Abstract
The observed diversity of protein coding sequences continues to increase far more rapidly than knowledge of their functions, making classification algorithms essential for assigning a function to proteins using only their sequence. Most pipelines for annotating proteins rely on searches for homologous sequences in databases of previously annotated proteins using BLAST or HMMER. Here, we develop a new approach for classifying proteins into a taxonomy of functions and demonstrate its utility for genome annotation. Our algorithm, IDTAXA, was more accurate than BLAST or HMMER at assigning sequences to KEGG ortholog groups. Moreover, IDTAXA correctly avoided classifying sequences with novel functions to existing groups, which is a common error mode for classification approaches that rely on E-values as a proxy for confidence. We demonstrate IDTAXA’s utility for annotating eukaryotic and prokaryotic genomes by assigning functions to proteins within a multi-level ontology and applied IDTAXA to detect genome contamination in eukaryotic genomes. Finally, we re-annotated 8604 microbial genomes with known antibiotic resistance phenotypes to discover two novel associations between proteins and antibiotic resistance. IDTAXA is available as a web tool (http://DECIPHER.codes/Classification.html) or as part of the open source DECIPHER R package from Bioconductor.
BACKGROUND
Classification is a fundamental task in bioinformatics where sequences are assigned to an ontology that is often hierarchical, such as a taxonomy of organisms or functions. Classifiers can be used for the purpose of gene annotation, where a coding sequence is assigned a name based on its putative function (1). These names are of great importance because they suggest the role a protein plays within the cell and provide context that links new protein sequences to the universe of known functions. Many gene annotation approaches involve finding the most similar sequence within a training set (i.e. database) of previously annotated sequences and inheriting a gene name or gene ontology (GO) term when sequence similarity is sufficiently high (2,3). In this way, protein classification is largely dependent upon fundamental search algorithms followed by curation of the resulting hits. Annotation software typically rely on BLAST (4–6) or HMMER (7) for homology searches and often differ in which databases they search. Due to the incredible diversity of proteins, this technique only allows for naming of about 25–75% of the proteins in bacterial genomes depending on the sequence databases employed (8). Gene annotation is such a challenging problem that the goal has historically been to assign names to the highest fraction of proteins. Less attention has been paid to false positive identifications, in part because the error rate of gene annotation is often difficult to establish.
Protein classifiers for gene annotation have been developed largely independently of the nucleotide classifiers that are commonly employed for taxonomic classification of organisms. In the nucleotide domain, robust approaches have been developed for quantifying classification error rates (9). Errors can be partitioned into misclassifications (MCs) and overclassifications (OCs), which have different importance depending on the training data. MCs occur when a classifier assigns a sequence to the wrong class when representatives of the correct class exist in the training set. This type of error is problematic but relatively rare for most nucleotide classifiers. In contrast, OCs occur when the classifier assigns the sequence to a group when the correct group is missing from the training set. OC errors are common in biological problems because training sets sparsely cover the repertoire of possibilities. For example, microbiome sequences contain a substantial fraction of ‘microbial dark matter’ that is unrepresented in nucleotide training sets composed of phylogenetic marker gene sequences (e.g., ribosomal RNA genes) (10,11). Similarly, bacterial genomes contain many ‘hypothetical proteins’ with low similarity to previously described sequences in curated protein databases.
We previously published IDTAXA (12), a classification algorithm for nucleotide sequences that outperforms existing approaches, including homology searching, for taxonomic classification in both MC and OC error rates. Our approach is a hybrid of traditional distance-based classifiers and machine learning approaches based on k-mers. In this study, we extended our IDTAXA algorithm to work on amino acid sequences and benchmarked it against fundamental approaches for label assignment to protein coding sequences. This required major changes to our algorithm to accommodate the wide breadth of protein sequences, including the incorporation of amino acid alphabet reduction. We demonstrate IDTAXA’s utility with the KEGG (13) database containing orthologs of experimentally characterized proteins. The extension of IDTAXA to protein sequences enables users to annotate protein coding genes with confidence and is a major step toward avoiding the propagation of errors (14–16) made by gene annotation approaches that simply match to the nearest known sequence. Therefore, IDTAXA offers an improvement over homology searches for classification of proteins into a sequence database.
MATERIALS AND METHODS
Extending the IDTAXA algorithm to amino acid sequences
We extended our previously published algorithm for nucleotide classification, IDTAXA (12), to incorporate features specific to amino acid classification. This required four substantial changes to the original algorithm:
First, IDTAXA now computes the equivalent entropy of a user-supplied alphabet and uses this to automatically calculate the size (k) of k-mers for classification. This enables the algorithm to accommodate any size of reduced amino acid alphabet provided as input without requiring the user to manually specify k. Given that the frequencies (f) of different characters (c) in the alphabet (a) are typically non-uniform, we calculate the size (x) of an alphabet having uniform character frequency and equivalent entropy:
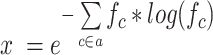 |
Then, k can be calculated using the formula:
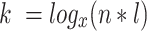 |
Where l is the length of sequences in the training set, and n is the number of random k-mers that must be drawn before one match is expected to occur by chance. We base l off of the upper 1-percentile of sequence lengths. The parameter n is user-specified with a default value of 500, meaning that k-mers are found by chance on average once per 500 k-mers sampled. In practice, we found that lower values of n (e.g. 100) selected sub-optimal values of k, whereas higher values offered no benefit.
Second, we modified IDTAXA to work with amino acid characters in the same way it works with nucleotide characters. We also enabled users to specify reduced amino acid alphabets as input rather than the standard 20-letter amino acid alphabet. Reduced alphabets are specified by the user as groupings of amino acids, and the optimized reduced alphabet is used by default (see Results).
Third, since amino acid sequences vary considerably in length, we implemented a pre-filter to subset relevant sequences within the training set to those within a multiple of the query sequence’s length. By default, the fold-difference in length is set by the 1st and 99th percentile of the distribution of full-length sequences in the training set. The fold-difference can be specified by the user via the fullLength parameter. This pre-filter improved both accuracy and speed when classifying full-length protein sequences; however, it may preclude rare correct matches that differ considerably in length from representatives of their group.
