ABSTRACT
Proteus mirabilis is a leading uropathogen of catheter-associated urinary tract infections (CAUTIs), which are among the most common health care-associated infections worldwide. A key factor that contributes to P. mirabilis pathogenesis and persistence during CAUTI is the formation of catheter biofilms, which provide increased resistance to antibiotic treatment and host defense mechanisms. Another factor that is important for bacterial persistence during CAUTI is the ability to resist reactive oxygen species (ROS), such as through the action of the catalase enzyme. Potent catalase activity is one of the defining biochemical characteristics of P. mirabilis, and the single catalase (katA) gene in strain HI4320 was recently identified as a candidate fitness factor for UTI, CAUTI, and bacteremia. Here, we show that disruption of katA results in increased ROS levels, increased sensitivity to peroxide, and decreased biofilm biomass. The biomass defect was due to a decrease in the production of extracellular polymeric substances (EPS) by the ΔkatA mutant and specifically due to reduced carbohydrate content. Importantly, the biofilm defect resulted in decreased antibiotic resistance in vitro and a colonization defect during experimental CAUTI. The ΔkatA mutant also exhibited decreased fitness in a bacteremia model, supporting a dual role for catalase in P. mirabilis biofilm development and immune evasion.
KEYWORDS: CAUTI, EPS, Proteus mirabilis, bacteremia, biofilms, catalase, catheter, hydrogen peroxide
INTRODUCTION
Proteus mirabilis is a Gram-negative bacterium that is present in numerous environments, such as soil, water, and the intestinal tracts of both humans and animals (1). However, P. mirabilis is also a leading uropathogen of catheter-associated urinary tract infections (CAUTIs), which are one the most common health care-associated infections worldwide (2–4). CAUTIs account for up to 80% of all nosocomial UTIs, and P. mirabilis CAUTIs are often complicated by bladder and kidney stone formation (urolithiasis), permanent renal damage, and progression to life-threatening bacteremia and sepsis (5–8). For instance, P. mirabilis is responsible for 12 to 31% of bacteremias in nursing home residents and is associated with a 1-year mortality rate ranging from 10% to 66% (9–13).
P. mirabilis CAUTIs have proven difficult to treat due to increasing antibiotic resistance and the formation of resistant biofilms on the catheter surface (14–17). P. mirabilis is particularly well adapted for biofilm formation, causing the encrustation and blockage of urethral catheters by the formation of crystalline biofilm structures (6, 18–22). Notably, biofilms containing P. mirabilis have been detected on catheters even after antibiotic treatment (23–25). As P. mirabilis biofilms play a significant role in both the pathogenesis and treatment of CAUTIs (4, 6, 26, 27), it is critical to investigate factors that influence biofilm development and antibiotic resistance in this species.
Biofilm formation is a multistep process, involving irreversible microbial attachment to a substrate, development of microcolonies, the production of extracellular polymeric substances (EPS), maturation, and dispersal, which is thought to contribute to persistent infection during CAUTIs and dissemination to the bloodstream (6, 28–30). The EPS matrix is highly hydrated (98% water) and comprised largely of proteins, carbohydrates, and extracellular DNA (eDNA); however, compositions vary depending on the bacterial strain, environmental conditions, and the maturation stage of the biofilm (31–34). EPS is considered to be a critical component of bacterial biofilms, as it defines the biofilm structure and is essential for the overall integrity and function of the biofilm (19, 28, 33, 35–37).
The EPS matrix also plays a substantial role in the increased antimicrobial resistance exhibited by bacterial biofilms (17, 37, 38). There are several ways in which biofilm formation impacts antimicrobial susceptibility. First, the antimicrobial agents must diffuse through the EPS matrix in order to act on the organisms within the biofilm. The EPS effectively acts as a shield that prevents diffusion of antimicrobials either by binding and chemically inhibiting the antimicrobial molecules or by limiting their rate of infiltration (39, 40). The biofilm-associated bacteria also experience reduced growth rates and are less metabolically active, which limits the efficacy of many antimicrobial agents (41). Lastly, the environment that immediately surrounds the biofilm may provide conditions that further protect the biofilm-associated bacteria (41). As a result, biofilm formation on indwelling medical devices poses a serious challenge due to the increased resistance of biofilm-associated organisms to antimicrobial agents (28, 29, 42, 43).
Another factor that contributes to bacterial persistence and pathogenicity is the ability to resist reactive oxygen species (ROS) (44). For instance, many bacterial species produce a catalase enzyme which reduces hydrogen peroxide (H2O2) to water (H2O) and oxygen (O2) and has been shown to protect biofilm-associated Pseudomonas aeruginosa by preventing full penetration of H2O2 through the biofilm matrix (44). Hydrogen peroxide is the longest-lived reactive oxygen species, and it can cause critical damage to both the bacterial cell membrane and essential cellular macromolecules, such as proteins, lipids, and nucleic acids (45, 46). ROS can also contribute to the effectiveness of antibiotic-mediated killing. For example, antibiotic exposure induces oxidative stress in P. aeruginosa, and antibiotic susceptibility was increased by disruption of a gene involved in reducing H2O2 (ahpC), indicating that ROS generation contributed to drug lethality (47). It has therefore been proposed that interfering with bacterial protection against ROS may improve antibiotic treatment, even for biofilms (47–50).
Potent catalase activity is one of the defining biochemical characteristics of P. mirabilis (51), and there are several indications that the single catalase (katA) gene of P. mirabilis is important for both fitness and pathogenicity during infection. For instance, katA was found to be upregulated during infection in a mouse model of UTI (52), it was identified as a candidate fitness factor during both single-species and polymicrobial CAUTI (53), and it was detected as an infection-specific fitness factor for survival in the bloodstream (54). Despite this evidence, the exact role of catalase in P. mirabilis pathogenesis had yet to be experimentally validated. In this study, we disrupted the katA gene in P. mirabilis strain HI4320 and investigated the contribution of catalase to bacterial growth, motility, ROS tolerance, biofilm formation, antibiotic resistance, pathogenicity within the urinary tract, and fitness within the bloodstream. We show that the loss of P. mirabilis catalase activity does not alter in vitro growth or planktonic antibiotic susceptibility, but peroxide detoxification is critical for production of a mature biofilm. The absence of catalase severely alters the composition of the EPS matrix, resulting in the increased antibiotic susceptibility of biofilm-associated bacteria. Additionally, we demonstrate that catalase contributes to the pathogenesis of P. mirabilis during CAUTI and fitness during bacteremia.
RESULTS
Generation and characterization of a P. mirabilis ΔkatA mutant.
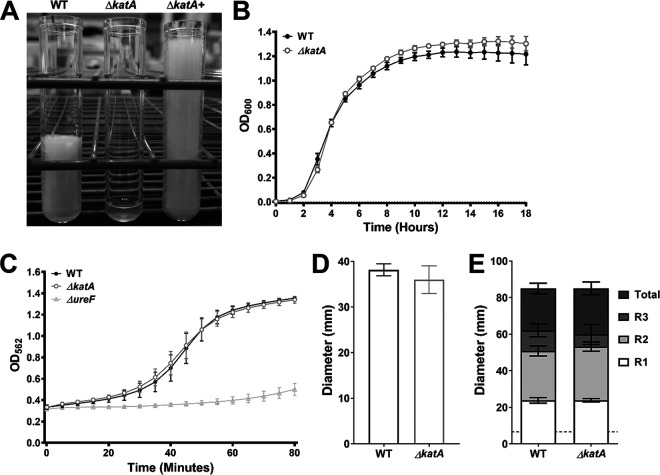
In order to investigate the importance of catalase activity to P. mirabilis growth, biofilm formation, and pathogenesis, the catalase (katA) gene of P. mirabilis strain HI4320 was disrupted by insertion of a kanamycin resistance cassette. Disruption of the katA gene and the resulting catalase activity were confirmed by PCR and a catalase foam height assay (Fig. 1A). As expected, the ΔkatA mutant was catalase negative when exposed to hydrogen peroxide, and providing the katA gene on a plasmid restored catalase activity. The complemented activity was more potent than that of the parental strain, which is likely due to plasmid copy number. The ΔkatA mutant grew similarly to wild-type (WT) P. mirabilis in LB broth and did not exhibit any defects in swimming motility, swarming motility, or urease activity (Fig. 1B to E), indicating that disruption of the katA gene did not affect the standard laboratory behaviors of P. mirabilis. Disruption of katA also had no impact on the expression of either adjacent gene by quantitative reverse transcription-PCR (qRT-PCR) (see Fig. S1 in the supplemental material). Taken together, these data indicate that targeted disruption of katA abrogates catalase activity without obvious polar effects.
FIG 1.
Characterization of a P. mirabilis ΔkatA mutant. The ΔkatA mutant was assessed for catalase activity (A), growth in LB (B), urease activity (C), swimming motility (D), and swarming motility (E) in comparison to those of wild-type P. mirabilis. (A) An SDS/peroxide foam height assay was conducted to provide phenotypic validation of successful generation of a P. mirabilis ΔkatA mutant and plasmid-generated ΔkatA+ complemented strain. (B) Bacterial growth in LB at 37°C was assessed by measurement of OD600 at hourly intervals for 18 h. Error bars represent means ± standard deviations (SD) from 6 independent experiments with 6 technical replicates each (representative graph shown). No significant differences in growth were determined by two-way ANOVA with Dunnett’s test for multiple comparisons. (C) Urease activity in human urine was assessed by measurement of phenol red color change (OD562) at 5-min intervals for 80 min, and a P. mirabilis ΔureF mutant was included as a negative control. Error bars represent means ± SD from 2 independent experiments with 6 technical replicates each. No significant differences between the WT and ΔkatA mutant were identified by two-way ANOVA. (D and E) Bacteria were cultured in LB broth overnight and stab inoculated into motility agar (D) or inoculated onto the surface of swarm agar plates (E). Motility diameter was measured in millimeters after 16 h of incubation at 30°C (D) or 37°C (E). R1 to R3 indicate individual swarm ring diameters. Error bars represent means ± SD from 3 independent experiments with at least 2 replicates each. No significant differences in motility diameter were determined by Student's t test (D) or two-way ANOVA (E).
Disrupting katA increases sensitivity to hydrogen peroxide and steady-state ROS levels.
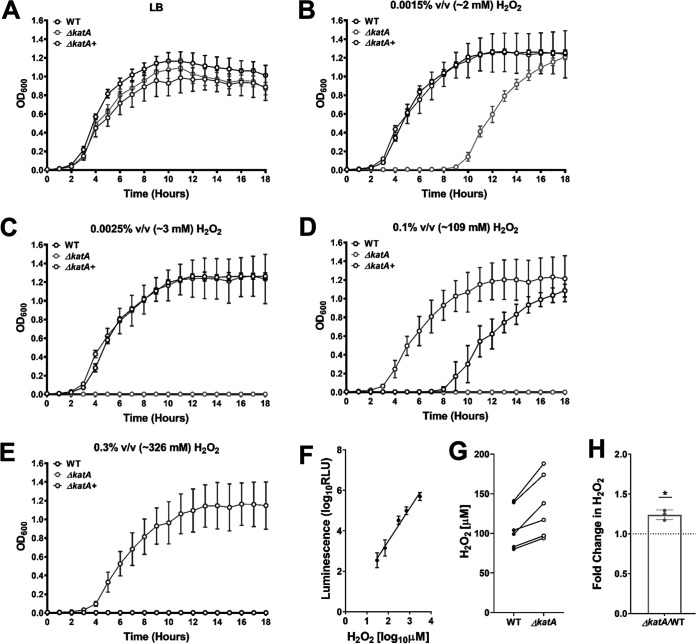
In order to define the limits of hydrogen peroxide tolerance in P. mirabilis, wild-type P. mirabilis and the ΔkatA mutant were cultured in LB broth with increasing concentrations of H2O2, and growth was assessed over the course of 18 h. The ΔkatA mutant was susceptible to ∼2 mM H2O2 and completely inhibited by ∼3 mM (Fig. 2B and C). In contrast, ∼109 mM H2O2 was needed to impair the growth of wild-type P. mirabilis, and ∼326 mM H2O2 was required to achieve complete growth inhibition (Fig. 2D and E). Importantly, providing the katA gene on a plasmid allowed the ΔkatA mutant to tolerate higher H2O2 concentrations than the wild-type strain (Fig. 2E), again likely due to a high copy number.
