ABSTRACT
Microbial penetration of the blood-brain barrier, a prerequisite for the development of central nervous system (CNS) infection, involves microbial invasion, intracellular traversal, and exocytosis. Microbial invasion of the blood-brain barrier has been investigated, but the molecular basis for microbial traversal and exit from the blood-brain barrier remains unknown. We performed transcriptome analysis of human brain microvascular endothelial cells (HBMEC) infected with Escherichia coli and Cryptococcus neoformans, representative bacterial and fungal pathogens common in CNS infections. Among the targets upregulated in response to E. coli and C. neoformans infection, PDLIM2 was knocked down by small hairpin RNA (shRNA) in HBMEC for further investigation. We demonstrated that Pdlim2 specifically regulated microbial traversal and exit from HBMEC by assessing microbial invasion, transcytosis, intracellular multiplication, and egression. Additionally, the defective exocytosis of internalized E. coli cells from the PDLIM2 shRNA knockdown cells was restored by treatment with a calcium ionophore (ionomycin). Moreover, we performed proximity-dependent biotin labeling with the biotin ligase BioID2 and identified 210 potential Pdlim2 interactors. Among the nine Pdlim2 interactors enriched in response to both E. coli and C. neoformans infection, we selected MPRIP and showed that HBMEC with knockdown of MPRIP mimicked the phenotype of PDLIM2 knockdown cells. These results suggest that the CNS-infecting microbes hijack Pdlim2 and Mprip for intracellular traversal and exocytosis in the blood-brain barrier.
KEYWORDS: Escherichia coli, Cryptococcus neoformans, blood-brain barrier, human brain microvascular endothelial cell, exocytosis, BioID
INTRODUCTION
Central nervous system (CNS) infection continues to be an important cause of mortality and morbidity (1, 2). Bacterial meningitis is an infection with high mortality and morbidity in the neonatal population (1). In developed countries, the incidence of bacterial meningitis is estimated at 0.3 per 1,000 live births, with a mortality of 10 to 15% (1). In developing countries, the reported incidence of bacterial meningitis is much higher, at 0.8 to 6.1 per 1,000 live births, and the mortality is 40 to 58% (1). Escherichia coli represents the second most common pathogen of bacterial meningitis and is isolated in 30% of all early-onset infections (1). In addition, E. coli has emerged as the most common cause of early-onset sepsis and meningitis among very low birth weight (<1,500 g) infants (1). Cryptococcus neoformans represents the most common pathogen of fungal meningitis and causes life-threating CNS infections in immunocompromised individuals, such as HIV-1-infected patients, resulting in over 180,000 deaths annually (2). C. neoformans is distributed globally and is particularly abundant in avian excreta (3). Infection with C. neoformans starts with inhalation of the fungal cells from the environment, followed by extrapulmonary spread, leading to hematogenous dissemination to the target organs, most commonly resulting in CNS infection (3).
E. coli and C. neoformans exhibit the ability to penetrate the blood-brain barrier, which is a prerequisite for the development of CNS infection (4), but the underlying mechanisms remain incompletely understood. Transcellular, paracellular, and Trojan-horse penetrations represent the known mechanisms for microbial penetration of the blood-brain barrier (4). Several CNS-infecting microbes, including E. coli and C. neoformans, exploit transcellular penetration of the blood-brain barrier (4). Transcellular penetration of the blood-brain barrier includes microbial invasion, intracellular traversal, and exocytosis (5). How microbes invade the blood-brain barrier has been investigated previously (6–9). For example, the FimH-CD48-RhoA, CNF1-37LRP-RhoA-FAK, OmpA-gp96-cPLA2α-CysLT-PKC, OmpA-gp96-STAT3-Rac1, IbeA-Caspr1-FAK-Rac1, and FimH/OmpA/NlpI-SphK2-EGFR-cSrc signaling pathways are exploited by E. coli, resulting in host cell cytoskeleton rearrangement and microbial internalization in human brain microvascular endothelial cells (HBMEC) (6). C. neoformans cell surface hyaluronic acid contributes to the invasion of HBMEC via its interaction with CD44 (7). C. neoformans phospholipase B1 (Plb1) was reported to activate the host cell Rac1 for traversal of an HBMEC monolayer (8). C. neoformans metalloprotease 1 (Mpr1) was shown to exploit the host cell annexinA2 (AnxA2) to facilitate transcytosis of the blood-brain barrier (9). However, the mechanism exploited by the CNS-infecting pathogens for intracellular traversal and exocytosis from HBMEC remains unknown.
In this report, we performed transcriptome sequencing (RNA-seq) analysis of HBMEC challenged with E. coli and C. neoformans. Among the upregulated genes, we identified PDLIM2, and we demonstrated that Pdlim2 specifically regulated the microbial traversal of and exit from HBMEC. In addition, we observed that the defective exit of internalized E. coli cells from PDLIM2 small hairpin RNA (shRNA) knockdown cells was restored by treatment with ionomycin. Moreover, we performed proximity-dependent biotin labeling and identified 210 potential Pdlim2 interactors. Among the nine Pdlim2 interactors enriched in response to both E. coli and C. neoformans infection, we identified MPRIP (myosin phosphatase Rho interacting protein gene), and we showed that MPRIP shRNA knockdown cells mimicked the phenotype of the PDLIM2 shRNA knockdown cells. Collectively, these results suggest that the CNS-infecting microbes hijack Pdlim2 and Mprip for intracellular traversal and exocytosis in the blood-brain barrier.
RESULTS
RNA-seq analysis of HBMEC in response to microbe infection.
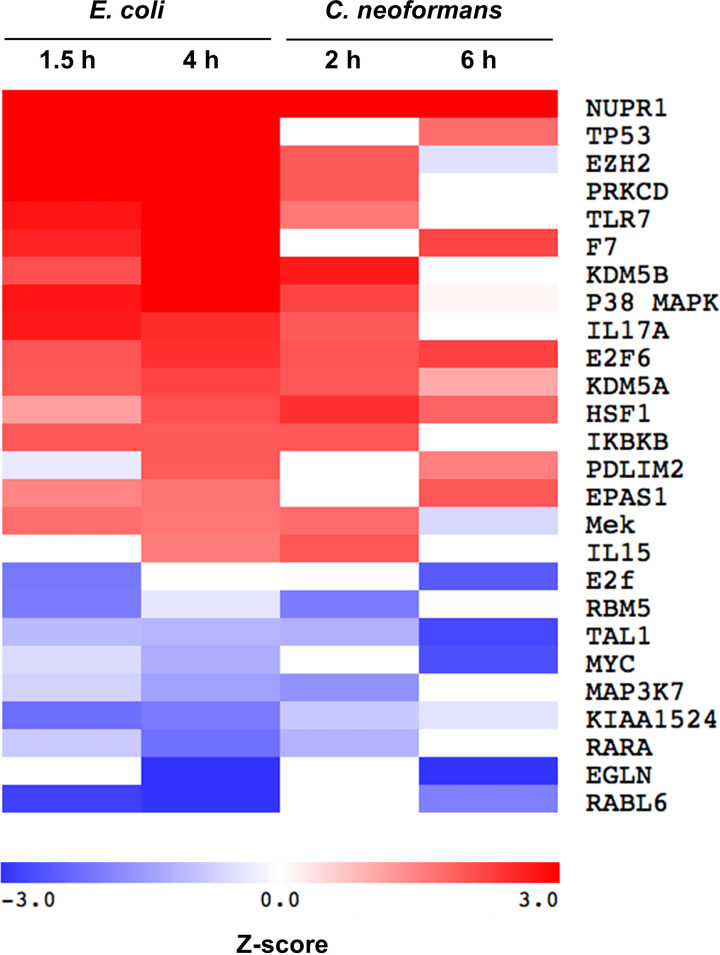
To investigate the microbial interaction with the blood-brain barrier, we analyzed the transcriptional response of HBMEC to infection with either E. coli or C. neoformans. HBMEC were infected with E. coli strain RS218 for 1.5 h or 4 h or with C. neoformans strain H99 for 2 h or 6 h. The uninfected HBMEC as a control were incubated for 6 h. Total RNA was purified from HBMEC for RNA-seq analysis. We defined the differentially expressed genes as those with a minimum 2-fold change in gene expression (P < 0.05) by comparing the infected group with the uninfected control. We observed that 277 genes were upregulated and 27 genes were downregulated for the 1.5-h E. coli infection and that 418 genes were upregulated and 48 genes were downregulated for the 4-h E. coli infection (Table S1 in the supplemental material). There were 228 upregulated genes and 327 downregulated genes for the 2-h C. neoformans infection, and there were 328 upregulated genes and 304 downregulated genes for the 6-h C. neoformans infection (Table S1). The Ingenuity Pathway Analysis (IPA) software (Ingenuity Systems) was used to identify the signaling pathways that are potentially regulated during infection (10, 11). Our analysis predicted the modulation of a total of 185 different signaling proteins from the HBMEC infected with E. coli and 34 different signaling proteins from the HBMEC infected with C. neoformans during at least one time point of the experiments. There were 17 activated signaling proteins and 9 repressed signaling proteins in response to both E. coli and C. neoformans (Fig. 1). The Z-score was used to determine whether an upstream transcription regulator had significantly more “activated” predictions than “inhibited” predictions (Z > 0) or vice versa (Z < 0) (12). This approach was validated by the predicted upregulation of epidermal growth factor receptor (EGFR) signaling during E. coli infection (Table S2). Previously, we have demonstrated that E. coli penetration of the blood-brain barrier exploits the EGFR signaling (13). We selected one new significantly upregulated molecule, Pdlim2, for functional analysis. Pdlim2 has not previously been recognized in connection with E. coli or C. neoformans infection and, in our experiments, is predicted to be activated at both 4 h of E. coli infection (1 of 13 genes; P value of overlap, 5.97 × 10−4) and 6 h of C. neoformans infection (1 of 11 genes; P value of overlap, 2.78 × 10−2) (Fig. 1, Tables S1 and S2).
