Abstract
Oral squamous cell carcinoma (OSCC) is one of the most common types of cancer worldwide. Accumulating evidence has shown that long noncoding RNAs (lncRNAs) serve important roles in the development of OSCC. The purpose of this study was to investigate the biological function and underlying regulatory mechanism of lncRNA homeobox A cluster antisense RNA2 (HOXA-AS2) in OSCC. RT-qPCR was performed to analyze the HOXA-AS2 expressions in human immortalized oral epithelial cell (HIOEC) line, human OSCC cell lines, and plasma. The expression of HOXA-AS2 and enhancer of zeste 2 polycomb repressive complex 2 subunit (EZH2) in Tca-8113 cells were knocked down or overexpressed by transfection with shRNA-HOXA-AS2 or pcDNA-EZH2, respectively. The interaction between HOXA-AS2 and EZH2 was validated by RNA immunoprecipitation assay. In addition, cell proliferation was assessed by CCK-8 and EdU assays. Cell cycle distribution was analyzed by flow cytometry. Cell migration and invasion were detected using wound healing and Transwell assays, respectively. Apoptosis was detected by TUNEL staining. The protein expression levels of cell cycle and apoptosis-related proteins were measured by western blot analysis. Compared with HIOEC cells, HOXA-AS2 expression in OSCC cells was upregulated. HOXA-AS2 knockdown significantly inhibited Tca-8113 cell proliferation, blocked the cell cycle by arresting cells in the G0/G1 phase, promoted apoptosis, and suppressed migration and invasion. In addition, HOXA-AS2 was predicted to directly target EZH2 and positively regulate EZH2 expression. EZH2 overexpression could reverse the inhibitory effect of HOXA-AS2 knockdown on the proliferation, migration, and invasion of Tca-8113 cells. In summary, the findings suggested that HOXA-AS2 may inhibit cell proliferation, invasion, and migration, induce cell cycle arrest in the G0/G1 phase, and increase cell apoptosis by targeting EZH2. The research indicated that HOXA-AS2/EZH2 axis may play a key role in the development of OSCC.
Keywords: oral squamous cell carcinoma, HOXA-AS2, EZH2, proliferation, invasion, migration
Introduction
Oral squamous cell carcinoma (OSCC) is a common malignant tumor of the head and neck, accounting for ∼3% of all systemic malignant tumors.1 At present, although most OSCC cases are treated by surgical resection, radiotherapy and chemotherapy, the prognosis remains poor and serious side effects from the treatment often occur. Previous studies have reported that metastasis is a major problem hindering in the treatment of OSCC and is, therefore, considered to be the primary factor affecting the prognosis of patients with OSCC.2 Once metastasis occurs in patients with OSCC, the cancer is difficult to effectively treat, resulting in a 5-year survival rate of 10% to 50%.3 Therefore, it remains an urgent priority to investigate the underling mechanisms of metastasis in OSCC to identify novel therapeutic targets and diagnostic markers.
Long noncoding RNAs (lncRNAs) are a type of nonprotein coding transcript of >200 nucleotides in length. Accumulating evidence has indicated that the biological functions of lncRNAs are complex but are closely associated with the proliferation, invasion, metastasis, and recurrence of tumors.4,5 For instance, lncRNA TMPO-AS1 facilitates OSCC metastases via activating GLI1 by targeting LAP2a.6 Gao et al7 demonstrated that the expression levels of lncRNAs were associated with the TNM stage and lymph node metastasis in patients with tongue squamous cell carcinoma (TSCC), which verified that the abnormal regulation of lncRNAs was associated with the biological activity of TSCC. An increasing number of studies have also reported that the expression levels of homeobox A cluster antisense RNA2 (HOXA-AS2) in various malignant tumors, which either regulated the expression levels of related genes.8 In hepatocellular carcinoma, HOXA-AS2 promoted the epithelial–mesenchymal transition (EMT) of HCC cells by regulating the microRNA-520c-3p/glypican 3 regulatory axis.9 In gallbladder carcinoma (GBC), HOXA-AS2 overexpression significantly enhanced the proliferation and EMT of GBC cells.10 In bladder cancer, HOXA-AS2 promoted the proliferation and invasion of bladder cancer 5637 and T24 cells by downregulating miR-125b expression.11 The overexpression of HOXA-AS2 significantly enhanced nasopharyngeal carcinoma progression, including the cell proliferative, migratory, and invasive abilities.12 However, to the best of our knowledge, little is known concerning the effects of HOXA-AS2 on the proliferation, migration, and invasion of OSCC cells. To gain a more comprehensive understanding of the role of HOXA-AS2 in all kinds of tumors, it is necessary to analyze the regulatory functions of HOXA-AS2 in OSCC cells.
Enhancer of zeste 2 polycomb repressive complex 2 subunit (EZH2) is a novel human gene that was discovered in 1996 and located on human chromosome 21q22.2, where it is associated with Down’s syndrome. As an important oncogene, EZH2 has been widely studied in various tumor types. For example, upregulated expression levels of EZH2 were found to be closely associated with cell proliferation and prognosis in OSCC.13 Another previous study demonstrated that HOXA-AS2 promoted colorectal cancer cell proliferation via downregulating P27 expression levels by targeting EZH2.14 However, whether HOXA-AS2 could regulate the progression of OSCC cells through targeting EZH2 has not been investigated to date.
Based on these aforementioned findings, the present study aimed to investigate HOXA-AS2 expression levels in OSCC cells and to determine the effects of HOXA-AS2 on the proliferation, migration, invasion, and apoptosis of OSCC cells.
