Abstract
Mass mapping using high-resolution mass spectrometry has been applied to identify and rapidly distinguish SARS-CoV-2 coronavirus strains across five major variants of concern. Deletions or mutations within the surface spike protein across these variants, which originated in the UK, South Africa, Brazil and India (known as the alpha, beta, gamma and delta variants respectively), lead to associated mass differences in the mass maps. Peptides of unique mass have thus been determined that can be used to identify and distinguish the variants. The same mass map profiles are also utilized to construct phylogenetic trees, without the need for protein (or gene) sequences or their alignment, in order to chart and study viral evolution. The combined strategy offers advantages over conventional PCR-based gene-based approaches exploiting the ease with which protein mass maps can be generated and the speed and sensitivity of mass spectrometric analysis.
Graphical abstract

Supplementary Information
The online version contains supplementary material available at 10.1007/s00216-021-03649-1.
Keywords: SARS-CoV-2, Coronavirus, Virus, Variants, Evolution, Mass spectrometry
Introduction
While the emergence of SARS-CoV-2 in late 2019 was followed by a period of relative evolutionary stasis [1], new variants caused by mutations in the viral proteins of the SARS-CoV-2 coronavirus are now taking hold as the virus spreads throughout the world’s population [2]. Many mutations are deleterious or neutral in terms of the virus’ transmissibility and infectivity, yet other non-synonymous mutations in genes that encode viral proteins have helped the virus to spread and cause more sustained and greater disease severity [3]. In the past 12 months, the emergence of sets of mutations in “variants of concern” strains [4] have been identified. These impact the virus’ transmissibility and antigenicity in response to a changing immune profile within the human population post-vaccination. Identifying and understanding the evolution of such variants is of paramount importance to control the virus through patient isolation and for the development of effective new vaccines and therapies [5].
Particular focus has concerned the surface or spike protein (S-protein) given its role in binding to the host’s angiotensin-converting enzyme 2 (ACE2) receptors to initiate infection. Following the early emergence of the D614G [6], the N501Y mutation was among the first identified within the receptor-binding domain (RBD) that allowed the virus to bind more tightly to ACE2 receptors, in cells and animal models, to improve its transmissibility [7].
Variants of concern possess a range of such mutations in the spike protein. Epidemiological data suggest that the Alpha B.1.1.7 variant, a descendant of the lineage containing the D614G mutation first identified in the UK that spread to other parts of the world, has heightened transmissibility. It also contains Δ69–70, an amino-terminal domain (NTD) deletion, which is predicted to alter the conformation of an exposed NTD loop region associated with increased infectivity [8].
Of all the RBD residues which have affected immune recognition, the mutation of E484 first identified in the South African beta B.1.351 variant is of principal importance. Changes at this residue to K, Q or P all have been shown to reduce neutralization titres by more than an order of magnitude [9]. The more recent Indian delta B.1.617 variant also contains this mutation in addition to L452R and T478K, the latter improving viral entry. The delta variant is considered to be 55% more transmissible (WHO) and twice as infectious as earlier alpha variants. Consequently, within only a few months since May 2021, the delta variant has rapidly spread around the world and is now the dominant strain in many countries.
Methods to rapidly detect and monitor the evolution of virus strains are of vital importance. Mass spectrometry is particularly suited to the analysis of viral proteins and their peptide segments and offers a viable and complementary alternative [10] to conventional gene-based sequencing strategies [11]. It has been demonstrated that MALDI-MS approaches, in particular, offer advantages in terms of the speed and sensitivity of analysis where viral proteins are best first isolated and then digested [12]. Subsequent mass maps can then be used to confidently identify SARS-CoV-2 coronavirus, given that direct swab analyses of specimens detect a whole range of host contaminants and residuals which both hamper and can even prevent virus detection [10].
Here, we employ high-resolution mass spectrometry to study and distinguish strains for the major variants of concern stains using isolates or viral proteins thereof using mass signatures. This work stems from our previous work to detect, type and subtype and distinguish respiratory viruses [10, 12–17] including SARS-CoV-2 [12], employing high-resolution mass spectrometry using signature peptides. This current study also demonstrates how such mass spectrometry data can also be used in the construction of phylogenetic trees [18, 19], analogous to those derived using gene sequence data, to chart viral evolution [20].
