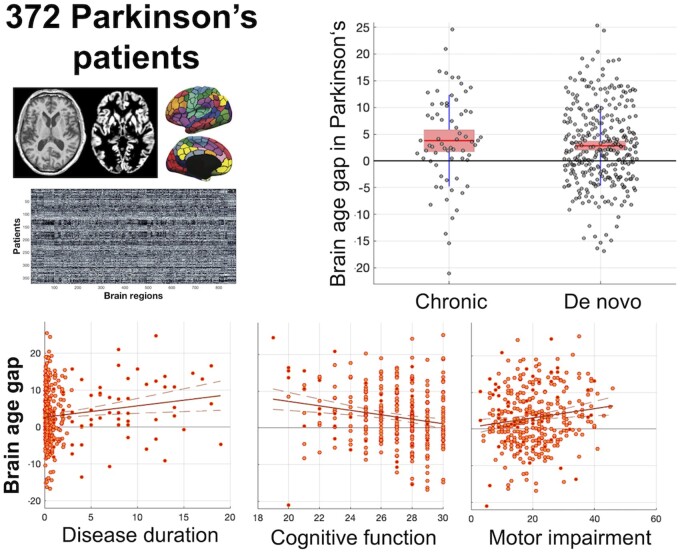
Abstract
Machine learning can reliably predict individual age from MRI data, revealing that patients with neurodegenerative disorders show an elevated biological age. A surprising gap in the literature, however, pertains to Parkinson’s disease. Here, we evaluate brain age in two cohorts of Parkinson’s patients and investigated the relationship between individual brain age and clinical characteristics. We assessed 372 patients with idiopathic Parkinson’s disease, newly diagnosed cases from the Parkinson’s Progression Marker Initiative database and a more chronic local sample, as well as age- and sex-matched healthy controls. Following morphometric preprocessing and atlas-based compression, individual brain age was predicted using a multivariate machine learning model trained on an independent, multi-site reference sample. Across cohorts, healthy controls were well predicted with a mean error of 4.4 years. In turn, Parkinson’s patients showed a significant (controlling for age, gender and site) increase in brain age of ∼3 years. While this effect was already present in the newly diagnosed sample, advanced biological age was significantly related to disease duration as well as worse cognitive and motor impairment. While biological age is increased in patients with Parkinson’s disease, the effect is at the lower end of what is found for other neurological and psychiatric disorders. We argue that this may reflect a heterochronicity between forebrain atrophy and small but behaviourally salient midbrain pathology. Finally, we point to the need to disentangle physiological ageing trajectories, lifestyle effects and core pathological changes.
Keywords: Parkinson’s, machine learning, age, atrophy, prediction
Eickhoff et al., analysed ‘brain age’, in two samples of Parkinson’s patients (de-novo and chronic). They found a significant increase in biological compared to chronological age of ∼3 years, which was already present in de-novo patients and significantly related to disease duration as well as worse cognitive and motor impairment.
Graphical Abstract
Graphical Abstract.
Introduction
Notwithstanding individual variability, the human brain ages in a systematic fashion, allowing machine learning models to robustly predict individual age from in-vivo imaging data with an accuracy of 3–5 years [mean absolute error (MAE)] for new subjects.1 The individual deviation of the biological ‘brain age’ from chronological age, the brain age gap (BAG), is now widely recognized as a summary marker of brain health.2 A negative BAG implies that a person’s brain appears younger than expected given the chronological age. In turn, a positive BAG, i.e. older appearing brains, are observed in patients with mild cognitive impairment (MCI), Alzheimer’s disease3–5 or healthy subjects with poorer cognitive performance.6–8 A recent large-scale analysis of ∼11 000 patients then showed elevated brain age in a broad range of neuro-psychiatric disorders.9 This suggests that BAG may be a sensitive albeit likely non-specific marker of not only neurodegeneration but brain pathology per se.
A surprising gap in the literature, however, pertains to the second most common neurodegenerative disorder10: Parkinson’s disease, with the only available investigation reporting a BAG of 1.5 years11 by a model that yielded a BAG of >9 years for patients with Alzheimer’s disease. While macroscopic brain atrophy in Parkinson’s disease has been discussed controversially and is a criterion for individual diagnosis,12 a recent meta-analysis of morphometric studies provided evidence for robust atrophy in Parkinson’s disease patients.13 Mirroring other neurodegenerative disorders, we would thus expect a positive BAG and, considering the progressive nature of Parkinson’s disease, a relationship to disease duration and severity.
In this study, we tested these hypotheses using a cohort of 372 Parkinson’s disease patients stemming from two sources, the Parkinson’s Progression Marker Initiative (PPMI)14 and a local sample. This not only allows us to test replicability of any findings, but also yields complementary insight. The PPMI provides a large multi-site sample of recently diagnosed Parkinson’s disease patients, which should provide a robust basis for generalized conclusions on potential brain age advancement in de-novo Parkinson’s patients. In addition, the PPMI provides a consistent set of clinical covariates across all subjects. Yet, owing to the focus on early biomarkers, more chronically ill patients are missing in this database. Hence, we aimed to replicate and extend the findings obtained from the PPMI in a local dataset reflecting a substantially broader range of progression.
We thus (i) assessed significant (univariate) differences in local grey matter volume (GMV) between patients with Parkinson’s disease and controls, (ii) predicted individual brain age by a model trained on an independent multi-site dataset and (iii) computed the BAG. The ensuing scores were then (iv) compared between patients with Parkinson’s disease and controls, and (v) correlated with individual disease duration as well as motor [Unified Parkinson’s Disease Rating Scale (UPDRS-III) motor scores] and cognitive impairment [Montreal Cognitive Assessment (MoCA)].
Materials and methods
Study participants
We analysed structural, T1 weighted MRI scans (standard MPRAGE, 3 T scanners) from 372 patients with idiopathic Parkinson’s disease (304 from PPMI and 68 from local samples) and 172 age- and sex-matched healthy controls (HCs, 101 from PPMI and 71 local) as summarized in Table 1. All patients were examined by an experienced movement disorders specialist. Diagnosis of idiopathic Parkinson’s disease was based on the UK Parkinson’s Disease Society Brain Bank Clinical Diagnostic Criteria15 following a thorough diagnostic work-up, including neurological examination, medical history and family anamnesis, neuropsychiatric assessment and positive levodopa response. In turn, non-idiopathic Parkinsonism was an exclusion criterion. Patients from the local sample were under their long-term dopaminergic treatment with individual drug regimens optimized by the attending neurologist. In turn, PPMI patients were unmedicated and not expected to require Parkinson’s disease medication within at least 6 months per the study protocol. Exclusion criteria for all subjects (patients and controls) were clinical dementia, major depression as well as any other physical, psychological, or medical condition that would interfere with the conduction of the study. Finally, standard contraindications for MRI such as pacemakers, metal implants, pregnancy, extreme obesity or claustrophobia likewise applied to all subjects. All subjects gave informed consent after approval of the study by the local ethics committees. Joint reanalysis was approved by the ethics committee of the University of Düsseldorf.