FIG 1.
Upstream regulator analysis of the infection-induced gene expression. Each regulator was predicted by Ingenuity Pathway Analysis (IPA) to be activated (red, Z-score > 1.5) or repressed (blue, Z-score < −1.5) during infection of HBMEC with either E. coli strain RS218 or C. neoformans strain H99. All the predicted pathways achieved a Z-score of ≥|1.5| in at least one time point for both organisms. White indicates no predicted activation or repression.
Microbes exploit Pdlim2 for intracellular traversal of HBMEC.
Pdlim2, containing a PDZ domain and a LIM domain, was reported to colocalize with cytoskeleton in the cytoplasm of the human epithelial cell line MCF-7 and monkey kidney fibroblast-like cell line COS-7 (14, 15). In contrast, other reports indicated that Pdlim2 was in the nucleus of mouse embryonic fibroblasts and macrophages (16, 17). Pdlim2 was also reported to be in both the cytoplasm and nucleus of DU145 cells and Huh7.5 cells (18, 19). To investigate its location in HBMEC, Pdlim2-green fluorescent protein (GFP) fusion protein was stably expressed in HBMEC via lentivirus-mediated transfection (Fig. 2A). We showed that Pdlim2-GFP was colocalized with F-actin in the cytoplasm (Fig. 2A, arrowhead in top panel). To exclude the artifact from the additional GFP tag, we stained HBMEC with Pdlim2 antibody and demonstrated that endogenous Pdlim2 colocalized with F-actin in the cytoplasm (Fig. S1).
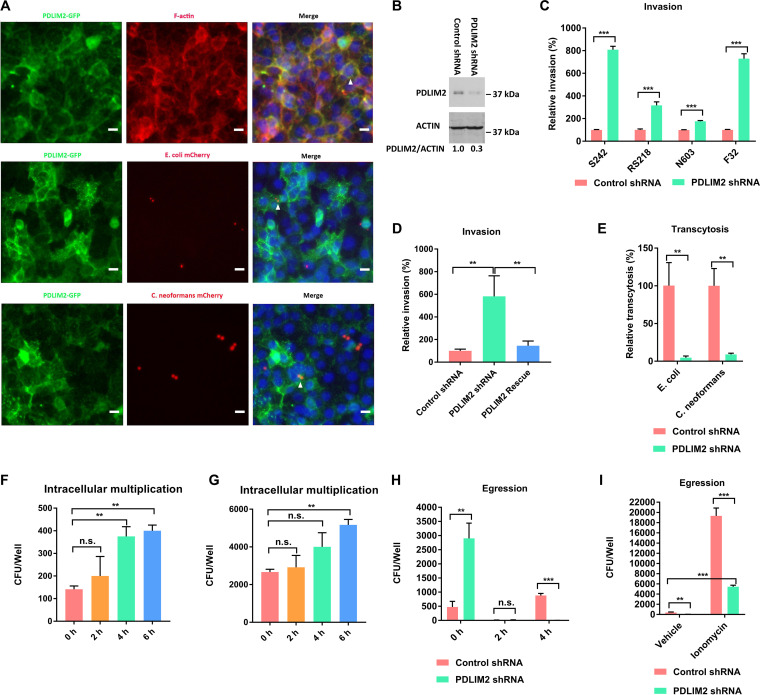
FIG 2.
Pdlim2 regulated microbial exocytosis from HBMEC. (A) Colocalization of Pdlim2-GFP with F-actin, E. coli S242 mCherry, or C. neoformans KN99 mCherry. F-actin was labeled with Alexa Fluor 594 phalloidin (red). DNA was labeled with DAPI (blue). Colocalization is highlighted by the arrowheads. Scale bars, 10 μm. (B) Pdlim2 expression in control shRNA cells and PDLIM2 shRNA knockdown cells. The assay was performed in duplicate. The results from one of the duplicates are presented. Student’s t test was performed to determine statistical significance. P = 0.016. (C) Relative invasion frequencies of E. coli strains S242, RS218, N603, and F32. The results were calculated as the percentages of the initial inoculum and are presented as the relative percentages of invasion compared with the invasion in the control shRNA cells. Student’s t test was performed to determine statistical significance. (D) Rescue experiment of PDLIM2 shRNA knockdown cells. The results were calculated as the percentages of the initial inoculum and are presented as the relative percentages of invasion compared with the invasion in the control shRNA cells. One-way analysis of variance (ANOVA) and multiple-comparison tests were performed to determine statistical significance. (E) Relative transcytosis frequencies of E. coli S242 and C. neoformans. The results were calculated as the percentages of the initial inoculum and are presented as the relative percentages of transcytosis compared with the transcytosis in the control shRNA cells. Student’s t test was performed to determine statistical significance. (F) Intracellular multiplication of E. coli S242 in control shRNA cells. ANOVA and multiple-comparison tests were performed to determine the statistical significance. (G) Intracellular multiplication of E. coli S242 in PDLIM2 shRNA knockdown cells. ANOVA and multiple-comparison tests were performed to determine statistical significance. (H) Egression of E. coli S242 from HBMEC. 0 h represents the initial internalized E. coli CFU. Student’s t test was performed to determine statistical significance. (H) Egression of E. coli S242 from HBMEC in the presence of ionomycin for 4 h. Student’s t test was performed to determine statistical significance. Each assay was performed in triplicate. Error bars indicate mean values ± standard deviations (SD). *, P < 0.05; **, P < 0.01; ***, P < 0.001; n.s., no significant difference.
To investigate its role in HBMEC-microbe interaction, we generated PDLIM2 shRNA knockdown cells, which were validated by Western blotting (Fig. 2B). An invasion assay was used to quantify the internalized E. coli cells (20). The HBMEC monolayer was incubated with E. coli for 1.5 h and then treated with 100 μg/ml gentamicin for 1 h to kill extracellular E. coli cells. The HBMEC monolayer was then lysed for quantification of internalized E. coli cells. The invasion assay showed that knockdown of PDLIM2 resulted in a significant increase in internalization into HBMEC by four meningitis isolates of E. coli (RS218, S242, N603, and F32) (Fig. 2C). E. coli strain S242 showed the highest increase in internalization into HBMEC and was used for further investigation (Fig. 2C).
To exclude any off-target effect of the PDLIM2 shRNA, we designed an shRNA-resistant cDNA of PDLIM2 by introducing five silent mutations within the PDLIM2 shRNA targeting sequence, as described previously (21), and stably expressed it in PDLIM2 knockdown cells to generate PDLIM2 rescue cells via lentivirus-mediated transfection. The increased invasion in the PDLIM2 shRNA knockdown cells was effectively rescued by the shRNA-resistant cDNA of PDLIM2 (Fig. 2D). The increased internalization in the PDLIM2 shRNA knockdown cells likely stems from increased invasion, decreased exocytosis, and/or increased multiplication of the internalized E. coli cells.
A transcytosis assay was utilized to quantify the microbes that traversed from the HBMEC monolayer (22). HBMEC were seeded on transwell inserts to form monolayers mimicking the blood-brain barrier in vitro. E. coli strain S242 was loaded into the upper chamber and incubated with HBMEC for 1.5 h. E. coli cells recovered from the lower chamber represented the traversed E. coli. The integrity of the HBMEC monolayer was assessed by measuring the transendothelial electrical resistance (TEER) with a World Precision Instruments epithelial voltohmmeter (WPI EVOM) before and after the assay. The transcytosis assay showed that cells with knockdown of PDLIM2 exhibited significantly reduced traversal of E. coli (Fig. 2E). These results indicated that the increased internalization of E. coli cells in the PDLIM2 shRNA knockdown cells was likely due to decreased exocytosis of the intracellular E. coli. In addition, knockdown of PDLIM2 led to significantly decreased traversal of C. neoformans cells (Fig. 2E). An internalization assay with C. neoformans was not feasible, as there is no drug to kill the extracellular C. neoformans. Internalization of E. coli and C. neoformans cells, however, was demonstrated by colocalization of mCherry-expressing strains E. coli S242 mCherry and C. neoformans KN99 mCherry with Pdlim2-GFP (Fig. 2A, arrowheads in middle and bottom panels).
