ABSTRACT
Upregulated in inflammation, calprotectin (complexed S100A8 and S100A9; S100A8/A9) functions as an innate immune effector molecule, promoting inflammation, and also as an antimicrobial protein. We hypothesized that antimicrobial S100A8/A9 would mitigate change to the local microbial community and promote resistance to experimental periodontitis in vivo. To test this hypothesis, S100A9−/− and wild-type (WT; S100A9+/+) C57BL/6 mice were compared using a model of ligature-induced periodontitis. On day 2, WT mice showed fewer infiltrating innate immune cells than S100A9−/− mice; by day 5, the immune cell numbers were similar. At 5 days post ligature placement, oral microbial communities sampled with swabs differed significantly in beta diversity between the mouse genotypes. Ligatures recovered from molar teeth of S100A9−/− and WT mice contained significantly dissimilar microbial genera from each other and the overall oral communities from swabs. Concomitantly, the S100A9−/− mice had significantly greater alveolar bone loss than WT mice around molar teeth in ligated sites. When the oral microflora was ablated by antibiotic pretreatment, differences disappeared between WT and S100A9−/− mice in their immune cell infiltrates and alveolar bone loss. Calprotectin, therefore, suppresses emergence of a dysbiotic, proinflammatory oral microbial community, which reduces innate immune effector activity, including early recruitment of innate immune cells, mitigating subsequent alveolar bone loss and protecting against experimental periodontitis.
KEYWORDS: innate immunity, calprotectin, microbiome, inflammation, experimental periodontitis
INTRODUCTION
Periodontitis is a chronic oral inflammatory disease leading to the destruction of gingival tissues and the underlying alveolar bone that supports the teeth. Although distributed globally, the prevalence of periodontitis in U.S. dentate adults (≥30 years) is 42.2%, and 7.8% of patients present with severe periodontitis (1). Disease initiates when a perturbation in the healthy oral periodontal microbial community promotes dysbiosis, altering the relative abundance of commensal species (2). In the dysbiotic community, some commensals with pathogenic potential overgrow their neighbors to emerge as pathobionts (3–5). Whereas single species (and strains) are studied to establish their virulence, pathobionts are not acquired exogenous microbes, but are emergent members of a dysbiotic community (6, 7). Over time, the imbalanced dysbiotic community elicits a hyperinflammatory host response, leading to the soft tissue destruction and alveolar bone loss that characterize periodontitis (8, 9).
In periodontitis, the role played by innate antimicrobial proteins, such as calprotectin, has not been established. Calprotectin is abundant in the cytoplasm of neutrophils (10, 11) and epithelial cells (12). Calprotectin is active intracellularly (13) and extracellularly within tissues (14–16) during infection and inflammation. In epithelial cells, calprotectin thwarts the growth of oral and enteric invasive bacteria, including Porphyromonas gingivalis, Salmonella enterica serovar Typhimurium, and Listeria monocytogenes (12, 17–19). A calcium-binding hetero-oligomer of S100A8 and S100A9 (S100A8/A9), whereby each S100 subunit contains two Ca2+ binding sites (19, 20), has antimicrobial activity involving the chelating of a subset of transition metals (i.e., Fe, Mn, Zn, and Ni), which serve as cofactors for many bacterial enzymes (16, 20, 21). By sequestering and restricting bacterial access to nutrient divalent cations, calprotectin promotes “nutritional” immunity, which suppresses bacterial growth (22) in species as diverse as Borrelia burgdorferi (23) and Staphylococcus aureus (24). By chelating Mn++, calprotectin inactivates superoxide dismutase to increase the effectiveness of bacterial killing by neutrophils (24, 25). Conversely, mechanisms to circumvent nutritional immunity by using high-metal-affinity metallophores have evolved, for example, in Pseudomonas aeruginosa (26) and Staphylococcus aureus (27), or likewise through flavin recovery during iron deprivation in Acinetobacter baumanii (28). During gastrointestinal inflammation, Salmonella enterica serovar Typhimurium, in the presence of elevated levels of S100A8/A9, upregulates high-affinity transport systems for Mn++ and Zn++ to outcompete neighboring commensals and activate superoxide-detoxifying enzymes to evade neutrophil killing (29–31).
Increased severity of periodontitis has been linked with a single nucleotide polymorphism (SNP) in a noncoding region of S100A8 in a family-based association study (32), in which gene associations are recognized by using relatives of case patients as control subjects (33). During the innate cellular response in acute periodontitis (8), S100A8/A9 is released from neutrophil cytoplasm into the surrounding environment (15, 34, 35), increasing extracellular levels in the interstitial fluid (36–41). Extracellular S100A8/A9 can complex in neutrophil extracellular traps, providing antimicrobial nutritional immunity in tissue spaces (14, 16, 21, 42–47). In the extracellular space, S100A8/A9 shows innate immune effector activity by acting as an alarmin, stimulating neutrophil chemotaxis (48) and cytokine release (49–51), and activating the proinflammatory TLR4 complex (48, 52) and the receptor for advanced glycation end products (RAGE) (49, 53, 54). The proinflammatory functions notwithstanding, calprotectin is protective against tissue damage and accumulation of bacteria in the lungs (55–57) and gastrointestinal tract (58) in animal models. Hence, the antimicrobial effects of S100A8/A9 may not be evident in all species and the proinflammatory roles difficult to predict in a complex community.
To study the underlying biological plausibility that S100A8/A9 contributes to resistance against experimental periodontitis, we used an established murine model (59). Experimental periodontitis is induced by a ligature that is tied around the maxillary right second molar tooth. Wild-type (WT) C57BL/6J mice and isogenic mice lacking S100A9 (S100A9−/−) were compared. S100A9−/− mice are deficient in S100A8 at the protein level and lack the calprotectin complex (60). Our findings demonstrate for the first time that S100A8/A9 mitigates dysbiotic changes in the periodontal microflora, protecting against an acute inflammatory cell infiltrate and the loss of alveolar bone that characterizes experimental periodontitis.
RESULTS
S100A8/A9 alters the microbiota during ligature-induced periodontitis.
Healthy unligated WT and S100A9−/− mice had similar alpha and beta diversities in their oral swab samples (Fig. 1; Fig. S2 in the supplemental material). After 5 days of ligature-induced periodontitis, however, the alpha diversities of oral swab samples differed significantly between ligated and unligated (healthy) sites in WT (P = 0.00062) mice and S100A9−/− (P = 0.0205) mice (Fig. 1). Swab sample alpha diversities from ligated WT and S100A9−/− mice did not differ significantly (P = 0.072) (Fig. 1). In contrast, the microbial diversity of communities recovered from the ligature loops (with knot removed) of WT and S100A9−/− mice differed significantly (P = 0.00016) (Fig. 1).
FIG 1.
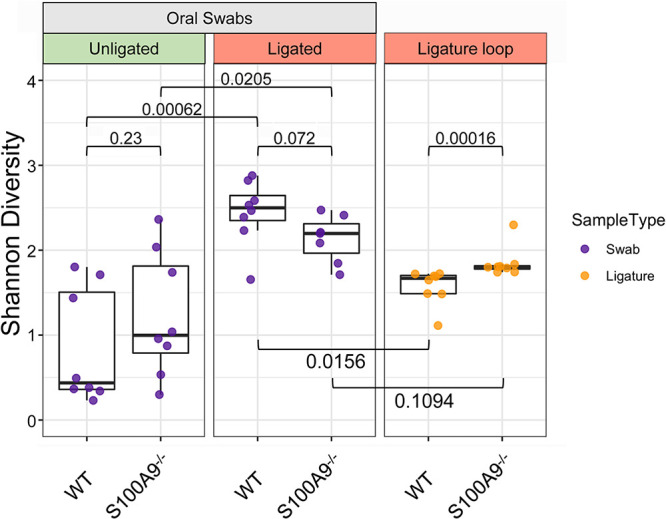
Microbial alpha diversity (i.e., within samples) during inflammation (5 days postligation) is greater in S100A9−/− than WT mice. Alpha diversity is represented by the Shannon index for taxonomic richness and evenness. Greater diversity within the sample constitutes a higher Shannon index value. Significant differences between group means were determined using the Mann-Whitney U test. Swab/ligature comparisons using the Mann-Whitney U test were made pairwise within-mouse. Between-group comparisons are indicated by bars with P values. P values of <0.05 were considered significant (n = 7 to 8 for each treatment group).
Oral swab microbial communities differed significantly between ligated WT and S100A9−/− mice based on Bray-Curtis dissimilarity (P = 0.018, permutational multivariate analysis of variance [PERMANOVA]), but not by unweighted or weighted UniFrac phylogenetic distance (Fig. S2). Microbial communities in samples recovered from the ligatures also showed significant differences using weighted UniFrac measures (P = 0.003, PERMANOVA) and trended toward difference with Bray-Curtis dissimilarity (P = 0.052, PERMANOVA) (Fig. S2). Since weighted UniFrac distance weighs distances by phylogenetic relationships, disparity in significance between microbial community distance measures may be influenced by phylogenetically related microbes. These data provide support that ligation differentially changes the composition of the healthy oral microbiota depending on the presence or absence of S100A8/A9.
