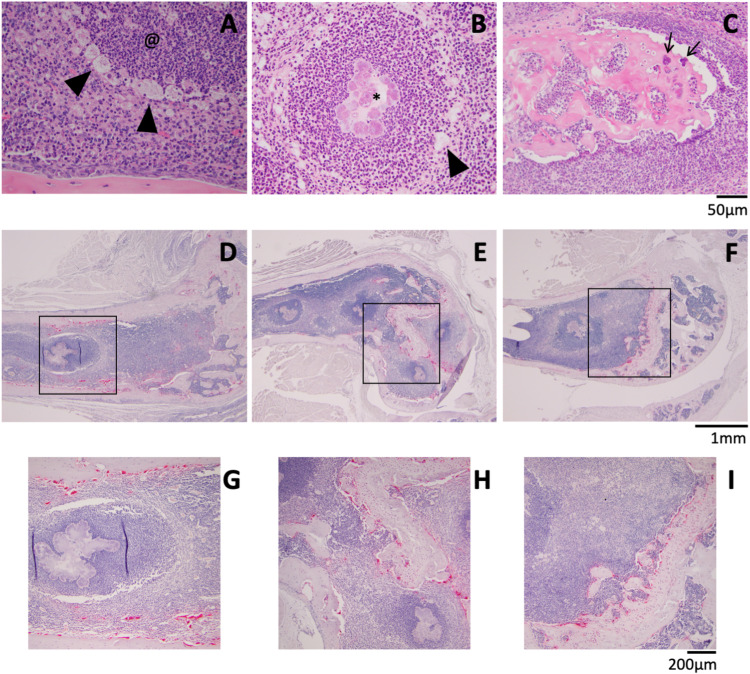
Focusing on OM lesions, higher-magnification images demonstrate characteristic abscesses with neutrophil infiltration and large macrophages surrounding the lesions (Fig. 6A and B, black arrowheads), some with central bacterial colonies (Fig. 6B, *). We observed only one example of a sequestrum, in a tibia from a LAC infection (Fig. 5F, black box). In the higher-magnification image, collections of cocci on the bone surface (Fig. 6C, black arrows) as well as empty lacunae can be seen. Since we saw dramatic bone loss, we looked for osteoclasts (OCs) by staining for tartrate-resistant acid phosphatase (TRAP) (an osteoclast marker), finding that many bone surfaces close to abscesses were covered by these resorptive cells (Fig. 6D to I).
FIG 6.
Detailed histological characteristics of HOM. (A and B) Abscesses without (@) (A) and with (*) (B) visible bacterial colonies were surrounded by enlarged macrophages (black arrowheads). (C) Magnified image of the sequestrum from Fig. 5F showing bacteria (black arrows). (D to I) TRAP staining (red) for osteoclast activity. Panels G to I depict higher magnifications of the black boxes indicated in panels D to F, respectively. Scale bars are presented for each row of images.
While it was not uncommon to find damage to the tibial epiphysis in cases with SA, an involvement of the tibial metaphysis or diaphysis was rare in all strains used except for LAC. One TI3-injected mouse had abscesses with bacterial colonies throughout the diaphyseal marrow space and a focus of periosteal reaction surrounding an abscess that appears to be outside the normal cortical shell (Fig. S7A). In addition to displaying markedly more tibial involvement than all the other strains, LAC-injected mice also had two cases of inflammation with reactive bone formation on the periosteal surface (Fig. S7B to D). In one case, the metaphysis of a tibia from a TI2-injected mouse was fibrotic, with a rim of new bone formation, just distal to the growth plate (Fig. S7E and F). While not prevalent, these instances of new bone formation provide further evidence that our murine HOM model shares many features with the human disorder. Altogether, this histology reveals significant pathological differences of OM infections between the clinical isolates.
Varying effects of clinical isolates on osteoblasts and osteoclasts.
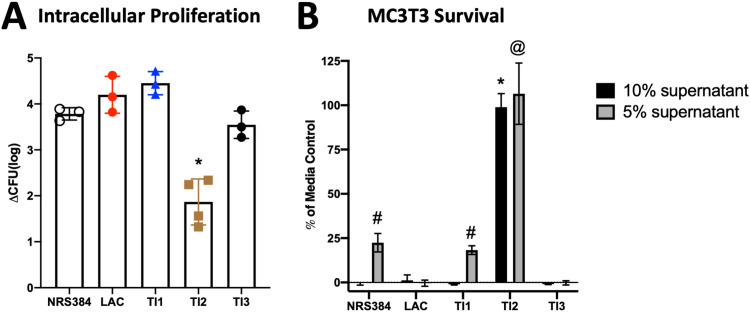
Next, we investigated whether these different strains exhibited virulence differences using our osteoclast intracellular proliferation (29) and osteoblast cytotoxicity (16) assays. First, osteoclasts differentiated for 3 days in culture were separately infected with each strain for 30 min, and all uninternalized bacteria were then killed by the addition of antibiotic to the culture for 1 h. Osteoclasts were lysed immediately thereafter (1.5 h postinfection), indicative of internalization, or cultured until 18 h postinfection, to allow proliferation, and CFU were enumerated. TI2 produced a significantly lower increase in the CFU between 1.5 and 18 h than all other strains (Fig. 7A). Input CFU and CFU measured 1.5 h after infection were similar among all strains (Fig. S8A and B), indicating that the observed difference in TI2 CFU at 18 h was related to decreased intracellular proliferation within osteoclasts and not internalization. Next, cytotoxicity was assessed by adding concentrated bacterial supernatants to cultured MC3T3 osteoblastic cells for 22 h (Fig. 7B). While all other strains showed complete cytotoxicity with the addition of 10% supernatant, the TI2 supernatant had no effect on osteoblast cell survival. LAC and TI3 were the most cytotoxic, with complete killing with 5% supernatant. Thus, the lower incidence of HOM with TI2, but not LAC, was associated with decreased direct effects on bone cells. These data, in addition to our histology, further characterize TI2 as functionally distinct from the other isolates.
