ABSTRACT
Riboflavin is an essential micronutrient, but its transport and utilization have remained largely understudied among pathogenic spirochetes. Here, we show that Borrelia burgdorferi, the zoonotic spirochete that causes Lyme disease, is able to import riboflavin via products of its rfuABCD-like operon as well as synthesize flavin mononucleotide and flavin adenine dinucleotide despite lacking canonical genes for their synthesis. Additionally, a mutant deficient in the rfuABCD-like operon is resistant to the antimicrobial effect of roseoflavin, a natural riboflavin analog, and is attenuated in a murine model of Lyme borreliosis. Our combined results indicate not only that are riboflavin and the maintenance of flavin pools essential for B. burgdorferi growth but also that flavin utilization and its downstream products (e.g., flavoproteins) may play a more prominent role in B. burgdorferi pathogenesis than previously appreciated.
KEYWORDS: Borrelia burgdorferi, Lyme disease, RfuABCD, riboflavin, roseoflavin
INTRODUCTION
Microorganisms have evolved elaborate mechanisms to acquire the essential micronutrient riboflavin (RF) (vitamin B2) (1). The RF biosynthetic pathway is an energetically costly process that requires the expression of enzymes encoded by the rib operon (ribDEABH) and metabolic precursors originating from purine biosynthesis (GTP) (2, 3) and the pentose phosphate pathway (ribulose-5 phosphate) (4). Additionally, the synthesis of one molecule of RF may require up to 25 molecules of ATP, whereas the uptake of RF requires only a few molecules of ATP (5). Despite the high energy cost, many bacteria, such as Vibrio cholerae (6), Clostridioides (formerly Clostridium) difficile (7), and methicillin-resistant Staphylococcus aureus (8), maintain both RF biosynthesis and uptake mechanisms, underscoring the importance of acquiring a sufficient supply of RF. Some other pathogenic bacteria, such as Listeria monocytogenes, Enterococcus faecalis, and Treponema pallidum, lack RF biosynthesis genes and thus must acquire RF from their environment. It is unknown why these pathogenic bacteria have dispensed with the ability to synthesize RF, but it likely is predicated on their ability to acquire sufficient quantities of exogenous RF through efficient import mechanisms. To date, nine different families of bacterial RF transporters have been identified. They are ImpX, RibM, RibN, RibU, RibV, RibXY, RibZ, RfnT (1), and RfuABCD, an ATP binding-cassette (ABC)-type uptake system recently reported by us for Treponema pallidum (9).
Treponema pallidum, the syphilis spirochete, relies heavily on flavin-dependent processes to satisfy a number of its physiological demands (10, 11), engendering what we have termed a “flavin-centric” metabolic lifestyle (12). Ideally, it would be advantageous to investigate the role of the rfuABCD gene products in RF transport in T. pallidum. However, despite a recent advance in the in vitro cultivation of T. pallidum (13), the spirochete remains genetically intractable. Thus, investigating the treponemal RfuABCD system for RF transport remains unachievable at this time. However, Borrelia burgdorferi, the causative agent of Lyme disease, also encodes a putative RfuABCD-like system (bb0319–bb0316) (14) and can be readily cultivated and genetically manipulated in vitro. As such, in this study, we utilized B. burgdorferi as a representative pathogenic spirochete to examine salient features of the putative RfuABCD transport system (9).
Borrelia burgdorferi, like T. pallidum, lacks many of the biosynthetic pathways involved in the de novo synthesis of amino acids, fatty acids, nucleotides, and other cofactors (14, 15). Bioinformatic analysis suggests that B. burgdorferi does not carry genes for an RF biosynthetic pathway or genes involved in the synthesis of flavin mononucleotide (FMN) and flavin adenine dinucleotide (FAD) (14, 15). Not only do FMN and FAD serve as cofactors for various enzymes and flavoproteins involved in oxidation-reduction reactions, but they also are necessary for cellular metabolism and energy production (16, 17). Thus, it is puzzling why B. burgdorferi would encode an RF uptake mechanism but lack the bifunctional FMN/FAD synthetase found in other bacteria (18, 19). Nevertheless, the operon containing bb0319 to bb0316 likely encodes B. burgdorferi’s RfuABCD-like RF uptake machinery, inasmuch as previous investigations in our laboratory have indicated that recombinant BB_0319 is able to bind RF in vitro (9). However, it is still unknown whether B. burgdorferi transports RF or what role this operon plays in borrelial growth in vitro and in vivo. Additionally, direct investigations of RF transport and RF interconversion in B. burgdorferi thus far have not been conducted. In this study, we focused on examining the presumed role of the RfuABCD-like operon in RF transport and assessed the synthesis of FMN and FAD in B. burgdorferi. We also examined the extent to which the RfuABCD-like operon might influence the infectivity phenotype of pathogenic B. burgdorferi in the murine model of Lyme borreliosis.
RESULTS
Borrelia burgdorferi imports RF.
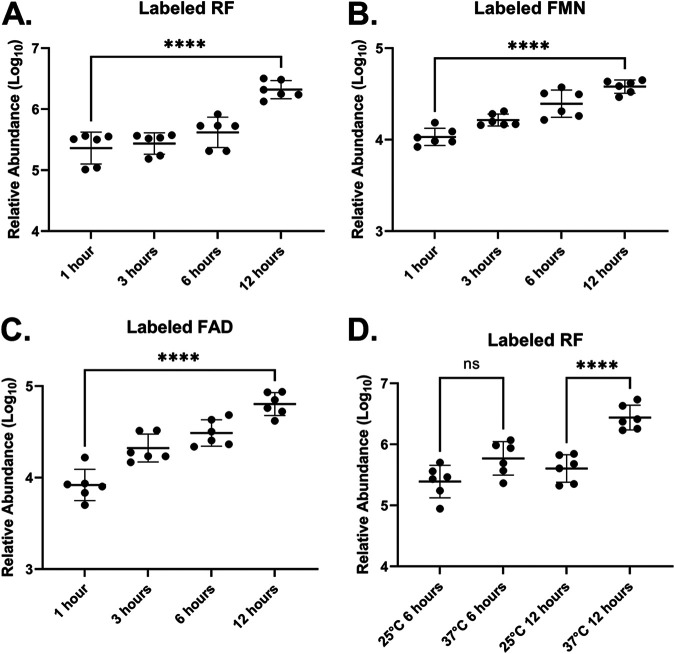
We postulated that B. burgdorferi could import RF because it carries an rfuABCD-like transport operon (bb0319–bb0316) and because we previously demonstrated that recombinant BB_0319 (RfuA) binds RF in vitro (9). To first assess RF uptake by B. burgdorferi, spirochetes were incubated at 37°C in Barbour-Stoenner-Kelly II (BSK-II) medium in the presence of 100 μM labeled RF (riboflavin-[13C4,15N2]dioxopyrimidine). After sequential collection of borreliae and subsequent analysis by liquid chromatography-mass spectrometry (LC-MS), labeled RF increased in a time-dependent manner among sampled spirochetes (Fig. 1A), indicating that B. burgdorferi imports RF from its environment.
FIG 1.
Borrelia burgdorferi B31 imports RF and synthesizes FMN and FAD. B. burgdorferi was grown to late log phase and then incubated at 37°C (A to C) or the indicated temperatures (D) in medium supplemented with labeled RF for the indicated times. Cell pellets were washed twice with BSK-II medium and then frozen. Labeled RF, FMN, and FAD concentrations were determined by LC-MS. Each data point represents a biological replicate (n = 6) from three independent experiments. Bars represent the averages of the displayed data points, and error bars indicate standard deviations (SD). One-way ANOVA with Sidak’s post hoc test was used for multiple comparisons. ns, not significant; ****, P < 0.0001.
