Abstract
Collectively referred to as the epitranscriptome, RNA modifications play important roles in gene expression control regulating relevant cellular processes. In the last few decades, growing numbers of RNA modifications have been identified not only in abundant ribosomal (rRNA) and transfer RNA (tRNA) but also in messenger RNA (mRNA). In addition, many writers, erasers and readers that dynamically regulate the chemical marks have also been characterized. Correct deposition of RNA modifications is prerequisite for cellular homeostasis, and its alteration results in aberrant transcriptional programs that dictate human disease, including breast cancer, the most frequent female malignancy, and the leading cause of cancer-related death in women. In this review, we emphasize the major RNA modifications that are present in tRNA, rRNA and mRNA. We have categorized breast cancer-associated chemical marks and summarize their contribution to breast tumorigenesis. In addition, we describe less abundant tRNA modifications with related pathways implicated in breast cancer. Finally, we discuss current limitations and perspectives on epitranscriptomics for use in therapeutic strategies against breast and other cancers.
Graphical Abstract
Graphical Abstract.
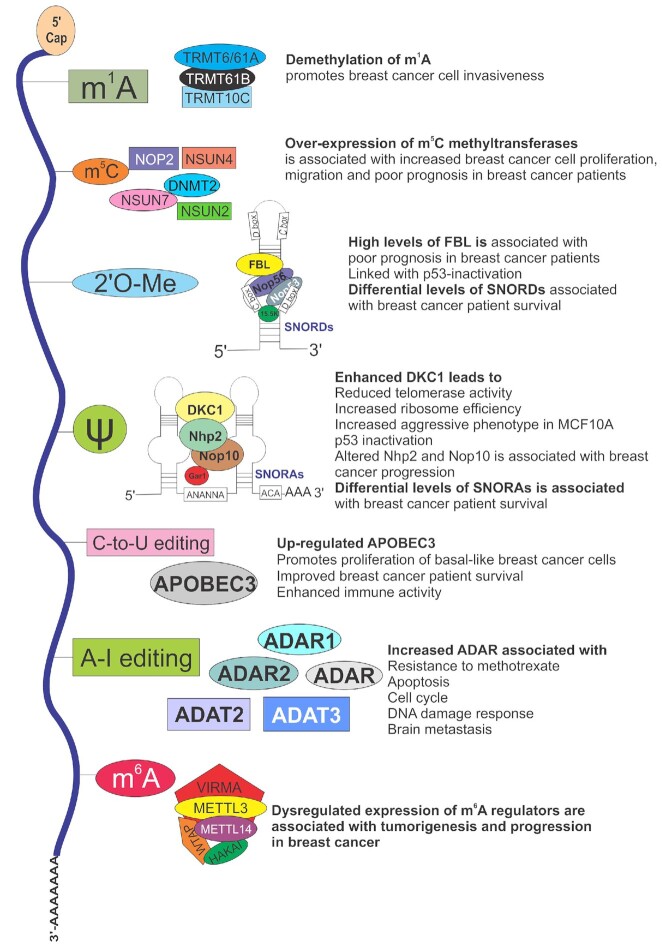
Modifications of RNA known in breast cancer.
INTRODUCTION
RNA modifications, collectively termed the epitranscriptome, are crucial regulators of temporal and spatial gene expression programs. Currently, over 170 RNA modifications decorating all RNA species and in all three kingdoms of life have been described (1,2). Although modification to RNA has been documented for over 50 years, the functions of most of these modifications are largely unknown. Recent development of more sensitive and specific technologies, such as high-throughput sequencing and improved mass spectrometry, shed light on to the exciting new field of RNA epitranscriptomics.
In general, the fate of modified RNAs is determined by the coordinated actions of writers, erasers and readers that impose, remove and recognize the chemical mark. Some of the writers are stand-alone enzymes, whereas others act as multiprotein writer complexes that also comprise accessory subunits. In addition, some of the RNA modifications are reversible i.e. removed by the erasers, whereas others are irreversible. Most of the known RNA modifications map to abundant RNAs such as transfer RNA (tRNA) and ribosomal RNA (rRNA), tRNA being the most extensively modified RNA type in the cell (3) (Figure 1).
Figure 1.
Chemical structures of common RNA modifications known to have a role in breast cancer. The modified groups are highlighted in red. The type of RNA in which the modification is detected is indicated using the symbols of rRNA, tRNA and mRNA.
In eukaryotes, the nuclear-encoded tRNAs contain on average 11–13 modifications per molecule whereas mitochondrial tRNAs are modified to a lesser extent, with an average of 5 modifications per molecule (4). The molecular consequence of the modification depends on the type of chemical modification and the location within a tRNA. Hence, modifications occurring at the wobble position (position 34) and to the residue adjacent to the anticodon loop (position 37) are highly conserved and lead to the strongest effects in optimization of codon usage, regulating translational efficiency and fidelity (5–7). However, modifications along the whole L-shape affect tRNA stability, localization and functional folding (8,9). Such modifications include but are not limited to 5-methylcytosine (m5C), N1-methyladenosine (m1A), pseudouridine (Ψ), 5-methyluridine (m5U), 1-methylguanosine and 7-methylguanosine (m1G and m7G, respectively), and inosine (I), and complex multistep chemical modifications, such as N6-threonylcarbamoyladenosine (t6A) and 5-methoxycarbonylmethyl-2-thiouridine (mcm5s2U) (10) (Figure 2).
Figure 2.
tRNA modifications. Clover leaf model representing the structure of a human tRNA. Nucleotides that undergo modification are shown in pink. Distinct chemical modifications are represented in different colors. The enzymes known to mediate tRNA modifications linked to breast cancer are capitalized and highlighted in blue.
Human ribosomes contain four rRNA types, i.e. 28S, 5S and 5.8S rRNAs in the 60S subunit, and 18S rRNA in the 40S subunit. In each subunit, the rRNAs form the core of the translation machinery whereas ribosomal proteins stabilize the structure and fine-tune the function of the ribosome (11,12). During ribosome biogenesis, rRNAs are extensively modified, expanding the topological properties of RNAs and optimizing the ribosome functionality (13). Recently, >130 individual rRNA modifications have been visualized in the three-dimensional structure of the human ribosome, being several of the modifications associated with degenerate states in cancer (14). In eukaryotic ribosomes, the most abundant rRNA modifications are ribose 2′-O-methylation (2′O-Me or Nm) and Ψ catalyzed by box C/D and box H/ACA ribonucleoprotein (RNP) enzymes, respectively, using small nucleolar RNAs (snoRNAs) for the recognition of specific rRNA target sites (15–17). Conversely, base modifications, such as methylations and acetylations, are catalyzed by conventional protein enzymes, most of which have only recently been identified (18–21), and thereby, their function is largely unknown (22).
Shortly after the discovery of the 5′ cap and 3′ polyadenylation, N6-methyladenosine (m6A) was identified in mRNA (18–21). m6A is the most abundant internal mRNA modification with on average 3–5 adenines methylated per mRNA (23,24). Other less abundant modifications within eukaryotic mRNA include m1A, N6,2′-O-dimethyladenosine (m6Am), m5C, 5-hydroxymethylcytosine (hm5C) and Ψ (Figure 3) (3). These modifications are not randomly distributed in the mRNA, and depending on the modification type and the deposition site, they can virtually affect all aspects of RNA fate including RNA processing, RNA export, mRNA translation and degradation. All these molecular events shape the transcriptome in a spatiotemporal manner to tightly regulate gene expression programs (25).
Figure 3.
Internal mRNA modifications. Schematic representation of internal mRNA modifications and the predicted location on the mRNA are shown. The main writers of the respective modifications and their functions in breast cancer are depicted.
The deposition of chemical modifications into RNA is dynamic allowing the rapid adaptation to changing environmental cues and to various stresses. Such adaptation is crucial for cellular homeostasis. Hence, alterations in the expression levels of RNA modifiers and thereby, dysregulated RNA modification pathways, have been linked to tumorigenesis as well as other human diseases (26). In this review, we describe the current understanding on how these epitranscriptomic marks are implicated in breast tumorigenesis, the most prevalent cancer among women worldwide. Specifically, we highlight several critical modifications, namely m6A, m1A, m5C, 2′O-Me, RNA editing and Ψ (Table 1). We describe which RNA species they have been identified in, their molecular and cellular functions (where known) and the evidence linking them to the development, maintenance and progression of breast cancer.
Table 1.
List of RNA modifications in breast cancer. RNA modifications, enzymes and associated effect in breast cancer
| Type of RNA | Enzymes | Function in breast cancer | Ref |
|---|---|---|---|
| m6A | |||
| mRNA | ALKBH5, IGF2BP1 | Promoted BCSC phenotype | (69,70,73) |
| mRNA | METTL3 | Promoted BCSC phenotype, induces metastasis | (201) |
| mRNA | IGF2BP2 | Promoted BCSC phenotype | (72) |
| mRNA | METTL3, METTL14, WTAP, FTO, ALKBH5 | Inhibited colony formation and migration | (202) |
| mRNA | YTHDF3 | Promoted breast cancer, induces brain metastasis and angiogenesis | (203) |
| mRNA | METTL14, ALKBH5, YTHDF3 | Promoted growth, proliferation and angiogenesis | (56) |
| mRNA | METTL14, ZC3H13 | Correlation with unfavorable prognosis in breast cancer patients | (49) |
| mRNA | METTL3 | Increased cell proliferation and tumor progression | (57) |
| mRNA | METTL3 | Increased proliferation and decreased apoptosis in breast cancer cells in vitro and in vivo | (58) |
| mRNA | METTL3, FTO, IGF2BP1, YTHDF1 | Promoted lung metastasis and clinical progression in breast cancer, induction of EMT | (59) |
| mRNA | METTL3 | Inhibited migration and invasive capacities of the cells in TNBC | (60) |
| mRNA | METTL14 | Promoted breast cancer initiation and progression | (62) |
| mRNA | FTO | Promoted cell proliferation, colony formation, tumor growth and metastasis | (63) |
| mRNA | METTL3 | Enhanced expression of oncogenes, induces acquired chemoresistance | (74) |
| mRNA | METTL3 | Induced breast cancer cell proliferation, associated with drug sensitivity | (76) |
| mRNA | YTHDF2 | Promoted breast cancer progression | (204) |
| mRNA | ALKBH3 | Increased breast cancer cell invasiveness | (83) |
| miRNA | METTL14 | Promoted migration and invasion in breast cancer cells | (50) |
| miRNA | FTO | Promoted cell invasion and migration | (52) |
| miRNA | METTL3 | Associated with acquired chemoresistance | (75) |
| circRNA | METTL3 | Promoted cell proliferation | (61) |
| rRNA | METTL5 | p70-S6K activation and translation initiation, increased breast cancer cell growth | (77) |
| m5C | |||
| Type of RNA | Enzymes | Function in breast cancer | Ref |
| mRNA | NSUN2-NSUN7, DNMT1, DNMT3A, DNMT3B, ALYREF TET2 | Affected tumor development, tumor immune microenvironment and potential markers for TNBC patients | (110) |
| 2’-O-Me | |||
| Type of RNA | Enzymes | Function in breast cancer | Ref |
| rRNA | Fibrillarin | Promoted BCSC phenotype | (128) |
| rRNA | Undefined | Associated with breast cancer subtypes and tumor grade, linked with overall patient survival | (127) |
| Ψ | |||
| Type of RNA | Enzymes | Function in breast cancer | Ref |
| rRNA | DKC1 | Telomerase activity, linked with better clinical outcome | (143) |
| A-to-I | |||
| Type of RNA | Enzymes | Function in breast cancer | Ref |
| mRNA | ADAR | Resistance to methotrexate | (167) |
| mRNA | ADAR | Cell viability, drug sensitivity clinically relevant editing events in breast tumors than normal tissues | (161) |
| mRNA | ADAR1 | Cell cycle control, DNA damage response, increased breast cancer cell progression | (163) |
| mRNA | ADAR1 | DNA damage, immunity, DNA replication, increased breast cancer cell progression | (159) |
| mRNA | ADAR1p110 | Increased proliferation and breast cancer metastasis | (164) |
| C-to-U | |||
| Type of RNA | Enzymes | Function in breast cancer | Ref |
| mRNA | APOBEC3 | Improved breast cancer survival | (173) |
| mRNA | Apobec-1 complementation factor (A1CF) | Increased breast cancer progression | (174) |
Breast cancer
According to a global cancer statistics study, breast cancer is the most commonly diagnosed cancer in women and among the leading cause of cancer-related death in females (27) (Figure 4). Although there is an evident increase in breast cancer incidence and mortality among pre- and post-menopausal women, females of younger age are also at risk every year, with higher emphasis in developing countries (28). Hence, although advanced therapies and early detection have improved the survival rate (29), the causal factors of breast carcinoma still remain elusive.
Figure 4.
Global cancer statistics. Pie chart depicting the percentage of new cases in each tumor type in 2020 for women globally. The data used to prepare the chart were taken from GLOBOCAN 2020 (27).