Materials and methods
Recovery of S-protein from virus specimen
Clinical specimens collected from infected patients containing SARS-CoV-2 were grown in cell culture using Vero E6 cells following a reported procedure [21] and, as used in a previous study [12], were the source of an originating-like strain. Following chemical and heat inactivation and filtration, the virus was precipitated with polyethylene glycol precipitation of virus was performed after filtration through a 300-K molecular weight cut-off (MWCO) filter (Pall Corporation, Cheltenham, Victoria). The retentate was reconstituted in buffer (50 mM ammonium bicarbonate), sonicated (3 × 30 min) and then deglycosylated following the addition of 1.2 units of recombinant peptide-N-glycosidase F (PNGaseF) (Roche Diagnostics, North Ryde, Sydney, Australia) and 5 mM octyl β-D-glucopyranoside (Sigma Aldrich–Merck, Castle Hill, Sydney, Australia). The released viral proteins were separated by SDS-PAGE and the S-protein (at some 150 kDa) was excised from the gel. The gel plug was transferred into 25 mM ammonium bicarbonate solution containing 10% v/v acetonitrile (ACN) and 10 mM dithiothreitol (DTT) (10 mM) and heated for 30 min at 60 °C. The gel plug was washed three times with 25 mM ammonium bicarbonate in 50% acetonitrile and then dried in a vacuum concentrator (Labconco Corporation, Kansas City, MI, USA).
S-protein digestion
Gel recovered S-protein or recombinant forms for several SARS-CoV-2 variants (UK, South Africa, India and Brazil) (Acro Biosystems, Newark, DE USA) were reconstituted in 100 μL digestion buffer (50 mM ammonium bicarbonate, 10% acetonitrile, 2 mM dithiothreitol) incubated for 2 h at 37 °C and digested overnight following the successive addition of 1 μL each of proteomics-grade trypsin and sequencing-grade endoproteinase GluC (Merck, Bayswater VIC, Australia) 4 h apart.
High-resolution MALDI-FT-ICR mass spectrometry
Solutions of viral peptides (1 μL) were diluted with a solution (5 μL) of matrix (5 mg/mL α-cyano-4-hydroxycinnaminic acid in 50% acetonitrile with 0.1% trifluoroacetic acid). Solution volumes of 1 μL were spotted onto a matrix-assisted laser desorption ionization (MALDI) sample plate and analysed on a Bruker (Bruker Daltonics, Preston Victoria, Australia) Fourier-transform ion cyclotron resonance (FT-ICR) 7 Tesla mass spectrometer [12, 17]. Spectra were acquired over a mass-to-charge ratio range of m/z 400–4000 using a broadband excitation. The instrument was calibrated externally with a standard peptide mixture and the S-protein tryptic + GluC peptides were identified based on the reported sequences for the S-proteins obtained from the NCBI protein database (QHD43416.1) or supplier (Acro Biosystems, Newark, DE USA). Peptides were matched to predicted proteolytic products generated in silico using the Expasy PeptideMass tool (https://web.expasy.org/peptide_mass/).
Mass tree construction
A mass tree was built from the masses for the proteolytic peptides generated upon digestion of the S-protein for the originating-like strain and major variants (UK, South Africa, India and Brazil) using the modified version [20] of the original algorithm [18]. The MassTree algorithm identifies mass values that are indistinguishable (within a mass error of 5 ppm) across the sets. A distance matrix is then generated through pairwise comparison of mass values across all datasets adopting a relaxed neighbour joining (NJ) approach [22] using the Clearcut algorithm [23]. The tree was visualized using the FigTree algorithm v1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/) and rooted to the originating-like strain.
Sequence tree construction
The sequence tree was built from residues 16–1213 derived for the originating strain (NCBI protein database entry QHD43416.1) and those for the recombinant variants (Acro Biosystems, Newark, DE USA) using the online Phylogeny.fr algorithm in the a la carte mode adopting a ClustalW alignment, Gblocks curation and a common neighbour joining (NJ) tree building approach to the MassTree algorithm. The tree was rooted to the originating strain and visualized with the FigTree algorithm v1.4.4 as above.
Results and discussion
The full-length spike protein sequence from the original strain of SARS-CoV-2 (NCBI protein database entry QHD43416.1) is over 141 kDa. It comprises both S1 receptor-binding and S2 fusion subunits that are formed by cleavage of the polyprotein at residues 682–685 with furin-like enzymes. Given that large segments exceeding 6 kDa are generated from its digestion with trypsin alone, the protein was digested in silico with both trypsin and endoproteinase GluC (pH 8). Segments across the S1 and S2 subunits, which contain mutations present in each of the five major variants of concern, are shown in Table 1. Their mass-to-charge (m/z) range from 134 to 2868 are within the acquisition range of most MALDI-based instruments, and most peptides contain only a single mutation site which is also desirable for mass-based phyogenetics [24].
Table 1.