FIG 7.
Effects of S. aureus strains on bone cells in vitro. (A) Intracellular proliferation within cultured osteoclasts measured by the change in CFU between 1.5 and 18 h postinfection. *, P ≤ 0.0015 compared to all other strains as measured by one-way ANOVA with Tukey’s post hoc test. (B) MC3T3 survival after 22 h of exposure to medium with either 5% or 10% concentrated S. aureus culture supernatant, relative to a medium-only control. Bars represent the means, and error bars are SD (n = 10). # and @, P < 0.0001 compared to all other 5% supernatants; *, P < 0.0001 compared to all other 10% supernatants (by two-way ANOVA with Tukey’s post hoc test).
Genetic sequence typing of the clinical isolates.
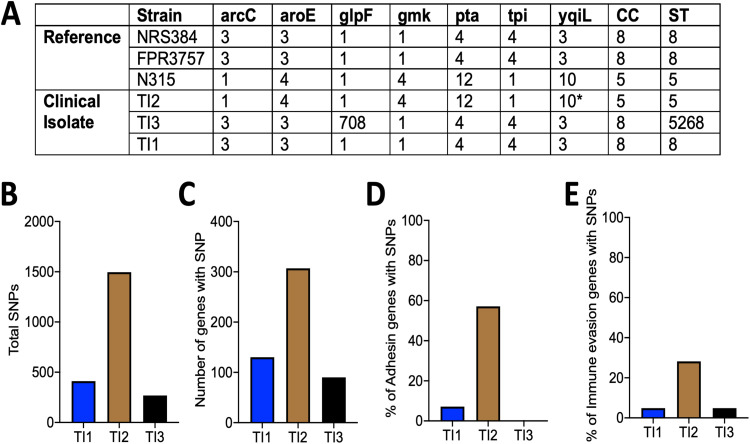
In order to begin investigating the inherent genetic differences underlying the observed differences in pathogenicity, the genomes of three clinical isolates (TI1, TI2, and TI3) were sequenced, and multilocus sequence typing (MLST) was performed as previously described (30). This analysis showed that TI2 belongs to clonal complex 5 (CC5), indicative of a USA100 clonal lineage, while TI1 and TI3 are both of CC ID8, consistent with a USA300 clonal lineage (Fig. 8A). When each isolate was compared with its respective clonal lineage reference strain (FPR3757 for USA300 and N315 for USA100), TI2 had more than 3 times the number of total polymorphisms, as well as genes containing at least one polymorphism, detected compared to TI1 and TI3 (Fig. 8B and C). Next, 173 virulence-associated genes within each isolate were grouped by function and examined for the presence of polymorphisms, compared to their respective reference strains (31). TI2 shows more polymorphisms in genes related to adhesins and immune evasion than Tl1 or TI3 (Fig. 8D and E; Fig. S9, red squares). However, polymorphisms in the other categories were relatively infrequent in all pediatric strains.
FIG 8.
Genetic differences between clinical isolates of S. aureus. (A) Multilocus sequence typing (MLST) of the clinical isolates (TI1, TI2, and TI3) and the reference strains NRS384 (USA300), FPR3757 (USA300), and N315 (USA100) listed with the allele identifiers (* denotes the closest match [no exact allele identification]) for each MLST locus, clonal complex (CC), and strain type (ST). (B to E) Enumerated single nucleotide polymorphisms (SNPs) compared to the reference strain (B), total number of genes with at least one SNP (C), and percentage of adhesin genes (D) or immune evasion genes (E), as designated in Fig. S8 in the supplemental material, with at least one SNP.
DISCUSSION
Using multiple clinical isolates of S. aureus from children with osteomyelitis as well as two community-acquired strains from adult patients (NRS384 and LAC, originally isolated from skin [32, 33]), we achieved reproducible development of HOM in mice. By generating stably bioluminescent bacteria, we were able to track overt infection occurrence and location over time. Important for their utility in the study of HOM, the strains and clinical isolates tested produced frequent hindlimb infections that did not rely upon direct bone inoculation or prior bone manipulation. Furthermore, mice did not become obviously septic immediately following injection and mostly survived for the duration of the experiment, except for occasional hindlimb lameness or paralysis necessitating euthanasia after several weeks. The ease of tracking, early onset of infection, associated bone loss, and formation of characteristic abscesses all stand to make this a very effective model for the further study of HOM pathogenesis. Because the BLI signal in bone correlated well with the bacterial load, both in vivo and in vitro, destructive CFU assays are not required to assess the effects of manipulations of either the host or pathogen on the bacterial burden in this model, and the bacteria can be tracked longitudinally in each mouse. Additionally, our HOM model mimics many of the hallmarks of juvenile HOM. Clinically, the most frequent site of infection is the metaphysis of the femur or tibia (12), a trend recapitulated in our studies. We also found several examples of abscesses located within the diaphysis of the femur and tibia, although this was not found in the majority of samples. We observed rapid trabecular bone destruction, as early as 2 weeks postinjection. However, significant cortical bone changes were rare, with only a few examples of periosteal reactive bone formation along with one sequestrum. These infections persisted over the duration of our 5-week follow-up, but longer periods of study are warranted to fully characterize the chronic effects of these infections, as persistence and recurrence are some of the most challenging aspects of treating HOM patients.