The clinical behavior and the treatment outcomes in breast cancer are highly influenced by tumor heterogeneity consisting in increased morphological variability and fluctuating therapy response (30). Based on the expression of the hormone receptors (estrogen (ER) and progesterone (PR)) and the human epidermal growth factor receptor 2 (HER2), there are four molecular subtypes of breast cancer, namely luminal A, luminal B, HER2 positive and basal-like or triple-negative (TNBC) (31). Although both luminal A and B are ER and PR positive, the later one has a worst prognosis and is either HER positive or negative with high levels of the proliferation marker Ki-67 (30,32). The HER2 subtype is ER and PR negative but positive for HER2 and the TNBC presents a triple negative immunophenotype (ER, PR and HER2 negative), increased proliferation rate and the highest incidence of relapse (30,33,34). Hence, in addition to providing prognostic information, the molecular subtypes can be used to evaluate clinical behaviors and response to treatments. However, the intrinsic heterogeneity of breast cancer impedes the full characterization based on the aforementioned histopathologic parameters. Thus, recent evidence supports the presence of multiple subtypes within a tumor (32) and the existence of even more than four subtypes (30), manifesting the complex molecular landscape of breast cancer cells. In addition, it has been shown that the microenvironment can dictate plasticity of breast cancer cells (35). For instance, the mammary stroma can induce basal differentiation in MCF7 cells, a luminal cellular model, and the site of injection of these cells into mice models can confer different tumor phenotypes, with injection in the milk ducts leading to an increased mimicry of the original tumor compared with the injection into the fat pads (35). Therefore, finding an effective cure for this heterogeneous and multifactorial disease is still a major challenge.
Such complexity and heterogeneity of breast cancer cells can be better understood through integration of multi-omics approaches that provide resourceful information of the different layers of gene expression regulation. Although genomic, epigenomic and transcriptomic datasets have added new insights into the true biological landscape of breast cancer, epitranscriptomic analysis are still in their infancy. Given that many of the RNA modifications are dysregulated in human cancers, the epitranscriptome represents a hot-spot of interest in the quest of elucidating the transition from a normal physiological to a pathological state, hoping that its study will enable the development of more efficient and effective therapies against cancer. For breast cancer, recognition of the function of RNA modifications in its development may represent the missing piece of the puzzle in deciphering the complex pathogenesis of these tumors.
N6-methyladenosine
m6A is the most studied and abundant internal modification on eukaryotic mRNA. It also appears on tRNAs and other non-coding RNAs (ncRNAs) and more recently, it has been found on rRNA (2,36,37). Due to its large impact on RNA metabolism at multiple levels i.e. splicing, nuclear export, mRNA stability, translation and even RNA and protein interactions, it plays a critical role not only in proper functioning of all sorts of biological processes but also in acquired pathologies such as cancer (26,38,39).
m6A is co-transcriptionally deposited on mRNA by a stable methyltransferase complex consisting of two core components, i.e. methyltransferase-like 3 (METTL3) and 14 (METTL14), and other proteins that ensure m6A specificity (40,41). The demethylases include fat mass and obesity-associated protein (FTO) and α-ketoglutarate dependent dioxygenase alk B homolog 5 (ALKBH5) (42,43). The most representative readers are members of the YTH domain-containing protein family (YTHDF1/2/3 and YTHDC1) (44,45).
In recent years, the expression of m6A regulators has been correlated with hallmarks of cancer and it was shown that breast cancer cells exhibited higher m6A methylation levels compared to healthy mammary epithelial cells (Table 1) (46,47). However, most of the studies seem controversial as a defined gene expression pattern of writers, erasers and readers is missing, reflecting the complexity and heterogeneity of breast pathogenesis. For instance the writer METTL14 was downregulated in TNBC, with METTL14 expression being correlated with favorable outcomes (48). Similarly, Gong et al. reported that low expression levels of METTL14 led to poor prognosis and tumor progression in TNBC (49). On the contrary, overexpression of METTL14 was observed in studies that compared breast cancer patients with healthy controls, METTL14 acting as an oncogene that promoted invading and migrating capacities of the cells (46,50). Dong et al. have attributed METTL14 overexpression to luminal A and B subtypes, being negatively correlated with tumor grade and not exerting influence over prognosis (51). In addition, the expression of the erasers of m6A has also been shown to be dysregulated in breast cancer patients, being the increased expression of ALKBH5 correlated with poor prognosis in TNBC (47), whereas the expression of the demethylase FTO appeared to be decreased in breast cancer patients compared to normal controls (47). However, Xu et al. observed upregulation of FTO in HER2+ subtype and a strong connection with unfavorable prognosis in this subtype (52). The expression of readers correlated as well with different outcomes. For instance, increased expression of YTHDF3 was linked with unfavorable survival, nodal metastasis and poor prognosis in breast cancer patients (53,54). Other readers such as YTHDF1 and YTHDF1 displayed increased expression in various analyses of breast cancer patient samples (48,54,55), while YTHDC1 and YTHDC2 were shown to be downregulated in TNBC (48). These studies reflect that the same writer protein, for example METTL14, can display distinct expression patters depending on the breast cancer subtype. They also show how the expression of distinct erasers or readers can follow opposite patterns of gene expression. Therefore, dysregulated expression of m6A writer, eraser and reader, either high or low, might lead to aberrant gene expression programs that promote breast tumorigenesis. Indeed, it has been shown that the interplay among m6A regulators determines the m6A levels and, consequently, the stability of several transcripts that are known to play a critical role in cell cycle, epithelial–mesenchymal transition (EMT) and angiogenesis in breast cancer (56).
Several mechanisms involving METTL3 in breast cancer progression have been described (Figure 5). METTL3 was found to participate in a feedback loop with Hepatitis B virus X-interacting protein (HBXIP) and the microRNA (miRNA) let-7g, in which METTL3 expression was positively regulated by HBXIP through inhibiting let-7g miRNA, while HBXIP expression was increased by METTL3. This positive feedback loop resulted in increased cell proliferation and, ultimately, in cancer progression (57). In addition, METTL3-mediated increased methylation levels promoted the translation of Bcl-2, a major regulator of cell death modulating cellular proliferation and apoptosis of cancerous cells (58). Furthermore, it has been shown that m6A can induce breast cancer lung metastasis by increasing the stability of a mRNA duplex formed from the keratin 7 (KRT7) transcript, encoding a regulator of EMT, cytoskeleton programming and cellular transformation, and its antisense long noncoding RNA (lncRNA) KRT7-AS (59). On the contrary, in TNBC cell lines, METTL3 halts metastatic progression by hypermethylation of the Collagen Type III Alpha 1 Chain (COL3A1) transcript which trigger its degradation (60). Recently, it was shown that METTL3 is the host gene of a circRNA, so called circMETTL3 whose expression was regulated via a m6A-dependent mechanism (61). This circRNA was upregulated and impacted cell proliferation, migration and invasion in breast cancer through upregulation of cyclin-dependent kinases (CDK1), recently identified as a breast cancer prognosis indicator (61). Moreover, METTL14 was reported to enhance cell proliferation by facilitating the deposition of m6A on the transcripts of the oncogenes CXCR4 and CYP1B1 (62). Mechanisms showing the implications of FTO in cell proliferation, colony formation and metastasis of breast cancer both in vitro and in vivo were also described. Hence, FTO-mediated demethylation of Bcl-2/adenovirus E1B mRNA (BNIP3), a pro-apoptotic tumor suppressor gene, favored tumor progression (63). Recently, the reader of m6A YTHDF2 was found to sustain MYC-driven cell growth and survival in TNBC cell lines by facilitating the turnover of mRNAs belonging to MAPK/ERK signaling pathways. Hence, YTHDF2 was limiting the endoplasmic reticulum stress response and therefore contributed to EMT and breast tumorigenesis (64).
Figure 5.
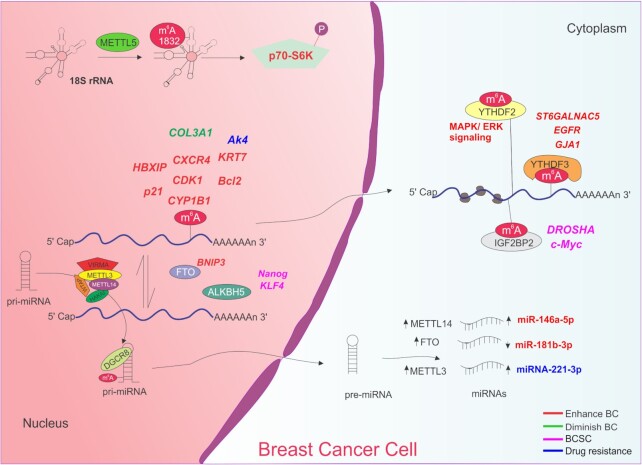
Functions of m6A in breast cancer. m6A modification is dynamically regulated by its writers (METTL3, METTL14, WTAP, HAKAI and VIRMA) erasers (FTO and ALKBH5) and readers (YTHDFC3 and IGF2BP2). Only enzymes with reported function in breast cancer are depicted for simplicity. Deposition of m6A on distinct transcripts can enhance (HBXIP, p21, CXCR4, CDK1, CYP1B1, KRT7, Bcl2; red) or diminish (COL3A1; green) breast cancer initiation and progression and affect therapy outcome (AK4; blue). Demethylation of BNIP3 also enhances breast cancer (red) whereas demethylation of Nanog, KLF4 and Sox2 promotes the breast cancer stem cell phenotype. Similarly, recognition of m6A-modified DROSHA and c-Myc mRNAs leads to the acquisition of breast cancer stem cell characteristics. YTHDFC3 binds to ST6GALNAC5, EGFR and GJA1 to induce breast cancer metastasis. YTHDF2 recognizes mRNAs involved in MAPK/ERK signaling. Deposition of m6A on pri-miRNA allows DGCR8 recognition and further processing. Increased expression of METTL14, FTO and METTL3 alters the expression of miRNA, enhancing the breast cancer phenotype (red) or induces drug resistance (blue) in breast cancer cells. Enhanced METTL5 mediated methylation at adenosine 1832 of mammalian 18S rRNA promotes p70-S6K activation and an increased translation initiation thus stimulating breast cancer cell growth.
Another way by which the m6A machinery can contribute to breast cancer formation and development is through its roles in miRNA biogenesis. METTL3 has been shown to deposit m6A on primary miRNAs (pri-miRNAs) enabling their recognition by the RNA binding protein DGCR8 and thereby facilitating miRNAs biogenesis (65). Particularly, in breast cancer, METTL14 reshaped the miRNA profile (50). Modulation of the expression of miR-146a-5p by METTL14 promoted cell migration and invasion underlying the predominant control that m6A may have over multiple aspects of breast tumorigenicity. In addition, inhibition of miR-181b-3p by FTO resulted in upregulation of ADP ribosylation factor like GTPase 5B (ARL5B) (52). ARL5B is responsible for promoting lysosome motility facilitating cell migration. Therefore, through this mechanism FTO enhanced the invasive and migratory capabilities of the breast cancer cells (52). Considering the data obtained in studying other cancer types, further studies will most certainly reveal an even higher impact of m6A on miRNA in breast carcinogenesis.
m6A plays a significant role in the formation of cancer stem cells (CSCs), tumorigenic cells with stem cell properties, which are known for their capabilities of facilitating the carcinogenic process and the resistance to therapy (66). Hypoxic environments, through the action of hypoxia-inducible factors (HIF), have been linked with EMT and thus with acquisition of stem cell-like properties that favor migration and invasion in breast cancer (67,68). Expression levels of ALKBH5 and the oncogenic factor Zinc-Finger Protein 217 (ZNF217) are increased in these environments (69,70). Notably, the mouse ortholog ZFP217 has been shown to interact with METTL3 and restrict the m6A deposition on pluripotency factors (71), increasing their mRNA stability. Thus, ZNF217- and ALKBH5-mediated demethylation of pluripotency factors, i.e. Nanog and KLF4, induced the acquisition of stem cell-like properties and therefore, BCSC specification (69,70). IGF2BP2, a reader of m6A, was also linked to mechanisms enhancing BCSC by stabilization of the mRNA of two oncogenes, namely DROSHA and c-Myc (72,73). All these results point toward a significant role of this mark in elucidating means that can provide new therapeutic strategies in both drug resistant and refractory hypoxic breast tumors.