Fourth, due to the wide diversity of protein sequences, the algorithm now detects the minimum number of k-mers (S) that must be sampled in each bootstrap replicate to avoid spurious hits in large training sets. In our previous study (12) S was set to L0.47, where L is the number of k-mers in the query sequence. This is problematic for very short sequences because a few k-mers can often be found by chance in large databases. We now impose the constraint that the probability of observing at least half of the sampled k-mers (≥ S/2) in one or more sequences is <1% per bootstrap replicate. In practice, this sets a lower bound on S of about 10 k-mers per bootstrap replicate, which excludes extremely small sequences (L < S + k) from testing.
IDTAXA is implemented in the LearnTaxa and IdTaxa functions within the DECIPHER (17) package for the R programming language (18). Users first train the classifier with LearnTaxa by supplying a training set of nucleotide or protein sequences and their associated classifications. The resulting classifier object can be given along with query sequences to IdTaxa to obtain classifications and their associated confidences at each hierarchical level of a taxonomy. All tests were performed using R v4.1.0 and DECIPHER v2.19.0 available from Bioconductor v3.13 (19) (https://bioconductor.org/packages/release/bioc/html/DECIPHER.html). We used the non-default parameters maxChildren = 1 in LearnTaxa and fullLength = 0.99 in IdTaxa. The argument maxChildren is set to avoid the tree descent algorithm described in our prior publication (12) because amino acid training sets are typically far more diverse than the nucleotide training sets composed of a single gene for which tree descent was originally designed. These non-default parameters are recommended for classifying protein sequences. All tests were performed on either a 2.6 GHz Intel i7 processor with 64 GB of RAM or run on Open Science Grid compute nodes with at least six processors, 32 GB of RAM, and 4 GB of available disk.
Optimizing a reduced amino acid alphabet
Reduced amino acid alphabets, where residues in the same group are viewed as interchangeable, can facilitate detection of distantly related sequences. Due to the immense space of possible alphabet reductions, it is challenging to determine an optimal reduced alphabet for protein classification. The number of possible alphabets is a Stirling number of the second kind, totaling more than 51 trillion possibilities. Searching through all possible alphabets is infeasible since testing each alphabet’s performance can take minutes. However, it is feasible to search through only a small fraction of possible alphabets because many reductions are unlikely to improve classification. We used a directed acyclic graph to query the space of possible reduced alphabets, using only high performing alphabets to seed new candidate alphabets for the next reduction level. This approach tested all size 20, 19 and 18 alphabets, after which point, only the top 500 size 18 alphabets were used to generate size 17 alphabets for testing. We repeated this process of selecting the top 500 from each reduction level down to an alphabet of size 2, testing a total of 419 130 unique alphabets.
To construct a test set for alphabet optimization, all single function, non-provisional and unambiguous enzyme classifications (EC) numbers were extracted from Swiss-Prot (20), including both eukaryotes and prokaryotes. This resulted in an initial set of 251 944 sequences. Long (>5000 amino acids) and short (<100 amino acids) sequences were removed from the training set, and gene names (if present) were trimmed of special characters and converted to lower case. The EC numbers and gene names were concatenated to create a group label for each sequence (e.g. ‘1.14.14.1.cyp2h1’ – a cytochrome p450). Since some groups contained many nearly identical sequences, we reduced this set to up to 10 randomly selected sequences per group. This final set contained 58 887 sequences assigned to one of 27 863 unique groups.
We randomly selected 2000 sequences for use as a hold-out set for testing alphabet performance. Hold-out sequences were split into two subsets: 1000 singletons (only member of their group) for determining the OC rate and 1000 non-singletons for determining the MC rate. Singleton sequences cannot be assigned to a group in the training data and, therefore, the only correct option is to assign them a very low confidence classification. As described in our previous publication (12), OC and MC error rates can be compared at a given fraction of sequences classified (based on non-singletons) to calculate accuracy. This benchmarking approach offers the advantage of being independent of an algorithm’s reported confidence, which allows comparing classification approaches having different confidence scales. Reduced alphabets were judged by the combined area under their OC and MC curves (AUC), where lower AUC corresponds to better accuracy.
Comparing against standard approaches for gene annotation
We performed benchmarking using the KEGG (v95.1) database containing functionally orthologous groups of experimentally characterized proteins (13). The complete KEGG database (including eukaryotes and prokaryotes) was randomly subsampled to maximize species diversity while enforcing a limit of 100 sequences per KEGG Orthology (KO) category. Only KEGG entries with both a nucleotide and amino acid sequence were allowed, and those with ambiguous positions (e.g., ‘X’ for amino acids or ‘N’ for nucleotides) were removed. The resulting set of 1 672 354 sequences were labeled according to KEGG’s four-level BRITE hierarchical classification (13) appended with a lineage (taxonomic) classification for increased resolution. For example, the KEGG classification ‘09100 Metabolism; 09101 Carbohydrate metabolism; 00010 Glycolysis / Gluconeogenesis [PATH:ko00010]; K00844 HK, hexokinase [EC:2.7.1.1]’ might be appended with ‘; Eukaryotes; Animals; Vertebrates; Reptiles’ if the training sequence originated from Gekko japonicus (Schlegel’s Japanese gecko). This process resulted in 21 157 unique groups at the KO-level and 568 912 groups at the lineage-level (i.e. after appending lineage information). KEGG training sets are available on Zenodo (https://doi.org/10.5281/zenodo.5057026).