Both BLI and CFU assays support the primary sites of infection after intravenous inoculation of all strains examined to be bone/joint and kidney/bladder. However, CFU assays also demonstrate low, but detectable, levels of live bacteria in femurs without distinct foci of bioluminescence or histological changes such as abscess formation. Interestingly, in mice inoculated with the pediatric isolates, these BLI− femurs had trabecular bone volumes that were higher than those of the BLI+ femurs but lower than those of control noninjected mice. The lack of bone volume differences between BLI+ and BLI− femurs in NRS384- and LAC-injected mice may indicate a limitation of BLI in predicting bone loss, but this could also indicate inherent differences in the abilities of certain strains to cause bone loss. Additionally, the presence of measurable bacteria within BLI− bones could suggest a limitation in the sensitivity of BLI as a readout for infection. However, most of the BLI− hindlimbs appeared completely normal by histology, with only 1 having signs of minor inflammation. Also, all samples from mice where low-level BLI positivity at early time points resolved, falling below our BLI threshold before the end of imaging, showed only low levels of inflammation but not abscesses or bacterial colonies. Thus, assessment of BLI over time correlates well with both the bacterial load and the histological features of OM, although latent bacteria present in bone are not detected by either BLI or standard histology. Ultimately, more detailed assessments, including examination of other skeletal sites and imaging of bacteria in situ at high resolution, are needed to determine whether bone loss at BLI-negative sites is driven by systemic inflammation, direct effects of sparsely distributed bacteria in bone, or a combination of both.
Interestingly, by using multiple clinical isolates of S. aureus, we found overt differences in their pathogenicities ranging from infection incidence to predilection toward specific infection types (osteomyelitis versus septic arthritis). There were also more subtle differences between the isolates, such as the intensity of the BLI signal in the BLI+ legs and the lesser drop in the bone volume of IT2-injected mice than for the other two isolates, although this could be a consequence of the lower percentage of bone-centered infections, as we were able to use microCT to analyze only two BLI+ legs. LAC also produced relatively few HOM infections, but those that occurred tended to involve the tibia rather than the femur. However, only TI2, and not LAC, displayed lower osteoblast cytotoxicity and intraosteoclast proliferation. These findings coincide with previous work establishing that S. aureus clinical isolates differ in their virulence, pathogenicity, and toxicity and that lower cytotoxicity correlates with increased persistence in vivo (34, 35). We found major differences in specific tissue tropisms among isolates. TI2 displayed a predilection toward promoting SA compared to the other strains, especially TI3, while LAC predominantly targeted the tibia over the femur. However, our knowledge of the clinical behavior of these isolates is limited to only one patient per isolate, so it is currently unclear how these findings translate to a broader population. Further work is needed to determine specific bacterial factors that may promote differences in infection incidence and tropism as well as if the lower cytotoxicity of TI2 contributes to chronicity as seen with other isolates (34, 35). It will be interesting to see if the incidence of sequestra or periosteal reactions, characteristic changes of chronic osteomyelitis, increases over time past the 5 weeks that we examined here.
Using genomic sequencing of the new clinical isolates, we have begun to explore how genomic content shapes the observed clinical differences. When the isolates were sequence typed, the TI2 isolate was shown to be of the USA100 clonal lineage, whereas the other isolates were of the USA300 lineage. USA100 has a higher prevalence in hospital-acquired infections, while USA300 predominates in community-acquired infections (36, 37). While none of the patients from whom these isolates were taken had hospital-acquired infections, a family member of the TI2 isolate patient was reported to be a health care worker. Although we do not yet know the traits of TI2 that underlie its pathogenic characteristics, this study highlights the importance of examining strains of other lineages besides USA300 as they are still circulating, are able to cause OM and SA, and could have differences in their pathogenicities. Sequencing these isolates is the first step in understanding these differences and gives us a resource to identify other genetically related strains. TI2 was found to have a number of polymorphisms in specific genes related to adhesins and immune evasion. This could explain the lower infection incidence as both surface adhesion and the ability to evade immune destruction are critical steps in infection initiation and propagation. Also, differences in adhesin expression may help explain the predilection of TI2 to promote SA as opposed to OM. Despite the different clades, TI2 and LAC have similar overall infection incidences in our model. However, LAC displayed osteoblast cytotoxicity and intraosteoclast proliferation similar to those of the other USA300 strains, whereas strain TI2 was significantly less cytotoxic. These results, taken together with infection incidence differences, suggest that the strain of bacteria could be important to note when comparing results from different models of osteomyelitis.
It is apparent in our study that many mice in our model develop kidney infections as well as OM in long bones, so it is difficult to determine with certainty whether OM is primary or secondary. However, some mice develop infections only in the tibia or femur, suggesting this to be the primary infection location, and we have not seen any examples of mice that develop OM after first having a kidney/bladder infection identified by BLI. Another limitation is that our study has focused only on MRSA isolates. Methicillin-susceptible strains of S. aureus are also a significant cause of hospital- and community-acquired infections, which will require investigation. Also, this study utilized only male mice in order to minimize variations between cohorts, as we were primarily interested in elucidating differences between the strains of S. aureus used. In the future, studies to determine host-specific influences, including sex differences, on HOM development and progression will need to be performed in order to assess the generalizability of this model.
