Abstract
Streptococcus pneumoniae (Spn) is a leading respiratory tract pathogen that colonizes the nasopharynx (NP) through adhesion to epithelial cells and immune evasion. Spn actively interacts with other microbiota in NP but the nature of these interactions are incompletely understood. Using 16S rRNA gene sequencing, we analyzed the microbiota composition in the NP of children with or without Spn colonization. 96 children were included in the study cohort. 74 NP samples were analyzed when children were 6 months old and 85 NP samples were analyzed when children were 12 months old. We found several genera that correlated negatively or positively with Spn colonization, and some of these correlations appeared to be influenced by daycare attendance or other confounding factors such as upper respiratory infection (URI) or Moraxella colonization. Among these genera, Corynebacterium showed a consistent inverse relationship with Spn colonization with little influence by daycare attendance or other factors. We isolated Corynebacterium propinquum and C. pseudodiphtheriticum and found that both inhibited the growth of Spn serotype 22F strain in vitro.
Introduction
Streptococcus pneumoniae (Spn) causes a variety of illnesses, including pneumonia, otitis media, bacteremia, and meningitis [1, 2]. Despite availability of pneumococcal vaccines, Spn remains the most common cause of bacterial infection in the developing world and most frequently infects children under 5 years old or elderly over 65. It was included as one of 12 priority pathogens by WHO in 2017 [3]. Spn colonizes the nasopharynx (NP) of children in the first month of life and 27–65% of children carry Spn asymptomatically [3]. It is incompletely understood how Spn progresses from a commensal state to a pathogenic state and eventually invades tissues and blood stream to cause local invasiveness and systemic infections. We hypothesize that commensal bacteria in NP may influence this process and promote or prevent Spn-related diseases in children.
The NP microenvironment harbors commensal flora, which maintain immune homeostasis and suppress pathogenic progression and/or colonization of respiratory tract pathobionts. Based on studies in the gut and in the NP, protective microbiota compete with pathogens in the microenvironment, through nutrient deprivation, production of anti-microbial molecules, and modulating the innate and adaptive immune system [3–12]. One dominant commensal genus in the NP is Corynebacterium. There are over 100 species of Corynebacterium [13], and at least 23 are present in NP [14]. Whether the different species exert distinct effects on Spn pathogenesis is not clear, highlighting the limit of our understanding of the influences of Corynebacterium on Spn-related illnesses. An inverse correlation of Corynebacterium detection in the NP to Spn colonization had been reported [15–18]. Some Corynebacterium spp. showed direct inhibition on in vitro Spn growth or in vivo colonization. For example, C. accolens was shown to inhibit growth of Spn in vitro [15]. C. pseudodiphtheriticum was reported to prevent Spn-induced pneumonia in mice but this effect appeared to be specific to particular C. pseudodiphtheriticum strains [19, 20].
In this study, we examined the NP microbiome of children at age 6 and 12 months with or without Spn colonization detected by bacterial culture. We found that detection of Corynebacterium spp. was inversely correlated with Spn colonization, consistent with previous reports [15–18]. We isolated two species of Corynebacterium: C. pseudodiphtheriticum and C. propinquum. We then investigated the effects of these Corynebacterium spp.on Spn growth in vitro and found that both exhibited inhibitory functions.
Materials and methods
The Rochester General Hospital IRB approved the study and written informed consent was obtained from parents before enrollment. All methods were performed in accordance with the IRB’s relevant guidelines and regulations.
Subject information and sample collection
NP washes were obtained during a prospective cohort study conducted in Rochester NY from 2006–2018 involving healthy children of 6 and 12 months old for study of respiratory illnesses, in particular acute otitis media (AOM), supported by the National Institutes of Deafness and Communication Disorders. Details of the cohort have been described previously [21–23]. Briefly, these children were recruited from middle-class suburban sociodemographic pediatric practices in Rochester, NY. Informed consent was obtained in writing at enrollment from the child’s parents or legally authorized representative. The parent/guardian agreed to provide follow-up information and arrange for all scheduled visits. Because this study was designed to investigate the impact of microbiome on AOM in children, individuals with uncertain diagnosis of AOM were excluded from the study. Additional exclusion criteria included diagnosis of otorrhea, presence of tympanostomy tube, or diagnosis of Down syndrome, cleft palate, craniofacial disorders, cystic fibrosis/mucoviscidosis immotile cilia syndrome, congenital immunodeficiency or HIV/AIDS, or other medical conditions that may interfere with implementation of the protocol or interpretation of study results. All children were vaccinated with pneumococcal conjugate vaccines (either PCV13 or PCV7 according to availability).
Definitions: Viral URI: Diagnosis of viral URI was made when children presented symptoms of nasal congestion, rhinorrhea, cough, and/or sore throat with or without fever, following established guidelines [24–27]. AOM: AOM was diagnosed using pneumatic otoscopy by validated otoscopists according to American Academy of Pediatrics guidelines [28]. The children presented acute onset of symptoms consistent with AOM and had tympanic membranes (TMs) that were: 1) bulging or full, with a cloudy or purulent effusion, or 2) completely opacified, and 3) with reduced or absent mobility.
NP washes from the children were cultured in vitro on Chocolate agar and tryptic soy agar (TSA) with 5% sheep blood (BD) for identification of Spn and other common respiratory bacterial pathogens using microbiology methods previously described [29, 30]. Spn was identified based on their colony morphology, alpha hemolytic activity on TSA blood plates, and sensitivity to optochin disc. Haemophilus influenzae (Hi) was identified based on their colony morphology, gram negative staining, growth on Chocolate agar plates but not on TSA blood plates, and their presence or absence of growth in specific quadrants on hemo ID quad plates (BD), depending on X- and V-factor requirements for the various Haemophilus species. We did not confirm every Hi isolates but our recent study found that more than 95% of Hi strains were non-typable (Fuji N et al, in press). Moraxella catarrahlis (Mcat) was identified based on colony morphology, gram-negative stain, positive oxidase reactivity, and positive reactivity to Remel Catarrahlis Test disc (Thermofisher). Staphylococcus aureus (SA) was identified based on its beta-hemolytic activity on blood plates and its being coagulase-positive and catalase-negative. Based on these microbiology methods, samples were split into Spn+ or Spn- groups for microbiome comparisons. The demographic information of the patient samples is listed in Table 1. The samples were stored at -80°C in Virus Transfer Media (VTM) after collection and then sent for 16S rRNA gene sequence analysis at the Microbiome Core Facility, University of North Carolina, Chapel Hill (https://www.med.unc.edu/microbiome/). All the above information on the samples could be accessed in S1 Table.