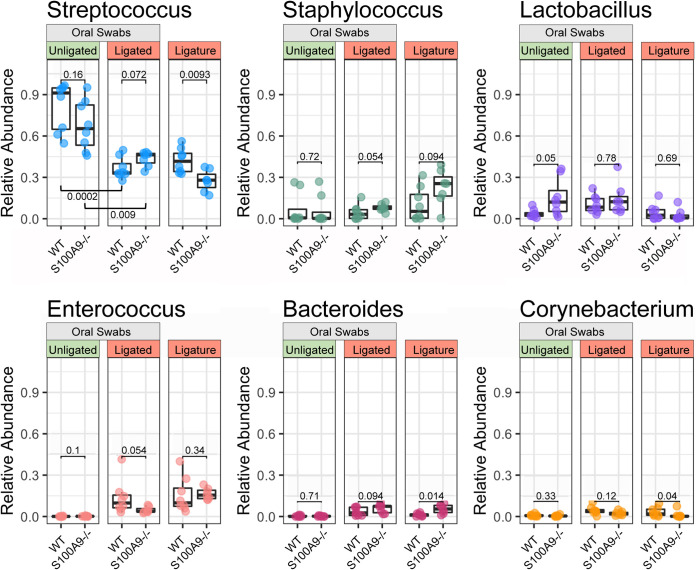
To determine the basis for the significant differences in bacterial communities associated with ligatures from WT and S100A9−/− mice, the genera associated with the community alterations were identified. After ligation, the genus Streptococcus was significantly depleted in both genotypes (WT, P = 0.0002; S100A9−/−, P = 0.009; Mann-Whitney U test) (Fig. 2). Comparing ligatures recovered from WT and S100A9−/− mice, the abundance of Streptococcus differed significantly (P = 0.0093), but this genotype-specific difference was not distinct (P = 0.072) in the full oral cavity (swab samples). Genotype-driven differences are seen in the abundance of Bacteroides (P = 0.014) and Corynebacterium (P = 0.04) in ligature communities. These trends and key genera were confirmed using relative abundance profiles for all samples and visualized using a stacked bar plot (Fig. S3) and a comparison of genotype group mean relative abundances (Table S1).
FIG 2.
WT and S100A9−/− mice have distinct oral microbes in response to ligation. The relative overall abundances of the most prevalent genera are represented by beta diversity. Genotype differences were evaluated using a Mann-Whitney U test. P values of <0.05 were considered significant (n = 7 to 8 for each treatment group).
Expression of S100A8/A9 is upregulated in ligature-induced periodontal disease.
After 5 days of ligature-induced periodontitis, gingival S100A8 and S100A9 were analyzed using immunohistochemistry and Western blot analysis. In WT unligated contralateral side gingiva, S100A8 and S100A9 were observed at low levels, and staining was confined to the upper spinous and keratin layers (Fig. S4A). In the gingiva of ligated teeth, S100A8 and S100A9 levels were substantially greater than from unligated contralateral sites, with intense staining throughout the spinous and keratin layers. The proteins were also observed in the suprabasal cells and occasionally in the basal cell layer. As expected, neither S100A8 nor S100A9 protein staining was seen in the ligated S100A9−/− mouse (Fig. S4A), although anti-S100A8 (Fig. S4A) and IgG and secondary antibody-only negative controls produced some artifactual staining in the keratinized layer in some sections (data not shown).
Protein levels in gingival tissue were also confirmed using Western blotting (Fig. S4B). S100A9−/− mice did not show observable S100A8 or S100A9 expression, corroborating the immunohistology results. Ligation increased S100A8 and S100A9 expression in the proximal gingiva by 36 and 34%, respectively, likely accompanied by dimers and multimers as suggested by the bands at ∼24 and ∼63 kDa. In sequential sections, immunoreactive S100A8 and S100A9 levels appeared greater in gingiva adjacent to the ligated tooth (Fig. S5A) than in the contralateral sites proximal to unligated teeth (Fig. S5B). Overall, the mean fluorescence intensity of anti-S100A8 and anti-S100A9 MAbs in gingival sections proximal to ligated teeth was significantly greater (*P < 0.05) than in the contralateral gingiva proximal to unligated teeth (Fig. S5C), whereas the levels of both proteins were similar by each mode of analysis.
S100A8/A9 reduces the early immune cell infiltrate in ligature-induced periodontitis.
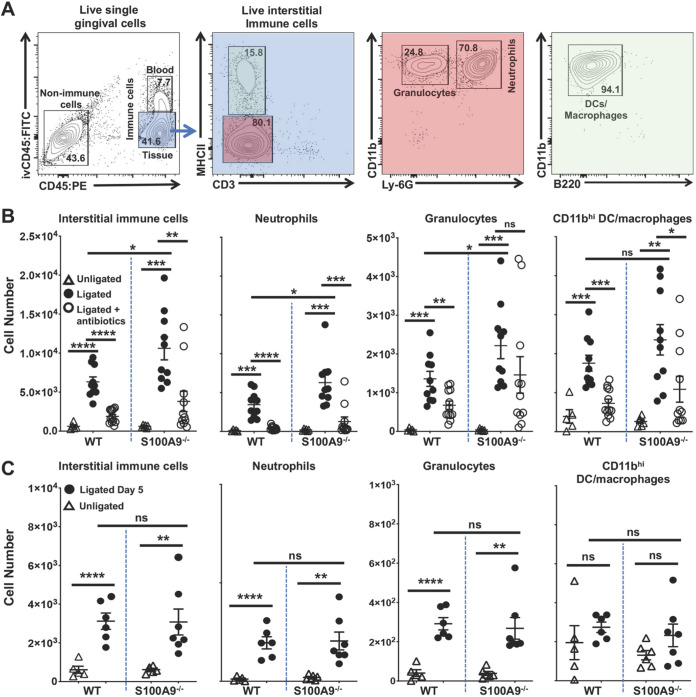
Alveolar bone destruction in ligature-induced periodontitis mouse models is driven primarily by an influx of innate immune cells, principally neutrophils (8, 61). Using the gating strategy shown (Fig. 3A), ligation induced an influx of interstitial immune cells into the gingiva, including neutrophils, granulocytes, CD11bhi dendritic cells, and macrophages on day 2 (Fig. 3B) and day 5 (Fig. 3C). The gingiva proximal to the ligature showed significantly greater numbers of infiltrating interstitial immune cells than unligated control sites in both S100A9−/− and WT mice. On day 2, gingiva proximal to the ligated tooth in S100A9−/− mice showed significantly more infiltrating interstitial immune cells than WT mice, including neutrophils and nonneutrophil granulocytes (P < 0.05) (Fig. 3B). However, the number of CD11bhi dendritic cells and macrophages was similar in WT and S100A9−/− mice. Antibiotic suppression of the oral microbiome (Fig. S1) inhibited the infiltration of interstitial immune cells in gingival sites proximal to ligated teeth (Fig. 3B), confirming that the immune cell infiltrate is driven by the microbial community (62, 63). On day 5, both genotypes showed similar numbers of interstitial immune cells and fewer than on day 2 (Fig. 3C).
FIG 3.
Interstitial immune cell infiltrates differ in S100A9−/− and WT mice following ligature placement. At 2 or 5 days post ligature placement, anesthetized mice were given 1.25 μg of FITC-conjugated anti-CD45 (CD45:FITC) MAb in a retro-orbital injection and euthanized 3 min later. Gingival cells were processed, stained, and counted by flow cytometry as reported in the Materials and Methods. Live immune cell numbers were normalized to 10,000 live nonimmune cells to account for variations in tissue size and sample lost during processing. (A) Flow cytometry gating strategy used to identify specific immune cell types of interest. Live interstitial immune cells were identified as Zombie Aqualo, CD45:FITC−, CD45:PE+; live neutrophils as Zombie Aqualo, CD45:FITC−, CD45:PE+, CD11bhi, Ly-6G+, MHCII−, CD3−; live nonneutrophil granulocytes as Zombie Aqualo, CD45:FITC−, CD45:PE+, CD11bhi, Ly-6G−, MHCII-, CD3−; and live CD11bhi dendritic cells and tissue macrophages as Zombie Aqualo, CD45:FITC−, CD45:PE+, CD11bhi, Ly-6G−, MHCII+, CD3−. (B) Effects of ligation and antibiotic suppression of the oral microbiota on the inflammatory cell infiltrate in WT and S100A9−/− mice at day 2. Summary data of at least three independent experiments are shown. Gingival tissues were harvested from sites in mice that were nonligated (open triangles), ligated (filled circles), or ligated after prophylactic antibiotic treatment (open circles). Cells were quantified using flow cytometry. Individual data points and means ± SEM are plotted. Data were analyzed by two-way ANOVA; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant. (C) At day 5 post ligature placement, S100A9−/− and WT mice show similar levels of infiltrating gingival immune cells. Individual data points from at least two independent experiments are plotted as means ± SEM. Data are for nonligated (open triangles) and ligated (filled circles) mice. Mean cell numbers were compared using two-tailed Student’s t test; **, P < 0.01; ****, P < 0.0001; ns, not significant.
S100A8/A9 mitigates alveolar bone loss.
After 5 days of ligature-induced periodontal disease, the ligated maxillary right second molar and the proximal third molar were mobile (in response to palpation) and exudate was present. The maxillae of co-caged WT and S100A9−/− mice were then harvested and scanned using micro-computed tomography (micro-CT). Loss of alveolar bone (i.e., reduced volume) within a defined volume of interest (VOI) was calculated in groups of experimental mice as described in the Materials and Methods. Ligated WT mice showed significantly less (P = 0.007) bone loss than S100A9−/− mice (Fig. 4). Maxillary bone was also visualized using 3D volume micro-CT reconstructions. In horizontal and vertical sections through the maxilla, voids appeared in the alveolar bone and periodontal ligament space surrounding the roots of the ligated and proximal teeth; voids were not observed on the unligated contralateral sides (Fig. S6A and B). In ligated sites, bone loss was greater in S100A9−/− mice than in WT mice (Fig. S6C). After treatment with antibiotics that ablated the oral microbiota (Fig. S1), ligature-induced bone loss was similar in S100A9−/− and WT mice (Fig. 4). In the absence of ligation and antibiotics, healthy WT and S100A9−/− mice showed similar bone volumes (P = 0.76).
FIG 4.
