Abstract
mRNAs that contain premature stop codons are selectively degraded in all eukaryotes tested, a phenomenon termed “nonsense-mediated mRNA decay” (NMD) or “mRNA surveillance.” NMD may function to eliminate aberrant mRNAs so that they are not translated, because such mRNAs might encode deleterious polypeptide fragments. In both yeasts and nematodes, NMD is a nonessential system. Mutations affecting three yeast UPF genes or seven nematode smg genes eliminate NMD. We report here the molecular analysis of smg-2 of Caenorhabditis elegans. smg-2 is homologous to UPF1 of yeast and to RENT1 (also called HUPF1), a human gene likely involved in NMD. The striking conservation of SMG-2, Upf1p, and RENT1/HUPF1 in both sequence and function suggests that NMD is an ancient system, predating the divergence of most eukaryotes. Despite similarities in the sequences of SMG-2 and Upf1p, expression of Upf1p in C. elegans does not rescue smg-2 mutants. We have prepared anti-SMG-2 polyclonal antibodies and identified SMG-2 on Western blots. SMG-2 is phosphorylated, and mutations of the six other smg genes influence the state of SMG-2 phosphorylation. In smg-1, smg-3, and smg-4 mutants, phosphorylation of SMG-2 was not detected. In smg-5, smg-6, and smg-7 mutants, a phosphorylated isoform of SMG-2 accumulated to abnormally high levels. In smg-2(r866) and smg-2(r895) mutants, which harbor single amino acid substitutions of the SMG-2 nucleotide binding site, phosphorylated SMG-2 accumulated to abnormally high levels, similar to those observed in smg-5, smg-6, and smg-7 mutants. We discuss these results with regard to the in vivo functions of SMG-2 and NMD.
Modulating the rates of mRNA degradation is an important control point for both regulated and constitutive gene expression. An understanding of the molecular mechanisms of selective mRNA turnover, however, is only beginning to emerge. mRNAs are selectively degraded by the interplay of specific cis-acting elements and trans-acting factors (for reviews, see references 35 and 57). cis-acting elements, which affect the stability of mRNAs in which they reside, have been defined in considerable detail. Examples include the poly(A) tail, AU-rich elements, the iron-responsive element, and translated instability elements (19, 29, 35, 71). Our understanding of trans-acting factors involved in mRNA turnover, however, is much less complete. Numerous proteins that interact with specific cis-acting elements have been identified, but how these factors regulate stability is unknown in most cases.
Several cis-acting elements function as part of deadenylation-dependent mRNA turnover, an important general system of mRNA turnover that has been elaborated in recent years. The components of this system have been most thoroughly described for yeast (17). Several tested wild-type mRNAs of yeast are degraded following the shortening of their poly(A) tails and subsequent Dcp1p-dependent removal of the 5′ m7G cap (12, 24, 33, 50). Decapped mRNAs are subsequently degraded by the 5′-to-3′ exoribonuclease Xrn1p (33, 50) and, to a lesser degree, by 3′-to-5′ exoribonucleases (3, 51). Like those of yeast, the poly(A) tails of several tested mammalian mRNAs are shortened prior to decay (reviewed in reference 57). Thus, a similar mechanism may operate in mammalian cells, although not all components of the yeast system have yet been described for mammals.
Many systems of mRNA turnover are coupled to translation. One of the clearest examples of translation-dependent mRNA turnover is nonsense-mediated mRNA decay (NMD), a system that is present in all eukaryotes tested. NMD selectively degrades mRNAs that contain premature stop codons (reviewed in references 30, 44, 46, and 59) and may serve as a surveillance system to prevent the expression of deleterious polypeptide fragments (56). NMD requires ongoing translation (13, 65, 78). Yeast nonsense-mutant mRNAs are decapped in a Dcp1p-dependent manner and degraded from their 5′ ends by Xrn1p (52). In this respect, NMD is similar to the deadenylation-dependent turnover described above. Nonsense-mutant mRNAs, however, are degraded without prior deadenylation (52). Thus, NMD appears to feed into the constitutive pathway of mRNA turnover at the decapping step.
Both cis-acting and trans-acting factors necessary for NMD have been identified. cis-acting factors include downstream elements of yeast (60, 77) and spliceable introns of mammalian mRNAs (18, 65, 75, 76). Such cis-acting elements must be located 3′ to a stop codon in order to elicit NMD. trans-acting factors include three yeast genes (UPF1, NMD2 [UPF2], and UPF3) (21, 28, 41, 42), seven Caenorhabditis elegans genes (smg-1 through smg-7) (16, 56), and a human ortholog of UPF1 that likely functions in mammalian NMD (5, 55, 64).
Loss-of-function mutations affecting any of the yeast UPF/NMD or C. elegans smg genes eliminate NMD without affecting viability or other systems of mRNA turnover. UPF1 (also known as NAM7, SAL1, IFS2, and MOF4) encodes a superfamily I RNA helicase, while NMD2 (also called UPF2, IFS1, and SUA1) and UPF3 (also called SUA6) encode novel proteins (21, 28, 41, 43, 69). Both the substrates of NMD and the yeast UPF/NMD proteins are associated with cytoplasmic polysomes (6, 78). UPF proteins interact with each other (27) and may be part of larger posttermination complexes that include translation release factors eRF1 and eRF3 (22). Such surveillance complexes may scan downstream of stop codons, inspecting mRNAs for the presence of downstream elements, and, if they find them, trigger decapping.
Mutations affecting the seven C. elegans smg genes were identified as allele-specific but non-gene-specific informational suppressors (16, 31). Genetic analysis of smg mutants and of smg-suppressible alleles demonstrates that smg mutations are loss-of-function alleles and that smg genes function in all tissues of the animal at all times of development. smg mutants exhibit mild morphogenetic defects (smg, for suppressor with morphogenetic effect on genitalia) and have smaller brood sizes than normal, but they are otherwise quite vigorous animals. Molecular analyses of mRNAs that contain premature nonsense mutations demonstrate that smg mutations eliminate NMD (49, 56). Thus, smg genes encode the C. elegans components of NMD, and as in yeast, C. elegans NMD is a nonessential system. We report here the molecular analysis of smg-2 and its encoded protein.
MATERIALS AND METHODS
Cloning of smg-2.
unc-54(r293) was introduced into a mut-2(r459) genetic background by being crossed twice with strain TR679 (20). mut-2(r259) was monitored in the cross by its Him and mutator phenotypes. smg-2(r920) was identified in this strain as a spontaneous suppressor of unc-54(r293). The smg-2(r920) strain was outcrossed eight times with the wild type prior to its molecular analysis. dpy-5(e61), which is located 18 centimorgans from smg-2, was recombined onto and off of the r920-containing chromosome three independent times during the outcrossing series. We then hybridized Southern blots of r920 with probes of Tc1, Tc3, Tc4, and Tc5. A copy of a transposon that is (i) absent in the wild type, (ii) present in the smg-2(r920) strain, (iii) tightly linked to smg-2, and (iv) absent in spontaneous Smg(+) revertants of r920 is a strong candidate for being located within smg-2 itself.
The Tc4 probe identified a novel 4.0-kb SacI restriction fragment that cosegregated with r920 and was absent in each of three revertants. A 2.5-kb SacI/BglII restriction fragment representing a junction of this novel Tc4 element was cloned as plasmid TR#178. TR#178 proved to contain repetitive DNA unrelated to Tc4. When used as a hybridization probe on genomic Southern blots, TR#178 hybridized to an unexpectedly large number of bands in the wild type. An ∼500-bp SacI-ClaI subclone of TR#178 (plasmid TR#179) proved to contain only unique genomic DNA (see Fig. 2 for its location). When used as a hybridization probe on a Southern blot, plasmid TR#179 detected a Tc4 insertion in smg-2(r920) that was absent in the wild type and each of three independent Smg(+) revertants. We screened 50 genome equivalents of a bacteriophage lambda genomic library by using plasmid TR#179 as a hybridization probe but were unable to identify genomic clones of the region. We screened a mixed-stage cDNA library (10) and identified one positive clone, plasmid TR#192.