Ultimately, our work demonstrates that HOM can be reproducibly initiated in immunocompetent mice using clinical isolates and that utilizing multiple isolates is likely necessary to capture the range of human diseases. This panel of bioluminescent strains will be critical for further studies to elucidate biological mechanisms underpinning pediatric HOM development and propagation, eventually aiding in more efficacious clinical treatments and therapies.
MATERIALS AND METHODS
Animals.
For these studies, we used 6-week-old male C57BL/6 mice, with a mutation in the tyrosinase gene [B-6 albino; B6(Cg)-Tyrc-2J/J; Lane, 1973] (JAX stock number 000058; The Jackson Laboratory, Bar Harbor, ME) (38) that renders their fur albino and allows improved bioluminescence imaging. For hematogenous osteomyelitis modeling, 2.5 × 106 bacteria were injected in 100 μl of PBS via the tail vein. Control, noninjected mice used for microCT analysis were age and sex matched. Mice were housed 5 per cage with food and water ad libitum and closely monitored throughout the experiments for signs of morbidity until they were humanely euthanized by asphyxiation with CO2.
Clinical isolates.
Clinical isolates were obtained from the St. Louis Children’s Hospital and Barnes Jewish Hospital clinical microbiology laboratories by Stephanie A. Fritz and transferred to Deborah J. Veis in accordance with Washington University Institutional Review Board approval (IRB number 201805091). None of the patients required pediatric intensive care unit admission or intubation. Antibiotic susceptibility profiles can be found in Table 3.
TABLE 3.
Antibiotic susceptibility profilesa
| Isolate | Drugs |
|
|---|---|---|
| Resistant | Susceptible | |
| TI1 | FOX, ERY, CRO | CLI, VAN, SXT, LZD, TET, DAP |
| TI2 | PEN, FOX, ERY | CLI, VAN, SXT |
| TI3 | PEN, FOX, ERY | CLI, VAN, SXT |
| TI4 | PEN, FOX, ERY | CLI, VAN, SXT |
| TI5 | PEN, FOX, ERY, CLI | VAN, SXT, TET, RIF |
Abbreviations: FOX, cefoxitin; CRO, ceftriaxone; CLI, clindamycin; DAP, daptomycin; ERY, erythromycin; LZD, linezolid; PEN, penicillin; RIF, rifampin; TET, tetracycline; SXT, trimethoprim-sulfamethoxazole; VAN, vancomycin.
Bacterial growth and preparation of inocula.
All strains of S. aureus were routinely grown on tryptic soy agar (TSA) or in tryptic soy broth (TSB) (Fisher Scientific, Hampton, NH) at 1:200 with shaking at 220 rpm overnight and subcultured at 1:100 for 2 h before injection. Bacteria were subsequently pelleted and resuspended in PBS to a final optical density at 600 nm (OD600) of 1. Injections were prepared by further dilution of bacteria in PBS to a final count of 2.5 × 106 bacteria in 100 μl of PBS.
The NRS384 S. aureus strain used was already stably bioluminescent (27). The LAC strain of S. aureus used was a dual-fluorescence luminescent strain of S. aureus. To generate the dual-fluorescence luminescent strain of S. aureus, the attC::PsarA-sodRBS-mCherry allele from S. aureus LAC (a gift from Victor Torres) was first cloned into PJC1306 (39) using Gibson assembly. PJC1306 containing this allele was electroporated into RN9011 and integrated into the genome with tetracycline selection for successful integration. The attC::PsarA-sodRBS-mCherry allele was transduced with phi-85 into an erythromycin-sensitized USA300 lineage S. aureus strain, AH1263 (28). Next, the construct PgapA-luxBADCE was transduced with phi-85 into LAC attC::PsarA-sodRBS-mCherry.
In order to produce stable bioluminescence in the clinical isolates, electrocompetent cells of each isolate were prepared and electroporated with the pRP1195 plasmid, as previously described (27). Clones containing the plasmid were selected for by chloramphenicol resistance and bioluminescence. Plasmid integration was then driven by a temperature shift to 43°C for 6 to 7 h, as previously described (27), after which bacteria were serially diluted onto TSA plates containing 5 μg/ml and grown at 43°C overnight. Clones with bioluminescence were again selected, and 20% glycerol stocks were made.
Planktonic growth assay.
Bacteria were streaked onto TSA plates and grown at 37°C overnight. Single colonies from clinical isolates were selected and cultured in TSB with shaking at 220 rpm overnight at 37°C. The culture grown overnight was then diluted in TSB at 1:100 in fresh TSB. The OD600 was measured every 30 min for 5 h.
Bioluminescence imaging.
In vivo bioluminescence imaging was performed on an Ivis 50 instrument (Living Image 4.3.1; PerkinElmer, Waltham, MA) using a 1-min exposure, bin 8, Field of View (FOV)12cm, Focal length (f/stop)1, and open filter. Mice were imaged under isoflurane anesthesia (2% vaporized in O2). Bacterial cultures grown in vitro were imaged using the same Ivis 50 instrument, using a 1-s exposure, bin 8, FOV12cm, f/stop1, and open filter. The total photon flux (photons per second) was measured from fixed regions of interest (ROIs) set over the hindlimbs, as depicted in Fig. S2 in the supplemental material, or from individual wells using Living Image 2.6.