We did an intracellular multiplication assay to test whether the internalized E. coli cells multiplied in the PDLIM2 shRNA knockdown cells, resulting in increased numbers of intracellular bacteria. HBMEC monolayers were incubated with E. coli strain S242 for 1.5 h and then treated with 100 μg/ml gentamicin for 1 h to kill extracellular E. coli. HBMEC monolayers were then cultured in the presence of 10 μg/ml gentamicin (to suppress any egressed E. coli) for 2 h, 4 h, and 6 h, separately. HBMEC monolayers were lysed for quantification of viable intracellular E. coli cells. The intracellular multiplication assay showed that the doubling time of intracellular E. coli within the control shRNA cells was about 4 h (Fig. 2F), while the doubling time of intracellular E. coli within the PDLIM2 knockdown cells was about 6 h (Fig. 2G), indicating that the increased internalization of E. coli within the PDLIM2 shRNA knockdown cells did not stem from increased multiplication of intracellular E. coli.
We then performed an egression assay to investigate whether knockdown of PDLIM2 would block the exit of intracellular E. coli cells from HBMEC. HBMEC monolayers on transwell inserts were incubated with E. coli strain S242 for 1.5 h and then treated with 100 μg/ml gentamicin for 1 h to kill extracellular E. coli. The HBMEC monolayers on the transwell inserts were then cultured in the absence of gentamicin for 2 h and 4 h separately. E. coli cells recovered from the lower chamber represented the egressed E. coli. In the 2-h egression assay, a few CFU (on control shRNA cells, 15 ± 2 CFU, and on PDLIM2 knockdown cells, 10 ± 13 CFU) were recovered from the lower chambers (Fig. 2H). In the 4-h egression assay, 880 ± 75 CFU were egressed from the control shRNA cells, whereas only 2 ± 3 CFU were egressed from the PDLIM2 knockdown cells, even though the initial number of internalized CFU (0 h in the egression assay) from the PDLIM2 knockdown cells was six times greater than that of the control shRNA cells (Fig. 2H). These results indicated that knockdown of PDLIM2 hindered the exit of intracellular E. coli cells from HBMEC.
We attempted to restore the defective exocytosis of intracellular E. coli from the PDLIM2 knockdown cells with four drugs (ionomycin, ambroxol, forskolin, and rapamycin) that were reported to promote exocytosis (23–26). Forskolin was reported to increase intracellular cAMP and induce exocytosis of E. coli from infected bladder epithelial cells (23). Rapamycin was reported to be an inducer of autophagy and to enhance bacterial expulsion from bladder epithelial cells (24). Ambroxol triggers lysosomal exocytosis by releasing the acidic Ca2+ stores (25). Ionomycin is a calcium ionophore that raises the intracellular calcium level to trigger calcium-regulated exocytosis (26). HBMEC monolayers on transwell inserts were incubated with E. coli strain S242 for 1.5 h, followed by treatment with 100 μg/ml gentamicin for 1 h to kill the extracellular E. coli. The HBMEC monolayers on the transwell inserts were then cultured in the presence of ionomycin, ambroxol, forskolin, or rapamycin for 4 h. The egression assay showed that only ionomycin restored the defective exocytosis of intracellular E. coli from the PDLIM2 knockdown cells (Fig. 2I). Ionomycin led to a 52-fold increase of egressed E. coli cells from the control shRNA cells and a 360-fold increase from the PDLIM2 knockdown cells (Fig. 2I). These data suggested that the microbial exit from HBMEC was likely to be dependent on the intracellular Ca2+.
BioID reveals involvement of Pdlim2 interactor Mprip in intracellular traversal of HBMEC.
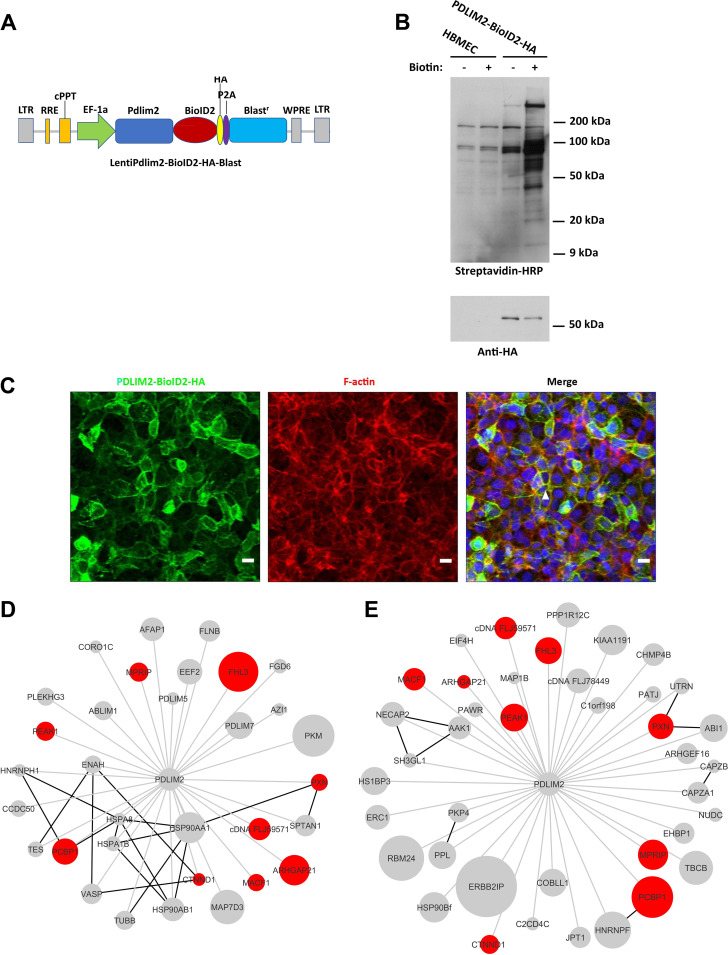
To further investigate the role of Pdlim2 in microbial traversal and exocytosis, we performed a proximity-dependent biotin ligation assay with the biotin ligase BioID2 to map the interactome of Pdlim2 during HBMEC-microbe interactions (27). The biotin ligase BioID2 is fused to a protein of interest and expressed in a cell, where it biotinylates the proximal endogenous proteins (27). The Pdlim2-BioID2-hemagglutinin (HA) fusion protein was stably expressed in HBMEC via lentivirus-mediated transfection (Fig. 3A). The stable expression of Pdlim2-BioID2-HA in HBMEC was validated by Western blotting (Fig. 3B, bottom). We observed some basal biotinylation in HBMEC, representing the endogenous biotinylated proteins (Fig. 3B, top). The cells expressing Pdlim2-BioID2-HA showed a drastic increase in the biotinylated proteins in the presence of 10 μM biotin (Fig. 3B, top). In addition, the Pdlim2-BioID2-HA was colocalized with F-actin (Fig. 3C, arrowhead). These data indicated that the Pdlim2-BioID2-HA fusion protein was functionally expressed in HBMEC.
FIG 3.
BioID revealed host proteins interacting with Pdlim2 in response to microbial infection. (A) Pdlim2 was fused with BioID2-HA and stably expressed in HBMEC. LTR, long terminal repeat; RRE, Rev response element; cPPT, central polypurine tract; P2A, 2A self-cleaving peptides; WPRE, woodchuck hepatitis virus posttranscriptional regulatory element. (B) The proteins biotinylated by Pdlim2-BioID2 were detected with HRP-conjugated streptavidin after SDS-PAGE separation. The expression of Pdlim2-BioID2 in HBMEC in the presence of biotin led to a drastic increase in biotinylation of the endogenous proteins (top). Fusion protein Pdlim2-BioID2-HA was detected with anti-HA antibody (bottom). (C) Pdlim2-BioID2-HA colocalized with F-actin. Pdlim2-BioID2-HA was labeled with anti-HA antibody (green). F-actin was labeled with Alexa Fluor 594 phalloidin (red). DNA was labeled with DAPI (blue). Colocalization is highlighted by the arrowhead. Scale bars, 10 μm. (D) Biotinylated proteins enriched in HBMEC infected with E. coli. Gray lines, BioID interactions; black lines, known protein interactions from the String networks; node sizes, normalized log2 fold enrichment. Proteins enriched in response to both E. coli and C. neoformans are highlighted in red. (E) Biotinylated proteins enriched in HBMEC infected with C. neoformans. Gray lines, BioID interactions; black lines, known protein interactions from the String networks; node sizes, normalized log2 fold enrichment. Proteins enriched in response to both E. coli and C. neoformans are highlighted in red.
The Pdlim2-BioID2-HA cells were incubated with E. coli S242 or C. neoformans for 20 h in the presence of biotin. The biotinylated proteins were purified with streptavidin beads and then submitted for mass spectrometry analysis. Mass spectrometry identified 210 biotinylated proteins, including some known Pdlim2 interactors, such as filamin A, actinin-1, and actinin-4 (Table S3) (14). Most of the 210 biotinylated proteins were cytoskeleton-related proteins, which reflected the colocalization of Pdlim2 with cytoskeleton (Fig. 3C, Table S3). Pdlim2 was reported to be an E3 ubiquitin ligase that acts to stimulate the degradation of STAT2, STAT3, and NF-κB in the nucleus (16, 17, 19). However, we did not detect biotinylated STAT2, STAT3, and NF-κB in HBMEC. The spectral counts were normalized with the following equation as previously described (28): normalized count per protein = [(spectral count per protein/predicted trypsin sites within a protein)]/total counts for all proteins in a sample × 106. There were 33 biotinylated proteins enriched in Pdlim2-BioID2-HA cells infected with E. coli S242 and 41 biotinylated proteins enriched in Pdlim2-BioID2-HA cells challenged with C. neoformans H99 compared to the biotinylated proteins in uninfected Pdlim2-BioID2-HA cells (Fig. 3D and E). In total, nine biotinylated proteins were enriched in Pdlim2-BioID2-HA cells in response to both E. coli and C. neoformans (Fig. 3D and E).