Table 1.
Demographic and clinical summary data
| PD | HC | P (PD vs HC) | ||
|---|---|---|---|---|
| Age | All | 62.7 ± 8.4 | 62.8 ± 8.3 | 0.861 |
| PPMI | 62.4 ± 8.2 | 63.6 ± 8.7 | 0.202 | |
| Local | 63.9 ± 9.0 | 61.7 ± 7.7 | 0.118 | |
| P (PPMI vs Local) | 0.191 | 0.124 | ||
| M/F | All | 243/129 | 108/64 | 0.566 |
| PPMI | 199/105 | 70/31 | 0.478 | |
| Local | 44/24 | 38/33 | 0.180 | |
| P (PPMI vs Local) | 0.954 | 0.108 | ||
| Disease duration (years) | All | 1.75 ± 3.39 | ||
| PPMI | 0.55 ± 0.54 | |||
| Local | 7.30 ± 5.09 | |||
| P (PPMI vs Local) | <0.001 | |||
| Hoehn and Yahr stage | All | 1.71 ± 0.67 | ||
| PPMI | 1.58 ± 0.51 | |||
| Local | 2.28 ± 0.97 | |||
| P (PPMI vs Local) | <0.001 | |||
| MoCA (adjusted for education) | All | 26.88 ± 2.44 | ||
| PPMI | 27.15 ± 2.19 | |||
| Local | 24.51 ± 3.14 | |||
| P (PPMI vs Local) | <0.001 | |||
| UPDRS-III | All | 20.34 ± 8.75 | ||
| PPMI | 20.75 ± 8.53 | |||
| Local | 18.19 ± 9.59 | |||
| P (PPMI vs Local) | 0.041 | |||
| LED | PPMI | None | ||
| Local | 679 ± 488 |
LED, Levodopa-equivalent dose; MoCA, montreal cognitive assessment; PD: Parkinson’s disease; UPDRS, Unified Parkinson’s Disease Rating Scale.
Motor scores were assessed in the pharmacological ‘ON’ state with patients on their regular medication. Disease duration corresponds to time since diagnosis. Data shown are number of subjects or mean ± standard deviation, respectively. For relationships between clinical covariates and chronological age, please refer to Supplementary Table 1.
Data availability statement
The PPMI dataset may be downloaded following application for access under http://PPMI-info.org/. The local data cannot be openly shared due to ethical restrictions and missing consent for sharing from the participants.
Data processing and representation
Structural MRI imaging for the local sample was performed using a T1-weighted MPRAGE sequence (3 T Siemens Trio scanner; TR = 2.3 s, TE = 2.96 ms, TI = 900 ms, flip-angle = 9°, FoV = 240 × 256 mm2 sagittal plane, slices = 160, voxel size = 1 × 1 × 1 mm³). The parameters for the local sample were thus in line with the PPMI protocol requiring an isotropic in-plane resolution of 1 × 1 mm³ and a maximum slice-thickness of 1.2 mm without gap while other parameters including repetition and echo time should follow the manufacturer’s recommendations for a T1-weighted, 3D sequence at each site.
After visual inspection for quality assurance, T1 scans were processed using the Computational Anatomy Toolbox (CAT12.5, http://www.neuro.uni-jena.de/cat/), including bias-field correction, noise removal and skull stripping, normalization to MNI space and tissue segmentation. Modulation by the amount of volume changes needed to match the individual brain to the template by non-linear registration then provided per-subject maps of local GMV adjusted for head size. We note that our approach of using only the Jacobian of the non-linear deformations for modulation is in effect very similar to modulating by the affine and non-linear deformations and subsequently adjusting for total intracranial volume but may be more robust as it does not rely on a full and correct estimation of the CSF component during segmentation. One of the challenges for machine learning on brain imaging is the high (nominal) dimensionality of voxel-wise data yielding a poor feature-to-sample ratio. Capitalizing on the topographic organization of the brain into distinct areas,16 we thus followed an atlas-based strategy for biologically informed compression based on 800 cortical,17 36 subcortical18 and 37 cerebellar19 grey matter regions. For all analyses, individual grey matter anatomy was hence represented by 873 features, each reflecting the winsorized mean of the voxel-wise GMV values for a given area.