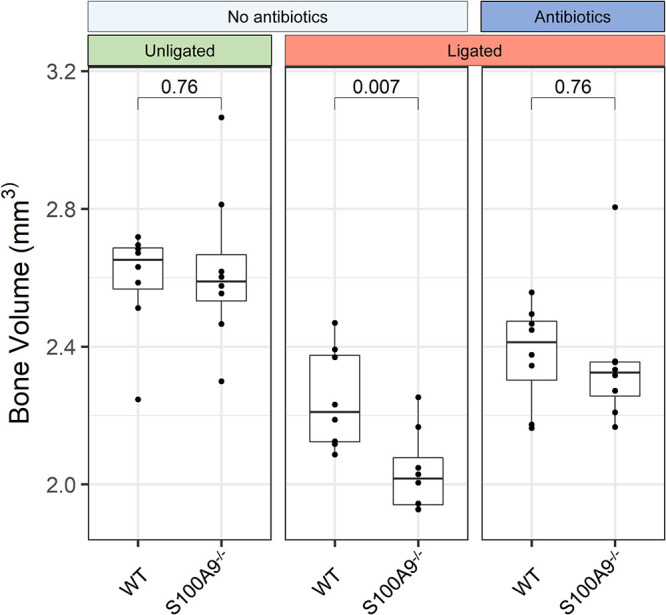
S100A8/A9 expression protects against alveolar bone loss in ligature-induced periodontitis. Harvested maxillae were imaged using micro-CT and reconstructed in 3D as described in the Materials and Methods. A volume of interest encompassing ligated and proximal molars and their roots was set using fixed anatomical landmarks. Bone volume was converted into cubic millimeters by reconstruction from the voxel size. Bone volume differences were analyzed by an unpaired independent Student’s t test. P values are indicated (n = 7 to 8 for each treatment group).
DISCUSSION
S100A8/A9 is a well-documented antimicrobial (10, 19, 22, 28, 64) and potentially immunomodulatory molecule (e.g., PMN chemotaxis [10], activation of TLR4 [48, 53] and the receptor for advanced glycation end products (RAGE) [54], neonatal sepsis [65–67], and CD69 engagement [68]). Using experimental periodontitis as a model, we analyzed the contributions of S100A8/A9 as an innate immune effector molecule and as an antimicrobial protein complex. Our data show that in the ligature-induced model of periodontal disease, loss of alveolar bone is greater in S100A9−/− mice than in WT mice. After mice were given a cocktail of antibiotics for 2 weeks before ligature placement, the resulting oral microbial read counts were virtually undetectable (Fig. S3), and the gingival inflammatory cell infiltrate at day 2 postligature placement and bone loss at day 5 were inhibited. In response to the pathobionts, immune cells infiltrate the gingiva and alveolar bone is lost; “true” acquired pathogens appear to play no role (6, 7). In experimental periodontitis, therefore, S100A8/A9 appeared to function primarily as a protective antimicrobial protein rather than as a directly immunomodulatory or proinflammatory agent. Based on our experiments, S100A8/A9 mitigates the emergence of pathobionts in the dysbiotic community.
With dysbiosis, lesions are impacted less by the relative abundance of specific taxa than the overall disturbance to community stability, which is manifested by changes in diversity in the commensal microbiota. Indeed, this mouse model of ligature-induced periodontitis has been performed in several laboratories with consistent pathological findings (7, 69). At the different centers, however, the community compositions of the oral microbiota at ligated sites are remarkably different; consistent, unique pathogens have not been identified and lesions result from the development of dysbiosis (5, 69). The composition of the oral microflora in C57BL6 mice reflects the vendor more than any mouse-strain-specific community structure and is generally stable over time (up to 52 weeks) (70). Horizontal transfer can occur, where mice from Taconic destabilize the oral microbiota of mice from Jackson Laboratory, whereas the converse is not as obvious (70). Since the amount of microbial DNA recoverable from a single ligature (one per mouse) is modest, investigators rely on 16S sequencing to characterize community composition at the family and genus level. Species-level characterization in this model is unreliable.
At the start of our experiments, we virtually eliminated neonatal differences in the oral microbiota between the WT and S100A9−/− mice by cohousing both genotypes together for 5 to 6 weeks before ligation. This step was crucial to the interpretation of the data, since the S100A9−/− mice were bred in-house and WT C57BL6/J mice were purchased from Jackson Laboratory, each with their acquired microflora. After cohousing, oral swab microbiomes of WT and S100A9−/− mice were statistically indistinguishable using both alpha diversity and beta diversity measurements (Fig. 1, Fig. 2, Fig. S2, and Fig. S3), as reported previously (70). Whereas the oral microbial composition was indistinguishable between the two genotypes at the start of ligation, the alpha diversity (i.e., Shannon index) of the microbiomes recovered from ligature loops was significantly higher in S100A9−/− than WT mice (Fig. 1). The abundance of Streptococcus was significantly lower in S100A9−/− than WT mice, whereas the abundance of Bacteroides was greater (Fig. 2). Indeed, the rank order abundances of the major genera differed between the WT and S100A9−/− mice (Table S1). These genotype-specific differences manifest as dysbiotic changes in the disease-causing community composition, rather than the acquisition of “true” pathogens.
How might S100A8/A9 mitigate the emergence of a dysbiotic community? Many species are sensitive to nutritional immunity (20), in which S100A8/A9 sequesters essential metals from bacteria to inhibit their growth (20, 71, 72); well-studied sensitive species include group B streptococcus (73), Pseudomonas aeruginosa (74), Staphylococcus aureus (75), Acinetobacter baumannii (76), and Helicobacter pylori (77). When nutritional immunity persists, many species, including Staphylococcus aureus (27) and Enterococcus faecalis (78), can evade or overcome nutritional immunity (as reviewed in reference 79). In our model, mouse gingival S100A8/A9 increases after ligation and appears to simulate the expected antimicrobial activity of human calprotectin, the species-specific structural differences notwithstanding.
During periodontitis, the oral environment would appear to favor calprotectin to function as an antimicrobial rather than as an innate immune effector. In the human oral salivary environment, the fluid-phase calcium concentration is estimated at 1.1 mM (80), whereas the concentration of S100A8/A9 in healthy individuals is about 20 μg/ml (81). In healthy gingiva, calcium concentrations in the extracellular environment are lower than in inflamed tissue and the heterodimeric form is likely to predominate (82). In the absence of Ca2+ ions, S100A8/A9 exists as a heterodimer (83, 84). Calprotectin heterodimers self-assemble to form heterotetramers, which promote nutritional immunity (20) and increased resistance to protease activity (83). In gingival crevicular fluid (GCF), a serum transudate from the periodontal tissues, calprotectin levels are estimated as 3.33 ± 0.92 μg/ml in chronic periodontitis patients compared to 2.82 ± 0.93 μg/ml in healthy controls (85). Calcium concentrations are likely to approximate those of serum. During inflammation, calcium is released from immune and epithelial cells into the extracellular environment, increasing the concentration to approximately 2 mM or greater, and thus favoring tetramer formation (82, 86, 87). Murine S100A8/A9 differs from its human orthologue by requiring 10-fold more Ca2+ equivalents to form a heterotetramer (84).
The heterotetramer is unable to bind TLR4/MD2, restricting the proinflammatory effects (88). In our ligature model, calcium and S100A8/A9 levels in the inflamed gingiva are likely to increase the formation of tetramers in comparison to heterodimers. Although we have not characterized tetramer formation directly, WT mice do appear to provide a gingival environment that limits the emergence of a dysbiotic cohort of microbes, mitigating inflammation and bone loss. Without attenuation by S100A8/A9, the ligatures recovered from S100A9−/− mice carry a dysbiotic community that causes increased inflammation and alveolar bone loss (Fig. 3 and 4). The inflamed gingival epithelium (36, 89) and transmigrating neutrophils (10, 35) are copious sources of S100A8/A9, greater than in the saliva or on the mucosal surfaces (10). The neutrophils (64) and squamous epithelial cells contribute to innate immunity (12, 17, 19). S100A8/A9 appears constitutively expressed (90) and is upregulated in the gingiva during periodontitis (89, 91), as we observed in our mouse model.
Our model appears to robustly simulate these important features of the human infection. The effects of the microbiota on the inflammatory cell infiltrate are also time- and S100A8/A9-dependent. In WT mice, the gingival inflammatory cell infiltrate was less abundant at 2 days of ligation than in S100A9−/− mice (Fig. 3B). By day 5 of ligation, levels of inflammatory cells in the S100A9−/− mice decreased to WT numbers, and levels in both strains of mice were reduced from day 2 (Fig. 3C). S100A8/A9 levels also change over time. S100A8/A9 and gingival cytokines increased in ligated sites (Fig. S4 and S5) as previously reported (92), whereas levels were low in the control nonligated, contralateral gingival sites (Fig. S4 and S5). The increase in S100A8/A9 in ligated sites reflects the numbers of infiltrating PMNs and the gingival epithelial cells, which also function as innate immune cells in response to microbial challenge, as we (93) and others (94) have reported. Furthermore, during ligature-induced periodontitis in WT mice, expression of innate immune response-related genes also appears to increase in the affected gingiva (40).
Our results are similar to some other models of inflammatory disease. Tobacco smoking is an important contributory etiologic factor in human periodontitis (95, 96) and is accompanied by an increase in S100A8/A9 in gingival crevicular fluid (i.e., a transudate) (36), as we see in our model. Similarly, in a mouse model of lung disease, smoking causes S100A9 increases in bronchial lavage fluid (97). In the mouse lung disease model, S100A9 appeared to signal through ERK and c-RAF to induce production of matrix metalloproteinase 3 and 9, monocyte chemoattractant protein 1 (MCP-1), interleukin 6 (IL-6), and IL-8, which contribute to the pathogenesis of chronic obstructive pulmonary disease. By inhibiting S100A9 engagement of TLR4, RAGE, or extracellular matrix metalloproteinase inducer (EMMPRIN), proinflammatory signaling pathways were blocked. Similar pathways are activated in the lungs of patients with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection (54). In models of lung infection, the absence of S100A9 is accompanied by a time-dependent increase and then reduction in PMN infiltration into the tissue parenchyma and fluid (56). In our model of periodontitis, absence of S100A9, which causes a dysbiotic microbiota, also results in a significantly more abundant inflammatory cell infiltrate early (2 days postligation) and greater alveolar bone loss later (5 days postligation). Several factors may explain the effects of S100A9, including the composition and dysbiosis of the proximal microbiota, site-specific differences in the anatomy, and the duration of inflammation when lesions were observed.