Table 1. Demographic factors associated with Spn colonization.
| # of samples | Race (White: non-white) | Female: Male | Breast-feeding (Yes:No) | Exposure to smoke (Yes:No) | Atopy (Yes:No) | Abx Treatment (Yes:No) | Daycare Attendance (Yes:No) | Siblings (Yes:No) | |
|---|---|---|---|---|---|---|---|---|---|
| Spn+ | |||||||||
| 6 m | 25 | 22:3 [0.53] | 10:15 [0.46] | 10:15 [0.46] | 0:24 [0.17] | 5:18 [0.57] | 1:20 [0.41] | 9:15 [0.0006]** | 3:22 [1] |
| 12 m | 28 | 24:4 [0.77] | 12:16 [0.17] | 10:16 [0.47] | 1:26 [0.26] | 6:19 [0.60] | 8:13 [0.060] | 10:17 [0.0078]* | 4:24 [1] |
| total | 53 | ||||||||
| Spn- | |||||||||
| 6 m | 48 | 39:9 [0.53] | 24:24 [0.46] | 24:23 [0.46] | 6:42 [0.17] | 13:29 [0.57] | 6:35 [0.41] | 2:45 [0.0006]** | 6:42 [1] |
| 12 m | 56 | 46:10 [0.77] | 33:23 [0.17] | 27:28 [0.47] | 8:47 [0.26] | 15:34 [0.60] | 7:38 [0.060] | 6:49 [0.0078]* | 9:47 [1] |
| total | 104 |
Note: Demographic factors are listed as column names. The number of children who were female or male, or who were Yes or No for a particular demographic factor is shown for each age group (6-month or 12-month) and for each Spn colonization phenotype (Spn+ or Spn-). In each age group, the proportion of children carrying a demographic trait was compared between Spn+ and Spn- samples and statistical significance assessed by Fisher’s Exact test. The p values are listed in square brackets.
*: p < 0.05;
**: p < 0.005.
16S rRNA gene sequencing analyses
Sample submission and sequencing analyses have been previously described [23]. Briefly, the V4 region of 16S rRNA gene was sequenced via Illumina sequencing, which were processed by Illumina Bcl2Fastq 2.18.0.12 and Cutadapt, and DADA2 [31]. The fastq sequences were deposited at Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) with accession number PRJNA720045. Bar plots were created by ggplot2 in R (version 3.6.1, r-project.org), after the conglomerate of data files at phylum, class, and family level. Alpha diversity indices of the samples were measured via estimate_richness and graphed by the plot_richness command in Phyloseq in R. To measure beta diversity of samples, Bray-Curtis distance and weighted unifrac between samples were calculated via phyloseq::distance command and NMDS plots were created by ordination method. For differential abundance analyses, 30 genera with the highest abundance, or genera present in at least 50% of the samples, were analyzed through DESeq2 (version 1.24.0) in R and the outcomes were corrected for batch effects. Adjusted p values were calculated after correction for multiple hypothesis testing using Holm-Bonferroni method and those less than 0.05 were graphed in ggplot2.
Isolation of Corynebacterium species
Cultures from NP swabs were plated on blood agar plates. Colonies with an appearance consistent with Corynebacterium spp. were randomly selected for PCR analyses [32] and for culturing in brain heart infusion (BHI) media at 37°C. Primers targeting the rpoB gene were used in the PCR analyses, which effectively distinguish species in the Corynebacterium genus [33]. The primer sequences are: C2700F: 5’-CGTATGAACATCGGCCAGGT– 3’, and C3130R: 5’-TCCATTTCGCCGAAGCGCTG-3’. The PCR program used was: 95°C 5min then 40 cycles of 95°C 15s, 55°C 15s, 72°C 15s. PCR products were separated on 1.5% agarose gel with an expected size of ~450bp and were subsequently isolated and extracted for DNA sequencing. Sequencing results were searched against GenBank database (blast.ncbi.nlm.nih.gov/Blast.cgi) for matches. C. propinquum and C. pseudodiphtheriticum were scored as the top candidates.
In vitro co-culture experiments
C. propinquum and C. pseudodiphtheriticum were cultured in BHI media to reach OD600 = 0.5. 5 μl of the culture was spotted on a blood agar plate and grown at 37°C for one day (for C. propinquum) or two days (for C. pseudodiphtheriticum), before 5 μl of Spn 22F strain (grown in THBY media to OD600 = 0.5) was spotted next to the Corynebacterium at different distances. Images were taken every 24 hours and analyzed by ImageJ. To correct for the variations in images taken on different days, the diameter of each image for the same plate was measured on each day and the ratio of its square over the square of the diameter measured on the first day was used as a normalizing factor. The area covered by each colony was measured via ImageJ, which was divided by the normalizing factor before being imported for graphing in Microsoft Excel. In the second approach to visualize how Corynebacterium might affect Spn growth, 600 μl of Spn 22F (OD600 = 0.5) or non-Spn alpha-hemolytic Streptococcus (AHS) as a control was spread onto 10 cm blood agar plates to form a lawn. The AHS strain was an alpha-hemolytic Streptococcus isolate from a pediatric patient that showed resistance to optochin, in contrast to Streptococcus pneumoniae. Cultures of C. propinquum and C. pseudodiphtheriticum were concentrated by centrifugation. 5 μl of the pellet was spotted onto the Spn22F or AHS lawn and incubated at 37°C. Images were taken the next day.
Statistics
Demographic factors and other factors that may influence Spn colonization were compared between Spn+ and Spn- samples by Fisher’s Exact test (https://www.socscistatistics.com/tests/fisher/default2.aspx). The difference in abundance of taxa, as shown in bar plots and in alpha diversity between Spn+ and Spn- samples, was calculated by Wilcoxon signed-rank test with Holm-Bonferroni adjustment. Statistical significance of differences between two populations based on beta diversities was calculated by Permanova via Adonis in R.
Results
Demographic and risk factors that associate with Spn colonization
96 children were included in the study cohort. 73 NP samples were analyzed when children were 6 months old and 84 NP samples were analyzed when children were 12 months old (Table 1). Gender, race, breast-feeding history, exposure to smoke, history of atopy, antibiotic treatment (30 days prior to sample collection), daycare attendance, and presence of siblings were variables that had frequently been examined for their association with Spn colonization and some were reported as risk factors [34–39]. We evaluated these factors in our cohort. As previously reported [34], daycare attendance correlated significantly with Spn colonization in the child’s NP, but none of the other demographic factors did in this study cohort (Table 1).
We investigated the association between Spn colonization and common respiratory pathogens, clinically-diagnosed viral upper respiratory infection (URI), and proness to acute otitis media (AOM) (Table 2). Spn colonization in NP has been reported to positively correlate with colonization of other otopathogens and with URI [36, 40]. In our study, Moraxella catarrhalis detected by culture was significantly associated with Spn carriage at 6 months, although not at 12 months (Table 2). Haemophilus influenzae (Hi) did not show significant association with Spn colonization although our group has previously reported an association [40]. This lack of significance in association could be due to the fact that few samples were positive for Hi in this cohort. Staphylococcus aureus did not show correlation with Spn colonization in our cohort (Table 2), although a negative association had been reported in other studies [41, 42]. URI was significantly associated with Spn carriage in 12-month samples but not so in 6-month samples (Table 2). AOM is a common disease in children and is frequently caused by Spn infection [43]. We have previously reported that while some children never develop AOM (AOM-free), some have frequent occurrences [44]. Those who develop at least 3 episodes of AOM (confirmed by tympanocentesis) within 6 months or 4 episodes in 12 months were categorized as sOP (stringently-defined Otitise-Prone). Elevation of Spn colonization was reported in sOP children compared with AOM-free children [36]. Similarly, in our cohort, sOP children had more frequent Spn colonization at both 6 months and 12 months of age compared to AOM-free children (Table 2).