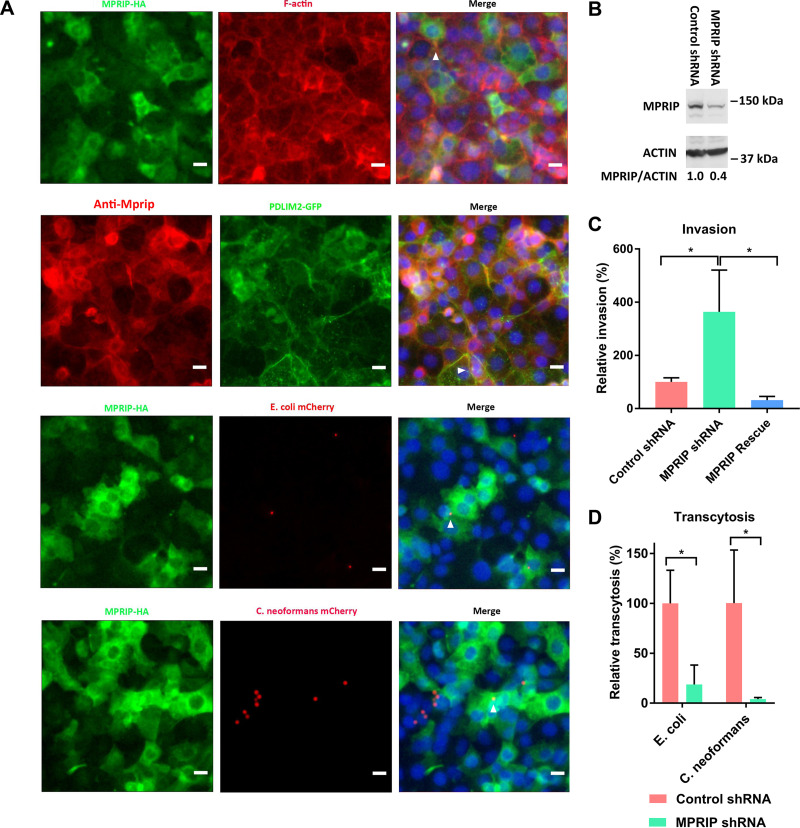
We selected Mprip from the nine overlapping proteins for functional validation. An Mprip-HA fusion protein was stably expressed in HBMEC via lentivirus-mediated transfection. Mprip colocalized with F-actin (Fig. 4A, arrowhead in top panel) as well as with Pdlim2 (Fig. 4A, arrowhead in second panel down). MPRIP shRNA knockdown cells were generated and validated by Western blotting (Fig. 4B). An invasion assay demonstrated that the MPRIP shRNA knockdown cells exhibited a significant increase in internalized E. coli, and the increased invasion in the MPRIP shRNA knockdown cells was rescued by the shRNA-resistant cDNA of MPRIP (Fig. 4C). A transcytosis assay showed that the MPRIP shRNA knockdown cells exhibited significantly decreased traversal by E. coli and C. neoformans cells (Fig. 4D). The integrity of the HBMEC monolayers was assessed by measuring the TEER with the WPI EVOM before and after the assay. In addition, E. coli S242 mCherry and C. neoformans KN99 mCherry were colocalized with Mprip (Fig. 4B, arrowheads in third-down and bottom panels). The phenotype of the MPRIP shRNA knockdown cells, therefore, mimicked that of the PDLIM2 shRNA knockdown cells. These results suggested that Mprip and Pdlim2 contributed to the microbial intracellular traversal and exocytosis in HBMEC.
FIG 4.
Mprip was involved in intracellular traversal and exocytosis in HBMEC. (A) Colocalization of Mprip with F-actin, Pdlim2, E. coli S242 mCherry, or C. neoformans KN99 mCherry. Mprip-HA was labeled with anti-HA antibody (green). F-actin was labeled with Alexa Fluor 594 phalloidin (red). Mprip was labeled with anti-MPRIP polyclonal antibody (red). DNA was labeled with DAPI (blue). Colocalization is highlighted by the arrowheads. Scale bars, 10 μm. (B) Mprip expression in control shRNA cells and MPRIP shRNA knockdown cells. The assay was performed in duplicate. The results for one of the duplicates are presented. Student’s t test was performed to determine statistical significance. P = 0.048. (C) Relative invasion frequencies in MPRIP shRNA knockdown and MPRIP rescue cells. The results were calculated as the percentages of the initial inoculum and are presented as the relative percentages of invasion compared with the invasion in the control shRNA cells. ANOVA and multiple-comparison tests were performed to determine statistical significance. (D) Relative transcytosis frequencies in MPRIP shRNA knockdown cells. The results were calculated as the percentages of the initial inoculum and are presented as the relative percentages of transcytosis compared with the transcytosis in the control shRNA cells. Student’s t test was performed to determine statistical significance. Error bars indicate mean values ± SD. Each assay was performed in triplicate. *, P < 0.05.
DISCUSSION
Microbial penetration of the blood-brain barrier includes microbial invasion, intracellular traversal, and exocytosis. Previous studies elucidated the mechanisms involved in microbial invasion, but the basis for microbial intracellular traversal and exocytosis remains unknown. In this report, we performed transcriptome analysis of HBMEC in response to infection with E. coli and C. neoformans, the common CNS-infecting pathogens. We did network analysis of the RNA-seq data to reveal the host signaling pathways affected by E. coli and C. neoformans infection and identified Pdlim2 as a key regulator of microbial intracellular traversal and exocytosis in HBMEC, essential steps for successful penetration of the blood-brain barrier.
We demonstrated that Pdlim2 specifically regulated the exit of internalized microbes from HBMEC through assessing microbial invasion, transcytosis, intracellular multiplication, and egression. In addition, we observed that the exocytosis of internalized microbes from HBMEC was enhanced by treatment with ionomycin. The calcium ionophore ionomycin, which could raise the intracellular calcium level, restored the defective exocytosis of internalized E. coli cells from the PDLIM2 shRNA knockdown cells, suggesting that the microbial exocytosis was triggered by the intracellular calcium. Unlike the exocytosis occurring in bladder epithelial cells, forskolin (which stimulates adenylate cyclase to raise the cyclic AMP level) and rapamycin (an inducer of autophagy) failed to induce the exocytosis of E. coli cells from HBMEC (23, 24). Ambroxol (which neutralizes the lamellar bodies’ pH, followed by an intracellular calcium release to induce lysosomal exocytosis) also failed to trigger the exocytosis of intracellular E. coli cells from HBMEC (25). Our previous study demonstrated that the internalized E. coli cells were localized within a membrane-bound vacuole in HBMEC and that the E. coli-containing vacuole did not fuse with the lysosome (29). Therefore, it appears that cyclic AMP-regulated exocytosis, autophagy-mediated exocytosis, and lysosomal exocytosis are not directly involved in the microbial exocytosis from HBMEC. We previously showed that E. coli invasion resulted in increased intracellular calcium in HBMEC and that chelating of the intracellular calcium reduced the bacterial invasion (30). These findings suggest that intracellular calcium plays a critical role in microbial invasion, intracellular traversal, and exocytosis. Additional studies are needed to elucidate the molecular basis for how the intracellular calcium triggers microbial exocytosis from HBMEC, particularly in connection with Pdlim2.
BioID has emerged as a powerful tool to screen for the physiologically relevant protein interactions that occur in living cells (31). We performed BioID using Pdlim2 as the bait and identified 210 potential proteins that physiologically interacted with Pdlim2. Of interest, Mprip was enriched in the HBMEC infected with E. coli and C. neoformans and phenocopied Pdlim2 in regulating microbial intracellular traversal and exocytosis in the blood-brain barrier. There are some contradictory reports about the roles of Pdlim2. In some reports, Pdlim2 is in the cytoplasm and acts as a cytoskeleton adaptor that promotes cell attachment and migration (14, 15). In other reports, Pdlim2 is in the nucleus and functions as a ubiquitin E3 ligase, negatively regulating the activities of STAT2, STAT3, and NF-κB (16, 17, 19). In this study, we showed that Pdlim2 was associated with the cytoskeletal factors in the cytoplasm and regulated the intracellular traversal and exocytosis of microbes in HBMEC. Mprip (myosin phosphatase Rho interacting protein) targets myosin phosphatase to the actin cytoskeleton to regulate myosin light-chain phosphorylation (32). Mprip was colocalized with Pdlim2 and appeared to coregulate the intracellular traversal and exocytosis of the CNS-infecting microbes in the blood-brain barrier with Pdlim2. Efficient microbial intracellular traversal and exocytosis exploit both Pdlim2 and Mprip, as knockdown of either gene resulted in defective exocytosis. Additional studies are needed to elucidate how Pdlim2 and Mprip exhibit similar functions in microbial intracellular traversal and exocytosis.