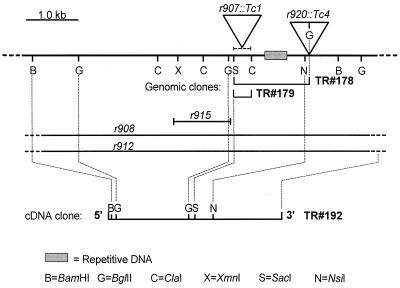
FIG. 2.
smg-2 genomic region. The genomic sequence of the smg-2 region is incomplete, but we deduced a partial restriction map of the region from genomic Southern blots by using either cDNA clone TR#192 or genomic clones TR#178 and TR#179 as hybridization probes. smg-2(r915) is an approximately 1.0-kb deletion. smg-2(r908) and smg-2(r912) delete all sequences contained on cDNA clone TR#192. smg-2(r907) and smg-2(r920) are insertions of Tc1 and Tc4, respectively.
Expression constructs.
For expression of SMG-2 in body wall muscle cells, the cDNA insert of plasmid TR#192 was excised with EcoRI, filled in with Klenow fragment, and ligated to EcoRV-digested plasmid pPD30.38. For expression of Upf1p in body wall muscle cells, a BamHI fragment containing the complete UPF1 open reading frame was removed from plasmid pBM272-UPF1 (provided by A. Atkin and M. Culbertson) and cloned into the BamHI site of pBluescript II KS(−), yielding plasmid TR#250. A SalI-SacI fragment of TR#250 was then cloned into SalI- and SacI-digested pPD30.38. The resulting plasmid, TR#253, was predicted to express full-length, native Upf1p from the unc-54 promoter contained on pPD30.38. Transforming DNAs were microinjected into the syncytial gonad as previously described (47) at either 1 μg/ml (TR#239) or 100 μg/ml (pRF4). We confirmed that the UPF1 fragment used for these experiments contained a functional UPF1 gene by removing it from plasmid TR#253 and cloning it into the URA3+, galactose-inducible expression vector pBM272 (36). We transformed the resulting UPF1 plasmid into yeast strain PLY38 (upf1-2 SUF1-1 his4-38 ura3-52) and selected URA3+ transformants. The transformants were phenotypically Upf− when grown on glucose and Upf+ when grown on galactose, as judged by temperature-sensitive suppression of his4-38. Transformants obtained with the vector alone were Upf− on both media.
Anti-SMG-2 antibodies.
We expressed two different His-tagged SMG-2 fusion proteins in Escherichia coli by using a pET-15b protein expression system (32). Fusion proteins SMG-2A and SMG-2B contain SMG-2 amino acids 20 to 726 and 52 to 522, respectively. Fusion proteins were purified as inclusion bodies, solubilized in 10% sodium dodecyl sulfate (SDS), electrophoresed through SDS–7% polyacrylamide gels, and eluted from gel slices (72). Approximately 500 μg of SMG-2B was injected into a New Zealand White rabbit; boosters were administered every 4 weeks. Sera were collected 2 weeks after the third booster and affinity purified with an Actigel column to which SMG-2A fusion protein had been coupled according to the manufacturer’s instructions (Sterogene Bioseparations, Inc.). Anti-E. coli antibodies were removed by adsorption to an acetone powder of E. coli not expressing a fusion protein (48). For Western blot analysis, either whole worms or worm soluble extracts (see below) were boiled in 2× sample buffer and electrophoresed on SDS–7% polyacrylamide gels. Proteins were transferred to polyvinylidene difluoride membranes (Millipore) in 48 mM Tris-Cl (pH 7.2)–39 mM glycine–20% methanol for 30 min at 15 V with a semidry protein blotter apparatus. Western blots were developed with an ECL kit (Amersham Life Sciences Corp.) as recommended by the manufacturer.
Phosphatase treatment of SMG-2.
Packed worms (floated on sucrose and washed in cold M9 buffer) were mixed with an equal volume of 10 mM morpholinepropanesulfonic acid (MOPS), pH 7.2, containing 1 mM phenylmethylsulfonyl fluoride and 1 μM Microcystin-LR (Calbiochem); they were then passed once through a French pressure cell at 14,000 lb/in2 and centrifuged three times at 14,000 × g, retaining the supernatant each time. Soluble extracts containing 1 mg of protein in 50 mM Tris-Cl (pH 7.5)–0.1 mM EDTA–5 mM dithiothreitol–0.01% Brij 35 were treated with 1,000 U of lambda protein phosphatase (an amount sufficient to overcome inhibition by Microcystin-LR; obtained from New England Biolabs) or buffer alone and incubated for 30 min at 30°C.
Human homolog of smg-2.
Four degenerate primers, corresponding to SMG-2 amino acids 478 to 484, 513 to 519, 735 to 741, and 798 to 809, were used to amplify a human homolog of SMG-2 via PCR by using a bacteriophage lambda placental cDNA library as a template. The resulting fragment (681 bp) was used as a hybridization probe to identify cDNA clone TR#314, which contains the 3′ end of a human SMG-2 homolog. TR#314 contains a 41-nucleotide (nt) poly(A) tail at its 3′ end but is incomplete at its 5′ end. The 5′ end of this transcript was identified as clone TR#329 by screening a heart cDNA library with portions of a human expressed sequence tag (GenBank accession no. H13971), known to be homologous to Upf1p, as a probe. cDNA clones TR#314 and TR#329 overlap to yield a 5,302-bp contiguous cDNA sequence that encodes a predicted protein of 1,118 amino acids.
Nucleotide sequence accession number.
The sequence of smg-2 cDNA has been assigned accession no. AF074017.
RESULTS
Cloning of smg-2.
We cloned smg-2 by transposon tagging. Spontaneous mutations that occur in a mutator genetic background are frequently caused by the insertion of transposable elements (4). In order to isolate transposon-induced alleles of smg-2, we first constructed an unc-54(r293) mut-2(r459) double mutant. unc-54(r293) is a smg-suppressible allele of the unc-54 myosin heavy chain gene. unc-54(r293) animals are paralyzed in smg(+) genetic backgrounds but have normal motility in smg(−) backgrounds (31). The mutator mutation mut-2(r459) activates several families of C. elegans transposable elements (4, 20). Among 30 spontaneous motile revertants of unc-54(r293) mut-2(r459), we identified six alleles of smg-2. One allele, smg-2(r920), was noteworthy, because upon passage it reverted spontaneously to Smg(+) five independent times. When we crossed smg-2(r920) into a background that did not contain mut-2(r459), we detected no Smg(+) revertants. Such mutator-dependent instability is characteristic of many C. elegans transposon insertions.
As described in Materials and Methods, we identified a copy of the transposable element Tc4 that cosegregated with smg-2(r920), cloned an insertional junction fragment of this Tc4 element (plasmid TR#179), and established that this novel Tc4 insertion was present in smg-2(r920::Tc4) animals but absent in the wild type and in the Smg(+) revertants of r920. The genomic region defined by plasmid TR#179 was not contained on any existing C. elegans cosmids (68), nor were we able to isolate bacteriophage lambda genomic clones from the region. smg-2 is located on YAC Y48G8 in a region of the genomic sequence that at present has not been completely assembled. Using TR#179 as a probe, we identified cDNA clone TR#192, which spans the insertion site of r920::Tc4.