Brain age prediction
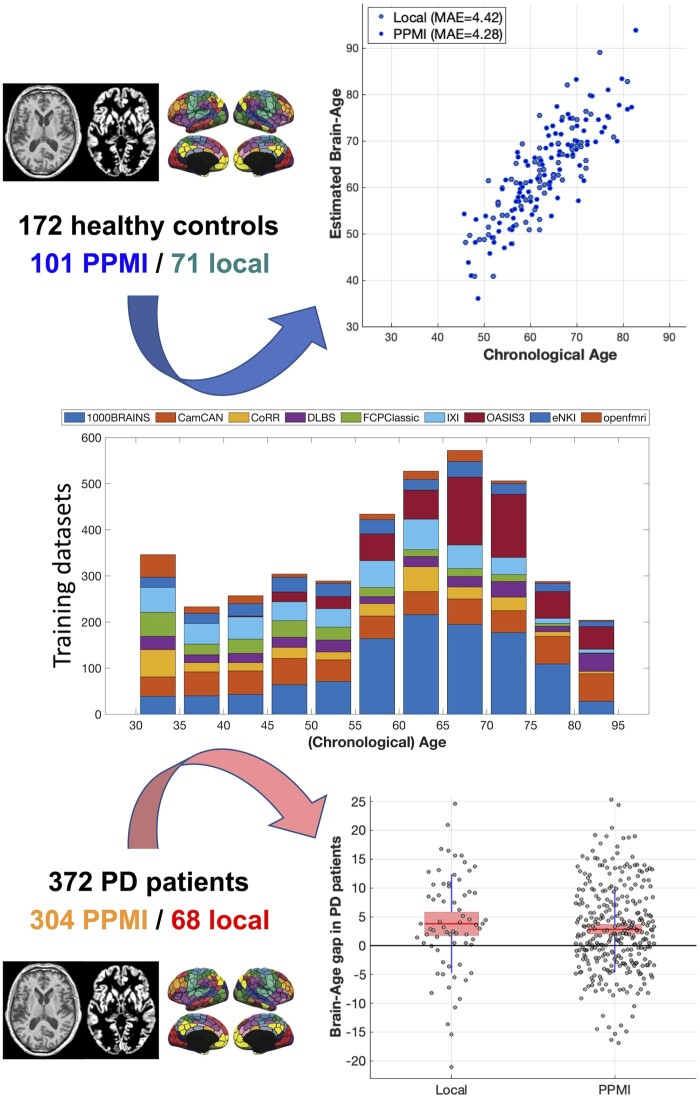
Individual age was estimated using an ensemble of linear support vector regression (SVR) with stratified subsampling.20 An independent dataset consisting of 3960 subjects (cf. Fig. 1) was compiled from several sources: 1000Brains,21 Cam-CAN,22 OpenfMRI,23 the Dallas Lifespan Brain Study,24 CoRR,25 IXI (http://brain-development.org/ixi-dataset) and the NKI-Rockland sample.26 We note that the age-range of the training dataset exceeds that of the Parkinson’s disease patients and HC by almost 20 years on the younger and older side. The higher sample size and broader age-range should provide a robust estimation of age-trajectories5 and increase stability of the models due to a more favourable feature-to-sample ratio.27 Given the imbalanced age- and sex-distribution across datasets, we performed stratified subsampling20 to ensure that the training set for each individual model within the ensemble was balanced in terms of age, site and sex distribution. At each iteration, 1562 subjects were independently drawn in a stratified manner without replacement and used to train an individual linear SVR with feature selection, providing a weak learner for the ensemble. Fitting of the SVR models was performed using LibSVM toolbox (https://www.csie.ntu.edu.tw/∼cjlin/libsvm/). This process was repeated 25 000 times, with all individual models being applied to the Parkinson’s disease patients and HCs. The thereby provided predictions were averaged (‘bagging’) to yield the final age-prediction, followed by a stacked model for bias-adjustment without leakage.28,29 Finally, individual BAGs were calculated as the difference between predicted (biological) and chronological age.
Figure 1.
Prediction of individual ‘brain age’ by an ensemble predictor trained on a heterogeneous database of ∼4000 subjects. This approach yielded a mean absolute error (MAE) of 4.4 years for HCs and a BAG increase of about 3 years in Parkinson’s disease patients. The middle plot shows the age-distribution of the training set for brain age prediction as a histogram (using 5-year age bins) stacked by site as denoted by the colours and named above the bars. The top right panel shows a scatter-plot of chronological age (x-axis) versus ‘brain age’ (as predicted from the model trained on the data summarized in the central panel) for the HCs from the PPMI cohort and the local sample. The mean absolute error (MAE) is computed as the average of the absolute individual deviations between brain and chronological age, i.e. the mean BAG ignoring the sign. The bottom right plot shows the distribution of the (signed) BAG for the Parkinson’s disease patients from both samples. The pink line denotes the average BAG in that sample, the shaded pink box the mean ± standard deviation.
Statistical analysis
All statistical analyses were performed independently for (i) the PPMI dataset and (ii) the local cohort as well as (iii) pooled across all patients. Differences between Parkinson’s disease patients and controls were assessed by an ANOVA including the factors ‘Diagnosis’ as well as ‘Gender’, ‘Age’ and ‘Site’. Significance was tested in a non-parametric (label-exchange) approach30 to accommodate the confound structure of the data without the need for distributional assumptions.
First, we analysed the areal-wise GMV to characterize the presence of regional brain atrophy accounting for gender, age and site. Inference was performed at p < 0.05, correcting for the false discovery rate (FDR) across parcels. Next, we compared biological ageing as reflected by the BAG (estimated—true age) between Parkinson’s disease and controls using the same approach. While the analysis of (area-wise) GMV differences was corrected for the number of assessed brain areas, no correction was necessary for the comparison of BAG values between groups, as the BAG collapses the multivariate pattern of atrophy into a single number.
Finally, we investigated associations between Parkinson’s disease patients’ BAG and individual disease duration, Hoehn and Yahr stage, cognitive (MoCA score) as well as motor (UPRDS-III score) affection using Pearson correlation analysis, again separately by cohort and in the overall sample. For these analyses, information on disease duration was missing for 2/372 patients, Hoehn and Yahr stage for 6/372, UPDRS-III scores for 10/372 and MoCA scores for 35/372. Given overlap among missing values, at least one covariate was missing for 40 patients, i.e. full information was available for 332. Individually for each analysis, i.e. covariate, cases with missing data for this covariate were excluded from the computation of the correlations, i.e. we did not perform any imputation.
