Abstract
Orphan genes are characteristic genomic features that have no detectable homology to genes in any other species and represent an important attribute of genome evolution as sources of novel genetic functions. Here, we identified 445 genes specific to Populus trichocarpa. Of these, we performed deeper reconstruction of 13 orphan genes to provide evidence of de novo gene evolution. Populus and its sister genera Salix are particularly well suited for the study of orphan gene evolution because of the Salicoid whole-genome duplication event which resulted in highly syntenic sister chromosomal segments across the Salicaceae. We leveraged this genomic feature to reconstruct de novo gene evolution from intergenera, interspecies, and intragenomic perspectives by comparing the syntenic regions within the P. trichocarpa reference, then P. deltoides, and finally Salix purpurea. Furthermore, we demonstrated that 86.5% of the putative orphan genes had evidence of transcription. Additionally, we also utilized the Populus genome-wide association mapping panel, a collection of 1,084 undomesticated P. trichocarpa genotypes to further determine putative regulatory networks of orphan genes using expression quantitative trait loci (eQTL) mapping. Functional enrichment of these eQTL subnetworks identified common biological themes associated with orphan genes such as response to stress and defense response. We also identify a putative cis-element for a de novo gene and leverage conserved synteny to describe evolution of a putative transcription factor binding site. Overall, 45% of orphan genes were captured in trans-eQTL networks.
Keywords: orphan genes, de novo gene evolution, genome evolution, whole-genome duplication, gene regulation, synteny
Significance
De novo gene evolution is an important source of new genes and has been shown to be important in adaptive processes and phenotypic novelty. In this study, we identified 445 Populus trichocarpa orphan genes which lack identifiable homology in other species and provided evidence of de novo evolution for 13 of these genes. Additionally, we performed a population-scale analysis of these orphan genes and identify putative regulators associated with their expression. Our study highlights the utility of whole-genome duplications in understanding de novo gene evolution and provides a foundation for future molecular validation of orphan genes in P. trichocarpa.
Introduction
To date, each new species sequenced has contained a cadre of orphan genes (Tautz and Domazet-Lošo 2011). Although detection of orphan genes is highly dependent on the group of species being searched against, they can be defined as any gene that lacks identifiable homologs in any other species. Additionally, some orphan genes when examined in the context of closely related species may show evidence of de novo evolution. De novo genes must arise from ancestrally noncoding sequence (Arendsee et al. 2014).
Initially, there was considerable speculation surrounding the possibility of de novo gene evolution. Both Susumu Ohno and François Jacob supported the duplication and divergence model and thought that de novo gene origination from noncoding sequence was highly unlikely, if not impossible (Ohno 1970; Jacob 1977). It was not until the early 2000s when empirical evidence for de novo evolved genes was available. The first example being five genes that evolved from noncoding sequence in Drosophila (Levine et al. 2006). Following this study, several additional examples of de novo genes were described in humans, plants, and primates (Knowles et al. 2009; Toll-Riera et al. 2009; Xiao et al. 2009).
Here, we provide evidence of de novo-evolved orphan genes in Populus trichocarpa via intragenomic, interspecific, and intergenera syntenic analyses. This novel analysis is possible as a result of the Salicoid whole-genome duplication (WGD) event that all members of the Salicaceae family share (Tuskan et al. 2006; Dai et al. 2014). The Salicoid WGD occurred 58 Ma in the ancestor of Populus and Salix, which was followed by the divergence of Populus and Salix 6 Myr after this WGD event (Dai et al. 2014). Additionally, we used transcriptomic and proteomic analysis across multiple experiments and tissue types to provide evidence of functionality. We also analyzed polymorphism of orphan genes throughout the Populus genome-wide association study (GWAS) mapping population and provide range-wide features of P. trichocarpa orphan genes. Lastly, we performed eQTL mapping to identify putative regulatory networks surrounding orphan genes. From these analyses, we propose novel insights into the mechanisms of de novo gene evolution, evidence of functionality, and characterization at the population scale.
Results
Identification and Curation of De Novo Genes in P. trichocarpa
Orphan genes represent an important aspect of genome evolution and their accurate identification allows for insights into adaptive processes. We first identified orphan genes in P. trichocarpa v3.1 reference genome assembly using several filtering processes, primarily utilizing tools such as BLAST (Altschul 1997) to exclude genes with homologs in the NCBI database. Following the steps described in supplementary figure S1, Supplementary Material online, we identified 445 putative orphan genes in P. trichocarpa, which had no detectable homology to any known genes based on the following analyses. For the initial BLASTP step, we used a total of 63 plant genomes available on the Phytozome database (supplementary table S1, Supplementary Material online). Next, we compared the remaining genes against the conserved domain database and the nonredundant protein database (nr database) (Marchler-Bauer et al. 2015). Finally, we removed candidates with missing open reading frames (ORFs) and then removed genes that had a copy number greater than one in P. trichocarpa to simplify downstream syntenic searches. Next, genes that had assigned gene models or transcriptomic evidence in P. deltoides or Salixpurpurea genomes were removed (supplementary table S11, Supplementary Material online). The remaining 445 genes were classified as specific to P. trichocarpa (supplementary table S2, Supplementary Material online). Within this set of 445, 386 (86.5%) met our threshold for expression based on five RNA-Seq data sets. These included 533 xylem, 529 root, and 470 leaf transcriptomes from the Populus GWAS panel, 438 xylem transcriptomes from a P. trichocarpa x P. deltoides pseudobackcross mapping population and 37 transcriptomes from the Joint Genome Institute (JGI) Plant Gene Atlas database representing various tissue types (supplementary table S3, Supplementary Material online). We also found proteomic evidence for 16 putative orphan genes based on a limited scan of leaf tissue from six P. trichocarpa genotypes from the GWAS mapping panel (Zhang et al. 2018). Next, for each gene, we attempted to locate the nongenic syntenic sequence in P. trichocarpa, P. deltoides, and S. purpurea and only retained genes that had alignments to all syntenic regions. From this analysis, we selected 13 genes that had tell-tale evidence of de novo gene evolution as illustrative cases. All 13 candidates had expression evidence, and two of them had proteomic evidence (supplementary fig. S2, Supplementary Material online).