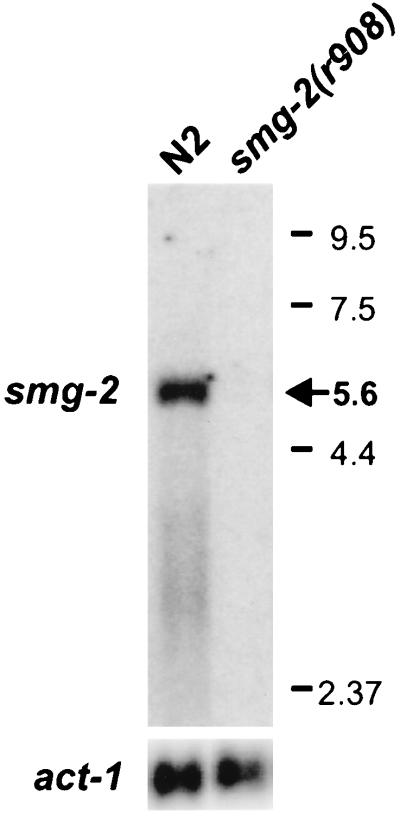
The cDNA insert of plasmid TR#192 is 3,388 bp long and contains a single open reading frame beginning with an ATG at nucleotide 12 and terminating with a UGA at nucleotide 3219. Using TR#192 as a hybridization probe on a Northern blot, we estimated that smg-2 mRNA was 5.6 kb long (Fig. 1). Thus, cDNA clone TR#192 is not full-length. TR#192 contains at its 5′ end nine nucleotides of the SL1 trans-spliced leader sequence (39), demonstrating that TR#192 is essentially full-length at its 5′ end. Assuming a poly(A) tail length of 200 nt, TR#192 is missing about 2.0 kb of 3′ untranslated region (UTR) material. Five additional smg-2 cDNA clones isolated from a mixed-stage cDNA library added only 124 nt of further 3′ UTR sequence. The sequence of smg-2 cDNA is discussed below. Although the smg-2 cDNA sequence is incomplete at its 3′ end, it contains the entire smg-2 coding region, because (i) the size of SMG-2 on Western blots agrees with that predicted by TR#192 (see below) and (ii) the cDNA insert of TR#192 provides smg-2(+) activity when introduced into smg-2(−) mutants (see below).
FIG. 1.
Northern blot hybridized with smg-2 cDNA clone TR#192. smg-2 mRNA is approximately 5.6 kb and is eliminated in smg-2(r908), a deletion that removes all smg-2 sequences. Numbers on the right are molecular sizes, in kilobases.
Transformation rescue of smg-2 mutants.
To test whether clone TR#192 contains the complete smg-2 coding region, we expressed the insert of clone TR#192 and tested whether it rescues smg-2 mutants. We ligated the cDNA insert of TR#192 to plasmid pPD30.38, a C. elegans expression vector driven by the unc-54 promoter and expressed in body wall muscle (47). We microinjected the resulting plasmid (TR#239) together with plasmid pRF4, which carries a dominant allele of rol-6 and serves as a marker for successful transformation (38), into unc-54(r293) smg-2(r863) hermaphrodites. Heritable transformants were identified by their roller phenotype and subsequently tested for their Smg phenotype based on suppression of unc-54(r293). Smg(+) animals were expected to be paralyzed, while Smg(−) animals were expected to have normal motility. We recovered three independent heritable transformants, all of which were paralyzed and had phenotypes indistinguishable from the unc-54(r293) phenotype. One transformant, TR2116 {unc-54(r293) smg-2(r863); rEx63[smg-2(+) rol-6(su1006dm)]}, was analyzed further. TR2116 exhibited a motility phenotype that was indistinguishable from the unc-54(r293) phenotype. Nonroller offspring of TR2116 (those that did not retain rEx63) had wild-type motility, indicating that they were smg-2(−). When crossed into an otherwise wild-type genetic background, rEx63 did not confer a dominant paralysis phenotype. These results demonstrate that the paralyzed phenotype of TR2116 was due to expression of a smg-2(+) transgene located on rEx63 and, hence, plasmid TR#192.
Identifying smg-2 null alleles.
As further proof that TR#192 represents smg-2 cDNA and to establish the smg-2 null phenotype, we investigated whether additional spontaneous smg-2 alleles contain alterations of the genomic region surrounding cDNA clone TR#192. We performed genomic Southern blot analysis of four additional spontaneous smg-2 alleles by using plasmid TR#192 or TR#179 as a hybridization probe. By comparing single and double digests of genomic DNA, we deduced a partial restriction map of the smg-2 genomic region and the structures of four mutations. These results are summarized in Fig. 2. All four tested alleles have alterations of the smg-2 genomic region. smg-2(r907) is a Tc1 insertion. smg-2(r915) is a deletion of approximately 1.0 kb. smg-2(r908) and smg-2(r912) are deletions of more than 7 kb that remove all sequences contained on cDNA clone TR#192. As described above, smg-2(r908) expressed no detectable smg-2 mRNA (Fig. 1). The phenotypes of smg-2(r908) and smg-2(r912) are indistinguishable from that of the canonical allele smg-2(e2008), establishing that the previously described smg-2 phenotype (31) represents the null phenotype.
smg-2 is homologous to UPF1 and RENT1/HUPF1.
The sequence of cDNA clone TR#192 predicted that SMG-2 is 120 kDa and has strong similarity to Upf1p of yeast and RENT1/HUPF1 of humans (Fig. 3). A 781-amino-acid central portion of SMG-2 is 50% identical to Upf1p, with particularly high conservation in the cysteine-rich region (92 amino acids; 58% identity), previously suggested to be zinc or zinc ring fingers (1, 43, 70), and in the superfamily I DNA-RNA helicase domain (400 amino acids; 61% identical) (37). The amino and carboxyl termini of SMG-2 and Upf1p are not similar, although the amino termini of both proteins are strongly acidic. SMG-2 is more similar to RENT1/HUPF1, a human ortholog of UPF1 (5, 55), than it is to Upf1p. The central 781 amino acids of SMG-2 are 59% identical to RENT1/HUPF1, as opposed to 50% identical to Upf1p. The amino-terminal 46 amino acids of SMG-2 (a region not similar to Upf1p) are 35% identical to RENT1/HUPF1. The carboxyl termini of SMG-2 and RENT1/HUPF1 are rich in serine-glutamine (SQ) dipeptides. SQ dipeptides are not concentrated at the carboxyl terminus of Upf1p.
FIG. 3.
Sequence of SMG-2. The sequence of SMG-2, deduced from cDNA clone TR#192, is aligned with those of Upf1p and RENT1/HUPF1. Amino acid residues identical in two or three of these three proteins are shaded with light gray and dark gray, respectively. The highly homologous zinc finger and RNA helicase domains are boxed with dotted and solid lines, respectively. The carboxyl termini of SMG-2 and RENT1/HUPF1 are rich in SQ dipeptides, which are underlined.
Based on sequence conservation between the helicase domains of SMG-2 and Upf1p, we synthesized degenerate PCR primers and amplified the helicase domain of a human homolog of SMG-2 from a cDNA library. We used this PCR product and a human expressed sequence tag homologous to Upf1p as hybridization probes to identify and sequence two overlapping cDNA clones that define a 5,302-nt transcript (see Materials and Methods). The sequence of this cDNA is essentially identical to the RENT1/HUPF1 sequence (5, 55). Our cDNA sequence contains a few minor differences from the RENT1/HUPF1 sequence and adds 1,613 nt of 3′ UTR material not previously described. We discuss the similarities of Upf1p-related proteins below.
Expression of Upf1p in C. elegans.