Recent evidence indicates that m6A also plays important roles in treatment outcomes, being either a vector of acquired resistance or a perfect target for more efficient therapies. On one hand, resistance to tamoxifen and doxorubicin were modulated by METTL3 through methylation of adenylate kinase 4 (AK4) transcripts, and miRNA-221–3p, respectively (74,75). AK4 is a mitochondrial matrix protein involved in energy metabolism homeostasis whose expression is linked to the progression of multiple cancers. In tamoxifen resistant MCF-7 cells, m6A deposition on AK4 mRNA led to increased expression which in turn resulted in higher reactive oxygen species and p38 levels while depletion of METTL3 and AK4 resensitized the cells to tamoxifen (74). METTL3 regulation of miRNA-221–3p through methylation of pri-miRNA-221–3p resulted in chemoresistance to doxorubicin in breast cancer cells, due to the suppression of the tumor suppressor HIPK2 that led to overexpression of Che-1, a transcription regulator known for its role in development of anticancer drug resistance (75). On the other hand, metformin, a drug used in the treatment of type 2 diabetes mellitus, has been shown to suppress m6A through downregulation of METTL3 (76). Metformin inhibited METTL3 and thus the m6A deposition on p21 (also known as cyclin-dependent kinase inhibitor 1 (CDKN1A)) exhibiting an antiproliferative effect in breast cancer cells (76).
Lastly, m6A on 18S rRNA at position 1832 has also been linked to breast cancer cell growth. This modification is catalyzed by METTL5 which must form a heterodimeric complex with TRMT112 to perform its methyltransferase activity (77). METTL5-mediated 1832 methylation at 18S rRNA fine-tuned the conformation of the ribosome decoding center, increasing its interaction with mRNAs (78). METTL5 has been shown to be overexpressed in breast cancer samples and its loss led to a reduction in proliferation of different breast cancer cell lines and of the S6K phosphorylation needed for the cells to initiate translation and to undergo growth (42). Similarly, enhanced expression of ZCCHC4, which catalyzes the m6A deposition of human 28S rRNA at position A4220 (79), is also evidenced in breast tumor tissues although the function of this modification in breast tumorigenesis is yet to be investigated (47). Taken together, this research topic is still in its infancy and more studies are needed to contour a better picture of the role of m6A on rRNA in breast cancer.
N1-methyladenosine
Methylation of adenosine at position N1 or m1A has the capacity to disrupt the Watson–Crick base-pairing specificity affecting RNA structure and protein–RNA interaction (80–82). It was originally discovered in tRNA and rRNA, and later studies detected its presence in mRNA although its ubiquity is still controversial. Hence, whereas original studies suggested that thousands of mRNAs showed m1A enrichment in the 5′UTR, usually in GC-rich regions and that m1A modification played an important role in translation and in environmental stress response (80,83), later studies revealed few transcripts with this modification (84). In addition, the original authors themselves conceded that m1A distribution showed no 5′UTR bias (85).
Therefore, although there is not enough evidence to suggest that m1A is present at appreciable levels on mRNA, one study has explored its role in breast cancer. Cytokine macrophage colony-stimulating factor (CSF-1) is an oncogene that promotes metastatic dispersion in breast and ovarian carcinomas. It has been shown that CSF-1 transcripts decorated with m1A were targeted for mRNA decay. Hence, ALKBH3-mediated demethylation increased the stability of CSF-1 mRNA, thereby increasing its expression. Given that ALKBH3 is overexpressed in many types of tumors, it is plausible that this is a general mechanism and not exclusively for breast or ovarian cancers (86).
5-methylcytosine
The methylation of the carbon 5 in cytosine (m5C) was originally found in rRNA and tRNA; however, recent studies have also detected m5C on mRNA (87,88). In eukaryotes, the large ribosomal subunit contains two m5C residues (22) that are essential to maintain ribosomal structure and fidelity during translation, playing a significant role in lifespan and stress resistance (89). On tRNAs, m5C protected from angiogenin-mediated endonucleolytic cleavage, and thereby avoided the biogenesis of tRNA-derived small RNA fragments (tRFs) (90,91). Such tRFs play a key role in regulating gene expression programs participating in various physiological processes such as cell stress, cell growth and cell differentiation (92). tRFs also play significant roles in various human diseases, including cancer (93). On mRNA, m5C promotes export (87), stabilization (88) and translation (94,95).
Depending on the RNA species, distinct m5C methyltransferase from the NOL1/NOP2/SUN domain (NSUN) family of proteins (NSUN1 to NSUN7) and the DNA methyltransferase member 2 (DNMT2) catalyze the m5C modification (95). Hence, NOP2 (NSUN1) and NSUN5 methylate 28S rRNA, while NSUN4 modify mitochondrial rRNA (96–99). DNMT2, NSUN2, NSUN3 and NSUN6 all methylate cytoplasmic tRNAs, with different specificity and at different residues, and NSUN3 targets mitochondrial tRNA (100–104). Moreover, NSUN6 also targets site-specific deposition of m5C in mRNA (105), NSUN2 in ncRNAs and mRNA (87,106), and NSUN7 targets enhancer RNAs (eRNAs) (107).
Several m5C methyltransferases, including NOP2, NSUN2 and NSUN4, are up-regulated in breast cancer although the molecular consequences of this overexpression are not well characterized (108–110). In addition, the genomic region containing NSUN2 (5p15.31–33) was associated with a strong risk for the development of breast cancer (111). Hence, both amplification and overexpression of NSUN2 has therapeutic potential as a drug target (109). Additionally, overexpression of NSUN2 by DNA hypomethylation has been associated with proliferation, migration and invasion while NSUN2 knockdown inhibited these processes in vitro and in vivo (112). These results were corroborated in another pan-cancer study showing that NSUN2, among other RNA methyltransferases, was amplified or mutated in breast cancer, and its expression was associated with poor prognosis in these patients (113). Notably, as NSUN2 can catalyze the deposition of m5C on distinct RNA species, i.e. of tRNAs, mRNAs and ncRNAs (87,106,114) additional work will be required to address the molecular mechanism that leads to breast carcinogenesis. Recently, Huang et al. reported that all eleven m5C regulators (NSUN2-NSUN7, DNMT1, DNMT3A, DNMT3B, ALYREF and TET2) were differentially expressed in TNBC and can potentially predict clinical prognostic risk in patients. Whereas upregulated expression of NSUN2 was found to be closely associated with cell cycle signaling pathways, RNA degradation and RNA polymerase, reduced expression of NSUN6 was linked with cell adhesion, metabolism and extracellular matrix receptor interaction (115).
Y-box binding protein 1 (YBX1) is a specific reader of m5C in mRNA which stabilizes the oncogene HDGF in urothelial carcinoma of the bladder (88). YBX1 is a multifunctional protein that is frequently overexpressed in breast cancer regardless of the subtype and its expression was correlated with poor survival, drug resistance and relapse (116,117). Therefore, these studies suggest that dysregulated YBX1-mediated decoding of m5C can lead to not only urothelial carcinoma but also to other cancers, including breast cancer.
2′-O-methylation
2′O-Me is the addition of a methyl group at the 2′ hydroxyl of the ribose moiety of all four nucleosides. This mark can be catalyzed by stand-alone enzymes (118) or by the methyltransferase fibrillarin in association with the C/D box family snoRNAs and the conserved proteins Nop56p, Nop58p and 15.5K or NHP2L1, collectively known as box C/D RNP complex (119). 2′O-Me is a predominant mark in rRNA, where >100 sites exist (120), but it has also been found in tRNA, mRNA (121–126) and other small RNAs (127).
Internal modification of 2′O-Me at 3′-terminal protects terminal ribose, inhibits the function of the RNA ligase and negatively impacts the efficiency of the polyA-polymerase (128,129), directly reducing the translational capacities of ribosomes (130). A Pan-cancer analysis highlighted that several RNA methyltransferases are either amplified or mutated in different cancers (68). Among these, FtsJ RNA 2′-O-methyltransferase 3 (FTSJ3), that modifies both rRNA and mRNA is among the stand-alone enzymes that has been correlated with cell growth and survival of breast cancer cells (113).
2′O-Me is linked to ribosome biogenesis and deregulated ribosome biogenesis is found to be associated with breast cancer cell progression (131). Therefore, studying the signature of 2′O-Me sites in both patient samples or cell lines is of therapeutic importance. This can be performed by RiboMethSeq which maps 2′O-Me sites based on the principle that methylation of ribose 2′OH makes 3′-adjacent phosphodiester bond resistance to alkaline hydrolysis and nuclease cleavage (132). Indeed, RiboMethSeq in 195 primary breast tumor samples showed that rRNA 2′O-Me levels differed between breast cancer subtypes and tumor grades (133). Particularly, TNBC patients displayed a signature where 2′O-Me levels at 18S-Am576 and at 18S-Gm1447 sites were increased and decreased, respectively. In addition, modification at the 18S-Gm1447 site was found to be guided by SNORD127 at 18S of rRNA, suggesting that studies evaluating the mechanism of SNORD127 in TNBC might be of therapeutic importance.
Recently, a distinct 2′O-Me rRNA pattern was observed upon exposing normal breast epithelial and breast cancer cell lines, i.e. MCF10A and T47D cells, to hypoxic conditions (134). In hypoxic conditions both cell lines displayed high 2′O-Me levels at 1858 and 4436 sites and reduced methylation at 390, 1612, 2848, 4806 and 1803 sites, compared to normoxia. Noteworthy, in low oxygen conditions genes such as vascular endothelial growth factor (VEGF-C) are translated in a cap-independent manner through internal ribosome entry site (IRES). Hence, a specific pool of ribosomes with a distinct 2′O-Me pattern facilitated the IRES recognition enhancing the translation of VEGF-C and most probably, other oncogenes (135). Although mechanistic evaluation of 2′O-Me in breast tumorigenesis is lacking, several groups have shown that deregulated expression of components of 2′O-Me RNP complex, such as C/D box snoRNAs and fibrillarin, led to breast tumorigenesis (136,137). For instance, increased fibrillarin expression was associated with aberrant 2′O-Me rRNA pattern and thus impaired translational fidelity. In addition, Marcel et al. showed that p53 acted as a safeguard of protein synthesis by repressing the expression of fibrillarin in breast cancer (138). Moreover, 58 differentially expressed snoRNAs were identified in 26 TNBC cell lines (139) being SNORD78, SNORD93, SNORD62A, SNORD2 and SNORD57 among the most highly upregulated snoRNAs in the invasive breast cancer MDA-MB-231 cell line. Small RNA sequencing in normal and primary breast tumor tissues discovered thirteen snoRNAs which were associated with overall survival and relapse free survival of breast cancer patients (140). In another study, Kothari et al. performed a transcriptome array in patient samples from different breast cancer subtypes and found SNORD114 and SNORD115 as important regulators of breast tumorigenesis (141).
The snoRNA host genes can be of great therapeutic importance as they mostly show altered expression in multiple cancers, and are known to regulate cell growth, tumor progression, metastasis and chemoresistance (142–144). ZFas1, host RNA for SNORD12, SNORD12b and SNORD12c (ZNFX1 Antisense RNA 1) regulated alveolar development and epithelial cell differentiation during normal mammary development and thus is a potential biomarker acting as a tumor suppressor in breast cancer (145). However, not all snoRNAs are involved in 2′O-Me. A combined analysis of high-throughput sequencing protocols and computational methods (146) suggested alternative roles for snoRNAs beyond 2′O-Me. Therefore, for a better understanding of correlation between snoRNAs expression and 2’O-Me, more research is needed in this area.
Pseudouridylation
Pseudouridylation is the isomerization of 1-ribosyluracil (uridine) to 5-ribosyluracil (pseudouridine, ψ) within the ribonucleoside. Known to be present in all kinds of RNA species, pseudouridylation is the second most common RNA modification of rRNA and has important roles in protein translation and pre-mRNA splicing (37,147). The mechanism of isomerization of uridine to pseudouridine can be either RNA independent or RNA dependent. RNA independent pseudouridylation is performed by enzymes having both substrate recognition and catalytic properties known as pseudouridine synthases (148). However, RNA dependent pseudouridylation is carried out by the RNP complex that consists of catalytic protein dyskerin (DKC1), box H/ACA snoRNAs (SNORAs) (substrate recognition), non-histone protein 2 (Nhp2), nucleolar protein 10 (Nop10) and glycine-arginine-rich protein-1 (Gar1) (149).
In breast cancer patients and cell lines, reduced expression of DKC1 was found to be associated with reduced telomerase activity, rRNA pseudouridylation and better clinical outcome (150,151). Similarly, depletion of DKC1 led to p53-inactivation and increased IRES-mediated VEGF mRNA translation in breast cancer cell lines (152,153). Moreover, enhanced DKC1 expression increased ribosome efficiency and stimulated the aggressive phenotype in the normal breast epithelial MCF10A cell line (154). In addition, expression of Nop10 was found to be associated with aggressive breast cancer (155) and coding missense variants of Nhp2 gene were detected in hereditary breast cancer patients although no pathogenic role was elucidated (156). Levels of small nucleolar RNAs (SNORNAs) in cancer are deregulated (157), being SNORA3, SNORA18, SNORA7B, SNORA13 SNORA2A among others highly expressed in metastatic breast cancer patients and cell lines (139,158,159). Inhibition of oncogenic SNORA7B impaired cell growth, migration and apoptosis in breast cancer cells (160). Relative pattern of pseudouridylation in normal and breast cancerous cases is yet to be evaluated and hence needs more research to elucidate its direct role in breast tumor initiation and progression.