Methods
Cell Culture
The human immortal oral epithelial cells (HIOEC) and human oral squamous cell carcinoma cells (Tca-8113) were purchased from Type Culture Collection of Chinese Academy of Sciences. All other OSCC cells (CAL-27, SCC-4, SCC-9, and SCC-15) were obtained from the American Type Culture Collection (ATCC). HIOEC cells were cultured in a defined keratinocyte-SFM medium (Gibco), while the other cells were cultured in DMEM supplemented with 10% FBS, 100 U/mL penicillin, and 100 U/mL streptomycin (Sigma). Cells were maintained in a fully humidified atmosphere containing 5% CO2 at 37 °C. The medium was changed regularly to observe the proliferation of cells over time. Cells were passaged when the cell confluence reached 90%. Cells in the logarithmic phase were used for subsequent experiments.
Cell Transfection
The short-hairpin RNA (shRNA) against HOXA-AS2 (shRNA-HOXA-AS2-1 and shRNA-HOXA-AS2-2) and corresponding empty vector (shRNA-NC) were purchased from GenePharma. The pcDNA-EZH2 and the empty vector pcDNA-NC were constructed from Sangon Biotech. Tca-8113 cells (1 × 106 cells/well) were inoculated into 12-well plates and then transfected with shRNA-NC, shRNA-HOXA-AS2, pcDNA-NC, and pcDNA-EZH2 using Lipofectamine® 3000 reagent (Thermo Fisher Scientific) following the manufacturer’s instruction. After 48 h, RT-qPCR was used to assess the transfection efficiencies. The transfected cells were collected for the next experiments.
RT-qPCR Analysis
Total RNA was extracted from cells using Trizol reagent (Invitrogen). The purity and quality of RNA and cDNA were assessed by the optical density (OD) at 260 and 280 nm, respectively, using an ultraviolet spectrophotometer (Thermo). Total RNA was reverse transcribed into cDNA and subsequently underwent qPCR using an All-in-One™ qRT-PCR Detection Kit (GeneCopoeia), according to the manufacturer’s protocol. qPCR was performed on an ABI Prism 7900HT instrument (Applied Biosystems). The following thermocycling conditions were used for the qPCR: Initial denaturation at 95 °C for 5 s; followed by 45 cycles of denaturation at 95 °C for 20 s, annealing at 58 °C for 20 s, and a final extension at 72 °C for 30 s. The primer sequences are as follow: HOXA-AS2: forward, 5′-AACCCATCTTTGCCTTCTGC-3′, reverse, 5′-CGGAGGAGTTTGGAGTTGG-3′; EZH2: forward, 5′-TGCACATCCTGACTTCTGTG-3′, reverse, 5′-AAGGGCATTCACCAACTCC-3′; GAPDH: forward, 5′-GAAGAGAGAGACCCTCACGCTG-3′, reverse, 5′-ACTGTGAGGAGGGGAGATTCAGT-3′. The relative expression of target gene was calculated by the 2−ΔΔCT method15 and normalized to GAPDH.
CCK-8 Assay
Transfected cells in the logarithmic phase were seeded into 96-well plates at the density of 2 × 103 cells/well for routine culture. Following 24, 48, or 72 h of culture, 10 μL of CCK-8 solution (Dojindo Molecular Technologies, Inc.) was added to each well and incubated for another 2 h at room temperature. The absorbance of each well was measured using a microplate reader (Bio-Rad) at a wavelength of 450 nm.
EdU Staining Assay
Transfected cells were seeded into 96-well plates at a density of 1 × 105 cells/mL and cultured overnight with 5% CO2 at 37 °C. EdU assay was performed by the EdU immunofluorescence staining kit (Ribobio). In short, cells of each group were first incubated with 100 μL of EdU reagent for 4 h. Following incubation, the culture medium was discarded, and cells were washed twice with PBS. Next, cells were fixed with 4% paraformaldehyde for 15 min and stained with 5 μg/mL Hoechst 33342 for 30 min. Stained cells were visualized and imaged under a fluorescence microscope (Olympus BX 60 fluorescence microscope).
TUNEL Staining Assay
Tca-8113 cells were inoculated into sterilized slides. Apoptotic cells were stained using a TUNEL staining kit (Nanjing KeyGen Biotech) following the manufacturer’s protocol. Cells in the section were observed by inverted microscopy (Nikon TS100) at 400× magnification and photographed. Cells with brown-yellow nuclei were identified as positive apoptotic cells.
Cell Cycle Assay
Cell cycle distribution was detected by the cell cycle detection kit (KeyGen). Transfected cells were inoculated into 6-well plates at the density of 1 × 106 cells/mL and cells were fixed with 70% ethanol at 4 °C overnight. Next, cells were washed with PBS for 3 times and co-incubated with 0.25 mg/mL RNaseA for 30 min at 37 °C. Then 50 μg/mL PI was then added to the cell suspension and continue to incubate for 30 min in the dark. The cells were analyzed in FL2 channel of flow cytometry (FACScan, Becton 4 Dickinson).
Cell Migration Assay
Cell migration was analyzed by the wound healing assay. Cells were inoculated into 24-well plates at the density of 2 × 105 cells/mL. After the cells were close to full confluency, the layer of cells was scratched to form wounds with a sterile 200 µL pipette tip and cultured with serum-free medium for 24 h. The image of the scratched area was captured after 0 and 24 h. The %age of cell migration was analyzed by ImageJ software (NIH).
Cell Invasion Assay
A Transwell assay was used to evaluate cell invasion using an 8 µm pore Transwell chamber (Corning) precoated with 5 µL Matrigel (Becton Dickinson) at room temperature for 24 h. Briefly, 5 × 104 cells/well were plated into the upper chambers of Transwell plates. Meanwhile, the complete medium supplemented with 10% FBS was added into the lower chambers. After 24 h of routine culture, cells remaining in the upper chamber were removed by washing with PBS and invasive cells in the lower chamber were fixed and stained with methanol and crystal violet. Invasive cells were observed and counted using an inverted microscope (Nikon).