Original coronavirus reference strain and tryptic and GluC proteolytic segments that contain sites of mutations in surface spike protein within five major variants of concern
| Lineage | Strainorigin | Sites of mutations in major variants | Tryptic + GluC segmenta | Sequencea | Mass [M + H]+ mono. |
|---|---|---|---|---|---|
| Reference | Wuhan, China | L18 or T19 or T20 | 1–21 | MFVFLVLLPLVSSQCVNLTTR | 2380.3132 |
| P26 | 22–34 | TQLPPAYTNSFTR | 1495.7540 | ||
| HV del 69–70 | 54–77 | LFLPFFSNVTWFHAIHVSGTNGTK | 2720.3984 | ||
| D80 | 79–80 | FD | 281.1132 | ||
| T95 | 89–96 | GVYFASTE | 873.3990 | ||
| D138 | 133–138 | FQFCND | 773.2923 | ||
| G142 or Y del 144 | 139–147 | PFLGVYYHK | 1123.5935 | ||
| E154 | 151–154 | SWME | 552.2123 | ||
| R158 or del 156–157 | 155–158 | SEFR | 538.2620 | ||
| R190 | 188–190 | NLR | 402.2460 | ||
| D215 | 215 | D | 134.0448 | ||
| 242–244 del. or R246I | 238–246 | FQTLLALHR | 1098.6419 | ||
| K417N | 409–417 | QIAPGQTGK | 899.4946 | ||
| L452 | 429–453 | FTGCVIAWNSNNLD | 1553.7053 | ||
| T478 + E484 | 472–484 | IYQAGSTPCNGVE | 1338.5995 | ||
| N501 | 499–504 | STNLVK | 661.3880 | ||
| A570 | 569–571 | IAD | 318.1660 | ||
| D614 | 587–614 | ITPCSFGGVSVITPGTNTSNQVAVLYQD | 2868.4084 | ||
| H655 | 655–661 | HVNNSYE | 862.3690 | ||
| P681 | 664–682 | IPIGAGICASYQTQTNSPR | 1976.9859 | ||
| A701 | 686–702 | SVASQSIIAYTMSLGAE | 1727.8521 | ||
| T716 | 703–725 | NSVAYSNNSIAIPTNFTISVTTE | 2443.1988 | ||
| D950 | 948–950 | LQD | 375.1875 | ||
| S982 | 979–982 | ILSR | 488.3192 | ||
| T1027 | 1019–1028 | ASANLAATK | 846.4680 | ||
| D1118 | 1112–1118 | PQIITTD | 787.4197 | ||
| V1176 | 1169–1181 | ISGINASVVNIQK | 1342.7689 |
aBased on NCBI protein database sequence QHD43416.1
The same sequence was modified to insert (or delete) mutations for the five major variants of concern and similar proteolytic segments containing these mutations are shown in Table 2. It is clear from Table 2 that many peptide segments are unique to each variant and accordingly have distinct masses. This enables the variants to be distinguished when any one or more of them are detected within a mass spectrum. In this regard, the few with mass values below m/z 500 that are frequently detected among a matrix ion background in the low mass region of a MALDI spectrum, or that are deflected using a cut-off filter during such analyses, are shown bracketed and not considered further. All others are distinguishable within a mass error of 83 ppm or greater from all both trypsin and endoproteinase GluC peptides of the original spike protein sequence across all segments even when missed cleaved sites and the oxidation of methionine residues are considered (see Table 2 footnote). This mass error is easily achieved with high and even mid-resolution mass spectrometers.
Table 2.
Major coronavirus variants of concern, mutation sites in surface spike protein and unique peptide masses that distinguish such strains
| Lineage | Origin | Mutations | Tryptic + GluC segmenta | Sequence (mutations shown underlined, except deletions)b | Mass [M + H]+ mono. | Strain distinguishing peptide massesc |
|---|---|---|---|---|---|---|
| B.1.17 (Alpha) | UK | HV69–70 del. | 54–77 minus 69–70 | LFLPFFSNVTWFHAISGTNGTK | 2484.2711 | 2484.2711 |
| Y144 del. | 139–147 minus 144 | PFLGVYHK | 960.5302 | 960.5302 | ||
| N501Y | 499–504 | STYLVK | 710.4084 | |||
| A570D | 569–570 | ID | 247.1289 | (247.1289) | ||
| D614G | 587–619 | ITPCSFGGVSVITPGTNTSNQVAVLYQGVNCTE | 3356.6138 | |||
| P681H | 664–682 | IPIGAGICASYQTQTNSHR | 2016.9920 | 2016.9920 | ||
| T716I | 703–725 | NSVAYSNNSIAIPINFTISVTTE | 2455.2352 | 2455.2352 | ||
| S982A | 979–982 | ILAR | 472.3242 | (472.3242) | ||