MC and OC error rates were computed using cross-validation (CV) by removing 10 unique hold-out sets of up to 1000 singleton and 1000 non-singleton sequences. Sequences were randomly selected such that no more than one non-singleton sequence was removed per group when constructing hold-out sets. For each fold of CV, IDTAXA was trained on the KEGG sequences minus the hold-out using LearnTaxa and the hold-out set was tested with IdTaxa. We compared IDTAXA’s results to those of BLAST (2.10.1) (21) by constructing a BLAST database from the KEGG sequences minus the hold-out set and querying the hold-out set using blastp or blastn with an E-value (EVL) cutoff of 10–3 and Smith–Waterman traceback enabled in blastp. To compare against HMMER (3.3.1) (22), protein multiple sequence alignments were constructed for each group of sequences using DECIPHER (v2.19.0) (23). Individual hidden Markov models (HMMs) were constructed using hmmbuild for each group without including the hold-out sequences, which required re-training HMMs for each fold of CV. The set of HMMs were combined with hmmpress to create a library of HMMs and the hold-out sequences were tested with hmmscan using default arguments.
Error rates for IDTAXA were contrasted to those of choosing the BLAST and HMMER top hit. We compared two proxies for BLAST confidence: EVL (-log10(EVL)) and local percent identity (PID) output by BLAST. In the case of PID, the top hit was selected based on PID rather than BLAST’s default ranking by EVL. In cases where BLAST or HMMER did not return any hits, we assigned the sequence to an unclassified placeholder at 0% confidence (i.e. a 0% PID or the maximum EVL). Scripts for reproduction of all cross-validation results are available on GitHub (https://github.com/npcooley/AAClassification), and cross-validation results are available on Zenodo (https://doi.org/10.5281/zenodo.5071173). The training set used to identify contamination is present on the DECIPHER website (http://DECIPHER.codes/Downloads.html).
Discovering novel antibiotic resistance associations with KO categories
To uncover associations between KEGG categories and antibiotic resistance, the NCBI pathogens Isolates Browser (https://www.ncbi.nlm.nih.gov/pathogens/isolates/) was used to download the proteome of all assemblies with corresponding antibiotic susceptibility test (phenotype) data. These data were subset to antibiotics with at least 1000 test results, species with 30 or more genomes, and antibiotics that were tested against >10% of the strains in a species. This resulted in seven testable species: Staphylococcus aureus (43 assemblies), Pseudomonas aureuginosa (148), Klebsiella pneumoniae (398), Escherichia coli (585), Campylobacter jejuni (928), Acinetobacter baumanii (1085) and Salmonella enterica (5417). Susceptibility tests with intermediate results were considered resistant. Protein sequences were classified with IDTAXA at 60% confidence. The program treeWAS (24) was used to determine statistically significant associations between the antibiotic resistance phenotypes and KO categories. Default settings were used with the exception of adjusting the P-value threshold (0.01) for multiple testing (Bonferroni correction) and supplying a neighbor-joining tree generated from a distance matrix based on the presence/absence matrix of classifications in each species. Only ‘simultaneous scores’ from treeWAS were evaluated, and treeWAS’s reported contingency tables were used to generate log-odds ratios for associations reported as significant. KO categories that were never associated with an antibiotic at a log-odds of 3.5 or greater were omitted.
RESULTS
Optimization of a reduced amino acid alphabet for classification
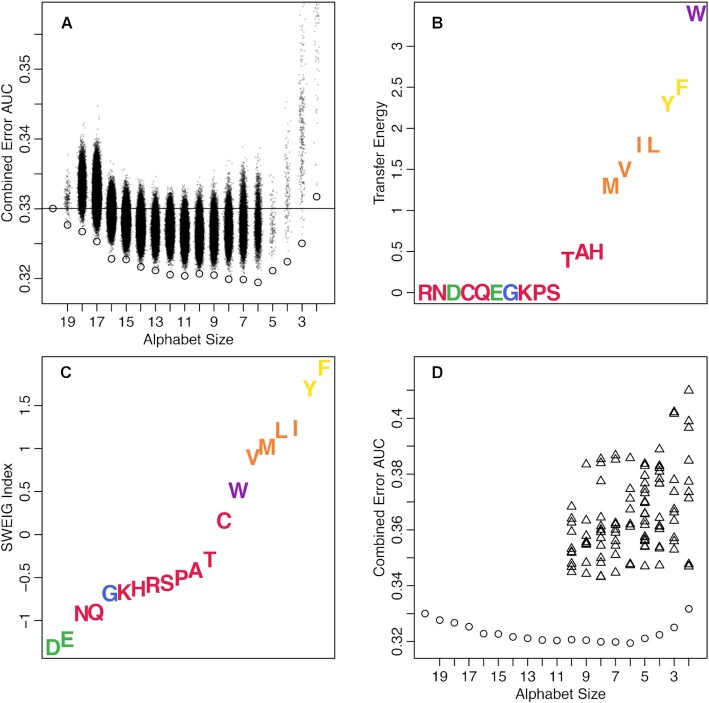
A major difference between amino acid and nucleotide sequences is that amino acids can be reduced by merging letters of the standard 20-letter alphabet into a lower dimensional space. For example, the amino acids isoleucine and valine are reducible to a single letter because the two are frequently substituted during protein evolution. This permits related k-mers to match in the reduced alphabet that would be mismatched in the standard 20-letter alphabet. We employed an iterative procedure to optimize a reduced alphabet for assigning enzyme classifications to Swiss-Prot sequences (see Materials and Methods). IDTAXA’s accuracy steadily improved from the standard 20 letter alphabet down to a 6 letter alphabet and then became worse with further reduction (Figure 1A). The optimal reduced alphabet consisted of ‘ACHKNPQRST’, ‘DE’, ‘FY’, ‘G’, ‘ILMV’ and ‘W’. This alphabet merges amino acids with similar values of hydrophobicity quantified in previous studies (Figure 1B,C) (25–28).
Figure 1.