Dysbiosis can also occur in the presence of elevated local levels of S100A8/A9 and concomitant inflammation such as produced by SARS-CoV-2 in the intestines of patients and in animal models (98). In other diseases, diminished S100A8/A9 levels and associated microbial changes are inconsistent with health. For example, locally produced S100A8/A9 in the gastrointestinal (GI) tracts of newborn humans and C57BL6 mice promote GI health and communities with abundant commensal Actinobacteria and Bifidobacteriaceae and suppressed Enterococcus faecalis (66). With diminished S100A8/A9 levels, however, abundance of endogenous Enterobacteriaceae increases in the complex GI community (66). Emerging as pathobionts, Enterobacteriaceae become proinflammatory, elicit a mucosal immune response, and cause sepsis and obesity.
Our work is the first to show that a single antimicrobial protein complex, calprotectin, can mitigate emergence of localized dysbiotic oral microflora, suppressing the early innate hyperinflammatory cell phenotype in the gingiva. Given the many antimicrobial mechanisms in the oral cavity, it is remarkable that an effect of calprotectin in limiting dysbiosis was found.
Suppression of commensal species by calprotectin, as we show, likely contributes to the emergence of pathobionts that are less sensitive to metal sequestration. Calprotectin appears to play a crucial indirect role, therefore, in the innate immune response to periodontal infection. How the early suppression of infiltrating innate inflammatory cells affects the later loss of alveolar bone remains to be elucidated.
MATERIALS AND METHODS
Mice.
C57BL/6J (stock number 000664) control mice (WT) were purchased from the Jackson Laboratory. S100A9−/− mice generated in a C57BL/6 background (60) were acquired from Daniel I. Simon, Case Western Reserve University (by material transfer agreement), backcrossed for three generations, and genotyped as C57BL6/J by Jackson Laboratories. All mice were bred and housed in specific-pathogen-free cages. Experimental procedures were approved for ethical considerations by the University of Minnesota IACUC.
Induction of periodontal inflammation.
Age- and sex-matched mice of both genotypes were co-caged immediately after weaning at 3 to 4 weeks to equilibrate the oral microbiota. At 10 weeks of age, a 5-0 silk suture (Roboz Surgical, Gaithersburg, MD) was ligated proximal to the gingiva around the maxillary right second molar of each mouse (59). Ligatures were left in place for 2 or 5 days.
To determine the influence of the microbiota on ligature-induced periodontal disease, some WT (n = 8) and S100A9−/− (n = 7) mice were given antibiotics in the drinking water ad libitum from 2 weeks before ligature placement until the conclusion of the experiment (at 10 weeks of age plus 2 days to analyze the inflammatory infiltrate or 5 days of ligature placement to quantify bone loss) (99). The antibiotic cocktail contained 1 mg/ml each of ampicillin (WG Critical Care, LLC, Paramus, NJ, USA), cefoperazone sodium salt (Sigma-Aldrich Co., St. Louis MO, USA), and clindamycin hydrochloride (Fagron, Inc., St. Paul, MN, USA). Control mice received drinking water without antibiotics. Efficacy of the antibiotic treatment was confirmed by sampling the oral microbiota and evaluating recovered operational taxonomic unit (OTU) counts (Fig. S3).
Immunohistochemistry of gingival tissue.
At 5 days postligature, gingival tissues from ligated and contralateral unligated sites were harvested from WT and S100A9−/− mice, placed in 2% paraformaldehyde solution in phosphate-buffered saline (PBS) for 2 h, washed 3 times in PBS for 10 min each, and incubated in 30% sucrose in PBS overnight, all at 4°C. The gingival specimens were placed in cryomolds with optimal cutting compound (OCT) and frozen using dry ice in 2-methylbutane. From each specimen, 8 sections (7-μm thickness) were cut at least 70 μm apart using a Leica CM1860 UV cryostat, washed with ice cold acetone for 8 min, and dried overnight to effectively survey the entire tissue.
Staining was performed essentially as described (100), modified for the gingiva and our proteins of interest. Tissue sections were rehydrated with 1× PBS for 10 min, permeabilized with 0.2% Triton X-100 in PBS for 15 min, and blocked for 1 h using 5% goat serum in 5% bovine serum albumin (BSA) in PBS. Primary antibodies were diluted in 5% BSA in PBS, incubated with tissue sections, and rinsed with 1× PBS for 5 min. The secondary antibodies were also diluted in 5% BSA in PBS, incubated, rinsed with 1× PBS for 5 min, and sections were counterstained with DAPI (4′,6-diamidino-2-phenylindole; 1:50,000) (Thermo Fisher Scientific, Waltham, MA, USA) in PBS for 10 min. The sections were rinsed again with 1× PBS and coverslips mounted. The primary antibodies included goat anti-mouse S100A8 polyclonal antibody (1:100 dilution, 1 h) (R&D Systems, Minneapolis, MN, USA; AF3059), goat anti-mouse S100A9 polyclonal antibody (1:100 dilution, 3 h) (R&D Systems; AF2065), and goat IgG control antibody (1:100 dilution, 1 h for S100A8, 3 h for S100A9) (R&D Systems; AB108C). Secondary antibody used was donkey anti-goat IgG (H+L) secondary antibody labeled with Alexa Fluor 647 (1:100 dilution for S100A9, 1:500 dilution for S100A9; 1 h) (Thermo Fisher Scientific; A-21447). Sections were counterstained using DAPI. Sections were imaged using a Leica DM6 B microscope. The images were formatted within ImageJ (NIH) and mean fluorescence intensities (MFI) of S100A8 and S100A9 were calculated using the integrated density measurements of the epithelia.
Western blotting of gingival S100A8 and S100A9.
Protein expression was also confirmed using Western blotting. Harvested gingival specimens of approximately equal wet weights were rinsed with cold PBS, snap-frozen in liquid nitrogen, suspended in 300 μl ice cold radioimmunoprecipitation assay (RIPA) buffer containing Halt Protease Inhibitor Cocktail (Thermo Fisher), and homogenized with a mortar and pestle (Corning). Homogenized tissue solutions were centrifuged at 4°C. Protein content of soluble supernatants was quantified by bicinchoninic acid (BCA) assay and samples were stored at −80°C. An equal mass (10 μg) of each protein extract was separated by SDS-PAGE and transferred to a nitrocellulose membrane using a Bio-Rad semidry transfer cell. Total protein in the blot was determined using the Revert 700 total protein kit (Li-Cor, Lincoln, NE) according to the manufacturer’s protocol. After removal of the Revert total protein stain, the membrane was blocked with Licor Odyssey (PBS) blocking buffer and treated overnight at 4°C with 1:200 dilutions of goat anti-mouse S100A8 polyclonal antibody (R&D Systems, AF3059) and goat anti-mouse S100A9 polyclonal antibody (R&D Systems, AF2065). After primary antibody treatment, the membrane was rinsed with PBS-Tween and treated with a 1:5,000 dilution of IRDye 800CW donkey anti-goat secondary antibody (Li-Cor). The membrane was incubated at room temperature for 1 h, rinsed, and visualized on a Li-Cor Odyssey imaging system. Integrated densities of S100A8 and S100A9 bands were normalized to total protein (per Revert stain) for each sample. The relative expressions of S100A8 and S100A9 were quantified using ImageJ.
Microbiota sampling.
The oral microbiota was sampled at 10 weeks of age plus 5 days (± ligature) using a PurFlock Ultra swab (25-3318-U; Puritan Medical Products Company LLC, Guilford, ME, USA) premoistened in PBS; oral tissues were swabbed circumferentially to include all four quadrants of the mouth for at least 20 s. Mice were then euthanized. The microbiota associated with periodontitis was recovered from the silk suture loop surrounding the ligated tooth (suture knots were removed). DNA was extracted from swabs and loops by enzyme digestion (lysozyme 20 mg/ml and mutanolysin 150 units/ml) (Sigma-Aldrich) followed by bead-based isolation using the DNeasy Powerlyzer Powersoil kit (Qiagen, Hilden, Germany).
Bioinformatic analyses.
Illumina MiSeq sequencing of the V4 region of 16S RNA gene was performed by the University of Minnesota Genomics Center. Raw FASTQ files were quality controlled using the self-learning short-read DNA pipeline SHI7 (101). After Nextera adaptor removal and paired-end read stitching, nucleotide sequences with a mean quality score greater than 36 were retained for downstream analyses. For analyses of bacterial community structure, operational taxonomic unit (OTU) picking against the NCBI RefSeq82 16S targeted loci reference database was performed using the taxonomy-aware exhaustive optimal alignment software BURST (102). Preprocessed sequences bearing a 93.5% nucleotide sequence similarity were clustered for production of OTU tables. Alpha diversity was determined using the Shannon index from OTU tables, representing the evenness and richness of OTU abundances within each sample. Differences in microbial composition between samples were represented by various measures for beta diversity, including relative genera abundances and dimensional scaling for dissimilarity of OTUs. All trends in diversity analyses were confirmed using the open-reference software package DADA2, to ensure confidence in results (103). Samples were further filtered by a minimum read count of 30,000 reads to eliminate low-confidence samples, including all controls, antibiotic-treated samples, and a single S100A9−/− ligated swab that was omitted from further analyses due to low quality (Fig. S1). Raw OTU counts obtained from the reads were normalized to convert to relative abundances per sample (104). OTUs were assigned genus level taxonomic assignments, where known, based upon 95% similarity with the RefSeq82 database. Unassigned OTUs were retained for applicable analyses.
Identification and enumeration of interstitial immune cells within gingival tissues.