| D1118H | 1112–1127 | PQIITTHNTFVSGNCD | 1746.8116 | 1746.8116 | ||
| B.1.351 (Beta) | South Africa | L18F | 1–21 | MFVFLVLLPLVSSQCVNFTTR | 2414.2975 | |
| D80A | 79–88 | FANPVLPFND | 1133.5626 | 1133.5626 | ||
| D215G | 215–224 | GLPQGFSALE | 1018.5204 | 1018.5204 | ||
| LAL 242–244 del. | 238–246 minus 242–244 | FQTLHR | 801.4366 | 801.4366 | ||
| R246I | 238–253 | FQTLLALHISYLTPGD | 1788.9531 | 1788.9531 | ||
| K417N | 409–419 | QIAPGQTGNIAD | 1184.5906 | 1184.5906 | ||
| E484K | 472–484 | IYQAGSTPCNGVK | 1337.6519 | 1337.6519 | ||
| N501Y | 499–504 | STYLVK | 710.4084 | |||
| D614G | 587–619 | ITPCSFGGVSVITPGTNTSNQVAVLYQGVNCTE | 3356.6138 | |||
| A701V | 686–702 | SVASQSIIAYTMSLGVE | 1755.8834 | 1755.8834 | ||
| B.1.617 (Delta) | India | T95I | 89–96 | GVYFASIE | 885.4353 | 885.4353 |
| G142D | 139–142 | PFLD | 491.2501 | |||
| E154K | 151–154 | SWMK | 551.2647 | 551.2647 | ||
| L452R | 429–452 | FTGCVIAWNSNNR | 1481.6955 | |||
| E484Q | 472–509 | IYQAGSTPCNGVQGFNCYFPLQSYGFQPTNGVGYQPYR | 4221.9222 | 4221.9222 | ||
| D614G | 587–619 | ITPCSFGGVSVITPGTNTSNQVAVLYQGVNCTE | 3356.6138 | |||
| P681R | 664–681 | IPIGAGICASYQTQTNSR | 1879.9331 | |||
| B.1.617.2 (Delta plus) | India | T19R | 1–19 | MFVFLVLLPLVSSQCVNLR | 2178.2178 | 2178.2178 |
| G142D | 139–142 | PFLD | 491.2501 | |||
| EF156–157 del. | 155–158 minus 156–157 | SR | 262.1510 | (262.1510) | ||
| R158G | 154–169 | SEFGVYSSANNCTFE | 1654.6690 | 1654.6690 | ||
| L452R | 429–452 | FTGCVIAWNSNNR | 1481.6955 | |||
| T478K | 472–478 | IYQAGSK | 766.4094 | 766.4094 | ||
| D614G | 587–619 | ITPCSFGGVSVITPGTNTSNQVAVLYQGVNCTE | 3356.6138 | |||
| P681R | 664–681 | IPIGAGICASYQTQTNSR | 1879.9331 | |||
| D950N | 948–964 | LQNVVNQNAQALNTLVK | 1867.0396 | 1867.0396 | ||
| P.1 (Gamma) | Brazil | L18F | 1–21 | MFVFLVLLPLVSSQCVNFTTR | 2414.2975 | |
| T20N | 1–21 | MFVFLVLLPLVSSQCVNLTNR | 2393.3084 | 2393.3084 | ||
| P26S | 22–34 | TQLPSAYTNSFTR | 1485.7333 | 1485.7333 | ||
| D138Y | 133–147 | FQFCNYPFLGVYYHK | 1925.9043 | 1925.9043 | ||
| R190S | 188–191 | NLSE | 462.2195 | (462.2195) | ||
| K417T | 409–420 | QIAPGQTGTIAD | 1171.5954 | 1171.5954 | ||
| E484K | 472–484 | IYQAGSTPCNGVK | 1337.6519 | 1337.6519 | ||
| N501Y | 499–504 | STYLVK | 710.4084 | 710.4084 | ||
| D614G | 587–619 | ITPCSFGGVSVITPGTNTSNQVAVLYQGVNCTE | 3356.6138 | |||
| H655Y | 655–661 | YVNNSYE | 888.3734 | 888.3734 | ||
| T1027I | 1019–1028 | ASANLAAIK | 858.5044 | 858.5044 | ||
| V1176F | 1169–1181 | ISGINASFVNIQK | 1390.7689 | 1390.7689 |
aResidue numbering is based on the originating strain and may differ in some variants due to the presence of deletion sites
bAll strain distinguishing peptides do not contain proline (F817P, A892P, A899P, A942P, K986P, V987P) or alanine substitutions (R683A and R685A) added to the recombinant forms for the variants introduced to stabilize the S-protein trimer
cThose with masses lower than 500 are bracketed since they typically appear among matrix background ions in MALDI mass spectra. All other peptides differ in mass by at least 83 ppm, as is the case for mass 1133.5626 and that of 1133.6565 for missed cleaved peptide 821–830 (of sequence LLFNKVTLAD) for the spike protein of the original reference strain
High-resolution MALDI mass spectra were recorded for the doubly digested protein extricated from laboratory-grown virus or recombinantly expressed in the case of highly transmissible variants given that the strains themselves require specialized containment facilities. The latter protein variants all contain a ten residue C-terminal histidine tag and additional proline and alanine substitutions to stabilize the trimeric prefusion state of the protein. All of the substitutions fall outside of the variant-specific peptides (see Table 2 footnote).