The similarity of SMG-2 and Upf1p prompted us to test whether UPF1 of yeast could rescue smg-2 mutants of C. elegans. In experiments analogous to those described above for expression of the smg-2 cDNA clone in body wall muscle, we cloned a complete UPF1 open reading frame into the expression vector pPD30.38. The resulting plasmid, TR#253 (see Materials and Methods for its construction), is predicted to express a transcript of 4.2 kb in which the first AUG of the transcript is the normal UPF1 translational initiation codon. We microinjected TR#253 and pRF4 into unc-54(r293) smg-2(r863) hermaphrodites and recovered three heritable roller transformants, all of which were phenotypically Smg(−). We confirmed by Northern blot analysis that transformants expressed an abundant UPF1 mRNA whose size is that predicted for the transgene. We removed the UPF1 fragment from plasmid TR#253 and demonstrated that it supplies UPF1+ function in yeast (see Materials and Methods). Thus, expression of UPF1 does not provide detectable smg-2(+) function in C. elegans. However, as we have no means to verify that UPF1 mRNA is translated in C. elegans, this conclusion must remain tentative.
SMG-2 is phosphorylated.
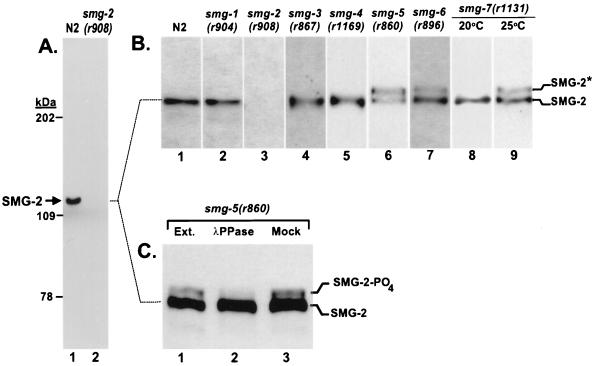
We prepared affinity-purified polyclonal antibodies against a His-tagged fusion protein containing SMG-2 amino acids 52 to 522. On Western blots of total C. elegans proteins, this antibody detected a single protein whose relative mobility (120 kDa) was that predicted for SMG-2 (Fig. 4A, lane 1). The 120-kDa protein, hereafter called SMG-2, was absent in the smg-2(r908) strain (Fig. 4A, lane 2) and in animals containing 7 of 13 tested smg-2 alleles (r865, r868, r870, r876, r890, r893, and r908). Mutants containing the remaining smg-2 alleles tested (r863, r866, r881, r882, r895, and r898) expressed an approximately full-length version of SMG-2. These alleles, therefore, likely represent missense alleles of smg-2. SMG-2 was present in approximately normal abundance in smg-1(r904), smg-3(r867), and smg-4(r1169) mutants (Fig. 4B, lanes 2, 4, and 5). In smg-5(r860), smg-6(r896), and smg-7(r1131) mutants grown at the nonpermissive temperature, SMG-2 was present, but we detected two distinct isoforms (Fig. 4B, lanes 6 to 9). One isoform corresponds to SMG-2 of the wild type. A second isoform, SMG-2* (Fig. 4B), migrates more slowly than SMG-2 and was detected only in smg-5, smg-6, and smg-7 mutants.
FIG. 4.
Phosphorylation of SMG-2. All panels display total protein Western blots reacted with anti-SMG-2 polyclonal antibodies. (A) Anti-SMG-2 antibodies react with a single protein estimated to be 120 kDa that is absent in smg-2(r908) mutants. (B) Expression of SMG-2 in wild-type animals (N2) and in mutants of all smg genes. All strains were grown at 20°C except the smg-7(r1131) strain, which was grown at both 20 and 25°C because it is temperature sensitive. Parallel experiments demonstrated that the wild type grown at 25°C did not accumulate detectable quantities of the more slowly migrating SMG-2 isoform. (C) Crude extracts (Ext.) of the smg-5(r860) mutant (lane 1) were incubated for 30 min with recombinant lambda protein phosphatase (λPPase) (lane 2) or in phosphatase buffer without added enzyme (lane 3) prior to electrophoresis.
Two lines of evidence demonstrate that the more slowly migrating isoform is a phosphorylated derivative of SMG-2. First, Microcystin-LR, an inhibitor of protein phosphatases 1 and 2A (74), had to be included in crude extracts in order to detect the more slowly migrating SMG-2 isoform. When it was omitted, only the more rapidly migrating SMG-2 isoform was detected in all strains tested. We presume that, in the absence of an inhibitor, one or more endogenous C. elegans protein phosphatases dephosphorylated SMG-2 after preparation of the crude extracts. Second, treatment of smg-5 mutant extracts with purified lambda protein phosphatase, which is active on phosphorylated serine, threonine, tyrosine, and histidine residues of many proteins (79), converted the more slowly migrating SMG-2 isoform into the more rapidly migrating isoform (Fig. 4C, lanes 1 and 2). We conclude that the more slowly migrating isoform of SMG-2 is phosphorylated. The phosphorylated isoform of SMG-2 was most consistently detected when living animals were immersed and boiled in sample loading buffer immediately prior to electrophoresis. Under such conditions, phosphorylated SMG-2 was readily detected in smg-5, smg-6, and smg-7 mutants, even when inhibitors of protein phosphatases were omitted, presumably due to the extremely rapid sample preparation. SMG-2 and phosphorylated SMG-2 were approximately equally abundant in smg-5 mutants, whereas SMG-2 was consistently more abundant than phosphorylated SMG-2 in smg-6 and smg-7 mutants. The relative proportions of SMG-2 and phosphorylated SMG-2 shown in Fig. 4 are typical of several independent Western blots. We did not detect phosphorylated SMG-2 in the wild type (Fig. 4B, lane 1), presumably because its abundance was below our threshold for detection. We believe that phosphorylation of SMG-2 is important to the in vivo roles of SMG-2 and not an aberrant event that occurs only in certain smg mutants, because functions of three other smg genes are required for SMG-2 phosphorylation and because certain smg-2 missense mutations accumulated the phosphorylated isoform (see below).
Activities of smg-1, smg-3, and smg-4 are required for SMG-2 phosphorylation.
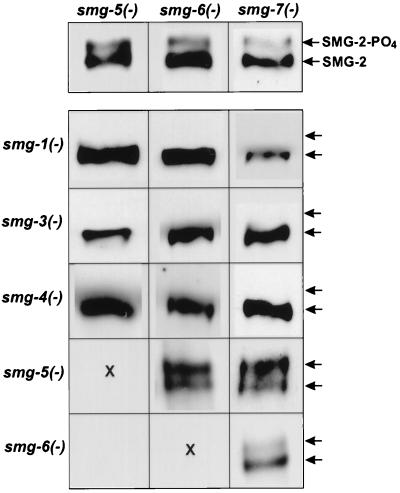
We tested whether functions of other smg genes are required for SMG-2 phosphorylation by constructing appropriate double mutants. If, for example, activity of smg-1 is needed to phosphorylate SMG-2, then the phosphorylated isoform of SMG-2 that we detected in smg-5, smg-6, and smg-7 single mutants should not accumulate in smg-1 smg-5, smg-1 smg-6, and smg-1 smg-7 double mutants. We constructed all relevant smg smg double mutant combinations and examined them for accumulation of SMG-2 and its phosphorylated isoform. The results are shown in Fig. 5. In smg-5, smg-6, and smg-7 single mutants, both SMG-2 and phosphorylated SMG-2 were detected (Fig. 5, row 1). In all double mutant combinations involving an allele of smg-5, smg-6, or smg-7 and an allele of smg-1, smg-3, or smg-4, only SMG-2 (not phosphorylated SMG-2) was detected (Fig. 5, rows 2 to 4). We concluded that activities of smg-1, smg-3, and smg-4 are required for phosphorylation of SMG-2.
FIG. 5.