Amino acid alphabet reduction leads to reduced error. (A) Reduced alphabets (dots) led to lower combined area under the MC and OC curves on the Swiss-Prot test set. The best performing alphabet of each size is circled and performance of the standard 20 letter amino acid alphabet is shown as a horizontal line. Lower error results in a smaller area under the curve with the best reduced alphabet identified at size 6. The best performing reduced alphabet (color groups) correlated with two measures of hydrophobicity: (B) transfer energy (28) and (C) SWEIG index (26). (D) Optimized alphabets (circles) outperformed 103 previously published literature alphabets (triangles) (29) on the Swiss-Prot test set.
We compared our optimized amino acid alphabet to a set of 103 previously published (29) reduced amino acid alphabets. However, none of the previously published alphabets outperformed the standard 20-letter amino acid alphabet on the Swiss-Prot test set (Figure 1D). The best performing alphabet from literature was ‘AST’, ‘CFWY’, ‘DEQ’, ‘G’, ‘HN’, ‘ILMV’, ‘KR’ and ‘P’. Notably, none of these published alphabets were constructed specifically for the purpose of amino acid classification.
Benchmarking IDTAXA for protein sequence classification
Having optimized a reduced amino acid alphabet for classification on the Swiss-Prot test set, we sought to compare IDTAXA’s performance to alternative classification approaches on an independent training set. We calculated MC and OC error rates for predicting the KO categories (from the BRITE hierarchy) using cross-validation with the KEGG orthology database. As expected, amino acid classification of the protein sequences resulted in far fewer errors than nucleotide classification of the equivalent coding sequences (Figure 2A). However, there was minimal difference on the KEGG training set among the optimized reduced alphabet, the 20-letter amino acid alphabet and the best performing reduced alphabet from literature. We observed similar results for classifying down to the lineage-level in the KEGG training set (Figure 2B). Here, the standard 20-letter alphabet slightly outperformed the other alphabets, which we attributed to the higher-resolution offered by use of all amino acids for sequences belonging to different lineages within the same KO functional category. This reflects the fact that different groups at the lineage-level can contain k-mers that are identical in the reduced amino acid space but different in the standard 20-letter amino acid space.
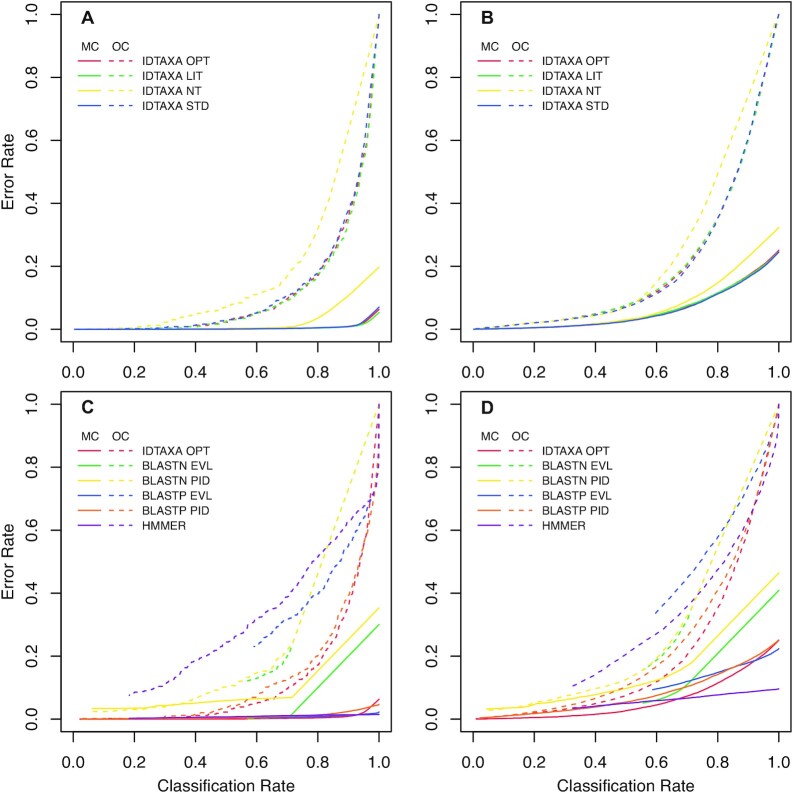
Figure 2.
Error rates for classification on the KEGG test set. Plots show the MC and OC error rates versus the fraction of classified non-singleton sequences as confidence is adjusted from 0% (right) to 100% (left). Classification of amino acid sequences yielded lower error rates than classifying the equivalent nucleotide (coding) sequences with IDTAXA on the KEGG test set at the (A) KO-level and (B) lineage-level. (C) IDTAXA outperformed BLAST and HMMER for classification of amino acid sequences on the KEGG test set at the KO-level. BLASTP PID as a proxy for confidence offered substantially lower error rates than using EVL. (D) At the lineage-level, IDTAXA even more substantially outperformed BLAST in both MC and OC error rates.
IDTAXA outperformed BLAST and HMMER in MC and OC error (Figure 2C and Table 1), although both BLASTP and IDTAXA both had exceptionally low error rates (Figure 2C). With BLASTP, E-value (EVL) offered lower MC error rates at the expense of much higher OC error rates than using percent identity (PID) as a proxy for confidence. At the lineage-level, IDTAXA outperformed BLAST in MC error rate by a larger margin than at the KO-level (Figure 2D). HMMER exhibited the lowest MC error rates at high fractions of sequences classified but had very high OC error rates. Nevertheless, OC error rates were too high for practical application above the point where 60% of test (hold-out) sequences were classified with the KEGG training set. At this point, IDTAXA’s confidence was 51% at the KO-level and 40% at the lineage-level, corresponding to a BLAST PID of ∼80% (Table 1). Such a high PID threshold suggests that sequences must be very similar to ensure membership in the same KO functional category.
Table 1.