RNA Immunoprecipitation Assay
A RIP assay was performed using the Magna RIP RNA-Binding Protein Immunoprecipitation kit (EMD Millipore) according to the manufacturer’s protocols. Cells were incubated with anti-EZH2 antibody (#5246, Cell Signaling Technology) or anti-IgG antibody as a control. Co-precipitated RNAs were analyzed using RT-qPCR.
Western Blot Analysis
Total proteins were extracted with RIPA lysis and protein concentrations were determined with a Pierce® BCA Protein Assay Kit (Thermo Fisher). Total proteins were separated via 12% SDS-PAGE, and the separated proteins were transferred onto PVDF membranes (Millipore) and blocked with 5% skimmed milk powder for 1 h at 37 °C. These membranes were subsequently treated with the following primary antibodies (all from Cell Signaling Technology Inc.) against CDK2 (1:1000; cat. no. #2546), cyclin E1 (1:1000; cat. no. #20808), P27 (1:1000; cat. no. #3686), MMP2 (1:1000; cat. no. #40994), MMP9 (1:1000; cat. no. #13667), Bcl-2 (1:1000; cat. no. #3498), Bax (1:1000; cat. no. #5023), Cleaved-caspase3 (1:1000; cat. no. #9661), caspase3 (1:1000; cat. no. #9662), and GAPDH (1:1000; cat. no. #5174) at 4 °C overnight, respectively. Following the primary antibody incubation, membranes were washed in TBST and then incubated with HRP-conjugated goat antirabbit secondary antibody (1:5000; cat. no. A0208; Beyotime Institute of Biotechnology) at room temperature for 2 h. Protein bands were visualized using an ECL kit (GE Healthcare Inc.). GAPDH was taken as an internal reference. Protein expression analysis was performed using Image J software (National Institute of Health) to calculate the relative protein expression.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism6.0 software (GraphPad Software Inc.). Data are presented as the mean ± standard deviation (SD) and statistical differences between groups were analyzed using a 1-way ANOVA followed by a Tukey’s post hoc test. P < .05 was considered to indicate a statistically significant difference.
Results
HOXA-AS2 Knockdown Inhibited the Proliferation of Tca-8113 Cells
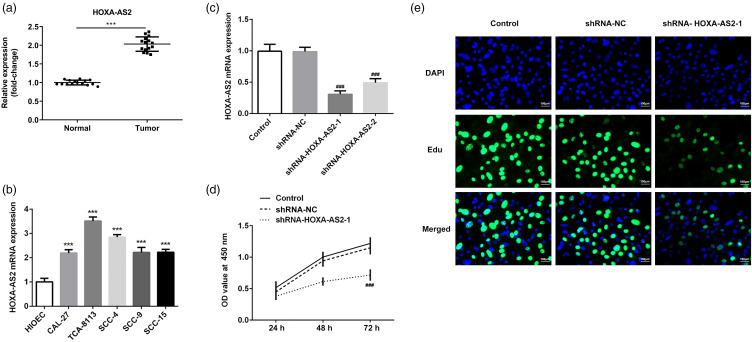
The expression levels of HOXA-AS2 in the serum of patients with OSCC are significantly higher than that of normal patients (Figure 1a). Besides, HOXA-AS2 expression in 5 OSCC cell lines (CAL-27, Tca-8113, SCC-4, SCC-9, and SCC-15) and one normal oral epithelial cell line (HIOEC) were then first determined by RT-qPCR. As shown in Figure 1b, compared with the HIOEC cells, HOXA-AS2 expression in OSCC cells was significantly upregulated. In particular, HOXA-AS2 expression was highest in Tca-8113 cells. Therefore, Tca-8113 cells were selected for use in subsequent experiments. Tca-8113 cells were subsequently transfected with shRNA-NC, shRNA-HOXA-AS-1, and shRNA-HOXA-AS-2. RT-qPCR analysis revealed that HOXA-AS2 knockdown significantly downregulated HOXA-AS2 expression compared with the control group, with the expression levels of HOXA-AS2 downregulated to the greatest extent in the shRNA-HOXA-AS2 to 1 group (Figure 1c). Cell viability and proliferation were determined using CCK-8 and EdU assays, respectively. The results of the CCK-8 and EdU assays demonstrated that HOXA-AS2 knockdown significantly reduced the viability and proliferation of Tca-8113 cells (Figure 1d and e). These results indicated that HOXA-AS2 knockdown may inhibit the proliferation of Tca-8113 cells.
Figure 1.
HOXA-AS2 knockdown inhibits the proliferation of Tca-8113 cells: (a) HOXA-AS2 mRNA expression in the serum of patients with OSCC. (b) HOXA-AS2 mRNA expression in HIOEC and human oral squamous cell carcinoma cells as analyzed by RT-qPCR. ***P < .001 versus HIOEC. (c) Knockdown efficiency of HOXA-AS2 expression was detected by RT-qPCR. (d) Cell viability in each group following transfection was detected using CCK-8 assay. (e) Cell proliferation was assessed by EdU assay. ###P < .001 versus shRNA-NC.
Abbreviations: HOXA-AS2, homeobox A cluster antisense RNA2; OSCC, oral squamous cell carcinoma; HIOEC, human immortal oral epithelial cells.