Results
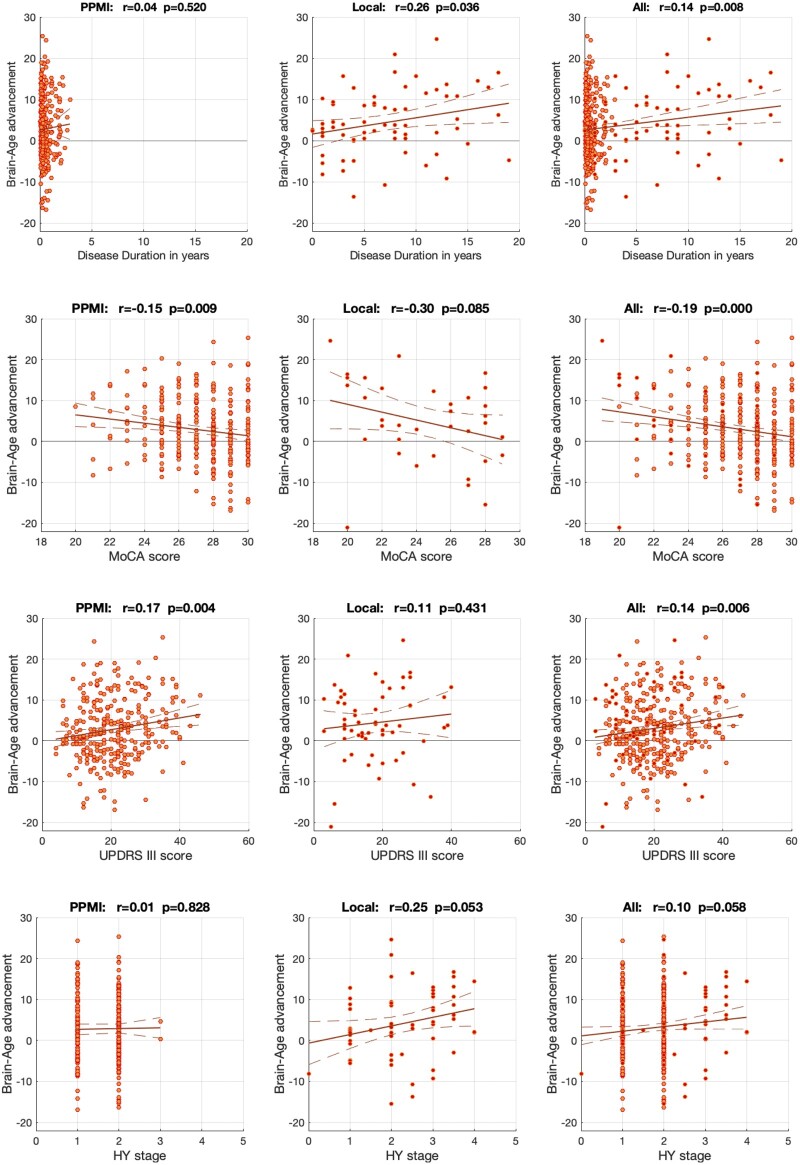
Age of the HCs was well predicted in both cohorts (overall r = 0.84, MAE 4.4 years; PPMI r = 0.85/MAE 4.3 years, local r = 0.81/MAE 4.5 years) using the SVR ensemble (Fig. 1, cf. Supplementary Fig. 1). When biological age was estimated by the same model for the Parkinson’s disease patients, we found an increased BAG of 2.9 years. In more details, local patients were estimated on average 3.3 years older than their true age, while the early-stage patients from the PPMI showed a mean advancement of 2.8 years (all p < 0.05 adjusting for age, sex and site). Convergently, we found a correlation between disease duration and BAG (r = 0.14, p < 0.008) in the pooled sample, mainly driven by the more chronic local patients (r = 0.26 / p = 0.036 versus r = 0.04 / p = 0.52 in the PPMI).
Advanced biological relative to chronological age was moreover related to clinical impairment (Fig. 2). There was a trend towards a negative correlation with Hoehn and Yahr stages in the pooled and local (chronic) sample, though (presumably due to a lack of variance) not in the PPMI data. More cognitively impaired patients showed advanced brain ageing as evident by a negative correlation between MoCA scores and BAG (r = −0.19, p < 0.001) in the overall sample as well as PPMI (r = −0.15, p < 0.01) and the local cohort (r = −0.30, p = 0.085). Likewise, patients with stronger Parkinson’s disease motor symptoms as reflected by UPDRS-III scores featured a higher BAG in the overall sample (r = 0.14, p < 0.006) and PPMI (r = 0.17, p < 0.004), though not the local cohort (r = 0.11, P = 0.43). In sum, we thus observed that individual BAG reflects disease duration, cognitive and motor affection.
Figure 2.
Relationship between individual brain age gap (BAG) and clinical covariates that were available for both cohorts. The BAG was significantly related to disease duration in the local and pooled samples, though not in the PPMI dataset comprising only early-stage patients. There was a significant negative relation to cognitive functioning (MoCA scores, lower values indicate stronger impairment) in the pooled and PPMI data and a trend towards such association in the subsample of the local cohort. Finally, correlation with motor impairments (UPDRS-III scores, higher values indicating stronger impairment) was evident in the PPMI and pooled sample. In turn, correlations between BAG and Hoehn and Yahr stages were not statistically significant, though showed a trend towards a positive correlation in the local and pooled samples.
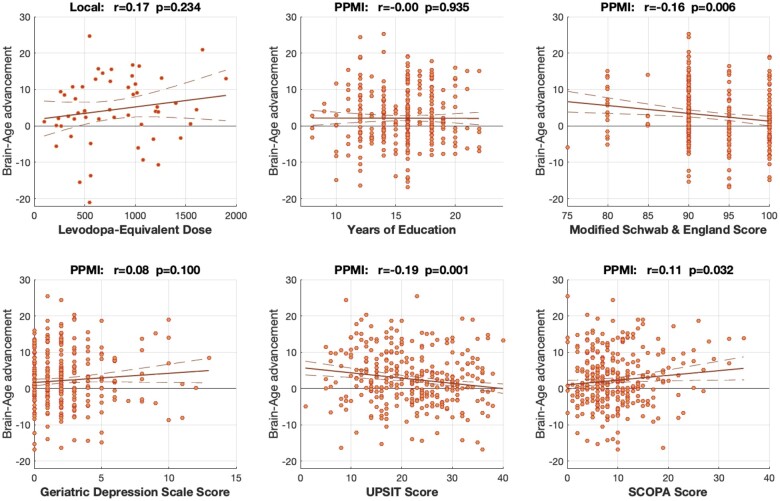
When testing for additional relationships between individual BAG and further clinical covariates in the PPMI sample as well as levodopa-equivalent doses (LEDs) in the local sample (as the PPMI patients were unmedicated), we found that higher BAG was associated with compromised independence in daily life (Schwab and England score), impaired smell (UPSIT scores) and more pronounced autonomous dysfunction (SCOPA-AUT scores). In turn, we found no significant correlation of BAG with education, LED or depressivity (Fig. 3).
Figure 3.
Correlations between individual brain age gap (BAG) and covariates that were only available in one of the two cohorts (PPMI or local sample). As shown, BAG showed a significant negative correlation to the individual Schwab & England and UPSIT scores (indicating an advanced biological age in patients with compromised independence or sense of smell) and a positive correlation with the SCOPA-AUT indicating a higher brain age in patients with more pronounced autonomous dysfunction. Importantly, we found no significant correlation of individual BAG with education, levodopa-equivalent dose or depressivity, rendering it unlikely that these covariates may have driven the main effects in Fig. 1.