Micro-computed tomography analysis.
Hindlimb bones were fixed for 48 h in 10% neutral buffered formalin and then stored in 70% ethanol. Bones were scanned ex vivo using a μCT40 instrument (Scanco Medical, Switzerland) (10 μm, 55 kilovoltage peak (kVp), 145 μA, 8 W, and 300-ms integration time) and analyzed with μCT Evaluation Program V6.6. For trabecular bone analyses, regions of interest were defined as 100 slices below the femoral growth plate. Outcome variables are reported in accordance with published consensus guidelines (40). Mice sacrificed earlier than the desired endpoint were not included in the micro-computed tomography (microCT) analysis.
Histology.
After microCT analysis was performed, bones were decalcified for 14 days in 14% EDTA-free acid and then stored in 70% ethanol. For tissue processing and staining, samples were submitted to the Washington University Musculoskeletal Histology and Morphometry Core for hematoxylin and eosin (H&E) or tartrate-resistant acid phosphatase (TRAP) staining. Two to three sections were examined per mouse. Images were taken using an Olympus BX-51 microscope equipped with an Olympus DP27 camera and cellSens Standard software.
Enumeration of CFU.
At 3 weeks postinjection, tissues were harvested using a Bullet-Blender and Navy lysis tubes (Next Advance, Inc., Averill Park, NY) at 4°C. To enumerate bacterial CFU, the whole organ (tibia, femur, heart, kidney, or liver) was homogenized in TSB, serially diluted in TSB, and plated on TSA plates at 37°C overnight for bacterial colony enumeration.
Intracellular proliferation assay.
Osteoclasts (OCs) were generated from enriched bone marrow macrophages (BMMs) harvested from the long bones of 10- to 12-week-old wild-type C57BL/6 mice and cultured in alpha-minimal essential medium (MEM) with 10% fetal bovine serum (FBS) and a 1:10 dilution of the CMG 14-12 cell supernatant (containing the equivalent of 100 ng/ml of macrophage colony-stimulating factor [M-CSF]) for 4 days to expand BMMs. BMMs were seeded into 12-well tissue culture-treated plates at 1.5 × 105 cells per well and differentiated into OCs for 3 days in alpha-MEM with 10% FBS, a 1:50 dilution of the CMG 14-12 cell supernatant (20 ng/ml M-CSF), and 60 ng/ml GST (glutathione S-transferase)-Receptor activator of nuclear factor kappa-B ligand (RANKL).
CFU were enumerated after a gentamicin protection assay, as previously described (29). Briefly, OCs were infected with S. aureus strains at a multiplicity of infection (MOI) of 10:1 for 30 min. Cells were then washed with PBS and cultured in medium containing antibiotic (0.3 mg/ml gentamicin) for 1 h to kill extracellular bacteria. Next, cells were again washed in PBS, and cells at the 1.5-h time point were lysed in sterile, cold, molecular-grade H2O. Cells at the 18-h time point were replenished with medium with M-CSF and RANKL until hypotonic lysis, as described above. Lysates were 10-fold serially diluted and plated on TSA plates for incubation overnight at 37°C.
S. aureus culture supernatant concentration.
Three colonies from each S. aureus strain were inoculated into 50 ml of RPMI 1640 supplemented with 1% Casamino Acids in a 250-ml Erlenmeyer flask. Flasks were stoppered and incubated at 37°C with shaking at 180 rpm for 15 h. Bacterial cultures were centrifuged at 4,000 × g for 10 min at 4°C, and culture supernatants were removed and filter sterilized using a 0.22-μm filter. The supernatants were concentrated with an Amicon Ultra 3-kDa nominal molecular weight limit centrifugal filter unit (Millipore), according to the manufacturer’s instructions. Concentrated supernatants were filter sterilized again with a 0.22-μm filter, and aliquots of each supernatant were stored at −80°C until use in the cytotoxicity assay.
Cytotoxicity assay.
MC3T3-E1 cells obtained from the American Type Culture Collection (ATCC) were propagated according to ATCC recommendations. Cells were grown at 37°C with 5% CO2, with replacement of the medium every 2 to 3 days. For cytotoxicity assays, MC3T3 cells were seeded in 96-well tissue culture plates at a density of 5,000 cells in 200 μl medium per well. After 24 h, the medium was removed, and fresh medium containing either 5% or 10% concentrated S. aureus culture supernatant or RPMI 1640 was added to the cell monolayers. The MC3T3 cells were incubated for an additional 22 h, and cell viability was then assayed using a CellTiter 96 AQueous one solution cell proliferation assay (Promega), according to the manufacturer’s instructions. Survival was calculated for MC3T3 cells exposed to each supernatant condition as a percentage of survival relative to MC3T3 cells exposed to medium containing RPMI 1640.
S. aureus clinical isolate sequencing.
DNA was isolated from bacterial cultures grown overnight using the DNeasy blood and tissue kit (catalog number 69504; Qiagen) according to the manufacturer’s instructions. Whole-genome sequencing was performed at the Genome Technology Access Center at the Washington University in St. Louis School of Medicine using an Illumina NovaSeq 6000 instrument with an S4 flow cell and 2-by-150 paired-end reads. Sequence reads were aligned to the reference genome using NovoAlign. USA300_FPR3757 (NCBI accession number NC_007793.1) was used for the USA300 reference genome, and N315 (NCBI accession number BA000018.3) was used for the USA100 reference genome. The alignments were sorted with SAMtools. Picard Tools was used to remove duplicate reads. Variant files and BAM alignments for each clinical isolate were analyzed using Geneious Prime v. 2021.0.1.