In summary, we linked Pdlim2 and Mprip to microbial exocytosis in HBMEC, an essential but unexplored step in microbial penetration of the blood-brain barrier. We also provided insight that the microbial exit from HBMEC was affected by a chemical affecting intracellular calcium. These findings are likely to facilitate the development of novel strategies for prevention of microbial penetration of the blood-brain barrier, the essential step in the development of CNS infection. More importantly, we showed that Pdlim2 and Mprip contribute to microbial exocytosis in HBMEC for two important CNS-infecting pathogens, a bacterium and a fungus, prompting an interesting concept that the findings shown in this report might be relevant to other CNS-infecting microbes for their penetration of the blood-brain barrier.
MATERIALS AND METHODS
Microbial strains.
E. coli strains S242, N603, F32, and RS218 are derived from the cerebrospinal fluid of neonates with E. coli meningitis. Strain S242 produces the extended-spectrum β-lactamase CTX-M-15, belonging to the worldwide-distributed O25b:H4-ST131 clone (33). An S242 strain expressing mCherry was generated by introducing pON.mCherry (84821; Addgene) into strain S242 via electroporation. Strain N603 is an E. coli meningitis isolate producing the TEM-type extended-spectrum β-lactamases (34). Strain F32 belongs to the O18:K1:H7 serotype. Strain RS218 is a prototype meningitis isolate (O18:K1:H7) (20). Strain H99 is a C. neoformans var. grubii strain causing fatal meningitis in humans (35). C. neoformans KN99 mCherry was a kind gift from Jennifer Lodge (36, 37).
Isolation of RNA from HBMEC.
HBMEC were isolated and characterized as previously described (38). HBMEC (3.5 × 106 cells) were seeded in a 100-mm petri dish and then cultured for 4 days until confluence in RPMI medium supplemented with 10% fetal bovine serum (FBS), 10% Nu-serum, 1% minimal essential medium (MEM) vitamins, 1% MEM nonessential amino acids, 1 mM sodium pyruvate, and 100 U/ml penicillin-streptomycin. The HBMEC monolayer was incubated for 1 h in the experimental medium, composed of medium 199 and Ham’s F-12 (1:1) supplemented with 5% FBS, before the infection assay. E. coli strain RS218 (5 × 107 cells/well, multiplicity of infection [MOI] = 100) was loaded and incubated for 1.5 h and 4 h, separately. C. neoformans (5 × 106 cells/well, MOI = 10) was loaded and incubated for 2 h and 6 h, separately. RNA was purified from HBMEC using the Quick-RNA microprep kit (catalog number R1050; Zymo Research).
RNA-seq and gene expression analysis.
RNA-seq was performed as described previously (10, 11). Briefly, all the RNA-seq libraries (non-strand specific, paired end) were prepared using the TruSeq RNA sample preparation kit (Illumina). Seventy-five nucleotides of the sequence were determined from each end of each cDNA fragment using the HiSeq 4000 platform (Illumina). Each time point was performed in triplicate. About 100 to 240 million reads were obtained for each sample (Table S4). The sequencing reads were annotated and aligned to Ensembl GRCh38 of the human reference genome (39) using TopHat2 (40). The alignment files from TopHat2 were used to generate the read counts for each gene. Statistical analysis of differential gene expression was performed using the EdgeR package from Bioconductor (41). The cutoffs included a P value of less than 0.05 and an absolute log2 fold change of 1 (infected/uninfected). We used the Ingenuity Pathway Analysis software (Ingenuity Systems) to identify the signaling proteins that were potentially activated or repressed during infection. This software assesses the overlap between lists of differentially expressed genes and an extensively curated database of regulator-target gene relationships. It then considers the direction of the gene expression changes to make predictions about the activation or repression of specific pathways.
Construction of the shRNA knockdown cell.
The shRNA lentiviral plasmids (PDLIM2 shRNA [TRCN0000154950] and MPRIP shRNA [TRCN0000048651]) were purchased from Sigma-Aldrich. HEK293T cells (CRL-3216; ATCC) (1.5 × 106 cells/well) were cultured in 6-well tissue culture plates overnight in 5 ml Opti-MEM I reduced-serum medium (catalog number 31985070; Thermo Fisher Scientific). The HEK293T cells were transfected with 3 μg lentiviral plasmids containing the target DNA sequence, 2 μg packaging plasmid psPAX2 (12260; Addgene), and 1 μg envelope plasmid pCMV-VSV-G (8454; Addgene) using FuGENE 6 transfection reagent (product number 2691; Promega). The lentivirus from the cell culture supernatant of the HEK293T cells was filtered with a 0.22-μm syringe filter and incubated with HBMEC for 72 h. The shRNA knockdown cells were selected with 1 μg/ml puromycin.
Western blot analysis.
HBMEC monolayers in 100-mm petri dishes were washed one time with cold phosphate-buffered saline (PBS) and then lysed in modified radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris HCl, pH 7.4, 150 mM NaCl, 2 mM EGTA, 1% Triton X-100) supplemented with protease/phosphatase inhibitor cocktail (product number 5872S; Cell Signaling).
For Western blot analysis, 25 μg of cell lysate was loaded for each well. Pdlim2 was immunoblotted with anti-PDLIM2 antibody (1:1,000, ab68220; Abcam). Mprip was immunoblotted with anti-MPRIP polyclonal antibody (1:1,000, PA5-54631; Invitrogen). Actin was immunoblotted with antiactin (20-33) antibody (1:5,000, A5060; Sigma). Anti-rabbit IgG horseradish peroxidase (HRP)-linked antibody (1:5,000, 7074S; Cell Signaling) and anti-mouse IgG HRP-linked antibody (1:5,000, 7076S; Cell Signaling) were purchased from Cell Signaling.
Bacterial invasion assay in HBMEC.
The bacterial invasion assay was carried out as described previously (20). E. coli strains were grown overnight in brain heart infusion broth and resuspended in the experimental medium composed of medium 199 and Ham’s F-12 (1:1) supplemented with 5% FBS. E. coli cells (1 × 107 cells/well) were added into confluent HBMEC monolayers in 24-well plates at an MOI of 100. The plates were incubated in a 37°C incubator with 5% CO2 for 90 min. The HBMEC monolayers were washed with the experimental medium and incubated in the experimental medium supplemented with 100 μg/ml gentamicin for 1 h to kill the extracellular bacteria. The HBMEC monolayers were washed three times with the experimental medium and then lysed with 0.025% Triton X-100 for 20 min at room temperature. The lysis suspension was diluted and plated on blood ager plates for quantification of the internalized E. coli. The result was calculated as the percentage of the initial inoculum and is presented as the relative percentage of invasion compared with the invasion in the control shRNA cells. The control shRNA cells contained enhanced green fluorescent protein (eGFP) shRNA (SHC005; Sigma-Aldrich). Each assay was performed in triplicate.
Microbial transcytosis assay in HBMEC.
The transcytosis assay was carried out as described previously (22, 42). HBMEC (6 × 104 cells/insert) were seeded in Millicell cell culture inserts with a pore size of 12 μm and diameter of 12 mm (EMD Millipore) and then cultured for 4 days. The HBMEC monolayers were incubated for 1 h in the experimental medium composed of medium 199 and Ham’s F-12 (1:1) supplemented with 5% FBS before the transcytosis assay. E. coli cells (3 × 106 cells/insert) were loaded into the inserts and incubated for 90 min. C. neoformans cells (1 × 106 cells/insert) were loaded into the inserts and incubated for 4 h. Samples from the lower chambers of inserts were collected and plated on agar plates for CFU determination. The integrity of the HBMEC monolayers was assessed by measuring the TEER with the WPI EVOM before and after the assay.
Bacterial multiplication assay in HBMEC.
HBMEC monolayers were infected with E. coli cells (1 × 107 cells/well) for 90 min, followed by gentamicin treatment as described for the invasion assay. The HBMEC monolayers were washed with the experimental medium and then incubated in the experimental medium containing 10 μg/ml gentamicin for 2 h, 4 h, and 6 h, separately. The HBMEC monolayers were washed three times with the experimental medium and then lysed with 0.025% Triton X-100 for 20 min at room temperature. The lysis suspensions were diluted and plated on blood ager plates for quantification of the intracellular E. coli cells.
Bacterial egression assay in HBMEC.
The HBMEC monolayers on the inserts were infected with E. coli cells (3 × 106 cells/well) for 90 min, followed by gentamicin treatment as described for the transcytosis assay. The HBMEC monolayers were washed three times with the experimental medium and then incubated in the experimental medium with or without a drug (1 μM ionomycin, 10 μM ambroxol, 10 μM forskolin, or 200 nM rapamycin) for 2 h and 4 h, separately. Samples from the lower chambers of the inserts were collected and plated on agar plates for CFU determination. The integrity of the HBMEC monolayers was assessed by measuring the TEER with the WPI EVOM before and after the assay.
Construction of cells expressing Pdlim2-BioID2-HA.
