Abstract
The RIKEN BioResource Research Center (BRC) was established in 2001 as a comprehensive biological resource center in Japan. The Experimental Animal Division, one of the BRC infrastructure divisions, has been designated as the core facility for mouse resources within the National BioResource Project (NBRP) by the Japanese government since FY2002. Our activities regarding the collection, preservation, quality control, and distribution of mouse resources have been supported by the research community, including evaluations and guidance on advancing social and research needs, as well as the operations and future direction of the BRC. Expenditure for collection, preservation, and quality-control operations of the BRC, as a national core facility, has been funded by the government, while distribution has been separately funded by users’ reimbursement fees. We have collected over 9000 strains created mainly by Japanese scientists including Nobel laureates and researchers in cutting-edge fields and distributed mice to 7000 scientists with 1500 organizations in Japan and globally. Our users have published 1000 outstanding papers and a few dozen patents. The collected mouse resources are accessible via the RIKEN BRC website, with a revised version of the searchable online catalog. In addition, to enhance the visibility of useful strains, we have launched web corners designated as the “Mouse of the Month” and “Today’s Tool and Model.” Only high-demand strains are maintained in live colonies, while other strains are cryopreserved as embryos or sperm to achieve cost-effective management. Since 2007, the RIKEN BRC has built up a back-up facility in the RIKEN Harima branch to protect the deposited strains from disasters. Our mice have been distributed with high quality through the application of strict microbial and genetic quality control programs that cover a globally accepted pathogens list and mutated alleles generated by various methods. Added value features, such as information about users’ publications, standardized phenotyping data, and genome sequences of the collected strains, are important to facilitate the use of our resources. We have added and disseminated such information in collaboration with the NBRP Information Center and the NBRP Genome Information Upgrading Program. The RIKEN BRC has participated in international mouse resource networks such as the International Mouse Strain Resource, International Mouse Phenotyping Consortium, and Asian Mouse Mutagenesis and Resource Association to facilitate the worldwide use of high-quality mouse resources, and as a consequence it contributes to reproducible life science studies and innovation around the globe.
Foundation of the RIKEN BioResource Research Center
The RIKEN BioResource Research Center (BRC) was established in January 2001 (https://web.brc.riken.jp/en/about). During these past 20 years, under the leadership of Dr. Toshihiko Shiroishi (current Director), Dr. Yuichi Obata (former Director and current senior adviser), and the late Dr. Kazuo Moriwaki (founding Director), the RIKEN BRC has successfully established its operational structure as a global biological resource center. The RIKEN BRC has been operating since its foundation according to three principles: trust, sustainability, and leadership. Our activities have been strongly supported by the scientific community as well as by the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of the Japanese government and RIKEN, the founding institution. Our collection, preservation, and quality-control (QC) operations have been funded by the government, while the distribution service has been funded separately by income generated by reimbursement fees collected from users.
To solve problems in the 21st Century related to health, food, the environment, and sustainable development, the importance of bioresources has grown exponentially. Among the many bioresources used in life science and biotechnology, the RIKEN BRC has specifically focused on the mouse, the most important experimental animal; Arabidopsis, the most important experimental plants; cultured cell lines of humans and animals; genetic materials from humans, animals, and microorganisms; and microorganisms such as bacteria, fungi, and archaea. These biological resources have been chosen according to criteria such as their importance to Japanese society and science, their original creation or derivation by Japanese scientists, the presence of a critical mass of users, and their merits for large-scale and intensive management. The RIKEN BRC has actively collected, preserved, conducted QC of, and distributed these biological resources with associated information and played a crucial role as a comprehensive biological resource hub to promote the life sciences and innovation. The RIKEN BRC has earned its status as one of the state-of-the-art research infrastructure centers of RIKEN.
Participation in the National BioResource Project
The National BioResource Project (NBRP) was launched by MEXT in FY2002, one year after the foundation of the RIKEN BRC (Yamazaki et al. 2010; Yokoyama et al. 2010; Yoshiki et al. 2009). The main program of the NBRP is the Core Facility Upgrading Program which aims to collect, preserve, and provide essential bioresources for life science studies strategically and systematically by the national government. Four divisions of the RIKEN BRC were selected at the start of the program as national core facilities of respective bioresources including the Experimental Animal Division as the NBRP core facility for mice (Yoshiki et al. 2009). Our activities as a national core facility have been evaluated extensively by the NBRP evaluation committee and designated over the course for 20 years during four 5-year programs.
Meeting social and research needs
Communication with the scientific community and comprehension of its advancing needs are essential to execute the mission of the research infrastructure. To achieve this objective, the RIKEN BRC has asked the domestic Resource Committee annually and the international RIKEN BRC Advisory Council (BRAC) every two or three years, each composed of several leading scientists and specialists in the biomedical sciences and global research infrastructures, for advice and recommendations regarding research needs and future directions. Moreover, the RIKEN Advisory Council provides recommendations on general research activities and management of the entire RIKEN institute, including the BRC, for guiding future research strategies and improving management structures. These recommendations form an integral part of the sustainable operation of the RIKEN BRC as a key element of global research infrastructure. The reports of previous Resource Committee and BRAC meetings are posted on the RIKEN BRC website (https://web.brc.riken.jp/en/reports) to ensure transparent operation. To earn trust in our activities for mouse resources, the RIKEN BRC has been supported by the Animal Experiments Committee and Recombinant DNA Committee which involve external scientists and non-scientists and include representatives of local residents and the municipal office of the city of Tsukuba to ensure animal welfare and safety measures in recombinant DNA experiments.