Reconstruction of De Novo Gene Evolution in P. trichocarpa
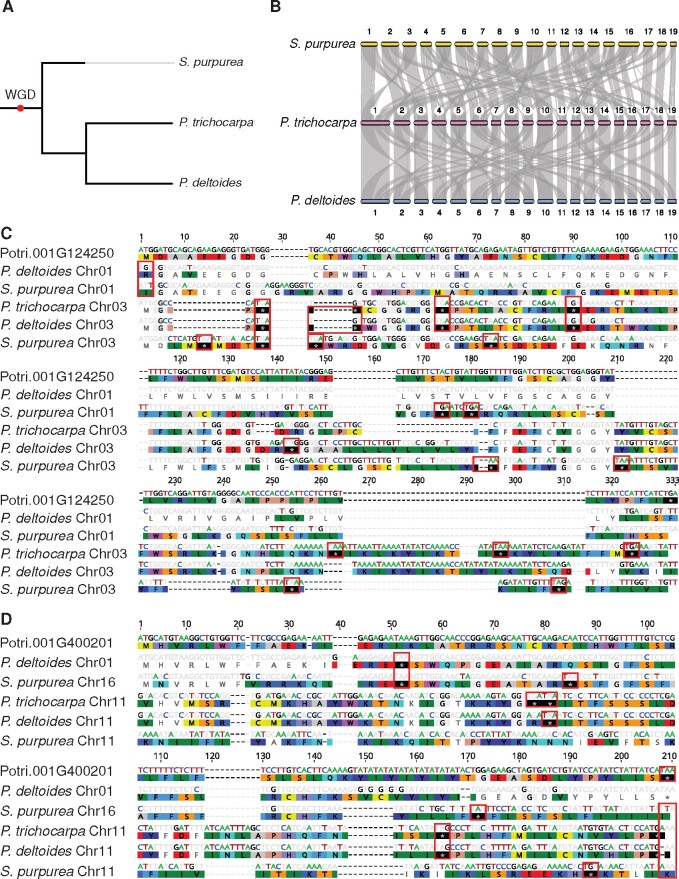
We used the common Salicoid WGD and resulting conserved macrosynteny to explore de novo gene evolution in the ancestral and extant states in Populus and Salix (fig. 1A and B). De novo gene reconstruction relies on identifying enabling mutations in noncoding sequence leading to the formation of the functional ORF. For example, figure 1C profiles a putative de novo gene on chromosome 1 (Chr01) of the P. trichocarpa reference genome. Syntenic blocks of genes exist between chromosome 1 and 3 and these syntenic regions were extracted and aligned. Two hypotheses arose from this alignment. The first is that after the WGD, there were functional ORFs, and over time the ORFs on the syntenic chromosomes accumulated mutations and eroded away, rendering them nonfunctional pseudogenes. The alternative, and more probable hypothesis, is that intergenic sequence was duplicated, and as such, the multiple sister chromosomes or secondary syntenic sequences represent noncoding ancestral states and outgroups that allow for a stepwise analysis of the enabling mutations leading to the evolution of a functional de novo gene. That is, the primary syntenic regions to Potri.001G124250 (P. deltoides Chr01 and S. purpurea Chr01) both have mutations resulting in a nonfunctional start codon, and all five syntenic regions, including P. trichocarpa Chr03, P. deltoides Chr03, and S. purpurea Chr03, have a nonfunctional stop codon (fig. 1C). This hypothesis is further supported by the high number of shared disabling mutations near residue 25, residue 62, and residue 89 as well as the high level of sequence conservation shared between the sister syntenic chromosomes of P. trichocarpa, P. deltoides, and S. purpurea (fig. 1C).
Fig. 1.
A species tree and macrosynteny relationships between P. trichocarpa, P. deltoides, and S. purpurea and alignments of de novo genes Potri.001G124250 and Potri.001G400201 against their respective syntenic intergenic regions in P. deltoides and S. purpurea. Highlighted bases and residues indicate disagreements to the de novo gene in P. trichocarpa. Red boxes indicate disabling mutations in the intergenic syntenic sequence. Both alignments follow the sequential structure of P. trichocarpa (de novo gene), P. deltoides (primary syntenic region), S. purpurea (primary syntenic region), P. trichocarpa (secondary syntenic region), P. deltoides (secondary syntenic region), and S. purpurea (secondary syntenic region). (A) A species tree of the phylogeny of P. trichocarpa, P. deltoides, and S. purpurea, the red point on the root of the tree is indicative of the WGD which occurred in the ancestor of Populus and Salix. (B) Macrosynteny relationships between P. trichocarpa and S. purpurea, and P. trichocarpa and P. deltoides. (C) Alignment of Potri.001G124250 against intergenic syntenic regions. (D) Alignment of Potri.001G400201 against intergenic syntenic regions.
Figure 1D profiles another example, Potri.001G400201, and depicts an alternative example where the gain of a start and stop codon can be observed along with a deletion at base 38 that places a potential stop codon out of frame therefore leading to a de novo gene in P. trichocarpa. We identified similar trends across most of the putative de novo gene alignments. Specifically, percent identity to the de novo gene of interest was highest for genes on the primary noncoding syntenic region (supplementary fig. S3A, Supplementary Material online) and noncoding secondary syntenic sequences were most similar to each other (supplementary fig. S3B, Supplementary Material online). The high similarity in the sister syntenic sequences serves as a useful comparator when examining the steps of de novo gene evolution. Finally, alignments to S. purpurea consistently had even lower identity values compared with P. deltoides, which can be explained by the earlier divergence of Salix and Populus (Dai et al. 2014). For example, average identity across all P. trichocarpa de novo genes compared with the P. deltoides primary syntenic chromosome alignments was 93.1% compared with 59.4% for S. purpurea alignments. Overall, it is evident that the Salicoid WGD provides a valuable resource through highly conserved sister syntenic sequence which allows for a clear understanding of ancestral sequence level changes essential to the evolution of a de novo gene. Alignments and genomic coordinates for other putative de novo genes against their noncoding syntenic regions are available in supplementary figures S4–S14 and table S12, Supplementary Material online.
Direction of selection of the 13 de novo genes in the GWAS mapping panel was assessed with the ratio of piN to piS to infer molecular function (supplementary table S4, Supplementary Material online). Two de novo genes showed evidence of purifying selection (piN/piS < 1) and one showed evidence of neutral selection (piN = piS). The remaining 10 had piN/piS values greater than one, or where piN was greater than piS, which may indicate positive or balancing selection.
De Novo-Evolved Genes Are Polymorphic within the P. trichocarpa Genome-Wide Association Study Population
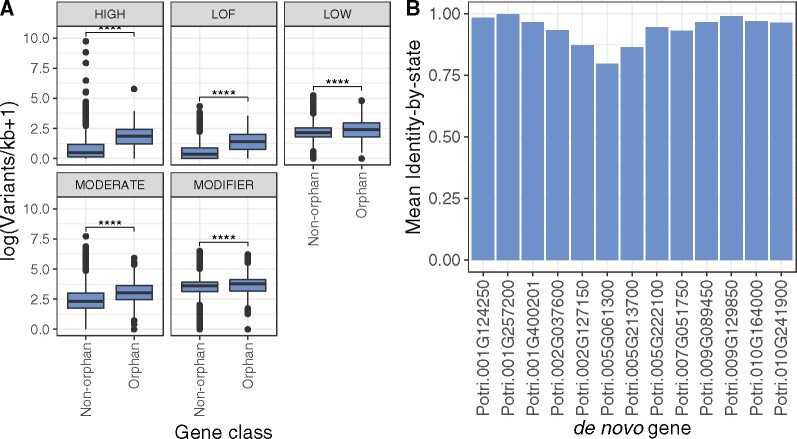
Genomic variant profiles were generated for all 42,950 P. trichocarpa genes and frequency of mutations were compared between nonorphan and orphan genes (fig. 2A). Nonorphan genes are defined as genes excluded as orphan genes as a result of our curation pipeline. For all mutation impact classes, orphan genes had higher frequencies of mutations when compared with nonorphan genes. Orphan genes were not significantly correlated with regions of high or low nucleotide diversity across 1 Mb windows (supplementary fig. S20, Supplementary Material online). Additionally, the subset of 13 de novo genes were inspected in more detail for variants affecting their coding potential. This analysis was performed with 917 unrelated P. trichocarpa individuals (Zhang et al. 2018). Although all 13 genes had high impact mutations, 7 of 13 did not have mutations impacting their ORF suggesting that they were fixed in the population (supplementary table S5, Supplementary Material online). Two examples that deviated substantially from the Nisqually-1 reference genome based on variant profile and had deleterious variants in their coding region were Potri.002G127150 and Potri.005G061300 (fig. 2B). For example, Potri.002G127150 had three nonsynonymous coding single-nucleotide polymorphisms (SNPs) that were homozygous for the alternate allele in greater than 75% of individuals. This same gene also had a small proportion of individuals (0.5%) where the stop codon was lost. Additionally, Potri.005G061300 had a nonsynonymous coding SNP that was homozygous for the alternate allele in 86% of individuals and a homozygous nonsense mutation that resulted in a gained stop codon in 90% of the individuals. On the other hand, de novo genes that more closely matched Nisqually-1’s variant profile included Potri.001G124250, Potri.001G257200, and Potri.009G129850 (fig. 2B). Specifically, nearly all variants in the coding region of Potri.001G124250 were homozygous for the reference allele, with the exception of 5% of individuals that had a mutation resulting in the loss of a stop codon. Additionally, Potri.001G257200, and Potri.009G129850, closely matched the Nisqually-1 variants and at the population level had small numbers of individuals that were homozygous for nonsynonymous or other deleterious mutations. In summary, based on the high similarity of genotype profiles in the GWAS mapping panel to the Nisqually-1 reference genotype profile (the P. trichocarpa genotype where de novo genes were identified) and low frequency of high impact or other deleterious variants, these three genes are relatively homogenous in the GWAS mapping panel.