FIG 2.
Disruption of katA increases sensitivity to hydrogen peroxide and steady-state ROS levels. (A to E) Growth at 37°C was assessed by measurement of OD600 at hourly intervals for 18 h for the WT (black), ΔkatA mutant (light gray), and plasmid-generated ΔkatA+ complemented strain (dark gray) in plain LB (A) or in the presence of increasing concentrations of hydrogen peroxide (H2O2) as follows: 0.0015% vol/vol (∼2 mM) (B); 0.0025% vol/vol (∼3 mM) (C); 0.1% vol/vol (∼109 mM) (D); and 0.3% vol/vol (∼326 mM) (E). Error bars represent means ± standard deviations (SD) from 3 independent experiments with 6 technical replicates each. (F) Peroxide standard curve for measurement of reactive oxygen species in bacterial cultures. (G) H2O2 concentrations present in cultures of the WT (dark gray) and ΔkatA mutant (white) after a 4-h incubation in LB broth. Individual data points represent results from 3 independent experiments with technical duplicates, and the WT and ΔkatA mutant values from each experiment are connected with a black line. (H) Average fold change in ΔkatA mutant H2O2 levels compared to those of the WT. *, P < 0.05 (by one-sample t test).
A luminescence-based H2O2 assay was next used to determine the extent of H2O2 accumulation in wild-type P. mirabilis and the ΔkatA mutant during growth in broth culture in order to gauge the contribution of catalase activity to the detoxification of endogenously produced ROS. For these experiments, a standard curve was generated by supplementing ΔkatA cultures with increasing concentrations of peroxide at the time of ROS measurement (Fig. 2F). After 4 h of culture in LB broth, samples from wild-type P. mirabilis contained an average of 109 ± 26.9 μM H2O2, while the ΔkatA samples contained an average of 135 ± 39.7 μM H2O2 (1.24-fold greater than for the wild type) (Fig. 2G and H). Notably, the concentration of H2O2 in the ΔkatA samples is ∼15-fold lower than the concentration at which the growth of the ΔkatA mutant was perturbed (∼2 mM, as seen in Fig. 2B), suggesting that the loss of catalase activity likely does not induce a significant state of oxidative stress during growth in LB broth. However, it may cause the ΔkatA mutant to be more susceptible to additional ROS insult or other stressors.
The ΔkatA mutant exhibits a defect in biofilm development.
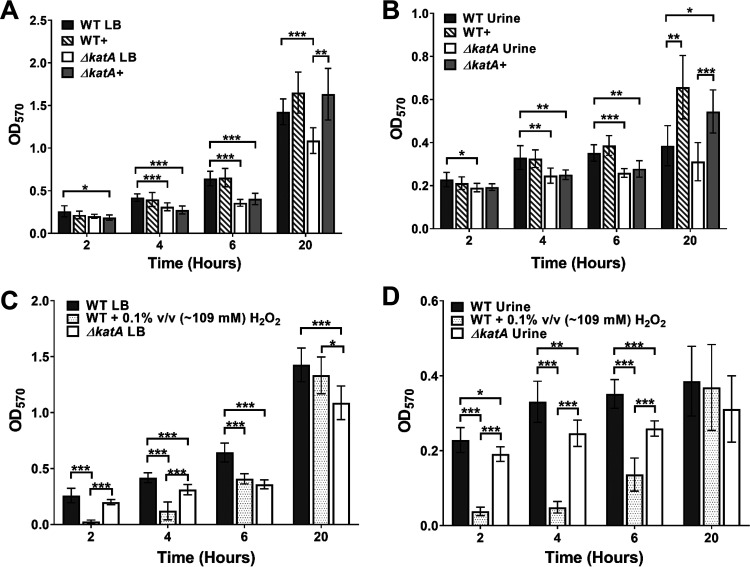
As biofilm formation is a critical component of P. mirabilis pathogenesis during CAUTI, we next assessed biofilm formation by the ΔkatA mutant at 2, 4, 6, and 20 h (Fig. 3). As determined by crystal violet staining, wild-type P. mirabilis established a robust biofilm over the 20-h incubation in LB broth, but the ΔkatA mutant exhibited a significant decrease in total biomass that was detectable at 4 h and remained evident for the rest of the time course (Fig. 3A). Importantly, assessment of viable bacteria within the biofilms at each time point revealed that the defect was not due to a decrease in numbers of bacterial CFU and therefore likely represents a defect in biofilm architecture (Fig. S2). These results were also recapitulated for biofilms established in pooled human urine, a physiologically relevant growth medium for CAUTI (Fig. 3B).
FIG 3.
The ΔkatA mutant exhibits a defect in biofilm development. (A, B) Biofilms of the WT (gray), WT plus bovine catalase (WT+; gray stripes), the ΔkatA mutant (white), and the ΔkatA mutant plus bovine catalase (ΔkatA+, light gray) were formed over a time course of 20 h in LB (A) or pooled human urine (B). Biofilm biomass was assessed at 2, 4, 6, and 20 h via crystal violet staining and detected by determining the OD570. (C, D) Biofilms of the WT (gray), WT plus 0.1% vol/vol (∼109 mM) H2O2 (gray dots), and the ΔkatA mutant (white) were formed over a time course of 20 h in LB (C) or pooled human urine (D). (A to D) Error bars represent means and SD from 3 independent experiments with 3 replicates each. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (by two-way ANOVA with Sidak’s test for multiple comparisons).
To define the specific contribution of peroxide detoxification to P. mirabilis biofilm formation, wild-type P. mirabilis and the ΔkatA mutant were supplemented with an amount of bovine catalase that results in equivalent foam heights produced by 1 × 108 CFU/ml of wild-type P. mirabilis (∼0.7 mg, ∼40,000 units). The addition of bovine catalase had no impact on biofilm development by wild-type P. mirabilis in LB but fully restored biofilm biomass for the ΔkatA mutant in both LB and human urine, albeit not until the 20-h time point (Fig. 3A and B). These data suggest that catalase is important for later stages of biofilm development and maturation rather than the initial stages of attachment and microcolony formation. Importantly, the contribution of bovine catalase was confirmed to specifically derive from enzymatic activity, as both 10-kDa filtration and heat inactivation of the catalase solution abrogated complementation of the ΔkatA mutant biofilm defect (Fig. S3).
To verify that the contribution of catalase to P. mirabilis biofilm development pertains specifically to peroxide detoxification, we next sought to determine if the addition of a nonlethal concentration of peroxide (∼109 mM, as seen in Fig. 2D) could impair biofilm formation by wild-type P. mirabilis. The addition of peroxide at the time of inoculation significantly decreased the P. mirabilis biofilm biomass for the first 6 h of biofilm development, after which time enough of the peroxide was likely broken down to restore full biofilm development (Fig. 3C). Similar results were also recapitulated in pooled human urine (Fig. 3D). Taken together, these data confirm that the biofilm defect of the ΔkatA mutant is specifically due to the loss of catalase activity and peroxide detoxification and likely pertains to biofilm maturation.
We next sought to determine if the biofilm defect of the ΔkatA mutant may be the result of increased oxidative stress. Throughout the time course of biofilm development, samples were taken from the ΔkatA mutant and wild-type P. mirabilis to examine the relative expression levels of two additional peroxide-detoxifying enzymes (alkylhydroperoxidases ahpC PMI1213 and PMI0073) and three superoxide dismutases (sodA, sodB, sodC). The expression of each gene was normalized to that of rpoA, and relative expression over time was determined for the wild-type strain alone and the ΔkatA mutant in comparison to the wild-type (Fig. S4). Expression of genes involved in peroxide detoxification (katA PMI1213 and PMI0073) remained unaffected in the wild-type strain over the time course of biofilm development (Fig. S4A to C); however, levels of expression of genes involved in superoxide detoxification (sodA, sodB, and sodC) were significantly upregulated over time (Fig. S4D to F). This suggests that wild-type P. mirabilis experiences some degree of oxidative stress during biofilm formation, primarily due to superoxide. If disruption of catalase and the resulting accumulation of endogenous peroxide generated substantial oxidative stress, we would expect to see an increased expression of some or all of these genes in the ΔkatA mutant relative to that in wild-type P. mirabilis. However, none of the selected genes exhibited an expression pattern in ΔkatA biofilms that was significantly different from that of wild-type biofilms (Fig. S4H to L). Thus, while P. mirabilis does exhibit signs of oxidative stress during biofilm formation, loss of catalase activity does not cause a substantial increase in oxidative stress.
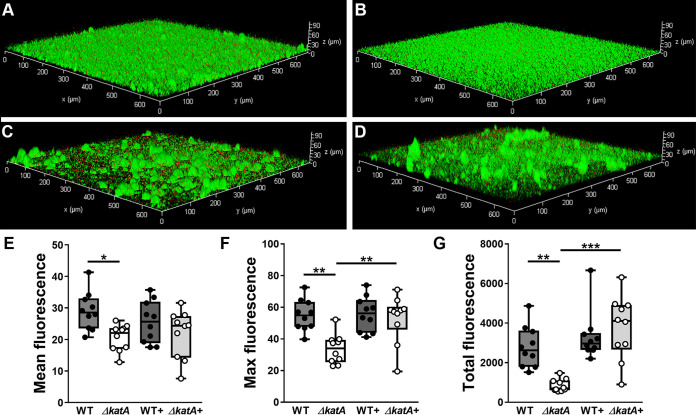
To better assess the contribution of catalase to biofilm maturation and architecture, biofilms were established for 20 h in LB broth, subjected to live/dead staining, and visualized by fluorescence microscopy (Fig. 4). Biofilms formed by wild-type P. mirabilis densely covered the surface substrate, with occasional tower-like structures (Fig. 4A), while those formed by the ΔkatA mutant were flatter and lacked visible three-dimensional structures (Fig. 4B). Interestingly, the addition of bovine catalase at the time of inoculation promoted the development of taller tower-like structures for wild-type P. mirabilis (Fig. 4C) as well as the ΔkatA mutant (Fig. 4D). Compared to wild-type biofilms, ΔkatA mutant biofilms exhibited a significant decrease in both the mean and maximum fluorescence per z-slice (Fig. 4E and F) as well as the total fluorescence of the entire z-stack (Fig. 4G), and the addition of bovine catalase restored maximum fluorescence and total fluorescence to wild-type levels. Taken together, these data indicate that catalase activity substantially impacts P. mirabilis biofilm architecture.
FIG 4.
Catalase contributes to P. mirabilis biofilm architecture. (A to D) Twenty-hour biofilms were formed by the WT (A), the ΔkatA mutant (B), the WT plus bovine catalase (C), and the ΔkatA mutant plus bovine catalase (D) and stained with Syto9 (green) and propidium iodide (red), and z-series images were acquired via fluorescence microscopy. Images are representative of two independent experiments. Total fluorescence (red plus green) was quantified for each z-slice from 10 z-stacks per condition for assessment of the average fluorescence per slice (E), the maximum fluorescence from any single slice in the stack (F), and a sum of the fluorescence from all slices in a stack (G). **, P < 0.01; ***, P < 0.001 (by one-way ANOVA with Tukey’s test for multiple comparisons).
The ΔkatA mutant exhibits a defect in biofilm EPS composition.