SMG-2 isoforms of smg smg double mutants. All panels display the SMG-2 region of Western blots reacted with anti-SMG-2 antibody. SMG-2 isoforms which accumulated in smg-5, smg-6, and smg-7 single mutants are shown in row 1. SMG-2 isoforms which accumulated in smg smg double mutants are shown at the intersections of rows and columns. For example, row 2, column 1, is a smg-1 smg-5 double mutant; row 3, column 1, is a smg-3 smg-5 double mutant, etc. Alleles used to construct the double mutants were smg-1(r861), smg-3(r867), smg-4(ma116), smg-5(r860), smg-6(r896), and smg-7(r1131). All strains were grown at 20°C except those containing smg-7(r1131). Because r1131 is temperature sensitive, all strains involving this mutation were grown at 25°C. Experiments not shown demonstrate that growth at 25°C does not influence the relative proportions of SMG-2 and phosphorylated SMG-2 in the wild type or smg-5 and smg-6 single mutants.
Mutations altering the SMG-2 nucleotide binding site affect SMG-2 phosphorylation.
As described above, containing mutants smg-2 with alleles r863, r866, r881, r882, r895, and r898 express approximately full-length SMG-2 polypeptides. These mutations, therefore, are likely to be missense alleles of smg-2. We examined on Western blots the SMG-2 protein(s) that accumulates in these mutants to determine whether SMG-2, phosphorylated SMG-2, or both proteins were present. Mutants containing one of four alleles (r863, r881, r882, and r898) accumulated only SMG-2 (Fig. 6, lanes 6 and 7, and data not shown), while mutants containing one of two alleles (r866 and r895) accumulated both SMG-2 and phosphorylated SMG-2 (Fig. 6, lanes 4 and 5). The doublet of SMG-2 polypeptides observed for smg-2(r866) and smg-2(r895) mutants is similar to that seen for smg-5, smg-6, and smg-7 mutants. These results suggest that the mutant SMG-2 of r866 and r895 is inefficiently dephosphorylated, although alternative interpretations are possible (see Discussion).
FIG. 6.
SMG-2 isoforms of smg-2 missense mutations. The SMG-2 region of a Western blot reacted with anti-SMG-2 antibody is shown. Phosphorylated SMG-2 was not detected in the wild type (lane 1) or in two mutants with smg-2 missense alleles (lanes 6 and 7) but was readily detected in smg-2(r866), smg-2(r895), and smg-5 (r860) (control) mutants. smg-2(r908) fully deletes smg-2. Amino acid substitutions predicted from the sequences of smg-2(r866) and smg-2(r895) are shown relative to the consensus sequence of the P-loop of mononucleotide binding proteins (14).
smg-2(r866) and smg-2(r895) mutants contain single amino acid substitutions of the SMG-2 nucleotide binding site. We amplified by PCR the helicase domains of r866 and r895 and determined their sequences. We detected a single nucleotide substitution in each case relative to the wild type. As shown in Fig. 6, both r866 and r895 cause single amino acid substitutions of invariant glycine residues of the P-loop of the SMG-2 nucleotide binding site. smg-2(r866) is a G→A transition of nt 1419 that changes glycine-470 to arginine. smg-2(r895) is a G→A transition of nt 1426 that changes glycine-472 to glutamic acid.
DISCUSSION
Genetic evidence demonstrates that both SMG-2 of C. elegans and Upf1p of yeast are required for NMD. In both organisms, loss-of-function mutations eliminate NMD. A central 781-amino-acid region of SMG-2 is 50% identical to the corresponding region of Upf1p. The proteins are particularly well conserved in the superfamily I helicase domain (400 amino acids; 61% identical) and in the cysteine-rich region (92 amino acids; 58% identical). The strong conservation of Upf1p and SMG-2, combined with genetic results indicating that these genes perform similar functions, suggests that NMD is an ancient system, one whose origins predate the divergence of yeast and nematodes.
SMG-2 is somewhat more similar to human RENT1/HUPF1 than to Upf1p (Fig. 3). The central 781 amino acids of SMG-2, which include the cysteine-rich and helicase domains, are 59% identical to RENT1/HUPF1 (compared to 50% identity for Upf1p). The amino terminus of SMG-2 is 35% identical to that of RENT1/HUPF1, whereas this region is not similar to Upf1p. The carboxyl termini of SMG-2 and RENT1/HUPF1, although not strongly similar, are both rich in SQ dipeptides. Although loss-of-function mutations are not available for RENT1/HUPF1, expression of a dominant negative form of RENT1/HUPF1 partially inhibits NMD in mammalian cells (64). Both Upf1p and RENT1/HUPF1 interact with translation release factors eRF1 and eRF3 (22). Thus, it seems likely that SMG-2, Upf1p, and RENT1/HUPF1 perform analogous functions in all cells and that the molecular mechanisms of NMD are similar in most eukaryotes.
Biochemical analysis of Upf1p demonstrates that it is a nucleic acid-dependent ATPase and helicase (23, 69). In the absence of ATP, Upf1p binds tightly to single-stranded nucleic acids, whereas ATP hydrolysis causes bound RNA to be released. The helicase and associated ATPase activities of Upf1p are required for NMD (69). Because of its similarity to Upf1p, we assume that the SMG-2 helicase has similar enzymatic properties. smg-2(r866) and smg-2(r895) are strongly predicted to eliminate the SMG-2 ATPase-helicase enzymatic function. Both r866 and r895 affect invariant glycine residues of the conserved P-loop of the SMG-2 nucleotide binding site. This flexible motif joins a β-strand and α-helix to form a pocket into which the phosphate groups insert. Mutation of glycine residues corresponding to those affected by r866 and r895 destroys nucleotide binding and/or hydrolysis in numerous proteins (for examples, see references 45, 54, 63, and 73; reviewed in reference 61). r866 and r895, therefore, almost certainly eliminate ATP binding and/or hydrolysis.
Upf1p functions not only in NMD but also in translation termination. In addition to defects in NMD, upf1 deletions exhibit elevated levels of translation through stop codons (43). Upf1p, furthermore, interacts with translation release factors eRF1 and eRF3 in vitro (22). upf1 mutations affecting the cysteine-rich region of Upf1p, which has been suggested to form zinc or zinc ring fingers, affect termination without affecting NMD (70). Certain mutations affecting the Upf1p helicase domain affect NMD without affecting termination (69). Does SMG-2 function in translation termination in a similar way? Certain smg-suppressible mutations of C. elegans are nonsense alleles, some of which are located near the 5′ end of the affected open reading frame (9, 40, 58). It is unlikely that such nonsense mutations are phenotypically suppressed solely by elevating mRNA levels, because the fragment polypeptides predicted by translation to the nonsense site are often quite short and lack functionally important domains of the affected proteins. A more likely explanation of such suppression is that SMG-2 is required for efficient translation termination, similar to the situation in yeast.
A phosphorylated isoform of SMG-2 accumulates to abnormally high levels in smg-2(r866), smg-2(r895), smg-5, smg-6, and smg-7 mutants (Fig. 4 and 6). We do not know at present which residue(s) of SMG-2 is phosphorylated or whether the more rapidly migrating SMG-2 isoform is completely free of phosphorylation. SQ dipeptides, which SMG-2 and RENT1/HUPF1 are rich in, have been shown to be phosphorylation sites for certain phosphatidylinositol 3-kinase-related kinases (2, 8, 11, 25, 53, 62). Two observations suggest that SQ dipeptides might be the sites of SMG-2 phosphorylation. First, SMG-2 is not phosphorylated in smg-1 mutants (Fig. 5), and second, smg-1 encodes a protein predicted to be a phosphatidylinositol 3-kinase-related kinase (53a). A simple interpretation of these observations is that SMG-1 directly phosphorylates SMG-2, possibly at SQ dipeptides.