Table 2. Other factors associated with Spn colonization.
| # of samples | Mcat | Hi | SA | URI | AOM-free: sOP | |
|---|---|---|---|---|---|---|
| Spn+ | ||||||
| 6 m | 26 | 15+, 11- [0.011]* | 3+, 23- [0.69] | 0+; 25- [0.088] | 5+, 21- [0.76] | 13:12 [0.036] * |
| 12 m | 29 | 10+, 19- [0.18] | 3+, 26- [0.69] | 1+, 27- [0.26] | 10+,19- [0.024]* | 16:12 [0.037]* |
| total | 55 | |||||
| Spn- | ||||||
| 6 m | 48 | 12+, 36- [0.011]* | 4+, 44- [0.69] | 6+, 42- [0.088] | 8+, 40- [0.76] | 37:11 [0.036] * |
| 12 m | 56 | 11+, 45- [0.18] | 4+, 52- [0.69] | 8+, 48- [0.26] | 7+, 48- [0.024] * | 45:11 [0.037]* |
| total | 104 |
Note: Specific factors are listed as column names. The number of children who were positive (+) or negative (-) for each potential bacterial respiratory pathogen or for URI, or were AOM-free or sOP, are indicated for each age group (6-month or 12-month) and for each Spn colonization phenotype (Spn+ or Spn-). In each age group, the proportion of children colonized with an otopathogen or URI, or designated as AOM-free or sOP, was compared between Spn+ and Spn- samples and statistical significance was assessed by Fisher’s Exact test. The p values are included in square brackets. SA: Staphylococcus aureus.
*: p < 0.05.
Microbiome composition during Spn colonization
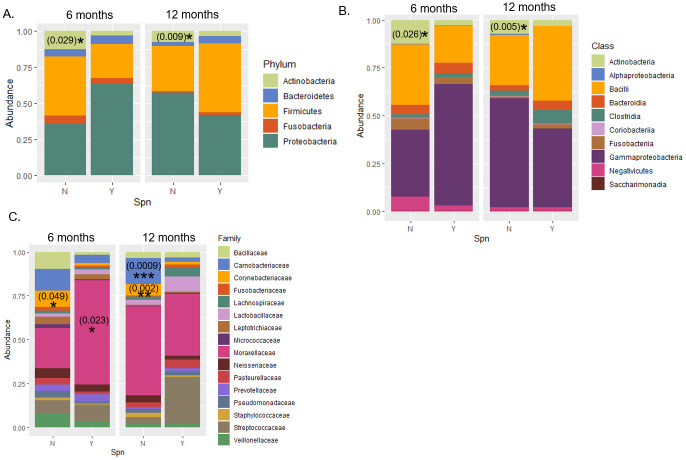
16S rRNA gene sequencing was performed to analyze the microbiome composition in each NP sample. Five phyla were most abundant: Actinobacteria, Bacteroidetes, Firmicutes, Fusobacteria, and Proteobacteria (Fig 1A). Among these, Actinobacteria was significantly reduced in Spn+ samples of both 6 (p = 0.029) and 12 months of ages (p = 0.009), relative to Spn- samples. This was also observed at the class level, where Actinobacteria was found inversely correlated with Spn colonization (Fig 1B; p = 0.026 in samples from 6-month olds and p = 0.005 in samples from 12-month olds). At the family level, Corynebacteriaceae was less abundant in Spn+ samples than in Spn- samples from children of 6 (p = 0.049) or 12 months old (p = 0.002), Moraxellaceae family was more abundant in Spn+ samples from 6 month old children (p = 0.023), and Carnobacteriaceae family was less abundant in Spn+ samples from 12 month old children (p = 0.0009).
Fig 1. Abundance of taxa in Spn+ and Spn- NP samples.
Proportion of taxa in Spn+ or Spn- NP samples at different ages were shown at the level of phylum (A), class (B), family (C) as bar plots. *: p < 0.05; **: p < 0.005; ***: p < 0.0005, Wilcoxon signed-rank test with Holm-Bonferroni adjustment. The absolute p values are included in parentheses on the graphs.
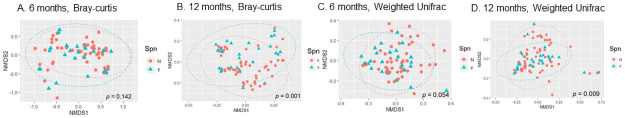
Four indices of alpha diversity were measured and no significant difference was observed between Spn+ and Spn- samples from children of either 6 or 12 months old (S1 Fig). In contrast, beta diversity measurements showed composition differences in the microbiome of Spn+ and Spn- samples. Bray-Curtis dissimilarity and weighted unifrac were measured among the samples and visualized in NMDS plots (Fig 2). Both measurements identified a significant difference in microbiome composition between Spn+ and Spn- samples from children at 12 months of age (p = 0.001 and 0.009), although only weighted unifrac identified a significant difference in microbiome composition between Spn+ and Spn- samples from 6-month old children (p = 0.054).
Fig 2. Difference in beta diversity of microbiome between Spn+ and Spn- NP samples in children at 12 months.
Bray-Curtis distance (A and B) or weighted unifrac (C and D) of samples collected at 6 months or 12 months were measured and visualized on NMDS plots. Permanova was performed for samples stratified by age to assess the difference between Spn+ and Spn- samples. The p values are shown at the right bottom corner of each plot.
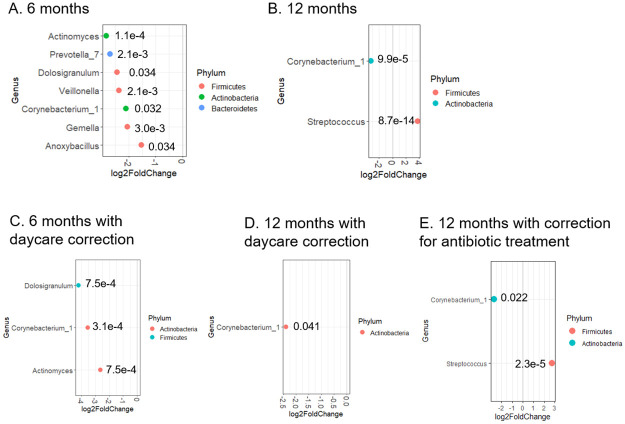
Individual bacteria genera that change abundance upon Spn colonization were identified (Fig 3). In samples from 6 month-old children, reduced abundance of Actinomyces (p = 0.0001), Prevotella_7 (p = 0.002), Dolosigranulum (p = 0.034), Veillonella (p = 0.002), Corynebacterium_1 (p = 0.032), Gemella (p = 0.003), and Anoxybacillus (p = 0.034) were identified in Spn+ samples compared to Spn- samples (Fig 3A). No genera, not even Streptococcus, were found in higher abundance in the Spn+ group. This observation could be explained if lower abundance of Streptococcus spp. other than S. pneumoniae were present in Spn+ samples relative to Spn- samples, as previously reported [27], so the genus of Streptococcus as a whole did not exhibit prominence in Spn+ samples. In 12 month-old children, the Streptococcus genus was more abundant (p = 8.7 x 10−14) and Corynebacterium less abundant (p = 9.9 x 10−5) in Spn+ samples compared with Spn- samples (Fig 3B). Because Spn colonization is influenced by demograhic factors such as daycare attendance (Table 1), we asked whether daycare attendance affects Spn-induced microbiome change. Indeed, fewer differentially abundant genera were observed between Spn+ and Spn- samples when daycare attendance was considered a covariate: among the genera listed in Fig 3A, only Dolosigranulum (p = 0.0007), Corynebacterium_1 (p = 0.0003), and Actinomyces (p = 0.0007) remained differentially abundant in Spn+ samples versus Spn- samples from 6-month old children (Fig 3C), and only Corynebacterium_1 remained differentially abundant in Spn+ samples compared with Spn- samples from 12-month old children (p = 0.041) (Fig 3D).