B. burgdorferi converts RF to FMN and FAD.
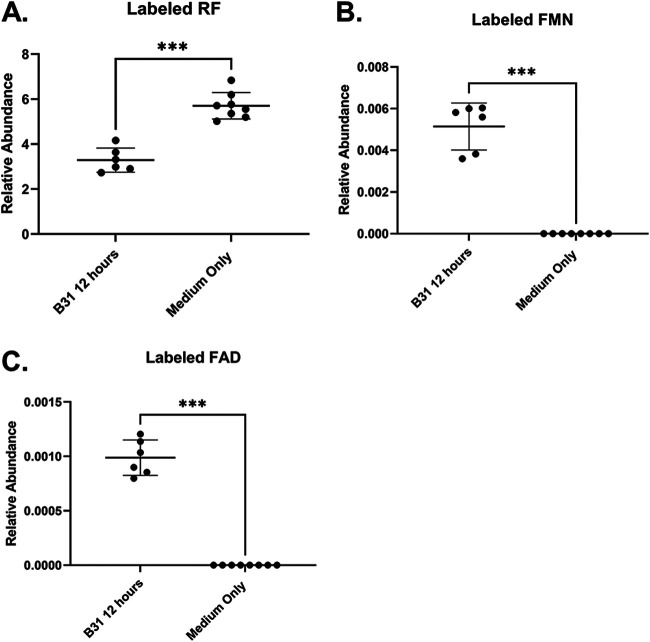
The most common biologically active flavins are FMN and FAD, which serve as cofactors in flavoproteins. These cofactors typically are generated in bacteria by a bifunctional FMN/FAD synthetase (20). However, bioinformatic analyses of the B. burgdorferi genome failed to identify enzymes required for the synthesis of FMN or FAD from RF (14). To investigate whether B. burgdorferi has the capacity to synthesize FMN and FAD, spirochete cultures grown in the presence of labeled RF were examined for the appearance of labeled FMN and FAD. Analysis by LC-MS detected increased concentrations of labeled FMN (Fig. 1B) and FAD (Fig. 1C) among the serially collected spirochete samples. Additionally, when uninoculated medium was compared with medium containing growing spirochetes, only samples from the active Borrelia cultures yielded labeled FMN and FAD (Fig. 2B and C), supporting the idea that FMN and FAD were synthesized by B. burgdorferi and were not somehow converted by the BSK-II medium alone. These results demonstrate that B. burgdorferi is capable of converting RF to FMN and FAD.
FIG 2.
Labeled FMN and FAD appear only in cultures containing B. burgdorferi. B. burgdorferi was grown at 37°C in BSK-II medium supplemented for 12 h with labeled RF. The supernatant from pelleted cultures was frozen and analyzed by LC-MS. A medium-only control was also analyzed to ensure that the labeled RF was not contaminated with labeled FMN or FAD. (A) Labeled RF was detected in the medium of the inoculated culture supernatants and in the medium-only controls. However, labeled FMN (B) and FAD (C) were detected only in the supernatants of the B. burgdorferi cultures. Each data point represents a biological replicate (n = 6) from three independent experiments. Data are normalized to an internal standard. Bars represent the averages of the displayed data points, and error bars indicate SD. One-way ANOVA with Sidak’s post hoc test was used for multiple comparisons. ***, P < 0.001.
RF transport by B. burgdorferi is influenced by temperature.
It is well documented that environmental temperature markedly influences both the growth rate and transcriptional profile of B. burgdorferi (21). In fact, the cultivation of B. burgdorferi at room temperature (25°C) is one parameter used to partially mimic B. burgdorferi’s residence within its tick host environment (22, 23). To investigate the potential influence of temperature on RF transport/utilization by B. burgdorferi, spirochetes were incubated in BSK-II medium at either 25°C or 37°C after the addition of 100 μM labeled RF. Consistent with previous results, labeled RF accumulated over time among spirochetes incubated at 37°C (Fig. 1D). However, labeled RF did not accumulate to similar levels when borreliae were incubated at 25°C (Fig. 1D). These results support the ideas that (i) RF import by B. burgdorferi is influenced, at least in part, by environmental temperature, with the implication that it is likely accomplished by an active uptake mechanism, and (ii) RF seems not to passively traverse the cytoplasmic membrane of B. burgdorferi. The latter conclusion is supported by the fact that RF uptake did not increase in cultures incubated at 25°C for up to 12 h.
Involvement of the rfuABCD-like gene cluster in RF transport.
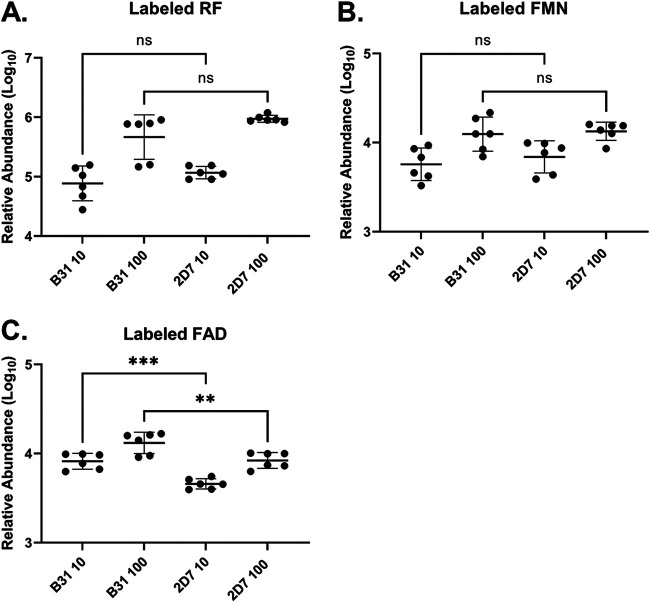
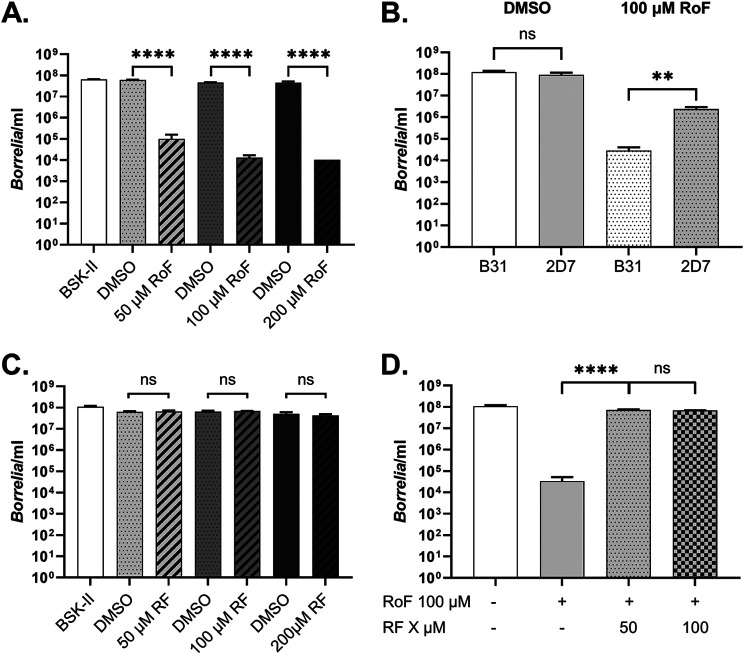
To examine whether the gene products of the B. burgdorferi rfuABCD operon collectively play a role in RF transport, we generated a B. burgdorferi mutant (2D7) lacking the entire rfuABCD-like operon (see Fig. S1A in the supplemental material). When we compared the progression of labeled RF uptake between wild-type B. burgdorferi and the operon-deficient mutant growing in vitro, there were no differences in either RF import kinetics (Fig. 3A) or the synthesis of FMN (Fig. 3B) or FAD (Fig. 3C) among the two strains. This result was perplexing in view of the homology between the T. pallidum and B. burgdorferi rfuABCD-like operons, because it implied that either the rfuABCD operon is not involved in RF transport (when B. burgdorferi is cultivated in vitro) or B. burgdorferi can acquire RF via an alternative import mechanism. To further probe the possibility that B. burgdorferi may use the rfuABCD operon for RF import, we employed roseoflavin (RoF), a natural RF structural analog that competes with RF transport (24, 25), as a potential inhibitor of spirochetal growth. We hypothesized that if the rfuABCD-like operon played a role in RF transport by B. burgdorferi, then the 2D7 mutant would be refractory to RoF-mediated growth inhibition. To test this possibility, spirochetes were cultured in vitro in BSK-II medium supplemented with graded concentrations of RoF. Cultures seeded with 104 bacteria were monitored for cell density up to 8 days postinoculation. As expected, wild-type B. burgdorferi was inhibited by RoF in a dose-dependent manner (0 to 200 μM) (Fig. 4A). In contrast, the 2D7 mutant was refractory to 100 μM RoF and reached cell densities ∼100-fold greater than those of wild-type B. burgdorferi when exposed to 100 μM RoF (Fig. 4B). These results imply that B. burgdorferi may exploit RfuABCD proteins to transport RF but may also acquire RF through an alternative mechanism when grown under the in vitro conditions employed.