Error rates and confidence levels at 60% of non-singleton sequences classified
| KEGG KO-level | KEGG lineage-level | |||||
|---|---|---|---|---|---|---|
| OC error rate | MC error rate | Confidence | OC error rate | MC error rate | Confidence | |
| IDTAXA − OPT | 0.051 | 0.001 | 51% | 0.134 | 0.051 | 40% |
| IDTAXA − LIT | 0.046 | 0.001 | 49% | 0.126 | 0.046 | 40% |
| IDTAXA − NT | 0.11 | 0.004 | 6% | 0.148 | 0.051 | 4% |
| IDTAXA − STD | 0.051 | 0.002 | 48% | 0.113 | 0.042 | 39% |
| BLASTP − EVL | 0.235 | 0.005 | EVL = 2e-177 | 0.340 | 0.096 | EVL = 9e-177 |
| BLASTP − PID | 0.071 | 0.006 | PID = 81% | 0.167 | 0.075 | PID = 82% |
| BLASTN − EVL | 0.130 | 0.004 | EVL = 2e-149 | 0.189 | 0.061 | EVL = 2e-140 |
| BLASTN − PID | 0.145 | 0.064 | PID = 83% | 0.195 | 0.125 | PID = 82% |
| HMMER | 0.320 | 0.009 | EVL = 4e-138 | 0.271 | 0.059 | EVL = 1e-197 |
Comparison of IDTAXA and BLAST assignments on a eukaryotic genome
Our benchmarking revealed that BLAST performed the best among previously existing approaches to classification. To investigate differences between BLAST and IDTAXA, we annotated the proteome of the yeast Brettanomyces bruxellensis using both classification approaches (Figure 3). We chose this genome because it was recently added to RefSeq and therefore was absent from our training dataset. IDTAXA’s assignment and BLAST’s top hit were largely in agreement when IDTAXA’s confidence was above 10%. BLAST’s PID and IDTAXA’s confidence were correlated, except for some proteins given low confidence assignments by IDTAXA despite high PID. This is expected to happen whenever there are competing assignments with high sequence homology and highlights the merits of assigning based on IDTAXA’s confidence rather than homology alone. In contrast to PID, EVL showed little correlation with IDTAXA’s confidence. There were many proteins with maximally low (1e-180) EVLs assigned low confidence by IDTAXA. This result partly explains why PID greatly outperformed EVL as a proxy for confidence in cross-validation.
Figure 3.
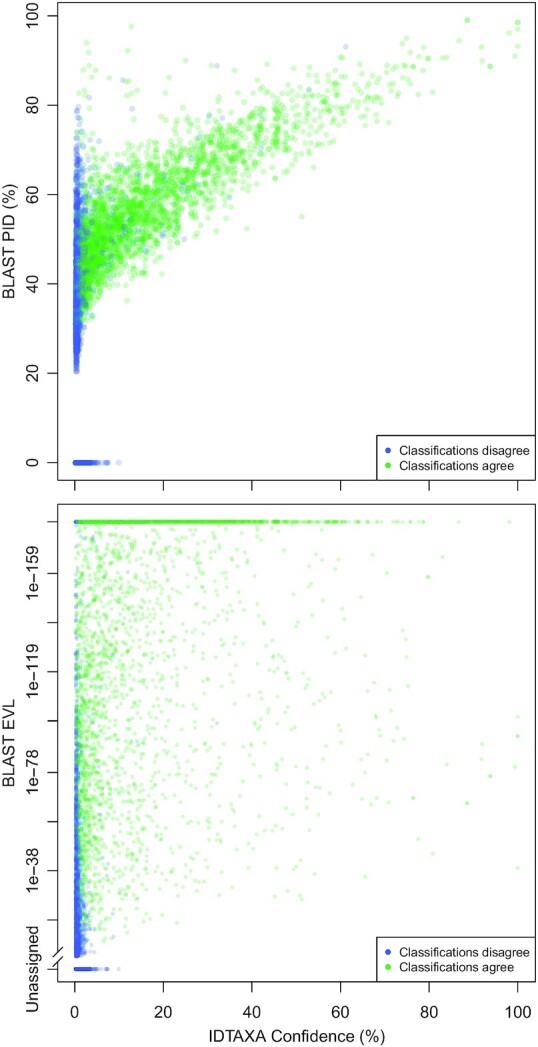
Comparison of eukaryotic protein assignments to KO categories. Each point represents one of 5243 protein sequence belonging to the genome of the yeast Brettanomyces bruxellensis (RefSeq accession number GCF_011074885.1). IDTAXA’s confidence was correlated with BLAST’s PID but largely uncorrelated with EVL. Most proteins that IDTAXA assigns a 40% or greater confidence were also found to have a BLASTP local PID above 60%, although many proteins with high PID were assigned low confidence classifications by IDTAXA (top). Proteins given low confidence classifications often had very low (∼0) BLAST EVLs (bottom). Note the inverted y-axis where higher EVLs are shown at the bottom.
Utility of IDTAXA for gene annotation and quality control
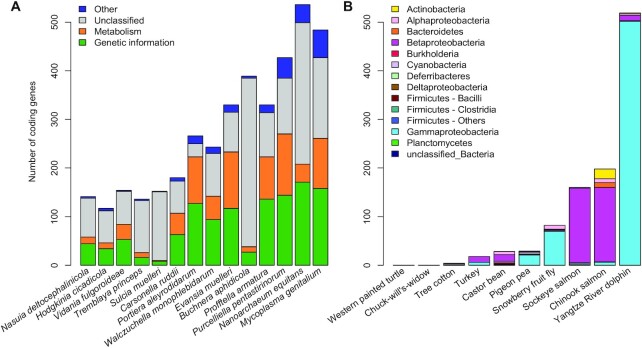
Since IDTAXA exhibited lower rates of incorrectly assigning annotations to novel proteins, we wished to re-annotate more genomes and visualize the resulting classifications. To this end, we trained IDTAXA on taxon-specific subsets of the complete KEGG database and then applied them to a diverse set of symbiont microbial genomes available from NCBI. Symbiont genomes are known to undergo genome reduction that results in maintenance of fewer genes. At 50% confidence, 7.2–90% of the genes in each genome were classifiable (Figure 4A). Notably, the Buchnera aphidicola genome, which belongs to the order Enterobacterales, had a low percentage (11%) of classifiable genes. This suggests that even members of well-annotated phylogenetic groups do not always yield high annotation coverage when the OC error rate is very low.