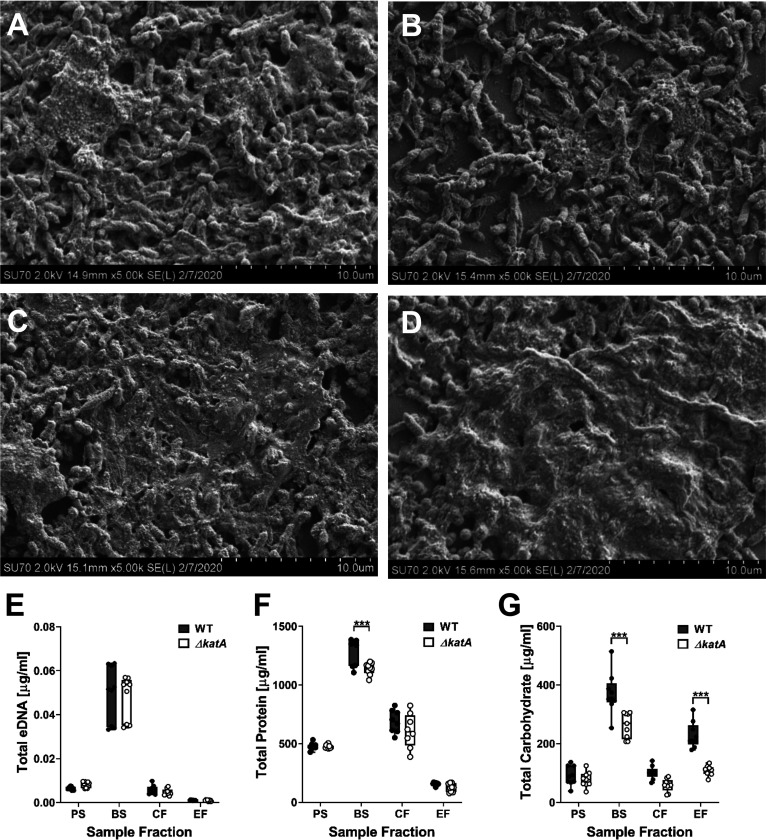
Since the ΔkatA mutant exhibited a defect in biofilm biomass that was independent of cell viability or oxidative stress, these data led us to investigate the overall composition of the biofilm. Aside from bacterial aggregates, the major component of bacterial biofilms is the matrix of extracellular polymeric substances (EPS), which coats and protects the bacterial cells from the surrounding environment. To qualitatively assess biofilm composition, we utilized scanning electron microscopy (SEM). Biofilms formed by wild-type P. mirabilis for 20 h were heavily coated with EPS (Fig. 5A), while biofilms formed by the ΔkatA mutant displayed a similar degree of aggregated clusters of bacteria but lacked the extensive EPS coating of the wild-type biofilms (Fig. 5B). Importantly, the addition of bovine catalase at the time of inoculation slightly enhanced EPS production by wild-type P. mirabilis (Fig. 5C) and fully restored EPS production by the ΔkatA mutant (Fig. 5D). These data indicate that the biomass defect of the ΔkatA mutant is likely due to a decrease in the production of EPS.
FIG 5.
The ΔkatA mutant exhibits a defect in biofilm EPS production. (A to D) Representative SEM images of 20-h biofilms formed by the WT (A), the ΔkatA mutant (B), the WT plus bovine catalase (C), and the ΔkatA mutant plus bovine catalase (D). (E to G) The WT (black circles) and ΔkatA mutant (white circles) were incubated for 20 h with aeration or kept stationary to generate planktonic suspensions (PS) and biofilm (BS) suspensions. EPS was further extracted from the biofilm suspension to generate a cellular fraction (CF) and EPS fraction (EF) for measurement of eDNA (E), protein (F), and carbohydrate (G). Box and whisker plots display the combined data from 3 independent experiments with 3 technical replicates each. ***, P < 0.001 (by two-way ANOVA with Sidak’s test for multiple comparisons).
The exact composition of P. mirabilis HI4320 biofilm EPS has yet to be fully elucidated; however, the EPS of bacterial biofilms is generally comprised of proteins, carbohydrates, and extracellular DNA (eDNA). To assess biofilm EPS composition, wild-type P. mirabilis and the ΔkatA mutant were incubated for 20 h in LB broth with aeration (planktonic suspension [PS]) or while stationary in wells of a 24-well plate from which a total biofilm sample was collected (biofilm suspension [BS]). The remaining biofilm material was then treated with formaldehyde to fix the bacterial cells and prevent lysis during subsequent steps of EPS extraction and NaOH addition to promote dissociation of the EPS from the biofilm and increase solubility. Following these treatments, a cellular fraction (CF) and dialyzed EPS fraction (EF) were collected. No differences in eDNA concentration were observed between strains in any of the tested fractions (Fig. 5E), and total protein measurement revealed only a slight decrease in the ΔkatA biofilm suspension compared to that in the wild-type suspension (Fig. 5F). However, total carbohydrate measurement revealed a substantial decrease in the biofilm suspension and EPS fraction of the ΔkatA mutant compared to those of wild-type P. mirabilis (Fig. 5G). Thus, the biofilm biomass defect of the ΔkatA mutant derives from a decrease in EPS carbohydrate content.
ΔkatA biofilms exhibit increased antibiotic susceptibility.
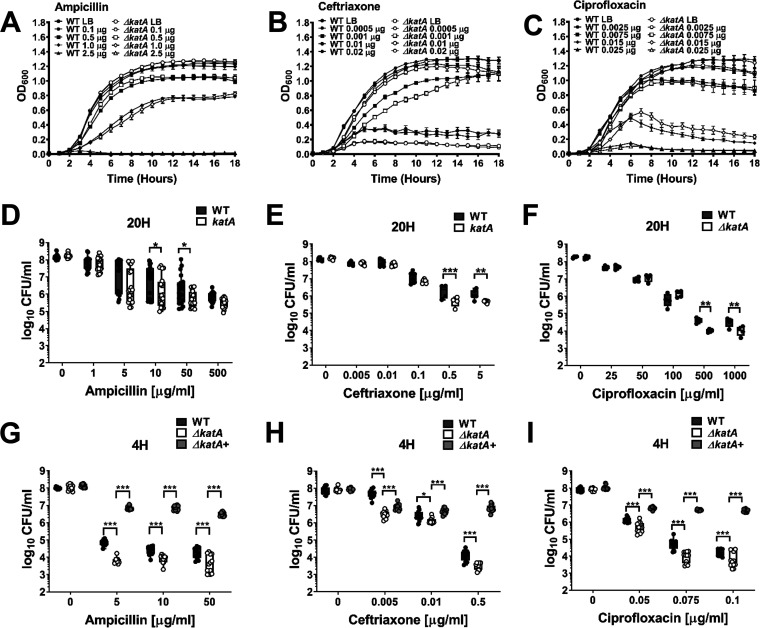
Due to the enhanced antimicrobial resistance of bacteria in a biofilm mode of growth, we sought to determine the impact of the ΔkatA biofilm biomass defect on antibiotic susceptibility. Wild-type P. mirabilis and the ΔkatA mutant were treated with two antibiotics that target the bacterial cell wall (ampicillin and ceftriaxone) and one that targets DNA replication (ciprofloxacin) during planktonic growth or after 20 h of biofilm growth. The ΔkatA mutant did not exhibit substantial differences in susceptibility to ampicillin or ciprofloxacin during planktonic growth, although a slight increase was detected for ceftriaxone at 0.02 and 0.01 μg/ml (Fig. 6A to C). However, 20-h ΔkatA biofilms were more susceptible to all three antibiotics, suggesting that the decrease in biofilm biomass impacts antibiotic susceptibility (Fig. 6D to F). To further explore the importance of the biofilm defect for antibiotic susceptibility, biofilms formed by the wild type and the ΔkatA mutant were also treated after only 4 h of development, as this was the earliest time point corresponding to a significant decrease in biofilm biomass for the ΔkatA mutant. Earlier exposure to antibiotics resulted in greater killing of both strains and further magnified the defect of the ΔkatA mutant (Fig. 6G to I). Importantly, the addition of bovine catalase restored the antibiotic resistance of the ΔkatA mutant biofilms to levels that were equivalent to or even greater than in the wild-type strain (Fig. 6G to I). Thus, we conclude that the loss of katA results in a biofilm-specific increase in antibiotic susceptibility due to disruption of EPS production and biofilm maturation.
FIG 6.
ΔkatA biofilms exhibit increased antibiotic susceptibility. (A to C) Growth at 37°C was assessed by measurement of OD600 at hourly intervals for 18 h for the WT (black symbols) and ΔkatA mutant (white symbols) in the presence of increasing concentrations (micrograms per milliliter) of ampicillin (A), ceftriaxone (B), or ciprofloxacin (C). Error bars represent means ± SD for 6 technical replicates, and graphs are representative of at least 3 independent experiments. Data were analyzed by two-way ANOVA with Tukey’s test for multiple comparisons. (D to I) Biofilms were established by the WT and ΔkatA mutant for 20 h (D to F) or 4 h (G to I), followed by treatment with increasing concentrations of ampicillin (D and G), ceftriaxone (E and H), or ciprofloxacin (F and I) for an additional 20-h period. Each black (WT), white (ΔkatA mutant), and light gray (ΔkatA mutant plus bovine catalase) circle represents the number of CFU per milliliter recovered from an individual biofilm, and box and whisker plots display combined results from at least 3 independent experiments with at least 3 technical replicates each. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (by two-way ANOVA with Sidak’s test for multiple comparisons).
The biofilm biomass defect of the ΔkatA mutant can be recapitulated on Foley catheters.
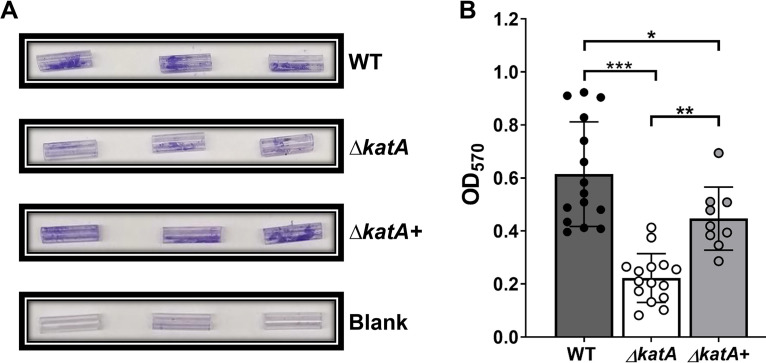
The bacterial biofilms that drive persistent infection in CAUTIs are most often formed on the surfaces and within the lumens of the catheters themselves. We therefore sought to determine if the ΔkatA biofilm biomass defect is similarly present when biofilms are formed directly on a silicone Foley catheter. Sterile Foley catheters (size 14 French) were cut into 15-mm segments, suspended in the wells of a 24-well plate, and inoculated with either wild-type P. mirabilis or the ΔkatA mutant in LB broth. Biofilms were established for 20 h, after which time the catheter segments were removed and air dried, and biofilm biomass was measured using an optimized crystal violet assay. Wild-type P. mirabilis established robust biofilms on the silicone catheter segments, and the ΔkatA mutant again exhibited a substantial biomass defect that could be complemented by the addition of bovine catalase at the time of inoculation (Fig. 7A and B). Thus, the ΔkatA biofilm biomass defect is present on a physiologically relevant biofilm substrate for CAUTI.
FIG 7.
The ΔkatA mutant exhibits a biofilm biomass defect on silicone catheters. (A) Representative images of crystal violet-stained 15-mm silicone Foley catheter segments after 20 h of incubation with the WT, the ΔkatA mutant, the ΔkatA mutant plus bovine catalase, or plain LB (blank). (B) Biomasses of 20-h WT (gray), ΔkatA mutant (white), or ΔkatA mutant plus bovine catalase (light gray) catheter biofilms as assessed by crystal violet staining at an OD570. Error bars represent means and SDs from at least 3 independent experiments with at least 3 technical replicates each. **, P < 0.01; ***, P < 0.001 (by one-way ANOVA with Tukey’s test for multiple comparisons).
Catalase contributes to both pathogenesis and fitness in vivo.