RNA editing
RNA editing refers to modifications of specific nucleotides in RNA sequence differentiating it from its corresponding DNA sequence that may lead to pathological consequences. The most common form of RNA editing is from adenosine (A) to inosine (I) (161) and is catalyzed by specific enzymes namely adenosine deaminases acting on RNA (ADARs). In mammals, three ADAR genes encode for four different isoforms known as ADAR1p150, ADAR1p110, ADAR2 and ADAR3 (162). A-to-I editing within the protein-coding region of an mRNA can result in an amino acid change in the encoded protein as the resulting inosine is interpreted as guanosine by the translational machinery leading to an amino acid substitution in the protein product (163). In addition, A-to-I can alter regulatory motifs, binding of RNA-binding proteins, or RNA secondary structures influencing pre-mRNA alternative splicing (164–166). Furthermore, RNA editing within 3′UTRs of target transcripts can alter miRNA targeting an perturb miRNA-mediated regulation of oncogenes and tumor-supressors (167). The enzymes involved in A-I editing on tRNAs are known as Adenosine deaminases that act on tRNAs (ADATs). Deregulated ADATs-mediated RNA editing is associated with diseases such as type 2 diabetes, neurological disorders, mitochondria-related disorders and cancer (168). Less frequent cytidine (C) to uridine (U) mRNA editing is carried out by cytidine deaminases belonging to the family of enzymes known as activation induced cytidine deaminase/apolipoprotein B editing complex (AID/APOBEC) or AADs (169), eleven of which are found in humans (170).
A-to-I editing in breast cancer
A-to-I editing is known to be the major source of mRNA variability in breast cancer (171), and it is controlled by type-I interferon response and high ADAR DNA copy number (172). Hence, >76 000 RNA editing sites were identified by using a bioluminescent reporter system and upon interferon treatment of the breast cancer MCF7 cell line (173). Breast tumors harbor amplified levels of ADAR (174) and inhibiting its activity has been shown to induce apoptosis. Indeed, A-to-I mRNA editing regulated breast cancer cell proliferation by modulating the stability and expression of genes involved in cell cycle and DNA damage response being ATM, MDM2, MDM4, CENPN and XPO1 among the affected transcripts (175). In another study, increased A-to-I mRNA editing was found on the mRNA of ATM, GINS4 and POLH affecting their stability and/ or expression in primary breast tumors when compared to the non-cancerous tissues (171). In addition, Gumireddy et al. reported that A-to-I mRNA editing of γ-amino butyric acid receptor alpha 3 (GABRA3), one of the subunits of GABA (A) receptor, changed its role from a metastasis promoter to a tumor suppressor by suppressing the ERK pathway activation (176). In an attempt to assess the role of A-to-I RNA editing on proteomic diversity, Peng et al. showed that A-to-I RNA editing contributed to protein heterogeneity in breast cancer (177). Hence, nine unique RNA editing sites with variant peptide evidence were identified such as COPA_I164V and IGFBP_R78G which were detected across 11 out of 36 BRCA mass spectrometry datasets (177). These peptide variants were shown to be presented as self-antigens by human leukocyte antigen (HLA) molecules, and thus recognized by the immune system. For instance, CD8+ effector T cells were evidenced in tumors in response to peptide variants generated from cyclin I (178). A-to-I RNA editing has also been linked to resistance to chemotherapy. Specifically, RNA editing at 26 sites of 3′UTR modulated the expression of dihydrofolate reductase resulting in highly proliferative breast cancer cells which were resistant to methotrexate (179). To sum up, existing data suggests an important role of A-to-I RNA editing in genes associated with breast cancer-relevant pathways and treatment outcomes, suggesting an important role of ADAR1 function in breast tumorigenesis.
C-to-U editing in breast cancer
APOBEC-dependent somatic mutation confers increased susceptibility for breast cancer (180) and is found to be enriched in the HER2 subtype (181,182). Overexpression of cytidine deaminase APOBEC3A in HEK293T cells caused mRNA alterations in several tumor associated genes including PTEN, KMT2A, ATM, BRCA1 and BRCA2 (183); however, their significance in breast tumorigenesis is yet to be evaluated. mRNA expression of APOBEC3B was evaluated in ductal carcinoma in situ (DCIS), invasive breast cancer (IBC) and normal breast cells (184). Normal breast cells expressed reduced levels of APOBEC3B, while increased expression was evident in DCIS and IBC. Recently, APOBEC3-mediated C-to-U RNA editing in breast cancer was shown to be associated with improved patient survival and enhanced immune activity (185). In another study, using a bioinformatics approach, C-to-U RNA editing levels were estimated in 1040 primary breast tumor tissues and 93 adjacent normal tissues. More than 5000 APOBEC3-mediated RNA editing sites were identified using TCGA sequencing data. For 440 sites, editing was found on 411 transcripts among which most prevalent were on GATA Zinc Finger Domain Containing 2B (GATAD2B), Serpin Family A Member 1 (SERPINA1) and Adenosine Monophosphate Deaminase 3 (AMPD3) (185). Moreover, Apobec-1 complementation factor (A1CF), another mRNA editing enzyme, promoted proliferation of basal-like breast cancer cells by targeting interleukin-6 (186). Overall, the existing data on C-to-U editing suggests important roles in breast cancer progression and further research may identify targets with therapeutic importance.
Other RNA modifications related to breast cancer
Growing evidence in recent years has pointed out that tRNAs and their derivatives are dysregulated in breast and other cancers (187). However, the functional role of tRNA modifications in tumorigenesis is still elusive. As mentioned before, several modifications are shared by the distinct RNA species but others are unique to tRNAs, as is the case of cm5U34 and mcm5U34 at the wobble position. These marks have been shown to influence the translational efficiency and the accuracy of the reading frame (188). In addition, the catalytic enzymes for mcm5s2 deposition are up-regulated in breast cancer sustaining metastasis (189). They enhanced the translation of DEK and LEF1, two oncogenes, enabling breast cancer cells to migrate and invade other tissues (189). Another modification that occurs at the wobble anticodon position is queuosine which is dependent on the gut microbiome (190). Thus, the microbiome product queuine is the substrate for the enzymes tRNA guanine transglycosylases (TGTs). In eukaryotes, the active TGT is a heterodimer formed of a catalytically active queuine tRNA-ribosyltransferase subunit 1 (QTRT1) and a catalytically inactive QTRT2 subunit (191). Knockout of QTRT1 in MCF7 and MDA-MB-231 cells produced changes in the functions of genes involved in cell proliferation, junction formation and migration (190). These results have been validated in mouse models, thus enforcing the significant role that this modification is playing in breast cancer development (190).
TRMT12, a tRNA methyltransferase that mediates posttranscriptional modifications on tRNAPhe has been reported to be consistently amplified and overexpressed in cell lines and breast cancer patient samples (192). In yeast, it catalyses the formation of wybutosine (yW) on the 37th residue of tRNAPhe, modification that helps in stabilization of the codon-anticodon interaction and in maintenance of the reading frame (192). Thus, disruption at the level of this modification can lead to translational errors and is therefore of great interest to establish the role of these enzymes, together with its effect on tRNAs, in tumorigenesis of breast cells.
It is noteworthy that many of the methyltransferases that catalyze the methylation of different types of RNAs have been shown to be deregulated in breast cancer. Although the lack of any association between the deposited marks and the expression of these proteins made us filter out these studies, there are some that are worth mentioning. For instance, tRNA methyltransferase 2 homolog A (TRMT2A) which catalyzes the methylation of the U5 of tRNAs and regulates cell cycle has been associated with increased recurrence risk in HER2+ breast cancer patients (193). Human tRNA methyltransferase 9-like (hTRM9L) protein, an another tRNA methyltransferase, is downregulated in breast cancer and was shown to suppress tumor growth in vivo by decreasing proliferation, cell cycle arrest in G0/G1, upregulation of LIN9, and blocking the hypoxia response (194) in colorectal cancer. Whether this mechanism is the same in breast cancer and if it is mediated though methylation remains to be further investigated.
Despite being less explored than other RNA marks, many of these modifications convey to be promising clues in deciphering the molecular enigma behind the development of breast cancer. Thus, it will be of increased interest to see how extensive studies of these marks on the various species of RNA can contribute to a more precise picture of breast cancer tumorigenic process.
Perspectives
In the last decade, and as a result of the advent of more reliable methodology, the number of epitranscriptomic studies has dramatically increased. The boost and excitement of this emerging field has led to a tremendous progress in identifying the machinery and the role of RNA modifications. Hence, RNA marks have emerged as important regulators of a variety of cellular processes and their roles in human disease, including cancer, have extensively been described. It has been shown that writers, erasers and readers can act as tumor suppressors or as oncogenes depending on the cellular context. Thus, a single enzyme can have opposing roles in distinct cancer types. However, as a consequence of this fast-growing discipline, many erroneous conclusions have been drawn, and a large number of controversial studies has emerged. In addition, most of the works have just focused on few RNA modifications, and thereby the functions of most RNA modifications remain to be characterized.
During these years, RNA modifications have been comprehensively mapped transcriptome-wide by coupling antibody immunoprecipitation or chemical probing with next-generation sequencing. Yet, tools for the simultaneous identification of distinct chemical marks in the same RNA are currently not available. Hence, it is unknown how the different RNA modifications interplay to influence cancer development. Aberrant expression of such RNA-modifying machinery has been acknowledged for most aspects of breast and other cancer types. One particular challenge is to attribute a phenotype to a particular chemical mark as there is a crosstalk between rRNA, tRNA and mRNA modifications which involves common modification factors. For example, NSUN2 can impose m5C on both mRNA and tRNA species (195). In addition, several previous conclusions might have to be revised as some of these key players are moonlighting proteins which perform multiple autonomous and often unrelated functions. Therefore, it is also challenging to distinguish between the canonical, i.e. directly related to RNA modifications and secondary protein functions. For instance, knockouts of IME4, the yeast homolog of METTL3, display more severe phenotypes than those derived by a catalytic mutant, suggesting that IME4 has methylation-independent functions (196,197). Indeed, it has been shown that in lung cancer, cytoplasmic METTL3 promotes the translation of oncogenes by a mechanism independent of its methyltransferase activity (198).
Because of the inherent reversibility of some of the RNA modifications, inhibitors targeting the RNA-modification machinery are promising therapeutic targets for breast and other tumors. As m6A is the most studied RNA modification, it is not surprising that lead biotech companies have developed drugs to target the writer complex, specifically METTL3 (199). Hence, STORM Therapeutics has developed the first catalytic inhibitor of METTL3. Pharmacological inhibition of METTL3 led to strong antitumor effects in vitro and in relevant mouse models of acute myeloid leukemia (AML) (200).
Overall, as highlighted in this review, considerable advance has been made in recent years in the research field of epitranscriptomics which has provided a link between RNA modifications and breast cancer. Yet, the epitranscriptome remains vastly unexplored. Thus, it appears that we just perceive the tip of the iceberg and that the number of RNA modifications is much higher than previously estimated. The exhaustive characterization of RNA modifications and the molecular mechanism in which they are involved holds a new avenue for novel cancer therapies.
ACKNOWLEDGEMENTS
We sincerely apologize to authors whose work could not be included due to space limitations.
Contributor Information
Kanchan Kumari, Department of Molecular Biology, Umeå University, SE-901 85 Umeå, Sweden; Wallenberg Centre for Molecular Medicine, Umeå University, SE-901 85 Umeå, Sweden.
Paula Groza, Department of Molecular Biology, Umeå University, SE-901 85 Umeå, Sweden; Wallenberg Centre for Molecular Medicine, Umeå University, SE-901 85 Umeå, Sweden.
Francesca Aguilo, Department of Molecular Biology, Umeå University, SE-901 85 Umeå, Sweden; Wallenberg Centre for Molecular Medicine, Umeå University, SE-901 85 Umeå, Sweden.
FUNDING
Knut and Alice Wallenberg Foundation (through the Wallenberg Centre for Molecular Medicine Umeå), Cancerfonden (Pj:190337), Cancer Research Foundation in Northern Sweden (AMP 21-1030).
Conflict of interest statement. None declared.