Statistical analysis.
All data are represented as means with standard deviations (SD). All data were analyzed by one-way analysis of variance (ANOVA) with Tukey’s multiple-comparison post hoc test (GraphPad InStat), except for correlation data, where the Pearson correlation coefficient was calculated, and Fig. 4A, where differences between BLI− and BLI+ legs for each isolate were calculated by Student’s t test (GraphPad InStat). A P value of <0.05 was taken as significant.
Ethics statement.
All animal procedures were approved by the Institutional Animal Care and Use Committee of Washington University (IACUC protocols 20170025 and 19-1059), according to established federal and state policies outlined in the Animal Welfare Act (AWA) and enforced by the U.S. Department of Agriculture (USDA), Animal and Plant Health Inspection Service (APHIS), USDA Animal Care. Clinical isolates were obtained under Washington University Institutional Review Board approval (IRB number 201805091).
Data availability.
Genomic sequences for TI1 (NCBI accession number SRX11116571), TI2 (NCBI accession number SRX11116572), and TI3 (NCBI accession number SRX11116573) are available at the NCBI Sequence Read Archive.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants R21 AR073507 and R01 AR070030 (D.J.V.) and by Shriners Hospitals for Children grant 85117 (D.J.V.). P.M.R. was supported by Skeletal Disorders Training Program grant T32 AR060719. J.E.C. was supported by grant R01AI132560 (NIAID), grant R01AI145992 (NIAID), and a career award for medical scientists from the Burroughs Wellcome Fund. K.R.E. was supported by Childhood Infection Research Program grant T32 AI095303. C.A.F. was supported through grant T32GM007347 (NIGMS) and is supported by grant F30AI138424 (NIAID). The bioluminescence imaging was performed at the Washington University School of Medicine Molecular Imaging Center, supported by NIH grant P50 CA094056 (Molecular Imaging Center) and NCI grant P30 CA091842 (Siteman Cancer Center Small Animal Cancer Imaging Shared Resource). The Washington University Musculoskeletal Research Center, supported by grant P30 R074992, provided resources for microCT through its Structure and Strength Core and histology via its Histology and Morphometry Core. Sequencing was performed by the Washington University Genome Technology Access Center.
We thank Crystal Idleburg and Samantha Coleman for expert histology, Roger Plaut for providing the lux operon plasmid pRP1195, Victor Torres for providing the mCherry S. aureus strain, Valeria Reyes Ruiz and Brittney Gimza for technical support with strain construction, and Julie Prior and Katie Duncan for their bioluminescence imaging assistance.
Footnotes
Supplemental material is available online only.
Contributor Information
Philip M. Roper, Email: proper@wustl.edu.
Deborah J. Veis, Email: dveis@wustl.edu.
Victor J. Torres, New York University School of Medicine
REFERENCES
- 1.Lew DP, Waldvogel FA. 2004. Osteomyelitis. Lancet 364:369–379. 10.1016/S0140-6736(04)16727-5. [DOI] [PubMed] [Google Scholar]
- 2.Calhoun JH, Manring MM, Shirtliff M. 2009. Osteomyelitis of the long bones. Semin Plast Surg 23:59–72. 10.1055/s-0029-1214158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goergens ED, McEvoy A, Watson M, Barrett IR. 2005. Acute osteomyelitis and septic arthritis in children. J Paediatr Child Health 41:59–62. 10.1111/j.1440-1754.2005.00538.x. [DOI] [PubMed] [Google Scholar]
- 4.Funk SS, Copley LA. 2017. Acute hematogenous osteomyelitis in children: pathogenesis, diagnosis, and treatment. Orthop Clin North Am 48:199–208. 10.1016/j.ocl.2016.12.007. [DOI] [PubMed] [Google Scholar]
- 5.Riise OR, Kirkhus E, Handeland KS, Flato B, Reiseter T, Cvancarova M, Nakstad B, Wathne KO. 2008. Childhood osteomyelitis—incidence and differentiation from other acute onset musculoskeletal features in a population-based study. BMC Pediatr 8:45. 10.1186/1471-2431-8-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peltola H, Paakkonen M. 2014. Acute osteomyelitis in children. N Engl J Med 370:352–360. 10.1056/NEJMra1213956. [DOI] [PubMed] [Google Scholar]
- 7.Street M, Puna R, Huang M, Crawford H. 2015. Pediatric acute hematogenous osteomyelitis. J Pediatr Orthop 35:634–639. 10.1097/BPO.0000000000000332. [DOI] [PubMed] [Google Scholar]