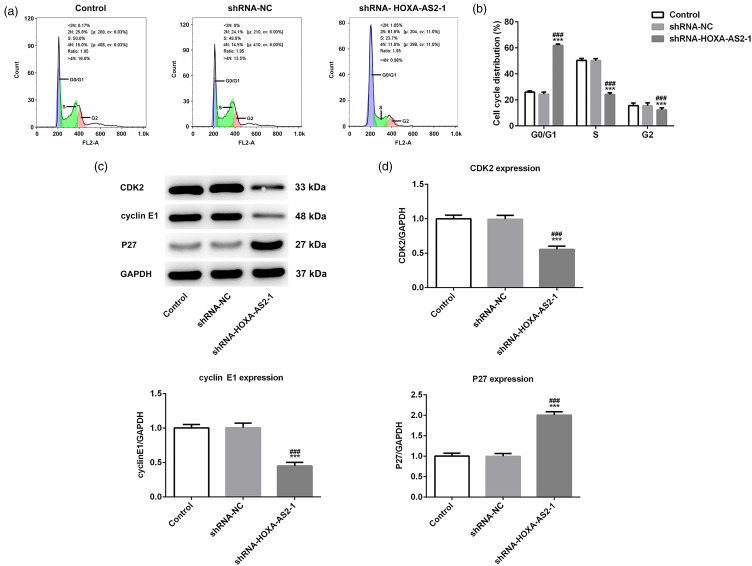
HOXA-AS2 Knockdown Induced Cell Cycle Arrest in Tca-8113 Cells
The cell cycle distribution and expression levels of cell cycle-related proteins were assessed by flow cytometry and western blotting, respectively. As shown in Figure 2a and b, compared with the control group, HOXA-AS2 knockdown increased the proportion of cells in the G0/G1 phase and decreased the proportion of cells in the S phase. It suggested that HOXA-AS2 knockdown could arrest Tca-8113 cells in the G0/G1 phase. Subsequently, the results of western blotting analysis showed that compared with the control group, HOXA-AS2 knockdown significantly downregulated the expression of CDK2 and cyclin E1, whereas the expression of P27 was upregulated (Figure 2c and d). These results indicated that HOXA-AS2 knockdown may induce cell cycle arrest in Tca-8113 cells.
Figure 2.
Homeobox A cluster antisense RNA2 (HOXA-AS2) knockdown-induced cell cycle arrest in Tca-8113 cells: (a) Cell cycle distribution was detected by flow cytometry. (b) Quantitative analysis of cell cycle distribution of Tca-8113 cells transfected with shRNA-HOXA-AS2 to 1. (c) Protein expression levels of CDK2, cyclin E1, and P27 were analyzed by western blotting. (d) Semiquantification of the expression levels of proteins presented in part (c), GAPDH was used as the control. ***P < .001 versus shRNA-NC.
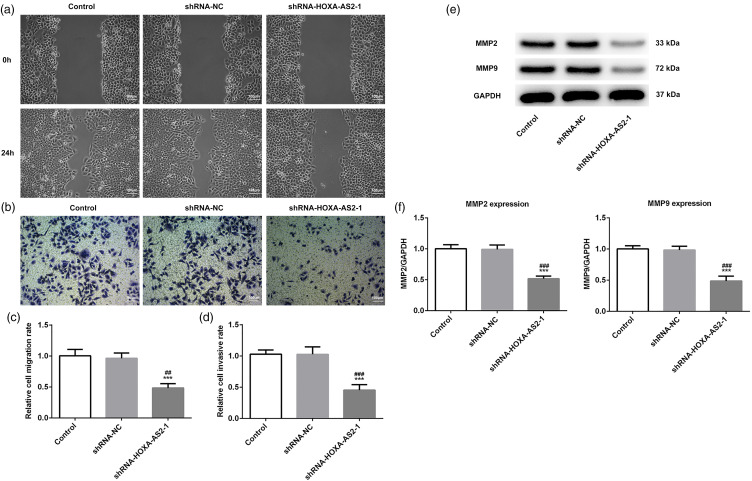
HOXA-AS2 Knockdown Inhibited the Migration and Invasion of Tca-8113 Cells
In view of the fact that metastasis is considered to be the primary factor affecting the prognosis of patients with OSCC. Cell migration and invasion were then detected by wound healing and Transwell assays, respectively. The results of the wound healing assay revealed that HOXA-AS2 knockdown significantly suppressed the migration of Tca-8113 cells compared with the control group (Figure 3a and c). Similarly, HOXA-AS2 knockdown could also significantly inhibited invasion compared with the control group (Figure 3b and d). In addition, the expressions of invasion-related proteins (MMP2 and MMP9) were measured by western blotting analysis.16 The results demonstrated that HOXA-AS2 knockdown significantly downregulated the expressions of MMP2 and MMP9 in Tca-8113 cells (Figure 3e and f). Taken together, these results indicated that HOXA-AS2 knockdown may inhibit migration and invasion of Tca-8113 cells.
Figure 3.
Homeobox A cluster antisense RNA2 (HOXA-AS2) knockdown inhibits the migration and invasion of Tca-8113 cells: (a) Cell migration was determined by wound healing assay. (b) Cell invasion was determined by Transwell assay. (c) Semiquantitative analysis of the migration rate. (d) Semiquantitative analysis of invasion rate. (e) Protein expression levels of MMP2 and MMP9 were analyzed by western blotting. (f) Semiquantitative analysis of the expression levels presented in part (e), GAPDH was used as the control. ***P < .001 versus shRNA-NC.