Collection of mouse strains developed in Japan
Mice are the premier mammalian model organism in terms of the availability of genetically defined inbred strains, high-quality genome information, advanced technologies for genomic alterations and phenotyping, and accumulated knowledge about their genetic diseases (Lloyd et al. 2020). Such model mammals are indispensable in an aging society and to achieve precision medicine by serving tools used to elucidate the molecular mechanisms of the onset of diseases such as cancer, dementia, and lifestyle-related diseases and to develop therapies as well. The Experimental Animal Division of the RIKEN BRC has collected useful mouse strains mainly developed by Japanese scientists to ensure that the collection does not overlap with those of other repositories around the world (Yoshiki et al. 2009).
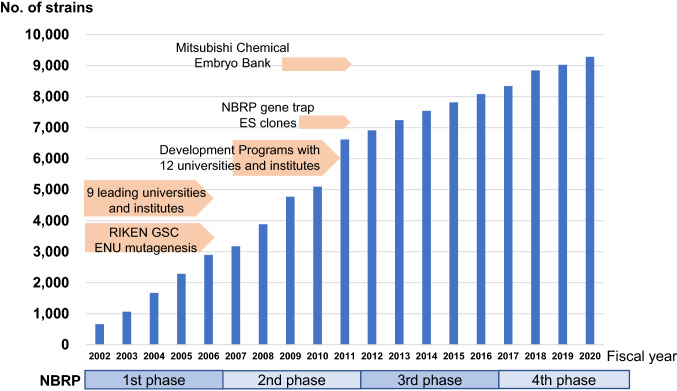
In the first phase of the NBRP, we collaborated with a large-scale ethylnitrosourea (ENU) mutagenesis project conducted by the Genome Science Center of RIKEN (Sakuraba et al. 2005) in addition to nine leading domestic universities and institutes. The collection has grown rapidly through collaborating facilities, with over 500 new strains added per year (Fig. 1). In the second phase of the NBRP, the archive expanded even more rapidly through the deposition of a large collection of frozen embryos from the Embryo Bank of the Mitsubishi Chemical Company and the gene-trap embryonic stem cell resources created by Dr. Yasumasa Ishida in the NBRP Fundamental Technology Upgrading Program (Shigeoka et al. 2005), as well as collaborative development programs for advanced genetically modified mouse models with 12 universities and institutes in Japan (Fig. 1). We have continued to ask individual scientists who have developed and published novel genetically modified strains to deposit their mice at the RIKEN BRC. In the third through fourth phases of the NBRP, we have focused on genetically modified strains to achieve an understanding of higher-order biological phenomena and advance research and development for overcoming diseases. Recently, genome-edited knockout and knock-in mice created using CRISPR/Cas9 have gradually accumulated in the collection since FY2014 and overtaken ES cell-based genetically modified mice since FY2019. The collection has steadily increased with the deposition of about 300 strains from 50 scientists on average each year, leading to the archiving of over 9,000 mouse models as of March 2021 (Fig. 1).
Fig. 1.
Number of mouse resources archived at the RIKEN BRC/NBRP
High-demand and high-impact strains
Our collection includes various gene knockouts, fluorescent reporters, conditional strains containing Cre–lox and Flp–FRT systems, and other useful congenic and inbred strains. Several mouse strains developed by Nobel laureates have also been deposited by the laureates and their colleagues, such as the stem cell reporter Nanog-EGFP knock-in mouse (RBRC02290) developed by Dr. Shinya Yamanaka (Okita et al. 2007), the autophagy reporter GFP-LC3#53 transgenic mouse (RBRC00806) developed by Dr. Noboru Mizushima (Kuma et al. 2004), and the PD-1 knockout mouse strain (RBRC02142) developed by Dr. Tasuku Honjo (Nishimura et al. 1999). Within the collection, there are a dozen unique strains with long-term periods of high demand that have been utilized in hundreds of publications by users (Table 1). These user publications have subsequently been cited in an outstanding number of studies (Table 1), indicating the large impact on the relevant research community.
Table 1.