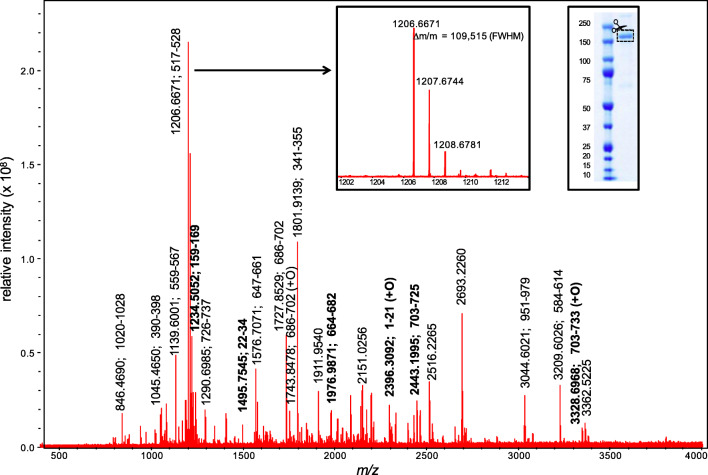
The high-resolution MALDI spectrum for the S-protein extricated from a laboratory grown strain [12] (Fig. 1) shows the presence of 17 proteolytic peptides with whose masses all fall within 1.5 ppm of those predicted sequence with a mass resolution of 109,515 (FWHM) measured at ions m/z 1206.6671 (Fig. 1 insert). The combined segments (Table 3) span 253 of a total of 1273 residues, or 20% of the protein, consistent with typical reported coverage levels [12]. Most peptides represent complete cleavage products and six (in bold in Table 3) contain regions that allow major SARS-CoV-2 variants to be distinguished based on the data of Table 2.
Fig. 1.
High-resolution MALDI mass spectra for the doubly digested (trypsin + GluC) S-protein extricated from laboratory grown virus. Peaks labelled in bold represent regions containing mutations in major variants of concern
Table 3.
Tryptic + GluC peptide ions detected for spike protein from lab grown specimen, their sequences and location
|
m/z
(mono.) experimental |
m/z
(mono.) theoretical |
Difference (ppm) | Residuesa | Sequence | Domainb |
|---|---|---|---|---|---|
| 846.4690 | 846.4680 | + 1.2 | 1020–1028 | ASANLAATK | S2 undefined |
| 1045.4650 | 1045.4659 | − 0.9 | 390–398 | LCFTNVYAD | S1 subunit receptor-binding domain (RBD) |
| 1139.6001 | 1139.5996 | + 0.4 | 559–567 | FLPFQQFGR | S1 undefined |
| 1206.6671 | 1206.6663 | + 0.7 | 517–528 | LLHAPATVCGPK | S1 subunit receptor-binding domain (RBD)—partial |
| 1234.5052 | 1234.5045 | + 0.6 | 159–169 | VYSSANNCTFE | S1 subunit N-terminal domain (NTD) |
| 1290.6985 | 1290.6974 | + 0.7 | 726–737 (1) | ILPVSMTKTSVD | S2 undefined |
| 1495.7545 | 1495.7540 | + 0.3 | 22–34 | TQLPPAYTNSFTR | S1 subunit N-terminal domain (NTD) |
| 1576.7071 | 1576.7060 | + 0.7 | 647–661 | AGCLIGAEHVNNSYE | S1 subunit C-terminal domain (CTD) |
| 1727.8529 | 1727.8520 | + 0.5 | 686–702 | SVASQSIIAYTMSLGAE | S2 subunit N-terminus at furin cleavage site |
| 1743.8478 | 1743.8469 | + 0.5 | 686–702 (+O) | SVASQSIIAYTMSLGAE | S2 subunit N-terminus at furin cleavage site |
| 1801.9139 | 1801.9133 | + 0.3 | 341–355 (1) | VFNATRFASVYAWNR | S1 subunit receptor-binding domain (RBD) |
| 1976.9871 | 1976.9858 | + 0.7 | 664–682 | IPIGAGICASYQTQTNSPR | S1 subunit C-terminus at furin cleavage site |
| 2396.3092 | 2396.3080 | + 0.5 | 1–21 (+O) | MFVFLVLLPLVSSQCVNLTTR | S1 subunit N-terminal domain (NTD) |
| 2443.1995 | 2443.1987 | + 0.3 | 703–725 | NSVAYSNNSIAIPTNFTISVTTE | Undefined |
| 3044.6021 | 3044.6011 | + 0.3 | 951–979 (1) | VVNQNAQALNTLVKQLSSNFGAISSVLND | HR1 domain—partial |
| 3209.6026 | 3209.6035 | − 0.3 | 584–614 (1) | ILDITPCSFGGVSVITPGTNTSNQVAVLYQD | S1 subunit receptor-binding domain (RBD)—partial |
| 3328.6968 | 3328.6981 | − 0.4 | 703–733 (+O) (1) | NSVAYSNNSIAIPTNFTISVTTEILPVSMTK | Undefined |
aBased on NCBI protein sequence QHD43416.1 where residues denoted (+O) are associated with an oxidized methionine residues and those with a (1) containing one missed cleavage site; all others contain no missed cleavage sites. Bolded entries represent regions that allow variants to be distinguished as identified in Table 2
bAs defined in UniPro knowledge base (uniprokb) at https://covid-19.uniprot.org/uniprotkb/ and ref. Acta Pharmacologica Sinica
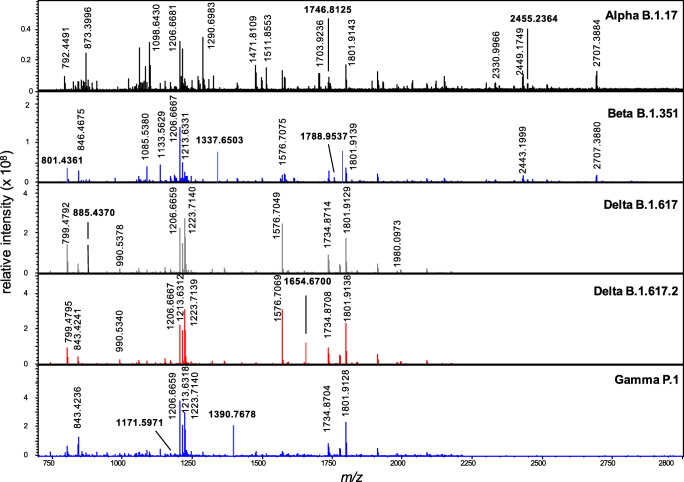