- 8.Wright JA, Nair SP. 2010. Interaction of staphylococci with bone. Int J Med Microbiol 300:193–204. 10.1016/j.ijmm.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Masters EA, Trombetta RP, de Mesy Bentley KL, Boyce BF, Gill AL, Gill SR, Nishitani K, Ishikawa M, Morita Y, Ito H, Bello-Irizarry SN, Ninomiya M, BrodellJD, Jr, Lee CC, Hao SP, Oh I, Xie C, Awad HA, Daiss JL, Owen JR, Kates SL, Schwarz EM, Muthukrishnan G. 2019. Evolving concepts in bone infection: redefining “biofilm”, “acute vs. chronic osteomyelitis”, “the immune proteome” and “local antibiotic therapy”. Bone Res 7:20. 10.1038/s41413-019-0061-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McNeil JC, Vallejo JG, Kok EY, Sommer LM, Hulten KG, Kaplan SL. 2019. Clinical and microbiologic variables predictive of orthopedic complications following Staphylococcus aureus acute hematogenous osteoarticular infections in children. Clin Infect Dis 69:1955–1961. 10.1093/cid/ciz109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clerc A, Zeller V, Marmor S, Senneville E, Marchou B, Laurent F, Lucht F, Desplaces N, Lustig S, Chidiac C, Ferry T. 2020. Hematogenous osteomyelitis in childhood can relapse many years later into adulthood: a retrospective multicentric cohort study in France. Medicine (Baltimore) 99:e19617. 10.1097/MD.0000000000019617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Popescu B, Tevanov I, Carp M, Ulici A. 2020. Acute hematogenous osteomyelitis in pediatric patients: epidemiology and risk factors of a poor outcome. J Int Med Res 48:300060520910889. 10.1177/0300060520910889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sukswai P, Kovitvanitcha D, Thumkunanon V, Chotpitayasunondh T, Sangtawesin V, Jeerathanyasakun Y. 2011. Acute hematogenous osteomyelitis and septic arthritis in children: clinical characteristics and outcomes study. J Med Assoc Thai 94(Suppl 3):S209–S216. [PubMed] [Google Scholar]
- 14.DeLeo FR, Otto M, Kreiswirth BN, Chambers HF. 2010. Community-associated meticillin-resistant Staphylococcus aureus. Lancet 375:1557–1568. 10.1016/S0140-6736(09)61999-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patel M, Rojavin Y, Jamali AA, Wasielewski SJ, Salgado CJ. 2009. Animal models for the study of osteomyelitis. Semin Plast Surg 23:148–154. 10.1055/s-0029-1214167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cassat JE, Hammer ND, Campbell JP, Benson MA, Perrien DS, Mrak LN, Smeltzer MS, Torres VJ, Skaar EP. 2013. A secreted bacterial protease tailors the Staphylococcus aureus virulence repertoire to modulate bone remodeling during osteomyelitis. Cell Host Microbe 13:759–772. 10.1016/j.chom.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harrasser N, Gorkotte J, Obermeier A, Feihl S, Straub M, Slotta-Huspenina J, von Eisenhart-Rothe R, Moser W, Gruner P, de Wild M, Gollwitzer H, Burgkart R. 2016. A new model of implant-related osteomyelitis in the metaphysis of rat tibiae. BMC Musculoskelet Disord 17:152. 10.1186/s12891-016-1005-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Mesy Bentley KL, Trombetta R, Nishitani K, Bello-Irizarry SN, Ninomiya M, Zhang L, Chung HL, McGrath JL, Daiss JL, Awad HA, Kates SL, Schwarz EM. 2017. Evidence of Staphylococcus aureus deformation, proliferation, and migration in canaliculi of live cortical bone in murine models of osteomyelitis. J Bone Miner Res 32:985–990. 10.1002/jbmr.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Cheng LI, Helfer DR, Ashbaugh AG, Miller RJ, Tzomides AJ, Thompson JM, Ortines RV, Tsai AS, Liu H, Dillen CA, Archer NK, Cohen TS, Tkaczyk C, Stover CK, Sellman BR, Miller LS. 2017. Mouse model of hematogenous implant-related Staphylococcus aureus biofilm infection reveals therapeutic targets. Proc Natl Acad Sci U S A 114:E5094–E5102. 10.1073/pnas.1703427114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Putnam NE, Fulbright LE, Curry JM, Ford CA, Petronglo JR, Hendrix AS, Cassat JE. 2019. MyD88 and IL-1R signaling drive antibacterial immunity and osteoclast-driven bone loss during Staphylococcus aureus osteomyelitis. PLoS Pathog 15:e1007744. 10.1371/journal.ppat.1007744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elasri MO, Thomas JR, Skinner RA, Blevins JS, Beenken KE, Nelson CL, Smeltzer MS. 2002. Staphylococcus aureus collagen adhesin contributes to the pathogenesis of osteomyelitis. Bone 30:275–280. 10.1016/S8756-3282(01)00632-9. [DOI] [PubMed] [Google Scholar]
- 22.Horst SA, Hoerr V, Beineke A, Kreis C, Tuchscherr L, Kalinka J, Lehne S, Schleicher I, Kohler G, Fuchs T, Raschke MJ, Rohde M, Peters G, Faber C, Loffler B, Medina E. 2012. A novel mouse model of Staphylococcus aureus chronic osteomyelitis that closely mimics the human infection: an integrated view of disease pathogenesis. Am J Pathol 181:1206–1214. 10.1016/j.ajpath.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 23.Tuchscherr L, Geraci J, Loffler B. 2017. Staphylococcus aureus regulator sigma B is important to develop chronic infections in hematogenous murine osteomyelitis model. Pathogens 6:31. 10.3390/pathogens6030031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corrado A, Donato P, Maccari S, Cecchi R, Spadafina T, Arcidiacono L, Tavarini S, Sammicheli C, Laera D, Manetti AG, Ruggiero P, Galletti B, Nuti S, De Gregorio E, Bertholet S, Seubert A, Bagnoli F, Bensi G, Chiarot E. 2016. Staphylococcus aureus-dependent septic arthritis in murine knee joints: local immune response and beneficial effects of vaccination. Sci Rep 6:38043. 