FIG 3.
Comparison of RF uptake by wild-type B. burgdorferi and the 2D7 mutant. B. burgdorferi was grown to late log phase and then incubated at 37°C in medium supplemented with labeled RF (10 or 100 μM) for 12 h. Cell pellets were washed twice with BSK-II medium and then frozen. Labeled RF (A), FMN (B), and FAD (C) concentrations were determined by LC-MS. Each data point represents a biological replicate (n = 6) from three independent experiments. Bars represent the averages of the displayed data points, and error bars indicate SD. One-way ANOVA with Sidak’s post hoc test was used for multiple comparisons. ns, not significant; **, P < 0.01; ***, P < 0.001.
FIG 4.
B. burgdorferi is inhibited by RoF. (A) B. burgdorferi was cultured in BSK-II medium alone or in medium supplemented with DMSO (RoF vehicle) or the indicated concentrations of RoF (0 to 200 μM). (B) B31 and 2D7 were grown in BSK-II medium supplemented with DMSO or 100 μM RoF. (C) B31 was cultured in BSK-II medium supplemented with DMSO or various concentrations of RF (0 to 200 μM). (D) B31 was cultured in BSK-II medium alone or in medium supplemented with an inhibitory concentration RoF (100 μM) and/or RF (50 or 100 μM). All cultures were grown for 8 days at 37°C. The concentration of DMSO added to medium reflected the amount of DMSO added for each respective RoF concentration. Bars represent the means from three independent experiments. Error bars indicate SD. Two-way ANOVA and Tukey’s post hoc test were used for multiple comparisons. ns, not significant; **, P < 0.01; ****, P < 0.0001.
Exogenous RF can overcome RoF-mediated growth inhibition of B. burgdorferi.
To investigate whether the RoF inhibition of borreliae by 50, 100, or 200 μM RoF (Fig. 4A) may have been due to nonspecific flavin toxicity, we grew B. burgdorferi in the presence of 50, 100, or 200 μM RF. B. burgdorferi growth was not inhibited when the bacteria were grown in BSK-II medium supplemented with these concentrations of RF (Fig. 4C), indicating that flavin-mediated toxicity likely was not responsible for the observed growth inhibition imparted by the RoF treatment (Fig. 4A). Additionally, to confirm that RoF competed with RF, B. burgdorferi was cultivated in BSK-II medium containing inhibitory concentrations of RoF and with various concentrations of exogenously added RF (as a competitor). When spirochetes were incubated in BSK-II medium containing 100 μM RoF and either 50 μM or 100 μM RF, spirochete cell densities increased by ∼1,000-fold and were similar to those in the untreated control (Fig. 4D). This observation suggests that RoF is able to compete with RF in the RF transport process. Furthermore, RF likely has a higher binding affinity for the transport protein(s) given that RF at a lower concentration than that of RoF was capable of reversing the RoF-mediated growth inhibition of B. burgdorferi.
RoF is bacteriostatic for B. burgdorferi.
There is a paucity of information regarding whether RoF is bacteriostatic or bactericidal for various bacteria (26). To assess this for B. burgdorferi, we took advantage of our observation that RoF inhibited the in vitro growth of B. burgdorferi and that inhibition could be alleviated by the addition of exogenous RF to the medium. If RoF is bactericidal for B. burgdorferi, then RF supplementation added at later intervals should not rescue borrelial growth. On the other hand, if RoF is bacteriostatic, then RF supplementation should restore borrelial growth to levels comparable to those of untreated control cultures. We first conducted growth curve analyses of B. burgdorferi incubated in the presence of RoF to assess whether B. burgdorferi replicated early on but may have succumbed by the time that we assessed borrelial numbers (8 days postinoculation). Following an initial burst in growth 2 days postinoculation, RoF-treated spirochete densities plateaued to 8 days postinoculation (Fig. 5A). When these growth experiments with RoF were repeated but modified by adding RF every 2 days (days 0, 2, 4, 6, and 8) over the 8-day period, borrelial cell densities initially remained in the 104- to 105-spirochetes/ml range but increased when RF was added to the cultures (Fig. 5B). This trend continued throughout the 8-day incubation period. These results support the observation that RoF is bacteriostatic for B. burgdorferi.
FIG 5.
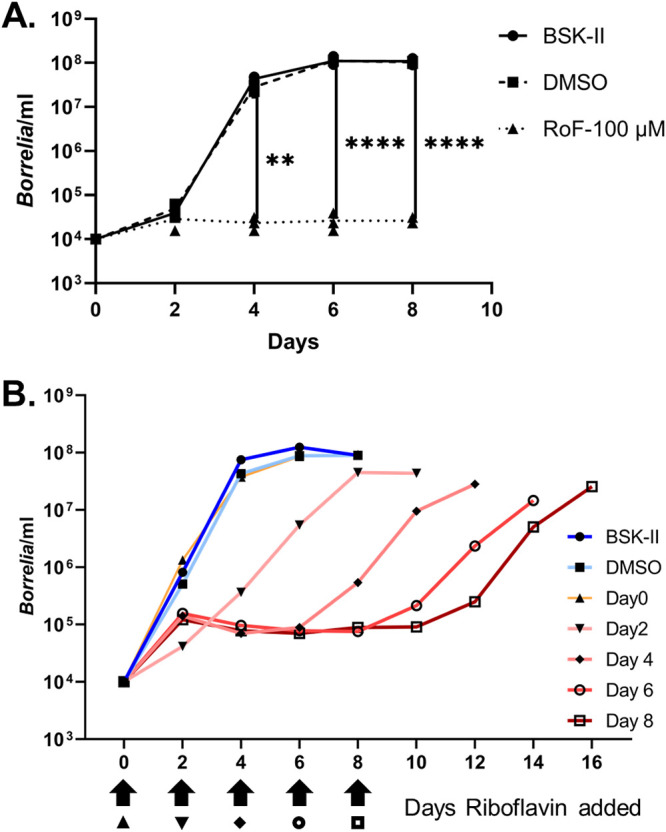
RoF-mediated growth inhibition of B. burgdorferi is bacteriostatic. (A) B. burgdorferi was cultured in BSK-II medium or in medium supplemented with RoF (100 μM) or DMSO (RoF vehicle). At the indicated times, a portion of each culture was taken and the concentration of spirochetes/ml was determined by dark-field microscopy. Spirochetes failed to replicate when cultured in the presence of 100 μM RoF. Lines represents the averages of the indicated data points from three independent experiments. The depicted comparison is between DMSO and RoF. Two-way ANOVA and Tukey’s post hoc test were used for multiple comparisons. **, P < 0.01; ****, P < 0.0001. (B) B31 was grown in BSK-II medium or medium supplemented with DMSO or RoF (100 μM). At the indicated times, RF (50 μM) was added to the cultures, and spirochetes were enumerated. Each data point represents an average for three biological replicates.