Fig. 2.
Orphan gene impact class frequency and variant profile similarity to Nisqually-1 (reference genome), (A) Frequency of variant impacts by mutation class (high, LOF = predicted loss of function, low, moderate, and modifier) in 445 orphan and 42,505 nonorphan genes. Nonorphan genes are defined as genes excluded as orphan genes as a result of our curation pipeline (**** = P < 1e−4, Wilcoxon signed rank test). (B) The 13 de novo genes and similarity to Nisqually-1’s (reference genome) genotype profile, represented as mean identity by state.
Populus trichocarpa Orphan Genes Exhibit a Narrow Expression Breadth, Lower Expression Levels, and a Subset Show Evidence of Translation
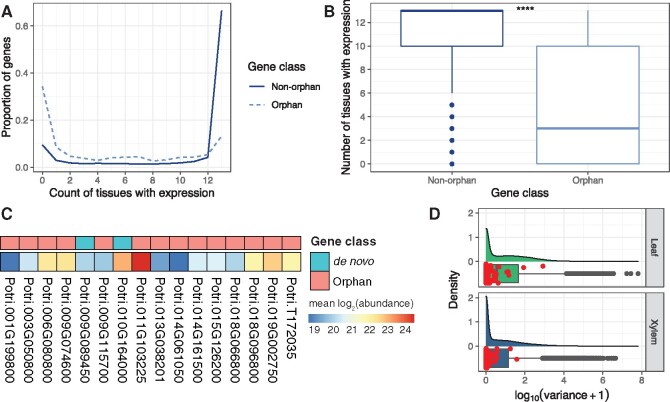
Orphan genes often exhibit lower expression levels and tend to have expression patterns that are relegated to specific tissues, commonly reported in male-biased and/or reproductive tissues (Levine et al. 2006; Cui et al. 2015). Thirteen expression libraries from various tissues (supplementary table S5, Supplementary Material online) were selected from the Populus Gene Atlas project and expression breadth (number of tissues in which transcription evidence is available) was determined for each of the 445 orphan genes. Expression breadth was assessed for both orphan and nonorphan genes. Results indicated that orphan genes were expressed in a smaller number of tissues and had narrower expression breadths (fig. 3A and B). Additionally, as shown in figure 3A, more than 40% of orphan genes were not expressed in the tissues analyzed, compared with less than 20% for nonorphan genes. Nearly 20% of orphan genes were expressed in all 13 tissues, in comparison to nearly 30% for nonorphan genes. To further complement the expression breadth analysis, the tissue specificity index (tau) was calculated using the same 13 tissues above and confirmed that orphan gene expression was more specific to particular tissues when compared with nonorphan genes (supplementary fig. S15, Supplementary Material online). Additionally, expression analysis across the five RNA-Seq data sets described above determined that orphan genes exhibited lower expression levels across all data sets when compared with nonorphan genes (supplementary fig. S16, Supplementary Material online). In addition to transcriptome data, proteomics data were generated from leaf tissue for six P. trichocarpa genotypes from the GWAS mapping panel (fig. 3C). It was determined that 16 orphan genes, including two de novo genes, showed evidence of translation.
Fig. 3.
Orphan gene expression breadth, translation evidence, and expression variation in the GWAS mapping panel. (A) Expression breadth across 13 Populus trichocarpa tissues. (B) Expression breadth distribution across 13 tissues derived from the JGI Gene Atlas project, which is publicly available RNA-Seq data (supplementary table S11, Supplementary Material online) (**** = P < 1e−4, Wilcoxon signed rank test). (C) A total of 16 orphan and de novo genes with proteomic data (MS/MS), shown is the mean of the log2 transformed abundance values across six P. trichocarpa genotypes. (D) Expression variation distribution in xylem (533 genotypes) and leaf (470 genotypes) from publicly available RNA-Seq data (supplementary table S11, Supplementary Material online) for all P. trichocarpa genes with expression evidence, orphan genes are highlighted as red points.
Cis-eQTL Analysis of Orphan Genes Provides Insight into Proximal Regulatory Features
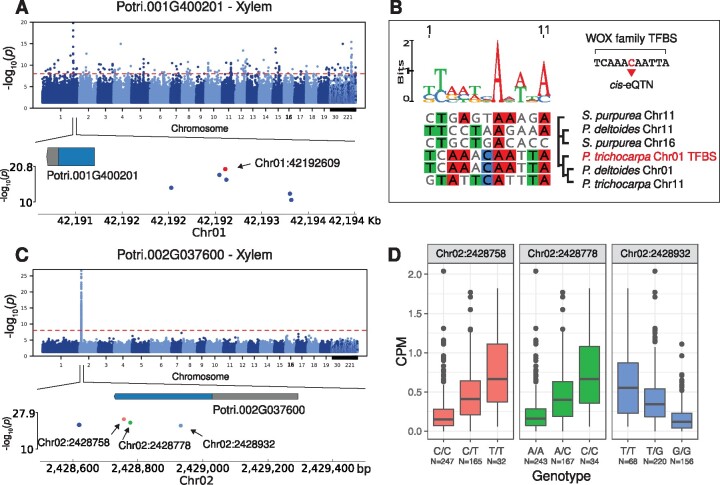
Even though orphan genes had relatively low expression, we observed significant expression variance across the GWAS population sufficient to facilitate expression quantitative trait loci (eQTL) mapping (fig. 3D and supplementary table S3, Supplementary Material online). To that end, eQTL mapping was performed using leaf and xylem transcriptomes to predict putative cis- and trans-regulatory elements underlying orphan gene expression. We identified putative cis-elements for 88 orphan genes that exhibited expression variation in the GWAS mapping panel (supplementary fig. S17A and table S6, Supplementary Material online). Of these 88, 19 had predicted cis-elements that could be aligned to all primary and secondary syntenic regions in P. trichocarpa, P. deltoides, and S. purpurea. Among these 19 was a de novo gene, Potri.001G400201 previously described above. The predicted cis-element of Potri.001G400201 occurred in a 3-kb interval upstream of the transcription start site. The flanking sequence of three SNPs were annotated as putative transcription factor binding site (TFBS) (Chr01:42192609, Chr01:42193303, and Chr01:42193318) based on motif searches. The most significant SNP in the cis-eQTL region of Potri.001G400201 was predicted to fall within a homeobox TFBS (fig. 4A and B). Additionally, after synteny-based reconstruction of this potential TFBS, the P. deltoides primary syntenic chromosome was the only syntenic sequence that had the same predicted homeobox TFBS. The other syntenic sequences either did not have any predicted TFBS (secondary P. deltoides, and primary/secondary S. purpurea) or had an entirely different predicted TFBS (secondary P. trichocarpa).