Fig 3. Bacterial genera that show differential abundance between Spn+ and Spn- samples.
Abundance of bacterial genera was compared between Spn+ and Spn- samples collected at 6 months (A), or at 12 months (B) without correction for daycare attendance, or with correction for daycare attendance (C, 6 months; D, 12 months) using DESeq2 software (see Materials and methods). The ones with adjusted p value less than 0.05 are shown as dots in the graphs. The adjusted p value is indicated next to each dot. The x axis exhibits the log2 ratio between abundance of bacterial genera in Spn+ samples and that in Spn- samples.
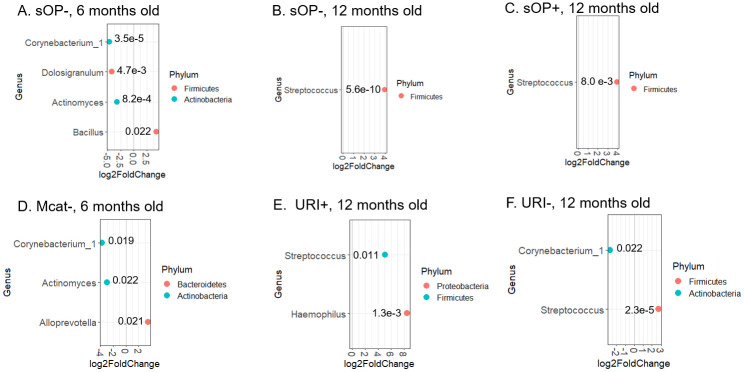
Among other factors associated with Spn colonization, sOP phenotype, Mcat colonization, and URI were significantly associated with Spn colonization in children (Table 2). To determine how these variables influence microbiome changes upon Spn colonization, we divided samples into variable+ and variable- subgroups and analyzed the differential abundance of genera between Spn+ and Spn- samples in each subgroup. We found that in the sOP- subgroup (i.e., AOM-free), Corynebacterium_1 (p = 3.5 x 10−5), Dolosigranulum (p = 0.0047), and Actinomyces (p = 8.2 x 10−4) remained at lower abundance in Spn+ samples relative to Spn- samples, but none of the other genera shown in Fig 3A did, and Bacillus became more abundant (p = 0.022) in Spn+ samples from children of 6-month old (Fig 4A). In sOP+ samples, however, no genus was found to differ in abundance between Spn+ and Spn- samples from 6-month old children. These differential findings from sOP+ and sOP- samples indicate that the sOP child phenotype may be associated with significant microbiome changes in response to Spn colonization when children were at 6 months of age. Among children of 12 months old, Corynebacterium was no longer at lower abundance but Streptococcus stayed at higher abundance in Spn+ samples relative to Spn- samples, once the population was split into AOM-free and sOP+ subgroups (Fig 4B and 4C; p = 5.5 x 10−10 and 0.008).
Fig 4. Mcat detection, URI and Otitis Proneness as factors that contribute to microbiome difference between Spn+ and Spn- NP samples.
Samples were divided into AOM-free/sOP, Mcat+/Mcat-, and URI+/URI- samples at each age group. Differential abundance of genera between Spn+ and Spn- samples was determined in each subgroup: AOM-free/sOP (A-C), Mcat (D), or URI (E-F), by DESeq2 software (see Materials and methods). The ones with adjusted p value less than 0.05 are shown as dots in the graphs. The adjusted p value is indicated next to each dot. The x axis exhibits the log2 ratio between abundance of bacterial genera in Spn+ samples and that in Spn- samples.
Mcat colonization detected by culture significantly segregated Spn+ and Spn- samples (Table 2) but this only occurred in 6-month old children. Consequently samples from 6-month old children were divided into Mcat+ and Mcat- subgroups to examine the effects of Mcat on microbiome differences when Spn colonization was detected. Within the Mcat- subgroup, Corynebacterium_1 (p = 0.019) and Actinomyces (p = 0.022) were at lower abundance in Spn+ samples, but none of the other genera shown in Fig 3A were, and Alloprevotella (p = 0.021) showed higher abundance in Spn+ samples, compared with Spn- samples (Fig 4D). Within the Mcat+ subgroup, however, no genus showed significant difference in abundance between Spn+ and Spn- samples, suggesting that Mcat colonization may override the effects of Spn colonization on microbiome changes.
The third factor we examined was URI, which showed a significant positive association with Spn colonization in children 12 months age but not children 6 months of age (Table 2). Consequently only samples from 12-month old children were divided into URI+ and URI- subgroups and investigated for the influence of URI on microbiome changes in association with Spn colonization. Withiin the URI+ samples, Haemophilus (p = 0.0013), in addition to Streptococcus (p = 0.011; as shown in Fig 3B), was found at higher abundance in Spn+ samples relative to Spn- samples (Fig 4E). Within the URI- subgroup, Corynebacterium_1 (p = 0.022) and Streptococcus (p = 2.3 x 10−5) remained at lower and higher abundance, respectively, in Spn+ samples than in Spn- samples (Fig 4F), as shown in the comparison among all samples from 12-month olds (Fig 3B).
Corynebacterium spp. inhibited Spn proliferation in vitro and reduced Spn colonization densities in vivo
An inverse relationship between Corynebacterium genus and Spn colonization was observed (Fig 3A and 3B), which was not affected by daycare attendance (Fig 3C and 3D) or other pathogens such as Moraxella or URI. The non-otitis-prone state was associated with the presence of Corynebacterium genus (Fig 4). Therefore, we hypothesized that Corynebacterium spp. interfered with Spn NP colonization. To test this, we first isolated Corynebacterium spp. from NP samples of children and, using PCR primers that target the rpoB gene in Corynebacterium [33], we identified C. propinquum and C. pseudodiphtheriticum (S2A–S2E Fig).