Mice were injected intravenously with 1.25 μg of fluorescein isothiocyanate (FITC)-conjugated rat anti-mouse CD45 monoclonal antibody (eBioscience San Diego, CA; clone 30-F11) 3 min before euthanasia. This procedure allowed immune cells within the blood or in blood contaminating the harvested tissue to be distinguished from the interstitial immune cells of interest (105). At 2 or 5 days after ligature placement, marginal gingival tissue (proximal to the maxillary right teeth) was harvested from ligated and unligated sites. Harvested gingival tissue was pooled from two cage mates with the same genetic background and treatment, and then placed in 1 ml of complete TexMacs medium (Miltenyi Biotec, San Diego, CA). Tissues were dispersed (105) and single-cell suspensions were stained with the cell viability dye Zombie Aqua (BioLegend, San Diego, CA). Immune cell subsets were identified using a panel of fluorochrome-conjugated anti-mouse MAbs against CD45 (conjugated to PE; eBioscience; clone 30-F11), CD3 (BioLegend; clone 17A2), CD4 (eBioscience; clone RM4-5), CD8 (eBioscience; clone 53-6.7), TCR γ/δ (BioLegend; clone GL3), TCR β (BioLegend; clone H57-597), CD11b (BioLegend; clone M1/70), B220 (eBioscience; clone RA3-6B2), MHC class II (I-A/I-E; BioLegend; clone M5/114.15-2), or Ly-6G (BioLegend; clone IA8). Cell fluorescence emissions were acquired on an LSR II flow cytometer (BD Biosciences, San Jose, CA) and data were analyzed using FlowJo software (Tree Star, Ashland, OR). Immune cell subsets were quantified and normalized to 10,000 live nonimmune cells in each sample. Normalization adjusts for sample-to-sample variation in processing, loss of cells during staining, and counting variation (105). The unligated contralateral left maxillary tissues of unligated and ligated mice served as the noninflammatory control site, since the interstitial immune cell infiltrates were similar in phenotype and numbers.
Micro-computed tomography imaging and analysis.
Harvested maxillae from day 5 mice (± ligature) were treated with 4% paraformaldehyde and stored in 0.1% sodium azide. Micro-computed tomography (Micro-CT) scans were obtained using an XT H 225 scanner (Nikon Metrology, Brighton, MI) with the following parameters: energy of 108 kV, 5.8 W, 708 ms of exposure, and 720 projections (4 frames per projection). Each image taken by the micro-CT scanner was converted to a BMP file format. To generate BMP data sets, each micro-CT scan was reconstructed using CT Pro 3D (Nikon Metrology). Three-dimensional reconstructed volumes were loaded into VG Studio MAX 2.1 (Volume Graphics GmbH, Heidelberg, Germany) to generate BMP data sets, which were imported into Data Viewer (Bruker-Micro-CT, Kontich, Belgium). The maxilla of each mouse was registered in 3D against an age-matched untreated reference mouse using fixed anatomical landmarks. The landmarks, which describe a volume of interest (VOI) included: (i) length from the distal aspect of the mesio-buccal root of the first molar to the orthogonal projection of the distal aspect of the third molar root (sagittal and horizontal planes); (ii) width from the lateral aspect of the palatine foramen to the orthogonal projection of the most buccal aspect of alveolar bone proximal to the mesio-buccal root (horizontal & frontal planes); and (iii) height from the most apical edge of alveolar bone of the first molar to the orthogonal projection of the most coronal aspect of the alveolar bone crest (sagittal and frontal planes) (described in Fig. S6). After registration, a VOI was determined for the right maxillary molar region, including roots of the teeth adjacent to the second molar, and imported into the CT analyzer (CTAN; Bruker). The lower- and upper-level thresholds were set at 100 and 255 greyscale units, respectively, to discriminate bone within the VOI. Voxel size from the digital reconstruction was used to convert bone volume into cubic millimeters.
Statistics.
Statistical tests and visual aids for microbiome data were generated using the R software package. Alpha diversity, computed independently within samples, was reported using the Shannon index. Treatment groups were tested for significant differences with the Mann-Whitney U test using the wilcox.test function from the R stats package. Beta diversity, measuring between-sample microbial community differences, was reported using the Bray-Curtis dissimilarity and UniFrac distance indices (106). Corresponding ordination plots were generated using nonmetric multidimensional scaling (NMDS) of the distance matrices. Group differences were evaluated with a PERMANOVA test, executed by the adonis function in the vegan package (107).
The frequency of interstitial immune cells was analyzed and plotted using Prism 6 software (GraphPad Software, San Diego, CA, USA) and expressed as mean ± standard error of the mean (SEM). Data were evaluated by two-way analysis of variance (ANOVA) and, where appropriate (comparison of two groups only), means were compared using unpaired two-tailed Student’s t tests. A P value of <0.05 was considered statistically significant. Mean fluorescence intensity (MFI) and bone volume differences were analyzed using an unpaired Student’s t test.
ACKNOWLEDGMENTS
This work was supported by grants NIH/NIDCR 1R21DE025711 and NIH/NIDCR 1 R01DE021206 (to M.C.H.), NIH/NIDCR 1 T90 DE022732 (to M.C.H. and W.S.B.), 5F32DE028174 (W.S.B.), DE025882 (P.D.B.-E.), DE022858 (M.C.), and DE026209 (M.C.), and the Schaffer Chair for Periodontal Research at the University of Minnesota. Additional support was provided by the Office of the Vice President for Research University of Minnesota and the Associate Dean for Research, School of Dentistry.
We thank the lab of Dan Knights, Department of Computer Science and Engineering at the University of Minnesota, for helpful discussions about microbiome analyses.
Footnotes
Supplemental material is available online only.
Contributor Information
Mark C. Herzberg, Email: mcherzb@umn.edu.
Marvin Whiteley, Georgia Institute of Technology School of Biological Sciences.
REFERENCES
- 1.Eke PI, Borgnakke WS, Genco RJ. 2020. Recent epidemiologic trends in periodontitis in the USA. Periodontol 2000 82:257–267. 10.1111/prd.12323. [DOI] [PubMed] [Google Scholar]
- 2.Gaffen SL, Moutsopoulos NM. 2020. Regulation of host-microbe interactions at oral mucosal barriers by type 17 immunity. Sci Immunol 5:eaau4594. 10.1126/sciimmunol.aau4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Costalonga M, Herzberg MC. 2014. The oral microbiome and the immunobiology of periodontal disease and caries. Immunol Lett 162:22–38. 10.1016/j.imlet.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiao Y, Hasegawa M, Inohara N. 2014. The role of oral pathobionts in dysbiosis during periodontitis development. J Dent Res 93:539–546. 10.1177/0022034514528212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kitamoto S, Nagao-Kitamoto H, Jiao Y, GillillandMG, 3rd, Hayashi A, Imai J, Sugihara K, Miyoshi M, Brazil JC, Kuffa P, Hill BD, Rizvi SM, Wen F, Bishu S, Inohara N, Eaton KA, Nusrat A, Lei YL, Giannobile WV, Kamada N. 2020. The intermucosal connection between the mouth and gut in commensal pathobiont-driven colitis. Cell 182:447–462. 10.1016/j.cell.2020.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nowicki EM, Shroff R, Singleton JA, Renaud DE, Wallace D, Drury J, Zirnheld J, Colleti B, Ellington AD, Lamont RJ, Scott DA, Whiteley M. 2018. Microbiota and metatranscriptome changes accompanying the onset of gingivitis. mBio 9:e00575-18. 10.1128/mBio.00575-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Curtis MA, Diaz PI, Van Dyke TE. 2020. The role of the microbiota in periodontal disease. Periodontol 2000 83:14–25. 10.1111/prd.12296. [DOI] [PubMed] [Google Scholar]
- 8.Hajishengallis G. 2020. New developments in neutrophil biology and periodontitis. Periodontol 2000 82:78–92. 10.1111/prd.12313. [DOI] [PubMed] [Google Scholar]
- 9.Maekawa T, Tamura H, Domon H, Hiyoshi T, Isono T, Yonezawa D, Hayashi N, Takahashi N, Tabeta K, Maeda T, Oda M, Ziogas A, Alexaki V, Chavakis T, Terao Y, Hajishengallis G. 2020. Erythromycin inhibits neutrophilic inflammation and mucosal disease by upregulating DEL-1. JCI Insight 5:e136706. 10.1172/jci.insight.136706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brandtzaeg P, Gabrielsen TO, Dale I, Muller F, Steinbakk M, Fagerhol MK. 1995. The leucocyte protein L1 (calprotectin): a putative nonspecific defence factor at epithelial surfaces. Adv Exp Med Biol 371A:201–206. 10.1007/978-1-4615-1941-6_41. [DOI] [PubMed] [Google Scholar]
- 11.Lusitani D, Malawista SE, Montgomery RR. 2003. Calprotectin, an abundant cytosolic protein from human polymorphonuclear leukocytes, inhibits the growth of Borrelia burgdorferi. Infect Immun 71:4711–4716. 10.1128/IAI.71.8.4711-4716.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zaia AA, Sappington KJ, Nisapakultorn K, Chazin WJ, Dietrich EA, Ross KF, Herzberg MC. 2009. Subversion of antimicrobial calprotectin (S100A8/S100A9 complex) in the cytoplasm of TR146 epithelial cells after invasion by Listeria monocytogenes. Mucosal Immunol 2:43–53. 10.1038/mi.2008.63. [DOI] [PubMed] [Google Scholar]