Fig. 4.
Cis-eQTL analysis facilitates assignment and reconstruction of probable proximal regulatory elements. (A) Potri.001G400201 Manhattan plot depicting the cis-eQTL interval, the red dot is the most significant SNP, with a significant match to a homeobox TFBS. (B) The reconstructed homeobox TFBS from the most significant SNP in Potri.001G400201’s cis-eQTL interval. (C) Potri.002G037600 Manhattan plot depicting the cis-eQTL interval, the three points within the exon are all nonsynonymous SNPs. (D) SNP effect on gene expression for Potri.002G037600, for three exonic SNPs, Chr02:2428758, Chr02:2428788, and Chr02:2428932.
Another de novo gene identified with a cis-eQTL signal was Potri.002G037600. There were three nonsynonymous SNPs within the gene body that were associated with its expression and are likely in linkage disequilibrium with the causal variant (fig. 4C). It is evident that there was a SNP effect on gene expression as the reference genotypes in the first two SNPs (Chr02:2428758 and Chr02:2428788) had lower expression when compared with the homozygous alternate. The opposite was true for the third SNP located in the gene body (Chr02:2428932), where the homozygous reference genotype had higher expression when compared with the homozygous alternate genotype (fig. 4D). Collectively, we identified cis-acting elements for a subset of orphan genes and profiled the proximal regulatory elements of two de novo evolved orphan genes.
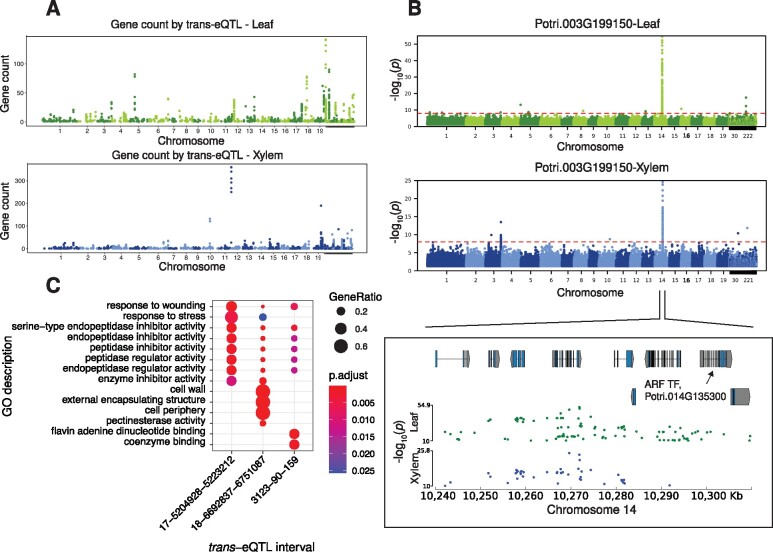
Orphan Genes Are Found in Leaf and Xylem Trans-eQTL Networks
After eQTL mapping, 128 and 136 orphan genes in leaf and xylem, respectively, were shown to be putatively regulated by one or more trans-eQTLs (supplementary tables S7–S9, Supplementary Material online). In these orphan gene sets, four de novo genes, used as illustrative cases above, could be associated with one or more trans-eQTLs. Interestingly, some orphan genes were found to be associated with trans-eQTLs that putatively regulate multiple genes (fig. 5A). Overall, 231 and 221 SNPs within leaf and xylem trans-eQTLs, respectively, had 2 or more orphan genes in their putative regulatory networks (supplementary tables S8 and S9, Supplementary Material online). Some orphan genes had trans-eQTLs that were exclusively found in either leaf (14.5%) or xylem (16.5%) (supplementary fig. S17B, Supplementary Material online). Lastly, a small cohort (2.6%) had overlapping eQTL regions in xylem and leaf (supplementary fig. S17B and table S10, Supplementary Material online). From this small cohort of orphan genes, Potri.003G199150 was an example where the same trans-eQTL on chromosome 14 was predicted in both xylem and leaf transcriptomes (fig. 5B). This example is also particularly interesting because of Potri.014G135300, an auxin response factor (ARF), transcription factor was present within the trans-eQTL interval. The closest homolog to Potri.014G135300 is ARF2 in Arabidopsis and has been shown to be a suppressor of auxin signaling and regulate many important developmental processes (fig. 5D) (Lim et al. 2010).
Fig. 5.
Trans-eQTL analysis of orphan genes in the GWAS mapping panel. (A) Count of genes regulated by putative trans-eQTL (SNPs) in leaf and xylem, all trans-eQTL shown putatively regulate one or more orphan gene. (B) Trans-eQTL analysis in leaf and xylem for Potri.003G199150. (C) Functional enrichment of three trans-eQTL intervals with similar functional roles, all three GO ontologies are represented in this figure.
A total of 37 and 47 trans-eQTL intervals that were associated with expression of more than 15 genes with at least 1 orphan gene in leaf and xylem, respectively, allowing for functional enrichment analyses using both the GO biological process and molecular function ontology (supplementary figs. S18 and S19, Supplementary Material online). Of those coregulated networks with significant enrichment, some shared biological processes across leaf and xylem were apparent. Most notably, those included homeostasis, cell wall-related processes, nucleoside and glycosyl metabolic process, response to wounding and stress, protein modification, ubiquitination, methylation, and alkylation functions. The corresponding GO molecular function processes included calcium ion binding, pectinesterase activity, endoribonuclease activity, enzyme inhibitor activity, serine-type endopeptidase activity, and ubiquitin-protein transferase activity.
It was evident from a comparative GO enrichment in two tissues that orphan genes were present in trans-eQTL-regulated networks that appear responsive to environmental stresses, although they were also present in networks representing diverse functions. One example of response to biotic stress in leaf was evident in three different coregulated networks targeted by trans-eQTLs (Chr17:5204928-5223212, Chr18:6692837-6751087, and scaffold_3123:90-159). These trans-eQTL intervals shared response to wounding as one of their enriched biological processes (fig. 5C). The same coregulated networks also had serine-type endopeptidase inhibitor activity as their most significantly enriched molecular function. The overrepresentation of serine-type endopeptidases likely represents an induced defense mechanism against pathogen proteases (Gottwald et al. 2012). The eQTL interval on Chr17 had a potential regulator, (Potri.017G057500) that is an alkaline ceramidase and has been shown to play an important role in defense response in Arabidopsis (Wu et al. 2015). The remaining two intervals were located in gene deserts and therefore putative regulator assignment was not possible during this study.
Discussion
In this work, we identified 13 orphan genes, which showed evidence of de novo evolution. All 13 of these putative de novo genes were expressed and two showed evidence of translation. Seven of the de novo genes were located on the antisense strand of an existing gene which is consistent with other studies where antisense de novo transcripts were identified and found to be functional (Ardern et al. 2020; Blevins et al. 2021). Additionally, 10 may be under positive or balancing selection, two were under purifying selection, and one was under neutral selection. Based on a recent study of 13 genomes of rice, gene age is positively correlated with the degree of purifying selection, which is also apparent in our results (Stein et al. 2018). In total, 445 orphan genes were identified representing 1% of genes in the P. trichocarpa genome that can be classified as species specific, which is lower than common estimates of 5–15%. However, these estimates vary considerably by species and methodology used for identification (Arendsee et al. 2014). One reason for this lower percentage may be that our curation pipeline was conservative (Vakirlis and McLysaght 2019). Specifically, two primary aspects contributing to this lower percentage include locating missing gene models in P. deltoides and S. purpurea, two close relatives to P. trichocarpa, as well as requiring there be no expression evidence after remapping P. deltoides and S. purpurea RNA-Seq data to the P. trichocarpa genome.