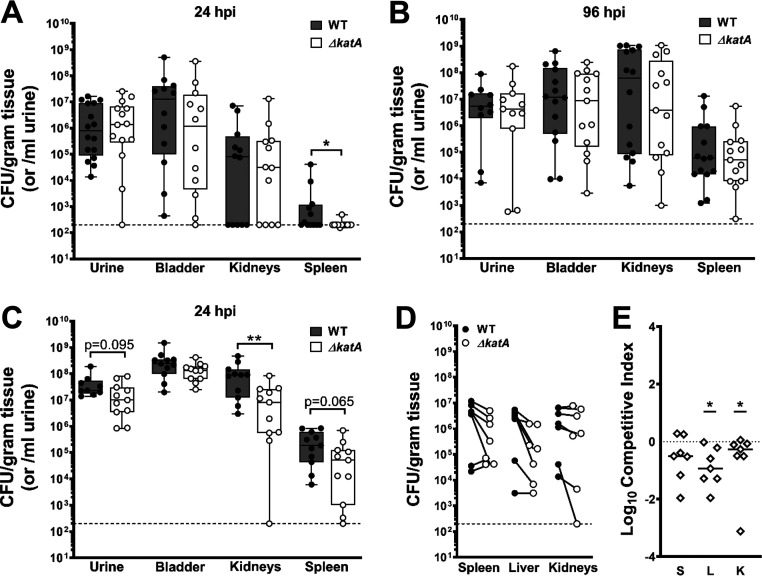
Considering the important contribution of bacterial biofilms to CAUTIs in humans, we next sought to determine the contribution of P. mirabilis catalase to colonization and pathogenicity during experimental CAUTI. To test this, we utilized a mouse model of CAUTI as previously described (55). Briefly, female CBA/J mice were transurethrally inoculated with 1 × 105 CFU of either wild-type P. mirabilis or the ΔkatA mutant, and a 4-mm segment of sterile silicone catheter tubing was inserted into the bladder at the time of inoculation to recapitulate CAUTI. Weight loss was monitored daily (Fig. S5), and mice were euthanized at 24 or 96 h postinoculation (hpi) to quantify bacterial burden in the urine, bladder, and kidneys, as well as the spleen as an indication of bacteremia. Infection with wild-type P. mirabilis resulted in slightly greater weight loss at 24 hpi than infection with the ΔkatA mutant, but no significant differences were detected at later time points (Fig. S5A). Similar colonization levels were observed between strains in the urine, bladder, and kidneys at 24 hpi. However, the ΔkatA mutant exhibited reduced spleen colonization at this time point (Fig. 8A), which may indicate a defect either in dissemination from the urinary tract to the bloodstream or decreased survival once within the bloodstream. Mice from both infection groups exhibited increased bacterial burdens and disease severity at 96 hpi compared to those at 24 hpi, although there were no significant differences between infection groups at this time point (Fig. 8B and Table 1). Thus, while the ΔkatA mutant exhibited reduced numbers of spleen CFU at 24 hpi, pathogenesis was equivalent to that in the wild type by 96 hpi.
FIG 8.
katA contributes to P. mirabilis CAUTI and bacteremia. (A, B) Female CBA/J mice were transurethrally inoculated with 50 μl of a bacterial suspension containing ∼2 × 106 CFU/ml of either the WT or the ΔkatA mutant, and a 4-mm segment of silicone catheter tubing was advanced into the bladder during inoculation. Mice were euthanized at 24 (A) and 96 (B) hpi for determination of numbers of bacterial CFU in the urine, bladder, kidneys, or spleen. Each black (WT) and white (ΔkatA mutant) circle represents the log10 number of CFU per milliliter of urine or gram of tissue from an individual mouse, and the dashed line indicates the limit of detection. *, P < 0.05; **, P < 0.01 (by the nonparametric Mann-Whitney test). (C) Female CBA/J mice were transurethrally inoculated via insertion of a 4-mm segment of silicone catheter tubing that had been precolonized by either the WT or the ΔkatA mutant for 12 h to establish a biofilm (∼2 × 106 CFU/ml). Mice were euthanized at 24 hpi for analysis of CFU as described above. (D, E) Female CBA/J mice were inoculated via tail vein injection of 100 μl of a bacterial suspension containing 1 × 108 CFU/ml of a 1:1 mixture of wild-type P. mirabilis and the ΔkatA mutant. Mice were euthanized at 24 hpi for determination of bacterial CFU in the spleen, liver, and kidneys. (D) Each black (WT) and white (ΔkatA mutant) circle connected with a solid line represents the log10 number of CFU per gram of tissue from an individual mouse. (E) A competitive index (CI) was calculated for the ΔkatA mutant on a per-mouse basis for the spleen (S), liver (L), and kidneys (K) from the ratio of mutant to wild-type cells recovered from the organ divided by the ratio of mutant to wild-type cells in the inoculum. Each data point represents the log10 CI from an individual mouse. Solid lines represent the median, and dashed lines indicate a competitive index of 1 or a 1:1 ratio of mutant to wild-type cells. *, P < 0.05 (by the Wilcoxon signed-rank test).
TABLE 1.
CAUTI disease severitya
| CAUTI disease severity | 24-h standard CAUTI model |
96-h standard CAUTI model |
24-h precolonized CAUTI model |
24-h standard CAUTI model vs 24-h precolonized CAUTI model (WT comparison) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WT | ΔkatA mutant | P value | WT | ΔkatA mutant | P value | WT | ΔkatA mutant | P value | CAUTI | Precolonized | P value | |
| Bladder hematoma and/or blood in urine | 0/12 | 0/12 | N/A | 0/14 | 0/13 | N/A | 8/11 | 0/11 | 0.0004 | 0/12 | 8/11 | 0.0003 |
| Kidney hematoma | 0/12 | 0/12 | N/A | 0/14 | 0/13 | N/A | 2/11 | 0/11 | 0.1380 | 0/12 | 2/11 | 0.1221 |
| Kidney color change and/or mottling | 0/12 | 0/12 | N/A | 3/14 | 3/13 | N/A | 8/11 | 5/11 | 0.1930 | 0/12 | 8/11 | 0.0003 |
| Kidney stone | 0/12 | 0/12 | N/A | 5/14 | 2/13 | 0.2280 | 4/11 | 0/11 | 0.0270 | 0/12 | 4/11 | 0.0215 |
| Any abnormality | 0/12 | 0/12 | N/A | 7/14 | 4/13 | 0.3090 | 9/11 | 5/11 | 0.0760 | 0/12 | 9/11 | 0.0001 |
All mice were assessed for the following parameters at the time of urine collection and euthanasia: bladder hematoma and/or blood in urine, kidney hematoma, kidney color change and/or mottling, and kidney stones. “Any abnormality” indicates whether at least one of these parameters was detected in a mouse. Data represent the number of mice positive for the above parameters out of the total number of mice infected for each strain during 24 h and 96 h of the standard CAUTI model and during 24 h of the precolonized CAUTI model, as well as a comparison of mice infected with the WT between the two models at 24 h. P values were determined by the chi-square test. N/A, not applicable.
A caveat to the CAUTI model is that the bacterial inoculum is introduced as a suspension, and biofilm formation occurs concurrently with defense against the host immune response. In order to isolate the contribution of the ΔkatA biofilm defect to P. mirabilis pathogenesis during CAUTI, we next inoculated mice with catheter segments that had been precolonized for 12 h by either wild-type P. mirabilis or the ΔkatA mutant to establish catheter biofilms. Importantly, precolonizing the catheter segments for 12 h resulted in an inoculum similar to that used for the injection of a bacterial suspension (∼1 × 105 CFU) but caused a more robust infection, with pronounced weight loss and disease severity after just 24 hpi (Fig. S5B and Table 1), such that the experiment could not be extended past this time point. Mice infected with wild-type P. mirabilis-precolonized catheters exhibited more uniform numbers of urine and bladder CFU than those in the traditional CAUTI model (compare Fig. 8C and A), as well as a significantly higher incidence of hematuria and hematoma, kidney discoloration and mottling, and macroscopically visible kidney stones (Table 1). Infection with the ΔkatA mutant also displayed greater uniformity in numbers of urine and bladder CFU in the precolonized model than in the traditional CAUTI model; however, in comparison to wild-type P. mirabilis, the ΔkatA mutant exhibited a profound decrease in kidney colonization and a significantly lower incidence of hematuria, bladder hematoma, and kidney stones, as well as a trend toward reduced bacterial burden in the urine and spleen (Fig. 8C and Table 1). Based on these data, the wild type displays greater infection severity and the ΔkatA mutant displays a more pronounced defect when the infection is seeded from a precolonized catheter, which underscores the importance of P. mirabilis biofilm development and EPS composition for CAUTI pathogenesis.
While the precolonized-catheter model clearly demonstrates a pathogenesis contribution for catalase stemming from a role in biofilm development, these studies cannot exclude a role for catalase in the defense against ROS. Neutrophils are the first and most abundant immune cells recruited to the bladder in response to CAUTI, and a neutrophil oxidative burst represents a robust source of ROS (56–58). Neutrophils appear to be the major factor that limits bacterial growth in the urinary tract during the initial 24 h postinfection (59), and many of the defense strategies used by neutrophils rely on oxidative mechanisms involving ROS (58, 60, 61). The oxidative burst of a single neutrophil has been shown to generate 1 to 10 μM hydrogen peroxide, and neutrophils in suspension (∼106/ml) are capable of producing steady-state concentrations of ∼10 μM (56). Thus, the decreased numbers of kidney and spleen CFU for the ΔkatA mutant support an additional role for catalase in protection against host defenses during dissemination and possibly survival within the bloodstream, in which biofilm formation is not likely to play a role.
To investigate the biofilm-independent fitness of the ΔkatA mutant, mice were inoculated via tail vein injection of a 1:1 mixture of wild-type and ΔkatA P. mirabilis. All mice exhibited high bacterial burdens in the spleen, liver, and kidneys 24 hpi (Fig. 8D), and the ΔkatA mutant was significantly outcompeted by wild-type P. mirabilis in the liver and kidneys, indicating a biofilm-independent fitness defect (Fig. 8E). As a neutrophil oxidative burst represents a potent source of hydrogen peroxide during CAUTI, we next investigated the contribution of P. mirabilis catalase to its defense against opsonophagocytic killing by neutrophils. Interestingly, the ΔkatA mutant was no more susceptible to opsonophagocytic killing than wild-type P. mirabilis (Fig. S6). These data indicate that the contribution of catalase to fitness within the bloodstream is likely due to factors other than the defense against neutrophil opsonophagocytosis.
DISCUSSION
The high degree of resistance against antimicrobials and host defenses that is conferred by bacterial biofilms represents a substantial challenge for the effective treatment of medical device-related infections, including CAUTIs. Catheter biofilms containing P. mirabilis pose an additional challenge, as they are typically comprised of bacterial aggregates, EPS, and crystalline deposits as a result of urease activity, and these crystalline biofilms can result in complete obstruction of urine flow (6). In general, P. mirabilis clinical isolates also exhibit a high degree of antimicrobial resistance, including intrinsic tolerance to tetracycline and polymyxin class drugs, acquisition of resistance to aminoglycosides and fluoroquinolones, and the spread of extended-spectrum beta-lactamases and carbapenemases (62–65). Combined with the added resistance provided by the biofilm mode of growth, these properties collectively contribute to the high mortality rate for P. mirabilis CAUTI and bacteremia (66–70). Investigation of the processes that contribute to P. mirabilis biofilm development and antimicrobial resistance is therefore critical to uncover new potential targets for prevention and treatment.