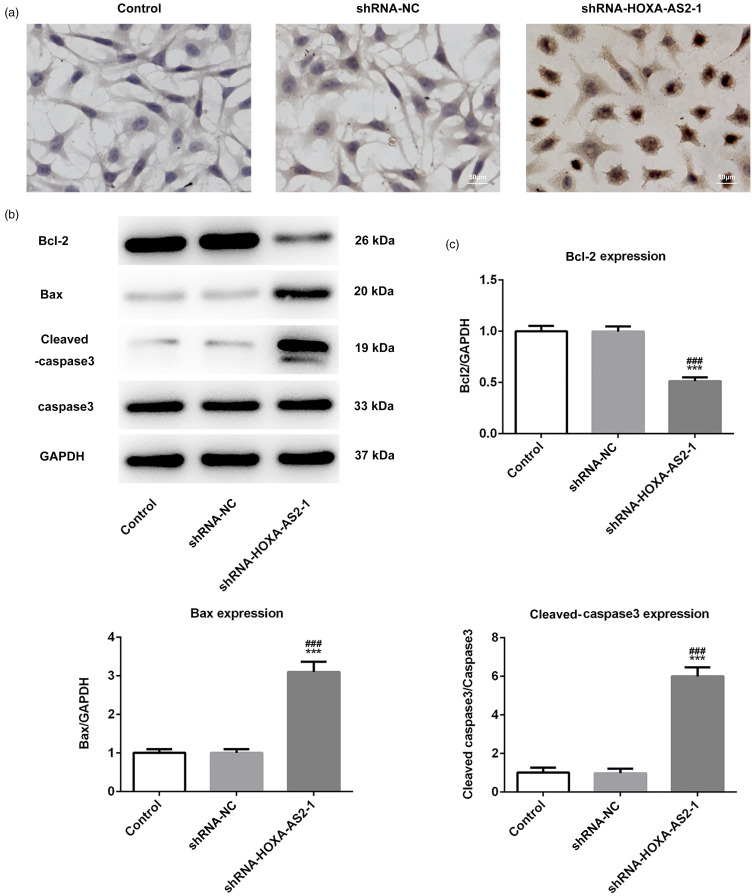
HOXA-AS2 Knockdown Promotes the Apoptosis of Tca-8113 Cells
Cell apoptosis was detected by TUNEL staining assay and western blotting analysis. Positive TUNEL staining was indicated by a brownish-yellow staining of cells, and most of these cells were also discovered to be apoptotic in morphology. Compared with the control group, the number of TUNEL-positive cells was significantly increased in the shRNA-HOXA-AS2 to 1 group, indicating that HOXA-AS2 knockdown significantly promoted the apoptosis of Tca-8113 cells (Figure 4a). Moreover, the pro- and antiapoptotic protein expressions in Tca-8113 cells were measured by western blotting analysis. The results demonstrated that HOXA-AS2 knockdown significantly downregulated the expression of Bcl-2, while the expression levels of Bax and Cleaved-caspase 3 were upregulated (Figure 4b and c). These results suggested that HOXA-AS2 knockdown may promote the apoptosis of Tca-8113 cells.
Figure 4.
Homeobox A cluster antisense RNA2 (HOXA-AS2) knockdown promotes the apoptosis of Tca-8113 cells: (a) Apoptosis was detected by TUNEL staining assay. (b) Protein expression levels of Bcl-2, Bax, Cleave-caspase3, and caspase3 were analyzed by western blotting. (c) Semiquantitative analysis of the expression levels of Bcl-2, Bax, Cleaved-caspase3, and caspase-3 in different groups from part (b), GAPDH was used as the control. ***P < .001 versus shRNA-NC.
HOXA-AS2 Directly Target EZH2
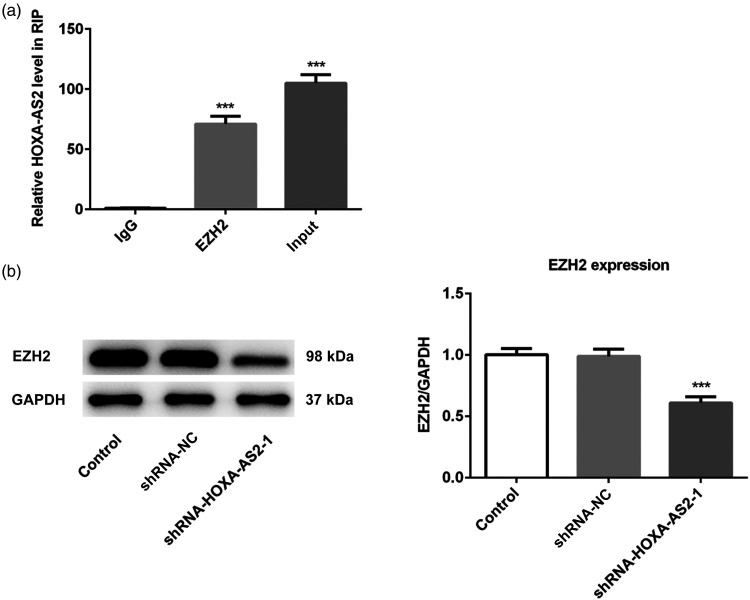
A previous study reported that HOXA-AS2 promoted colorectal cancer cells proliferation by targeting EZH2.14 Therefore, RIP assays were performed to validate the interaction between HOXA-AS2 and EZH2 in OSCC. The results of the RIP assay identified an association between HOXA-AS2 and EZH2 in Tca-8113 cells (Figure 5a). Moreover, compared with the control group, HOXA-AS2 knockdown significantly downregulated EZH2 expression (Figure 5b). These results indicated that HOXA-AS2 may target and positively regulate EZH2 expression.
Figure 5.
HOXA-AS2 directly target EZH2: (a) RIP assay was performed to determine the binding relationship between HOXA-AS2 and EZH2. (b) Relative protein expression levels of EZH2 in different groups were determined by western blotting, GAPDH was used as the control. ***P < .001 versus shRNA-NC.