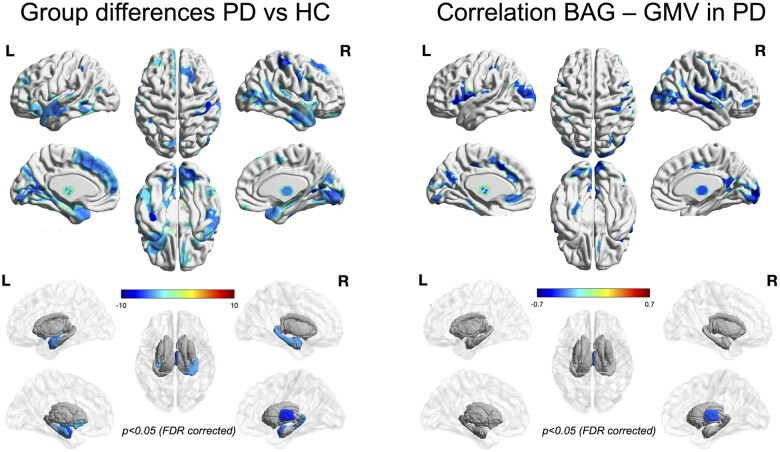
On the univariate level (P < 0.05, corrected for multiple comparisons), Parkinson’s disease patients featured widespread cortical atrophy, in particular in the right central region, medial frontal but also the visual and temporal cortices, as well as the thalamus and basal ganglia (Fig. 4, left panel). Interestingly, there was only a moderate (r = 0.22) correlation between the parcel-wise atrophy estimates of the (early stage) PPMI cohort and the (more advanced) local cohort, though both were represented well by the overall analysis (pooled versus PPMI: r = 0.85; pooled versus local r = 0.67). As illustrated in Supplementary Fig. 2, the most pronounced difference between the cohorts was the stronger prefrontal atrophy in the (chronically ill) local cohort as compared to the early-stage PPMI patients. Finally, noting both positive and negative BAG among the patients, we correlated the individual BAG of the Parkinson’s disease patients with the per-parcel GMV to explore, where atrophy was most strongly related to increased brain age (Fig. 4, right panel). Interestingly, the negative correlation between BAG and GMV, i.e. association between increased brain age and atrophy, was strongest in the Rolandic Operculum, the cingulate cortex and visual areas, showing a distinct pattern from the categorical differences between Parkinson’s disease patients and controls. We take this finding as an indication that atrophy patterns in Parkinson’s disease may only partially map onto differences in brain age, even though we must note that brain age as a multivariate prediction will inevitably depend on the entire pattern in the feature space rather than local features in isolation.
Figure 4.
Left: Significant grey matter volume differences between Parkinson’s disease patients and HCs accounting for effects of age, sex and site. Analyses were performed on the parcel level, i.e. using the same data that was used for brain age prediction. Values show percent atrophy in PD relative to controls, solid coloured parcels survive statistical inference at p < 0.05 corrected for multiple comparisons by controlling the False Discovery Rate (FDR). Right: Significant correlations between parcel-wise GMV and individual BAG in PD patients accounting for the effects of age, site and sex. Values show Fisher-Z transformed correlation coefficients, solid coloured parcels survive statistical inference at p < 0.05 corrected for multiple comparisons by controlling the False Discovery Rate (FDR).
Discussion
We investigated grey matter loss in two independent, large cohorts of Parkinson’s patients and matched controls. Mass-univariate comparison revealed distributed atrophy in fronto-occipito-temporal cortices of Parkinson’s disease patients when controlling for age, gender and site. Our results resonate well with findings from a recent meta-analysis13 regarding effects in the basal ganglia as well as superior and medial frontal cortex, but we note that the current analysis also robustly highlighted ventral occipital and temporal areas. In addition, the more extensive frontal affection of the more chronic local patients matches the increased prevalence of executive symptoms with increased disease progression.31
Training and applying a multivariate model for individual age prediction then allowed the assessment of discrepancies between biological and chronological age in patients with Parkinsons’s disease and matched controls. While the age of the HCs was expectedly well predicted, we found brain ageing in Parkinson’s disease patients to be advanced by about 3 years and related to disease duration as well as cognitive and motor affection.
The concept of ‘brain age’
To estimate a summary marker of individual neurodegeneration, we employed the ‘brain age’ concept.32 The ensuing ‘BAG’ has the advantage of absorbing physiological changes, rendering it conceptually independent from chronological age. That is, given that the BAG reflects the difference between estimated ‘brain age’ and chronological age, it should be comparable across the lifespan as long as underfitting due to regression dilution and distribution imbalance are corrected for.29,33 Moreover, from the current literature i appears that BAG dispersion does not systematically change across the (adult) lifespan, i.e. the distribution of BAG values seems to be similarly wide in younger and older adults. On the one hand, this may be expected when balanced training sets are used, as in that case, the model will be optimized to capture the entire age-range represented in the training data equally well (while often failing to extrapolate beyond the training range). On the other hand, we would like to note that the same BAG should represent the same relative (in terms of years of further ageing) rather than an absolute deviation (in terms of percent volume loss) of atrophy patterns across the lifespan. Given that changes in brain structure are much smaller in, e.g. the 30s are compared to the 60s,34,35 this implies that a similar BAG of 4 years would correspond to more substantial absolute atrophy later in life but a similar shift relative to the reference cohort.
We note that the MAE of 4.4 years for our HCs compares well with the current literature reporting MAEs of 3–5 years for healthy subjects1,32 but acknowledge, that the BAG should represent a mixture of true biological deviation and generalization error, i.e. noise. While their relative contribution is unknowable, recent reports suggest that even with massive training datasets and complex models, the lower bound of prediction accuracy may be in the range of 2–3 years, likely reflecting biological variability. Deep learning on almost 12 000 subjects from a multi-site dataset resulted in an MAE of 3.7 years in cross-validation and 4.1 years for an independent test-sample.36 In turn, a deep learning ensemble trained on ∼13 000 subjects from the UKbiobank achieved an MAE of 2.14 years on held-out subjects from the same dataset.37
Comparison to other disorders
While we found a significantly elevated BAG in both cohorts, the absolute increase of ∼3 years is rather moderate relative to other neurodegenerative disorders.9,36,38 As recently reviewed,2,32 BAGs of up to 10 years and Cohen’s d values > 1 have been reported for Alzheimer’s disease. MCI patients also show substantially increased brain ages, though the findings are more variable (mean BAG: 3–8 years, Cohen’s d: 0.4–0.7). Effects at the lower end of that spectrum have been shown for schizophrenia and psychotic disorders39–41 whereas BAG values of patients with multiple sclerosis even exceed those for Alzheimer’s disease.9 A mean BAG of ∼3 years (Cohen’s d ∼ 0.4) thus places Parkinson’s disease at the lower end of brain disorders featuring broad impairments of mental functions.