The rfuABCD mutant is growth deficient at room temperature.
Wild-type B. burgdorferi and the 2D7 mutant were cultivated in BSK-II medium at 25°C to compare their growth patterns. Borrelial cell densities were assessed at 14, 21, and 28 days postinoculation. The mutant displayed significant growth defects at 25°C in comparison to its wild-type parent (Fig. 6A). However, growth of the mutant was not impaired when it was cultivated at 37°C (Fig. 6B).
FIG 6.
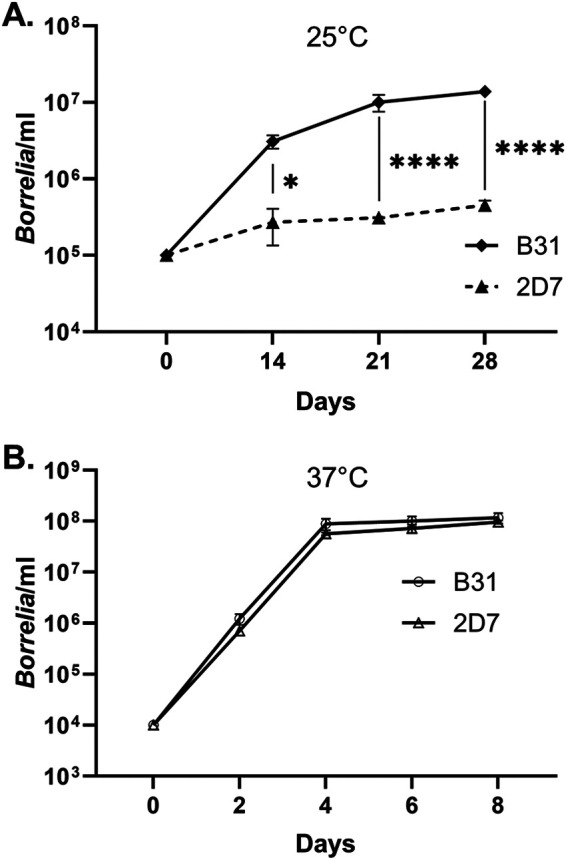
Growth of the 2D7 mutant is impaired at 25°C but not at 37°C. BSK-II medium was inoculated with B. burgdorferi and the cultures were incubated at either (A) 25°C or (B) 37°C. The initial inoculum for the 25°C cultures was 105 spirochetes/ml, whereas the initial inoculum for the 37°C cultures was 104 spirochetes/ml. The concentration of spirochetes was determined at the indicated times. Each data point represents an average for three biological replicates. Two-way ANOVA and Tukey’s post hoc test were used for multiple comparisons. *, P < 0.05; ****, P < 0.0001.
The rfuABCD-like operon influences B. burgdorferi mammalian infectivity and dissemination.
Prior to murine infection experiments, we first confirmed that the 2D7 mutant maintained all mammalian infection-associated plasmids (Fig. S1B). C3H/HeN mice were infected intradermally with either 103 or 104 spirochetes of either the wild-type parent or the 2D7 mutant. Twenty-one days postinoculation, mice were euthanized and ear, heart, and joint tissues were cultured for 21 days in BSK-II medium. In the case of the wild-type parent, as expected, spirochetes were observed growing in all cultures of all tissues harvested from mice inoculated with either 103 or 104 bacteria. However, in the case of the 2D7 mutant, none of the cultures of tissues harvested from mice inoculated with 103 spirochetes were positive, and only 3 cultures, all from the same mouse, inoculated with 104 borreliae yielded spirochetes (Table 1). These results establish that mammalian infection and dissemination by the 2D7 mutant are markedly attenuated. Of note, attenuation of the 2D7 mutant was unlikely due to disruption of the virulence-promoting RpoN-RpoS regulatory pathway (27), because both upstream (BosR) (28) and downstream (OspC) (29) transcripts (key components of the pathway) were similarly expressed by wild-type B. burgdorferi and the 2D7 mutant (data not shown).
TABLE 1.
Mammalian infectivity and dissemination are attenuated for the 2D7 mutanta
| Strain | No. positive/total |
|||||
|---|---|---|---|---|---|---|
| Joint |
Heart |
Skin (ear) |
||||
| 103 | 104 | 103 | 104 | 103 | 104 | |
| B31 | 10/10 | 9/9 | 10/10 | 9/9 | 10/10 | 9/9 |
| 2D7 | 0/10 | 1*/9 | 0/10 | 1*/9 | 0/10 | 1*/9 |
Mice were intradermally inoculated with 103 or 104 spirochetes of either wild-type B31 or the 2D7 mutant. Twenty-one days postinoculation, mice were euthanized, and joint, heart, and ear tissues were collected for culture. Tissues were incubated for 21 days at 37°C in BSK-II medium supplemented with BAM (antibiotic cocktail). Cultures were observed every 3 days, up to 21 days, for the presence of spirochetes. *, same mouse.
DISCUSSION
RF is an essential micronutrient utilized by all organisms. Many bacteria are able to synthesize RF utilizing genes similar to the ribGBAHT operon of Bacillus subtilis (30). However, some bacteria have lost this capability and rely on exogenous sources and uptake mechanisms to acquire RF (1). One of these groups comprises the closely related pathogenic spirochetes Treponema pallidum and Borrelia burgdorferi. Until recently, it was unknown what genes pathogenic spirochetes such as T. pallidum may utilize to import RF (9), as many bacteria that take up RF seem to have evolved independent RF uptake mechanisms (1). Our laboratory demonstrated that recombinant RfuA of T. pallidum binds RF and proposed that this protein was part of a larger ABC-type transporter complex that likely mediates the uptake of RF. Additionally, we demonstrated that BB_0319 of B. burgdorferi was also capable of binding RF in vitro, suggesting that these two pathogenic spirochete species may acquire RF via a similar mechanism(s) (9).
To investigate whether the rfuABCD-like operon encoded by B. burgdorferi may be involved in the transport of RF, we generated a mutant lacking this operon and demonstrated that it is refractory to the inhibitory action of the RF structural analog RoF (Fig. 4). RoF is known to compete with RF during RF transport. Once within cells, RoF can be converted into toxic forms of FMN and FAD (31), which further inhibit proper functioning of target flavoproteins (32, 33). The observation that the inhibition of B. burgdorferi by RoF could be alleviated when RF was added to the medium supported the idea that the mechanism of RoF inhibition in B. burgdorferi involves limiting sufficient RF internalization and synthesis of FMN and FAD. However, because we were able to generate an rfuABCD-deficient mutant, it is reasonable to conclude that B. burgdorferi likely is capable of acquiring RF through an alternative unknown uptake mechanism, at least when growing in vitro. This hypothesis is bolstered by the fact that the import of labeled RF by wild-type B31 or the 2D7 mutant was similar (Fig. 3). Although believed to be uncommon, some bacteria such as Clostridioides difficile encode multiple RF uptake mechanisms. C. difficile encodes a RibU (ypaA) (7), an RF transporter commonly found in many bacteria such as Bacillus subtilis, Lactococcus lactis, and Staphylococcus aureus. C. difficile also encodes an exclusive RF transporter designated RibZ (1). Thus, it is plausible that B. burgdorferi encodes more than one RF uptake mechanism, especially when one considers its disparate zoonotic life cycle and its consequent need to cycle between the diverse tick and mammalian host environments. However, one enigmatic feature of this multiple transporter hypothesis concerns why wild-type B. burgdorferi was sensitive to the action of RoF whereas the 2D7 mutant was not. Although there is no clear explanation for the discordance at this time, this property may serve as a strategic tool for the identification of genes encoding the alternative RF transport mechanism(s).