First-strand cDNAs were synthesized from RNA with the OneTaq reverse transcription (RT)-PCR kit (E5310S; NEB). The PDLIM2 cDNA was amplified from the first-strand cDNAs with primers PDLIM2-F (CCGCCAGAACACAGGACCGGTTCTAGATAGCCACCATGGCGTTGACGGTG) and PDLIM2-R (GGCCCGAGAGCTGAGGGTGG). BioID2-HA was amplified from plasmid MCS-BioID2-HA (74224; Addgene) with primers BioID2-HA-F (CTGCCACCCTCAGCTCTCGGGCCCGCTTAAGGCCTGTTAACCGG) and BioID2-HA-R (GCAGAGAGAAGTTTGTTGCGCCGGATCCTGCGTAATCCGGTACATCGTAAGGG) (27). To generate LentiPdlim2-BioID2-HA cells, the PDLIM2 cDNA and BioID2-HA PCR fragments were assembled into XbaI- and BamHI-linearized lentiCas9-Blast (52962; Addgene) using the NEBuilder hifi DNA assembly kit. The resulting LentiPdlim2-BioID2-HA plasmid was introduced into HBMEC to generate LentiPdlim2-BioID2-HA cells using Lipofectamine 2000 reagent (catalog number 11668-030; Invitrogen).
The resulting LentiPdlim2-BioID2-HA cells were seeded in 100-mm dishes and then cultured in biotin-free Dulbecco’s modified Eagle’s medium (DMEM) containing 5% FBS for 4 days until confluence. The LentiPdlim2-BioID2-HA cell monolayers were then cultured in DMEM containing 5% FBS with or without 10 μM biotin for 20 h. The LentiPdlim2-BioID2-HA cell monolayers were washed three time with cold PBS and lysed in buffer (50 mM Tris·Cl, pH 7.4, 500 mM NaCl, 0.2% SDS) supplemented with protease/phosphatase inhibitor cocktail (product number 5872S; Cell Signaling). For Western blot analysis, 25 μg of cell lysate was loaded for each well. Pdlim2-BioID2-HA was immunoblotted with anti-HA tag antibody (HA.C5) (1:1,000, ab18181; Abcam). The biotinylated proteins were immunoblotted with Pierce high-sensitivity streptavidin-HRP (1:1,000, catalog number 21130; Thermo Fisher Scientific).
The LentiPdlim2-BioID2-HA cells were fixed in 4% paraformaldehyde/PBS for 10 min. The fixed cells were permeabilized with 0.25% Triton X-100/PBS for 10 min. The cell samples were labeled with anti-HA tag antibody (HA.C5) (1:200, ab18181; Abcam), followed by goat anti-mouse IgG(H+L) highly cross-adsorbed secondary antibody, Alexa Fluor plus 488 (1:500, catalog number A32723; Invitrogen), and Alexa Fluor 594 phalloidin (1:500, catalog number A12381; Invitrogen). DNA was detected with DAPI (4′,6-diamidino-2-phenylindole). The cell samples were imaged using a Zeiss LSM700 confocal microscope and processed with ImageJ software.
Construction of cells expressing Pdlim2-GFP or Mprip-HA.
PDLIM2 cDNA was amplified from first-strand cDNAs with primers PDLIM2-F (CCGCCAGAACACAGGACCGGTTCTAGATAGCCACCATGGCGTTGACGGTG) and PDLIM2-R (GGCCCGAGAGCTGAGGGTGG). GFP was amplified from pEGFP-N1 (6085-1; Addgene) with primers GFP-F (ACCTGCCACCCTCAGCTCTCGGGCCCGCTTAAGGCCTGTTGTGAGCAAGGGCGAGGAGCT) and GFP-R (GCAGAGAGAAGTTTGTTGCGCCGGATCCCTTGTACAGCTCGTCCATGC). To generate LentiPdlim2-GFP cells, PDLIM2 cDNA and GFP PCR fragments were assembled into XbaI- and BamHI-linearized lentiCas9-Blast (52962; Addgene) using the NEBuilder hifi DNA assembly kit. The resulting LentiPdlim2-GFP plasmid was introduced into HBMEC using Lipofectamine 2000 reagent (catalog number 11668-030; Invitrogen).
MPRIP cDNA was amplified from the first-strand cDNAs with primers MPRIP-HA-F (CGCCAGAACACAGGACCGGTTCTAGATAGCCACCATGGCGTCGGCAGCCAAGGAGAACCC) and MPRIP-HA-R (GAGAGAAGTTTGTTGCGCCGGATCCTGCGTAATCCGGTACATCGTAAGGGTAGGTATCCCACGAGACCTGCT). To generate LentiMprip-HA cells, the MPRIP-HA PCR fragment was assembled into XbaI- and BamHI-linearized lentiCas9-Blast (52962; Addgene) using the NEBuilder hifi DNA assembly kit. The resulting LentiMprip-HA plasmid was introduced into HBMEC using Lipofectamine 2000 reagent (catalog number 11668-030; Invitrogen).
For colocalization analysis, the LentiPdlim2-GFP cells were fixed in 4% paraformaldehyde/PBS for 10 min. The cell samples were permeabilized with 0.25% Triton X-100/PBS for 10 min and then labeled with Alexa Fluor 594 phalloidin (1:500, catalog number A12381; Invitrogen).
The LentiPdlim2-GFP cells were labeled with anti-MPRIP polyclonal antibody (1:200, catalog number PA5-54631; Invitrogen), followed by goat anti-rabbit IgG(H+L) cross-adsorbed secondary antibody and Alexa Fluor 568 (1:500, A-11011; Invitrogen).
The LentiPdlim2-GFP cells were infected with strain S242 mCherry or KN99 mCherry and then fixed in 4% paraformaldehyde/PBS for 10 min. The cell samples were permeabilized with 0.25% Triton X-100/PBS for 10 min.
The LentiMprip-HA cells were fixed in 4% paraformaldehyde/PBS for 10 min. The cell samples were permeabilized with 0.25% Triton X-100/PBS for 10 min and then labeled with anti-HA tag antibody (HA.C5) (1:200, ab18181; Abcam), followed by goat anti-mouse IgG(H+L) highly cross-adsorbed secondary antibody, Alexa Fluor plus 488 (1:500, catalog number A32723; Invitrogen), and Alexa Fluor 594 phalloidin (1:500, catalog number A12381; Invitrogen).
The LentiMprip-HA cells were infected with S242 mCherry or KN99 mCherry and then fixed in 4% paraformaldehyde/PBS for 10 min. The cell samples were permeabilized with 0.25% Triton X-100/PBS for 10 min and then labeled with anti-HA tag antibody (HA.C5) (1:200, ab18181; Abcam), followed by goat anti-mouse IgG(H+L) highly cross-adsorbed secondary antibody and Alexa Fluor plus 488 (1:500, catalog number A32723; Invitrogen).
DNA was detected with DAPI. Cell samples were imaged using a Zeiss AxioImager.A1 inverted microscope with a DP80 charge-coupled-device (CCD) camera and processed with ImageJ software.
Construction of PDLIM2 rescue and MPRIP rescue cells.
PDLIM2 cDNA was amplified from first-strand cDNAs into two parts with primers PDLIM2-rescue_1 F (CCGCCAGAACACAGGACCGGTTCTAGATAGCCACCATGGCGTTGACGGTG), PDLIM2-rescue_1 R (GGACTGGCTCTCGGTGTATGTTCTCACGGAGCCCTGGAAGCGAGT), PDLIM2-rescue_2 F (CGTGAGAACATACACCGAGAGCCAGTCCTCCTTAAGGTCCTCCT), and PDLIM2-rescue_2 R (AGAGAGAAGTTTGTTGCGCCGGATCCTGCGTAATCCGGTACATCGTAAGGGTAGGCCCGAGAGCTGAGGGTGG). The PDLIM2 shRNA target sequence is underlined, and five silent mutations are highlighted in bold. To generate LentiPdlim2-rescue cells, the PDLIM2 rescue_1 and PDLIM2 rescue_2 PCR fragments were assembled into XbaI- and BamHI-linearized lentiCas9-Blast (52962; Addgene) using the NEBuilder hifi DNA assembly kit. The resulting LentiPdlim2-rescue plasmid was introduced into the PDLIM2 shRNA knockdown cells using Lipofectamine 2000 reagent (catalog number 11668-030; Invitrogen).
The MPRIP cDNA was amplified from first-strand cDNAs into two parts with primers MPRIP-rescue_1 F (CGCCAGAACACAGGACCGGTTCTAGATAGCCACCATGGCGTCGGCAGCCAAGGAGAACCC), MPRIP-rescue_1 R (GCTCGTACAGTATGAAGAATCGGCGCTGCCATTTCCGAGACCGGTG), MPRIP-rescue_2 F (GCGCCGATTCTTCATACTGTACGAGCACGGCCTCTTGCGC), and MPRIP-rescue_2 R (GAGAGAAGTTTGTTGCGCCGGATCCTGCGTAATCCGGTACATCGTAAGGGTAGGTATCCCACGAGACCTGCT). The MPRIP shRNA target sequence is underlined, and four silent mutations ae highlighted in bold. To generate LentiMprip-rescue cells, the MPRIP-rescue_1 and MPRIP-rescue_2 PCR fragments were assembled into XbaI- and BamHI-linearized lentiCas9-Blast (52962; Addgene) using the NEBuilder hifi DNA assembly kit. The resulting LentiMprip-rescue plasmid was introduced into the MPRIP shRNA knockdown cells using Lipofectamine 2000 reagent (catalog number 11668-030; Invitrogen).