P. mirabilis has long been characterized as having potent catalase activity, and catalase (katA) has been implicated as contributing to infection in three prior genome-wide screens. In these screens, (i) katA expression was upregulated during infection in a mouse model of UTI (52), (ii) katA was identified as a candidate fitness factor during both single-species and polymicrobial CAUTI (53), and (iii) katA was detected as an infection-specific fitness factor for survival in the bloodstream (54). We therefore endeavored to delineate the contribution of catalase to several stages of P. mirabilis pathogenesis. Here, we show that disruption of catalase results in a slight increase in ROS levels during growth in broth culture, which does not appear to induce oxidative stress in P. mirabilis but does increase sensitivity to additional peroxide insult. We also uncovered a critical role for peroxide detoxification via catalase activity in the production of a mature biofilm by P. mirabilis. Specifically, the loss of catalase activity decreased biofilm biomass due to a severe alteration of EPS composition, particularly the carbohydrate fraction. Importantly, this EPS defect rendered the ΔkatA biofilms more susceptible to antibiotics and also decreased colonization and dissemination in a mouse model of CAUTI.
The exact sugar moieties that comprise the EPS of P. mirabilis strain HI4320 have yet to be fully elucidated; however, hexose moieties previously identified in the LPS outer core of P. mirabilis (glucose, galactose, and N-acetylglucosamine) are likely involved (71, 72). Further exploration is needed to identify the specific carbohydrate moieties that are decreased in the ΔkatA mutant and to determine how loss of catalase activity impacts carbohydrate biosynthesis and EPS production. Based on the transcription profiles of select oxidative-stress response genes, the slight increase in hydrogen peroxide accumulation in ΔkatA biofilms does not appear to induce a more pronounced state of oxidative stress than what is experienced by the wild-type strain, but it may cause sufficient damage to perturb EPS production or linkages. In the absence of catalase, the accumulated hydrogen peroxide may become converted to hydroxyl radicals capable of stealing hydrogen atoms from the C–H bonds of aldoses, uronic acids, and other sites on carbohydrates (73). Ultimately, this process can damage glycosidic linkages and cause the breakdown of polysaccharide chains. The accumulation of hydrogen peroxide in the ΔkatA mutant and any resulting damage may also result in activation of other stress response genes, which may indirectly interfere with the regulation of carbohydrate biosynthesis and EPS production.
EPS defects have been demonstrated to impact the biofilm formation and antibiotic susceptibility of other Gram-negative bacteria. For instance, katA is important for P. aeruginosa biofilm development and biofilm-dependent resistance of treatment with high concentrations of H2O2 (74), and biofilms formed by P. aeruginosa Δpsl (polysaccharide synthesis locus) mutants exhibit increased antibiotic susceptibility (75, 76). Our data also fit within existing literature, which has revealed a contribution of catalase to biofilm architecture (77). In Mycoplasma pneumoniae (an organism that lacks superoxide dismutase and catalase), treatment with catalase enhanced biofilm formation and altered biofilm structure such that fewer and smaller tower structures were produced, and the resulting biofilms were smoother and more homogenous (3). Excess catalase appears to have the opposite effect on P. mirabilis, as the addition of bovine catalase promoted the development of tower structures. A full determination of P. mirabilis HI4320 EPS carbohydrate composition will be necessary to reveal which components are altered when catalase is disrupted and which (if any) are increased in the wild-type strain upon supplementation with excess catalase.
We also revealed that disruption of P. mirabilis catalase results in a biofilm-dependent defect in a mouse model of CAUTI and a biofilm-independent defect in fitness within the bloodstream, thereby demonstrating two separate contributions of the P. mirabilis catalase enzyme to pathogenesis. Precolonizing the catheter resulted in greater overall bacterial colonization during CAUTI than delivery of a bacterial suspension, which highlights the importance of biofilm formation in CAUTI pathogenesis. The precolonized catheter also resulted in greater infection severity overall, as seen by the increased incidence of hematuria and hematoma, kidney discoloration and mottling, and macroscopically visible kidney stones. These results are in agreement with other investigations concerning the pathogenic potential of biofilm-associated bacteria and bacterial populations that are newly dispersed from a biofilm (77). For instance, Streptococcus pneumoniae cells that have been dispersed from a biofilm have distinct phenotypic properties that differ from those of either biofilm-associated or broth-grown planktonic bacteria, including differential gene expression and an increased ability to disseminate and cause infection (78).
The colonization defect of the ΔkatA mutant that was observed in the spleen during both CAUTI models is also notable, as it may indicate a reduced capacity of the mutant to disseminate from the urinary tract to the spleen, a defect in resistance against host defenses and survival within the bloodstream or a combination thereof. Since the ΔkatA mutant also exhibited a fitness defect during direct competition with the wild-type strain in a bloodstream infection, the spleen colonization defect likely pertains to a reduced capacity to survive host immune defenses, such as a neutrophil oxidative burst. However, the ΔkatA mutant was no more susceptible to neutrophil opsonophagocytic killing than the parental strain. One possible explanation for this observation is that neutrophil-mediated opsonophagocytic killing of P. mirabilis may be due largely to nonoxidative mechanisms, such as granule fusion rather than oxidative burst (79–81). Alternatively, P. mirabilis catalase may be more critical for resistance to an oxidative burst from other cell types, such as macrophages (82, 83). A third possibility is that the slight increase in peroxide levels that occurs in the ΔkatA mutant may render it more sensitive to other oxidative and nonoxidative host defenses. While the ΔkatA mutant did not exhibit any differences in expression of select genes involved in oxidative stress, other stress responses may be induced in response to the slight increase in peroxide and resulting damage. If so, the heightened stress state may ultimately drive the biofilm defects and colonization defects that were observed for the ΔkatA mutant. Further exploration will be necessary to fully elucidate the transcriptional profile of the ΔkatA mutant and the role of P. mirabilis catalase in immune evasion.
In summary, peroxide detoxification via catalase activity is a critical component of P. mirabilis pathogenesis due to a role in biofilm development, antimicrobial resistance, and the resistance of host defenses. Elucidation of the underlying mechanism through which catalase impacts carbohydrate and EPS production may uncover new strategies for preventing P. mirabilis biofilm formation, disrupting existing biofilms, or increasing sensitivity to ROS and antimicrobial agents. Such strategies may be particularly beneficial for patient populations requiring long-term catheterization.
MATERIALS AND METHODS
Ethics statement.
All animal protocols were approved by the Institutional Animal Care and Use Committee (IACUC) at the State University of New York at Buffalo, Jacobs School of Medicine and Biomedical Sciences (MIC31107Y), and were in accordance with the Office of Laboratory Animal Welfare (OLAW), the U.S. Department of Agriculture (USDA), and the guidelines specified by the Association for Assessment and Accreditation of Laboratory Animal Care, International (AAALAC, Intl.).
Bacterial strains and culture conditions.
Proteus mirabilis strain HI4320 was previously isolated from the urine of a catheterized patient (84). The P. mirabilis ΔkatA mutant was constructed in strain HI4320 by the insertion of a kanamycin resistance cassette into the katA gene according to the Sigma TargeTron group II intron protocol, as previously described (85). Mutants were verified by selection on kanamycin, PCR, and a catalase foam height assay (see below). As insertion of a kanamycin cassette via TargeTron retrohoming can result in polar effects, the ΔkatA mutant was complemented by PCR amplification of the wild-type katA gene and 500 bp of flanking sequences, ligation into a linearized pGEN-MCS vector, electroporation, and selection on ampicillin. Primers used for the generation of the ΔkatA mutant and complemented mutant (ΔkatA+) are provided in Table S1 in the supplemental material. P. mirabilis HI4320 (wild type) and the ΔkatA mutant were routinely cultured at 37°C with aeration in 5 ml of LB broth (10 g/liter tryptone, 5 g/liter yeast extract, 0.5 g/liter NaCl) or on low-salt LB agar (10 g/liter tryptone, 5 g/liter yeast extract, 0.1 g/liter NaCl, 15 g/liter agar). Biofilm assays were performed using LB broth (10 g/liter tryptone, 5 g/liter yeast extract, 0.5 g/liter NaCl) or filter-sterilized human urine (Cone Bioproducts, Sequin, TX).
Catalase foam height assay.
One hundred microliters of an overnight culture of each strain was added to a 12- by 75-mm polystyrene tube. Subsequently, 100 μl of 1% Triton X-100 and 100 μl of 30%, vol/vol, hydrogen peroxide were added to the tubes, which were mixed thoroughly and incubated at room temperature for ∼5 min. Following completion of the reaction, the height of O2-forming foam in the test tube was observed.
Growth curves.
Bacterial overnight cultures were diluted 1:100 into fresh LB and grown in 96-well plates at 37°C with aeration via double orbital shaking in a BioTek Synergy H1 microplate reader. Growth was assessed by measurement of the optical density at 600 nm (OD600) at 15-min intervals for 18 h. For hydrogen peroxide sensitivity growth curves, the indicated concentrations of hydrogen peroxide were added to the media at the time of inoculation.
Urease assay.
Urease activity was measured as described previously (55). Briefly, overnight cultures of bacteria were washed once in sterile saline and adjusted to ∼5 × 109 CFU/ml. Cultures were then diluted 1:10 in filter-sterilized human urine supplemented with 0.001%, wt/vol, phenol red and 500 mM urea and dispensed into the wells of a clear-bottom 96-well plate. Absorbance (OD562) was measured every 30 s for a total of 90 min in a BioTek Synergy H1 reader.
Motility assays.
Swimming motility agar plates (10 g/liter tryptone, 0.5 g/liter NaCl, 3 g/liter agar) were stab inoculated with an overnight culture of wild-type P. mirabilis or the ΔkatA mutant and incubated without inversion at 30°C for 16 h prior to the measurement of swimming diameter. Swarming was assessed by inoculating 5 μl of an overnight culture of wild-type P. mirabilis or the ΔkatA mutant onto the surface of a swarm plate (LB agar with 5 g/liter NaCl); the inoculum was allowed to soak in for ∼10 min, and the plate was incubated at 37°C for 16 h prior to the measurement of the diameter of each swarm ring.
ROS quantification.
The ROS-Glo H2O2 assay kit (Promega) was used per the manufacturer’s specifications, with the following modifications, to measure the basal level of H2O2 present in the wild-type and ΔkatA mutant strain in broth culture. Overnight cultures of each strain were centrifuged, washed once with fresh LB, adjusted to the same optical density, diluted 1:100 in fresh LB, and incubated at 37°C with aeration for 4 h. After the 4-h incubation, 80 μl of each sample was added to a 96-well plate with 20 μl of derivatized luciferin substrate and incubated for 30 min at 37°C. To generate a standard curve, a series of ΔkatA samples were supplemented with increasing concentrations of H2O2 or an equivalent volume of distilled H2O. One hundred microliters of ROS-Glo detection solution was then added to each well, the plate was incubated at room temperature for 20 min, and the relative luminescence units (RLU) were read in a BioTek Synergy H1 microplate reader.
Biofilm formation for determination of viability.
Static biofilm formation was performed in tissue culture-treated 24-well plates (Falcon 353047) by introducing ∼2 × 107 CFU/ml of either wild-type P. mirabilis or the ΔkatA mutant into 1.5 ml LB broth per well and incubating the cultures at 37°C for 20 h. Blank wells for normalization contained only assay medium (LB broth or urine). Following incubation, supernatants were removed and biofilms were washed twice with 1× phosphate-buffered saline (PBS) to remove any remaining planktonic bacteria. A volume of 1.5 ml of sterile PBS was then added to each well, and biofilms were scraped with a sterile micropipette tip to resuspend the viable bacteria contained within each biofilm. Samples underwent serial 10-fold dilutions and were spiral plated (Eddy Jet 2; Neutec Group Inc., Farmingdale, NY) onto low-salt LB agar for enumeration of CFU using a ProtoCOL 3 automated colony counter (Synbiosis).
Biofilm biomass analysis.