We hypothesized that if B. burgdorferi indeed maintains an alternative RF uptake mechanism, then growth of the 2D7 mutant may be attenuated under conditions that somewhat mimic the unfed tick midgut, such as growth at ambient temperature. As such, we incubated wild-type B31 and the 2D7 mutant at room temperature and compared their growth kinetics. Replication of the mutant strain was significantly attenuated at 25°C, suggesting that one or more of the rfu genes are important for RF acquisition at environmental temperature when B. burgdorferi is within its tick host. However, it remains possible that the normal growth of the mutant at higher temperature is due to the induction of a second transporter that may not be expressed at lower temperature. Nonetheless, when we compared the infectivity profiles between wild-type B31 and the 2D7 mutant following murine infection, the mutant was highly attenuated for its mammalian infectivity phenotype; the results for the one mouse infected with an inoculum of 104 bacteria could have been due to spirochete clumping, which may have inadvertently resulted in a much higher intradermal dose of bacteria. Of note, it was somewhat counterintuitive that the 2D7 mutant grew normally in vitro at 37°C but displayed diminished infectivity for mice. Nevertheless, our mouse infection results are very consistent with a previous report showing that a mutant lacking the rfuB (bb0318) gene also was attenuated for murine infectivity (34). This prompts an extended hypothesis that B. burgdorferi may have evolved multiple mechanisms to acquire RF and that the competition for RF in mammalian hosts requires maintenance of the rfuABCD RF transport genes for proper growth and spirochetal tissue dissemination. It also remains possible that variable micronutrient availability in differing tissue compartments, as well as ensuing host immune responses, also contributed to clearance of the 2D7 mutant.
Although we have reported evidence supporting the idea that bb0319 (Bb_RfuA) likely is the RF-binding component of the rfuABCD operon (9), additional experiments are warranted to explore more completely the function and roles of the other three members of this operon in B. burgdorferi. One caveat relative to the interpretations in this study is that phenotypic characterizations of the 2D7 mutant are potentially limited by the lack of a genetically complemented strain; numerous attempts to generate a genetic complement (either in cis or in trans) to the 2D7 mutant thus far have been unsuccessful for unknown reasons.
To our knowledge, this is the first study to demonstrate that B. burgdorferi is capable of synthesizing FMN and FAD from exogenous RF. Unlike Treponema pallidum, which encodes a version of the bifunctional riboflavin kinase/FAD synthetase (TP0888) found in most bacteria (15, 35), the relevant enzyme(s) in B. burgdorferi remains elusive (14, 36). Alternatively, one could propose that B. burgdorferi may import FMN and/or FAD from external sources. Previous investigations into RF uptake in Lactococcus lactis indicate that RibU can facilitate the uptake of RF and, to a lesser extent, FMN (25). However, we believe that it is unlikely that FMN and FAD are imported by B. burgdorferi. Our combined experiments showed that prior to cultivation, we could not detect labeled FMN or labeled FAD in the medium, but after subsequent cultivation of B. burgdorferi in medium containing labeled RF, we detected labeled FMN and labeled FAD, supporting the idea that the spirochetes actively synthesized FMN and FAD from the labeled RF.
The RF derivatives FMN and FAD as coenzymes are of paramount importance to the cell, impacting many types of metabolic functions. FMN is generated from RF via riboflavin kinase, and FAD is made from FMN by the action of FAD synthetase (37, 38). FMN and FAD, bound either covalently (39, 40) or noncovalently (41, 42), subsequently serve as cofactors for many types of enzymes (flavoproteins) that impact cellular metabolism. Given the broad metabolic impact on the cell, it is thus not surprising that a finely controlled balance of intracellular flavin is requisite for maintaining proper cellular homeostasis, of which flavoprotein biogenesis is an important component. With respect to flavoproteins, KEGG genomic data predict at least six flavoprotein genes within the B. burgdorferi genome. These are bb0515 (trxB; FAD-dependent thioredoxin reductase), bba76 (thyX; FAD-dependent thymidylate synthase), bb0178 (gidA; FAD-dependent tRNA uridine 5-carboxymethylaminomethyl modification enzyme), bb0684 (fni; FMN-dependent isopentenyl-diphosphate delta-isomerase), bb0728 (cdr; FAD-dependent CoA-disulfide reductase), and bb0812 (dfp, coaBC; FMN-dependent coenzyme A [CoA] biosynthesis [bifunctional protein]).
Dfp (CoaBC) catalyzes two steps in the synthesis of CoA from pantothenate (43, 44). In B. burgdorferi, dfp (coaBC) has been shown to have increased expression at 35°C relative to growth at 25°C (21), and we have shown that this gene is regulated by BosR, which is an essential regulator of B. burgdorferi virulence (28). In Borrelia, CoA, which is an essential cofactor involved in many cellular processes in bacteria, also may serve to help protect spirochetes from reactive oxygen species. Specifically, reduced CoA is the major low-molecular-weight thiol in Borrelia and, in conjunction with CoADR (bb0728) (which regenerates oxidized CoA back to reduced CoA), is able to reduce H2O2. CoADR also is regulated by BosR (45), and is likely an important ROS protection mechanism during mammalian infection (46). Given the importance of all of these cellular processes, it is thus not surprising that B. burgdorferi growth was inhibited by RoF. Taking these results together, it is tempting to speculate how the broad metabolic vulnerability engendered by flavin essentiality may be exploited to develop additional structural analogs of RF; the application of contemporary principles of medicinal chemistry (8, 47) may engender other analogs inhibitory for B. burgdorferi, potentially representing new candidate therapeutics for Lyme borreliosis.
MATERIALS AND METHODS
Ethics statement.
The Institutional Animal Care and Use Committee at the University of Texas (UT) Southwestern Medical Center approved all experiments involving animals in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (48).
Bacterial strains and culture conditions.
Borrelia burgdorferi strain B31 and all described mutants were cultured and maintained in Barbour-Stoenner-Kelly II medium supplemented with 6% heat-inactivated rabbit serum (Pel-Freeze Biologicals, Rogers, AR) at a pH of 7.6 (BSK-II) (49). When needed, BSK-II medium was supplemented with kanamycin (300 μg/ml). Plasmid content of each B31 strain was evaluated using PCR primers as previously described (50, 51). BSK-II medium was inoculated with B. burgdorferi at a concentration of 104 spirochetes/ml and grown to stationary phase (108 spirochetes/ml) under an atmosphere of 5% CO2. All cultures were incubated at 37°C, except where otherwise noted.
Generation of the 2D7 mutant.
GeneArt seamless cloning and assembly enzyme mix (Thermo Fisher Scientific) was used to generate the suicide plasmid construct for removal of the RF transporting locus (bb0319–bb0316) in B. burgdorferi. Four DNA fragments were used in the simultaneous assembly: (i) the pUC origin was amplified from pUC19 (Thermo Fisher Scientific), (ii) the flgB-kan antibiotic resistance cassette was amplified from OY153 (52), and the flanking sequences (iii) upstream and (iv) downstream of bb0319–bb0316 were amplified from B31 genomic DNA. All PCRs were conducted using Phusion high-fidelity DNA polymerase (Thermo Fisher Scientific). The four PCR fragments were purified with a QIAquick gel extraction kit (Qiagen) according to the manufacturer’s protocol, mixed together in a 1:1:1:1 molar ratio, and ligated following the procedures provided with the GeneArt enzyme mix. The ligation product was then transformed into NEB 10-beta competent Escherichia coli cells (New England Biolabs). Bacteria carrying the suicide plasmid were selected using LB agar with 50 μg/ml of kanamycin. The suicide plasmid was extracted with a QIAprep Plasmid Plus maxikit (Qiagen) and confirmed by Sanger sequencing. B. burgdorferi strain B31 was transformed via electroporation with the suicide plasmid, and Δbb0319–bb0316 mutants were identified using the 96-well liquid culture method as previously described (53). Mutants were verified for loss of the operon and maintenance of essential virulence-associated plasmids (Fig. S1).