REFERENCES
- 1. Ashraf S.S., Guenther R.H., Ansari G., Malkiewicz A., Sochacka E., Agris P.F. Role of modified nucleosides of yeast tRNA(Phe) in ribosomal binding. Cell Biochem. Biophys. 2000; 33:241–252. [DOI] [PubMed] [Google Scholar]
- 2. Boccaletto P., Machnicka M.A., Purta E., Piatkowski P., Baginski B., Wirecki T.K., de Crecy-Lagard V., Ross R., Limbach P.A., Kotter A. et al. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018; 46:D303–D307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Roundtree I.A., Evans M.E., Pan T., He C. Dynamic RNA Modifications in Gene Expression Regulation. Cell. 2017; 169:1187–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pan T. Modifications and functional genomics of human transfer RNA. Cell Res. 2018; 28:395–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ranjan N., Rodnina M.V. tRNA wobble modifications and protein homeostasis. Translation (Austin). 2016; 4:e1143076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hanson G., Coller J. Codon optimality, bias and usage in translation and mRNA decay. Nat. Rev. Mol. Cell Biol. 2018; 19:20–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rezgui V.A., Tyagi K., Ranjan N., Konevega A.L., Mittelstaet J., Rodnina M.V., Peter M., Pedrioli P.G. tRNA tKUUU, tQUUG, and tEUUC wobble position modifications fine-tune protein translation by promoting ribosome A-site binding. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:12289–12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lorenz C., Lunse C.E., Morl M. tRNA Modifications: Impact on Structure and Thermal Adaptation. Biomolecules. 2017; 7:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vare V.Y., Eruysal E.R., Narendran A., Sarachan K.L., Agris P.F. Chemical and Conformational Diversity of Modified Nucleosides Affects tRNA Structure and Function. Biomolecules. 2017; 7:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Suzuki T. The expanding world of tRNA modifications and their disease relevance. Nat. Rev. Mol. Cell Biol. 2021; 22:375–392. [DOI] [PubMed] [Google Scholar]
- 11. Wilson D.N., Doudna Cate J.H. The structure and function of the eukaryotic ribosome. Cold Spring Harb. Perspect. Biol. 2012; 4:a011536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Voigts-Hoffmann F., Klinge S., Ban N. Structural insights into eukaryotic ribosomes and the initiation of translation. Curr. Opin. Struct. Biol. 2012; 22:768–777. [DOI] [PubMed] [Google Scholar]
- 13. Sloan K.E., Warda A.S., Sharma S., Entian K.D., Lafontaine D.L.J., Bohnsack M.T. Tuning the ribosome: the influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol. 2017; 14:1138–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Natchiar S.K., Myasnikov A.G., Kratzat H., Hazemann I., Klaholz B.P. Visualization of chemical modifications in the human 80S ribosome structure. Nature. 2017; 551:472–477. [DOI] [PubMed] [Google Scholar]
- 15. Kiss-Laszlo Z., Henry Y., Bachellerie J.P., Caizergues-Ferrer M., Kiss T. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell. 1996; 85:1077–1088. [DOI] [PubMed] [Google Scholar]
- 16. Ganot P., Bortolin M.L., Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 1997; 89:799–809. [DOI] [PubMed] [Google Scholar]
- 17. Lapinaite A., Simon B., Skjaerven L., Rakwalska-Bange M., Gabel F., Carlomagno T. The structure of the box C/D enzyme reveals regulation of RNA methylation. Nature. 2013; 502:519–523. [DOI] [PubMed] [Google Scholar]
- 18. Adams J.M., Cory S. Modified nucleosides and bizarre 5′-termini in mouse myeloma mRNA. Nature. 1975; 255:28–33. [DOI] [PubMed] [Google Scholar]
- 19. Desrosiers R., Friderici K., Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. U.S.A. 1974; 71:3971–3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dubin D.T., Taylor R.H. The methylation state of poly A-containing messenger RNA from cultured hamster cells. Nucleic Acids Res. 1975; 2:1653–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Perry R.P., Kelley D.E., Friderici K., Rottman F. The methylated constituents of L cell messenger RNA: evidence for an unusual cluster at the 5′ terminus. Cell. 1975; 4:387–394. [DOI] [PubMed] [Google Scholar]
- 22. Sharma S., Lafontaine D.L.J. ‘View From A Bridge’: a new perspective on eukaryotic rRNA base modification. Trends Biochem. Sci. 2015; 40:560–575. [DOI] [PubMed] [Google Scholar]
- 23. Meyer K.D., Saletore Y., Zumbo P., Elemento O., Mason C.E., Jaffrey S.R. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012; 149:1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., Cesarkas K., Jacob-Hirsch J., Amariglio N., Kupiec M. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012; 485:201–206. [DOI] [PubMed] [Google Scholar]
- 25. Nachtergaele S., He C. Chemical modifications in the life of an mRNA transcript. Annu. Rev. Genet. 2018; 52:349–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Destefanis E., Avsar G., Groza P., Romitelli A., Torrini S., Pir P., Conticello S.G., Aguilo F., Dassi E. A mark of disease: how mRNA modifications shape genetic and acquired pathologies. RNA. 2021; 27:367–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021; 71:209–249. [DOI] [PubMed] [Google Scholar]
- 28. He Z., Chen Z., Tan M., Elingarami S., Liu Y., Li T., Deng Y., He N., Li S., Fu J. et al. A review on methods for diagnosis of breast cancer cells and tissues. Cell Prolif. 2020; 53:e12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021; 71:7–33. [DOI] [PubMed] [Google Scholar]
- 30. Tsang J.Y.S., Tse G.M. Molecular classification of breast cancer. Adv. Anat. Pathol. 2020; 27:27–35. [DOI] [PubMed] [Google Scholar]
- 31. Perou C.M., Sorlie T., Eisen M.B., van de Rijn M., Jeffrey S.S., Rees C.A., Pollack J.R., Ross D.T., Johnsen H., Akslen L.A. et al. Molecular portraits of human breast tumours. Nature. 2000; 406:747–752. [DOI] [PubMed] [Google Scholar]
- 32. Yeo S.K., Guan J.L. Breast cancer: multiple subtypes within a tumor. Trends Cancer. 2017; 3:753–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Badve S., Dabbs D.J., Schnitt S.J., Baehner F.L., Decker T., Eusebi V., Fox S.B., Ichihara S., Jacquemier J., Lakhani S.R. et al. Basal-like and triple-negative breast cancers: a critical review with an emphasis on the implications for pathologists and oncologists. Mod. Pathol. 2011; 24:157–167. [DOI] [PubMed] [Google Scholar]
- 34. Yang X.R., Sherman M.E., Rimm D.L., Lissowska J., Brinton L.A., Peplonska B., Hewitt S.M., Anderson W.F., Szeszenia-Dabrowska N., Bardin-Mikolajczak A. et al. Differences in risk factors for breast cancer molecular subtypes in a population-based study. Cancer Epidemiol. Biomarkers Prev. 2007; 16:439–443. [DOI] [PubMed] [Google Scholar]
- 35. Sflomos G., Dormoy V., Metsalu T., Jeitziner R., Battista L., Scabia V., Raffoul W., Delaloye J.F., Treboux A., Fiche M. et al. A preclinical model for eralpha-positive breast cancer points to the epithelial microenvironment as determinant of luminal phenotype and hormone response. Cancer Cell. 2016; 29:407–422. [DOI] [PubMed] [Google Scholar]
- 36. Yue Y., Liu J., He C. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 2015; 29:1343–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Taoka M., Nobe Y., Yamaki Y., Sato K., Ishikawa H., Izumikawa K., Yamauchi Y., Hirota K., Nakayama H., Takahashi N. et al. Landscape of the complete RNA chemical modifications in the human 80S ribosome. Nucleic Acids Res. 2018; 46:9289–9298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Malla S., Melguizo-Sanchis D., Aguilo F. Steering pluripotency and differentiation with N(6)-methyladenosine RNA modification. Biochim. Biophys. Acta Gene Regul. Mech. 2019; 1862:394–402. [DOI] [PubMed] [Google Scholar]
- 39. Aguilo F., Walsh M.J. The N(6)-Methyladenosine RNA modification in pluripotency and reprogramming. Curr. Opin. Genet. Dev. 2017; 46:77–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lence T., Paolantoni C., Worpenberg L., Roignant J.Y. Mechanistic insights into m(6)A RNA enzymes. Biochim. Biophys. Acta Gene Regul. Mech. 2019; 1862:222–229. [DOI] [PubMed] [Google Scholar]
- 41. Shi H., Wei J., He C. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol. Cell. 2019; 74:640–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jia G., Fu Y., Zhao X., Dai Q., Zheng G., Yang Y., Yi C., Lindahl T., Pan T., Yang Y.G. et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011; 7:885–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zheng G., Dahl J.A., Niu Y., Fedorcsak P., Huang C.M., Li C.J., Vagbo C.B., Shi Y., Wang W.L., Song S.H. et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell. 2013; 49:18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Patil D.P., Pickering B.F., Jaffrey S.R. Reading m(6)A in the Transcriptome: m(6)A-Binding Proteins. Trends Cell Biol. 2018; 28:113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zaccara S., Ries R.J., Jaffrey S.R. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 2019; 20:608–624. [DOI] [PubMed] [Google Scholar]
- 46. Xiao H., Fan X., Zhang R., Wu G. Upregulated N6-methyladenosine RNA in peripheral blood: potential diagnostic biomarker for breast cancer. Cancer Res. Treat. 2021; 53:399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. He X., Tan L., Ni J., Shen G. Expression pattern of m(6)A regulators is significantly correlated with malignancy and antitumor immune response of breast cancer. Cancer Gene Ther. 2021; 28:188–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang S., Zou X., Chen Y., Cho W.C., Zhou X. Effect of N6-methyladenosine regulators on progression and prognosis of triple-negative breast cancer. Front Genet. 2020; 11:580036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gong P.J., Shao Y.C., Yang Y., Song W.J., He X., Zeng Y.F., Huang S.R., Wei L., Zhang J.W. Analysis of N6-methyladenosine methyltransferase reveals METTL14 and ZC3H13 as tumor suppressor genes in breast cancer. Front. Oncol. 2020; 10:578963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yi D., Wang R., Shi X., Xu L., Yilihamu Y., Sang J. METTL14 promotes the migration and invasion of breast cancer cells by modulating N6methyladenosine and hsamiR146a5p expression. Oncol. Rep. 2020; 43:1375–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dong X.F., Wang Y., Huang B.F., Hu G.N., Shao J.K., Wang Q., Tang C.H., Wang C.Q. Downregulated METTL14 expression correlates with breast cancer tumor grade and molecular classification. Biomed. Res. Int. 2020; 2020:8823270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Xu Y., Ye S., Zhang N., Zheng S., Liu H., Zhou K., Wang L., Cao Y., Sun P., Wang T. The FTO/miR-181b-3p/ARL5B signaling pathway regulates cell migration and invasion in breast cancer. Cancer Commun (Lond). 2020; 40:484–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang B., Gu Y., Jiang G. Expression and prognostic characteristics of m(6) A RNA methylation regulators in breast cancer. Front Genet. 2020; 11:604597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Liu L., Liu X., Dong Z., Li J., Yu Y., Chen X., Ren F., Cui G., Sun R. N6-methyladenosine-related genomic targets are altered in breast cancer tissue and associated with poor survival. J. Cancer. 2019; 10:5447–5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Anita R., Paramasivam A., Priyadharsini J.V., Chitra S. The m6A readers YTHDF1 and YTHDF3 aberrations associated with metastasis and predict poor prognosis in breast cancer patients. Am J Cancer Res. 2020; 10:2546–2554. [PMC free article] [PubMed] [Google Scholar]
- 56. Panneerdoss S., Eedunuri V.K., Yadav P., Timilsina S., Rajamanickam S., Viswanadhapalli S., Abdelfattah N., Onyeagucha B.C., Cui X., Lai Z. et al. Cross-talk among writers, readers, and erasers of m(6)A regulates cancer growth and progression. Sci. Adv. 2018; 4:eaar8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cai X., Wang X., Cao C., Gao Y., Zhang S., Yang Z., Liu Y., Zhang X., Zhang W., Ye L. HBXIP-elevated methyltransferase METTL3 promotes the progression of breast cancer via inhibiting tumor suppressor let-7g. Cancer Lett. 2018; 415:11–19. [DOI] [PubMed] [Google Scholar]
- 58. Wang H., Xu B., Shi J. N6-methyladenosine METTL3 promotes the breast cancer progression via targeting Bcl-2. Gene. 2020; 722:144076. [DOI] [PubMed] [Google Scholar]
- 59. Chen F., Chen Z., Guan T., Zhou Y., Ge L., Zhang H., Wu Y., Jiang G.M., He W., Li J. et al. N(6) -methyladenosine regulates mRNA stability and translation efficiency of KRT7 to promote breast cancer lung metastasis. Cancer Res. 2021; 81:2847–2860. [DOI] [PubMed] [Google Scholar]