Twenty-hour biofilm formation was performed as described above. Where indicated, biofilms were supplemented at the time of inoculation with either 0.7 mg of active bovine catalase liver enzyme (∼40,000 units) or 0.1% vol/vol (∼109 mM) H2O2. Supernatants were then carefully removed from wells of a 24-well plate using a VIAFLO Voyager automated pipette (Integra Biosciences, Hudson, NJ), inserted slowly at a 45° angle (with the technician making sure to avoid touching the sides and bottom of wells), and set to a speed of 2 for gentle aspiration. Biofilms were air dried for 5 min, stained with 1.5 ml of 0.1% crystal violet for 10 min, washed once with distilled H2O to remove excess stain, and solubilized in 2 ml of 95% ethanol for 10 min on an orbital shaker at 210 rpm. The wells of the 24-well plate containing the biofilms were then scraped and resuspended with a micropipette tip to ensure that all stained biofilm material was solubilized. Crystal violet absorbance (OD570) was measured in a BioTek Synergy H1 microplate reader and blanked using cell-free control wells for each biofilm assay.
Fluorescence microscopy.
Biofilms were grown in 24-well optically clear glass-bottom plates (MatTek) using the conditions described above. Supernatants were removed and replaced with 0.9% saline containing 5 μm Syto9 (ThermoFisher) and 30 μM propidium iodide (Cayman Chemical). Images were acquired on a Leica Dmi8 fluorescence microscope using the 20× air objective, and images were processed using Leica THUNDER computational clearing. Values for the mean, maximum, and total fluorescence of z-series images were determined using Fiji (ImageJ) via the multimeasure feature of the region-of-interest manager.
Catheter biofilm biomass analysis.
Biofilms were established in 24-well plates as described above, and a 15-mm segment of a silicone Foley catheter (Bard 57165814; size, 14 French) was placed in the wells at the time of inoculation. After 20 h, catheter segments were transferred into empty wells of a new 24-well plate, all of the residual LB supernatant was removed from the catheter lumen, and the segments were air dried for 15 min. The segments were then stained with 1.5 ml 0.1% crystal violet for 10 min, washed by gentle dunking in distilled H2O to remove excess stain, and solubilized in a microcentrifuge tube containing 1 ml of 95% ethanol. The tubes were then transferred to a water bath sonicator for a total of 10 min (with vortexing every 5 min) to ensure removal of the biofilms from the catheter segments. The tubes were then incubated at room temperature at 220 rpm for 10 min to ensure full solubilization. The now-clean catheter segments were removed from the solubilized solution, solubilized solutions received a final vortex, and crystal violet absorbance (OD570) was measured in a BioTek Synergy H1 reader. Absorbance was blanked using cell-free catheters for each biofilm assay.
SEM.
Biofilms were established on 12-mm circular micro cover glass (VWR) within 24-well plates by following the same protocol as described above. The supernatants were aspirated using a vacuum manifold fitted with a 300-μl tip and inserted slowly at a 45° angle without touching the coverslip. Coverslips were gently transferred via sterile forceps to a new/sterile well, and biofilms were fixed for 1 h with 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer containing 0.075% ruthenium red and 0.075 M lysine acetate, pH 7.2. Samples were rinsed three times with 0.2 M sodium cacodylate buffer containing 0.075% ruthenium red (pH 7.2) and then subjected to graded incubations in 30%, 50%, 75%, 95%, and 100% ethanol. Samples were submerged twice in 100% hexamethyldisilazane and air dried. Scanning electron microscopy (SEM) images were captured at the University at Buffalo South Campus Instrument Center using a Hitachi SU-70 microscope equipped with a tilt stage for side-angle views.
EPS isolation and analysis.
The following protocol was adapted from the work of Bales et al. (86). Biofilms were established in 24-well plates for 20 h as described above, and 20-h planktonic samples of each strain were also grown as described above. At the 20-h time point, the planktonic samples were normalized by optical density, 1 ml of each was centrifuged, the supernatants were removed, the cell pellets were resuspended in sterile Milli-Q H2O, and the resulting planktonic suspension (PS) was aliquoted and frozen until future use. Supernatants were removed from the biofilm samples, and all biofilm material from the entire 24-well plate was scraped and resuspended into 3 ml sterile Milli-Q H2O. A portion of the resulting biofilm suspension (BS) was aliquoted and frozen until future use, and the rest was transferred to sterile microcentrifuge tubes and treated with 6 μl of 37% formaldehyde solution per 1 ml to fix the cells and prevent lysis during subsequent extraction steps. The mixture was incubated at room temperature in a chemical hood with gentle shaking (100 rpm) for 1 h. Four hundred microliters of 1 M NaOH was then added for each 1 ml of biofilm suspension and incubated at room temperature with gentle shaking (100 rpm) for 3 h to extract EPS. Cell suspensions were then centrifuged (20,000 relative centrifugal force [rcf]) for 1 h at 4°C. The supernatant containing soluble EPS was transferred to a new sterile microcentrifuge tube, the cellular pellet was resuspended in 1 ml of sterile Milli-Q H2O, and the resulting cellular fraction (CF) was aliquoted and frozen until future use. The EPS was then filtered through a Spin-X 0.22-μm centrifuge tube filter (Costar 8160) and dialyzed against sterile Milli-Q H2O (∼300 times the volume of the sample) using a 3,500-molecular-weight-cutoff (MWCO) Slide-A-Lyzer dialysis cassette (Thermo Scientific). Samples were dialyzed for ∼18 h at room temperature, with H2O changes at 2 and 4 h. The resulting dialyzed EPS fractions (EF) were aliquoted and frozen until future use.
For EPS component analyses, all isolated fractions (PS, BS, CF, and EF) were thawed and analyzed for characterization of their chemical compositions using commercially available kits. Total carbohydrate was determined by the Dubois phenol-sulfuric acid method (87), adapted to a 96-well plate assay with a total carbohydrate assay kit (MAK 104; Sigma-Aldrich), using d-glucose as the standard, and absorbance was read at a wavelength of 490 nm. Total protein content was determined by the Lowry method (88) adapted to a 96-well plate assay with the Microplate BCA protein assay kit (23252; Thermo Scientific), using bovine serum albumin (BSA) as the standard, and absorbance was read at an OD595. Total eDNA was determined by the Quant-iT PicoGreen double-stranded DNA assay kit (P11496; Invitrogen) adapted to a 96-well plate, using double-stranded lambda DNA as the standard, and relative fluorescence units (RFU) were read with 470-nm excitation and a 525-nm detection. All readings for chemical composition were performed on a BioTek Synergy H1 reader.
Antibiotic susceptibility assay.
The following antibiotics were diluted as per manufacturer specifications and made fresh before each experiment: ampicillin sodium salt (Research Products International; A40040), ceftriaxone sodium salt (Cayman Chemical; 18866), and ciprofloxacin (Tokyo Chemical Industry; C2510). For planktonic bacteria, antibiotic susceptibility was determined by assessing growth in increasing concentrations of each antibiotic via determining the OD600, as described above. Antibiotic susceptibilities of biofilms were determined by establishing biofilms as described above. After either 4 h or 20 h (as indicated in the text), supernatants were carefully removed and replaced with fresh LB containing increasing concentrations of each antibiotic. Biofilms were then incubated for an additional 20 h, and numbers of viable CFU per milliliter were determined as described above.
Mouse model of CAUTI.
CAUTI studies were carried out as previously described (53). Briefly, the inoculum was prepared by washing overnight cultures of wild-type P. mirabilis and the ΔkatA mutant in PBS, adjusting each to an OD600 of 0.2 (∼2 × 108 CFU/ml), and diluting cultures 1:100 to achieve an inoculum of 2 × 106 CFU/ml. Female CBA/J mice aged 6 to 8 weeks (Jackson Laboratory) were anesthetized with a weight-appropriate dose (0.1 ml for a mouse weighing 20 g) of ketamine-xylazine (100 mg/kg of body weight for ketamine and 10 mg/kg for xylazine) by intraperitoneal (i.p.) injection and inoculated transurethrally with 50 μl of the diluted suspension (1 × 105 CFU/mouse). A 4-mm segment of sterile silicone tubing (0.64-mm outside diameter [o.d.], 0.30-mm inside diameter [i.d.]; Braintree Scientific, Inc.) was carefully advanced into the bladder during inoculation and retained for the duration of the study as described previously (55, 89). At both 24 and 96 h postinfection (hpi), urine was collected, the mice were euthanized, and bladders, kidneys, and spleens were harvested into 5-ml Eppendorf tubes containing 1 ml PBS. Tissues were homogenized using a Bullet Blender 5 Gold (Next Advance) and plated using an Eddy Jet 2 spiral plater (Neutec Group) for determination of CFU numbers using a ProtoCOL 3 automated colony counter (Synbiosis).
Mouse model of CAUTI using a precolonized catheter.
Biofilms of wild-type P. mirabilis and the ΔkatA mutant were formed over a 12-h period on 4-mm segments of sterile silicone tubing (0.64-mm o.d., 0.30-mm i.d.; Braintree Scientific, Inc.) and transferred to PBS to remove nonadherent bacteria, resulting in an inoculum of ∼2 × 106 CFU/ml of biofilm-associated bacteria. Female CBA/J mice aged 6 to 8 weeks (Jackson Laboratory) were anesthetized as described above, and precolonized catheter segments were then carefully advanced into the bladder. At 24 hpi, urine was collected, the mice were euthanized, and bladders, kidneys, and spleens were harvested and processed as described above.
Mouse model of bacteremia.
Bacteremia studies were carried out as previously described (90). Briefly, the inoculum was prepared by washing overnight cultures of wild-type P. mirabilis and the ΔkatA mutant in PBS and diluting them in PBS to an OD600 of 0.1 (1 × 108 CFU/ml). Female CBA/J mice aged 6 to 8 weeks (Jackson Laboratory) were inoculated by tail vein injection of 100 μl (1 × 107 CFU/mouse) of a 1:1 mixture of the P. mirabilis HI4320 and ΔkatA mutant suspensions. Mice were euthanized 24 hpi, and organs were harvested into 5-ml Eppendorf tubes containing PBS (1 ml for spleens and kidneys, 2 ml for livers). Tissues were homogenized as described above and plated onto plain LB agar (total CFU) and LB with kanamycin (ΔkatA CFU) for determination of CFU. A competitive index (CI) was calculated as follows for all samples in which bacterial burden was above the limit of detection: CI = (ΔkatA mutant output/WT output)/(ΔkatA mutant input/WT input). A log10 CI of 0 indicates that the ratio of the strains in the output is similar to that in the input, and neither strain had an advantage. A log10 CI of >0 indicates that the ΔkatA mutant has a competitive advantage over the wild type.
Statistical analysis.
Normalcy was assessed for all data sets by the Shapiro-Wilk and D’Agostino-Pearson normality tests. Significance was assessed using two-way analysis of variance (ANOVA), one-way ANOVA, the nonparametric Mann-Whitney test, the unpaired t test, a one-sample t test, the chi-square test, or the Wilcoxon signed-rank test, as indicated in the figure legends. All P values are two tailed at a 95% confidence interval. All analyses were performed using Prism version 7.03 (GraphPad Software, San Diego, CA).
ACKNOWLEDGMENTS
We thank members of the Department of Microbiology & Immunology in the Jacobs School of Medicine and Biomedical Sciences at the University at Buffalo for helpful comments and critiques. SEM analyses were performed with training and assistance from Peter Bush and the University of Buffalo South Campus Instrument Center, and fluorescence microscopy was performed with training and assistance from Wade Sigurdson and the Confocal Microscope and Flow Cytometry Core Facility.
This work was supported by the National Institutes of Health (grants R00 DK105205 and R01 DK123158 to C.E.A.). The sponsors were not involved in the study design, methods, data collections, analysis, or preparation of the paper. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funders.
We have no financial or nonfinancial competing interests to declare.
Footnotes
Supplemental material is available online only.
Contributor Information
Chelsie E. Armbruster, Email: chelsiea@buffalo.edu.