Expression of orphan genes is often relegated to particular tissues when compared with more broadly expressed nonorphan genes. Most commonly, these include male reproductive tissues such as the testis, as well as the brain in humans (Begun et al. 2007; Li et al. 2010; Zhao et al. 2014; Cui et al. 2015). Consistent with previous findings, we also observe significantly lower expression, expression breadth, and higher tissue specificity compared with nonorphan genes. We also examine population-level expression variation of orphan genes in leaf and xylem. Although, orphan genes have considerably lower expression variation when compared with nonorphan genes, we were able to identify strong associations with putative cis-elements and trans-regulatory factors. Future work could further examine the genomic context underlying variation in the P. trichocarpa GWAS population, possibly through epigenetic and pan-genome approaches. In summary, through our extensive use of multitissue and population-level transcriptome data sets, we are able to confirm previously accepted orphan gene expression trends in the context of P. trichocarpa and show there was sufficient expression variation for association studies. We also provide evidence of translation for 16 orphan genes (including two orphan genes, which showed evidence of de novo evolution) based on a subset of six P. trichocarpa genotypes. Although this is a much lower percentage compared with recent studies, a more thorough sampling of the GWAS mapping panel for proteomics analysis and ribosome profiling would allow for further discovery of orphan genes with evidence of translation (Zhang et al. 2019).
Regulatory networks play essential roles in the control of transcription, signaling, and development (Prud’homme et al. 2007). Recently, the process of de novo gene integration into existing networks has been explored in more depth (Majic and Payne 2020). Several studies have examined the integration of de novo genes into existing regulatory networks and rely on sequence homology of known cis-elements (Carvunis et al. 2012; Li et al. 2016). In our study, we expand upon homology-based detection of known regulatory elements through the use cis-eQTL analysis. Through the identification of probable causal SNPs that were associated with gene expression, we identified potential TFBS and provided evidence of network rewiring for the addition of de novo genes. The use of cis-eQTL analysis adds an additional layer of evidence in addition to homology-guided detection of binding sites that the TFBS is likely functional and provides a foundation for future molecular validations.
In addition to gene reconstruction, synteny can also be leveraged to reconstruct the evolution of cis-elements of de novo genes. Cis-elements have been shown to be essential to de novo gene evolution by creating an appropriate context for transcription and integrating new genes into existing networks (Werner et al. 2018; Majic and Payne 2020). Our findings provide clear examples of the evolution of a TFBS in the context of a cis-eQTL study. Although, we do not provide experimental evidence of transcription factor binding, future work could validate these predictions through molecular assays.
Thus far, insight into orphan gene functional repertoire has primarily been limited to examples of molecular validation. To expand and confirm upon known functions provided by molecular studies, we used functional enrichment of putative trans-eQTL regulatory networks that contain orphan genes to assign putative functional roles to orphan genes found in those networks. Our functional enrichment results aligned with well-known functional niches of orphan genes such as response to environmental stress and host–pathogen interaction and uncovered additional functional diversity as well. Collectively, this study captured 45% of P. trichocarpa orphan genes in trans-eQTL networks.
Forest trees are keystone species and have significant environmental importance. P.trichocarpa has an extensive species range which spans considerable abiotic and biotic diversity (Evans et al. 2014). From previously described functional validation in other systems, orphan genes have been shown play essential roles in adaptation and phenotypic novelty (Xiao et al. 2009; Qi et al. 2019). Moreover, the identification of orphan genes in P. trichocarpa may provide a platform for future studies interested in their roles in adaptive processes. By exploring de novo gene evolution through the lens of a WGD event which serves as an outgroup that has extensive syntenic conservation, we were able to concretely provide evidence of a noncoding ancestral state. Furthermore, the methodology developed here may enable future de novo gene studies in species, which lack closely related outgroups but retain highly conserved WGDs. Additionally, through the use of multiomics data available to P. trichocarpa, we could accurately describe orphans’ primary functional niches and regulatory origins. Future work will place an emphasis on empirically validating their function and regulation.
Materials and Methods
Phylogenetics, Synteny, and Selection Analysis
The species tree was constructed with Orthofinder v2.2.6 with default parameters using the primary protein isoform sequences of P. trichocarpa v3.1, P. deltoides v2.1, and S. purpurea v1.1 (Emms and Kelly 2019). Macrosynteny relationships between P. trichocarpa, P. detloides, and S. purpurea were constructed with MCScan using default parameters (JCVI utility libraries v0.8.12) with the primary transcripts of P. trichocarpa v3.1, P. deltoides v2.1, and S. purpurea v1.1 (Tang et al. 2008). Selection analysis (piN/piS) was performed with SNPGenie with 917 individuals, using biallelic SNPs found in the CDS of the de novo gene with a minimum allele frequency of 0.01 (Nelson et al. 2015).
Orphan Gene Curation
The P. trichocarpa v.3.1 genome using the primary transcript from all genes was searched against 63 proteomes (supplementary table S1, Supplementary Material online) available in Phytozome 12 using BLASTP 2.6.0+ with an e-value cutoff of 0.001 (Altschul 1997). This resulted in 1,079 genes that were found to be exclusive to P. trichocarpa. These 1,079 genes were then analyzed with BLASTP 2.6.0+ with an e-value threshold of 0.001 against the NCBI nr database excluding Populus spp., and 32 genes had hits within the NCBI nr database, reducing the count to 1,047. The remaining genes were then analyzed within the Conserved Domain Database, which contains 50,369 PSSM, and resulted in 4 additional hits, and reduced the gene count to 1,043. Genes were then analyzed for their coding intactness, which resulted in 68 orphan genes that had missing start or stop codons, which resulted in 977 total genes. These genes were further analyzed for missing homologous gene models in S. purpurea v.1.0 and P. deltoides v.2.1 genomes with genblastG v1.0.138 (She et al. 2011). An e-value cutoff of 1e−5, coverage threshold of 90%, and identity threshold of 50% was used. This excluded 329 candidate orphan genes that had missing homologous gene models, and resulted in 648 genes. A script was also used to verify the validity of the genblastG gene models and predicted models that did not have start or stop codons, had internal stop codons, or were not divisible by 3 were removed from consideration. Next, 55 genes that were duplicated in clusters of 2 or more removed, which resulted in 593 genes. The remaining 593 genes were then analyzed for expression evidence in P. deltoides D124 and across six tissues in S. purpurea 94006 (supplementary table S11, Supplementary Material online). These RNA-Seq data sets were aligned to P. trichocarpa v3.1 reference genome following the methods described below. Genes that had expression evidence of counts per million (CPM) greater than one in one replicate or greater than zero in two replicates in P. deltoides D124 or across six tissues in S. purpurea were removed, which resulted in 445 genes.