- 60. Shi Y., Zheng C., Jin Y., Bao B., Wang D., Hou K., Feng J., Tang S., Qu X., Liu Y. et al. Reduced expression of METTL3 promotes metastasis of triple-negative breast cancer by m6a methylation-mediated COL3A1 up-regulation. Front. Oncol. 2020; 10:1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Li Z., Yang H.Y., Dai X.Y., Zhang X., Huang Y.Z., Shi L., Wei J.F., Ding Q. CircMETTL3, upregulated in a m6A-dependent manner, promotes breast cancer progression. Int. J. Biol. Sci. 2021; 17:1178–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sun T., Wu Z., Wang X., Wang Y., Hu X., Qin W., Lu S., Xu D., Wu Y., Chen Q. et al. LNC942 promoting METTL14-mediated m(6)A methylation in breast cancer cell proliferation and progression. Oncogene. 2020; 39:5358–5372. [DOI] [PubMed] [Google Scholar]
- 63. Niu Y., Lin Z., Wan A., Chen H., Liang H., Sun L., Wang Y., Li X., Xiong X.F., Wei B. et al. RNA N6-methyladenosine demethylase FTO promotes breast tumor progression through inhibiting BNIP3. Mol. Cancer. 2019; 18:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Einstein J.M., Perelis M., Chaim I.A., Meena J.K., Nussbacher J.K., Tankka A.T., Yee B.A., Li H., Madrigal A.A., Neill N.J. et al. Inhibition of YTHDF2 triggers proteotoxic cell death in MYC-driven breast cancer. Mol. Cell. 2021; 81:3048–3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Alarcon C.R., Lee H., Goodarzi H., Halberg N., Tavazoie S.F. N6-methyladenosine marks primary microRNAs for processing. Nature. 2015; 519:482–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Celia-Terrassa T., Jolly M.K. Cancer stem cells and epithelial-to-mesenchymal transition in cancer metastasis. Cold Spring Harb. Perspect. Med. 2020; 10:a036905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Vivanco Mdel M. Mammary Stem Cells. Preface. Methods Mol. Biol. 2015; 1293:v–vi. [DOI] [PubMed] [Google Scholar]
- 68. Yang M.H., Wu M.Z., Chiou S.H., Chen P.M., Chang S.Y., Liu C.J., Teng S.C., Wu K.J. Direct regulation of TWIST by HIF-1alpha promotes metastasis. Nat. Cell Biol. 2008; 10:295–305. [DOI] [PubMed] [Google Scholar]
- 69. Zhang C., Samanta D., Lu H., Bullen J.W., Zhang H., Chen I., He X., Semenza G.L. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m(6)A-demethylation of NANOG mRNA. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E2047–E2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zhang C., Zhi W.I., Lu H., Samanta D., Chen I., Gabrielson E., Semenza G.L. Hypoxia-inducible factors regulate pluripotency factor expression by ZNF217- and ALKBH5-mediated modulation of RNA methylation in breast cancer cells. Oncotarget. 2016; 7:64527–64542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Aguilo F., Zhang F., Sancho A., Fidalgo M., Di Cecilia S., Vashisht A., Lee D.F., Chen C.H., Rengasamy M., Andino B. et al. Coordination of m(6)A mRNA methylation and gene transcription by ZFP217 regulates pluripotency and reprogramming. Cell Stem Cell. 2015; 17:689–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Peng F., Xu J., Cui B., Liang Q., Zeng S., He B., Zou H., Li M., Zhao H., Meng Y. et al. Oncogenic AURKA-enhanced N(6)-methyladenosine modification increases DROSHA mRNA stability to transactivate STC1 in breast cancer stem-like cells. Cell Res. 2021; 31:345–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zhu P., He F., Hou Y., Tu G., Li Q., Jin T., Zeng H., Qin Y., Wan X., Qiao Y. et al. A novel hypoxic long noncoding RNA KB-1980E6.3 maintains breast cancer stem cell stemness via interacting with IGF2BP1 to facilitate c-Myc mRNA stability. Oncogene. 2021; 40:1609–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Liu X., Gonzalez G., Dai X., Miao W., Yuan J., Huang M., Bade D., Li L., Sun Y., Wang Y. Adenylate kinase 4 modulates the resistance of breast cancer cells to tamoxifen through an m(6)A-based epitranscriptomic mechanism. Mol. Ther. 2020; 28:2593–2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Pan X., Hong X., Li S., Meng P., Xiao F. METTL3 promotes adriamycin resistance in MCF-7 breast cancer cells by accelerating pri-microRNA-221-3p maturation in a m6A-dependent manner. Exp. Mol. Med. 2021; 53:91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Cheng L., Zhang X., Huang Y.Z., Zhu Y.L., Xu L.Y., Li Z., Dai X.Y., Shi L., Zhou X.J., Wei J.F. et al. Metformin exhibits antiproliferation activity in breast cancer via miR-483-3p/METTL3/m(6)A/p21 pathway. Oncogenesis. 2021; 10:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. van Tran N., Ernst F.G.M., Hawley B.R., Zorbas C., Ulryck N., Hackert P., Bohnsack K.E., Bohnsack M.T., Jaffrey S.R., Graille M. et al. The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res. 2019; 47:7719–7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Rong B., Zhang Q., Wan J., Xing S., Dai R., Li Y., Cai J., Xie J., Song Y., Chen J. et al. Ribosome 18S m(6)A methyltransferase METTL5 promotes translation initiation and breast cancer cell growth. Cell Rep. 2020; 33:108544. [DOI] [PubMed] [Google Scholar]
- 79. Pinto R., Vagbo C.B., Jakobsson M.E., Kim Y., Baltissen M.P., O’Donohue M.F., Guzman U.H., Malecki J.M., Wu J., Kirpekar F. et al. The human methyltransferase ZCCHC4 catalyses N6-methyladenosine modification of 28S ribosomal RNA. Nucleic Acids Res. 2020; 48:830–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Li X., Xiong X., Wang K., Wang L., Shu X., Ma S., Yi C. Transcriptome-wide mapping reveals reversible and dynamic N(1)-methyladenosine methylome. Nat. Chem. Biol. 2016; 12:311–316. [DOI] [PubMed] [Google Scholar]
- 81. Haruehanroengra P., Zheng Y.Y., Zhou Y., Huang Y., Sheng J. RNA modifications and cancer. RNA Biol. 2020; 17:1560–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Oerum S., Degut C., Barraud P., Tisne C. m1A Post-Transcriptional Modification in tRNAs. Biomolecules. 2017; 7:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Dominissini D., Nachtergaele S., Moshitch-Moshkovitz S., Peer E., Kol N., Ben-Haim M.S., Dai Q., Di Segni A., Salmon-Divon M., Clark W.C. et al. The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature. 2016; 530:441–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Safra M., Sas-Chen A., Nir R., Winkler R., Nachshon A., Bar-Yaacov D., Erlacher M., Rossmanith W., Stern-Ginossar N., Schwartz S. The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature. 2017; 551:251–255. [DOI] [PubMed] [Google Scholar]
- 85. Zhou H., Rauch S., Dai Q., Cui X., Zhang Z., Nachtergaele S., Sepich C., He C., Dickinson B.C. Evolution of a reverse transcriptase to map N(1)-methyladenosine in human messenger RNA. Nat. Methods. 2019; 16:1281–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Woo H.H., Chambers S.K. Human ALKBH3-induced m(1)A demethylation increases the CSF-1 mRNA stability in breast and ovarian cancer cells. Biochim. Biophys. Acta Gene Regul. Mech. 2019; 1862:35–46. [DOI] [PubMed] [Google Scholar]
- 87. Yang X., Yang Y., Sun B.F., Chen Y.S., Xu J.W., Lai W.Y., Li A., Wang X., Bhattarai D.P., Xiao W. et al. 5-methylcytosine promotes mRNA export - NSUN2 as the methyltransferase and ALYREF as an m(5)C reader. Cell Res. 2017; 27:606–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Chen X., Li A., Sun B.F., Yang Y., Han Y.N., Yuan X., Chen R.X., Wei W.S., Liu Y., Gao C.C. et al. 5-methylcytosine promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nat. Cell Biol. 2019; 21:978–990. [DOI] [PubMed] [Google Scholar]
- 89. Schosserer M., Minois N., Angerer T.B., Amring M., Dellago H., Harreither E., Calle-Perez A., Pircher A., Peter Gerstl M., Pfeifenberger S. et al. Corrigendum: Methylation of ribosomal RNA by NSUN5 is a conserved mechanism modulating organismal lifespan. Nat. Commun. 2016; 7:11530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Blanco S., Dietmann S., Flores J.V., Hussain S., Kutter C., Humphreys P., Lukk M., Lombard P., Treps L., Popis M. et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J. 2014; 33:2020–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Schaefer M., Pollex T., Hanna K., Tuorto F., Meusburger M., Helm M., Lyko F. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010; 24:1590–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Guzzi N., Bellodi C. Novel insights into the emerging roles of tRNA-derived fragments in mammalian development. RNA Biol. 2020; 17:1214–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Zhu W., Wang J.Z., Xu Z., Cao M., Hu Q., Pan C., Guo M., Wei J.F., Yang H. Detection of N6methyladenosine modification residues (Review). Int. J. Mol. Med. 2019; 43:2267–2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Tang H., Fan X., Xing J., Liu Z., Jiang B., Dou Y., Gorospe M., Wang W. NSun2 delays replicative senescence by repressing p27 (KIP1) translation and elevating CDK1 translation. Aging (Albany NY). 2015; 7:1143–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Bohnsack K.E., Hobartner C., Bohnsack M.T. Eukaryotic 5-methylcytosine (m(5)C) RNA methyltransferases: mechanisms, cellular functions, and links to disease. Genes (Basel). 2019; 10:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Janin M., Ortiz-Barahona V., de Moura M.C., Martinez-Cardus A., Llinas-Arias P., Soler M., Nachmani D., Pelletier J., Schumann U., Calleja-Cervantes M.E. et al. Epigenetic loss of RNA-methyltransferase NSUN5 in glioma targets ribosomes to drive a stress adaptive translational program. Acta Neuropathol. 2019; 138:1053–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Heissenberger C., Liendl L., Nagelreiter F., Gonskikh Y., Yang G., Stelzer E.M., Krammer T.L., Micutkova L., Vogt S., Kreil D.P. et al. Loss of the ribosomal RNA methyltransferase NSUN5 impairs global protein synthesis and normal growth. Nucleic Acids Res. 2019; 47:11807–11825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Heissenberger C., Rollins J.A., Krammer T.L., Nagelreiter F., Stocker I., Wacheul L., Shpylovyi A., Tav K., Snow S., Grillari J. et al. The ribosomal RNA m(5)C methyltransferase NSUN-1 modulates healthspan and oogenesis in Caenorhabditis elegans. Elife. 2020; 9:e56205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Metodiev M.D., Spahr H., Loguercio Polosa P., Meharg C., Becker C., Altmueller J., Habermann B., Larsson N.G., Ruzzenente B. NSUN4 is a dual function mitochondrial protein required for both methylation of 12S rRNA and coordination of mitoribosomal assembly. PLos Genet. 2014; 10:e1004110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Tuorto F., Liebers R., Musch T., Schaefer M., Hofmann S., Kellner S., Frye M., Helm M., Stoecklin G., Lyko F. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat. Struct. Mol. Biol. 2012; 19:900–905. [DOI] [PubMed] [Google Scholar]
- 101. Khoddami V., Cairns B.R. Identification of direct targets and modified bases of RNA cytosine methyltransferases. Nat. Biotechnol. 2013; 31:458–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Goll M.G., Kirpekar F., Maggert K.A., Yoder J.A., Hsieh C.L., Zhang X., Golic K.G., Jacobsen S.E., Bestor T.H. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science. 2006; 311:395–398. [DOI] [PubMed] [Google Scholar]
- 103. Frye M., Watt F.M. The RNA methyltransferase Misu (NSun2) mediates Myc-induced proliferation and is upregulated in tumors. Curr. Biol. 2006; 16:971–981. [DOI] [PubMed] [Google Scholar]
- 104. Haag S., Warda A.S., Kretschmer J., Gunnigmann M.A., Hobartner C., Bohnsack M.T. NSUN6 is a human RNA methyltransferase that catalyzes formation of m5C72 in specific tRNAs. RNA. 2015; 21:1532–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Selmi T., Hussain S., Dietmann S., Heiss M., Borland K., Flad S., Carter J.M., Dennison R., Huang Y.L., Kellner S. et al. Sequence- and structure-specific cytosine-5 mRNA methylation by NSUN6. Nucleic Acids Res. 2021; 49:1006–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Sajini A.A., Choudhury N.R., Wagner R.E., Bornelov S., Selmi T., Spanos C., Dietmann S., Rappsilber J., Michlewski G., Frye M. Loss of 5-methylcytosine alters the biogenesis of vault-derived small RNAs to coordinate epidermal differentiation. Nat. Commun. 2019; 10:2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Aguilo F., Li S., Balasubramaniyan N., Sancho A., Benko S., Zhang F., Vashisht A., Rengasamy M., Andino B., Chen C.H. et al. Deposition of 5-methylcytosine on enhancer RNAs enables the coactivator function of PGC-1alpha. Cell Rep. 2016; 14:479–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Freeman J.W., McGrath P., Bondada V., Selliah N., Ownby H., Maloney T., Busch R. K., Busch H. Prognostic significance of proliferation associated nucleolar antigen p120 in human breast carcinoma. Cancer Res. 1991; 51:1973–1978. [PubMed] [Google Scholar]