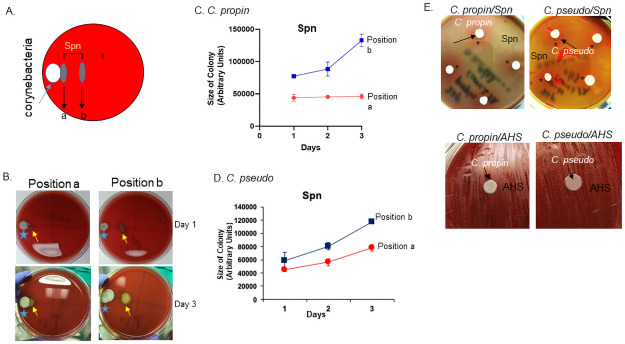
To evaluate the interaction between Corynebacterium and Spn in vitro, we inoculated a colony of Spn22F at different distances to C. propinquum or C. pseudodiphtheriticum colonies and monitored their growth over three days (Fig 5A and 5B). Spn colonies farther away from the Corynebacterium colonies grew faster than those closer (Fig 5C and 5D), indicating an inhibitory effect of C. propinquum and C. pseudodiphtheriticum on Spn proliferation. An alternative approach was employed to confirm this inhibitory effect. Concentrated C. propinquum or C. pseudodiphtheriticum was added to a lawn of Spn22F on blood agar plates and the growth of Spn22F was visualized the next day. A pink ring (indicatng minimal hemolysis) of Spn22F was observed around C. propinquum or C. pseudodiphtheriticum colony (Fig 5E, *), in contrast to the yellow/green lawn (indicating complete hemolysis) of Spn22F, suggesting inhibited growth of Spn22F by Corynebacterium. This inhibition was not observed when C. propinquum or C. pseudodiphtheriticum was inoculated on a lawn of non-Spn strain of alpha-hemolytic Streptococcus (AHS) as a control (Fig 5E) suggesting that the inhibitory effect was not achieved by competition for nutrients in the agar.
Fig 5. Growth inhibition of Corynebacterium by Spn.
A) Schematic representation of co-culture experiment for Corynebacterium spp. and Spn. Spn22F was inoculated right next to a Corynebacterium colony (position a) or with some distance (position b). B) Representative images of Corynebacterium spp. co-cultured with Spn22F at position a or b (as shown in A) on day 1 and day 3. C, D) Measurement of size changes in Spn colonies placed at positions a and b next to C. propinquum (C) or next to C. pseudodiphtheriticum (D) over 3 days. E) Images of culturing C. propinquum (left) or C. pseudodiphtheriticum (right) on top of a lawn of Spn22F (top panels) or AHS (bottom panels).
Discussion
The intranasal colonization of Spn is a prerequisite step for its pathogenesis, which occurs via interactions with host as well as other microorganisms in NP [3]. We show here that several genera of bacteria in the NP microbiome correlated negatively or positively with Spn colonization, and some of these correlations appeared to be influenced by daycare attendance or other factors such as upper respiratory infection (URI), Moraxella co-colonization, and propensity to develop AOM among young children. Among these genera, Coryenbacterium showed a consistent inverse relationship with Spn colonization with little influence by daycare attendance or other factors. We isolated C. propinquum and C. pseudodiphtheriticum and found that both inhibited the growth of Spn serotype 22F strain in vitro.
We first evaluated the distribution of demographic and other factors that associate with Spn colonization. We found that daycare attendance, Mcat colonization, URI, and AOM recurrence were significantly associated with Spn colonization, but breastfeeding, race, gender, exposure to smoke, symptoms of atopy, presence of siblings were not (Table 1). These findings were generally consistent with previous reports [34–38]. Some inconsistencies were observed, in particular in the effects of race and siblings, which had been reported as risk factors for Spn colonization and infection [37, 39]. This disparity could be due to sampling difference, in that the majority of samples in our study were collected from children in families that were mostly middle class and caucacian. We did not observe influence of antibiotic treatment on colonization of Spn, inconsistent with what was reported [37]. Perhaps the antibiotic regimens (types of antibiotics, duration and intervals) in our cohorts differ from the previous report.
Microbiota in NP consists of pathogens and commensals [4–6, 45], both of which influence Spn colonization and pathogenesis [3, 45, 46]. We focused on the roles of commensals on Spn colonization, because they are relatively less studied and impose novel therapeutic potentials for treating Spn-related diseases. The Actinobacteria phyla had an inverse relationship with Spn colonization in our chorts, consistent with previous reports on microbiome distribution in human nostrils [47]. A dominant genus of Actinobacteria phylum in nostrils is the Corynebacterium [48], which was reported to negatively correlate with Spn colonization in pediatric samples [15, 17, 18] and was confirmed in our study (Fig 3). In URI+ samples however, Corynebacterium no longer exhibited inverse relationship with Spn colonization. Perhaps, the condition of URI supports the colonization of Corynebacterium, counteracting the effects of Spn. This notion is consistent with the report by Edouard et al that C. propinquum was elevated in patients with symptoms of viral respiratory tract infections, compared with healthy controls [49].
We also found an inverse relationship between Dolosigranulum and Spn, as previously reported [18]. Our study revealed a few new taxa that correlated negatively with Spn colonization in NP of 6-month olds—Actinomyces, Prevotella, Veillonella, Gemella, and Anoxybacillus. Among these, Prevotella, Veillonella, and Gemella were found inversely correlated with pneumonia in adults and elderly people [50], who had elevated Spn in oropharyngeal samples. Prevetella was reported to correlate with a reduced risk of hospital-acquired pneumonia in ICU patients [51]. Notably, these anaerobic genera were most pronouncedly influenced by daycare attendance and other confounding factors (Figs 3C and 4), suggesting their sensitivity to these factors. They were no longer found differentially abundant between the Spn+ and Spn- NPs of 12-month old children (Fig 3B). Perhaps nasal microbiome in young children is more susceptible to changes induced by Spn colonization than older children. It is known that microbiome composition undergoes profound changes within the first year of a child’s life [50, 52], with bacteria density increasing and gained dominance of certain taxa such as Moraxella spp. Spn colonization may thus not be sufficient to alter the abundance of taxa like Moraxella in the NP of 12-month olds. In contrast, the abundance of Corynebacterium spp. decreases as a child ages, making them remain susceptible to depletion upon Spn colonization at the age of 12 months, as was observed (Fig 3B).
Among human commensals, the Corynebacterium is one of the most studied and has been extensively reported to correlate with a healthy state in nasopharynx [53–59]. Its function in Spn pathology was also reported. C. accolens, for example, was found to inhibit Spn in vitro via releasing fatty acid; its functions in vivo however were not reported [15]. On the other hand, C. pseudodiphtheriticum 090104 was shown to inhibit secondary infection of S. pneumoniae in vivo probably by inducing elevated TNF-alpha and interferon gamma levels [19]. The immune effect of Corynebacterium was reported to be strain-specific, since a different C. pseudodiphtheriticum strain did not exhibit this function [20].
Our study showed the in vitro effects of two Corynebacterium species, C. propinquum and C. pseudodiphtheriticum, on Spn proliferation. We do not know the nature of this inhibition, but we suspect that it would differ from aforementioned inhibition by C. accolens [15], since C. accolens is a lipophilic species and requires lipid for its growth but C. propinquum and C. pseudodiphtheriticum are not [13]. It was reported recently that C. propinquum released siderophores to inhibit the growth of coagulase-negative Staphylococcus species but not coagulase-positive Staphylococcus species such as S. aureus [60]. This inhibition was not observed from C. pseudodiphtheriticum so likely differs from the inhibition we observed in this study.