The current P. trichocarpa v4.1 Nisqually-1 reference genome assembly used a homology-based annotation method, which excluded the majority of orphan genes because our curation pipeline specifically eliminated genes that had any detectable homology with genic features in all existing genomic databases. The exception was 24 orphan genes, which were annotated based on homology to gene models in another P. trichocarpa genome assembly, Stettler 14 v1.1 (https://phytozome-next.jgi.doe.gov/info/PtrichocarpaStettler14_v1_1). These 24 did not have homology to any other genes outside of the 2 P. trichocarpa genome assemblies, further supporting our conclusions of species specificity. Regardless of exclusion by annotation methodology, we used genblastG and were able to identify 437 of 445 orphan genes in the v4.1 reference genome suggesting a 98% transfer rate across assemblies.
De Novo Gene Identification
First, MCScanX with default parameters (Wang et al. 2012), was used to generate a collinearity map in the following way, P. trichocarpa versus P. trichocarpa, P. trichocarpa versus P. deltoides, and P. trichocarpa versus S. purpurea (Wang et al. 2012). In order to identify the primary and secondary syntenic chromosomes for the 445 orphan genes, five genes flanking the orphan gene were used to search each collinearity map. Some genes were missing their primary and secondary syntenic regions and following this step only 250 genes had syntenic regions in P. trichocarpa, P. deltoides, and S. purpurea. Next, whole-genome alignments were generated with nucmer from the Mummer4 package with the same species comparisons as above (Marçais et al. 2018). The boundary coordinates of the five flanking genes were then used to extract the orphan gene region and using the identified primary and secondary chromosomes, the synteny map was split into primary and secondary syntenic maps. Next, Synder 0.28.0 was used to identify the expected syntenic region of the candidate de novo gene within the primary and secondary syntenic map (Arendsee et al. 2019). The resulting region was then filtered to include the highest scoring interval that was at least the size of the candidate de novo gene and less than 125 kb, which resulted in 202 genes, which had intervals in the expected syntenic regions. The regions were extracted and compiled with the candidate de novo gene and aligned with MAFFT linsi v7.407 with default parameter settings (Katoh and Standley 2013). To ensure that the primary and secondary syntenic regions were nongenic, we utilized multiple lines of evidence: the syntenic region was required to not overlap a gene model (annotated by JGI) on the same strand second, the use of genblastG did not result in a reasonable gene model based on the thresholds above, the absence of expression of the query de novo gene after remapping RNA-Seq data from both P. deltoides and S. purpurea after applying the thresholds above, and identification of shared disabling mutations in the syntenic alignments. This analysis resulted in 13 de novo genes that had high-quality alignments. Upon further inspection of the nongenic syntenic sequence with BLASTN 2.6.0 + (Altschul 1997), seven de novo genes could be classified as having their origins in overlapping existing gene features on the antisense strand whereas the remaining six are from intergenic regions.
SNP Effect and Polymorphism Analysis in 917 P. trichocarpa Individuals
A vcf with the variant calls from 917 P. trichocarpa individuals was annotated with SnpEff 4.3t using the P. trichocarpa v.3.1 GFF3 file (available at https://phytozome.jgi.doe.gov/pz/portal.html) (Cingolani et al. 2012). To determine the variant frequency by mutation class, gene regions were extracted with bcftools v1.9, each mutation class count was divided by the gene length, and then multiplied by 1000. For similarity to Nisqually-1, gene regions were extracted with bcftools v1.9 (Li 2011). Then, Plink v1.90 was used to calculate pairwise identity by state with default parameter settings, multiallelic SNPs were excluded (Purcell et al. 2007). Identity by state was calculated for each of the 13 de novo genes. Nucleotide diversity was calculated with the same vcf as above with VCFtools v0.1.16 with a window size of 1 Mb (Danecek et al. 2011).
Mass Spectrometry of Six P. trichocarpa Genotypes
Protein Extraction and Digestion
Six genotypes, BESC-377, BESC-907, BESC-901, BESC-886, BESC-900 and Nisqually-1, from the P. trichocarpa GWAS population were selected for analysis by mass spectrometry. Each genotype had three technical replicates. One hundred milligrams of leaf tissue was ground with two 5-mm stainless steel grinding beads in the Qiagen TissueLyser twice for 30 s at 30 Hz. Ground tissue pellets were suspended in sodium dodecyl sulfate lysis buffer (2% in 100 mM of NH4HCO3, 10 mM DTT). Samples were physically disrupted by bead beating (0.15 mm) at 8000 rpm for 5 min. Crude lysates were boiled for 5 min at 90 °C and then samples were adjusted to 30 mM IAA and incubated in the dark for 15 min at room temperature to avoid disulfide bridge reformation. Proteins were precipitated using a chloroform/methanol/water extraction. Dried protein pellets were resolubilized in 2% (w/v) sodium deoxycholate (SDC) (100 mM NH4HCO3) and protein amounts were estimated by performing a BCA assay (Pierce Biotechnology). For each sample, an aliquot of approximately 500 µg of protein was digested via two aliquots of sequencing-grade trypsin (Promega, 1:75 [w: w]) at two different sample dilutions, (overnight) followed by incubating 3 h at 37 °C. The peptide mixture was adjusted to 0.5% formaldehyde to precipitate SDC. Hydrated ethyl acetate was added to each sample at a 1:1 [v:v] ratio three times to effectively remove SDC. Samples were then placed in a SpeedVac Concentrator (Thermo Fischer Scientific) to remove ethyl acetate and further concentrate the sample. The peptide-enriched flow through was quantified using the BCA assay, desalted on RP-C18 stage tips (Pierce Biotechnology) and then stored at −80 °C prior to liquid chromatography (LC)-MS/MS analysis.
LC-MS/MS Analysis
All samples were analyzed on a Q Exactive Plus mass spectrometer (Thermo Fisher Scientific) coupled with a Proxeon EASY-nLC 1200 LC pump (Thermo Fisher Scientific). Peptides were separated on a 75-μm inner diameter microcapillary column packed with 25 cm of Kinetex C18 resin (1.7 μm, 100 Å, Phenomenex). For each sample, a 2-μg aliquot was loaded in buffer A (0.1% formic acid, 2% acetonitrile) and eluted with a linear 150 min gradient of 2–20% of buffer B (0.1% formic acid, 80% acetonitrile), followed by an increase in buffer B to 30% for 10 min, another increase to 50% buffer for 10 min and concluding with a 10-min wash at 98% buffer A. The flow rate was kept at 200 nl/min. MS data were acquired with the Thermo Xcalibur software v2.2, a topN method where N could be up to 15. Target values for the full-scan MS spectra were 1 × 106 charges in the 300–1,500 m/z range with a maximum injection time of 25 ms. Transient times corresponding to a resolution of 70,000 at m/z 200 were chosen. A 1.6-m/z isolation window and fragmentation of precursor ions was performed by higher-energy C-trap dissociation with a normalized collision energy of 27 eV. MS/MS sans were performed at a resolution of 17,500 at m/z 200 with an ion target value of 1 × 106 and a maximum injection time of 50 ms. Dynamic exclusion was set to 30 s to avoid repeated sequencing of peptides.
Peptide Identification and Protein Inference
MS raw data files were searched against the P. trichocarpa v3.0 reference FASTA database to which mitochondrial and chloroplast-encoded proteins had been added. A list of common protein contaminants (e.g., keratin) were appended to the reference database. A decoy database, consisting of the reversed sequences of the target database, was appended to discern the false-discovery rate (FDR) at the spectral level. For standard database searching, the peptide fragmentation spectra (MS/MS) were analyzed by the Crux pipeline v3.0 (McIlwain et al. 2014). The MS/MS were searched using the Tide algorithm and was configured to derive fully tryptic peptides using default settings except for the following parameters: allowed clip nterm-methionine, a precursor mass tolerance of 10 ppm, a static modification on cysteines (iodoacetamide; +57.0214 Da), and dynamic modifications on methionine (oxidation; 15.9949). The results were processed by Percolator to estimate q values. Peptide spectrum matches and peptides were considered identified at a q-value <0.01. Across the entire experimental data set, proteins were required to have at least two distinct peptide sequences and two minimum spectra per protein. All proteomics spectral data in this study were deposited at ProteomeXchange Consortium via the MASSIVE repository (https://massive.ucsd.edu/). The data can be reviewed under the username “MSV000087050_reviewer” and password “muchero.”