- 109. Frye M., Dragoni I., Chin S.F., Spiteri I., Kurowski A., Provenzano E., Green A., Ellis I.O., Grimmer D., Teschendorff A. et al. Genomic gain of 5p15 leads to over-expression of Misu (NSUN2) in breast cancer. Cancer Lett. 2010; 289:71–80. [DOI] [PubMed] [Google Scholar]
- 110. Kar S.P., Beesley J., Amin Al Olama A., Michailidou K., Tyrer J., Kote-Jarai Z., Lawrenson K., Lindstrom S., Ramus S.J., Thompson D.J. et al. Genome-wide meta-analyses of breast, ovarian, and prostate cancer association studies identify multiple new susceptibility loci shared by at least two cancer types. Cancer Discov. 2016; 6:1052–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Vachon C.M., Sellers T.A., Carlson E.E., Cunningham J.M., Hilker C.A., Smalley R.L., Schaid D.J., Kelemen L.E., Couch F.J., Pankratz V.S. Strong evidence of a genetic determinant for mammographic density, a major risk factor for breast cancer. Cancer Res. 2007; 67:8412–8418. [DOI] [PubMed] [Google Scholar]
- 112. Yi J., Gao R., Chen Y., Yang Z., Han P., Zhang H., Dou Y., Liu W., Wang W., Du G. et al. Overexpression of NSUN2 by DNA hypomethylation is associated with metastatic progression in human breast cancer. Oncotarget. 2017; 8:20751–20765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Manning M., Jiang Y., Wang R., Liu L., Rode S., Bonahoom M., Kim S., Yang Z.Q. Pan-cancer analysis of RNA methyltransferases identifies FTSJ3 as a potential regulator of breast cancer progression. RNA Biol. 2020; 17:474–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Shinoda S., Kitagawa S., Nakagawa S., Wei F.Y., Tomizawa K., Araki K., Araki M., Suzuki T., Suzuki T. Mammalian NSUN2 introduces 5-methylcytidines into mitochondrial tRNAs. Nucleic Acids Res. 2019; 47:8734–8745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Huang Z., Pan J., Wang H., Du X., Xu Y., Wang Z., Chen D Prognostic Significance and Tumor Immune Microenvironment Heterogenicity of m5C RNA Methylation Regulators in Triple-Negative Breast Cancer. Front. Cell Dev. Biol. 2021; 9:657547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Huang J., Tan P.H., Li K.B., Matsumoto K., Tsujimoto M., Bay B.H. Y-box binding protein, YB-1, as a marker of tumor aggressiveness and response to adjuvant chemotherapy in breast cancer. Int. J. Oncol. 2005; 26:607–613. [PubMed] [Google Scholar]
- 117. Maciejczyk A., Szelachowska J., Ekiert M., Matkowski R., Halon A., Lage H., Surowiak P. Elevated nuclear YB1 expression is associated with poor survival of patients with early breast cancer. Anticancer Res. 2012; 32:3177–3184. [PubMed] [Google Scholar]
- 118. Somme J., Van Laer B., Roovers M., Steyaert J., Versees W., Droogmans L. Characterization of two homologous 2′-O-methyltransferases showing different specificities for their tRNA substrates. RNA. 2014; 20:1257–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Yang Z., Wang J., Huang L., Lilley D.M.J., Ye K. Functional organization of box C/D RNA-guided RNA methyltransferase. Nucleic Acids Res. 2020; 48:5094–5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Sharma S., Marchand V., Motorin Y., Lafontaine D.L.J. Identification of sites of 2′-O-methylation vulnerability in human ribosomal RNAs by systematic mapping. Sci. Rep. 2017; 7:11490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Ayadi L., Galvanin A., Pichot F., Marchand V., Motorin Y. RNA ribose methylation (2′-O-methylation): Occurrence, biosynthesis and biological functions. Biochim. Biophys. Acta Gene Regul. Mech. 2019; 1862:253–269. [DOI] [PubMed] [Google Scholar]
- 122. Wu H., Qin W., Lu S., Wang X., Zhang J., Sun T., Hu X., Li Y., Chen Q., Wang Y. et al. Long noncoding RNA ZFAS1 promoting small nucleolar RNA-mediated 2′-O-methylation via NOP58 recruitment in colorectal cancer. Mol. Cancer. 2020; 19:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Bizarro J., Deryusheva S., Wacheul L., Gupta V., Ernst F.G.M., Lafontaine D.L.J., Gall J.G., Meier U.T. Nopp140-chaperoned 2′-O-methylation of small nuclear RNAs in Cajal bodies ensures splicing fidelity. Genes Dev. 2021; 35:1123–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Marchand V., Pichot F., Thuring K., Ayadi L., Freund I., Dalpke A., Helm M., Motorin Y. Next-Generation Sequencing-Based RiboMethSeq Protocol for Analysis of tRNA 2′-O-Methylation. Biomolecules. 2017; 7:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Angelova M.T., Dimitrova D.G., Da Silva B., Marchand V., Jacquier C., Achour C., Brazane M., Goyenvalle C., Bourguignon-Igel V., Shehzada S. et al. tRNA 2′-O-methylation by a duo of TRM7/FTSJ1 proteins modulates small RNA silencing in Drosophila. Nucleic Acids Res. 2020; 48:2050–2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Elliott B.A., Ho H.T., Ranganathan S.V., Vangaveti S., Ilkayeva O., Abou Assi H., Choi A.K., Agris P.F., Holley C.L. Modification of messenger RNA by 2′-O-methylation regulates gene expression in vivo. Nat. Commun. 2019; 10:3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Kurth H.M., Mochizuki K. 2′-O-methylation stabilizes Piwi-associated small RNAs and ensures DNA elimination in Tetrahymena. RNA. 2009; 15:675–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Wang N., Qu S., Sun W., Zeng Z., Liang H., Zhang C.Y., Chen X., Zen K. Direct quantification of 3′ terminal 2′-O-methylation of small RNAs by RT-qPCR. RNA. 2018; 24:1520–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Munafo D.B., Robb G.B. Optimization of enzymatic reaction conditions for generating representative pools of cDNA from small RNA. RNA. 2010; 16:2537–2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Erales J., Marchand V., Panthu B., Gillot S., Belin S., Ghayad S.E., Garcia M., Laforets F., Marcel V., Baudin-Baillieu A. et al. Evidence for rRNA 2′-O-methylation plasticity: Control of intrinsic translational capabilities of human ribosomes. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:12934–12939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Belin S., Beghin A., Solano-Gonzalez E., Bezin L., Brunet-Manquat S., Textoris J., Prats A.C., Mertani H.C., Dumontet C., Diaz J.J. Dysregulation of ribosome biogenesis and translational capacity is associated with tumor progression of human breast cancer cells. PLoS One. 2009; 4:e7147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Marchand V., Blanloeil-Oillo F., Helm M., Motorin Y. Illumina-based RiboMethSeq approach for mapping of 2′-O-Me residues in RNA. Nucleic Acids Res. 2016; 44:e135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Marcel V., Kielbassa J., Marchand V., Natchiar K.S., Paraqindes H., Nguyen Van Long F., Ayadi L., Bourguignon-Igel V., Lo Monaco P., Monchiet D. et al. Ribosomal RNA 2′O-methylation as a novel layer of inter-tumour heterogeneity in breast cancer. NAR Cancer. 2020; 2:zcaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Metge B.J., Kammerud S.C., Pruitt H.C., Shevde L.A., Samant R.S. Hypoxia re-programs 2′-O-Me modifications on ribosomal RNA. iScience. 2021; 24:102010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Samanta D., Gilkes D.M., Chaturvedi P., Xiang L., Semenza G.L. Hypoxia-inducible factors are required for chemotherapy resistance of breast cancer stem cells. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E5429–E5438. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 136. Gong J., Li Y., Liu C.J., Xiang Y., Li C., Ye Y., Zhang Z., Hawke D.H., Park P.K., Diao L. et al. A Pan-cancer Analysis of the Expression and Clinical Relevance of Small Nucleolar RNAs in Human Cancer. Cell Rep. 2017; 21:1968–1981. [DOI] [PubMed] [Google Scholar]
- 137. Shubina M.Y., Musinova Y.R., Sheval E.V. Proliferation, cancer, and aging-novel functions of the nucleolar methyltransferase fibrillarin. Cell Biol. Int. 2018; 42:1463–1466. [DOI] [PubMed] [Google Scholar]
- 138. Marcel V., Ghayad S.E., Belin S., Therizols G., Morel A.P., Solano-Gonzalez E., Vendrell J.A., Hacot S., Mertani H.C., Albaret M.A. et al. p53 acts as a safeguard of translational control by regulating fibrillarin and rRNA methylation in cancer. Cancer Cell. 2013; 24:318–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Guo Y., Yu H., Wang J., Sheng Q., Zhao S., Zhao Y.Y., Lehmann B.D. The landscape of small non-coding RNAs in triple-negative breast cancer. Genes (Basel). 2018; 9:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Krishnan P., Ghosh S., Wang B., Heyns M., Graham K., Mackey J.R., Kovalchuk O., Damaraju S. Profiling of small nucleolar RNAs by next generation sequencing: potential new players for breast cancer prognosis. PLoS One. 2016; 11:e0162622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Kothari C., Ouellette G., Labrie Y., Jacob S., Diorio C., Durocher F. Identification of a gene signature for different stages of breast cancer development that could be used for early diagnosis and specific therapy. Oncotarget. 2018; 9:37407–37420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Mourtada-Maarabouni M., Pickard M.R., Hedge V.L., Farzaneh F., Williams G.T. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009; 28:195–208. [DOI] [PubMed] [Google Scholar]
- 143. Zimta A.A., Tigu A.B., Braicu C., Stefan C., Ionescu C., Berindan-Neagoe I. An Emerging Class of Long Non-coding RNA With Oncogenic Role Arises From the snoRNA Host Genes. Front. Oncol. 2020; 10:389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Qin Y., Sun W., Wang Z., Dong W., He L., Zhang T., Zhang H. Long non-coding small nucleolar RNA host genes (SNHGs) in endocrine-related cancers. Onco Targets Ther. 2020; 13:7699–7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Askarian-Amiri M.E., Crawford J., French J.D., Smart C.E., Smith M.A., Clark M.B., Ru K., Mercer T.R., Thompson E.R., Lakhani S.R. et al. SNORD-host RNA Zfas1 is a regulator of mammary development and a potential marker for breast cancer. RNA. 2011; 17:878–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Gumienny R., Jedlinski D.J., Schmidt A., Gypas F., Martin G., Vina-Vilaseca A., Zavolan M. High-throughput identification of C/D box snoRNA targets with CLIP and RiboMeth-seq. Nucleic Acids Res. 2017; 45:2341–2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Chen C., Zhao X., Kierzek R., Yu Y.T. A flexible RNA backbone within the polypyrimidine tract is required for U2AF65 binding and pre-mRNA splicing in vivo. Mol. Cell. Biol. 2010; 30:4108–4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. De Zoysa M.D., Yu Y.T. Posttranscriptional RNA pseudouridylation. Enzymes. 2017; 41:151–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Nombela P., Miguel-Lopez B., Blanco S. The role of m(6)A, m(5)C and Psi RNA modifications in cancer: novel therapeutic opportunities. Mol. Cancer. 2021; 20:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Montanaro L., Brigotti M., Clohessy J., Barbieri S., Ceccarelli C., Santini D., Taffurelli M., Calienni M., Teruya-Feldstein J., Trere D. et al. Dyskerin expression influences the level of ribosomal RNA pseudo-uridylation and telomerase RNA component in human breast cancer. J. Pathol. 2006; 210:10–18. [DOI] [PubMed] [Google Scholar]
- 151. Rocchi L., Barbosa A.J., Onofrillo C., Del Rio A., Montanaro L. Inhibition of human dyskerin as a new approach to target ribosome biogenesis. PLoS One. 2014; 9:e101971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Montanaro L., Calienni M., Bertoni S., Rocchi L., Sansone P., Storci G., Santini D., Ceccarelli C., Taffurelli M., Carnicelli D. et al. Novel dyskerin-mediated mechanism of p53 inactivation through defective mRNA translation. Cancer Res. 2010; 70:4767–4777. [DOI] [PubMed] [Google Scholar]
- 153. Rocchi L., Pacilli A., Sethi R., Penzo M., Schneider R.J., Trere D., Brigotti M., Montanaro L. Dyskerin depletion increases VEGF mRNA internal ribosome entry site-mediated translation. Nucleic Acids Res. 2013; 41:8308–8318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Guerrieri A.N., Zacchini F., Onofrillo C., Di Viggiano S., Penzo M., Ansuini A., Gandin I., Nobe Y., Taoka M., Isobe T. et al. DKC1 overexpression induces a more aggressive cellular behavior and increases intrinsic ribosomal activity in immortalized mammary gland cells. Cancers (Basel). 2020; 12:3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Elsharawy K.A., Althobiti M., Mohammed O.J., Aljohani A.I., Toss M.S., Green A.R., Rakha E.A. Nucleolar protein 10 (NOP10) predicts poor prognosis in invasive breast cancer. Breast Cancer Res. Treat. 2021; 185:615–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Yanowsky K., Barroso A., Osorio A., Urioste M., Benitez J., Martinez-Delgado B. Mutational analysis of telomere genes in BRCA1/2-negative breast cancer families with very short telomeres. Breast Cancer Res. Treat. 2012; 134:1337–1343. [DOI] [PubMed] [Google Scholar]