Abbreviations: HOXA-AS2, homeobox A cluster antisense RNA2; EZH2: enhancer of zeste 2 polycomb repressive complex 2 subunit; RIP: RNA immunoprecipitation.
HOXA-AS2 Promoted the Proliferation, Migration, and Invasion of Tca-8113 Cells by Upregulating EZH2 Expression
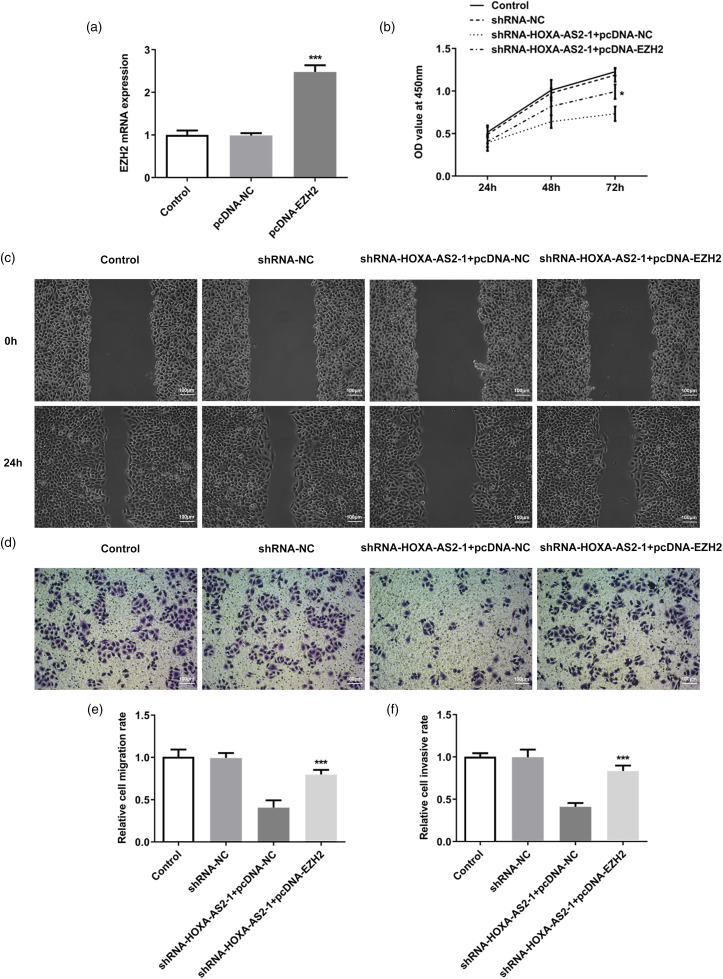
Finally, CCK-8, wound healing, and invasion assays were performed to determine the role of EZH2 in the effect of HOXA-AS2 knockdown. Tca-8113 cells were transfected with pcDNA-NC and pcDNA-EZH2, and RT-qPCR analysis revealed that EZH2 overexpression significantly upregulated EZH2 expression compared with the control group (Figure 6a). The results of the CCK-8 assay demonstrated that, compared with the shRNA-HOXA-AS2 to 1 + pcDNA-NC group, EZH2 overexpression alleviated the inhibitory effect of HOXA-AS2 knockdown on cell viability (Figure 6b). The results of the migration and invasion assays found that, compared with the shRNA-HOXA-AS2 to 1 + pcDNA-NC group, EZH2 overexpression could significantly reversed the inhibitory effect of HOXA-AS2 knockdown on cell migration and invasion (Figure 6c to f). In general, these results suggested that HOXA-AS2 may promote the proliferation, migration, and invasion of Tca-8113 cells by upregulating EZH2 expression.
Figure 6.
HOXA-AS2 promotes the proliferation, migration, and invasion of Tca-8113 cells by upregulating EZH2 expression: (a) Transfection efficiency of EZH2 expression was detected by qRT-PCR. (b) Cell viability in each group following transfection was detected using CCK-8 assay. (c) Cell migration was determined by wound healing assay. (d) Cell invasion was determined by Transwell assay. (e) Semiquantitative analysis of the migration rate from part (c). (f) Semiquantitative analysis of the invasion rate from part (d). *P < .05 and ***P < .001 versus shRNA-HOXA-AS2 to 1 + pcDNA-NC.
Abbreviations: HOXA-AS2, homeobox A cluster antisense RNA2; EZH2: enhancer of zeste 2 polycomb repressive complex 2 subunit.
Discussion
The results of the present study revealed that HOXA-AS2 expression levels were upregulated in OSCC cells compared with human oral epithelial cells. HOXA-AS2 knockdown effectively inhibited the proliferation, migration, and invasion of Tca-8113 cells, and induced apoptosis and cell cycle arrest in Tca-8113 cells. In addition, HOXA-AS2 was found to directly bind to the oncogenic factor EZH2, thereby suggesting the existence of a positive association between the expression levels of HOXA-AS2 and EZH2. EZH2 overexpression alleviated the inhibitory effects of HOXA-AS2 knockdown on the proliferation, migration, and invasion of Tca-8113 cells. The findings of the present study may provide a greater understanding of the underlying mechanism of HOXA-AS2 in OSCC metastasis and provide a novel potential therapeutic target for the treatment of OSCC.