Protein Quantification
For label-free quantification, MS1-level precursor intensities were derived from MOFF (Argentini et al. 2016) using the following parameters: 10 ppm mass tolerance, retention time window for extracted ion chromatogram was 3 min, time window to get the apex for MS/MS precursor was 30 s. Protein intensity-based values, which were calculated by summing together quantified peptides, normalized by dividing by protein length and then LOESS and median central tendency procedures were performed on log2-transformed data using the freely available software Perseus (http://www.perseus-framework.org). Missing values were replaced by random numbers drawn from a normal distribution (width = 0.3 and downshift = 2.8).
RNA-Seq and Data Analysis
For RNA extraction procedures, refer to Zhang et al. (2018). Raw RNA-Seq reads were filtered and trimmed using the JGI QC pipeline. BBDuk (https://sourceforge.net/projects/bbmap/) was used to evaluate raw reads were for sequence artifacts by kmer matching (kmer = 25) allowing 1 mismatch and detected artifacts were trimmed from the 3′ end of the reads. RNA spike-in reads, PhiX reads, and reads containing any Ns were removed. Quality trimming was performed using the phred trimming method set at Q6. Following trimming, reads under the length threshold were removed (minimum length 25 bases or 1/3 of the original read length; whichever was longer). Raw reads from each library were aligned to the P. trichocarpa v3.1 reference genome using STAR v2.6.1b (Dobin et al. 2013). FeatureCounts 1.6.3 in stranded mode was used to generate raw gene counts, excluding multimapping reads (Liao, et al. 2014). For the QTL mapping pedigree, eQTL xylem, root, and leaf samples, and the P. trichocarpa GeneAtlas data set, EdgeR 3.24.3 was used to generate scaling factors and was subsequently converted to CPM to account for RNA composition and library size between samples (Robinson et al. 2010). Orphan genes were considered to be expressed if CPM was greater than one in one data set or greater than zero in two data sets. The JGI Plant Gene Atlas can be found at https://phytozome.jgi.doe.gov/phytomine/aspect.do?name=Expression. The Salix data set was processed with the same JGI QC pipeline, the raw reads were then mapped against the P. trichocarpa v3.1 reference with STAR v2.6.1b, and raw counts were generated with featureCounts in unstranded mode, excluding multimapping reads, and converted to CPM. For population-level expression variation, the statistic used is variance. For the tissue specificity index (tau), at least one tissue was required to have a CPM value of greater than 1, the script used for this analysis is available in Le Béguec et al. (2018). Refer to supplementary table S11, Supplementary Material online for all SRA identifiers for RNA-Seq data used.
eQTL Analysis
Whole-genome sequencing, short variant discovery, and functional annotation of 545 P. trichocarpa individuals are described in Evans et al. (2014). The exact same analysis pipeline was used in this study, with the exception being that 917 individuals were used. This SNP data set is available at http://bioenergycenter.org/besc/gwas/. A total of 390 and 444 RNA-Seq samples in leaf and xylem, respectively, were used to perform eQTL analysis with EMMAX v20120210 with default parameters (Kang et al. 2010). All genes with expression evidence were used in the eQTL analysis, which is 40,301 in leaf and 39,380 in xylem. Association results were then filtered with a threshold of P value less than or equal to 1e-10, followed by bedtools merge with a distance of 100 kb to extract the eQTL interval (Quinlan and Hall 2010). Additionally, in order for the eQTL interval to be considered significant, at least five SNPs needed to be present in the peak. eQTLs on different chromosomes than the target gene were considered to be trans-eQTL, and eQTLs on the same chromosome within 1 Mb of the target gene were classified as cis-eQTLs. TFBS were reconstructed with synteny via same methods described above. TFBS were identified by adding a flanking sequence (5 bp) to target SNPs of interest, which was then searched with FIMO v4.11.2 using default parameters (Grant et al. 2011) against the PlantPAN v3.0 position weight matrix (Chow et al. 2019). Functional enrichment of the networks with orphan genes was performed with ClusterProfiler v3.14.3 enrichGO function, P values were adjusted with Benjamini–Hochberg correction (Yu et al. 2012).
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This research was supported by the United States Department of Energy’s Office of Science Early Career Research Program under the Biological and Environmental Research office and, in part, by the Plant-Microbe Interfaces Scientific Focus Area in the Genomic Science Program, and by the Center for Bioenergy Innovation at Oak Ridge National Laboratory. Oak Ridge National Laboratory is managed by UT-Battelle, LLC for the U.S. Department of Energy under Contract Number DE-AC05-00OR22725. Part of this work was performed at the Oak Ridge Leadership Computing Facility (OLCF) including resources of the Compute and Data Environment for Science (CADES) at Oak Ridge National Laboratory. The work conducted by the U.S. Department of Energy Joint Genome Institute was supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. We thank Dr Lawrence Smart and collaborators for prepublication use of the Salix purpurea v1.0 genome and Salix purpurea RNA-Seq data sets. We thank the Department of Energy Joint Genome Institute and colleagues at CBI for prepublication access to RNA-seq data sets and Populus deltoides WV94 v2.1. We thank collaborators at the Oak Ridge National Laboratory, Duke University, and the DOE Joint Genome Institute for access to the prepublication access to the genome and annotation of Sphagnum fallax v0.5. We thank Xiaohan Yang and the Department of Energy Joint Genome Institute for prepublication access to Kalanchoe laxiflora v1.1. We thank the Department of Energy Joint Genome Institute and collaborators for prepublication access to Phaseolus vulgaris v2.1.
Author Contributions
T.B.Y. and W.M. designed this study. T.B.Y conducted data analysis and wrote the manuscript. V.S., K.B., and A.L. analyzed and curated the RNA-Seq and WGS data. S.J.S. and L.G. generated data. K.F. performed variant calling and performed data analysis. T.B.Y and J.Z. performed eQTL analysis. C.P. and P.R. contributed to data analysis. P.E.A. generated and analyzed proteomic data. T.B.Y, P.E.A., J.S., J.-G.C, G.A.T., and W.M. revised the manuscript. All authors read and approved the final manuscript.
Data Availability
The data underlying this article are available in its Supplementary Material online.
Notice: This manuscript has been authored by UT-Battelle, LLC under Contract No. DE-AC05-00OR22725 with the U.S. Department of Energy. The United States Government retains and the publisher, by accepting the article for publication, acknowledges that the United States Government retains a nonexclusive, paid-up, irrevocable, worldwide license to publish or reproduce the published form of this manuscript, or allow others to do so, for United States Government purposes. The Department of Energy will provide public access to these results of federally sponsored research in accordance with the DOE Public Access Plan (http://energy.gov/downloads/doe-public-access-plan).