Our study has several limitations. First, 16S rRNA gene sequencing at the V4 region does not provide resolution to the species level, therefore we do not know what bacterial species correlate with Spn colonization. This limitation significantly reduces the number of taxa to be uncovered and imposes difficulty in isolating relevant commensals for follow-up studies. Full-length 16S rRNA gene sequencing or shotgun metagenomics with genome reconstruction would need to be performed to overcome this shortcoming. Second, our in vitro assays imply that the Corynebacterium species inhibit the proliferation of Spn22F. This interpretation needs to be considered cautiously, as only one isolate of each species was tested in our study. Additionally, the in vitro effects we observed may not directly translate to their effects on Spn colonization in patients, since Spn colonization involves more steps (survival, adhesion, etc.) than proliferation. Furthermore, we only used one Spn strain (Spn22F) in our study whereas in human more than 90 strains of Spn have been identified. More Spn strains would need to be tested to evaluate whether the Corynebacterium species inhibit Spn proliferation in general, but not specific to Spn22F. We also do not know whether the inhibitory effects of Corynebacterium in vitro could be translated to their effects in vivo. C. pseudodiphtheriticum had been shown recently to induce a pro-immune response in mice [19], which in turn may imped the colonization of Spn. In contrast, C. propinquum was reported to correlate with anti-flammatory mediators in human nasopharynx [61]. Whether and what type of immune responses the Corynebacterium species may elicit in vivo to imped Spn colonization thus await future investigation. Finally, our study population is from middle-class, suburban pediatric practices and thus may not represent the general population which are more socioeconomically diverse.
Supporting information
Alpha diversity indices of nasal microbiome from Spn+ or Spn- children of 6 and 12 months olds were calculated and graphed in box plots.
(TIF)
A) Agarose gel electrophoresis of PCR products from rpoB gene. Lanes 1 and 3: PCR products from colonies grown on chocolate plates; lanes 2 and 4: PCR products from colonies grown on blood agar plates. The growth of Corynebacterium on chocolate plates was not as robust as on blood agar plates, so the colonies grown on chocolate plates were found to differ from Corynebacterium and served as a negative control. B,C) Sequences of PCR products from two Corynebacterium species that were later identified as C. propinquum and C. pseudodiphtheriticum. D) Alignment of PCR sequences from C. propinquum against GeneBank database. E) Alignment of PCR sequences from C. pseudodiphtheriticum against GenBank database.
(TIF)
(XLSX)
(TIF)
Acknowledgments
We would like to thank Eduardo Gonzalez for technical assistance and University of North Carolina Microbiome Core for processing samples and sequencing the 16S rRNA gene.
Data Availability
The sequence and sample data have been submitted to NCBI Sequence Read Archive, accession number (PRJNA720045).
Funding Statement
This work was supported by the Rochester General Hospital Kidd Fund grant awarded to LX. Sponsors did not play a role in study design, data collection and analysis, decision to publish or preparation of manuscript.
References
- 1.Käyhty H NA, Soininen A, Väkeväinen M. The Immunological Basis for Immunization Series, Module 12: Pneumococcal Vaccines. Publications of the World Health Organization, Department of Immunization, Vaccines and Biologicals, World Health Organization. 2009.
- 2.Feldman C, Anderson R. Epidemiology, virulence factors and management of the pneumococcus. F1000Research. 2016;5:2320. doi: 10.12688/f1000research.9283.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weiser JN, Ferreira DM, Paton JC. Streptococcus pneumoniae: transmission, colonization and invasion. Nature reviews Microbiology. 2018Jun;16(6):355–67. doi: 10.1038/s41579-018-0001-8 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biesbroek G, Tsivtsivadze E, Sanders EA, Montijn R, Veenhoven RH, Keijser BJ, et al. Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am J Respir Crit Care Med. 2014Dec1;190(11):1283–92. doi: 10.1164/rccm.201407-1240OC . [DOI] [PubMed] [Google Scholar]
- 5.Bomar L, Brugger SD, Lemon KP. Bacterial microbiota of the nasal passages across the span of human life. Curr Opin Microbiol. 2018Feb;41:8–14. doi: 10.1016/j.mib.2017.10.023 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Man WH, de Steenhuijsen Piters WA, Bogaert D. The microbiota of the respiratory tract: gatekeeper to respiratory health. Nature reviews Microbiology. 2017May;15(5):259–70. doi: 10.1038/nrmicro.2017.14 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Planer JD, Peng Y, Kau AL, Blanton LV, Ndao IM, Tarr PI, et al. Development of the gut microbiota and mucosal IgA responses in twins and gnotobiotic mice. Nature. 2016Jun9;534(7606):263–6. doi: 10.1038/nature17940 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Badal VD, Wright D, Katsis Y, Kim HC, Swafford AD, Knight R, et al. Challenges in the construction of knowledge bases for human microbiome-disease associations. Microbiome. 2019Sep5;7(1):129. doi: 10.1186/s40168-019-0742-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Round JL, O’Connell RM, Mazmanian SK. Coordination of tolerogenic immune responses by the commensal microbiota. Journal of autoimmunity. 2010May;34(3):J220–5. doi: 10.1016/j.jaut.2009.11.007 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Atarashi K, Honda K. Microbiota in autoimmunity and tolerance. Curr Opin Immunol. 2011Dec;23(6):761–8. doi: 10.1016/j.coi.2011.11.002 . [DOI] [PubMed] [Google Scholar]
- 11.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009Oct30;139(3):485–98. doi: 10.1016/j.cell.2009.09.033 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kriegel MA, Sefik E, Hill JA, Wu HJ, Benoist C, Mathis D. Naturally transmitted segmented filamentous bacteria segregate with diabetes protection in nonobese diabetic mice. Proc Natl Acad Sci U S A. 2011Jul12;108(28):11548–53. doi: 10.1073/pnas.1108924108 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Funke G, von Graevenitz A, Clarridge JE 3rd, Bernard KA. Clinical microbiology of coryneform bacteria. Clinical microbiology reviews. 1997Jan;10(1):125–59. doi: 10.1128/CMR.10.1.125 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaspar U, Kriegeskorte A, Schubert T, Peters G, Rudack C, Pieper DH, et al. The culturome of the human nose habitats reveals individual bacterial fingerprint patterns. Environ Microbiol. 2016Jul;18(7):2130–42. doi: 10.1111/1462-2920.12891 . [DOI] [PubMed] [Google Scholar]