Number of user publications and citations associated with research using mouse strains distributed by the RIKEN BRC
| RBRC no | Strain name | Category: description | No. of publications | Total no. of citations |
|---|---|---|---|---|
| 00806 | C57BL/6-Tg(CAG-EGFP/LC3)53NmzRbrc | TG: GFP-tagged reporter for autophagosome LC3 | 108 | 18,891 |
| 00267 | C57BL/6-Tg(CAG-EGFP)C14-Y01-FM131OsbRbrc | TG: Green mice, EGFP expression in all tissues | 53 | 2484 |
| 01390 | B6.129P2-Nfe2l2tm1Mym/MymRbrc | Null-KO: Nfe2l2, nuclear factor, erythroid derived 2, like 2 gene null deficient mice | 46 | 2277 |
| 01834 | C57BL/6-Tg(CAG-flpe)36Ito/ItoRbrc | TG: Ubiquitous deleter for Flp-FRT recombination system | 30 | 1092 |
| 00858 | B6;129S6- Bcl2l12/Irf3tm1Ttg/TtgRbrc | Null-KO: Interferon regulatory factor 3 gene | 24 | 964 |
| 01420 | B6;129P2-Irf7tm1Ttg/TtgRbrc | Null-KO: Interferon regulatory factor 7 gene, exons 2–3 replaced by the neo cassette | 23 | 888 |
| 01828 | B6.Cg-Tg(CAG-Cre)CZ-MO2OsbRbrc | TG: Ubiquitous deleter for Cre-loxP recombination system | 23 | 657 |
| 02975 | B6.129S-Atg5tm1Myok/Rbrc | cKO: Autophagy-related 5 gene floxed knockout mouse | 23 | 4576 |
| 01361 | B6.Cg-Trp53tm1Sia/Rbrc | Null-KO: Cancer related p53 deficient mice | 21 | 275 |
| 00144 | C57BL/6-Ly5.1/Rbrc | B6 congenic strain carrying Ly5.1: Useful for tissue/cell and B cell markers in transplantation studies | 19 | 371 |
| 00209 | MSM/MsRbrc | Japanese wild-derived inbred | 19 | 577 |
| 00639 | JF1/MsRbrc | Japanese wild-derived inbred | 18 | 363 |
Two autophagy-related strains, RBRC00806 and 02975 which are given in bold showed highest impact in regard to publications with out standing number of citations
TG transgenic mouse, KO; knockout mouse, KI knock-in mouse, cKO conditional knockout mouse
It is very important for the resource repository to continuously collect novel strains created in cutting-edge research for sustainable operations to meet the needs of advancing life science research and innovation. Notably, the C57BL/6-Apptm3(NL−G−F)Tcs/TcsRbrc (RBRC06344) strain, a next-generation Alzheimer's disease (AD) model created by Dr. Takaomi C. Saido and his colleagues at the RIKEN Center for Brain Science, has become the strain with the highest demand since FY2015 soon after its deposition. This novel AD knock-in model contains humanized sequences and clinical mutations in the endogenous mouse App gene, exhibiting typical Aβ pathology, neuroinflammation, and memory impairment in an age-dependent manner (Saito et al. 2014; Sasaguri et al. 2017). The next-generation AD model has been further advanced by crossing the model strain with human MAPT knock-in mice (Saito et al. 2019) for future preclinical studies.
Accessibility and distribution
Mouse strains deposited at the RIKEN BRC can be accessed via the BRC home page (https://mus.brc.riken.jp/en/), or browsed or searched from the web catalog (https://mus.brc.riken.jp/en/search_for_mouse_strain). Mice are distributed in the form of live animals, frozen embryos or sperm, and recovered litters from frozen embryos or sperm. Organs, tissues, and genomic DNA have been provided on request. We use a material transfer agreement to protect the intellectual property (IP) right of the depositor and other IP rights associated with the strain, while clarifying for users the terms and conditions set forth by the depositor. The total distribution increased in proportion to the size of the collection during the first phase of the NBRP. Through the second to fourth phases of the NBRP, 2500–3000 items of mouse resources per year were stably distributed (Fig. 2). We have so far distributed our mouse resources to 4377 users at 627 domestic organizations and 3225 users at 917 overseas organizations in 41 countries, resulting in 1047 user publications and 41 patents. Among these resources, autophagy reporter GFP-LC3 mice (RBRC00806) have been used since FY2005 at 272 organizations in 24 countries, resulting in 108 publications. Since FY2015, the knock-in AD model has been requested most frequently and distributed to 175 organizations in 16 countries.
Fig. 2.
Number of mouse resource items distributed by the RIKEN BRC/NBRP
Dissemination of information
Dissemination of information associated with mouse strains has a vital role in facilitating active use of large-scale mouse resources by the biomedical community. We have recently renewed the RIKEN BRC home page and developed a revised version of the searchable online catalog of mouse strains that functions as a part of the BRC-integrated database (Masuya et al. 2021).
Monthly email newsletters are sent to announce newly deposited mouse strains, users’ publications, relevant news, and coming events in both Japanese and English. To complement the functions of the integrated database and email news, we have launched proactive outreach corners focusing on specific strains, such as “Today’s Tool for Functional Analysis,” “Today’s Model for Human Disease” and (https://mus.brc.riken.jp/en/todays_tool_and_model), and “Mouse of the Month” (https://mus.brc.riken.jp/en/mouse_of_month) to provide researchers with information about advanced strains and facilitate the use of such strains for cutting-edge research. For the Mouse of the Month, strains are selected based on the potential impact of relevant publications as well as access log data for strains from the web catalog. The articles are designed in a mini-review format that includes representative images or figures and a summary of the strain including gene details, use applications, the latest research results, and references. The strain name with its repository unique identifier (RBRC number) is linked to each strain data sheet page of the online catalog supported by the BRC-integrated database. The Today's Tool and Model series are presented in a list format to disseminate more genetic tools and disease models; the list provides the title of the strain, its strain ID with a link to its strain data sheet, brief descriptions, and references with PubMed links.