We would hypothesize that this surprising observation may reflect the usual time of diagnosis in the progression of Parkinson’s disease. Clinical diagnosis is usually attained in Braak stages 3/4,42 when the disease has entered the substantia nigra and striatal function is disturbed through loss of dopaminergic innervation. In turn, substantial cortical atrophy corresponding to Braak-stages 5/6 may trail by several years.43 While the Braak hypothesis has been questioned,44 it stands to reason that patients recently diagnosed with Parkinson’s disease due to striatal dysfunction may not yet feature substantial forebrain atrophy, resulting in a low BAG. In contrast, relevant cortical atrophy in disorders like AD, MCI and schizophrenia should precede the onset of the clinical symptoms, resulting in more substantial BAG increases for recently diagnosed patients. That is, in comparison to other disorders, the behaviourally salient disturbance of striatal function in Parkinson’s disease may prepone the time of diagnosis relative to substantial forebrain degeneration.
Comparison to previous work on brain age in Parkinson’s disease
The to date only existing study on brain age in Parkinson’s disease,11 reported a BAG of 1.5 ± 6.0 years for PPMI patients based on a model using grey matter features and 2.5 ± 5.9 for one based on WM features. While both the work by Beheshti and the current estimate of a 2.8 year BAG for the PPMI thus point to a only very moderate increase in brain age compared to other disorders, the two investigations differed in multiple aspects that may explain the divergence in the reported BAG. First, we not only included a second cohort in order to replicate the findings from the PPMI data and extend them to more chronically ill patients, but notably also almost twice as many patients from the PPMI dataset (304 versus 160). From the methodological perspective, the current age-prediction model was trained on a substantially larger training set, which allowed us to train each weak (but unbiased) learner using more subjects than were available to Beheshti et al. Combined with the use of a biologically informed data-compression by a state-of-the-art brain atlas relative to low-resolution resampling, we would argue that the current model should have a much higher capacity for robust, unbiased and precise estimation of individual age. Yet, it may be noted that the age-prediction in the PPMI controls was only slightly better using our model (4.28 versus 4.38 year MAE). We would attribute this observation to the fact that the current paper performed a true out-of-sample validation, i.e. no PPMI subject was part of the training set whereas Beheshti et al. performed 10-fold cross-validation. Hence information on the PPMI data, though not on the test-subjects themselves, were available to the previous but not the current model.
Variance and specificity of the BAG in Parkinson’s disease
While the BAG was significantly increased in both Parkinson’s disease cohorts, we noted a substantial variability between individuals with about one-third of the patients featuring negative BAG values, i.e. brains appearing younger than their chronological age. While this may be partially attributed to imperfect algorithm-generalization, it must be remembered that a particular diagnosis such as Parkinson’s disease is just one of many factors potentially influencing ‘brain age’. To exemplify, effects on BAG have been reported for education and physical activity,45 obesity46 and metabolic syndrome,46,47 childbirth,48 meditation practice,49 negative life events50 as well as alcohol and nicotine consumption.51 The observed variance could thus reflect influences of such factors. In turn, recording (during data collection) and accommodating (during analysis) more lifestyle and health related information will be a critical step in the further development of ‘brain age’ approaches. Ultimately, we would then envision an ‘adjusted’ BAG reflecting the part of the deviation between biological and chronological age that is not attributable to known influences.
Whether such adjustment would lead to an increase or decrease in the mean BAG, however, is up for conjecture, as unmeasured and hence unknown confounds may bias results either way. To illustrate, most neurological and psychiatric conditions entail a reduced physical activity.52,53 Advanced brain age may hence partially reflect general patterns of involution due to a more reclusive lifestyle rather than true neurodegeneration. In turn, adjusting may unmask differences that are obscured by confounding lifestyle and physical health factors.
This leads to a final but critical consideration that also relates back to the rather small BAG advancement in Parkinson’s disease. As noted, brain age models describe the multivariate patterns of physiological changes in the brain, i.e. the normative trajectory of age-related changes.1 The observation that the BAG is increased in patients with neurodegenerative disorders has hence been conceptualized as an ‘acceleration’ of normal ageing.54 This view, however, is at odds with observations of distinct disease-related patterns for Alzheimer’s and Parkinson’s disease, Schizophrenia, multiple sclerosis, etc.; each confirmed by meta-analysis.13,55–57 In the current brain age framework, multivariate patterns of atrophy due to neurodegenerative disorders that differ from physiological changes would then be projected onto the normal trajectory. Consequently, a disorder featuring atrophy patterns that closely resemble physiological changes would result in a higher BAG than one evoking atrophy that strongly differs from normal age-related involution. In summary, we would argue that the brain age concept offers a unique opportunity to objectively quantify the deviation of high-dimensional morphometric patterns from their age-related norm in a single number. Yet, further work is needed to disentangle the effects of lifestyle features and the alignment between physiological and pathological patterns from raw BAG scores to obtain sensitive, specific and clinically useful58 biomarkers of neurodegeneration and their progression, understand patient heterogeneity and finally develop tools for individual prognosis.
Limitations
This study investigated ‘brain age’ in Parkinson’s disease based on two well-sized cohorts of de-novo and chronic patients (304 and 68 patients, respectively) using a large, heterogeneous training database and true out-of-sample predictions. Yet, we need to acknowledge that a deeper, more consistent clinical and neurocognitive phenotyping may have allowed further insight into the behavioural correlates of inter-individual differences in BAG across patients. Most importantly, only core clinical features (disease duration and HY stage as well as MoCA and UPDRS-III scores) were available for both the PPMI cohort as well as the local sample. This limited analysis of covariates such as the Geriatric Depression Scale, UPSIT, Modified Schwab and England scale, SCOPA-AUT and unfortunately also education (which was used in the local sample to correct the MoCA scores but not systematically recorded for further analysis) to the PPMI lacking more chronically ill patients. There is hope, however, that future releases of the PPMI will allow to track the relation of these variables to brain ageing, while we encourage the design of future cohorts to include a more detailed cognitive and affective test battery.
Conclusions and outlook
We demonstrated that Parkinson’s disease leads to a moderate but replicable increase in predicted brain age that reflects both disease duration and severity. These findings are well in line with the neurodegenerative nature of Parkinson’s disease. However, more longitudinal work on populations with detailed social and medical history is needed to disentangle physiological ageing trajectories, lifestyle effects and core pathological changes.
Supplementary material
Supplementary material is available at Brain Communications online.
Funding
This study was supported by the Deutsche Forschungsgemeinschaft, the Helmholtz Portfolio Theme ‘Supercomputing and Modeling for the Human Brain’, the European Union’s Horizon 2020 Research and Innovation Programme under Grant Agreement No. 945539 (HBP SGA3) and No. 826421 (VirtualBrainCloud).
Competing interests
The authors report no competing interests.