Quantification of B. burgdorferi spirochetes.
B. burgdorferi cultures were vortexed to ensure an even distribution of spirochetes. Wet mounts were then prepared by spotting 10 μl of each culture onto glass slides. Dark-field microscopy was used to enumerate B. burgdorferi spirochetes from 32 random fields per culture (Olympus BX41 microscope, 40× objective). The average number of spirochetes per field was used to determine the density of spirochetes/ml in each culture.
Detection of RF, FMN, and FAD. (i) Growth conditions.
Fifty-milliliter cultures of B. burgdorferi strain B31 or the 2D7 mutant were grown to stationary phase (∼108 spirochetes/ml) and pelleted at 3,000 × g for 30 min at 25°C. Cell pellets were resuspended in 1 ml of BSK-II medium supplemented with heavy-isotope-labeled riboflavin (riboflavin-[13C4,15N2]dioxopyrimidine; Santa Cruz Biotechnology) to a final concentration of 100 μM. Cultures were incubated at either 25°C or 37°C for 12 h in a 5% CO2 incubator. Bacteria were pelleted at 16,000 × g for 15 min at 4°C and washed three times in BSK-II medium. The supernatant was saved for LC-MS analysis. The final pellet was snap-frozen using liquid nitrogen and stored at −80°C.
(ii) Sample preparation.
Cell metabolites were extracted by suspending bacterial pellets in acetonitrile-methanol-water (40:40:20) with 0.1% formic acid and incubating at −20°C for 20 min. Insoluble materials were removed via centrifugation at 16,000 × g for 10 min. Resulting supernatants were transferred to an autosampler vial and stored at −80°C until LC-MS analyses (54). The cellular protein content of the pelleted insoluble material was determined using the Pierce bicinchoninic acid (BCA) protein assay kit (Thermo Fisher Scientific) following solubilization in 1% SDS and 0.1 M NaOH. Sample contents were adjusted based on total protein to account for differences in bacterial numbers among the samples. Metabolites were extracted from spent medium by adding methanol containing d5-phenylalanine (100 ng/ml) as an internal standard to a final concentration of 75% methanol. Samples were incubated at −20°C for 1 h and insoluble materials removed by centrifugation at 1,500 × g for 10 min. The resulting supernatant was dried under vacuum and stored at −80°C. Prior to LC-MS analyses, the dried extract was suspended in 50% methanol (55).
(iii) LC-MS analyses.
Metabolite extracts were applied to a XBridge BEH C18 XP column (2.5-μm particle size, 2.1 mm by 100 mm) (Waters, Milford, MA, USA) and eluted with an 11-min linear gradient of 4% to 90% methanol in 5 mM ammonium acetate using a Waters Acquity H-class ultraperformance liquid chromatography (UPLC) system (Waters, Milford, MA, USA) (32). Extracts were analyzed using a Bruker maXis quadrupole time-of-flight mass spectrometer equipped with an electrospray ionization source operated in positive ion polarity. Source parameters were as follows: endplate offset, 500 V; capillary voltage, 3,500 V; nebulizer, 3.0 × 105 Pa; drying gas, 10 liters/min; and dry temperature, 300°C.
Labeled and unlabeled versions of RF, FAD, and FMN, as well as d5-phenylalanine, were quantified using Skyline (56). Specifically, molecular ions m/z 383.1531, m/z 792.1719, and m/z 463.1194 for labeled RF, FAD, and FMN, respectively, and m/z 377.1456, m/z 786.1644, and m/z 457.1119 for unlabeled RF, FAD, and FMN, respectively, were identified in the chromatographic spectra. The molecular ion m/z 171.1103 was used for d5-phenylalanine. The peak area of each molecular ion was determined in Skyline and normalized by the total protein content (in milligrams) for the metabolites extracted from cells or the peak area of d5-phenylalanine for the metabolites extracted from the medium.
Flavin treatment of Borrelia cultures.
RF or RoF was solubilized in dimethyl sulfoxide (DMSO) at a concentration of 10 mg/ml following overnight agitation at room temperature. One milliliter of BSK-II medium supplemented with the indicated concentrations of RF and/or RoF was inoculated with 104 spirochetes/ml and incubated at 37°C in a 5% CO2 incubator for 8 days. DMSO controls correspond to the amount of DMSO added to each culture minus the indicated compound. For the RoF bacteriostatic growth curve, the appropriate volume of RF stock (10 mg/ml) was added to the indicated cultures at the indicated times to a final concentration of 50 μM RF.
Murine infectivity and dissemination.
Four-week-old female C3H/HeN mice (C3H/HeNCrl; Charles River Laboratories) were anesthetized with a ketamine/xylazine cocktail (30 mg/ml and 4 mg/ml, respectively), shaved, and then inoculated with 103 or 104 organisms of wild-type B. burgdorferi strain B31 or the 2D7 mutant intradermally on the lower right back quadrant. Twenty-one days postinfection, infected mice were euthanized, and the left ear, left joint, and apex heart tissue were collected and incubated in BSK-II medium supplemented with BAM cocktail (sulfamethoxazole, fosfomycin, rifampin, trimethoprim, and amphotericin) for 21 days at 37°C in a 5% CO2 incubator. Mice were considered infected if spirochetes were detected in any of these tissues.
Statistical analysis.
Data were analyzed and graphs generated using GraphPad Prism 9. An unpaired two-tailed Student's t test was used when two groups were compared, and when multiple comparisons were made, an analysis of variance (ANOVA) and Tukey’s or Sidak’s post hoc test for multiple comparisons was utilized.
ACKNOWLEDGMENTS
This work was funded by HHS award from the National Institutes of Health (AI059062 and AI056305). We also thank the Global Lyme Alliance and the Deborah and Mark Blackman Postdoctoral Fellowship, which supported the efforts of M.K.M. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material is available online only.
Contributor Information
Michael V. Norgard, Email: Michael.Norgard@UTSouthwestern.edu.