The spectra for the recombinant forms for each of the 5 major variants are shown in Fig. 2. As expected, they contain a number of common ions including those at ions m/z 1206, associated with residues 517–228, and m/z 1801 resulting from a missed cleaved peptide comprising residues 341–355. Note that the actual numbering of these residues and other peptide segments will vary from the originating strain due to the presence of deletion sites in some variants (see Table 2). All of the vertically labelled masses have been assigned (see Supplementary Table 1) but residue segments are not shown on the spectra for clarity. Those labelled horizontally and in bold represent those peptides that can be used to distinguish the variants (Table 2). For example, the spectrum of the alpha variant exhibits two distinguishing peptides comprising residues 703–725 (at m/z 2455.2364) and 1112–1127 (at m/z 1746.8125) that contain the T716I and D1118H mutations respectively (Table 2). The beta variant is identified by three peptides at m/z 801.4361, 1337.6503 and 1788.9537 representing residues 238–246 (Δ242–244), 472–484 and 238–253 containing the 242–244 deletion, E484K and R246I mutations. The delta variants are distinguished from other variants of concern, and from each other, based upon the detection of the peptides at m/z 885.4370 (89–96) and 1654.6700 (154–169) containing the T95I and R158G mutations. The gamma variant is distinguished by two peptides at m/z 1171.5971 (409–420) and 1390.7678 (1169–1181) containing the unique K419T and V1176F mutations. Irrespective of the coverage and the ionization of particular peptides, the variants of concern can be identified, and distinguished from one another, based on the detection of any one of the peptides of unique mass (Table 2) in these maps.
Fig. 2.
High-resolution MALDI mass spectra for the doubly-digested (trypsin + GluC) recombinant S-protein for five major variants of concern. Peaks labelled horizontally containing mutations that distinguish the variants. Residue segments for all peaks are provided in Supplementary Table 1
The ability of the results to correctly chart the evolution of the variants was assessed using the MassTree algorithm. This algorithm builds phylogenetic-like trees from mass map data generated experimentally or theoretically, or using some combination of both. Mass trees have been found to highly congruent with sequence-based trees in a series of studies [18–20, 25].
A mass tree built from the labelled masses shown in Figs. 1 and 2, which represent the identified peptide segments of the S-protein from the originating strain and each major variant, is shown in Fig. 3. Even without the use of any sequence data or their alignment [19], the mass tree correctly predicts the evolution of the beta, delta and gamma variants from the alpha UK lineage as well as the close association of the two delta variants. Even though the mass datasets do not represent complete S-protein coverage, and despite the very different nature of the data itself, the tree closely resembles the topology of a sequence tree built following the alignment of protein sequence data (across a common span of residues 16 to 1213) (Supplementary Fig. 1). The latter is consistent with that reported elsewhere [26]. Where greater coverage is achieved, the MassTree algorithm has been shown to be able to correctly identify point mutations and display on them on the mass tree, providing each peptide segment contains a single mutation [24].
Fig. 3.