We did not detect phosphorylated SMG-2 in the wild type. While it is in principle possible that phosphorylation of SMG-2 is an abnormal event occurring only in certain smg(−) mutants (not in the wild type), we consider this unlikely for two reasons. First, not all smg mutants accumulate phosphorylated SMG-2. smg-5, smg-6, and smg-7 mutants do so, but smg-1, smg-3, and smg-4 mutants do not. Functions of smg-1, smg-3, and smg-4 are, in fact, required for SMG-2 phosphorylation. If SMG-2 phosphorylation were an artifact of being Smg(−), it is unlikely that other smg gene products would be required for the abnormal event. Second, smg-2(r866) and smg-2(r895) mutants, but not mutants with five other tested smg-2 missense alleles, accumulate phosphorylated SMG-2 in a manner similar to that seen in smg-5, smg-6, and smg-7 mutants. Such alleles appear to be blocked in the dephosphorylation (or further metabolism; see below) of phosphorylated SMG-2. r866 and r895 are mutations affecting the SMG-2 nucleotide binding site and are strongly predicted to eliminate ATP binding and/or hydrolysis. This suggests that conformational changes of the SMG-2 helicase domain, mediated via its ATPase activity, are important for dephosphorylation. Our inability to detect phosphorylated SMG-2 in the wild type suggests that this state is transient, causing the steady-state level to be below our threshold of detection.
How is metabolism of phosphorylated SMG-2 affected in smg-5, smg-6, and smg-7 mutants? With regard to SMG-2 phosphorylation, the phenotypes of smg-5, smg-6, and smg-7 mutants are similar to those of smg-2(r866) and smg-2(r895) mutants. Perhaps SMG-5, SMG-6, and SMG-7 are positive regulators of SMG-2 ATP binding and/or hydrolysis, such that in smg-5, smg-6, and smg-7 mutants, SMG-2 accumulates in a phosphorylated, inactive state. Alternatively, perhaps SMG-5, SMG-6, and SMG-7 are directly or indirectly required for a SMG-2 phosphatase. Both SMG-5 and SMG-7 are novel proteins (1a, 16). If this model is correct, it is unlikely that one isoform of SMG-2 is active while the other isoform is inactive. Mutations that block either SMG-2 phosphorylation (smg-1, smg-3, and smg-4) or dephosphorylation (smg-5, smg-6, and smg-7) fully eliminate NMD, suggesting that complete cycles of phosphorylation and dephosphorylation must occur for NMD. Alternatively, perhaps phosphorylation of SMG-2 targets it for proteolysis, with smg-5, smg-6, and smg-7 being required for degradation of phosphorylated SMG-2. smg-5, smg-6, and smg-7 mutants would accumulate phosphorylated SMG-2 because of their failure to degrade it.
What role does SMG-2 phosphorylation play in NMD? Perhaps the simplest would be to regulate enzymatic activity of the SMG-2 ATPase or helicase. The activity of human p68 RNA helicase, for example, is modulated by phosphorylation (15), as is the activity of human DNA helicase IV (66). Phosphorylation of SMG-2 might affect assembly or disassembly of a protein complex, such as posttermination scanning complexes that have been proposed to sense yeast downstream elements (26, 59). Phosphorylation of eIF4E and eIF4E binding proteins, for example, regulates the assembly of eukaryotic translation initiation complexes (34). Perhaps phosphorylation of SMG-2 alters the distribution of NMD proteins within the cell, such as occurs with NF-κB following phosphorylation of IκB (7), or targets SMG-2 for proteolysis, such as occurs with yeast Sic1p upon entry into S phase (67). Refinement of these or other models will require an understanding of the biochemical activities of SMG-2 and the role of SMG-2 phosphorylation in those processes.
ACKNOWLEDGMENTS
We thank Rock Pulak for isolating spontaneous smg-2 alleles and for assistance with yeast transformation.
This work was supported by a grant from the NIH (GM50933), by a Howard Hughes Predoctoral Fellowship to A.G., and by the University of Wisconsin Training Grant in Genetics.
REFERENCES
- 1.Altamura N, Groudinsky O, Dujardin G, Slonimski P P. NAM7 nuclear gene encodes a novel member of a family of helicases with a Zn-ligand motif and is involved in mitochondrial functions in Saccharomyces cerevisiae. J Mol Biol. 1992;224:575–587. doi: 10.1016/0022-2836(92)90545-u. [DOI] [PubMed] [Google Scholar]
- 1a.Anders, K., and P. Anderson. Unpublished observations.
- 2.Anderson C W, Lees-Miller S P. The nuclear serine/threonine protein kinase DNA-PK. Crit Rev Eukaryot Gene Expression. 1992;2:283–314. [PubMed] [Google Scholar]
- 3.Anderson J S J, Parker R. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson P. Mutagenesis. Methods Cell Biol. 1995;48:31–58. [PubMed] [Google Scholar]
- 5.Applequist S E, Selg M, Raman C, Jack H M. Cloning and characterization of HUPF1, a human homolog of the Saccharomyces cerevisiae nonsense mRNA-reducing UPF1 protein. Nucleic Acids Res. 1997;25:814–821. doi: 10.1093/nar/25.4.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Atkin A L, Schenkman L R, Eastham M, Dahlseid J N, Lelivelt M J, Culbertson M R. Relationship between yeast polyribosomes and upf proteins required for nonsense mRNA decay. J Biol Chem. 1997;272:22163–22172. doi: 10.1074/jbc.272.35.22163. [DOI] [PubMed] [Google Scholar]
- 7.Baldwin A S., Jr The NF-kappa B and I kappa B proteins: new discoveries and insights. Annu Rev Immunol. 1996;14:649–683. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 8.Bannister A J, Gottlieb T M, Kouzarides T, Jackson S P. c-Jun is phosphorylated by the DNA-dependent protein kinase in vitro; definition of the minimal kinase recognition motif. Nucleic Acids Res. 1993;21:1289–1295. doi: 10.1093/nar/21.5.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barnes T M, Hodgkin J. The tra-3 sex determination gene of Caenorhabditis elegans encodes a member of the calpain regulatory protease family. EMBO J. 1996;15:4477–4484. [PMC free article] [PubMed] [Google Scholar]
- 10.Barstead R J, Waterston R. The basal component of the nematode dense body is vinculin. J Biol Chem. 1989;264:10177–10185. [PubMed] [Google Scholar]
- 11.Baskaran R, Wood L D, Whitaker L L, Canman C E, Morgan S E, Xu Y, Barlow C, Baltimore D, Wynshaw-Boris A, Kastan M B, Wang J Y. Ataxia telangiectasia mutant protein activates c-Abl tyrosine kinase in response to ionizing radiation. Nature. 1997;387:516–519. doi: 10.1038/387516a0. [DOI] [PubMed] [Google Scholar]
- 12.Beelman C A, Stevens A, Caponigro G, LaGrandeur T E, Hatfield L, Fortner D M, Parker R. An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature. 1996;382:642–646. doi: 10.1038/382642a0. [DOI] [PubMed] [Google Scholar]
- 13.Belgrader P, Cheng J, Maquat L E. Evidence to implicate translation by ribosomes in the mechanism by which nonsense codons reduce the nuclear level of human triosephosphate isomerase mRNA. Proc Natl Acad Sci USA. 1993;90:482–486. doi: 10.1073/pnas.90.2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bossemeyer D. The glycine-rich sequence of protein kinases: a multifunctional element. Trends Biochem Sci. 1994;19:201–205. doi: 10.1016/0968-0004(94)90022-1. [DOI] [PubMed] [Google Scholar]
- 15.Buelt M K, Glidden B J, Storm D R. Regulation of p68 RNA helicase by calmodulin and protein kinase C. J Biol Chem. 1994;269:29367–29370. [PubMed] [Google Scholar]
- 16.Cali B M, Kuchma S L, Latham J, Anderson P. smg-7 is required for mRNA surveillance in C. elegans. Genetics. 1998;151:605–616. doi: 10.1093/genetics/151.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caponigro G, Parker R. Mechanisms and control of mRNA turnover in Saccharomyces cerevisiae. Microbiol Rev. 1996;60:233–249. doi: 10.1128/mr.60.1.233-249.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carter M S, Li S, Wilkinson M F. A splicing-dependent regulatory mechanism that detects translation signals. EMBO J. 1996;15:5965–5975. [PMC free article] [PubMed] [Google Scholar]