De'Broski R. Herbert, University of Pennsylvania
REFERENCES
- 1.Gutierrez-Preciado A, Torres AG, Merino E, Bonomi HR, Goldbaum FA, Garcia-Angulo VA. 2015. Extensive identification of bacterial riboflavin transporters and their distribution across bacterial species. PLoS One 10:e0126124. 10.1371/journal.pone.0126124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mac LJ. 1952. The effects of certain purines and pyrimidines upon the production of riboflavin by Eremothecium ashbyii. J Bacteriol 63:233–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mailander B, Bacher A. 1976. Biosynthesis of riboflavin. Structure of the purine precursor and origin of the ribityl side chain. J Biol Chem 251:3623–3628. 10.1016/S0021-9258(17)33390-2. [DOI] [PubMed] [Google Scholar]
- 4.Volk R, Bacher A. 1991. Biosynthesis of riboflavin. Studies on the mechanism of L-3,4-dihydroxy-2-butanone 4-phosphate synthase. J Biol Chem 266:20610–20618. 10.1016/S0021-9258(18)54753-0. [DOI] [PubMed] [Google Scholar]
- 5.Marsili E, Baron DB, Shikhare ID, Coursolle D, Gralnick JA, Bond DR. 2008. Shewanella secretes flavins that mediate extracellular electron transfer. Proc Natl Acad Sci U S A 105:3968–3973. 10.1073/pnas.0710525105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chao MC, Pritchard JR, Zhang YJ, Rubin EJ, Livny J, Davis BM, Waldor MK. 2013. High-resolution definition of the Vibrio cholerae essential gene set with hidden Markov model-based analyses of transposon-insertion sequencing data. Nucleic Acids Res 41:9033–9048. 10.1093/nar/gkt654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vitreschak AG, Rodionov DA, Mironov AA, Gelfand MS. 2002. Regulation of riboflavin biosynthesis and transport genes in bacteria by transcriptional and translational attenuation. Nucleic Acids Res 30:3141–3151. 10.1093/nar/gkf433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang H, Mann PA, Xiao L, Gill C, Galgoci AM, Howe JA, Villafania A, Barbieri CM, Malinverni JC, Sher X, Mayhood T, McCurry MD, Murgolo N, Flattery A, Mack M, Roemer T. 2017. Dual-targeting small-molecule inhibitors of the Staphylococcus aureus FMN riboswitch disrupt riboflavin homeostasis in an infectious setting. Cell Chem Biol 24:576–588.E6. 10.1016/j.chembiol.2017.03.014. [DOI] [PubMed] [Google Scholar]
- 9.Deka RK, Brautigam CA, Biddy BA, Liu WZ, Norgard MV. 2013. Evidence for an ABC-type riboflavin transporter system in pathogenic spirochetes. mBio 4:e00615-12. 10.1128/mBio.00615-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deka RK, Brautigam CA, Liu WZ, Tomchick DR, Norgard MV. 2013. The TP0796 lipoprotein of Treponema pallidum is a bimetal-dependent FAD pyrophosphatase with a potential role in flavin homeostasis. J Biol Chem 288:11106–11121. 10.1074/jbc.M113.449975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deka RK, Brautigam CA, Liu WZ, Tomchick DR, Norgard MV. 2015. Evidence for posttranslational protein flavinylation in the syphilis spirochete Treponema pallidum: structural and biochemical insights from the catalytic core of a periplasmic flavin-trafficking protein. mBio 6:e00519-15. 10.1128/mBio.00519-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Radolf JD, Deka RK, Anand A, Smajs D, Norgard MV, Yang XF. 2016. Treponema pallidum, the syphilis spirochete: making a living as a stealth pathogen. Nat Rev Microbiol 14:744–759. 10.1038/nrmicro.2016.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edmondson DG, Hu B, Norris SJ. 2018. Long-term in vitro culture of the syphilis spirochete Treponema pallidum subsp. pallidum. mBio 9:e01153-18. 10.1128/mBio.01153-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, White O, Ketchum KA, Dodson R, Hickey EK, Gwinn M, Dougherty B, Tomb JF, Fleischmann RD, Richardson D, Peterson J, Kerlavage AR, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams MD, Gocayne J, Weidman J, Utterback T, Watthey L, McDonald L, Artiach P, Bowman C, Garland S, Fuji C, Cotton MD, Horst K, Roberts K, Hatch B, Smith HO, Venter JC. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580–586. 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 15.Fraser CM, Norris SJ, Weinstock GM, White O, Sutton GG, Dodson R, Gwinn M, Hickey EK, Clayton R, Ketchum KA, Sodergren E, Hardham JM, McLeod MP, Salzberg S, Peterson J, Khalak H, Richardson D, Howell JK, Chidambaram M, Utterback T, McDonald L, Artiach P, Bowman C, Cotton MD, Fujii C, Garland S, Hatch B, Horst K, Roberts K, Sandusky M, Weidman J, Smith HO, Venter JC. 1998. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science 281:375–388. 10.1126/science.281.5375.375. [DOI] [PubMed] [Google Scholar]
- 16.Fraaije MW, Mattevi A. 2000. Flavoenzymes: diverse catalysts with recurrent features. Trends Biochem Sci 25:126–132. 10.1016/s0968-0004(99)01533-9. [DOI] [PubMed] [Google Scholar]
- 17.van Berkel WJ, Kamerbeek NM, Fraaije MW. 2006. Flavoprotein monooxygenases, a diverse class of oxidative biocatalysts. J Biotechnol 124:670–689. 10.1016/j.jbiotec.2006.03.044. [DOI] [PubMed] [Google Scholar]
- 18.Kearney EB, Goldenberg J, Lipsick J, Perl M. 1979. Flavokinase and FAD synthetase from Bacillus subtilis specific for reduced flavins. J Biol Chem 254:9551–9557. 10.1016/S0021-9258(19)83550-0. [DOI] [PubMed] [Google Scholar]
- 19.Manstein DJ, Pai EF. 1986. Purification and characterization of FAD synthetase from Brevibacterium ammoniagenes. J Biol Chem 261:16169–16173. 10.1016/S0021-9258(18)66693-1. [DOI] [PubMed] [Google Scholar]
- 20.Mack M, van Loon AP, Hohmann HP. 1998. Regulation of riboflavin biosynthesis in Bacillus subtilis is affected by the activity of the flavokinase/flavin adenine dinucleotide synthetase encoded by ribC. J Bacteriol 180:950–955. 10.1128/JB.180.4.950-955.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ojaimi C, Brooks C, Casjens S, Rosa P, Elias A, Barbour A, Jasinskas A, Benach J, Katona L, Radolf J, Caimano M, Skare J, Swingle K, Akins D, Schwartz I. 2003. Profiling of temperature-induced changes in Borrelia burgdorferi gene expression by using whole genome arrays. Infect Immun 71:1689–1705. 10.1128/IAI.71.4.1689-1705.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tokarz R, Anderton JM, Katona LI, Benach JL. 2004. Combined effects of blood and temperature shift on Borrelia burgdorferi gene expression as determined by whole genome DNA array. Infect Immun 72:5419–5432. 10.1128/IAI.72.9.5419-5432.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang X, Goldberg MS, Popova TG, Schoeler GB, Wikel SK, Hagman KE, Norgard MV. 2000. Interdependence of environmental factors influencing reciprocal patterns of gene expression in virulent Borrelia burgdorferi. Mol Microbiol 37:1470–1479. 10.1046/j.1365-2958.2000.02104.x. [DOI] [PubMed] [Google Scholar]
- 24.Otani S, Takatsu M, Nakano M, Kasai S, Miura R. 1974. Roseoflavin, a new antimicrobial pigment from Streptomyces. J Antibiot (Tokyo) 27:86–87. [PubMed] [Google Scholar]