Literature Cited
- Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25(17):3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardern Z, Neuhaus K, Scherer S.. 2020. Are antisense proteins in prokaryotes functional? Front Mol Biosci. 7:187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendsee Z, et al. 2019. Synder: inferring genomic orthologs from synteny maps. bioRxiv. 554501. doi: 10.1101/554501. [DOI]
- Arendsee ZW, Li L, Wurtele ES.. 2014. Coming of age: orphan genes in plants. Trends Plant Sci. 19(11):698–708. [DOI] [PubMed] [Google Scholar]
- Argentini A, et al. 2016. MoFF: a robust and automated approach to extract peptide ion intensities. Nat Methods. 13(12):964–966. [DOI] [PubMed] [Google Scholar]
- Begun DJ, Lindfors HA, Kern AD, Jones CD.. 2007. Evidence for de novo evolution of testis-expressed genes in the Drosophila yakuba/Drosophila erecta clade. Genetics 176(2):1131–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blevins WR, et al. 2021. Uncovering de novo gene birth in yeast using deep transcriptomics. Nat Commun. 12(1):604–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvunis A-R, et al. 2012. Proto-genes and de novo gene birth. Nature 487(7407):370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow CN, et al. 2019. Plantpan3.0: a new and updated resource for reconstructing transcriptional regulatory networks from chip-seq experiments in plants. Nucleic Acids Res. 47(D1):D1155–D1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, et al. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6(2):80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, et al. 2015. Young genes out of the male: an insight from evolutionary age analysis of the pollen transcriptome. Mol Plant. 8(6):935–945. [DOI] [PubMed] [Google Scholar]
- Dai X, et al. 2014. The willow genome and divergent evolution from poplar after the common genome duplication. Cell Res. 24(10):1274–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, et al. 2011. The variant call format and VCFtools. Bioinformatics 27(15):2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, et al. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S.. 2019. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20(1):1–4. [DOI] [PMC free article] [PubMed]
- Evans LM, et al. 2014. Population genomics of Populus trichocarpa identifies signatures of selection and adaptive trait associations. Nat Genet. 46(10):1089–1096. [DOI] [PubMed] [Google Scholar]
- Gottwald S, Samans B, Lück S, Friedt W.. 2012. Jasmonate and ethylene dependent defence gene expression and suppression of fungal virulence factors: two essential mechanisms of Fusarium head blight resistance in wheat? BMC Genomics. 13:369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, Noble WS.. 2011. FIMO: Scanning for occurrences of a given motif. Bioinformatics 27(7):1017–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F.1977. Evolution and tinkering. Science 196(4295):1161–1166. [DOI] [PubMed] [Google Scholar]
- Kang HM, et al. 2010. Variance component model to account for sample structure in genome-wide association studies. Nat Genet. 42(4):348–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles DG, Mclysaght A, Knowles DG, Mclysaght A.. 2009. Recent de novo origin of human protein-coding genes. Genome Res. 19(10):1752–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Béguec C, et al. 2018. Characterisation and functional predictions of canine long non-coding RNAs. Sci Rep. 8(1):13444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine MT, Jones CD, Kern AD, Lindfors HA, Begun DJ.. 2006. Novel genes derived from noncoding DNA in Drosophila melanogaster are frequently X-linked and exhibit testis-biased expression. Proc Natl Acad Sci USA. 103(26):9935–9939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CY, et al. 2010. A human-specific de novo protein-coding gene associated with human brain functions. PLoS Comput Biol. 6(3):e1000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27(21):2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z-W, et al. 2016. On the origin of de novo genes in Arabidopsis thaliana populations. Genome Biol Evol. 8(7):2190–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W.. 2014. FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30(7):923–930. [DOI] [PubMed] [Google Scholar]
- Lim PO, et al. 2010. Auxin response factor 2 (ARF2) plays a major role in regulating auxin-mediated leaf longevity. J Exp Bot. 61(5):1419–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majic P, Payne JL.. 2020. Enhancers facilitate the birth of de novo genes and gene integration into regulatory networks. Mol Biol Evol. 37(4):1165–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marçais G, et al. 2018. MUMmer4: a fast and versatile genome alignment system. PLoS Comput Biol. 14(1):e1005944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, et al. 2015. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 43(Database issue):D222–D226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIlwain S, et al. 2014. Crux: rapid open source protein tandem mass spectrometry analysis. J Proteome Res. 13(10):4488–4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CW, Moncla LH, Hughes AL.. 2015. SNPGenie: estimating evolutionary parameters to detect natural selection using pooled next-generation sequencing data. Bioinformatics 31(22):3709–3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S.1970. The enormous diversity in genome sizes of fish as a reflection of nature’s extensive experiments with gene duplication. Trans Am Fish Soc. 99(1):120–130. [Google Scholar]
- Prud’homme B, Gompel N, Carroll SB.. 2007. Emerging principles of regulatory evolution. Light Evol. 1:109–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 81(3):559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi M, et al. 2019. QQS orphan gene and its interactor NF-YC4 reduce susceptibility to pathogens and pests. Plant Biotechnol J. 17(1):252–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM.. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26(6):841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK.. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26(1):139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She R, et al. 2011. genBlastG: using BLAST searches to build homologous gene models. Bioinformatics 27(15):2141–2143. [DOI] [PubMed] [Google Scholar]
- Stein JC, et al. 2018. Genomes of 13 domesticated and wild rice relatives highlight genetic conservation, turnover and innovation across the genus Oryza. Nat Genet. 50(2):285–296. [DOI] [PubMed] [Google Scholar]
- Tang H, et al. 2008. Synteny and collinearity in plant genomes. Science 320(5875):486–488. [DOI] [PubMed] [Google Scholar]
- Tautz D, Domazet-Lošo T.. 2011. The evolutionary origin of orphan genes. Nat Rev Genet. 12(10):692–702. [DOI] [PubMed] [Google Scholar]
- Toll-Riera M, et al. 2009. Origin of primate orphan genes: a comparative genomics approach. Mol Biol Evol. 26(3):603–612. [DOI] [PubMed] [Google Scholar]
- Tuskan GA, et al. 2006. The genome of black cottonwood. Science 313(5793):1596–1605. [DOI] [PubMed] [Google Scholar]
- Vakirlis N, McLysaght A.. 2019. Computational prediction of de novo emerged protein-coding genes. In: Sikosek T, editor. Computational methods in protein evolution. New York (NY): Humana Press. p. 63–81. [DOI] [PubMed] [Google Scholar]
- Wang Y, et al. 2012. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40(7):e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner MS, et al. 2018. Young genes have distinct gene structure, epigenetic profiles, and transcriptional regulation. Genome Res. 28(11):1675–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JX, et al. 2015. The Arabidopsis ceramidase AtACER functions in disease resistance and salt tolerance. Plant J. 81(5):767–780. [DOI] [PubMed] [Google Scholar]
- Xiao W, et al. 2009. A rice gene of de novo origin negatively regulates pathogen-induced defense response. PLoS One. 4(2):e4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang LG, Han Y, He QY.. 2012. ClusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16(5):284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, et al. 2018. Genome-wide association studies and expression-based quantitative trait loci analyses reveal roles of HCT2 in caffeoylquinic acid biosynthesis and its regulation by defense-responsive transcription factors in Populus. New Phytol. 220(2):502–516. [DOI] [PubMed] [Google Scholar]
- Zhang L, et al. 2019. Rapid evolution of protein diversity by de novo origination in Oryza. Nat Ecol Evol. 3(4):679–690. [DOI] [PubMed] [Google Scholar]
- Zhao L, Saelao P, Jones CD, Begun DJ.. 2014. Origin and spread of de novo genes in Drosophila melanogaster populations. Science 343(6172):769–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in its Supplementary Material online.