Craig R. Roy, Yale University School of Medicine
REFERENCES
- 1.Wenner JJ, Rettger LF. 1919. A systematic study of the Proteus group of bacteria. J Bacteriol 4:331–353. 10.1128/jb.4.4.331-353.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hooton T, Bradley S, Cardenas D, Colgan R, Geerlings S, C Rice J, Saint S, Schaeffer A, Tambyah P, Tenke P, E Nicolle L. 2010. Diagnosis, prevention, and treatment of catheter-associated urinary tract infection in adults: 2009 International Clinical Practice Guidelines from the Infectious Diseases Society of America. Clin Infect Dis 50:625–663. 10.1086/650482. [DOI] [PubMed] [Google Scholar]
- 3.Warren JW. 2001. Catheter-associated urinary tract infections. Int J Antimicrob Agents 17:299–303. 10.1016/s0924-8579(00)00359-9. [DOI] [PubMed] [Google Scholar]
- 4.Jacobsen SM, Stickler DJ, Mobley HLT, Shirtliff ME. 2008. Complicated catheter-associated urinary tract infections due to Escherichia coli and Proteus mirabilis. Clin Microbiol Rev 21:26–59. 10.1128/CMR.00019-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li X, Zhao H, Lockatell CV, Drachenberg CB, Johnson DE, Mobley HLT. 2002. Visualization of Proteus mirabilis within the matrix of urease-induced bladder stones during experimental urinary tract infection. Infect Immun 70:389–394. 10.1128/IAI.70.1.389-394.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Armbruster CE, Mobley HLT, Pearson MM. 2018. Pathogenesis of Proteus mirabilis infection. EcoSal Plus 8:ESP-0009-2017. 10.1128/ecosalplus.ESP-0009-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Griffith DP, Musher DM, Itin C. 1976. Urease. The primary cause of infection-induced urinary stones. Invest Urol 13:346–350. [PubMed] [Google Scholar]
- 8.Lo E, Nicolle L, Classen D, Arias KM, Podgorny K, Anderson DJ, Burstin H, Calfee DP, Coffin SE, Dubberke ER, Fraser V, Gerding DN, Griffin FA, Gross P, Kaye KS, Klompas M, Marschall J, Mermel LA, Pegues DA, Perl TM, Saint S, Salgado CD, Weinstein RA, Wise R, Yokoe DS. 2008. Strategies to prevent catheter-associated urinary tract infections in acute care hospitals. Infect Control Hosp Epidemiol 29:S41–S50. 10.1086/591066. [DOI] [PubMed] [Google Scholar]
- 9.Setia U, Serventi I, Lorenz P. 1984. Bacteremia in a long-term care facility: spectrum and mortality. Arch Intern Med 144:1633–1635. 10.1001/archinte.1984.00350200143021. [DOI] [PubMed] [Google Scholar]
- 10.Nicolle L, McIntyre M, Hoban D, Murray D. 1994. Bacteremia in a long term care facility. Can J Infect Dis 5:130–132. 10.1155/1994/647804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mylotte JM, Tayara A, Goodnough S. 2002. Epidemiology of bloodstream infection in nursing home residents: evaluation in a large cohort from multiple homes. Clin Infect Dis 35:1484–1490. 10.1086/344649. [DOI] [PubMed] [Google Scholar]
- 12.Melzer M, Welch C. 2013. Outcomes in UK patients with hospital-acquired bacteraemia and the risk of catheter-associated urinary tract infections. Postgrad Med J 89:329–334. 10.1136/postgradmedj-2012-131393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daniels KR, Lee GC, Frei CR. 2014. Trends in catheter-associated urinary tract infections among a national cohort of hospitalized adults, 2001–2010. Am J Infect Control 42:17–22. 10.1016/j.ajic.2013.06.026. [DOI] [PubMed] [Google Scholar]
- 14.Weiner LM, Webb AK, Limbago B, Dudeck MA, Patel J, Kallen AJ, Edwards JR, Sievert DM. 2016. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2011–2014. Infect Control Hosp Epidemiol 37:1288–1301. 10.1017/ice.2016.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peterson LR. 2009. Bad bugs, no drugs: no ESCAPE revisited. Clin Infect Dis 49:992–993. 10.1086/605539. [DOI] [PubMed] [Google Scholar]
- 16.Pendleton JN, Gorman SP, Gilmore BF. 2013. Clinical relevance of the ESKAPE pathogens. Expert Rev Anti Infect Ther 11:297–308. 10.1586/eri.13.12. [DOI] [PubMed] [Google Scholar]
- 17.Stewart PS, Costerton JW. 2001. Antibiotic resistance of bacteria in biofilms. Lancet 358:135–138. 10.1016/s0140-6736(01)05321-1. [DOI] [PubMed] [Google Scholar]
- 18.Macleod SM, Stickler DJ. 2007. Species interactions in mixed-community crystalline biofilms on urinary catheters. J Med Microbiol 56:1549–1557. 10.1099/jmm.0.47395-0. [DOI] [PubMed] [Google Scholar]
- 19.Stickler DJ, Morgan SD. 2008. Observations on the development of the crystalline bacterial biofilms that encrust and block Foley catheters. J Hosp Infect 69:350–360. 10.1016/j.jhin.2008.04.031. [DOI] [PubMed] [Google Scholar]
- 20.Stickler D, Ganderton L, King J, Nettleton J, Winters C. 1993. Proteus mirabilis biofilms and the encrustation of urethral catheters. Urol Res 21:407–411. 10.1007/BF00300077. [DOI] [PubMed] [Google Scholar]
- 21.Stickler D, Morris N, Moreno MC, Sabbuba N. 1998. Studies on the formation of crystalline bacterial biofilms on urethral catheters. Eur J Clin Microbiol Infect Dis 17:649–652. 10.1007/BF01708349. [DOI] [PubMed] [Google Scholar]
- 22.Roshni Amalaradjou MA, Kumar V. 2013. Role of bacterial biofilms in catheter-associated urinary tract infections (CAUTI) and strategies for their control. IntechOpen 10.5772/55200. [DOI] [Google Scholar]
- 23.Muzzi-Bjornson L, Macera L. 2011. Preventing infection in elders with long-term indwelling urinary catheters. J Am Acad Nurse Pract 23:127–134. 10.1111/j.1745-7599.2010.00588.x. [DOI] [PubMed] [Google Scholar]
- 24.Walker JN, Flores-Mireles AL, Lynch AJL, Pinkner C, Caparon MG, Hultgren SJ, Desai A. 2020. High-resolution imaging reveals microbial biofilms on patient urinary catheters despite antibiotic administration. World J Urol 38:2237–2245. 10.1007/s00345-019-03027-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Talsma SS. 2007. Biofilms on medical devices. Home Healthc Nurse 25:589–594. 10.1097/01.NHH.0000296117.87061.14. [DOI] [PubMed] [Google Scholar]
- 26.Wasfi R, Hamed SM, Amer MA, Fahmy LI. 2020. Proteus mirabilis biofilm: development and therapeutic strategies. Front Cell Infect Microbiol 10:414–414. 10.3389/fcimb.2020.00414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacobsen SM, Shirtliff ME. 2011. Proteus mirabilis biofilms and catheter-associated urinary tract infections. Virulence 2:460–465. 10.4161/viru.2.5.17783. [DOI] [PubMed] [Google Scholar]
- 28.Donlan RM. 2001. Biofilm formation: a clinically relevant microbiological process. Clin Infect Dis 33:1387–1392. 10.1086/322972. [DOI] [PubMed] [Google Scholar]
- 29.Flemming HC, Wingender J, Szewzyk U, Steinberg P, Rice SA, Kjelleberg S. 2016. Biofilms: an emergent form of bacterial life. Nat Rev Microbiol 14:563–575. 10.1038/nrmicro.2016.94. [DOI] [PubMed] [Google Scholar]
- 30.Hall-Stoodley L, Costerton JW, Stoodley P. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol 2:95–108. 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 31.Lewandowski Z. 2000. Structure and function of biofilms, p 1–17. In Evans LV (ed), Biofilms: recent advances in their study and control. Harwood Academic Publishers, Amsterdam, Netherlands. [Google Scholar]
- 32.Charlton SGV, White MA, Jana S, Eland LE, Jayathilake PG, Burgess JG, Chen J, Wipat A, Curtis TP. 2019. Regulating, measuring, and modeling the viscoelasticity of bacterial biofilms. J Bacteriol 201:e00101-19. 10.1128/JB.00101-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Di Martino P. 2018. Extracellular polymeric substances, a key element in understanding biofilm phenotype. AIMS Microbiol 4:274–288. 10.3934/microbiol.2018.2.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kostakioti M, Hadjifrangiskou M, Hultgren SJ. 2013. Bacterial biofilms: development, dispersal, and therapeutic strategies in the dawn of the postantibiotic era. Cold Spring Harb Perspect Med 3:a010306. 10.1101/cshperspect.a010306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Donlan RM. 2002. Biofilms: microbial life on surfaces. Emerg Infect Dis 8:881–890. 10.3201/eid0809.020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Flemming H-C, Wingender J. 2010. The biofilm matrix. Nat Rev Microbiol 8:623–633. 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 37.Yan J, Bassler BL. 2019. Surviving as a community: antibiotic tolerance and persistence in bacterial biofilms. Cell Host Microbe 26:15–21. 10.1016/j.chom.2019.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pinto RM, Soares FA, Reis S, Nunes C, Van Dijck P. 2020. Innovative strategies toward the disassembly of the EPS matrix in bacterial biofilms. Front Microbiol 11:952–952. 10.3389/fmicb.2020.00952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stewart PS. 1996. Theoretical aspects of antibiotic diffusion into microbial biofilms. Antimicrob Agents Chemother 40:2517–2522. 10.1128/AAC.40.11.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stewart PS, Raquepas JB. 1995. Implications of reaction-diffusion theory for the disinfection of microbial biofilms by reactive antimicrobial agents. Chem Eng Sci 50:3099–3104. 10.1016/0009-2509(95)00143-S. [DOI] [Google Scholar]
- 41.Burmølle M, Webb JS, Rao D, Hansen LH, Sørensen SJ, Kjelleberg S. 2006. Enhanced biofilm formation and increased resistance to antimicrobial agents and bacterial invasion are caused by synergistic interactions in multispecies biofilms. Appl Environ Microbiol 72:3916–3923. 10.1128/AEM.03022-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bjarnsholt T. 2013. The role of bacterial biofilms in chronic infections. APMIS 121:1–58. 10.1111/apm.12099. [DOI] [PubMed] [Google Scholar]
- 43.Burmølle M, Thomsen TR, Fazli M, Dige I, Christensen L, Homøe P, Tvede M, Nyvad B, Tolker-Nielsen T, Givskov M, Moser C, Kirketerp-Møller K, Johansen HK, Høiby N, Jensen P, Sørensen SJ, Bjarnsholt T. 2010. Biofilms in chronic infections—a matter of opportunity—monospecies biofilms in multispecies infections. FEMS Immunol Med Microbiol 59:324–336. 10.1111/j.1574-695X.2010.00714.x. [DOI] [PubMed] [Google Scholar]
- 44.Stewart PS, Roe F, Rayner J, Elkins JG, Lewandowski Z, Ochsner UA, Hassett DJ. 2000. Effect of catalase on hydrogen peroxide penetration into Pseudomonas aeruginosa biofilms. Appl Environ Microbiol 66:836–838. 10.1128/AEM.66.2.836-838.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Staerck C, Gastebois A, Vandeputte P, Calenda A, Larcher G, Gillmann L, Papon N, Bouchara J-P, Fleury MJJ. 2017. Microbial antioxidant defense enzymes. Microb Pathog 110:56–65. 10.1016/j.micpath.2017.06.015. [DOI] [PubMed] [Google Scholar]
- 46.Cornelis P, Wei Q, Andrews SC, Vinckx T. 2011. Iron homeostasis and management of oxidative stress response in bacteria. Metallomics 3:540–549. 10.1039/c1mt00022e. [DOI] [PubMed] [Google Scholar]