OSCC is a multiple malignant tumor, which is prone to develop lymph node metastasis and leads a poor prognosis in patients. At present, surgical resection or lymph node dissection is the current treatments used for OSCC clinically, but the postoperative survival and quality of life of patients remains unsatisfactory.17 With the development of genomics, a set of lncRNAs with more than 200 nt is abnormally expressed in a variety of tumor cells, where they have been found to play important roles in regulating the biological behavior of tumor cells including proliferation, invasion, and metastasis.18,19 Therefore, the in-depth investigations of lncRNAs may help to provide novel treatment strategies for OSCC. As a subset of lncRNAs, HOXA-AS2 fulfills a carcinogenic role in many kinds of cancers.8 Notably, HOXA-AS2 has been reported to promote the metastasis in numerous tumor cells. For example, in nonsmall cell lung cancer (NSCLC), HOXA-AS2 knockdown significantly reduced the migration and invasion of NSCLC cells by targeting miR-520 to 3p.20 In prostate cancer, HOXA-AS2 knockdown inhibited tumor cell migration, invasion, and EMT progression by downregulating PBX3.21 In osteosarcoma, HOXA-AS2 promoted the EMT process in osteosarcoma cells by sponging miR-520 to 3p.22 In the present study, the expression levels of HOXA-AS2 were found to be upregulated in OSCC cells, which was consistent with the results of previous studies. Results of the in vitro experiments demonstrated that HOXA-AS2 knockdown significantly suppressed the cell proliferation, migration, and invasion, and induced cell cycle arrest and apoptosis. These results indicated that the upregulation of HOXA-AS2 may be associated with the malignant metastasis of OSCC, thereby demonstrating strong potential as a novel biomarker of OSCC progression.
To further determine the potential mechanism of HOXA-AS2 in OSCC, the present study used RIP assays; the results revealed that HOXA-AS2 could directly bind to EZH2, EZH2 is an oncogene that has been found to regulate the silencing of downstream genes through the trimethylation of histone 3 lysine 27 (H3K27me3). As an oncogenic growth factor, it was also demonstrated to play an important role in the regulation of tumor cell proliferation, differentiation, migration, and tumorigenesis.23 A previous study reported that the upregulated expression levels of EZH2 were associated with the poor prognosis of OSCC.13 Wu et al24 found that lncRNA HOX transcript antisense RNA promoted the invasion and metastasis of OSCC cells by targeting EZH2. Hong et al25 reported that lncRNA H1 sponged miR-138 to enhance the proliferative and invasive abilities of OSCC cells by upregulating EZH2. Thus, the findings of the aforementioned studies suggested that EZH2 may play a key role in the OSCC progression. It is also worthy to note that 1 study showed that HOXA-AS2 enhanced colorectal cancer cells proliferation by binding with EZH2.14 Consistent with the previous findings, the expression levels of EZH2 were upregulated in OSCC cells in the present study. In fact, HOXA-AS2 could directly target EZH2 and regulate the expression of EZH2 in OSCC cells. In addition, EZH2 overexpression reversed the effect of HOXA-AS2 knockdown on the proliferation, migration, and invasion of OSCC cells.
In summary, the results of the present study indicated that HOXA-AS2 may serve a role in OSCC progression as an oncogene. The expression levels of HOXA-AS2 were found to be upregulated in OSCC cells and HOXA-AS2 knockdown significantly decreased the proliferation, migration, and invasion of OSCC cells, and promoted cell cycle arrest and apoptosis. Notably, EZH2 overexpression effectively alleviated these effects. To the best of our knowledge, the present study was the first to report that HOXA-AS2 may affect the proliferation, invasion, and metastasis of OSCC cells through EZH2, which may provide a novel biological target for the treatment of OSCC.
Abbreviations
- EMT
epithelial–mesenchymal transition
- EZH2
enhancer of zeste 2 polycomb repressive complex 2 subunit
- GBC
gallbladder carcinoma
- HIOEC
human immortal oral epithelial cells
- HOXA-AS2
homeobox A cluster antisense RNA2
- lncRNAs
long noncoding RNAs
- NSCLC
nonsmall cell lung cancer
- OD
optical density
- OSCC
Oral squamous cell carcinoma
- RIP
RNA immunoprecipitation
- shRNA
short-hairpin RNA
- TSCC
tongue squamous cell carcinoma
Footnotes
Authors’ Contributions: ZZ and SJ made substantial contributions to the conception and design of the present study. ZZ and Y X designed the research; ZZ, YX, FY, ZZ, YS, and JS performed the research; ZZ and YX analyzed the data and wrote the paper.