10.1038/srep38043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilde AD, Snyder DJ, Putnam NE, Valentino MD, Hammer ND, Lonergan ZR, Hinger SA, Aysanoa EE, Blanchard C, Dunman PM, Wasserman GA, Chen J, Shopsin B, Gilmore MS, Skaar EP, Cassat JE. 2015. Bacterial hypoxic responses revealed as critical determinants of the host-pathogen outcome by TnSeq analysis of Staphylococcus aureus invasive infection. PLoS Pathog 11:e1005341. 10.1371/journal.ppat.1005341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sahukhal GS, Tucci M, Benghuzzi H, Wilson G, Elasri MO. 2020. The role of the msaABCR operon in implant-associated chronic osteomyelitis in Staphylococcus aureus USA300 LAC. BMC Microbiol 20:324. 10.1186/s12866-020-01964-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Plaut RD, Mocca CP, Prabhakara R, Merkel TJ, Stibitz S. 2013. Stably luminescent Staphylococcus aureus clinical strains for use in bioluminescent imaging. PLoS One 8:e59232. 10.1371/journal.pone.0059232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boles BR, Thoendel M, Roth AJ, Horswill AR. 2010. Identification of genes involved in polysaccharide-independent Staphylococcus aureus biofilm formation. PLoS One 5:e10146. 10.1371/journal.pone.0010146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krauss JL, Roper PM, Ballard A, Shih CC, Fitzpatrick JAJ, Cassat JE, Ng PY, Pavlos NJ, Veis DJ. 2019. Staphylococcus aureus infects osteoclasts and replicates intracellularly. mBio 10:e02447-19. 10.1128/mBio.02447-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol 38:1008–1015. 10.1128/JCM.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Collins A, Wakeland EK, Raj P, Kim MS, Kim J, Tareen NG, Copley LAB. 2018. The impact of Staphylococcus aureus genomic variation on clinical phenotype of children with acute hematogenous osteomyelitis. Heliyon 4:e00674. 10.1016/j.heliyon.2018.e00674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kennedy AD, Otto M, Braughton KR, Whitney AR, Chen L, Mathema B, Mediavilla JR, Byrne KA, Parkins LD, Tenover FC, Kreiswirth BN, Musser JM, DeLeo FR. 2008. Epidemic community-associated methicillin-resistant Staphylococcus aureus: recent clonal expansion and diversification. Proc Natl Acad Sci U S A 105:1327–1332. 10.1073/pnas.0710217105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen H, Akoda E, Zhang K. 2013. Methicillin-resistant Staphylococcus aureus carriage among students at a historically black university: a case study. Int J Microbiol 2013:979734. 10.1155/2013/979734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tuchscherr L, Pollath C, Siegmund A, Deinhardt-Emmer S, Hoerr V, Svensson CM, Thilo Figge M, Monecke S, Loffler B. 2019. Clinical S. aureus isolates vary in their virulence to promote adaptation to the host. Toxins (Basel) 11:135. 10.3390/toxins11030135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Musso N, Caruso G, Bongiorno D, Grasso M, Bivona DA, Campanile F, Caraci F, Stefani S. 2021. Different modulatory effects of four methicillin-resistant Staphylococcus aureus clones on MG-63 osteoblast-like cells. Biomolecules 11:72. 10.3390/biom11010072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Limbago B, Fosheim GE, Schoonover V, Crane CE, Nadle J, Petit S, Heltzel D, Ray SM, Harrison LH, Lynfield R, Dumyati G, Townes JM, Schaffner W, Mu Y, Fridkin SK, Active Bacterial Core surveillance MRSA Investigators. 2009. Characterization of methicillin-resistant Staphylococcus aureus isolates collected in 2005 and 2006 from patients with invasive disease: a population-based analysis. J Clin Microbiol 47:1344–1351. 10.1128/JCM.02264-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.See I, Mu Y, Albrecht V, Karlsson M, Dumyati G, Hardy DJ, Koeck M, Lynfield R, Nadle J, Ray SM, Schaffner W, Kallen AJ. 2020. Trends in incidence of methicillin-resistant Staphylococcus aureus bloodstream infections differ by strain type and healthcare exposure, United States, 2005-2013. Clin Infect Dis 70:19–25. 10.1093/cid/ciz158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Le Fur N, Kelsall SR, Mintz B. 1996. Base substitution at different alternative splice donor sites of the tyrosinase gene in murine albinism. Genomics 37:245–248. 10.1006/geno.1996.0551. [DOI] [PubMed] [Google Scholar]
- 39.Chen J, Yoong P, Ram G, Torres VJ, Novick RP. 2014. Single-copy vectors for integration at the SaPI1 attachment site for Staphylococcus aureus. Plasmid 76:1–7. 10.1016/j.plasmid.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bouxsein ML, Boyd SK, Christiansen BA, Guldberg RE, Jepsen KJ, Muller R. 2010. Guidelines for assessment of bone microstructure in rodents using micro-computed tomography. J Bone Miner Res 25:1468–1486. 10.1002/jbmr.141. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download IAI.00180-21-s0001.pdf, PDF file, 4.0 MB (4MB, pdf)
Data Availability Statement
Genomic sequences for TI1 (NCBI accession number SRX11116571), TI2 (NCBI accession number SRX11116572), and TI3 (NCBI accession number SRX11116573) are available at the NCBI Sequence Read Archive.