- 13.Ross KF, Herzberg MC. 2016. Autonomous immunity in mucosal epithelial cells: fortifying the barrier against infection. Microbes Infect 18:387–398. 10.1016/j.micinf.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Urban CF, Ermert D, Schmid M, Abu-Abed U, Goosmann C, Nacken W, Brinkmann V, Jungblut PR, Zychlinsky A. 2009. Neutrophil extracellular traps contain calprotectin, a cytosolic protein complex involved in host defense against Candida albicans. PLoS Pathog 5:e1000639. 10.1371/journal.ppat.1000639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Striz I, Trebichavsky I. 2004. Calprotectin—a pleiotropic molecule in acute and chronic inflammation. Physiol Res 53:245–253. [PubMed] [Google Scholar]
- 16.Corbin BD, Seeley EH, Raab A, Feldmann J, Miller MR, Torres VJ, Anderson KL, Dattilo BM, Dunman PM, Gerads R, Caprioli RM, Nacken W, Chazin WJ, Skaar EP. 2008. Metal chelation and inhibition of bacterial growth in tissue abscesses. Science 319:962–965. 10.1126/science.1152449. [DOI] [PubMed] [Google Scholar]
- 17.Nisapakultorn K, Ross KF, Herzberg MC. 2001. Calprotectin expression in vitro by oral epithelial cells confers resistance to infection by Porphyromonas gingivalis. Infect Immun 69:4242–4247. 10.1128/IAI.69.7.4242-4247.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nisapakultorn K, Ross KF, Herzberg MC. 2001. Calprotectin expression inhibits bacterial binding to mucosal epithelial cells. Infect Immun 69:3692–3696. 10.1128/IAI.69.6.3692-3696.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Champaiboon C, Sappington KJ, Guenther BD, Ross KF, Herzberg MC. 2009. Calprotectin S100A9 calcium-binding loops I and II are essential for keratinocyte resistance to bacterial invasion. J Biol Chem 284:7078–7090. 10.1074/jbc.M806605200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zygiel EM, Nolan EM. 2018. Transition metal sequestration by the host-defense protein calprotectin. Annu Rev Biochem 87:621–643. 10.1146/annurev-biochem-062917-012312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brophy MB, Hayden JA, Nolan EM. 2012. Calcium ion gradients modulate the zinc affinity and antibacterial activity of human calprotectin. J Am Chem Soc 134:18089–18100. 10.1021/ja307974e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zackular JP, Chazin WJ, Skaar EP. 2015. Nutritional immunity: S100 proteins at the host-pathogen interface. J Biol Chem 290:18991–18998. 10.1074/jbc.R115.645085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kehl-Fie TE, Skaar EP. 2010. Nutritional immunity beyond iron: a role for manganese and zinc. Curr Opin Chem Biol 14:218–224. 10.1016/j.cbpa.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Damo SM, Kehl-Fie TE, Sugitani N, Holt ME, Rathi S, Murphy WJ, Zhang Y, Betz C, Hench L, Fritz G, Skaar EP, Chazin WJ. 2013. Molecular basis for manganese sequestration by calprotectin and roles in the innate immune response to invading bacterial pathogens. Proc Natl Acad Sci U S A 110:3841–3846. 10.1073/pnas.1220341110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kehl-Fie TE, Chitayat S, Hood MI, Damo S, Restrepo N, Garcia C, Munro KA, Chazin WJ, Skaar EP. 2011. Nutrient metal sequestration by calprotectin inhibits bacterial superoxide defense, enhancing neutrophil killing of Staphylococcus aureus. Cell Host Microbe 10:158–164. 10.1016/j.chom.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gomez NO, Tetard A, Ouerdane L, Laffont C, Brutesco C, Ball G, Lobinski R, Denis Y, Plesiat P, Llanes C, Arnoux P, Voulhoux R. 2021. Involvement of the Pseudomonas aeruginosa MexAB-OprM efflux pump in the secretion of the metallophore pseudopaline. Mol Microbiol 115:84–98. 10.1111/mmi.14600. [DOI] [PubMed] [Google Scholar]
- 27.Grim KP, San Francisco B, Radin JN, Brazel EB, Kelliher JL, Parraga Solorzano PK, Kim PC, McDevitt CA, Kehl-Fie TE. 2017. The metallophore staphylopine enables Staphylococcus aureus to compete with the host for zinc and overcome nutritional immunity. mBio 8:e01281-17. 10.1128/mBio.01281-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J, Lonergan ZR, Gonzalez-Gutierrez G, Nairn BL, Maxwell CN, Zhang Y, Andreini C, Karty JA, Chazin WJ, Trinidad JC, Skaar EP, Giedroc DP. 2019. Multi-metal restriction by calprotectin impacts de novo flavin biosynthesis in Acinetobacter baumannii. Cell Chem Biol 26:745–755. 10.1016/j.chembiol.2019.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Behnsen J, Jellbauer S, Wong CP, Edwards RA, George MD, Ouyang W, Raffatellu M. 2014. The cytokine IL-22 promotes pathogen colonization by suppressing related commensal bacteria. Immunity 40:262–273. 10.1016/j.immuni.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diaz-Ochoa VE, Lam D, Lee CS, Klaus S, Behnsen J, Liu JZ, Chim N, Nuccio SP, Rathi SG, Mastroianni JR, Edwards RA, Jacobo CM, Cerasi M, Battistoni A, Ouellette AJ, Goulding CW, Chazin WJ, Skaar EP, Raffatellu M. 2016. Salmonella mitigates oxidative stress and thrives in the inflamed gut by evading calprotectin-mediated manganese sequestration. Cell Host Microbe 19:814–825. 10.1016/j.chom.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu JZ, Jellbauer S, Poe AJ, Ton V, Pesciaroli M, Kehl-Fie TE, Restrepo NA, Hosking MP, Edwards RA, Battistoni A, Pasquali P, Lane TE, Chazin WJ, Vogl T, Roth J, Skaar EP, Raffatellu M. 2012. Zinc sequestration by the neutrophil protein calprotectin enhances Salmonella growth in the inflamed gut. Cell Host Microbe 11:227–239. 10.1016/j.chom.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ren XY, Xu L, Meng HX, Zhao HS, Lu RF, Chen ZB, Feng XH, Shi D, Zhang L, Tian Y. 2009. Family-based association analysis of S100A8 genetic polymorphisms with aggressive periodontitis. J Periodontal Res 44:184–192. 10.1111/j.1600-0765.2008.01103.x. [DOI] [PubMed] [Google Scholar]
- 33.Gauderman WJ, Witte JS, Thomas DC. 1999. Family-based association studies. J Natl Cancer Inst Monogr 1999:31–37. 10.1093/oxfordjournals.jncimonographs.a024223. [DOI] [PubMed] [Google Scholar]
- 34.Rammes A, Roth J, Goebeler M, Klempt M, Hartmann M, Sorg C. 1997. Myeloid-related protein (MRP) 8 and MRP14, calcium-binding proteins of the S100 family, are secreted by activated monocytes via a novel, tubulin-dependent pathway. J Biol Chem 272:9496–9502. 10.1074/jbc.272.14.9496. [DOI] [PubMed] [Google Scholar]
- 35.Hetland G, Talgo GJ, Fagerhol MK. 1998. Chemotaxins C5a and fMLP induce release of calprotectin (leucocyte L1 protein) from polymorphonuclear cells in vitro. Mol Pathol 51:143–148. 10.1136/mp.51.3.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakamura T, Kido J, Kido R, Ohishi K, Yamauchi N, Kataoka M, Nagata T. 2000. The association of calprotectin level in gingival crevicular fluid with gingival index and the activities of collagenase and aspartate aminotransferase in adult periodontitis patients. J Periodontol 71:361–367. 10.1902/jop.2000.71.3.361. [DOI] [PubMed] [Google Scholar]
- 37.Berg-Hansen P, Vandvik B, Fagerhol M, Holmoy T. 2009. Calprotectin levels in the cerebrospinal fluid reflect disease activity in multiple sclerosis. J Neuroimmunol 216:98–102. 10.1016/j.jneuroim.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 38.Jensen LJ, Kistorp C, Bjerre M, Raymond I, Flyvbjerg A. 2012. Plasma calprotectin levels reflect disease severity in patients with chronic heart failure. Eur J Prev Cardiolog 19:999–1004. 10.1177/1741826711421078. [DOI] [PubMed] [Google Scholar]
- 39.Abildtrup M, Kingsley GH, Scott DL. 2015. Calprotectin as a biomarker for rheumatoid arthritis: a systematic review. J Rheumatol 42:760–770. 10.3899/jrheum.140628. [DOI] [PubMed] [Google Scholar]
- 40.Maekawa S, Onizuka S, Katagiri S, Hatasa M, Ohsugi Y, Sasaki N, Watanabe K, Ohtsu A, Komazaki R, Ogura K, Miyoshi-Akiyama T, Iwata T, Nitta H, Izumi Y. 2019. RNA sequencing for ligature induced periodontitis in mice revealed important role of S100A8 and S100A9 for periodontal destruction. Sci Rep 9:14663. 10.1038/s41598-019-50959-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wei L, Liu M, Xiong H. 2019. Role of calprotectin as a biomarker in periodontal disease. Mediators Inflamm 2019:3515026. 10.1155/2019/3515026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakashige TG, Zhang B, Krebs C, Nolan EM. 2015. Human calprotectin is an iron-sequestering host-defense protein. Nat Chem Biol 11:765–771. 10.1038/nchembio.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zackular JP, Moore JL, Jordan AT, Juttukonda LJ, Noto MJ, Nicholson MR, Crews JD, Semler MW, Zhang Y, Ware LB, Washington MK, Chazin WJ, Caprioli RM, Skaar EP. 2016. Erratum: Dietary zinc alters the microbiota and decreases resistance to Clostridium difficile infection. Nat Med 22:1502. 10.1038/nm1216-1502d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zackular JP, Moore JL, Jordan AT, Juttukonda LJ, Noto MJ, Nicholson MR, Crews JD, Semler MW, Zhang Y, Ware LB, Washington MK, Chazin WJ, Caprioli RM, Skaar EP. 2016. Dietary zinc alters the microbiota and decreases resistance to Clostridium difficile infection. Nat Med 22:1330–1334. 10.1038/nm.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakashige TG, Zygiel EM, Drennan CL, Nolan EM. 2017. Nickel sequestration by the host-defense protein human calprotectin. J Am Chem Soc 139:8828–8836. 10.1021/jacs.7b01212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Besold AN, Gilston BA, Radin JN, Ramsoomair C, Culbertson EM, Li CX, Cormack BP, Chazin WJ, Kehl-Fie TE, Culotta VC. 2017. Role of calprotectin in withholding zinc and copper from Candida albicans. Infect Immun 86:e00779-17. 10.1128/IAI.00779-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Urban CF, Nett JE. 2019. Neutrophil extracellular traps in fungal infection. Semin Cell Dev Biol 89:47–57. 10.1016/j.semcdb.2018.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ehrchen JM, Sunderkotter C, Foell D, Vogl T, Roth J. 2009. The endogenous Toll-like receptor 4 agonist S100A8/S100A9 (calprotectin) as innate amplifier of infection, autoimmunity, and cancer. J Leukoc Biol 86:557–566. 10.1189/jlb.1008647. [DOI] [PubMed] [Google Scholar]