BioID pulldown.
LentiPdlim2-BioID2-HA cells were cultured in a 100-mm dish in biotin-free DMEM containing 10% FBS for 4 days until confluence. The HBMEC monolayers were then incubated with E. coli cells (1 × 103 cells/dish) or C. neoformans cells (1 × 107 cells/dish) in DMEM containing 5% FBS and 10 μM biotin for 20 h. The HBMEC monolayers were washed three times with cold PBS and lysed in buffer (50 mM Tris·Cl, pH 7.4, 500 mM NaCl, 0.2% SDS) supplemented with protease/phosphatase inhibitor cocktail (product number 5872S; Cell Signaling). The biotinylated proteins were isolated with Pierce streptavidin magnetic beads (catalog number 88817; Thermo Fisher Scientific). The beads were washed following the BioID protocol (31). The biotinylated proteins were eluted from the streptavidin beads by heating at 100°C for 10 min in 1× SDS-PAGE running buffer and then precipitated with trichloroacetic acid (TCA). The pellets were washed twice with acetone, allowed to dry, and submitted to mass spectrometry analysis.
Mass spectrometry analysis and data analysis.
The protein pellets were reconstituted in 10 μl 500 mM triethylammonium bicarbonate (TEAB) plus 300 μl water and sonicated for 15 min. The protein samples were treated with a trypsin/Lys-C mixture, mass spectrometry grade (product number V5071; Promega), at 37°C overnight. The tryptic peptides were desalted and then analyzed with liquid chromatography-tandem mass spectrometry (LC–MS-MS) on a Q Exactive plus instrument in tandem Fourier transform (FT-FT) mode at the Mass Spectrometry and Proteomics Facility of Johns Hopkins University.
The MS-MS spectra were searched via PD2.2 with Mascot against the Uniprot human species (180915) database. The Mascot files were compiled in Scaffold and sent back to PD2.2 to identify the peptides. In total, 210 proteins were identified against the Uniprot human species database with at least one peptide at a 1% false discovery rate (FDR).
The spectral counts were converted into the normalized log2 protein ratios (28). The networks in Fig. 2 were generated with Cytoscape (version 3.2), with the nodes corresponding to the genes encoding the proteins identified by BioID and the node sizes proportional to fold enrichment over the values for uninfected LentiPdlim2-BioID2-HA cells. The bait Pdlim2 node size was set manually.
Data availability.
The raw sequencing reads from this study have been submitted to the NCBI sequence read archive (SRA) under BioProject accession no. PRJNA632824.
ACKNOWLEDGMENTS
We thank Weifeng She for preparing the RNA samples for RNA-seq, Ningyu Zhu, Donna Pearce, Wei Liu, and Chengxian Zhang for their assistance in the bacterial invasion assay, the Mass Spectrometry and Proteomics Facility (Johns Hopkins University) for the mass spectrometry analysis, Jeremy Nathans (Johns Hopkins University) for the confocal microscope, Ross Imaging Center (Johns Hopkins University) for the Zeiss AxioImager.A1 inverted microscope, Jennifer Lodge (Washington University in St. Louis) for the KN99 mCherry strain, Jun Liu (Johns Hopkins University) for providing the drugs (ionomycin, ambroxol, forskolin, and rapamycin), and Brendan Cormack (Johns Hopkins University) for comments on the manuscript.
Z.L. and K.S.K. conceived this project. Z.L. performed the experiments and analyzed the data. V.M.B. performed RNA-seq and analyzed the RNA-seq data. K.S.K. supervised the research. Z.L. wrote the original draft. Z.L., V.M.B., and K.S.K. reviewed and edited the manuscript.
This work was supported in part by NIH grants (https://www.nih.gov/) (grants number AI84894, NS94012, AI127570, and AI147699) to K.S.K. and (grant number U19AI110820) to V.M.B. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
We declare that we have no conflicts of interest with the contents of this article.
Footnotes
Supplemental material is available online only.
Contributor Information
Kwang Sik Kim, Email: kwangkim@jhmi.edu.
Denise Monack, Stanford University.
REFERENCES
- 1.Ku LC, Boggess KA, Cohen-Wolkowiez M. 2015. Bacterial meningitis in the infant. Clin Perinatol 42:29–45. 10.1016/j.clp.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rajasingham R, Smith RM, Park BJ, Jarvis JN, Govender NP, Chiller TM, Denning DW, Loyse A, Boulware DR. 2017. Global burden of disease of HIV-associated cryptococcal meningitis: an updated analysis. Lancet Infect Dis 17:873–881. 10.1016/S1473-3099(17)30243-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.May RC, Stone NRH, Wiesner DL, Bicanic T, Nielsen K. 2016. Cryptococcus: from environmental saprophyte to global pathogen. Nat Rev Microbiol 14:106–117. 10.1038/nrmicro.2015.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim KS. 2008. Mechanisms of microbial traversal of the blood-brain barrier. Nat Rev Microbiol 6:625–634. 10.1038/nrmicro1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim KS. 2010. Acute bacterial meningitis in infants and children. Lancet Infect Dis 10:32–42. 10.1016/S1473-3099(09)70306-8. [DOI] [PubMed] [Google Scholar]
- 6.Kim KS. May2016. Human meningitis-associated Escherichia coli. EcoSal Plus 2016. 10.1128/ecosalplus.ESP-0015-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jong A, Wu C-H, Shackleford GM, Kwon‐Chung KJ, Chang YC, Chen H-M, Ouyang Y, Huang S-H. 2008. Involvement of human CD44 during Cryptococcus neoformans infection of brain microvascular endothelial cells. Cell Microbiol 10:1313–1326. 10.1111/j.1462-5822.2008.01128.x. [DOI] [PubMed] [Google Scholar]
- 8.Maruvada R, Zhu L, Pearce D, Zheng Y, Perfect J, Kwon-Chung KJ, Kim KS. 2012. Cryptococcus neoformans phospholipase B1 activates host cell Rac1 for traversal across the blood-brain barrier. Cell Microbiol 14:1544–1553. 10.1111/j.1462-5822.2012.01819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Na Pombejra S, Salemi M, Phinney BS, Gelli A. 2017. The metalloprotease, Mpr1, engages annexinA2 to promote the transcytosis of fungal cells across the blood-brain barrier. Front Cell Infect Microbiol 7:296. 10.3389/fcimb.2017.00296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y, Shetty AC, Schwartz JA, Bradford LL, Xu W, Phan QT, Kumari P, Mahurkar A, Mitchell AP, Ravel J, Fraser CM, Filler SG, Bruno VM. 2015. New signaling pathways govern the host response to C. albicans infection in various niches. Genome Res 25:679–689. 10.1101/gr.187427.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watkins TN, Gebremariam T, Swidergall M, Shetty AC, Graf KT, Alqarihi A, Alkhazraji S, Alsaadi AI, Edwards VL, Filler SG, Ibrahim AS, Bruno VM. 2018. Inhibition of EGFR signaling protects from mucormycosis. mBio 9:e01384-18. 10.1128/mBio.01384-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krämer A, Green J, Pollard J, Tugendreich S. 2014. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 30:523–530. 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X, Maruvada R, Morris AJ, Liu JO, Wolfgang MJ, Baek DJ, Bittman R, Kim KS. 2016. Sphingosine 1-phosphate activation of EGFR as a novel target for meningitic Escherichia coli penetration of the blood-brain barrier. PLoS Pathog 12:e1005926. 10.1371/journal.ppat.1005926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Torrado M, Senatorov VV, Trivedi R, Fariss RN, Tomarev SI. 2004. Pdlim2, a novel PDZ-LIM domain protein, interacts with alpha-actinins and filamin A. Invest Ophthalmol Vis Sci 45:3955–3963. 10.1167/iovs.04-0721. [DOI] [PubMed] [Google Scholar]
- 15.Loughran G, Healy NC, Kiely PA, Huigsloot M, Kedersha NL, O’Connor R. 2005. Mystique is a new insulin-like growth factor-I-regulated PDZ-LIM domain protein that promotes cell attachment and migration and suppresses Anchorage-independent growth. Mol Biol Cell 16:1811–1822. 10.1091/mbc.e04-12-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanaka T, Grusby MJ, Kaisho T. 2007. PDLIM2-mediated termination of transcription factor NF-κB activation by intranuclear sequestration and degradation of the p65 subunit. Nat Immunol 8:584–591. 10.1038/ni1464. [DOI] [PubMed] [Google Scholar]