- 15.Bomar L, Brugger SD, Yost BH, Davies SS, Lemon KP. Corynebacterium accolens Releases Antipneumococcal Free Fatty Acids from Human Nostril and Skin Surface Triacylglycerols. MBio. 2016Jan5;7(1):e01725–15. doi: 10.1128/mBio.01725-15 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laufer AS, Metlay JP, Gent JF, Fennie KP, Kong Y, Pettigrew MM. Microbial communities of the upper respiratory tract and otitis media in children. mBio. 2011Feb1;2(1):e00245–10. doi: 10.1128/mBio.00245-10 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kelly MS, Surette MG, Smieja M, Pernica JM, Rossi L, Luinstra K, et al. The Nasopharyngeal Microbiota of Children With Respiratory Infections in Botswana. Pediatr Infect Dis J. 2017Sep;36(9):e211–e8. doi: 10.1097/INF.0000000000001607 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Steenhuijsen Piters WAA, Jochems SP, Mitsi E, Rylance J, Pojar S, Nikolaou E, et al. Interaction between the nasal microbiota and S. pneumoniae in the context of live-attenuated influenza vaccine. Nature communications. 2019Jul5;10(1):2981. doi: 10.1038/s41467-019-10814-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanmani P, Clua P, Vizoso-Pinto MG, Rodriguez C, Alvarez S, Melnikov V, et al. Respiratory Commensal Bacteria Corynebacterium pseudodiphtheriticum Improves Resistance of Infant Mice to Respiratory Syncytial Virus and Streptococcus pneumoniae Superinfection. Front Microbiol. 2017;8:1613. doi: 10.3389/fmicb.2017.01613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ortiz Moyano R, Raya Tonetti F, Tomokiyo M, Kanmani P, Vizoso-Pinto MG, Kim H, et al. The Ability of Respiratory Commensal Bacteria to Beneficially Modulate the Lung Innate Immune Response Is a Strain Dependent Characteristic. Microorganisms. 2020May13;8(5). doi: 10.3390/microorganisms8050727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pichichero ME, Kaur R, Casey JR, Sabirov A, Khan MN, Almudevar A. Antibody response to Haemophilus influenzae outer membrane protein D, P6, and OMP26 after nasopharyngeal colonization and acute otitis media in children. Vaccine. 2010Oct18;28(44):7184–92. doi: 10.1016/j.vaccine.2010.08.063 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaur R, Morris M, Pichichero ME. Epidemiology of Acute Otitis Media in the Postpneumococcal Conjugate Vaccine Era. Pediatrics. 2017Sep;140(3). doi: 10.1542/peds.2017-0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu L, Earl J, Bajorski P, Gonzalez E, Pichichero ME. Nasopharyngeal microbiome analyses in otitis-prone and otitis-free children. Int J Pediatr Otorhinolaryngol. 2021Jan21;143:110629. doi: 10.1016/j.ijporl.2021.110629. [DOI] [PubMed] [Google Scholar]
- 24.Kalu SU, Ataya RS, McCormick DP, Patel JA, Revai K, Chonmaitree T. Clinical spectrum of acute otitis media complicating upper respiratory tract viral infection. Pediatr Infect Dis J. 2011Feb;30(2):95–9. doi: 10.1097/INF.0b013e3181f253d5 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patel JA, Nair S, Revai K, Grady J, Chonmaitree T. Nasopharyngeal acute phase cytokines in viral upper respiratory infection: impact on acute otitis media in children. Pediatr Infect Dis J. 2009Nov;28(11):1002–7. doi: 10.1097/INF.0b013e3181aa5b13 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ren D, Xu Q, Almudevar AL, Pichichero ME. Impaired Proinflammatory Response in Stringently Defined Otitis-prone Children During Viral Upper Respiratory Infections. Clin Infect Dis. 2019Apr24;68(9):1566–74. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Friedel V, Chang A, Wills J, Vargas R, Xu Q, Pichichero ME. Impact of respiratory viral infections on alpha-hemolytic streptococci and otopathogens in the nasopharynx of young children. Pediatr Infect Dis J. 2013Jan;32(1):27–31. doi: 10.1097/INF.0b013e31826f6144 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lieberthal AS, Carroll AE, Chonmaitree T, Ganiats TG, Hoberman A, Jackson MA, et al. The diagnosis and management of acute otitis media. Pediatrics. 2013Mar;131(3):e964–99. doi: 10.1542/peds.2012-3488 . [DOI] [PubMed] [Google Scholar]
- 29.Xu Q, Casey JR, Chang A, Pichichero ME. When co-colonizing the nasopharynx haemophilus influenzae predominates over Streptococcus pneumoniae except serotype 19A strains to cause acute otitis media. Pediatr Infect Dis J. 2012Jun;31(6):638–40. doi: 10.1097/INF.0b013e31824ba6f7 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chapman TJ, Morris MC, Xu L, Pichichero ME. Nasopharyngeal colonization with pathobionts is associated with susceptibility to respiratory illnesses in young children. PLoS One. 2020;15(12):e0243942. doi: 10.1371/journal.pone.0243942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nature methods. 2016Jul;13(7):581–3. doi: 10.1038/nmeth.3869 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kong P, Richardson PA, Hong C. Direct colony PCR-SSCP for detection of multiple pythiaceous oomycetes in environmental samples. J Microbiol Methods. 2005Apr;61(1):25–32. doi: 10.1016/j.mimet.2004.10.019 . [DOI] [PubMed] [Google Scholar]
- 33.Khamis A, Raoult D, La Scola B. rpoB gene sequencing for identification of Corynebacterium species. J Clin Microbiol. 2004Sep;42(9):3925–31. doi: 10.1128/JCM.42.9.3925-3931.2004 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petraitiene S, Alasevicius T, Staceviciene I, Vaiciuniene D, Kacergius T, Usonis V. The influence of Streptococcus pneumoniae nasopharyngeal colonization on the clinical outcome of the respiratory tract infections in preschool children. BMC Infect Dis. 2015Sep30;15:403. doi: 10.1186/s12879-015-1149-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perez AC, Pang B, King LB, Tan L, Murrah KA, Reimche JL, et al. Residence of Streptococcus pneumoniae and Moraxella catarrhalis within polymicrobial biofilm promotes antibiotic resistance and bacterial persistence in vivo. Pathog Dis. 2014Apr;70(3):280–8. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu Q, Wischmeyer J, Gonzalez E, Pichichero ME. Nasopharyngeal polymicrobial colonization during health, viral upper respiratory infection and upper respiratory bacterial infection. J Infect. 2017Jul;75(1):26–34. doi: 10.1016/j.jinf.2017.04.003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koliou MG, Andreou K, Lamnisos D, Lavranos G, Iakovides P, Economou C, et al. Risk factors for carriage of Streptococcus pneumoniae in children. BMC pediatrics. 2018Apr26;18(1):144. doi: 10.1186/s12887-018-1119-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pettigrew MM, Gent JF, Revai K, Patel JA, Chonmaitree T. Microbial interactions during upper respiratory tract infections. Emerg Infect Dis. 2008Oct;14(10):1584–91. doi: 10.3201/eid1410.080119 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wortham JM, Zell ER, Pondo T, Harrison LH, Schaffner W, Lynfield R, et al. Racial disparities in invasive Streptococcus pneumoniae infections, 1998–2009. Clin Infect Dis. 2014May;58(9):1250–7. . [DOI] [PubMed] [Google Scholar]