Figure 4.
Using IDTAXA to re-annotate published genomes. (A) Microbial symbionts are known to undergo genome reduction, which results in smaller genome sizes and a skewed distribution of core gene functions. Bar heights show the number of genes in each symbiont genome annotated as belonging to KO categories involved in genetic information processing (e.g. transcription and translation), metabolism, other categories, or unclassified at 50% confidence. Notably, the fraction of unclassified proteins varies from species-to-species and can be high even for members of well-studied groups (e.g. Buchnera aphidicola, a member of Enterobacterales). (B) Using the full KEGG training set with lineage information, IDTAXA reports both a KO category and a taxonomic lineage for each query sequence. Bar heights show the number of taxonomic assignments from the set of protein sequences belonging to the top 10 contaminated genomes in RefSeq according to a recent study (30). The presence of prokaryotic proteins in eukaryotic genomes suggests contamination and may serve to alert the authors of the genome assembly. Most contaminated proteins are classified as originating from the Proteobacteria bacterial phylum.
Another advantage of IDTAXA is its ability to assign confidences at each level of a hierarchical classification. We applied this feature to identify contaminating sequences during genome annotation. Genome contamination is a major issue in public sequence databases (30), especially among eukaryotic genomes. Contamination has been deemed responsible for controversial claims of extensive horizontal gene transfer between bacteria and some eukaryotes (31–33). We used the KEGG database with lineage information to identify possible bacterial genes within the top 10 eukaryotic genomes in RefSeq reported to harbor (eukaryotic or prokaryotic) contaminants in a recent study (30). This training set enables us to classify proteins past the KO-level to the taxonomic lineage from which the training sequences originated. Proteins with annotations arising from bacterial training sequences represented a notable fraction of some genomes (Figure 4B). Most of these sequences were classified to proteins originating from Proteobacteria and likely represent contaminants.
Re-annotating proteomes to discover novel antibiotic resistance associations
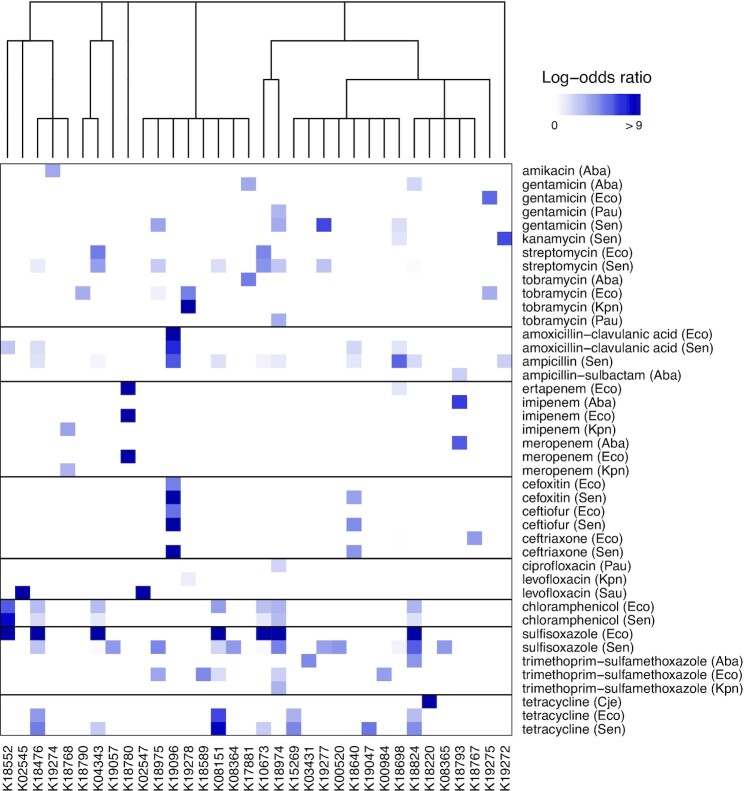
We next sought to apply IDTAXA’s high quality annotations to identify genotype–phenotype associations. We re-annotated the genomes of 8604 bacterial pathogens with known antibiotic resistance phenotypes using the KEGG training set. The resulting dataset contained assignments to 5215 KO categories and phenotypes specifying resistance or susceptibility to 38 possible antibiotics. We used treeWAS (24) to identify 151 statistically significant associations between an antibiotic and a KO category for each of seven bacterial pathogens and narrowed these results to the 69 with log-odds ratios of at least 3.5 (Figure 5). Of these, 32 (out of 34) KO categories were already documented in the literature as implicated in antibiotic resistance, validating our approach. We identified two novel associations in Salmonella: K15269 (PecM) associated with tetracycline resistance (log-odds = 4.8) and K18640 (ParM) associated with resistance to three cephalosporins (log-odds = 3.9 to 4.6). PecM is an efflux pump known to be associated with virulence in Proteobacteria, and is part of a two-component system with PecS, which belongs to the multiple antibiotic resistance (mar) family of regulatory proteins implicated in antibiotic resistance (34,35). PecM’s partner, PecS, was not detected because it is not represented by a KO category. ParM is involved in plasmid maintenance, which might be beneficial for maintaining resistance genes located on mobile elements (36). Similar to the PecM/PecS two-component system, ParM’s associated partner (ParR) is not included in a KO category. Overall, these results verified the utility of high-quality annotations produced by IDTAXA for discovering genotype–phenotype associations.
Figure 5.
Significant associations between KEGG categories and antibiotics. Proteomes belonging to pathogens were re-annotated with IDTAXA and assessed for statistically significant associations with antibiotic resistance. Antibiotics are grouped by class, with species abbreviations shown in parentheses. The dendrogram at the top shows the functional taxonomy of the 34 KO categories found to have genotype–phenotype associations. For example, K18589 and K19728 are both part of ‘01504 Antimicrobial resistance genes’. Only two of 34 KO categories were not previously implicated in antibiotic resistance: K15269 and K18640.