- 17.Li L, Sun F, Han L, Liu X, Xiao Y, Gregory AD, Shapiro SD, Xiao G, Qu Z. 2021. PDLIM2 repression by ROS in alveolar macrophages promotes lung tumorigenesis. JCI Insight 6:e144394. 10.1172/jci.insight.144394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bowe RA, Cox OT, Ayllón V, Tresse E, Healy NC, Edmunds SJ, Huigsloot M, O’Connor R. 2014. PDLIM2 regulates transcription factor activity in epithelial-to-mesenchymal transition via the COP9 signalosome. Mol Biol Cell 25:184–195. 10.1091/mbc.E13-06-0306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joyce MA, Berry-Wynne KM, Dos Santos T, Addison WR, McFarlane N, Hobman T, Tyrrell DL. 2019. HCV and flaviviruses hijack cellular mechanisms for nuclear STAT2 degradation: up-regulation of PDLIM2 suppresses the innate immune response. PLoS Pathog 15:e1007949. 10.1371/journal.ppat.1007949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang SH, Wass C, Fu Q, Prasadarao NV, Stins M, Kim KS. 1995. Escherichia coli invasion of brain microvascular endothelial cells in vitro and in vivo: molecular cloning and characterization of invasion gene ibe10. Infect Immun 63:4470–4475. 10.1128/iai.63.11.4470-4475.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Massengill MT, Young BM, Lewin AS, Ildefonso CJ. 2019. Co-delivery of a short-hairpin RNA and a shRNA-resistant replacement gene with adeno-associated virus: an allele-independent strategy for autosomal-dominant retinal disorders. Methods Mol Biol 1937:235–258. 10.1007/978-1-4939-9065-8_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stins MF, Badger J, Sik Kim K. 2001. Bacterial invasion and transcytosis in transfected human brain microvascular endothelial cells. Microb Pathog 30:19–28. 10.1006/mpat.2000.0406. [DOI] [PubMed] [Google Scholar]
- 23.Bishop BL, Duncan MJ, Song J, Li G, Zaas D, Abraham SN. 2007. Cyclic AMP-regulated exocytosis of Escherichia coli from infected bladder epithelial cells. Nat Med 13:625–630. 10.1038/nm1572. [DOI] [PubMed] [Google Scholar]
- 24.Miao Y, Li G, Zhang X, Xu H, Abraham SN. 2015. A TRP channel senses lysosome neutralization by pathogens to trigger their expulsion. Cell 161:1306–1319. 10.1016/j.cell.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fois G, Hobi N, Felder E, Ziegler A, Miklavc P, Walther P, Radermacher P, Haller T, Dietl P. 2015. A new role for an old drug: ambroxol triggers lysosomal exocytosis via pH-dependent Ca2+ release from acidic Ca2+ stores. Cell Calcium 58:628–637. 10.1016/j.ceca.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 26.Liu T, Sun L, Xiong Y, Shang S, Guo N, Teng S, Wang Y, Liu B, Wang C, Wang L, Zheng L, Zhang CX, Han W, Zhou Z. 2011. Calcium triggers exocytosis from two types of organelles in a single astrocyte. J Neurosci 31:10593–10601. 10.1523/JNEUROSCI.6401-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim DI, Jensen SC, Noble KA, Kc B, Roux KH, Motamedchaboki K, Roux KJ. 2016. An improved smaller biotin ligase for BioID proximity labeling. Mol Biol Cell 27:1188–1196. 10.1091/mbc.E15-12-0844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uezu A, Kanak DJ, Bradshaw TWA, Soderblom EJ, Catavero CM, Burette AC, Weinberg RJ, Soderling SH. 2016. Identification of an elaborate complex mediating postsynaptic inhibition. Science 353:1123–1129. 10.1126/science.aag0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim KJ, Elliott SJ, Di Cello F, Stins MF, Kim KS. 2003. The K1 capsule modulates trafficking of E. coli-containing vacuoles and enhances intracellular bacterial survival in human brain microvascular endothelial cells. Cell Microbiol 5:245–252. 10.1046/j.1462-5822.2003.t01-1-00271.x. [DOI] [PubMed] [Google Scholar]
- 30.Kim YV, Pearce D, Kim KS. 2008. Ca2+/calmodulin-dependent invasion of microvascular endothelial cells of human brain by Escherichia coli K1. Cell Tissue Res 332:427–433. 10.1007/s00441-008-0598-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roux KJ, Kim DI, Burke B, May DG. 2018. BioID: a screen for protein-protein interactions. Curr Protoc Protein Sci 91:19.23.1–19.23.15. 10.1002/cpps.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Surks HK, Riddick N, Ohtani K-I. 2005. M-RIP targets myosin phosphatase to stress fibers to regulate myosin light chain phosphorylation in vascular smooth muscle cells. J Biol Chem 280:42543–42551. 10.1074/jbc.M506863200. [DOI] [PubMed] [Google Scholar]
- 33.Pouillot F, Chomton M, Blois H, Courroux C, Noelig J, Bidet P, Bingen E, Bonacorsi S. 2012. Efficacy of bacteriophage therapy in experimental sepsis and meningitis caused by a clone O25b:H4-ST131 Escherichia coli strain producing CTX-M-15. Antimicrob Agents Chemother 56:3568–3575. 10.1128/AAC.06330-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moissenet D, Salauze B, Clermont O, Bingen E, Arlet G, Denamur E, Mérens A, Mitanchez D, Vu-Thien H. 2010. Meningitis caused by Escherichia coli producing TEM-52 extended-spectrum beta-lactamase within an extensive outbreak in a neonatal ward: epidemiological investigation and characterization of the strain. J Clin Microbiol 48:2459–2463. 10.1128/JCM.00529-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Janbon G, Ormerod KL, Paulet D, Byrnes EJ, Yadav V, Chatterjee G, Mullapudi N, Hon C-C, Billmyre RB, Brunel F, Bahn Y-S, Chen W, Chen Y, Chow EWL, Coppée J-Y, Floyd-Averette A, Gaillardin C, Gerik KJ, Goldberg J, Gonzalez-Hilarion S, Gujja S, Hamlin JL, Hsueh Y-P, Ianiri G, Jones S, Kodira CD, Kozubowski L, Lam W, Marra M, Mesner LD, Mieczkowski PA, Moyrand F, Nielsen K, Proux C, Rossignol T, Schein JE, Sun S, Wollschlaeger C, Wood IA, Zeng Q, Neuvéglise C, Newlon CS, Perfect JR, Lodge JK, Idnurm A, Stajich JE, Kronstad JW, Sanyal K, Heitman J, Fraser JA, et al. 2014. Analysis of the genome and transcriptome of Cryptococcus neoformans var. grubii reveals complex RNA expression and microevolution leading to virulence attenuation. PLoS Genet 10:e1004261. 10.1371/journal.pgen.1004261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Upadhya R, Lam WC, Maybruck BT, Donlin MJ, Chang AL, Kayode S, Ormerod KL, Fraser JA, Doering TL, Lodge JK. 2017. A fluorogenic C. neoformans reporter strain with a robust expression of m-cherry expressed from a safe haven site in the genome. Fungal Genet Biol 108:13–25. 10.1016/j.fgb.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nielsen K, Cox GM, Wang P, Toffaletti DL, Perfect JR, Heitman J. 2003. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and alpha isolates. Infect Immun 71:4831–4841. 10.1128/IAI.71.9.4831-4841.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stins MF, Gilles F, Kim KS. 1997. Selective expression of adhesion molecules on human brain microvascular endothelial cells. J Neuroimmunol 76:81–90. 10.1016/s0165-5728(97)00036-2. [DOI] [PubMed] [Google Scholar]
- 39.Cunningham F, Amode MR, Barrell D, Beal K, Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fitzgerald S, Gil L, Girón CG, Gordon L, Hourlier T, Hunt SE, Janacek SH, Johnson N, Juettemann T, Kähäri AK, Keenan S, Martin FJ, Maurel T, McLaren W, Murphy DN, Nag R, Overduin B, Parker A, Patricio M, Perry E, Pignatelli M, Riat HS, Sheppard D, Taylor K, Thormann A, Vullo A, Wilder SP, Zadissa A, Aken BL, Birney E, Harrow J, Kinsella R, Muffato M, Ruffier M, Searle SMJ, Spudich G, Trevanion SJ, Yates A, Zerbino DR, Flicek P. 2015. Ensembl 2015. Nucleic Acids Res 43:D662–D669. 10.1093/nar/gku1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14:R36. 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chang YC, Stins MF, McCaffery MJ, Miller GF, Pare DR, Dam T, Paul-Satyaseela M, Kim KS, Kwon-Chung KJ, Paul-Satyasee M. 2004. Cryptococcal yeast cells invade the central nervous system via transcellular penetration of the blood-brain barrier. Infect Immun 72:4985–4995. 10.1128/IAI.72.9.4985-4995.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download IAI.00128-21-s0001.xlsx, XLSX file, 0.3 MB (314.9KB, xlsx)
Supplemental material. Download IAI.00128-21-s0002.xlsx, XLSX file, 0.02 MB (19.9KB, xlsx)
Supplemental material. Download IAI.00128-21-s0003.xlsx, XLSX file, 0.1 MB (105.8KB, xlsx)
Supplemental material. Download IAI.00128-21-s0004.xlsx, XLSX file, 0.01 MB (11.7KB, xlsx)
Supplemental material. Download IAI.00128-21-s0005.pdf, PDF file, 0.1 MB (140.8KB, pdf)
Data Availability Statement
The raw sequencing reads from this study have been submitted to the NCBI sequence read archive (SRA) under BioProject accession no. PRJNA632824.