- 40.Xu Q, Almudervar A, Casey JR, Pichichero ME. Nasopharyngeal bacterial interactions in children. Emerg Infect Dis. 2012Nov;18(11):1738–45. doi: 10.3201/eid1811.111904 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chien YW, Vidal JE, Grijalva CG, Bozio C, Edwards KM, Williams JV, et al. Density interactions among Streptococcus pneumoniae, Haemophilus influenzae and Staphylococcus aureus in the nasopharynx of young Peruvian children. Pediatr Infect Dis J. 2013Jan;32(1):72–7. doi: 10.1097/INF.0b013e318270d850 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dunne EM, Smith-Vaughan HC, Robins-Browne RM, Mulholland EK, Satzke C. Nasopharyngeal microbial interactions in the era of pneumococcal conjugate vaccination. Vaccine. 2013May1;31(19):2333–42. doi: 10.1016/j.vaccine.2013.03.024 . [DOI] [PubMed] [Google Scholar]
- 43.Pichichero ME. Otitis media. Pediatric clinics of North America. 2013Apr;60(2):391–407. doi: 10.1016/j.pcl.2012.12.007 . [DOI] [PubMed] [Google Scholar]
- 44.Pichichero ME. Ten-Year Study of Acute Otitis Media in Rochester, NY. Pediatr Infect Dis J. 2016Sep;35(9):1027–32. doi: 10.1097/INF.0000000000001216 . [DOI] [PubMed] [Google Scholar]
- 45.Hardy BL, Merrell DS. Friend or Foe: Interbacterial Competition in the Nasal Cavity. J Bacteriol. 2021Feb8;203(5). doi: 10.1128/JB.00480-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bergenfelz C, Hakansson AP. Streptococcus pneumoniae Otitis Media Pathogenesis and How It Informs Our Understanding of Vaccine Strategies. Current otorhinolaryngology reports. 2017;5(2):115–24. doi: 10.1007/s40136-017-0152-6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lemon KP, Klepac-Ceraj V, Schiffer HK, Brodie EL, Lynch SV, Kolter R. Comparative analyses of the bacterial microbiota of the human nostril and oropharynx. mBio. 2010;1(3). doi: 10.1128/mBio.00129-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ibironke O, McGuinness LR, Lu SE, Wang Y, Hussain S, Weisel CP, et al. Species-level evaluation of the human respiratory microbiome. GigaScience. 2020Apr1;9(4). . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Edouard S, Million M, Bachar D, Dubourg G, Michelle C, Ninove L, et al. The nasopharyngeal microbiota in patients with viral respiratory tract infections is enriched in bacterial pathogens. Eur J Clin Microbiol Infect Dis. 2018Sep;37(9):1725–33. doi: 10.1007/s10096-018-3305-8 . [DOI] [PubMed] [Google Scholar]
- 50.de Steenhuijsen Piters WAA, Binkowska J, Bogaert D. Early Life Microbiota and Respiratory Tract Infections. Cell Host Microbe. 2020Aug12;28(2):223–32. doi: 10.1016/j.chom.2020.07.004 . [DOI] [PubMed] [Google Scholar]
- 51.Bousbia S, Papazian L, Saux P, Forel JM, Auffray JP, Martin C, et al. Repertoire of intensive care unit pneumonia microbiota. PLoS One. 2012;7(2):e32486. doi: 10.1371/journal.pone.0032486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mika M, Mack I, Korten I, Qi W, Aebi S, Frey U, et al. Dynamics of the nasal microbiota in infancy: a prospective cohort study. J Allergy Clin Immunol. 2015Apr;135(4):905–12 e11. doi: 10.1016/j.jaci.2014.12.1909 . [DOI] [PubMed] [Google Scholar]
- 53.Pettigrew MM, Laufer AS, Gent JF, Kong Y, Fennie KP, Metlay JP. Upper respiratory tract microbial communities, acute otitis media pathogens, and antibiotic use in healthy and sick children. Applied & Environmental Microbiology. 2012;78(17):6262–70. doi: 10.1128/AEM.01051-12 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lappan R, Imbrogno K, Sikazwe C, Anderson D, Mok D, Coates H, et al. A microbiome case-control study of recurrent acute otitis media identified potentially protective bacterial genera. BMC Microbiol. 2018Feb20;18(1):13. doi: 10.1186/s12866-018-1154-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Neumann RP, Hilty M, Xu B, Usemann J, Korten I, Mika M, et al. Nasal microbiota and symptom persistence in acute respiratory tract infections in infants. ERJ open research. 2018Oct;4(4). doi: 10.1183/23120541.00066-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McCauley K, Durack J, Valladares R, Fadrosh DW, Lin DL, Calatroni A, et al. Distinct nasal airway bacterial microbiotas differentially relate to exacerbation in pediatric patients with asthma. J Allergy Clin Immunol. 2019Nov;144(5):1187–97. doi: 10.1016/j.jaci.2019.05.035 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Santee CA, Nagalingam NA, Faruqi AA, DeMuri GP, Gern JE, Wald ER, et al. Nasopharyngeal microbiota composition of children is related to the frequency of upper respiratory infection and acute sinusitis. Microbiome. 2016Jul01;4(1):34. doi: 10.1186/s40168-016-0179-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brugger SD, Bomar L, Lemon KP. Commensal-Pathogen Interactions along the Human Nasal Passages. PLoS Pathog. 2016Jul;12(7):e1005633. doi: 10.1371/journal.ppat.1005633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Toivonen L, Hasegawa K, Waris M, Ajami NJ, Petrosino JF, Camargo CA Jr., et al. Early nasal microbiota and acute respiratory infections during the first years of life. Thorax. 2019Jun;74(6):592–9. doi: 10.1136/thoraxjnl-2018-212629 . [DOI] [PubMed] [Google Scholar]
- 60.Stubbendieck RM, May DS, Chevrette MG, Temkin MI, Wendt-Pienkowski E, Cagnazzo J, et al. Competition among Nasal Bacteria Suggests a Role for Siderophore-Mediated Interactions in Shaping the Human Nasal Microbiota. Appl Environ Microbiol. 2019May15;85(10). doi: 10.1128/AEM.02406-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Enoksson F, Ruiz Rodriguez A, Peno C, Balcazar Lopez C, Tjernstrom F, Bogaert D, et al. Niche- and Gender-Dependent Immune Reactions in Relation to the Microbiota Profile in Pediatric Patients with Otitis Media with Effusion. Infect Immun. 2020Sep18;88(10). doi: 10.1128/IAI.00147-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alpha diversity indices of nasal microbiome from Spn+ or Spn- children of 6 and 12 months olds were calculated and graphed in box plots.
(TIF)
A) Agarose gel electrophoresis of PCR products from rpoB gene. Lanes 1 and 3: PCR products from colonies grown on chocolate plates; lanes 2 and 4: PCR products from colonies grown on blood agar plates. The growth of Corynebacterium on chocolate plates was not as robust as on blood agar plates, so the colonies grown on chocolate plates were found to differ from Corynebacterium and served as a negative control. B,C) Sequences of PCR products from two Corynebacterium species that were later identified as C. propinquum and C. pseudodiphtheriticum. D) Alignment of PCR sequences from C. propinquum against GeneBank database. E) Alignment of PCR sequences from C. pseudodiphtheriticum against GenBank database.
(TIF)
(XLSX)
(TIF)
Data Availability Statement
The sequence and sample data have been submitted to NCBI Sequence Read Archive, accession number (PRJNA720045).