- 49.Chen B, Miller AL, Rebelatto M, Brewah Y, Rowe DC, Clarke L, Czapiga M, Rosenthal K, Imamichi T, Chen Y, Chang CS, Chowdhury PS, Naiman B, Wang Y, Yang D, Humbles AA, Herbst R, Sims GP. 2015. S100A9 induced inflammatory responses are mediated by distinct damage associated molecular patterns (DAMP) receptors in vitro and in vivo. PLoS One 10:e0115828. 10.1371/journal.pone.0115828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chernov AV, Dolkas J, Hoang K, Angert M, Srikrishna G, Vogl T, Baranovskaya S, Strongin AY, Shubayev VI. 2015. The calcium-binding proteins S100A8 and S100A9 initiate the early inflammatory program in injured peripheral nerves. J Biol Chem 290:11771–11784. 10.1074/jbc.M114.622316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Crowe LAN, McLean M, Kitson SM, Melchor EG, Patommel K, Cao HM, Reilly JH, Leach WJ, Rooney BP, Spencer SJ, Mullen M, Chambers M, Murrell GAC, McInnes IB, Akbar M, Millar NL. 2019. S100A8 & S100A9: alarmin mediated inflammation in tendinopathy. Sci Rep 9:1463. 10.1038/s41598-018-37684-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schenten V, Plancon S, Jung N, Hann J, Bueb JL, Brechard S, Tschirhart EJ, Tolle F. 2018. Secretion of the phosphorylated form of S100A9 from neutrophils is essential for the proinflammatory functions of extracellular S100A8/A9. Front Immunol 9:447. 10.3389/fimmu.2018.00447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gao H, Zhang X, Zheng Y, Peng L, Hou J, Meng H. 2015. S100A9-induced release of interleukin (IL)-6 and IL-8 through toll-like receptor 4 (TLR4) in human periodontal ligament cells. Mol Immunol 67:223–232. 10.1016/j.molimm.2015.05.014. [DOI] [PubMed] [Google Scholar]
- 54.Roy D, Ramasamy R, Schmidt AM. 2021. Journey to a receptor for advanced glycation end products connection in severe acute respiratory syndrome coronavirus 2 infection: with stops along the way in the lung, heart, blood vessels, and adipose tissue. ATVB 41:614–627. 10.1161/ATVBAHA.120.315527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Achouiti A, Vogl T, Urban CF, Rohm M, Hommes TJ, van Zoelen MA, Florquin S, Roth J, van 't Veer C, de Vos AF, van der Poll T. 2012. Myeloid-related protein-14 contributes to protective immunity in Gram-negative pneumonia derived sepsis. PLoS Pathog 8:e1002987. 10.1371/journal.ppat.1002987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Achouiti A, Vogl T, Van der Meer AJ, Stroo I, Florquin S, de Boer OJ, Roth J, Zeerleder S, van 't Veer C, de Vos AF, van der Poll T. 2015. Myeloid-related protein-14 deficiency promotes inflammation in staphylococcal pneumonia. Eur Respir J 46:464–473. 10.1183/09031936.00183814. [DOI] [PubMed] [Google Scholar]
- 57.Palmer LD, Maloney KN, Boyd KL, Goleniewska AK, Toki S, Maxwell CN, Chazin WJ, PeeblesRS, Jr, Newcomb DC, Skaar EP. 2019. The innate immune protein S100A9 protects from T-helper cell type 2-mediated allergic airway inflammation. Am J Respir Cell Mol Biol 61:459–468. 10.1165/rcmb.2018-0217OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zackular JP, Skaar EP. 2018. The role of zinc and nutritional immunity in Clostridium difficile infection. Gut Microbes 9:469–476. 10.1080/19490976.2018.1448354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abe T, Hajishengallis G. 2013. Optimization of the ligature-induced periodontitis model in mice. J Immunol Methods 394:49–54. 10.1016/j.jim.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hobbs JA, May R, Tanousis K, McNeill E, Mathies M, Gebhardt C, Henderson R, Robinson MJ, Hogg N. 2003. Myeloid cell function in MRP-14 (S100A9) null mice. Mol Cell Biol 23:2564–2576. 10.1128/MCB.23.7.2564-2576.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moutsopoulos NM, Konkel J, Sarmadi M, Eskan MA, Wild T, Dutzan N, Abusleme L, Zenobia C, Hosur KB, Abe T, Uzel G, Chen W, Chavakis T, Holland SM, Hajishengallis G. 2014. Defective neutrophil recruitment in leukocyte adhesion deficiency type I disease causes local IL-17-driven inflammatory bone loss. Sci Transl Med 6:229ra40. 10.1126/scitranslmed.3007696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rovin S, Costich ER, Gordon HA. 1966. The influence of bacteria and irritation in the initiation of periodontal disease in germfree and conventional rats. J Periodontal Res 1:193–204. 10.1111/j.1600-0765.1966.tb01860.x. [DOI] [PubMed] [Google Scholar]
- 63.Xiao E, Mattos M, Vieira GHA, Chen S, Correa JD, Wu Y, Albiero ML, Bittinger K, Graves DT. 2017. Diabetes enhances IL-17 expression and alters the oral microbiome to increase its pathogenicity. Cell Host Microbe 22:120–128. 10.1016/j.chom.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miyasaki KT, Bodeau AL, Murthy AR, Lehrer RI. 1993. In vitro antimicrobial activity of the human neutrophil cytosolic S-100 protein complex, calprotectin, against Capnocytophaga sputigena. J Dent Res 72:517–523. 10.1177/00220345930720020801. [DOI] [PubMed] [Google Scholar]
- 65.Ulas T, Pirr S, Fehlhaber B, Bickes MS, Loof TG, Vogl T, Mellinger L, Heinemann AS, Burgmann J, Schoning J, Schreek S, Pfeifer S, Reuner F, Vollger L, Stanulla M, von Kockritz-Blickwede M, Glander S, Barczyk-Kahlert K, von Kaisenberg CS, Friesenhagen J, Fischer-Riepe L, Zenker S, Schultze JL, Roth J, Viemann D. 2017. S100-alarmin-induced innate immune programming protects newborn infants from sepsis. Nat Immunol 18:622–632. 10.1038/ni.3745. [DOI] [PubMed] [Google Scholar]
- 66.Willers M, Ulas T, Vollger L, Vogl T, Heinemann AS, Pirr S, Pagel J, Fehlhaber B, Halle O, Schoning J, Schreek S, Lober U, Essex M, Hombach P, Graspeuntner S, Basic M, Bleich A, Cloppenborg-Schmidt K, Kunzel S, Jonigk D, Rupp J, Hansen G, Forster R, Baines JF, Hartel C, Schultze JL, Forslund SK, Roth J, Viemann D. 2020. S100A8 and S100A9 are important for postnatal development of gut microbiota and immune system in mice and infants. Gastroenterology 159:2130–2145. 10.1053/j.gastro.2020.08.019. [DOI] [PubMed] [Google Scholar]
- 67.Wirtz TH, Buendgens L, Weiskirchen R, Loosen SH, Haehnsen N, Puengel T, Abu Jhaisha S, Brozat JF, Hohlstein P, Koek G, Eisert A, Mohr R, Roderburg C, Luedde T, Trautwein C, Tacke F, Koch A. 2020. Association of serum calprotectin concentrations with mortality in critically ill and septic patients. Diagnostics 10:990. 10.3390/diagnostics10110990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lin CR, Wei TY, Tsai HY, Wu YT, Wu PY, Chen ST. 2015. Glycosylation-dependent interaction between CD69 and S100A8/S100A9 complex is required for regulatory T-cell differentiation. FASEB J 29:5006–5017. 10.1096/fj.15-273987. [DOI] [PubMed] [Google Scholar]
- 69.Dutzan N, Kajikawa T, Abusleme L, Greenwell-Wild T, Zuazo CE, Ikeuchi T, Brenchley L, Abe T, Hurabielle C, Martin D, Morell RJ, Freeman AF, Lazarevic V, Trinchieri G, Diaz PI, Holland SM, Belkaid Y, Hajishengallis G, Moutsopoulos NM. 2018. A dysbiotic microbiome triggers TH17 cells to mediate oral mucosal immunopathology in mice and humans. Sci Transl Med 10:eaat0797. 10.1126/scitranslmed.aat0797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Abusleme L, O’Gorman H, Dutzan N, Greenwell-Wild T, Moutsopoulos NM. 2020. Establishment and stability of the murine oral microbiome. J Dent Res 99:721–729. 10.1177/0022034520915485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brophy MB, Nolan EM. 2015. Manganese and microbial pathogenesis: sequestration by the mammalian immune system and utilization by microorganisms. ACS Chem Biol 10:641–651. 10.1021/cb500792b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hadley RC, Gagnon DM, Ozarowski A, Britt RD, Nolan EM. 2019. Murine calprotectin coordinates Mn(II) at a hexahistidine site with Ca(II)-dependent affinity. Inorg Chem 58:13578–13590. 10.1021/acs.inorgchem.9b00763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Burcham LR, Le Breton Y, Radin JN, Spencer BL, Deng L, Hiron A, Ransom MR, Mendonca JDC, Belew AT, El-Sayed NM, McIver KS, Kehl-Fie TE, Doran KS. 2020. Identification of zinc-dependent mechanisms used by group B Streptococcus to overcome calprotectin-mediated stress. mBio 11:e02302-20. 