Mass tree for the S-protein of an originating strain and five major variants of concern, constructed using the mass map data of Figs. 1 and 2
Conclusions
This study demonstrates that the detection of SARS-CoV-2 variants of concern is no longer reliant on genome sequencing. Peptide signatures of unique mass can be used to identify the presence of mutations associated with the evolution of the virus without the need for gene or protein sequences. Further, the evolution of the virus can be correctly charted from the mass maps as has been shown previously for other viruses [18–20] and a wider range of organisms [25] by this laboratory. Such a mass spectrometry-based strategy offers an alternative to conventional PCR-based genetic detection and analysis of the virus [27] where, after RNA extraction from viral specimens, studies of its evolutionary dynamics require the relatively time-consuming generation, interpretation and processing of large genome sequence datasets. The mass spectrometric approach does require the initial isolation, or at least partial purification, of the S-protein but if optimized, using procedures under development [28], this could be performed within a similar timeframe to the many steps needed to isolate, purify and amplify the virus’ genes or genome [29]. While mutations within the S-protein or its gene have been the focus of most studies of the variants of concern [4, 6–9] given the role the protein plays in host cell interactions [3], variant-specific mutations identified within other protein-coding regions [5] (e.g. for nucleocapsid) could be detected by the same MS approach.
The ease with which protein mass maps can be generated, once a viral protein is isolated, and the speed and sensitivity of mass spectrometric approaches afford benefits over gene-based approaches [10]. Protein-based approaches are further more transferable to studies in structural biology that identify antiviral drug targets [30] for therapeutic interventions, as well as vaccine candidates.
Supplementary information
(DOCX 23 kb)
(PDF 80 kb)
Funding
Author Downard acknowledges support from the Clinical Research Fund and donors to this study.
Data availability
The MassTree algorithm can be accessed for use by contacting the corresponding author.
Declarations
Ethics approval
All procedures for collection, preparation and transport of the samples were carried out in accordance with the Communicable Diseases Network Australia (CDNA) national guidelines for Coronavirus Disease 2019 and NSW Health restrictions and protocols with the virus cultured in Vero E6 cells within a physical containment laboratory [21].
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.MacLean OA, Orton RJ, Singer JB, Robertson DL. No evidence for distinct types in the evolution of SARS-CoV-2. Virus Evol. 2020; 6: veaa034. [DOI] [PMC free article] [PubMed]
- 2.Zhou HY, Ji CY, Fan H, Han N, Li XF, Wu A, Qin CF. Convergent evolution of SARS-CoV-2 in human and animals. Protein Cell. 2021;30:1–4. doi: 10.1007/s13238-021-00847-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banoun H. Evolution of SARS-CoV-2: review of mutations, role of the host immune system. Nephron. 2021;145:392–403. doi: 10.1159/000515417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanyaolu A, Okorie C, Marinkovic A, Haider N, Abbasi AB, Jaferi U, Prakash S, Balendra V. The emerging SARS-CoV-2 variants of concern. Therapeutic Advances in Infectious Disease. 2021;8:20499361211024372. doi: 10.1177/20499361211024372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang R, Hozumi Y, Yin C, Wei G-W. Decoding SARS-CoV-2 transmission and evolution and ramifications for COVID-19 diagnosis, vaccine, and medicine. J Chem Inf Model. 2020;60:5853–5865. doi: 10.1021/acs.jcim.0c00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, Hengartner N, Giorgi EE, Bhattacharya T, Foley B, Hastie KM, Parker MD, Partridge DG, Evans CM, Freeman TM, de Silva TI, Sheffield COVID-19 Genomics Group. McDanal C, Perez LG, Tang H, Moon-Walker A, Whelan SP, CC LB, Saphire EO, Montefiori DC. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ali F, Kasry A, Amin M. The new SARS-CoV-2 strain shows a stronger binding affinity to ACE2 due to N501Y mutant. Med Drug Discov. 2021;10:100086. doi: 10.1016/j.medidd.2021.100086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meng B, Kemp SA, Papa G, Datir R, Ferreira IATM, Marelli S, Harvey WT, Lytras S, Mohamed A, Gallo G, Thakur N, Collier DA, Mlcochova P, COVID-19 Genomics UK (COG-UK) Consortium. Duncan LM, Carabelli AM, Kenyon JC, Lever AM, De Marco A, Saliba C, Culap K, Cameroni E, Matheson NJ, Piccoli L, Corti D, James LC, Robertson DL, Bailey D, Gupta RK. Recurrent emergence of SARS-CoV-2 spike deletion H69/V70 and its role in the alpha variant B.1.1.7. Cell Rep. 2021;35:109292. doi: 10.1016/j.celrep.2021.109292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jangra S, Ye C, Rathnasinghe R, Stadlbauer D, Personalized Virology Initiative study group. Krammer F, Simon V, Martinez-Sobrido L, García-Sastre A, Schotsaert M. SARS-CoV-2 spike E484K mutation reduces antibody neutralisation. Lancet Microbe. 