Cryopreservation and secure back-up storage
Mice with high demand are maintained as live stocks for faster distribution, while mice with lower demand are preserved as frozen sperm or embryos in liquid nitrogen tanks for cost-effective management. Two-cell stage embryos of various strains have been successfully cryopreserved by using ethylene glycol-based vitrification (Mochida et al. 2011). Sperm samples were cryopreserved by the method reported by Nakagata and Takeshima (1993). Recent advances in assisted reproductive technologies have facilitated efficient preservation and recovery of various inbred strains and dozens of unique wild-derived strains belonging to different species and subspecies of Mus (Hasegawa et al. 2012, 2016, 2021; Mochida et al. 2014). Cryopreservation also plays a key role in protecting the deposited mouse strains by minimizing genetic drift, which may occur during continuous breeding periods (Wiles & Taft 2010). We cryopreserve embryos or sperm at the earliest stages after deposition and whenever possible with genomic DNA samples and breeding records in order to recover a live colony from earlier generations after extensive breeding periods or whenever undesired mutations are detected.
We have preserved approximately 95% of collected strains as frozen embryos and sperm. A rapidly increasing number of genetically modified strains developed by genome editing have efficiently been cryopreserved by sperm freezing. To protect the archived strains from disasters, the RIKEN BRC has built a back-up facility in the RIKEN Harima branch 700 km away, and every frozen strain has regularly been transferred there since 2007. Moreover, after the Great Eastern Japan Earthquake of 2011, we have installed an in-house water supply system, liquid nitrogen suppliers, and large fuel tanks for an emergency power supply to sufficiently handle a one-week outage to strengthen the security of the main collection in Tsukuba.
Microbial quality control
Ensuring the reproducibility of animal experiments in life science research is the most important issue for a resource repository. Microbial and genetic QC are two major pillars for the mouse resource repository to ensure reproducible animal experiments. In general, commercial breeders have established global standards and contributed to high-quality animal experiments in Japan. The RIKEN BRC, which regularly receives a number of original developed mouse strains from universities and research institutes, preserves and distributes the mice around Japan and globally, and also plays a critical role in ensuring the microbial quality of the mice made available by eradicating pathogenic agents as listed in Table 2.
Table 2.
Categorized list of pathogenic microbes to be excluded from the barrier facility at RIKEN BRC
| Class | A | B | C | D |
|---|---|---|---|---|
| Pathogenic agents | Clostridium piliforme, Ectromelia virus, Lymphocytic choriomeningitis virus (LCMV), Mouse hepatitis virus (MHV), Mycoplasma pulmonis, Salmonella typhimurium, Sendai virus (HVJ) | Filobacterium rodentium, Citrobacter rodentium, Corynebacterium kutscheri, Dermatophytes, Helicobacter bilis, Helicobacter hepaticus, Pasteurella pneumotropica, Ectoparasites, Intestinal protozoa, Pinworms, Salmonella spp. | Staphylococcus aureus, Pneumocystis murina, Pseudomonas aeruginosa |
Lactate dehydrogenase elevating virus (LDHEV), Mouse adenovirus (MAV), Mouse cytomegalovirus (MCMV), Mouse minute virus (MMV), Mouse norovirus (MNV), Mouse parvovirus (MPV), Mouse polyoma virus (Poly), Mouse rotavirus (EDIM), Pneumonia virus of mice (PVM), Reovirus type 3 (Reo 3), Theiler’s mouse encephalomyelitis virus (TMEV) |
Mice deposited by universities or institutes are received at the first quarantine facility, where they remain for one night and two days for microbial serology tests and initial external health observations. The mice are subsequently transferred to the second intermediate facility and housed in negative or positive pressure bioBUBBLE Clean Room Units (bioBUBBLE, Inc., Fort Collins, CO) according to the microbial test results. In the second facility, the deposited mice are maintained prior to rederivation procedures, such as breeding for cesarean section or sacrificing sperm donors for in vitro fertilization (IVF) followed by IVF and embryo transfer. After confirming negative results for all the listed pathogenic microbes, mice are introduced into a separate barrier facility, where approximately 10,000 breeding cages can be accommodated.
We have adopted the sentinel mouse program with a specialized protocol for individually ventilated cage units (CLEA Japan, Tokyo, Japan). The program uses dirty bedding sentinels to detect pathogenic microbes. Sentinel mice are exposed to dirty bedding collected from every cage of the unit at regular intervals (Ike et al. 2007). In FY2010, pathogenic agents such as mouse hepatitis virus, Pasteurella pneumotropica, and Helicobacter hepaticus were detected in 13% of the deposited strains in addition to intestinal protozoa and pinworms in 43% of the strains. Recently, the microbial status of deposited mice has been improved by intensive efforts of research communities to eradicate mouse hepatitis virus and other pathogens, while intestinal protozoa and pinworms have persistently been detected. The RIKEN BRC has also continued to clean-up newly deposited strains for pathogens, such as intestinal pathogens, detected in the mice received from research colonies of universities and institutes.
Genetic quality control
To set up appropriate genetic QC tests for ensuring reproducibility, we ask all depositor scientists to provide us with precise information including reference publications, primer sequences for genotyping, sequences of vectors and transgenes used for genetic modifications, breeding records, and any other relevant information. If there is any discrepancy between the depositor’s information and our test results, we communicate the results to the depositor and ask for a correction of the supplied information or resending correct mice. We use different genetic QC tests (Nakata et al. 2009, 2021), particularly PCR, depending on various genetic mutations (Table 3). The test results and protocols for each strain are posted in the strain data sheet in the web catalog when tested. When the depositor’s protocol includes methods other than PCR, we design a PCR test so that every user laboratory can easily set up the test based on the information provided by the depositor. Whenever PCR genotyping data from a mutant strain becomes available based on a published paper, we replicate or redesign the PCR genotyping protocol and post it to the web catalog.