- 19.Chen C Y, Shyu A B. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 20.Collins J, Saari B, Anderson P. Activation of a transposable element in the germ line but not the soma of Caenorhabditis elegans. Nature. 1987;328:726–728. doi: 10.1038/328726a0. [DOI] [PubMed] [Google Scholar]
- 21.Cui Y, Hagan K W, Zhang S, Peltz S W. Identification and characterization of genes that are required for the accelerated degradation of mRNAs containing a premature translational termination codon. Genes Dev. 1995;9:423–436. doi: 10.1101/gad.9.4.423. [DOI] [PubMed] [Google Scholar]
- 22.Czaplinski K, Ruiz-Echevarria M J, Paushkin S V, Han X, Weng Y, Perlick H A, Dietz H C, Ter-Avanesyan M D, Peltz S W. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev. 1998;12:1665–1677. doi: 10.1101/gad.12.11.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Czaplinski K, Weng Y, Hagan K W, Peltz S W. Purification and characterization of the Upf1 protein: a factor involved in translation and mRNA degradation. RNA. 1995;1:610–623. [PMC free article] [PubMed] [Google Scholar]
- 24.Decker C J, Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- 25.Giffin W, Kwast-Welfeld J, Rodda D J, Prefontaine G G, Traykova-Andonova M, Zhang Y, Weigel N L, Lefebvre Y A, Hache R J. Sequence-specific DNA binding and transcription factor phosphorylation by Ku autoantigen/DNA-dependent protein kinase. Phosphorylation of Ser-527 of the rat glucocorticoid receptor. J Biol Chem. 1997;272:5647–5658. doi: 10.1074/jbc.272.9.5647. [DOI] [PubMed] [Google Scholar]
- 26.Hagan K W, Ruiz-Echevarria M J, Quan Y, Peltz S W. Characterization of cis-acting sequences and decay intermediates involved in nonsense-mediated mRNA turnover. Mol Cell Biol. 1995;15:809–823. doi: 10.1128/mcb.15.2.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He F, Brown A H, Jacobson A. Upf1p, Nmd2p, and Upf3p are interacting components of the yeast nonsense-mediated mRNA decay pathway. Mol Cell Biol. 1997;17:1580–1594. doi: 10.1128/mcb.17.3.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He F, Jacobson A. Identification of a novel component of the nonsense-mediated mRNA decay pathway by use of an interacting protein screen. Genes Dev. 1995;9:437–454. doi: 10.1101/gad.9.4.437. [DOI] [PubMed] [Google Scholar]
- 29.Hentze M W, Kuhn L C. Molecular control of vertebrate iron metabolism: mRNA-based regulatory circuits operated by iron, nitric oxide, and oxidative stress. Proc Natl Acad Sci USA. 1996;93:8175–8182. doi: 10.1073/pnas.93.16.8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hentze M W, Kulozik A E. A perfect message: RNA surveillance and nonsense-mediated decay. Cell. 1999;96:307–310. doi: 10.1016/s0092-8674(00)80542-5. [DOI] [PubMed] [Google Scholar]
- 31.Hodgkin J, Papp A, Pulak R, Ambros V, Anderson P. A new kind of informational suppression in the nematode Caenorhabditis elegans. Genetics. 1989;123:301–313. doi: 10.1093/genetics/123.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoffmann A, Roeder R G. Purification of his-tagged proteins in non-denaturing conditions suggests a convenient method for protein interaction studies. Nucleic Acids Res. 1991;19:6337–6338. doi: 10.1093/nar/19.22.6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hsu C L, Stevens A. Yeast cells lacking 5′→3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol Cell Biol. 1993;13:4826–4835. doi: 10.1128/mcb.13.8.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jackson R J, Wickens M. Translational controls impinging on the 5′-untranslated region and initiation factor proteins. Curr Opin Genet Dev. 1997;7:233–241. doi: 10.1016/s0959-437x(97)80133-5. [DOI] [PubMed] [Google Scholar]
- 35.Jacobson A, Peltz S W. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu Rev Biochem. 1996;65:693–739. doi: 10.1146/annurev.bi.65.070196.003401. [DOI] [PubMed] [Google Scholar]
- 36.Johnston M, Davis R W. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koonin E V. A new group of putative RNA helicases. Trends Biochem Sci. 1992;17:495–497. doi: 10.1016/0968-0004(92)90338-a. [DOI] [PubMed] [Google Scholar]
- 38.Kramer J M, French R P, Park E, Johnson J J. The Caenorhabditis elegans rol-6 gene, which interacts with the sqt-1 collagen gene to determine organismal morphology, encodes a collagen. Mol Cell Biol. 1990;10:2081–2089. doi: 10.1128/mcb.10.5.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krause M, Hirsh D. A trans-spliced leader sequence on actin mRNA in C. elegans. Cell. 1987;49:753–761. doi: 10.1016/0092-8674(87)90613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuwabara P E, Okkema P G, Kimble J. tra-2 encodes a membrane protein and may mediate cell communication in the Caenorhabditis elegans sex determination pathway. Mol Biol Cell. 1992;3:461–473. doi: 10.1091/mbc.3.4.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee B S, Culbertson M R. Identification of an additional gene required for eukaryotic nonsense mRNA turnover. Proc Natl Acad Sci USA. 1995;92:10354–10358. doi: 10.1073/pnas.92.22.10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leeds P, Peltz S W, Jacobson A, Culbertson M R. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 1991;5:2303–2314. doi: 10.1101/gad.5.12a.2303. [DOI] [PubMed] [Google Scholar]
- 43.Leeds P, Wood J M, Lee B S, Culbertson M R. Gene products that promote mRNA turnover in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:2165–2177. doi: 10.1128/mcb.12.5.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li S, Wilkinson M F. Nonsense surveillance in lymphocytes? Immunity. 1998;8:135–141. doi: 10.1016/s1074-7613(00)80466-5. [DOI] [PubMed] [Google Scholar]
- 45.Liu Q, Summers W. Site-directed mutagenesis of a nucleotide-binding domain in HSV-1 thymidine kinase: effects on catalytic activity. Virology. 1988;163:638–642. doi: 10.1016/0042-6822(88)90308-x. [DOI] [PubMed] [Google Scholar]
- 46.Maquat L E. When cells stop making sense: effects of nonsense codons on RNA metabolism in vertebrate cells. RNA. 1995;1:453–465. [PMC free article] [PubMed] [Google Scholar]
- 47.Mello C, Fire A. DNA transformation. In: Epstein H F, Shakes D C, editors. Caenorhabditis elegans: modern biological analysis of an organism. San Diego, Calif: Academic Press; 1995. pp. 451–482. [Google Scholar]
- 48.Miller D M, Shakes D C. Immunofluorescence microscopy. Methods Cell Biol. 1995;48:365–394. [PubMed] [Google Scholar]
- 49.Morrison M, Harris K S, Roth M B. smg mutants affect the expression of alternatively spliced SR protein mRNAs in Caenorhabditis elegans. Proc Natl Acad Sci USA. 1997;94:9782–9785. doi: 10.1073/pnas.94.18.9782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muhlrad D, Decker C J, Parker R. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′→3′ digestion of the transcript. Genes Dev. 1994;8:855–866. doi: 10.1101/gad.8.7.855. [DOI] [PubMed] [Google Scholar]
- 51.Muhlrad D, Decker C J, Parker R. Turnover mechanisms of the stable yeast PGK1 mRNA. Mol Cell Biol. 1995;15:2145–2156. doi: 10.1128/mcb.15.4.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Muhlrad D, Parker R. Premature translational termination triggers mRNA decapping. Nature. 1994;370:578–581. doi: 10.1038/370578a0. [DOI] [PubMed] [Google Scholar]
- 53.Niu H, Erdjument-Bromage H, Pan Z Q, Lee S H, Tempst P, Hurwitz J. Mapping of amino acid residues in the p34 subunit of human single-stranded DNA-binding protein phosphorylated by DNA-dependent protein kinase and Cdc2 kinase in vitro. J Biol Chem. 1997;272:12634–12641. doi: 10.1074/jbc.272.19.12634. [DOI] [PubMed] [Google Scholar]
- 53a.O’Connor, S. L., and P. Anderson. Unpublished observations.