DISCUSSION
Modern approaches to annotation are based on similarity searches in databases of previously annotated proteins (6,37). Such approaches assign genes to their nearest neighbor if they are within a pre-specified similarity threshold, typically using HMMs or BLAST. Unfortunately, as we have shown, HMMER results in unacceptably high OC error rates. The widespread application of this approach propagates errors among genomes and encourages skepticism in automatic annotations (14–16). A major advantage of IDTAXA is that it assigns a percent confidence to each classification that supersedes ambiguous terms commonly prepended to gene annotations, such as possible, probable, predicted and putative. This confidence also makes it straightforward to annotate across multiple databases by selecting the label with highest confidence. We anticipate that these features will be appreciated by users who are concerned with the accuracy of their gene annotations.
Another major advantage of IDTAXA is that it can classify proteins into a hierarchical taxonomy with multiple levels of names. For example, KEGG’s BRITE hierarchy consists of four levels that are each assigned a confidence by IDTAXA. This allows higher-level functional comparisons to be drawn (Figure 4), analogous to examining the enrichment of gene ontology terms. Also, using multiple levels allows for an appropriate level of ambiguity in assignments where enzymes belong to several categories at the lowest level of a hierarchy (e.g. multi-functional or promiscuous enzymes). When a query sequence matches more than one category it is assigned a low confidence at the lowest-level but a greater confidence in upper levels. Unfortunately, only a subset of protein databases adhere to multi-level ontologies, such as the BRITE hierarchy or the EC numbering scheme. Nevertheless, these are some of the most comprehensive databases of classified proteins, so we expect multi-level hierarchies to be a useful feature in practice.
Our results led to several surprising conclusions. First, EVL is an unacceptable proxy for confidence with nucleotide or amino acid sequences. Even at the most stringent EVL, sequences belonging to novel functional categories were incorrectly assigned to a group in KEGG 23% of the time in amino acid space (blastp) and 11% in nucleotide space (blastn). Second, PID values at low error rates imply that commonly used PID thresholds are too lenient. At a PID threshold of 60% the OC error rate was 26% for blastp and 23% for blastn at the KO-level, meaning that a sequence belonging to a novel group would be incorrectly assigned to an existing KEGG category over 20% of the time. Third, HMMER’s high OC error rate in comparison to BLAST and IDTAXA was unanticipated given its frequent usage for gene annotation. We attributed this to HMMER’s high sensitivity, which results in detection of weak hits that lower its MC error rate at the expense of raising its OC error rate. We believe IDTAXA outperforms approaches based on homology searches because it not only accounts for the degree to which a protein matches the training data but also accounts for the number of competing candidate categories where a protein could be assigned.
IDTAXA was designed to be simple for users to apply to their own gene sequences using the DECIPHER R package and is also accessible as a web tool (http://DECIPHER.codes/Classification.html). Users only need to supply their protein or nucleotide (coding) sequences and choose an appropriate database for classification. Notably, we also decided to split the complete KEGG database into subsets by major taxonomic group to speed up classification, and because we noticed that assignments originated from organisms predominantly related to that of the query sequence when contamination was not present. We provide pre-trained classifiers for prokaryotic and eukaryotic groups, as well as the entire database for protein sets having multiple origins (e.g. metagenomes). Annotations with lineage information are provided when classifying with the entire KEGG database, which can offer insight into assembly quality when contamination might be present. We anticipate that users will find the ease of gene annotation with IDTAXA to be a major advantage in addition to its high accuracy classifications.
DATA AVAILABILITY
Scripts necessary to reproduce the results are available on GitHub (https://github.com/npcooley/AAClassification). Trained classifiers and the original training sequences are available on Zenodo (https://doi.org/10.5281/zenodo.5057026).
ACKNOWLEDGEMENTS
Compute resources for this study were provided by the Open Science Grid.
Contributor Information
Nicholas P Cooley, Department of Biomedical Informatics, University of Pittsburgh, Pittsburgh, PA 15206, USA.
Erik S Wright, Department of Biomedical Informatics, University of Pittsburgh, Pittsburgh, PA 15206, USA; Center for Evolutionary Biology and Medicine, Pittsburgh, PA 15219, USA.
FUNDING
NPC was supported by the NLM at the NIH (grant number 5T15LM007059-32). This study was funded by the NIAID at the NIH (grant number 1DP2AI145058-01 to ESW).
Conflict of interest statement. None declared.