2021;2:e283–e284. doi: 10.1016/S2666-5247(21)00068-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Griffin JH, Downard KM. Mass spectrometry analytical responses to the SARS-CoV2 coronavirus in review. Trends Anal Chem. 2021;142:116328. doi: 10.1016/j.trac.2021.116328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Udugama B, Kadhiresan P, Kozlowski HN, Malekjahani A, Osborne M, Li V, Chen H, Mubareka S, Gubbay JB, Chan W. Diagnosing COVID-19: the disease and tools for detection. ACS Nano. 2020;14:3822–3835. doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- 12.Dollman NL, Griffin JH, Downard KM. Detection, mapping, and proteotyping of SARS-CoV-2 coronavirus with high resolution mass spectrometry. ACS Infect Dis. 2020;6:3269–3276. doi: 10.1021/acsinfecdis.0c00664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwahn AB, Wong JWH, Downard KM. Subtyping of the influenza virus by high resolution mass spectrometry. Anal Chem. 2009;81:3500–3506. doi: 10.1021/ac900026f. [DOI] [PubMed] [Google Scholar]
- 14.Downard KM. Proteotyping for the rapid identification of pandemic influenza virus and other biopathogens. Chem Soc Rev. 2013;42:8584–8595. doi: 10.1039/c3cs60081e. [DOI] [PubMed] [Google Scholar]
- 15.Nguyen AP, Downard KM. Proteotyping of the parainfluenza virus with high resolution mass spectrometry. Anal Chem. 2013;85:1097–1105. doi: 10.1021/ac302962u. [DOI] [PubMed] [Google Scholar]
- 16.Fernandes ND, Downard KM. Incorporation of a proteotyping approach using mass spectrometry for the surveillance of the influenza virus in cell culture. J Clin Microbio. 2014;52:725–735. doi: 10.1128/JCM.02315-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uddin R, Downard KM. Subtyping of hepatitis C virus with high resolution mass spectrometry. Clin Mass Spectrom. 2017;4-5:19–24. doi: 10.1016/j.clinms.2017.08.003. [DOI] [Google Scholar]
- 18.Lun ATL, Swaminathan K, Wong JWH, Downard KM. Mass trees – a new phylogenetic approach and algorithm to chart evolutionary history with mass spectrometry. Anal Chem. 2013;85:5475–5482. doi: 10.1021/ac4005875. [DOI] [PubMed] [Google Scholar]
- 19.Downard KM. Sequence free phylogenetics with mass spectrometry. Mass Spectrom Rev. 2021, in press. 10.1002/mas.21658. [DOI] [PubMed]
- 20.Akand EH, Downard KM. Mutational analysis employing a phylogenetic mass tree approach in a study of the evolution of the influenza virus. Mol Phylogenet Evol. 2017;112:209–217. doi: 10.1016/j.ympev.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 21.Druce J, Tran T, Kostecki R, Chibo D, Morris M, Catton M, Birch C. SARS–associated coronavirus replication in cell lines. Emerg Infect Dis. 2006;12:128–33. [DOI] [PMC free article] [PubMed]
- 22.Evans J, Sheneman L, Foster JA. Relaxed neighbor joining: a fast distance-based phylogenetic tree construction method. J Mol Evol. 2006;62:785–792. doi: 10.1007/s00239-005-0176-2. [DOI] [PubMed] [Google Scholar]
- 23.Sheneman L, Evans J, Foster JA. Clearcut: a fast implementation of relaxed neighbor joining. Bioinformatics. 2006;22:2823–2834. doi: 10.1093/bioinformatics/btl478. [DOI] [PubMed] [Google Scholar]
- 24.Mann C, Downard KM. Evolution of SARS CoV-2 coronavirus surface protein investigated with mass spectrometry based phylogenetics. Anal Lett. 2021; in press. 10.1080/00032719.2021.1928685.
- 25.Downard KM. Darwin’s tree of life is numbered. Resolving the origins of species by mass. Evol Biol. 2020;47:325–333. doi: 10.1007/s11692-020-09517-7. [DOI] [Google Scholar]
- 26.Pattabiraman C. Tracking SARS-COV-2 variants of concern. Observer Research Foundation Special Report. 2021;144:1–13. [Google Scholar]
- 27.Udugama B, Kadhiresan P, Kozlowski HN, Malekjahani A, Osborne M, Li VYC, Chen H, Mubareka S, Gubbay JB, Chan WCW. Diagnosing COVID-19: the disease and tools for detection. ACS Nano. 2020;14:3822–3835. doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- 28.Ponce-Rojas JC, Costello MS, Proctor DA, Kosik KS, Wilson MZ, Arias C, Acosta-Alvear D. A fast and accessible method for the isolation of RNA, DNA, and protein to facilitate the detection of SARS-CoV-2. J Clin Microbiol. 2021;59:e02403–e02420. doi: 10.1128/JCM.02403-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ambrosi C, Prezioso C, Checconi P, Scribano D, Sarshar M, Capannari M, Tomino C, Fini M, Garaci E, Palamara AT, De Chiara G, Limongi D. SARS-CoV-2: comparative analysis of different RNA extraction methods. J Virol Methods. 2021;287:114008. doi: 10.1016/j.jviromet.2020.114008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang Y, Yang C, Xu X-F, Xu W, Liu S-W. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol Sinica. 2020;41:1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 23 kb)
(PDF 80 kb)
Data Availability Statement
The MassTree algorithm can be accessed for use by contacting the corresponding author.