- 54.Parsonage D, Wilke-Mounts S, Senior A. E. coli F1-ATPase: site-directed mutagenesis of the beta-subunit. FEBS Lett. 1988;232:111–114. doi: 10.1016/0014-5793(88)80397-1. [DOI] [PubMed] [Google Scholar]
- 55.Perlick H A, Medghalchi S M, Spencer F A, Kendzior R J, Jr, Dietz H C. Mammalian orthologues of a yeast regulator of nonsense transcript stability. Proc Natl Acad Sci USA. 1996;93:10928–10932. doi: 10.1073/pnas.93.20.10928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pulak R, Anderson P. mRNA surveillance by the C. elegans smg genes. Genes Dev. 1993;7:1885–1897. doi: 10.1101/gad.7.10.1885. [DOI] [PubMed] [Google Scholar]
- 57.Ross J. mRNA stability in mammalian cells. Microbiol Rev. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rougvie A E, Ambros V. The heterochronic gene lin-29 encodes a zinc finger protein that controls a terminal differentiation event in Caenorhabditis elegans. Development. 1995;121:2491–2500. doi: 10.1242/dev.121.8.2491. [DOI] [PubMed] [Google Scholar]
- 59.Ruiz-Echevarria M J, Czaplinski K, Peltz S W. Making sense of nonsense in yeast. Trends Biochem Sci. 1996;21:433–438. doi: 10.1016/s0968-0004(96)10055-4. [DOI] [PubMed] [Google Scholar]
- 60.Ruiz-Echevarria M J, Gonzalez C I, Peltz S W. Identifying the right stop: determining how the surveillance complex recognizes and degrades an aberrant mRNA. EMBO J. 1998;17:575–589. doi: 10.1093/emboj/17.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saraste M, Sibbald P R, Wittinghofer A. The P-loop—a common motif in ATP- and GTP-binding proteins. Trends Biochem Sci. 1990;15:430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- 62.Shafman T, Khanna K K, Kedar P, Spring K, Kozlov S, Yen T, Hobson K, Gatei M, Zhang N, Watters D, Egerton M, Shiloh Y, Kharbanda S, Kufe D, Lavin M F. Interaction between ATM protein and c-Abl in response to DNA damage. Nature. 1997;387:520–523. doi: 10.1038/387520a0. [DOI] [PubMed] [Google Scholar]
- 63.Shen H, Yao B, Mueller D M. Primary structural constraints of P-loop of mitochondrial F1-ATPase from yeast. J Biol Chem. 1994;269:9424–9428. [PubMed] [Google Scholar]
- 64.Sun X, Perlick H A, Dietz H C, Maquat L E. A mutated human homologue to yeast Upf1 protein has a dominant-negative effect on the decay of nonsense-containing mRNAs in mammalian cells. Proc Natl Acad Sci USA. 1998;95:10009–10014. doi: 10.1073/pnas.95.17.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thermann R, Neu-Yilik G, Deters A, Frede U, Wehr K, Hagemeier C, Hentze M W, Kulozik A E. Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J. 1998;17:3484–3494. doi: 10.1093/emboj/17.12.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tuteja N, Huang N W, Skopac D, Tuteja R, Hrvatic S, Zhang J, Pongor S, Joseph G, Faucher C, Amalric F, Falaschi A. Human DNA helicase IV is nucleolin, an RNA helicase modulated by phosphorylation. Gene. 1995;160:143–148. doi: 10.1016/0378-1119(95)00207-m. [DOI] [PubMed] [Google Scholar]
- 67.Verma R, Annan R S, Huddleston M J, Carr S A, Reynard G, Deshaies R J. Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S phase. Science. 1997;278:455–460. doi: 10.1126/science.278.5337.455. [DOI] [PubMed] [Google Scholar]
- 68.Waterston R, Sulston J. The genome of Caenorhabditis elegans. Proc Natl Acad Sci USA. 1995;92:10836–10840. doi: 10.1073/pnas.92.24.10836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weng Y, Czaplinski K, Peltz S W. Genetic and biochemical characterization of mutations in the ATPase and helicase regions of the Upf1 protein. Mol Cell Biol. 1996;16:5477–5490. doi: 10.1128/mcb.16.10.5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weng Y, Czaplinski K, Peltz S W. Identification and characterization of mutations in the UPF1 gene that affect nonsense suppression and the formation of the Upf protein complex but not mRNA turnover. Mol Cell Biol. 1996;16:5491–5506. doi: 10.1128/mcb.16.10.5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wickens M. In the beginning is the end: regulation of poly(A) addition and removal during early development. Trends Biochem Sci. 1990;15:320–324. doi: 10.1016/0968-0004(90)90022-4. [DOI] [PubMed] [Google Scholar]
- 72.Williams J A, Langeland J A, Thalley B S, Skeath J B, Carroll S B. Expression of foreign proteins in E. coli using plasmid vectors and purification of specific polyclonal antibodies. In: Hames B D, Glover D, editors. DNA cloning: a practical approach. II. Expression systems. Oxford, England: Oxford University Press; 1995. pp. 15–58. [Google Scholar]
- 73.Yoneya T, Tagaya M, Kishi F, Nakazawa A, Fukui T. Site-directed mutagenesis of Gly-15 and Gly-20 in the glycine-rich region of adenylate kinase. J Biochem. 1989;105:158–160. doi: 10.1093/oxfordjournals.jbchem.a122631. [DOI] [PubMed] [Google Scholar]
- 74.Yoshizawa S, Matsushima R, Watanabe M F, Harada K, Ichihara A, Carmichael W W, Fujiki H. Inhibition of protein phosphatases by microcystins and nodularin associated with hepatotoxicity. J Cancer Res Clin Oncol. 1990;116:609–614. doi: 10.1007/BF01637082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang J, Sun X, Qian Y, LaDuca J P, Maquat L E. At least one intron is required for the nonsense-mediated decay of triosephosphate isomerase mRNA: a possible link between nuclear splicing and cytoplasmic translation. Mol Cell Biol. 1998;18:5272–5283. doi: 10.1128/mcb.18.9.5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang J, Sun X, Qian Y, Maquat L E. Intron function in the nonsense-mediated decay of beta-globin mRNA: indications that pre-mRNA splicing in the nucleus can influence mRNA translation in the cytoplasm. RNA. 1998;4:801–815. doi: 10.1017/s1355838298971849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang S, Ruiz-Echevarria M J, Quan Y, Peltz S W. Identification and characterization of a sequence motif involved in nonsense-mediated mRNA decay. Mol Cell Biol. 1995;15:2231–2244. doi: 10.1128/mcb.15.4.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang S, Welch E M, Hogan K, Brown A H, Peltz S W, Jacobson A. Polysome-associated mRNAs are substrates for the nonsense-mediated mRNA decay pathway in Saccharomyces cerevisiae. RNA. 1997;3:234–244. [PMC free article] [PubMed] [Google Scholar]
- 79.Zhuo S, Clemens J C, Hakes D J, Barford D, Dixon J E. Expression, purification, crystallization, and biochemical characterization of a recombinant protein phosphatase. J Biol Chem. 1993;268:17754–17761. [PubMed] [Google Scholar]