Supplementary Material
Glossary
- BAG =
brain age gap
- GMV =
grey matter volume
- HCs =
healthy controls
- LED =
levodopa-equivalent dose
- MAE =
mean absolute error
- MoCA =
Montreal Cognitive Assessment
- PPMI =
Parkinson’s disease marker initiative
- SCOPA-AUT =
Scales for Outcomes in Parkinson’s Disease—Autonomic Dysfunction
- SVR =
support vector regression
- UPDRS =
Unified Parkinson’s Disease Rating Scale
- UPSIT =
University of Pennsylvania Smell Identification Test
References
- 1.Cole JH, Franke K.. Predicting age using neuroimaging: Innovative brain ageing biomarkers. Trends Neurosci. 2017;40(12):681–690. [DOI] [PubMed] [Google Scholar]
- 2.Cole JH, Marioni RE, Harris SE, Deary IJ.. Brain age and other bodily ‘ages’: Implications for neuropsychiatry. Mol Psychiatry. 2019;24(2):266–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davatzikos C, Xu F, An Y, Fan Y, Resnick SM.. Longitudinal progression of Alzheimer’s-like patterns of atrophy in normal older adults: The SPARE-AD index. Brain. 2009;132(Pt 8):2026–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moradi E, Pepe A, Gaser C, Huttunen H, Tohka J.. Machine learning framework for early MRI-based Alzheimer’s conversion prediction in MCI subjects. Neuroimage. 2015;104:398–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Varikuti DP, Genon S, Sotiras A, et al. Evaluation of non-negative matrix factorization of grey matter in age prediction. Neuroimage. 2018;173:394–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cole JH.Multimodality neuroimaging brain-age in UK biobank: Relationship to biomedical, lifestyle, and cognitive factors. Neurobiol Aging. 2020;92:34–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elliott ML, Belsky DW, Knodt AR, et al. Brain-age in midlife is associated with accelerated biological aging and cognitive decline in a longitudinal birth cohort. Mol Psychiatry. 2019. doi:10.1038/s41380-019-0626-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liem F, Varoquaux G, Kynast J, et al. Predicting brain-age from multimodal imaging data captures cognitive impairment. Neuroimage. 2017;148:179–188. [DOI] [PubMed] [Google Scholar]
- 9.Kaufmann T, van der Meer D, Doan NT, et al. Common brain disorders are associated with heritable patterns of apparent aging of the brain. Nat Neurosci. 2019;22(10):1617–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorsey ER, Bloem BR.. The Parkinson pandemic—A call to action. JAMA Neurol. 2018;75(1):9–10. [DOI] [PubMed] [Google Scholar]
- 11.Beheshti I, Mishra S, Sone D, Khanna P, Matsuda H.. T1-weighted MRI-driven brain age estimation in Alzheimer’s disease and Parkinson's disease. Aging Dis. 2020;11(3):618–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gelb DJ, Oliver E, Gilman S.. Diagnostic criteria for Parkinson disease. Arch Neurol. 1999;56(1):33–39. [DOI] [PubMed] [Google Scholar]
- 13.Xu X, Han Q, Lin J, Wang L, Wu F, Shang H.. Grey matter abnormalities in Parkinson’s disease: A voxel-wise meta-analysis. Eur J Neurol. 2020;27(4):653–659. [DOI] [PubMed] [Google Scholar]
- 14.Marek K, Chowdhury S, Siderowf A, et al. The Parkinson’s progression markers initiative (PPMI) - establishing a PD biomarker cohort. Ann Clin Transl Neurol. 2018;5(12):1460–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hughes AJ, Daniel SE, Kilford L, Lees AJ.. Accuracy of clinical diagnosis of idiopathic Parkinson’s disease: A clinico-pathological study of 100 cases. J Neurol Neurosurg Psychiatry. 1992;55(3):181–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eickhoff SB, Yeo BTT, Genon S.. Imaging-based parcellations of the human brain. Nat Rev Neurosci. 2018;19(11):672–686. [DOI] [PubMed] [Google Scholar]
- 17.Schaefer A, Kong R, Gordon EM, et al. Local-global parcellation of the human cerebral cortex from intrinsic functional connectivity MRI. Cereb Cortex. 2018;28(9):3095–3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan L, Li H, Zhuo J, et al. The Human Brainnetome Atlas: A new brain atlas based on connectional architecture. Cereb Cortex. 2016;26(8):3508–3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buckner RL, Krienen FM, Castellanos A, Diaz JC, Yeo BTT.. The organization of the human cerebellum estimated by intrinsic functional connectivity. J Neurophysiol. 2011;106(5):2322–2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohajer B, Abbasi N, Mohammadi E, et al. Gray matter volume and estimated brain age gap are not linked with sleep-disordered breathing. Hum Brain Mapp. 2020;41(11):3034–3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caspers S, Moebus S, Lux S, et al. Studying variability in human brain aging in a population-based German cohort-rationale and design of 1000BRAINS. Front Aging Neurosci. 2014;6:149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shafto MA, Tyler LK, Dixon M, et al. The Cambridge Centre for Ageing and Neuroscience (Cam-CAN) study protocol: A cross-sectional, lifespan, multidisciplinary examination of healthy cognitive ageing. BMC Neurol. 2014;14:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poldrack RA, Barch DM, Mitchell JP, et al. Toward open sharing of task-based fMRI data: The OpenfMRI project. Front Neuroinform. 2013;7:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park J, Carp J, Kennedy KM, et al. Neural broadening or neural attenuation? Investigating age-related dedifferentiation in the face network in a large lifespan sample. J Neurosci. 2012;32(6):2154–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zuo X-N, Anderson JS, Bellec P, et al. An open science resource for establishing reliability and reproducibility in functional connectomics. Sci Data. 2014;1:140049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nooner KB, Colcombe SJ, Tobe RH, et al. The NKI-rockland sample: A model for accelerating the pace of discovery science in psychiatry. Front Neurosci. 2012;6:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He T, Kong R, Holmes AJ, et al. Deep neural networks and kernel regression achieve comparable accuracies for functional connectivity prediction of behavior and demographics. Neuroimage. 2020;206:116276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang H, Zhang F, Niu X.. Investigating systematic bias in brain age estimation with application to post‐traumatic stress disorders. Hum Brain Mapp. 2019;40(11):3143–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith SM, Vidaurre D, Alfaro-Almagro F, Nichols TE, Miller KL.. Estimation of brain age delta from brain imaging. Neuroimage. 2019;200:528–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bludau S, Mühleisen TW, Eickhoff SB, Hawrylycz MJ, Cichon S, Amunts K.. Integration of transcriptomic and cytoarchitectonic data implicates a role for MAOA and TAC1 in the limbic-cortical network. Brain Struct Funct. 2018;223(5):2335–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alzahrani H, Venneri A.. Cognitive and neuroanatomical correlates of neuropsychiatric symptoms in Parkinson’s disease: A systematic review. J Neurol Sci. 2015;356(1-2):32–44. [DOI] [PubMed] [Google Scholar]