Table 3.
Genetic QC tests conducted at RIKEN BRC
| Genetic QC test | Explanations |
|---|---|
| Transgene-specific PCR | A PCR test that uses transgene-specific primers designed based on information provided by the depositor to detect the promoter and the structural gene driven by the promoter |
| Targeted gene-specific genotyping PCR | A PCR test that uses targeted gene-specific primers designed based on information provided by the depositor |
| Detection of marker genes for genetic modifications | PCR tests for TG, BAC-TG, cTG, KO, KI, CRISPR/Cas9, cKO and spontaneous/ENU-induced mutant strains to detect any contamination with other genetically modified mice by using primers for 10 frequently used marker genes, including neo, Pgk-neo, Tk-neo, IRES, lacZ, GFP, Cre, Flp, Puro, and Hyg. (Nakata et al. 2009) |
| loxP and FRT tests | PCR tests of cTG and cKO mice that examines the fragment length between loxPs or FRTs with primers designed based on information provided by the depositor. Confirmation of structure of conditional strains that carry loxP or FRT flanked alleles. (Nakata et al. 2021) |
| Detection of BAC-loxP | A PCR test for BAC-TG to detect loxPs derived from BAC vectors |
| Genetic background test | PCR tests for the genetic background of inbred and wild-derived strains that uses microsatellite or single nucleotide polymorphism (SNP) markers (Mekada et al. 2009, 2015). Test results are posted to the web catalog after testing. Prior to phenotyping at the Japan Mouse Clinic, the strains are subjected to high-speed genetic profiling using the TaqMan assay. (https://ja.brc.riken.jp/lab/jmc/mouse_clinic/en/assistive/index.html) |
| Sequencing | PCR products are sequenced when the product size is different from the expected size |
TG transgenic, BAC-TG bacterial artificial chromosome-transgenic, cTG conditional transgenic, KO knockout, KI knock-in, cKO conditional knockout
Genome editing technology using the CRISPR/Cas9 system (Wang et al. 2013) has facilitated production of genetically modified mouse models across the wider research community and transgenic core facilities around the world. There is a concern, however, that the importance of QC is likely to be overlooked and left behind during rapidly expanding production. Mouse resource organizations around the world that gathered in the INFRAFRONTIER IMPC workshop in Munich agreed to conduct strict QC of mouse resources, including genome-edited mice, in 2014 (Nature editorial 2014). The RIKEN BRC established a policy to accept deposition of genome-edited mouse lines only after confirmation of germline transmission and to not accept founder mice that are likely to be mosaic (Singh et al. 2015; Ayabe et al. 2019) and are, therefore, unsuitable for reproducible experiments. We request the depositor to provide all accurate information, including the results of sequence analyses of mutations induced by genome editing, as listed in Table 4, and preserve the lines by cost-effective sperm freezing. Recently, more attention has been paid to the sequences around the on-target sites to confirm whether the genome was indeed correctly edited as designed and does not contain unintentional genomic changes (Kosicki et al. 2018; Gurumurthy et al. 2019).
Table 4.
Information required for genetic quality control of CRISPR/Cas9 genome-edited mice
| Items | Descriptions |
|---|---|
| Reference Publications | PubMed ID or DOI |
| Editing target | Gene or genomic region |
| Design of editing | Endonuclease (wild-type Cas9, nickase, others), injection reagents (mRNA, DNA, protein), injection methods (Pronuclear, Cytoplasmic, Pronuclear + Cytoplasmic, electroporation, others) |
| Guide RNA | Sequence and vector IDs |
| Sequencing results | Wild-type and mutated genome sequences |
| Detection of the mutation | PCR (primer sequence, PCR condition, fragment size), sequencing, RFLP, TaqMan real-time PCR etc |
| Off-target analysis | Done or not done |
| Background strain | Background strain: C57BL/6 N, C57BL/6 J, or others |
| Generation | Filial or backcross generations |
| Pedigree | Breeding records and/or pedigree |
Adding value to collected mouse strains
Researchers generally access a specific mouse strain found in publications because the strain has been used by others and proved to be useful through providing relevant information for a particular research purpose. Therefore, information about publications by users is indispensable for enriching the value of each research resource. In addition to direct feedback from users, the RIKEN BRC systematically collects information about users’ publications mainly by using the alert function of Google Scholar and Stanford HighWire and by emails received whenever such publications appear on the website. After manually confirming RBRC strains in publications, we add the paper to the users’ publication list. Such publication information is regularly added to the strain data sheet of the RIKEN BRC Mouse web catalog and then transferred to the NBRP Information Center to update the Research Resource Circulation (RRC) database to enrich the value of NBRP resources (https://rrc.nbrp.jp/projects/1?lang=en).