10.1128/mBio.02302-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zygiel EM, Nelson CE, Brewer LK, Oglesby-Sherrouse AG, Nolan EM. 2019. The human innate immune protein calprotectin induces iron starvation responses in Pseudomonas aeruginosa. J Biol Chem 294:3549–3562. 10.1074/jbc.RA118.006819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kehl-Fie TE, Zhang Y, Moore JL, Farrand AJ, Hood MI, Rathi S, Chazin WJ, Caprioli RM, Skaar EP. 2013. MntABC and MntH contribute to systemic Staphylococcus aureus infection by competing with calprotectin for nutrient manganese. Infect Immun 81:3395–3405. 10.1128/IAI.00420-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hood MI, Mortensen BL, Moore JL, Zhang Y, Kehl-Fie TE, Sugitani N, Chazin WJ, Caprioli RM, Skaar EP. 2012. Identification of an Acinetobacter baumannii zinc acquisition system that facilitates resistance to calprotectin-mediated zinc sequestration. PLoS Pathog 8:e1003068. 10.1371/journal.ppat.1003068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gaddy JA, Radin JN, Loh JT, Piazuelo MB, Kehl-Fie TE, Delgado AG, Ilca FT, Peek RM, Cover TL, Chazin WJ, Skaar EP, Scott Algood HM. 2014. The host protein calprotectin modulates the Helicobacter pylori cag type IV secretion system via zinc sequestration. PLoS Pathog 10:e1004450. 10.1371/journal.ppat.1004450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Colomer-Winter C, Flores-Mireles AL, Baker SP, Frank KL, Lynch AJL, Hultgren SJ, Kitten T, Lemos JA. 2018. Manganese acquisition is essential for virulence of Enterococcus faecalis. PLoS Pathog 14:e1007102. 10.1371/journal.ppat.1007102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Antelo GT, Vila AJ, Giedroc DP, Capdevila DA. 2020. Molecular evolution of transition metal bioavailability at the host-pathogen interface. Trends Microbiol 29:441–457. 10.1016/j.tim.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Baumann T, Bereiter R, Lussi A, Carvalho TS. 2017. The effect of different salivary calcium concentrations on the erosion protection conferred by the salivary pellicle. Sci Rep 7:12999. 10.1038/s41598-017-13367-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Majster M, Almer S, Bostrom EA. 2019. Salivary calprotectin is elevated in patients with active inflammatory bowel disease. Arch Oral Biol 107:104528. 10.1016/j.archoralbio.2019.104528. [DOI] [PubMed] [Google Scholar]
- 82.Brini M, Ottolini D, Cali T, Carafoli E. 2013. Calcium in health and disease. Met Ions Life Sci 13:81–137. 10.1007/978-94-007-7500-8_4. [DOI] [PubMed] [Google Scholar]
- 83.Stephan JR, Nolan EM. 2016. Calcium-induced tetramerization and zinc chelation shield human calprotectin from degradation by host and bacterial extracellular proteases. Chem Sci 7:1962–1975. 10.1039/C5SC03287C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hadley RC, Gu Y, Nolan EM. 2018. Initial biochemical and functional evaluation of murine calprotectin reveals Ca(II)-dependence and its ability to chelate multiple nutrient transition metal ions. Biochemistry 57:2846–2856. 10.1021/acs.biochem.8b00309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gao H, Xu J, He L, Meng H, Hou J. 2021. Calprotectin levels in gingival crevicular fluid and serum of patients with chronic periodontitis and type 2 diabetes mellitus before and after initial periodontal therapy. J Periodontal Res 56:121–130. 10.1111/jre.12800. [DOI] [PubMed] [Google Scholar]
- 86.Kaslick RS, Chasens AI, Mandel ID, Weinstein D, Waldman R, Pluhar T, Lazzara R. 1970. Quantitative analysis of sodium, potassium and calcium in gingival fluid from gingiva in varying degrees of inflammation. J Periodontol 41:93–97. 10.1902/jop.1970.41.2.93. [DOI] [PubMed] [Google Scholar]
- 87.Olszak IT, Poznansky MC, Evans RH, Olson D, Kos C, Pollak MR, Brown EM, Scadden DT. 2000. Extracellular calcium elicits a chemokinetic response from monocytes in vitro and in vivo. J Clin Invest 105:1299–1305. 10.1172/JCI9799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vogl T, Stratis A, Wixler V, Voller T, Thurainayagam S, Jorch SK, Zenker S, Dreiling A, Chakraborty D, Frohling M, Paruzel P, Wehmeyer C, Hermann S, Papantonopoulou O, Geyer C, Loser K, Schafers M, Ludwig S, Stoll M, Leanderson T, Schultze JL, Konig S, Pap T, Roth J. 2018. Autoinhibitory regulation of S100A8/S100A9 alarmin activity locally restricts sterile inflammation. J Clin Invest 128:1852–1866. 10.1172/JCI89867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Eversole LR, Miyasaki KT, Christensen RE. 1993. Keratinocyte expression of calprotectin in oral inflammatory mucosal diseases. J Oral Pathol Med 22:303–307. 10.1111/j.1600-0714.1993.tb01077.x. [DOI] [PubMed] [Google Scholar]
- 90.Ross KF, Herzberg MC. 2001. Calprotectin expression by gingival epithelial cells. Infect Immun 69:3248–3254. 10.1128/IAI.69.5.3248-3254.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schlegel Gomez R, Langer P, Pelka M, von den Driesch P, Johannessen AC, SimonM, Jr. 1995. Variational expression of functionally different macrophage markers (27E10, 25F9, RM3/1) in normal gingiva and inflammatory periodontal disease. J Clin Periodontol 22:341–346. 10.1111/j.1600-051X.1995.tb00159.x. [DOI] [PubMed] [Google Scholar]
- 92.Nishii K, Usui M, Yamamoto G, Yajima S, Tsukamoto Y, Tanaka J, Tachikawa T, Yamamoto M. 2013. The distribution and expression of S100A8 and S100A9 in gingival epithelium of mice. J Periodontal Res 48:235–242. 10.1111/jre.12000. [DOI] [PubMed] [Google Scholar]
- 93.Sorenson BS, Khammanivong A, Guenther BD, Ross KF, Herzberg MC. 2012. IL-1 receptor regulates S100A8/A9-dependent keratinocyte resistance to bacterial invasion. Mucosal Immunol 5:66–75. 10.1038/mi.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rollenhagen C, Wollert T, Langford GM, Sundstrom P. 2009. Stimulation of cell motility and expression of late markers of differentiation in human oral keratinocytes by Candida albicans. Cell Microbiol 11:946–966. 10.1111/j.1462-5822.2009.01303.x. [DOI] [PubMed] [Google Scholar]
- 95.Leite FRM, Nascimento GG, Baake S, Pedersen LD, Scheutz F, Lopez R. 2019. Impact of smoking cessation on periodontitis: a systematic review and meta-analysis of prospective longitudinal observational and interventional studies. Nicotine Tob Res 21:1600–1608. 10.1093/ntr/nty147. [DOI] [PubMed] [Google Scholar]
- 96.Leite FRM, Nascimento GG, Scheutz F, Lopez R. 2018. Effect of smoking on periodontitis: a systematic review and meta-regression. Am J Prev Med 54:831–841. 10.1016/j.amepre.2018.02.014. [DOI] [PubMed] [Google Scholar]
- 97.Railwah C, Lora A, Zahid K, Goldenberg H, Campos M, Wyman A, Jundi B, Ploszaj M, Rivas M, Dabo A, Majka SM, Foronjy R, El Gazzar M, Geraghty P. 2020. Cigarette smoke induction of S100A9 contributes to chronic obstructive pulmonary disease. Am J Physiol Lung Cell Mol Physiol 319:L1021–L1035. 10.1152/ajplung.00207.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Guo M, Tao W, Flavell RA, Zhu S. 2021. Potential intestinal infection and faecal-oral transmission of SARS-CoV-2. Nat Rev Gastroenterol Hepatol 18:269–283. 10.1038/s41575-021-00416-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Staley C, Kaiser T, Beura LK, Hamilton MJ, Weingarden AR, Bobr A, Kang J, Masopust D, Sadowsky MJ, Khoruts A. 2017. Stable engraftment of human microbiota into mice with a single oral gavage following antibiotic conditioning. Microbiome 5:87. 10.1186/s40168-017-0306-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fiege JK, Stone IA, Dumm RE, Waring BM, Fife BT, Agudo J, Brown BD, Heaton NS, Langlois RA. 2019. Long-term surviving influenza infected cells evade CD8+ T cell mediated clearance. PLoS Pathog 15:e1008077. 10.1371/journal.ppat.1008077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Al-Ghalith GA, Hillmann B, Ang K, Shields-Cutler R, Knights D. 2018. SHI7 is a self-learning pipeline for multipurpose short-read DNA quality control. mSystems 3:e00202-17. 10.1128/mSystems.00202-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hillmann B, Al-Ghalith GA, Shields-Cutler RR, Zhu Q, Gohl DM, Beckman KB, Knight R, Knights D. 2018. Evaluating the information content of shallow shotgun metagenomics. mSystems 3:e00069-18. 10.1128/mSystems.00069-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.McMurdie PJ. 2018. Normalization of microbiome profiling data. Methods Mol Biol 1849:143–168. 10.1007/978-1-4939-8728-3_10. [DOI] [PubMed] [Google Scholar]
- 105.Bittner-Eddy PD, Fischer LA, Tu AA, Allman DA, Costalonga M. 2017. Discriminating between interstitial and circulating leukocytes in tissues of the murine oral mucosa avoiding nasal-associated lymphoid tissue contamination. Front Immunol 8:1398. 10.3389/fimmu.2017.01398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hamady M, Lozupone C, Knight R. 2010. Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J 4:17–27. 10.1038/ismej.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O'Hara RB, Simpson GL, Solymos P, Henry M, Stevens H, Szoecs E, Wagner H. 2019. Community Ecology Package. R package version 2.5-6. https://CRAN.R-project.org/package=vegan.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download IAI.00122-21-s0001.pdf, PDF file, 1.9 MB (1.9MB, pdf)