- 25.Burgess CM, Slotboom DJ, Geertsma ER, Duurkens RH, Poolman B, van Sinderen D. 2006. The riboflavin transporter RibU in Lactococcus lactis: molecular characterization of gene expression and the transport mechanism. J Bacteriol 188:2752–2760. 10.1128/JB.188.8.2752-2760.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou LF, Wu J, Li S, Li Q, Jin LP, Yin CP, Zhang YL. 2021. Antibacterial potential of termite-associated Streptomyces spp. ACS Omega 6:4329–4334. 10.1021/acsomega.0c05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hubner A, Yang X, Nolen DM, Popova TG, Cabello FC, Norgard MV. 2001. Expression of Borrelia burgdorferi OspC and DbpA is controlled by a RpoN-RpoS regulatory pathway. Proc Natl Acad Sci U S A 98:12724–12729. 10.1073/pnas.231442498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ouyang Z, Kumar M, Kariu T, Haq S, Goldberg M, Pal U, Norgard MV. 2009. BosR (BB0647) governs virulence expression in Borrelia burgdorferi. Mol Microbiol 74:1331–1343. 10.1111/j.1365-2958.2009.06945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang XF, Lybecker MC, Pal U, Alani SM, Blevins J, Revel AT, Samuels DS, Norgard MV. 2005. Analysis of the ospC regulatory element controlled by the RpoN-RpoS regulatory pathway in Borrelia burgdorferi. J Bacteriol 187:4822–4829. 10.1128/JB.187.14.4822-4829.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bacher A, Mailander B. 1978. Biosynthesis of riboflavin in Bacillus subtilis: function and genetic control of the riboflavin synthase complex. J Bacteriol 134:476–482. 10.1128/jb.134.2.476-482.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grill S, Busenbender S, Pfeiffer M, Kohler U, Mack M. 2008. The bifunctional flavokinase/flavin adenine dinucleotide synthetase from Streptomyces davawensis produces inactive flavin cofactors and is not involved in resistance to the antibiotic roseoflavin. J Bacteriol 190:1546–1553. 10.1128/JB.01586-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langer S, Hashimoto M, Hobl B, Mathes T, Mack M. 2013. Flavoproteins are potential targets for the antibiotic roseoflavin in Escherichia coli. J Bacteriol 195:4037–4045. 10.1128/JB.00646-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langer S, Nakanishi S, Mathes T, Knaus T, Binter A, Macheroux P, Mase T, Miyakawa T, Tanokura M, Mack M. 2013. The flavoenzyme azobenzene reductase AzoR from Escherichia coli binds roseoflavin mononucleotide (RoFMN) with high affinity and is less active in its RoFMN form. Biochemistry 52:4288–4295. 10.1021/bi400348d. [DOI] [PubMed] [Google Scholar]
- 34.Showman AC, Aranjuez G, Adams PP, Jewett MW. 2016. Gene bb0318 is critical for the oxidative stress response and infectivity of Borrelia burgdorferi. Infect Immun 84:3141–3151. 10.1128/IAI.00430-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smajs D, McKevitt M, Howell JK, Norris SJ, Cai WW, Palzkill T, Weinstock GM. 2005. Transcriptome of Treponema pallidum: gene expression profile during experimental rabbit infection. J Bacteriol 187:1866–1874. 10.1128/JB.187.5.1866-1874.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Casjens S, Palmer N, van Vugt R, Huang WM, Stevenson B, Rosa P, Lathigra R, Sutton G, Peterson J, Dodson RJ, Haft D, Hickey E, Gwinn M, White O, Fraser CM. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol Microbiol 35:490–516. 10.1046/j.1365-2958.2000.01698.x. [DOI] [PubMed] [Google Scholar]
- 37.Coquard D, Huecas M, Ott M, van Dijl JM, van Loon AP, Hohmann HP. 1997. Molecular cloning and characterisation of the ribC gene from Bacillus subtilis: a point mutation in ribC results in riboflavin overproduction. Mol Gen Genet 254:81–84. 10.1007/s004380050393. [DOI] [PubMed] [Google Scholar]
- 38.Efimov I, Kuusk V, Zhang X, McIntire WS. 1998. Proposed steady-state kinetic mechanism for Corynebacterium ammoniagenes FAD synthetase produced by Escherichia coli. Biochemistry 37:9716–9723. 10.1021/bi972817j. [DOI] [PubMed] [Google Scholar]
- 39.Singer TP, Kearney EB, Massey V. 1956. Observations on the flavin moiety of succinic dehydrogenase. Arch Biochem Biophys 60:255–257. 10.1016/0003-9861(56)90415-5. [DOI] [PubMed] [Google Scholar]
- 40.Willie A, Edmondson DE, Jorns MS. 1996. Sarcosine oxidase contains a novel covalently bound FMN. Biochemistry 35:5292–5299. 10.1021/bi952995h. [DOI] [PubMed] [Google Scholar]
- 41.Dym O, Eisenberg D. 2001. Sequence-structure analysis of FAD-containing proteins. Protein Sci 10:1712–1728. 10.1110/ps.12801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ryan KS, Howard-Jones AR, Hamill MJ, Elliott SJ, Walsh CT, Drennan CL. 2007. Crystallographic trapping in the rebeccamycin biosynthetic enzyme RebC. Proc Natl Acad Sci U S A 104:15311–15316. 10.1073/pnas.0707190104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kupke T, Uebele M, Schmid D, Jung G, Blaesse M, Steinbacher S. 2000. Molecular characterization of lantibiotic-synthesizing enzyme EpiD reveals a function for bacterial Dfp proteins in coenzyme A biosynthesis. J Biol Chem 275:31838–31846. 10.1074/jbc.M004273200. [DOI] [PubMed] [Google Scholar]
- 44.Strauss E, Kinsland C, Ge Y, McLafferty FW, Begley TP. 2001. Phosphopantothenoylcysteine synthetase from Escherichia coli. Identification and characterization of the last unidentified coenzyme A biosynthetic enzyme in bacteria. J Biol Chem 276:13513–13516. 10.1074/jbc.C100033200. [DOI] [PubMed] [Google Scholar]
- 45.Boylan JA, Hummel CS, Benoit S, Garcia-Lara J, Treglown-Downey J, CraneEJ, III, Gherardini FC. 2006. Borrelia burgdorferi bb0728 encodes a coenzyme A disulphide reductase whose function suggests a role in intracellular redox and the oxidative stress response. Mol Microbiol 59:475–486. 10.1111/j.1365-2958.2005.04963.x. [DOI] [PubMed] [Google Scholar]
- 46.Eggers CH, Caimano MJ, Malizia RA, Kariu T, Cusack B, Desrosiers DC, Hazlett KR, Claiborne A, Pal U, Radolf JD. 2011. The coenzyme A disulphide reductase of Borrelia burgdorferi is important for rapid growth throughout the enzootic cycle and essential for infection of the mammalian host. Mol Microbiol 82:679–697. 10.1111/j.1365-2958.2011.07845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pedrolli DB, Jankowitsch F, Schwarz J, Langer S, Nakanishi S, Frei E, Mack M. 2013. Riboflavin analogs as antiinfectives: occurrence, mode of action, metabolism and resistance. Curr Pharm Des 19:2552–2560. 10.2174/1381612811319140006. [DOI] [PubMed] [Google Scholar]
- 48.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed. National Academies Press, Washington, DC. [Google Scholar]
- 49.Pollack RJ, TelfordSR, 3rd, Spielman A. 1993. Standardization of medium for culturing Lyme disease spirochetes. J Clin Microbiol 31:1251–1255. 10.1128/jcm.31.5.1251-1255.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Purser JE, Norris SJ. 2000. Correlation between plasmid content and infectivity in Borrelia burgdorferi. Proc Natl Acad Sci U S A 97:13865–13870. 10.1073/pnas.97.25.13865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiang X, Yang Y, Du J, Lin T, Chen T, Yang XF, Lou Y. 2017. Investigation of ospC expression variation among Borrelia burgdorferi Strains. Front Cell Infect Microbiol 7:131. 10.3389/fcimb.2017.00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ouyang Z, Zhou J, Norgard MV. 2014. CsrA (BB0184) is not involved in activation of the RpoN-RpoS regulatory pathway in Borrelia burgdorferi. Infect Immun 82:1511–1522. 10.1128/IAI.01555-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang XF, Pal U, Alani SM, Fikrig E, Norgard MV. 2004. Essential role for OspA/B in the life cycle of the Lyme disease spirochete. J Exp Med 199:641–648. 10.1084/jem.20031960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rabinowitz JD, Kimball E. 2007. Acidic acetonitrile for cellular metabolome extraction from Escherichia coli. Anal Chem 79:6167–6173. 10.1021/ac070470c. [DOI] [PubMed] [Google Scholar]
- 55.Fitzgerald BL, Molins CR, Islam MN, Graham B, Hove PR, Wormser GP, Hu L, Ashton LV, Belisle JT. 2020. Host metabolic response in early Lyme disease. J Proteome Res 19:610–623. 10.1021/acs.jproteome.9b00470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Adams KJ, Pratt B, Bose N, Dubois LG, St John-Williams L, Perrott KM, Ky K, Kapahi P, Sharma V, MacCoss MJ, Moseley MA, Colton CA, MacLean BX, Schilling B, Thompson JW, Alzheimer's Disease Metabolomics Consortium. 2020. Skyline for small molecules: a unifying software package for quantitative metabolomics. J Proteome Res 19:1447–1458. 10.1021/acs.jproteome.9b00640. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download IAI.00307-21-s0001.pdf, PDF file, 0.6 MB (622.2KB, pdf)