- 157. Deogharia M., Majumder M. Guide snoRNAs: drivers or passengers in human disease. Biology (Basel). 2018; 8:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Schulten H.J., Bangash M., Karim S., Dallol A., Hussein D., Merdad A., Al-Thoubaity F.K., Al-Maghrabi J., Jamal A., Al-Ghamdi F. et al. Comprehensive molecular biomarker identification in breast cancer brain metastases. J. Transl. Med. 2017; 15:269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Liang F., Qu H., Lin Q., Yang Y., Ruan X., Zhang B., Liu Y., Yu C., Zhang H., Fang X. et al. Molecular biomarkers screened by next-generation RNA sequencing for non-sentinel lymph node status prediction in breast cancer patients with metastatic sentinel lymph nodes. World J. Surg. Oncol. 2015; 13:258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Sun Y., Chen E., Li Y., Ye D., Cai Y., Wang Q., Li Q., Zhang X. H/ACA box small nucleolar RNA 7B acts as an oncogene and a potential prognostic biomarker in breast cancer. Cancer Cell Int. 2019; 19:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Bazak L., Haviv A., Barak M., Jacob-Hirsch J., Deng P., Zhang R., Isaacs F.J., Rechavi G., Li J.B., Eisenberg E. et al. A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes. Genome Res. 2014; 24:365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Savva Y.A., Rieder L.E., Reenan R.A. The ADAR protein family. Genome Biol. 2012; 13:252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Gommans W.M., Mullen S.P., Maas S. RNA editing: a driving force for adaptive evolution?. Bioessays. 2009; 31:1137–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Rueter S.M., Dawson T.R., Emeson R.B. Regulation of alternative splicing by RNA editing. Nature. 1999; 399:75–80. [DOI] [PubMed] [Google Scholar]
- 165. Washburn M.C., Hundley H.A. Controlling the Editor: The Many Roles of RNA-Binding Proteins in Regulating A-to-I RNA Editing. Adv. Exp. Med. Biol. 2016; 907:189–213. [DOI] [PubMed] [Google Scholar]
- 166. Hsiao Y.E., Bahn J.H., Yang Y., Lin X., Tran S., Yang E.W., Quinones-Valdez G., Xiao X. RNA editing in nascent RNA affects pre-mRNA splicing. Genome Res. 2018; 28:812–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Zhang L., Yang C.S., Varelas X., Monti S. Altered RNA editing in 3′ UTR perturbs microRNA-mediated regulation of oncogenes and tumor-suppressors. Sci. Rep. 2016; 6:23226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Torres A.G., Batlle E., Ribas de Pouplana L. Role of tRNA modifications in human diseases. Trends Mol. Med. 2014; 20:306–314. [DOI] [PubMed] [Google Scholar]
- 169. Smith H.C., Bennett R.P., Kizilyer A., McDougall W.M., Prohaska K.M. Functions and regulation of the APOBEC family of proteins. Semin. Cell Dev. Biol. 2012; 23:258–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Conticello S.G. The AID/APOBEC family of nucleic acid mutators. Genome Biol. 2008; 9:229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Sagredo E.A., Blanco A., Sagredo A.I., Perez P., Sepulveda-Hermosilla G., Morales F., Muller B., Verdugo R., Marcelain K., Harismendy O. et al. ADAR1-mediated RNA-editing of 3′UTRs in breast cancer. Biol. Res. 2018; 51:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Fumagalli D., Gacquer D., Rothe F., Lefort A., Libert F., Brown D., Kheddoumi N., Shlien A., Konopka T., Salgado R. et al. Principles Governing A-to-I RNA Editing in the Breast Cancer Transcriptome. Cell Rep. 2015; 13:277–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Han L., Diao L., Yu S., Xu X., Li J., Zhang R., Yang Y., Werner H.M.J., Eterovic A.K., Yuan Y. et al. The genomic landscape and clinical relevance of A-to-I RNA editing in human cancers. Cancer Cell. 2015; 28:515–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Paz-Yaacov N., Bazak L., Buchumenski I., Porath H.T., Danan-Gotthold M., Knisbacher B.A., Eisenberg E., Levanon E.Y. Elevated RNA Editing Activity Is a Major Contributor to Transcriptomic Diversity in Tumors. Cell Rep. 2015; 13:267–276. [DOI] [PubMed] [Google Scholar]
- 175. Sagredo E.A., Sagredo A.I., Blanco A., Rojas De Santiago P., Rivas S., Assar R., Perez P., Marcelain K., Armisen R. ADAR1 Transcriptome editing promotes breast cancer progression through the regulation of cell cycle and DNA damage response. Biochim. Biophys. Acta Mol. Cell Res. 2020; 1867:118716. [DOI] [PubMed] [Google Scholar]
- 176. Gumireddy K., Li A., Kossenkov A.V., Sakurai M., Yan J., Li Y., Xu H., Wang J., Zhang P.J., Zhang L. et al. The mRNA-edited form of GABRA3 suppresses GABRA3-mediated Akt activation and breast cancer metastasis. Nat. Commun. 2016; 7:10715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Peng X., Xu X., Wang Y., Hawke D.H., Yu S., Han L., Zhou Z., Mojumdar K., Jeong K.J., Labrie M. et al. A-to-I RNA editing contributes to proteomic diversity in cancer. Cancer Cell. 2018; 33:817–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Zhang M., Fritsche J., Roszik J., Williams L.J., Peng X., Chiu Y., Tsou C.C., Hoffgaard F., Goldfinger V., Schoor O. et al. RNA editing derived epitopes function as cancer antigens to elicit immune responses. Nat. Commun. 2018; 9:3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Nakano M., Fukami T., Gotoh S., Nakajima M. A-to-I RNA editing up-regulates human dihydrofolate reductase in breast cancer. J. Biol. Chem. 2017; 292:4873–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Nik-Zainal S., Wedge D.C., Alexandrov L.B., Petljak M., Butler A.P., Bolli N., Davies H.R., Knappskog S., Martin S., Papaemmanuil E. et al. Association of a germline copy number polymorphism of APOBEC3A and APOBEC3B with burden of putative APOBEC-dependent mutations in breast cancer. Nat. Genet. 2014; 46:487–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Roberts S.A., Lawrence M.S., Klimczak L.J., Grimm S.A., Fargo D., Stojanov P., Kiezun A., Kryukov G.V., Carter S.L., Saksena G. et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat. Genet. 2013; 45:970–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Trevino V. Integrative genomic analysis identifies associations of molecular alterations to APOBEC and BRCA1/2 mutational signatures in breast cancer. Mol Genet Genomic Med. 2019; 7:e810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Sharma S., Patnaik S.K., Kemer Z., Baysal B.E. Transient overexpression of exogenous APOBEC3A causes C-to-U RNA editing of thousands of genes. RNA Biol. 2017; 14:603–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Sieuwerts A.M., Doebar S.C., de Weerd V., Verhoef E.I., Beauford C.M., Agahozo M.C., Martens J.W.M., van Deurzen C.H.M. APOBEC3B gene expression in ductal carcinoma in situ and synchronous invasive breast cancer. Cancers (Basel). 2019; 11:1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Asaoka M., Ishikawa T., Takabe K., Patnaik S.K. APOBEC3-Mediated RNA Editing in Breast Cancer is Associated with Heightened Immune Activity and Improved Survival. Int. J. Mol. Sci. 2019; 20:5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Zhou L., Hao J., Yuan Y., Peng R., Wang H., Ni D., Gu Y., Huang L., Mao Z., Lyu Z. et al. EIYMNVPV Motif is Essential for A1CF Nucleus Localization and A1CF (-8aa) Promotes Proliferation of MDA-MB-231 Cells via Up-Regulation of IL-6. Int. J. Mol. Sci. 2016; 17:811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Pavon-Eternod M., Gomes S., Geslain R., Dai Q., Rosner M.R., Pan T. tRNA over-expression in breast cancer and functional consequences. Nucleic Acids Res. 2009; 37:7268–7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Rozov A., Demeshkina N., Khusainov I., Westhof E., Yusupov M., Yusupova G. Novel base-pairing interactions at the tRNA wobble position crucial for accurate reading of the genetic code. Nat. Commun. 2016; 7:10457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Delaunay S., Rapino F., Tharun L., Zhou Z., Heukamp L., Termathe M., Shostak K., Klevernic I., Florin A., Desmecht H. et al. Elp3 links tRNA modification to IRES-dependent translation of LEF1 to sustain metastasis in breast cancer. J. Exp. Med. 2016; 213:2503–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Zhang J., Lu R., Zhang Y., Matuszek Z., Zhang W., Xia Y., Pan T., Sun J. tRNA queuosine modification enzyme modulates the growth and microbiome recruitment to breast tumors. Cancers (Basel). 2020; 12:628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Chen Y.C., Kelly V.P., Stachura S.V., Garcia G.A. Characterization of the human tRNA-guanine transglycosylase: confirmation of the heterodimeric subunit structure. RNA. 2010; 16:958–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Rodriguez V., Chen Y., Elkahloun A., Dutra A., Pak E., Chandrasekharappa S. Chromosome 8 BAC array comparative genomic hybridization and expression analysis identify amplification and overexpression of TRMT12 in breast cancer. Genes Chromosomes Cancer. 2007; 46:694–707. [DOI] [PubMed] [Google Scholar]
- 193. Hicks D.G., Janarthanan B.R., Vardarajan R., Kulkarni S.A., Khoury T., Dim D., Budd G.T., Yoder B.J., Tubbs R., Schreeder M.T. et al. The expression of TRMT2A, a novel cell cycle regulated protein, identifies a subset of breast cancer patients with HER2 over-expression that are at an increased risk of recurrence. BMC Cancer. 2010; 10:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Begley U., Sosa M.S., Avivar-Valderas A., Patil A., Endres L., Estrada Y., Chan C.T., Su D., Dedon P.C., Aguirre-Ghiso J.A. et al. A human tRNA methyltransferase 9-like protein prevents tumour growth by regulating LIN9 and HIF1-alpha. EMBO Mol. Med. 2013; 5:366–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Schumann U., Zhang H.N., Sibbritt T., Pan A., Horvath A., Gross S., Clark S.J., Yang L., Preiss T. Multiple links between 5-methylcytosine content of mRNA and translation. BMC Biol. 2020; 18:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Clancy M.J., Shambaugh M.E., Timpte C.S., Bokar J.A. Induction of sporulation in Saccharomyces cerevisiae leads to the formation of N6-methyladenosine in mRNA: a potential mechanism for the activity of the IME4 gene. Nucleic Acids Res. 2002; 30:4509–4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Agarwala S.D., Blitzblau H.G., Hochwagen A., Fink G.R. RNA methylation by the MIS complex regulates a cell fate decision in yeast. PLos Genet. 2012; 8:e1002732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Lin S., Choe J., Du P., Triboulet R., Gregory R.I. The m(6)A methyltransferase METTL3 promotes translation in human cancer cells. Mol. Cell. 2016; 62:335–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Cully M. Chemical inhibitors make their RNA epigenetic mark. Nat. Rev. Drug Discov. 2019; 18:892–894. [DOI] [PubMed] [Google Scholar]
- 200. Yankova E., Blackaby W., Albertella M., Rak J., De Braekeleer E., Tsagkogeorga G., Pilka E.S., Aspris D., Leggate D., Hendrick A.G. et al. Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature. 2021; 593:597–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Xie J., Ba J., Zhang M., Wan Y., Jin Z., Y. Yao. The m6A methyltransferase METTL3 promotes the stemness and malignant progression of breast cancer by mediating m6A modification on SOX2. J. BUON. 2021; 26:444–449. [PubMed] [Google Scholar]
- 202. Wu L., Wu D., Ning J., Liu W., Zhang D. Changes of N6-methyladenosine modulators promote breast cancer progression. BMC Cancer. 2019; 19:326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Chang G., Shi L., Ye Y., Shi H., Zeng L., Tiwary S., Huse J.T., Huo L., Ma L., Ma Y. et al. YTHDF3 Induces the Translation of m6A-Enriched Gene Transcripts to Promote Breast Cancer Brain Metastasis. Cancer Cell. 2020; 38:857–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Einstein J.M., Perelis M., Chaim I.A., Meena J.K., Nussbacher J.K., Tankka A.T., Yee B.A., Li H., Madrigal A.A., Neill N.J. et al. Inhibition of YTHDF2 triggers proteotoxic cell death in MYC-driven breast cancer. Mol. Cell. 2021; 81:3048–3064. [DOI] [PMC free article] [PubMed] [Google Scholar]