Phenotypic data for each strain is an important key to evaluating the strain as a potential human disease model or a tool to study disease mechanisms. The Integrated Bioresource Information Division has recently upgraded the mouse web catalog by manually adding 24,490 phenotype annotations to over 4300 strains from publications so that users can find a specific mouse strain based on phenotype ontology terms in addition to gene names (Masuya et al. 2021), thus increasing the findability of the strains. We also promote comprehensive phenotyping of deposited mouse strains by the Japan Mouse Clinic at the RIKEN BRC (Wakana et al. 2009). The broad-based phenotyping platform has been built in collaboration with the International Mouse Phenotyping Consortium (IMPC) to add useful phenotyping data, which should facilitate evaluation of the strain as a potential disease model as demonstrated by the IMPC (Dickinson et al. 2016; Meehan et al. 2017; Birling et al. 2021). Thus far, 69 domestic users have agreed to deposit their 121 mouse strains with phenotypic data generated at the Japan Mouse Clinic, which will be available to the public soon after publication.
Collaborations with the NBRP Genome Information Upgrading Program have contributed to the addition of genome sequence data for wild-derived strains of Mus musculus, including the Japanese wild-derived strains MSM and JF1 in the MoG+ mouse genomic variation database (https://molossinus.brc.riken.jp/mogplus/#JF1; Takada et al. 2013, 2015). The genome information is essential and useful for future functional studies based on comprehensive phenotyping and genetic modifications of genetically divergent unique mouse strains created by genome editing (Hirose et al. 2017).
International collaborations
The RIKEN BRC has participated in the International Mouse Strain Resource (IMSR), a one-stop shop database operated by the Jackson Laboratory (Epigg et al. 2015) to register mouse resources developed by Japanese scientists and share those resources with the global biomedical community. Scientists who find interesting strains or queries in the IMSR list can be directed to the holder repository with one click. This function has increased the visibility and accessibility of our mouse strains. By the further collaboration of the IMSR with the Resource Identification Initiative (Bandrowski et al. 2016), our mouse resources are given unique Research Resource Identifiers in addition to repository-specific RBRC numbers, which ensures the ability to identify the exact mouse strains used in publications as well as reproducibility.
Since 2006, mouse repositories and genetics institutes in Asia and Australia have established the Asian Mouse Mutagenesis & Resource Association (AMMRA) to strengthen communications and technical exchanges in the Asia and Australia regions (http://ammra.info/). We have collaborated with major global repository members such as the Jackson Laboratory and the Mutant Mouse Resource and Research Centers in USA, the European Mouse Mutant Archive in Europe, the Centre for Phenogenomics in Canada, and AMMRA members mutually to provide technical assistance in cryo-recovery to local investigators. Moreover, the RIKEN BRC has participated in the IMPC and contributed to the production, preservation, QC, phenotyping, and distribution of knockout mouse resources (Dickinson et al. 2016; Birling et al. 2021). The RIKEN BRC has joined with the IMPC and AMMRA members in the activities of the Global Mouse Models for COVID-19 Consortium and posted our own biological resources, including mouse strains, for respective studies. Thus, our international collaborations and networks function together to facilitate the active use of high-quality mouse resources around the world.
Conclusion
As progress in decoding the genome of patients continues, the need to verify causative gene candidates underlying genetic diseases in a model organism increases exponentially. Recent genome editing technology enables us to collaborate with clinical scientists in the generation of knock-in mice for variants or mutations found in patients, in addition to the development of null-knockout mice in the same production platform (Lloyd et al. 2020). The Next Generation Human Disease Model Team of the RIKEN BRC has started a new research and development program with the goals of promoting precise mouse modeling of human diseases and contributing to the realization of precision medicine. Advances in genome editing technology will be a major key to the expansion of such mouse resources, while developing technology for use in genome editing may inevitably generate new quality issues that need to be controlled. As a national and international hub of mouse resources and as a critical research resource infrastructure, we will continue expansion of the premier mammalian model organism, mice. We will do this in collaboration with global mouse repositories to make available genetics tools to decipher the molecular mechanisms of health and disease with the highest priority in order to ensure the reproducibility of life science experiments and innovation.
Acknowledgements
Special thanks to the late Dr. Kazuo Moriwaki, founding Director of RIKEN BRC, for his continuous encouragement and support. We thank all user scientists and advisors for their kind and generous support and all members of the Experimental Animal Division, Bioresource Engineering Division, Gene Engineering Division, Integrated Bioresource Information Division, and Technology and Development Team for Mouse Phenotype Analysis for their continuous support in the collection, preservation, quality control, phenotyping, and distribution of mouse resources in addition to providing various customer services. The Experimental Animal Division of the RIKEN BRC has participated in the NBRP supported by the MEXT, Japan and has been designated as the NBPR core facility for mouse resources.