- 47.Yeom J, Imlay JA, Park W. 2010. Iron homeostasis affects antibiotic-mediated cell death in Pseudomonas species. J Biol Chem 285:22689–22695. 10.1074/jbc.M110.127456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Acker H, Gielis J, Acke M, Cools F, Cos P, Coenye T. 2016. The role of reactive oxygen species in antibiotic-induced cell death in Burkholderia cepacia complex bacteria. PLoS One 11:e0159837. 10.1371/journal.pone.0159837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eason MM, Fan X. 2014. The role and regulation of catalase in respiratory tract opportunistic bacterial pathogens. Microb Pathog 74:50–58. 10.1016/j.micpath.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 50.Dariush M-T, Imana SA, Mina KM, Zahra SD, Elmira RZ, Tahmineh EP. 2014. Comparing inhibitory effect of tramadol on catalase of Pseudomonas aeruginosa and mouse liver. Curr Enzyme Inhibit 10:53–57. 10.2174/15734080113099990002. [DOI] [Google Scholar]
- 51.Taylor WI, Achanzar D. 1972. Catalase test as an aid to the identification of Enterobacteriaceae. Appl Microbiol 24:58–61. 10.1128/am.24.1.58-61.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pearson MM, Yep A, Smith SN, Mobley HLT. 2011. Transcriptome of Proteus mirabilis in the murine urinary tract: virulence and nitrogen assimilation gene expression. Infect Immun 79:2619–2631. 10.1128/IAI.05152-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Armbruster CE, Forsyth-DeOrnellas V, Johnson AO, Smith SN, Zhao L, Wu W, Mobley HLT. 2017. Genome-wide transposon mutagenesis of Proteus mirabilis: essential genes, fitness factors for catheter-associated urinary tract infection, and the impact of polymicrobial infection on fitness requirements. PLoS Pathog 13:e1006434. 10.1371/journal.ppat.1006434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Armbruster CE, Forsyth VS, Johnson AO, Smith SN, White AN, Brauer AL, Learman BS, Zhao L, Wu W, Anderson MT, Bachman MA, Mobley HLT. 2019. Twin arginine translocation, ammonia incorporation, and polyamine biosynthesis are crucial for Proteus mirabilis fitness during bloodstream infection. PLoS Pathog 15:e1007653. 10.1371/journal.ppat.1007653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Armbruster CE, Smith SN, Johnson AO, DeOrnellas V, Eaton KA, Yep A, Mody L, Wu W, Mobley HLT. 2017. The pathogenic potential of Proteus mirabilis is enhanced by other uropathogens during polymicrobial urinary tract infection. Infect Immun 85:e00808-16. 10.1128/IAI.00808-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jesaitis AJ, Franklin MJ, Berglund D, Sasaki M, Lord CI, Bleazard JB, Duffy JE, Beyenal H, Lewandowski Z. 2003. Compromised host defense on Pseudomonas aeruginosa biofilms: characterization of neutrophil and biofilm interactions. J Immunol 171:4329–4339. 10.4049/jimmunol.171.8.4329. [DOI] [PubMed] [Google Scholar]
- 57.Hirschfeld J. 2014. Dynamic interactions of neutrophils and biofilms. J Oral Microbiol 6:26102–26102. 10.3402/jom.v6.26102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rada B. 2017. Interactions between neutrophils and Pseudomonas aeruginosa in cystic fibrosis. Pathogens 6:10. 10.3390/pathogens6010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haraoka M, Hang L, Frendéus B, Godaly G, Burdick M, Strieter R, Svanborg C. 1999. Neutrophil recruitment and resistance to urinary tract infection. J Infect Dis 180:1220–1229. 10.1086/315006. [DOI] [PubMed] [Google Scholar]
- 60.Guiton PS, Hannan TJ, Ford B, Caparon MG, Hultgren SJ. 2013. Enterococcus faecalis overcomes foreign body-mediated inflammation to establish urinary tract infections. Infect Immun 81:329–339. 10.1128/IAI.00856-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hayes BW, Abraham SN. 2016. Innate immune responses to bladder infection. Microbiol Spectr 4:UTI-0024-2016. 10.1128/microbiolspec.UTI-0024-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang J-T, Chen P-C, Chang S-C, Shiau Y-R, Wang H-Y, Lai J-F, Huang IW, Tan M-C, Lauderdale T-LY, Hospitals T. 2014. Antimicrobial susceptibilities of Proteus mirabilis: a longitudinal nationwide study from the Taiwan surveillance of antimicrobial resistance (TSAR) program. BMC Infect Dis 14:486–486. 10.1186/1471-2334-14-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kanayama A, Kobayashi I, Shibuya K. 2015. Distribution and antimicrobial susceptibility profile of extended-spectrum β-lactamase-producing Proteus mirabilis strains recently isolated in Japan. Int J Antimicrob Agents 45:113–118. 10.1016/j.ijantimicag.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 64.Girlich D, Bonnin RA, Dortet L, Naas T. 2020. Genetics of acquired antibiotic resistance genes in Proteus spp. Front Microbiol 11:256. 10.3389/fmicb.2020.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Adamus-Bialek W, Zajac E, Parniewski P, Kaca W. 2013. Comparison of antibiotic resistance patterns in collections of Escherichia coli and Proteus mirabilis uropathogenic strains. Mol Biol Rep 40:3429–3435. 10.1007/s11033-012-2420-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ahn JY, Ann HW, Jeon Y, Ahn MY, Oh DH, Kim YC, Kim EJ, Song JE, Jung IY, Kim MH, Jeong W, Ku NS, Jeong SJ, Choi JY, Yong D, Song YG, Kim JM. 2017. The impact of production of extended-spectrum β-lactamases on the 28-day mortality rate of patients with Proteus mirabilis bacteremia in Korea. BMC Infect Dis 17:327–327. 10.1186/s12879-017-2431-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Endimiani A, Luzzaro F, Brigante G, Perilli M, Lombardi G, Amicosante G, Rossolini GM, Toniolo A. 2005. Proteus mirabilis bloodstream infections: risk factors and treatment outcome related to the expression of extended-spectrum beta-lactamases. Antimicrob Agents Chemother 49:2598–2605. 10.1128/AAC.49.7.2598-2605.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tumbarello M, Trecarichi EM, Fiori B, Losito AR, D'Inzeo T, Campana L, Ruggeri A, Di Meco E, Liberto E, Fadda G, Cauda R, Spanu T. 2012. Multidrug-resistant Proteus mirabilis bloodstream infections: risk factors and outcomes. Antimicrob Agents Chemother 56:3224–3231. 10.1128/AAC.05966-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Luzzaro F, Ortisi G, Larosa M, Drago M, Brigante G, Gesu G. 2011. Prevalence and epidemiology of microbial pathogens causing bloodstream infections: results of the OASIS multicenter study. Diagn Microbiol Infect Dis 69:363–369. 10.1016/j.diagmicrobio.2010.10.016. [DOI] [PubMed] [Google Scholar]
- 70.Korytny A, Riesenberg K, Saidel-Odes L, Schlaeffer F, Borer A. 2016. Bloodstream infections caused by multi-drug resistant Proteus mirabilis: epidemiology, risk factors and impact of multi-drug resistance. Infect Dis (Lond) 48:428–431. 10.3109/23744235.2015.1129551. [DOI] [PubMed] [Google Scholar]
- 71.Aquilini E, Azevedo J, Jimenez N, Bouamama L, Tomás JM, Regué M. 2010. Functional identification of the Proteus mirabilis core lipopolysaccharide biosynthesis genes. J Bacteriol 192:4413–4424. 10.1128/JB.00494-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vinogradov E. 2011. Structure of the core part of the lipopolysaccharide from Proteus mirabilis genomic strain HI4320. Biochemistry (Mosc) 76:803–807. 10.1134/S000629791107011X. [DOI] [PubMed] [Google Scholar]
- 73.Duan J, Kasper DL. 2011. Oxidative depolymerization of polysaccharides by reactive oxygen/nitrogen species. Glycobiology 21:401–409. 10.1093/glycob/cwq171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Elkins JG, Hassett DJ, Stewart PS, Schweizer HP, McDermott TR. 1999. Protective role of catalase in Pseudomonas aeruginosa biofilm resistance to hydrogen peroxide. Appl Environ Microbiol 65:4594–4600. 10.1128/AEM.65.10.4594-4600.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Moradali MF, Ghods S, Rehm BHA. 2017. Pseudomonas aeruginosa lifestyle: a paradigm for adaptation, survival, and persistence. Front Cell Infect Microbiol 7:39. 10.3389/fcimb.2017.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Matsukawa M, Greenberg EP. 2004. Putative exopolysaccharide synthesis genes influence Pseudomonas aeruginosa biofilm development. J Bacteriol 186:4449–4456. 10.1128/JB.186.14.4449-4456.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vestby LK, Grønseth T, Simm R, Nesse LL. 2020. Bacterial biofilm and its role in the pathogenesis of disease. Antibiotics (Basel) 9:59. 10.3390/antibiotics9020059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Marks LR, Davidson BA, Knight PR, Hakansson AP. 2013. Interkingdom signaling induces Streptococcus pneumoniae biofilm dispersion and transition from asymptomatic colonization to disease. mBio 4:e00438-13. 10.1128/mBio.00438-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Spitznagel JK, Shafer WM. 1985. Neutrophil killing of bacteria by oxygen-independent mechanisms: a historical summary. Rev Infect Dis 7:398–403. 10.1093/clinids/7.3.398. [DOI] [PubMed] [Google Scholar]
- 80.Standish AJ, Weiser JN. 2009. Human neutrophils kill Streptococcus pneumoniae via serine proteases. J Immunol 183:2602–2609. 10.4049/jimmunol.0900688. [DOI] [PubMed] [Google Scholar]
- 81.Segal AW. 2005. How neutrophils kill microbes. Annu Rev Immunol 23:197–223. 10.1146/annurev.immunol.23.021704.115653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Slauch JM. 2011. How does the oxidative burst of macrophages kill bacteria? Still an open question. Mol Microbiol 80:580–583. 10.1111/j.1365-2958.2011.07612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Iles KE, Forman HJ. 2002. Macrophage signaling and respiratory burst. Immunol Res 26:95–105. 10.1385/IR:26:1-3:095. [DOI] [PubMed] [Google Scholar]
- 84.Mobley HL, Warren JW. 1987. Urease-positive bacteriuria and obstruction of long-term urinary catheters. J Clin Microbiol 25:2216–2217. 10.1128/jcm.25.11.2216-2217.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pearson MM, Mobley HLT. 2007. The type III secretion system of Proteus mirabilis HI4320 does not contribute to virulence in the mouse model of ascending urinary tract infection. J Med Microbiol 56:1277–1283. 10.1099/jmm.0.47314-0. [DOI] [PubMed] [Google Scholar]
- 86.Bales PM, Renke EM, May SL, Shen Y, Nelson DC. 2013. Purification and characterization of biofilm-associated EPS exopolysaccharides from ESKAPE organisms and other pathogens. PLoS One 8:e67950. 10.1371/journal.pone.0067950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dubois M, Gilles K, Hamilton JK, Rebers PA, Smith F. 1951. A colorimetric method for the determination of sugars. Nature 168:167–167. 10.1038/168167a0. [DOI] [PubMed] [Google Scholar]
- 88.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. 1951. Protein measurement with the Folin phenol reagent. J Biol Chem 193:265–275. 10.1016/S0021-9258(19)52451-6. [DOI] [PubMed] [Google Scholar]
- 89.Guiton PS, Hung CS, Hancock LE, Caparon MG, Hultgren SJ. 2010. Enterococcal biofilm formation and virulence in an optimized murine model of foreign body-associated urinary tract infections. Infect Immun 78:4166–4175. 10.1128/IAI.00711-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Smith SN, Hagan EC, Lane MC, Mobley HLT. 2010. Dissemination and systemic colonization of uropathogenic Escherichia coli in a murine model of bacteremia. mBio 1:e00262-10. 10.1128/mBio.00262-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download IAI.00177-21-s0001.pdf, PDF file, 1.2 MB (1.2MB, pdf)