REFERENCES
- 1.Dong Y., Li C., Kim K., Cui L., Liu X.. Genome annotation of disease-causing microorganisms. Brief. Bioinform. 2021; 22:845–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ruiz-Perez C.A., Conrad R.E., Konstantinidis K.T.. MicrobeAnnotator: a user-friendly, comprehensive functional annotation pipeline for microbial genomes. BMC Bioinformatics. 2021; 22:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galperin M.Y., Koonin E.V.. Sources of systematic error in functional annotation of genomes: domain rearrangement, non-orthologous gene displacement and operon disruption. In Silico Biol. 1998; 1:55–67. [PubMed] [Google Scholar]
- 4.Cantalapiedra C.P., Hernández-Plaza A., Letunic I., Bork P., Huerta-Cepas J.. eggNOG-mapper v2: functional annotation, orthology assignments, and domain prediction at the metagenomic scale. 2021; bioRxiv doi:03 June 2021, preprint: not peer reviewed 10.1101/2021.06.03.446934. [DOI] [PMC free article] [PubMed]
- 5.Aziz R.K., Bartels D., Best A.A., DeJongh M., Disz T., Edwards R.A., Formsma K., Gerdes S., Glass E.M., Kubal M.et al.. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008; 9:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seemann T.Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014; 30:2068–2069. [DOI] [PubMed] [Google Scholar]
- 7.Jones P., Binns D., Chang H.Y., Fraser M., Li W., McAnulla C., McWilliam H., Maslen J., Mitchell A., Nuka G.et al.. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014; 30:1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lobb B., Tremblay B.J., Moreno-Hagelsieb G., Doxey A.C.. An assessment of genome annotation coverage across the bacterial tree of life. Microb. Genom. 2020; 6:e000341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edgar R.C.Accuracy of taxonomy prediction for 16S rRNA and fungal ITS sequences. PeerJ. 2018; 6:e4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rinke C., Schwientek P., Sczyrba A., Ivanova N.N., Anderson I.J., Cheng J.-.F., Darling A., Malfatti S., Swan B.K., Gies E.A.et al.. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013; 499:431–437. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Z., Wang J., Wang J., Wang J., Li Y.. Estimate of the sequenced proportion of the global prokaryotic genome. Microbiome. 2020; 8:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murali A., Bhargava A., Wright E.S.. IDTAXA: a novel approach for accurate taxonomic classification of microbiome sequences. Microbiome. 2018; 6:140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanehisa M., Sato Y., Furumichi M., Morishima K., Tanabe M.. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019; 47:D590–D595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richardson E.J., Watson M.. The automatic annotation of bacterial genomes. Brief. Bioinform. 2013; 14:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salzberg S.L.Next-generation genome annotation: we still struggle to get it right. Genome Biol. 2019; 20:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei X., Zhang C., Freddolino P.L., Zhang Y.. Detecting gene ontology misannotations using taxon-specific rate ratio comparisons. Bioinformatics. 2020; 36:4383–4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wright E.S.Using DECIPHER v2.0 to analyze big biological sequence data in R. R J. 2016; 8:352–359. [Google Scholar]
- 18.R Core Team R: A Language and Environment for Statistical Computing. 2019; Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 19.Huber W., Carey V.J., Gentleman R., Anders S., Carlson M., Carvalho B.S., Bravo H.C., Davis S., Gatto L., Girke T.et al.. Orchestrating high-throughput genomic analysis with bioconductor. Nat. Methods. 2015; 12:115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.The UniProt C UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017; 45:D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J.. Basic local alignment search tool. J. Mol. Biol. 1990; 215:403–410. [DOI] [PubMed] [Google Scholar]
- 22.Eddy S.R.Accelerated profile HMM searches. PLoS Comput. Biol. 2011; 7:e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wright E.S.DECIPHER: harnessing local sequence context to improve protein multiple sequence alignment. BMC Bioinformatics. 2015; 16:322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Collins C., Didelot X.. A phylogenetic method to perform genome-wide association studies in microbes that accounts for population structure and recombination. PLoS Comput. Biol. 2018; 14:e1005958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manavalan P., Ponnuswamy P.K.. Hydrophobic character of amino acid residues in globular proteins. Nature. 1978; 275:673–674. [DOI] [PubMed] [Google Scholar]
- 26.Sweet R.M., Eisenberg D.. Correlation of sequence hydrophobicities measures similarity in three-dimensional protein structure. J. Mol. Biol. 1983; 171:479–488. [DOI] [PubMed] [Google Scholar]
- 27.Zhou H., Zhou Y.. Quantifying the effect of burial of amino acid residues on protein stability. Proteins. 2004; 54:315–322. [DOI] [PubMed] [Google Scholar]
- 28.Nozaki Y., Tanford C.. The solubility of amino acids and two glycine peptides in aqueous ethanol and dioxane solutions. Establishment of a hydrophobicity scale. J. Biol. Chem. 1971; 246:2211–2217. [PubMed] [Google Scholar]
- 29.Solis A.D.Amino acid alphabet reduction preserves fold information contained in contact interactions in proteins. Proteins Struct. Funct. Genet. 2015; 83:2198–2216. [DOI] [PubMed] [Google Scholar]
- 30.Steinegger M., Salzberg S.L.. Terminating contamination: large-scale search identifies more than 2,000,000 contaminated entries in genbank. Genome Biol. 2020; 21:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boothby T.C., Tenlen J.R., Smith F.W., Wang J.R., Patanella K.A., Nishimura E.O., Tintori S.C., Li Q., Jones C.D., Yandell M.et al.. Evidence for extensive horizontal gene transfer from the draft genome of a tardigrade. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:15976–15981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koutsovoulos G., Kumar S., Laetsch D.R., Stevens L., Daub J., Conlon C., Maroon H., Thomas F., Aboobaker A.A., Blaxter M.. No evidence for extensive horizontal gene transfer in the genome of the tardigrade hypsibius dujardini. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:5053–5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Delmont T.O., Eren A.M.. Identifying contamination with advanced visualization and analysis practices: metagenomic approaches for eukaryotic genome assemblies. PeerJ. 2016; 4:e1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sulavik M.C., Gambino L.F., Miller P.F.. The MarR repressor of the multiple antibiotic resistance (mar) operon in escherichia coli: prototypic member of a family of bacterial regulatory proteins involved in sensing phenolic compounds. Mol. Med. 1995; 1:436–446. [PMC free article] [PubMed] [Google Scholar]
- 35.Praillet T., Reverchon S., Nasser W.. Mutual control of the PecS/PecM couple, two proteins regulating virulence-factor synthesis in erwiniachrysanthemi. Mol. Microbiol. 1997; 24:803–814. [DOI] [PubMed] [Google Scholar]
- 36.Gerdes K., Howard M., Szardenings F.. Pushing and pulling in prokaryotic DNA segregation. Cell. 2010; 141:927–942. [DOI] [PubMed] [Google Scholar]
- 37.Tatusova T., DiCuccio M., Badretdin A., Chetvernin V., Nawrocki E.P., Zaslavsky L., Lomsadze A., Pruitt K.D., Borodovsky M., Ostell J.. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016; 44:6614–6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Scripts necessary to reproduce the results are available on GitHub (https://github.com/npcooley/AAClassification). Trained classifiers and the original training sequences are available on Zenodo (https://doi.org/10.5281/zenodo.5057026).