Declarations
Conflict of interest
The authors declare no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- (2014) Still much to learn about mice. Nature 509:399. 10.1038/509399a [DOI] [PubMed]
- Ayabe S, Nakashima K, Yoshiki A. Off- and on-target effects of genome editing in mouse embryos. J Reprod Dev. 2019;65:1–5. doi: 10.1262/jrd.2018-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birling MC, Yoshiki A, Adams DJ, Ayabe S, Beaudet AL, Bottomley J, Bradley A, Brown SDM, Bürger A, Bushell W, et al. A resource of targeted mutant mouse lines for 5,061 genes. Nat Genet. 2021;53:416–419. doi: 10.1038/s41588-021-00825-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandrowski A, Brush M, Grethe JS, Haendel MA, Kennedy DN, Hill S, Hof PR, Martone ME, Pols M, Tan SS, et al. The resource identification initiative: a cultural shift in publishing. Neuroinformatics. 2016;14:169–182. doi: 10.1007/s12021-015-9284-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson ME, Flenniken AM, Ji X, Teboul L, Wong MD, White JK, Meehan TF, Weninger WJ, Westerberg H, Adissu H, et al. High-throughput discovery of novel developmental phenotypes. Nature. 2016;537(7621):508–514. doi: 10.1038/nature19356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppig JT, Motenko H, Richardson JE, Richards-Smith B, Smith CL. The International Mouse Strain Resource (IMSR): cataloging worldwide mouse and ES ell line resources. Mamm Genome. 2015;26:448–455. doi: 10.1007/s00335-015-9600-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurumurthy CB, O'Brien AR, Quadros RM, Adams J, Jr, Alcaide P, Ayabe S, Ballard J, Batra SK, Beauchamp MC, Becker KA, et al. Reproducibility of CRISPR-Cas9 methods for generation of conditional mouse alleles: a multi-center evaluation. Genome Biol. 2019;20:171. doi: 10.1186/s13059-019-1776-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa A, Mochida K, Matoba S, Yonezawa K, Ohta A, Watanabe G, Taya K, Ogura A. Efficient production of offspring from Japanese wild-derived strains of mice (Mus musculus molossinus) by improved assisted reproductive technologies. Biol Reprod. 2012;86(167):1–7. doi: 10.1095/biolreprod.111.098491. [DOI] [PubMed] [Google Scholar]
- Hasegawa A, Mochida K, Inoue H, Noda Y, Endo T, Watanabe G, Ogura A. High-yield superovulation in adult mice by anti-inhibin serum treatment combined with estrous cycle synchronization. Biol Reprod. 2016;94(1):21. doi: 10.1095/biolreprod.115.134023. [DOI] [PubMed] [Google Scholar]
- Hasegawa A, Mochida K, Matoba S, Inoue K, Hama D, Kadota M, Hiraiwa N, Yoshiki A, Ogura A. Development of assisted reproductive technologies for Mus spretus. Biol Reprod. 2021;104:234–243. doi: 10.1093/biolre/ioaa177. [DOI] [PubMed] [Google Scholar]
- Hirose M, Hasegawa A, Mochida K, Matoba S, Hatanaka Y, Inoue K, Goto T, Kaneda H, Yamada I, Furuse T, Abe K, Uenoyama Y, Tsukamura H, Wakana S, Honda A, Ogura A. CRISPR/Cas9-mediated genome editing in wild-derived mice: generation of tamed wild-derived strains by mutation of the a (nonagouti) gene. Sci Rep. 2017;14(7):42476. doi: 10.1038/srep42476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ike F, Bourgade F, Ohsawa K, Sato H, Morikawa S, Saijo M, Kurane I, Takimoto K, Yamada YK, Jaubert J, Berard M, Nakata H, Hiraiwa N, Mekada K, Takakura A, Itoh T, Obata Y, Yoshiki A, Montagutelli X. Lymphocytic choriomeningitis infection undetected by dirty-bedding sentinel monitoring and revealed after embryo transfer of an inbred strain derived from wild mice. Comp Med. 2007;57:272–281. [PubMed] [Google Scholar]
- Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- Kosicki M, Tomberg K, Bradley A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat Biotechnol. 2018;36:765–771. doi: 10.1038/nbt.4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd KCK, Adams DJ, Baynam G, Beaudet AL, Bosch F, Boycott KM, Braun RE, Caulfield M, Cohn R, Dickinson ME, Dobbie MS, et al. The deep genome project. Genome Biol. 2020;21:18. doi: 10.1186/s13059-020-1931-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuya H, Usuda D, Nakata H, Yuhara N, Kurihara K, Namiki Y, Iwase S, Takada T, Tanaka N, Suzuki K, Yamagata Y, Kobayashi N, Yoshiki A, Kushida T. Establishment and application of information resource of mutant mice in RIKEN BioResource Research Center. Lab Anim Res. 2021;37:6. doi: 10.1186/s42826-020-00068-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehan TF, Conte N, West DB, Jacobsen JO, Mason J, Warren J, Chen CK, Tudose I, Relac M, Matthews P, et al. Disease model discovery from 3328 gene knockouts by the international mouse phenotyping consortium. Nat Genet. 2017;49:1231–1238. doi: 10.1038/ng.3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekada K, Abe K, Murakami A, Nakamura S, Nakata H, Moriwaki K, Obata Y, Yoshiki A. Genetic differences among C57BL/6 substrains. Exp Anim. 2009;58:141–149. doi: 10.1538/expanim.58.141. [DOI] [PubMed] [Google Scholar]