Declaration of Conflicting Interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: Shanghua Jing https://orcid.org/0000-0003-4515-2094
References
- 1.Salute TS, Ali M, Mishra P, Kumar V, Singh SK. Prognostic value of cancer stem cell markers in potentially malignant disorders of oral mucosa: a meta-analysis. Cancer Epidemiol Prev Biomarkers. 2019;28(1):144-153. doi: 10.1158/1055-9965.EPI-18-0672. [DOI] [PubMed] [Google Scholar]
- 2.Ren ZH, Xu JL, Li B, Fan TF, Ji T, Zhang CP. Elective versus therapeutic neck dissection in node-negative oral cancer: evidence from five randomized controlled trials. Oral Oncol .2015;51(11):976-981. doi: 10.1016/j.oraloncology.2015.08.009. [DOI] [PubMed] [Google Scholar]
- 3.Ouyang SB, Wang J, Zhao SY, Zhang XH, Liao L. CircRNA_0109291 regulates cell growth and migration in oral squamous cell carcinoma and its clinical significance. Iran J Basic Med Sci .2018;21(11):1186-1191. 10.22038/ijbms.2018.30347.7313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huarte M. The emerging role of lncRNAs in cancer. Nat Med .2015;21(11):1253-1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 5.Li Y, Wang X. Role of long noncoding RNAs in malignant disease. Mol Med Rep .2016;13(2):1463-1469. doi: 10.3892/mmr.2015.4711. [DOI] [PubMed] [Google Scholar]
- 6.Luo XJ, Liu J, Ju HQ, Xu RH. IDDF2019-ABS-0307 Long non-coding RNA TMPO-AS1 regulates oesophageal squamous cell carcinoma metastases through activating GLI1 by maintaining LAP2a expression. Int Dig Dis Forum .2019;68(Suppl 1):36. doi: 10.1136/gutjnl-2019-IDDFAbstracts.69. [DOI] [Google Scholar]
- 7.Gao W, Chan JY, Wong TS. Long non-coding RNA deregulation in tongue squamous cell carcinoma. BioMed Res Int .2014;2014:1-10. doi: 10.1155/2014/405860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang J, Su Z, Lu S, et al. LncRNA HOXA-AS2 and its molecular mechanisms in human cancer. Clin Chim Acta .2018;485:229-233. doi: 10.1016/j.cca.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y, Xu J, Zhang S, et al. HOXA-AS2 promotes proliferation and induces epithelial-mesenchymal transition via the miR-520c-3p/GPC3 axis in hepatocellular carcinoma. Cell Physiol Biochem .2018;50(6):2124-2138. doi: 10.1159/000495056. [DOI] [PubMed] [Google Scholar]
- 10.Zhang P, Cao P, Zhu X, et al. Upregulation of long non-coding RNA HOXA-AS2 promotes proliferation and induces epithelial-mesenchymal transition in gallbladder carcinoma. Oncotarget .2017;8(20):33137-33143. doi: 10.18632/oncotarget.16561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang F, Wu D, Chen J, et al. Long non-coding RNA HOXA-AS2 promotes the migration, invasion and stemness of bladder cancer via regulating miR-125b/Smad2 axis. Exp Cell Res .2019;375(1):1-10. doi: 10.1016/j.yexcr.2018.11.005. [DOI] [PubMed] [Google Scholar]
- 12.Wang S, You H, Yu S. Long non-coding RNA HOXA-AS2 promotes the expression levels of hypoxia-inducible factor-1α and programmed death-ligand 1, and regulates nasopharyngeal carcinoma progression via miR-519. Oncol Lett .2020;20(5):245. doi: 10.3892/ol.2020.12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kidani K, Osaki M, Tamura T, et al. High expression of EZH2 is associated with tumor proliferation and prognosis in human oral squamous cell carcinomas. Oral Oncol .2009;45(1):39-46. doi: 10.1016/j.oraloncology.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 14.Ding J, Xie M, Lian Y, et al. Long noncoding RNA HOXA-AS2 represses P21 and KLF2 expression transcription by binding with EZH2, LSD1 in colorectal cancer. Oncogenesis. 2017;6(1):e288. doi: 10.1038/oncsis.2016.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402-408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Dofara SG, Chang SL, Diorio C. Gene polymorphisms and circulating levels of MMP-2 and MMP-9: a review of their role in breast cancer risk. Anticancer Res. 2020;40(7):3619-3631. doi: 10.21873/anticanres.14351. [DOI] [PubMed] [Google Scholar]
- 17.Bijina BR, Ahmed J, Shenoy N, Ongole R, Shenoy S, Baliga S. Detection of human papilloma virus in potentially malignant and malignant lesions of the oral cavity and a study of associated risk factors. South Asian J Cancer. 2016;5(4):179-181. 10.4103/2278-330X.195337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi X, Sun M, Wu Y, et al. Post-transcriptional regulation of long noncoding RNAs in cancer. Tumor Biol. 2015;36(2):503-513. doi: 10.1007/s13277-015-3106-y. [DOI] [PubMed] [Google Scholar]
- 19.Han X, Wang J, Wang J, et al. ScaPD: a database for human scaffold proteins. BMC Bioinformatics. 2017;18(Suppl 11):87-91. doi: 10.1186/s12859-017-1806-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y, Lin X, Zhou S, Zhang P, Shao G, Yang Z. Long noncoding RNA HOXA-AS2 promotes non-small cell lung cancer progression by regulating miR-520a-3p. Biosci Rep .2019;39(5):1-28. doi: 10.1042/BSR20190283. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Xiao S, Song B. LncRNA HOXA-AS2 promotes the progression of prostate cancer via targeting miR-509-3p/PBX3 axis. Biosci Rep .2020;40(8):1-10. doi: 10.1042/BSR20193287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Zhang R, Cheng G, Xu R, Han X. Long non-coding RNA HOXA-AS2 promotes migration and invasion by acting as a ceRNA of miR-520c-3p in osteosarcoma cells. Cell Cycle. 2018;17(13):1637-1648. doi: 10.1080/15384101.2018.1489174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu M, Chen X, Lin K, et al. lncRNA SNHG6 regulates EZH2 expression by sponging miR-26a/b and miR-214 in colorectal cancer. J Hematol Oncol. 2019;12(1):3. doi: 10.1186/s13045-018-0690-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu Y, Zhang L, Zhang L, et al. Long non-coding RNA HOTAIR promotes tumor cell invasion and metastasis by recruiting EZH2 and repressing E-cadherin in oral squamous cell carcinoma. Int J Oncol .2015;46(6):2586-2594. doi: 10.3892/ijo.2015.2976. [DOI] [PubMed] [Google Scholar]
- 25.Hong Y, He H, Sui W, Zhang J, Zhang S, Yang D. Long non-coding RNA H1 promotes cell proliferation and invasion by acting as a ceRNA of miR138 and releasing EZH2 in oral squamous cell carcinoma. Int J Oncol .2018;52(3):901-912. doi: 10.3892/ijo.2018.4247. [DOI] [PubMed] [Google Scholar]