- 32.Franke K, Gaser C.. Ten years of as a neuroimaging biomarker of brain aging: What insights have we gained? Front Neurol. 2019;10:789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beheshti I, Nugent S, Potvin O, Duchesne S.. Bias-adjustment in neuroimaging-based brain age frameworks: A robust scheme. Neuroimage Clin. 2019;24:102063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Battaglini M, Gentile G, Luchetti L, et al. Lifespan normative data on rates of brain volume changes. Neurobiol Aging. 2019;81:30–37. [DOI] [PubMed] [Google Scholar]
- 35.Ziegler G, Dahnke R, Jäncke L, Yotter RA, May A, Gaser C.. Brain structural trajectories over the adult lifespan. Hum Brain Mapp. 2012;33(10):2377–2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bashyam VM, Erus G, Doshi J, et al. MRI signatures of brain age and disease over the lifespan based on a deep brain network and 14 468 individuals worldwide. Brain. 2020;143(7):2312–2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peng H, Gong W, Beckmann CF, Vedaldi A, Smith SM.. Accurate brain age prediction with lightweight deep neural networks. Med Image Anal. 2021;68:101871- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Franke K, Ziegler G, Klöppel S, Gaser C.. Estimating the age of healthy subjects from T1-weighted MRI scans using kernel methods: Exploring the influence of various parameters. Neuroimage. 2010;50(3):883–892. [DOI] [PubMed] [Google Scholar]
- 39.Koutsouleris N, Davatzikos C, Borgwardt S, et al. Accelerated brain aging in schizophrenia and beyond: A neuroanatomical marker of psychiatric disorders. Schizophr Bull. 2014;40(5):1140–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nenadić I, Dietzek M, Langbein K, Sauer H, Gaser C.. BrainAGE score indicates accelerated brain aging in schizophrenia, but not bipolar disorder. Psychiatry Res Neuroimaging. 2017;266:86–89. [DOI] [PubMed] [Google Scholar]
- 41.Schnack HG, van Haren NEM, Nieuwenhuis M, Hulshoff Pol HE, Cahn W, Kahn RS.. Accelerated brain aging in schizophrenia: A longitudinal pattern recognition study. Am J Psychiatry. 2016;173(6):607–616. [DOI] [PubMed] [Google Scholar]
- 42.Braak H, Del Tredici K.. Neuropathological staging of brain pathology in sporadic Parkinson’s disease: Separating the wheat from the Chaff. J Parkinson’s Dis. 2017;7(s1):S71–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hawkes CH, Del Tredici K, Braak H.. A timeline for Parkinson’s disease. Parkinsonism Relat Disord. 2010;16(2):79–84. [DOI] [PubMed] [Google Scholar]
- 44.Brooks DJ.Examining Braak’s hypothesis by imaging Parkinson's disease. Mov Disord. 2010;25 (Suppl 1):S83–8. [DOI] [PubMed] [Google Scholar]
- 45.Steffener J, Habeck C, O'Shea D, Razlighi Q, Bherer L, Stern Y.. Differences between chronological and brain age are related to education and self-reported physical activity. Neurobiol Aging. 2016;40:138–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ronan L, Alexander-Bloch AF, Wagstyl K, et al. Obesity associated with increased brain age from midlife. Neurobiol Aging. 2016;47:63–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Franke K, Ristow M, Gaser C.. Gender-specific effects of health and lifestyle markers on individual BrainAGE [Internet]. 2013 International Workshop on Pattern Recognition in Neuroimaging. Philadelphia: IEEE; 2013. doi:10.1109/prni.2013.33. [Google Scholar]
- 48.de Lange A-MG, Kaufmann T, van der Meer D, et al. Population-based neuroimaging reveals traces of childbirth in the maternal brain. Proc Natl Acad Sci U S A. 2019;116(44):22341–22346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luders E, Cherbuin N, Gaser C.. Estimating brain age using high-resolution pattern recognition: Younger brains in long-term meditation practitioners. Neuroimage. 2016;134:508–513. [DOI] [PubMed] [Google Scholar]
- 50.Hatton SN, Franz CE, Elman JA, et al. Negative fateful life events in midlife and advanced predicted brain aging. Neurobiol Aging. 2018;67:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ning K, Zhao L, Matloff W, Sun F, Toga AW.. Association of relative brain age with tobacco smoking, alcohol consumption, and genetic variants. Sci Rep. 2020;10(1):10- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheval B, Orsholits D, Sieber S, Courvoisier D, Cullati S, Boisgontier MP.. Relationship between decline in cognitive resources and physical activity. Health Psychol. 2020;39(6):519–528. [DOI] [PubMed] [Google Scholar]
- 53.Wittwer JE, Webster KE, Menz HB.. A longitudinal study of measures of walking in people with Alzheimer’s disease. Gait Posture. 2010;32(1):113–117. [DOI] [PubMed] [Google Scholar]
- 54.Elliott ML.MRI-based biomarkers of accelerated aging and dementia risk in midlife: How close are we? Ageing Res Rev. 2020;61:101075. [DOI] [PubMed] [Google Scholar]
- 55.Chapleau M, Aldebert J, Montembeault M, Brambati SM.. Atrophy in Alzheimer’s disease and semantic dementia: An ALE meta-analysis of voxel-based morphometry studies. J Alzheimers Dis. 2016;54(3):941–955. [DOI] [PubMed] [Google Scholar]
- 56.Chiang FL, Wang Q, Yu FF, et al. Localised grey matter atrophy in multiple sclerosis is network-based: A coordinate-based meta-analysis. Clin Radiol. 2019;74(10):816.e19–816.e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goodkind M, Eickhoff SB, Oathes DJ, et al. Identification of a common neurobiological substrate for mental illness. JAMA Psychiatry. 2015;72(4):305–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hedderich DM, Eickhoff SB.. Machine learning for psychiatry: Getting doctors at the black box? Mol Psychiatry. 2021;26(1):23–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The PPMI dataset may be downloaded following application for access under http://PPMI-info.org/. The local data cannot be openly shared due to ethical restrictions and missing consent for sharing from the participants.