- Mekada K, Hirose M, Murakami A, Yoshiki A. Development of SNP markers for C57BL/6N-derived mouse inbred strains. Exp Anim. 2015;64:91–100. doi: 10.1538/expanim.14-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida K, Hasegawa A, Taguma K, Yoshiki A, Ogura A. Cryopreservation of mouse embryos by ethylene glycol-based vitrification. J Vis Exp. 2011;57:3155. doi: 10.3791/3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida K, Hasegawa A, Otaka N, Hama D, Furuya T, Yamaguchi M, Ichikawa E, Ijuin M, Taguma K, Hashimoto M, et al. Devising assisted reproductive technologies for wild-derived strains of mice: 37 strains from five subspecies of Mus musculus. PLoS ONE. 2014;9(12):e114305. doi: 10.1371/journal.pone.0114305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagata N, Takeshima T. Cryopreservation of mouse spermatozoa from inbred and F1 hybrid strains. Jikken Dobutsu. 1993;42:317–320. doi: 10.1538/expanim1978.42.3_317. [DOI] [PubMed] [Google Scholar]
- Nakata H, Hashimoto T, Seki Y, Mekada K, Obata Y, Yoshiki A. Simultaneous detection of multiple transgenes for genetically-modified mouse strains. Exp Anim. 2009;58:437–442. doi: 10.1538/expanim.58.437. [DOI] [PubMed] [Google Scholar]
- Nakata H, Hashimoto T, Yoshiki A. Quick validation of genetic quality for conditional alleles in mice. Genes Cells. 2021;26:240–245. doi: 10.1111/gtc.12834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura H, Nose M, Hiai H, Minato N, Honjo T. Development of lupus-like autoimmune diseases by disruption of the PD-1 gene encoding an ITIM motif-carrying immunoreceptor. Immunity. 1999;11:141–151. doi: 10.1016/s1074-7613(00)80089-8. [DOI] [PubMed] [Google Scholar]
- Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- Singh P, Schimenti JC, Bolcun-Filas E. A mouse geneticist's practical guide to CRISPR applications. Genetics. 2015;199:1–15. doi: 10.1534/genetics.114.169771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaguri H, Nilsson P, Hashimoto S, Nagata K, Saito T, De Strooper B, Hardy J, Vassar R, Winblad B, Saido TC. APP mouse models for Alzheimer's disease preclinical studies. EMBO J. 2017;36:2473–2487. doi: 10.15252/embj.201797397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Matsuba Y, Mihira N, Takano J, Nilsson P, Itohara S, Iwata N, Saido TC. Single App knock-in mouse models of Alzheimer's disease. Nat Neurosci. 2014;17:661–663. doi: 10.1038/nn.3697. [DOI] [PubMed] [Google Scholar]
- Saito T, Mihira N, Matsuba Y, Sasaguri H, Hashimoto S, Narasimhan S, Zhang B, Murayama S, Higuchi M, Lee VMY, Trojanowski JQ, Saido TC. Humanization of the entire murine Mapt gene provides a murine model of pathological human tau propagation. J Biol Chem. 2019;294:12754–12765. doi: 10.1074/jbc.RA119.009487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigeoka T, Kawaichi M, Ishida Y. Suppression of nonsense-mediated mRNA decay permits unbiased gene trapping in mouse embryonic stem cells. Nucleic Acids Res. 2005;33:e20. doi: 10.1093/nar/gni022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuraba Y, Sezutsu H, Takahasi KR, Tsuchihashi K, Ichikawa R, Fujimoto N, Kaneko S, Nakai Y, Uchiyama M, Goda N, et al. Molecular characterization of ENU mouse mutagenesis and archives. Biochem Biophys Res Commun. 2005;336:609–616. doi: 10.1016/j.bbrc.2005.08.134. [DOI] [PubMed] [Google Scholar]
- Takada T, Ebata T, Noguchi H, Keane TM, Adams DJ, Narita T, Shin-I T, Fujisawa H, Toyoda A, Abe K, et al. The ancestor of extant Japanese fancy mice contributed to the mosaic genomes of classical inbred strains. Genome Res. 2013;23:1329–1338. doi: 10.1101/gr.156497.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada T, Yoshiki A, Obata Y, Yamazaki Y, Shiroishi T. NIG_MoG: a mouse genome navigator for exploring intersubspecific genetic polymorphisms. Mamm Genome. 2015;26:331–337. doi: 10.1007/s00335-015-9569-8. [DOI] [PubMed] [Google Scholar]
- Wakana S, Suzuki T, Furuse T, Kobayashi K, Miura I, Kaneda H, Yamada I, Motegi H, Toki H, Inoue M, et al. Introduction to the Japan Mouse Clinic at the RIKEN BioResource Center. Exp Anim. 2009;58:443–450. doi: 10.1538/expanim.58.443. [DOI] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153(4):910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiles MV, Taft RA. The sophisticated mouse: protecting a precious reagent. Methods Mol Biol. 2010;602:23–36. doi: 10.1007/978-1-60761-058-8_2. [DOI] [PubMed] [Google Scholar]
- Yamazaki Y, Akashi R, Banno Y, Endo T, Ezura H, Fukami-Kobayashi K, Inaba K, Isa T, Kamei K, Kasai F, et al. NBRP databases: databases of biological resources in Japan. Nucleic Acids Res. 2010;38:D26–32. doi: 10.1093/nar/gkp996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama KK, Murata T, Pan J, Nakade K, Kishikawa S, Ugai H, Kimura M, Kujime Y, Hirose M, Masuzaki S, et al. Genetic materials at the gene engineering division, RIKEN BioResource Center. Exp Anim. 2010;59:115–124. doi: 10.1538/expanim.59.115. [DOI] [PubMed] [Google Scholar]
- Yoshiki A, Ike F, Mekada K, Kitaura Y, Nakata H, Hiraiwa N, Mochida K, Ijuin M, Kadota M, Murakami A, Ogura A, Abe K, Moriwaki K, Obata Y. The mouse resources at the RIKEN BioResource center. Exp Anim. 2009;58(2):85–96. doi: 10.1538/expanim.58.85. [DOI] [PubMed] [Google Scholar]