Abstract
Lung cancer is listed as the most common reason for cancer-related death all over the world despite diagnostic improvements and the development of chemotherapy and targeted therapies. MicroRNAs control both physiological and pathological processes including development and cancer. A microRNA-9 to 1 (miR-9 to 1) overexpression model in lung cancer cell lines was established and miR-9 to 1 was found to significantly suppress the proliferation rate in lung cancer cell lines, colony formation in vitro, and tumorigenicity in nude mice of A549 cells. Ubiquitin-like containing PHD and RING finger domains 1 (UHRF1) was then identified to direct target of miR-9 to 1. The inhibition of UHRF1 by miR-9 to 1 causes G1 arrest and p15, p16, and p21 were re-expressed in miR-9 to 1 group in mRNA level and protein level. Silence of UHRF1 expression in A549 cells resulted in the similar re-expression of p15, p16, p21 which is similar with miR-9 to 1 infection. Therefore, we concluded that UHRF1 is a new target for miR-9 to 1 to suppress cell proliferation by re-expression of tumor suppressors p15, p16, and p21 mediated by UHRF1.
Keywords: miR-9 to 1, UHRF1, lung cancer, proliferation, apoptosis
Introduction
In higher eukaryotes, MicroRNAs (miRNAs) can be used as a posttranscriptional regulator of gene expression.1-3 Recent estimates have found that 20%–30% of all genes are targets of miRNA. Therefore, miRNAs are considered to play a significant role in organisms.4-6 MiRNAs control both physiological and pathological processes such as development and cancer.7-10 MicroRNA-9 to 1 (miR-9-1) is initially identified as an important regulator which plays a pivotal role in nervous system development.11-13 In insects and humans, the mature miR-9 to 1 sequence is found to be widespread.14 In addition, except for Xenopus laevis, the mature sequence of miR-9 to 1 in all vertebrates was completely consistent, suggesting that the role of miR-9 to 1 is widely conservative.15 MiR-9 to 1 is expressed almost in the brain and is a mediator in mammalian neurogenesis.16 The function of miR-9 to 1 in nerve system development and differentiation has been extensively identified in several model organisms such as mice and zebrafish.17
An increasing number of literatures suggested that miR-9 to 1 were also involved in tumorigenesis.18-21 In different types of cancer cells, miR-9 to 1 can have different effects on apoptosis and proliferation of cancer cells through targeting different mRNA targets.22-24 miR-9 to 1 is found to over-express in human Hodgkin's lymphoma cells,25 primary brain tumors,26 cervical cancer,27 and colorectal cancer.28 However, miR-9 to 1 is identified to down-regulate in the ovarian tumor,29 gastric adenocarcinomas,30 breast cancer,31 oral carcinomas.32 Over-expression of miR-9 to 1 suppresses the proliferation in ovarian cancer,33 human medulloblastoma cells.34-36 Therefore, the abnormal expression of miR-9 to 1 in many types of cancer makes it possible to become a disease diagnosis and prognosis marker with important clinical significance.37-40 However, little is known about the pathological function and mechanisms of miR-9 to 1 in lung cancer.
Ubiquitin-like, containing PHD and RING finger domains 1 (UHRF1) is important to maintain the methylation status of DNA.41-43 UHRF1 is a ubiquitin E3 ligase and contains at least 4 domains including a plant homeodomain domain, a ubiquitin-like domain, a RING domain, and a SET and RING associated (SRA) domain.44-47 It was confirmed that UHRF1 could co-locate with DNA methyltransferase protein DNMT1 in the S phase.48 The SRA domain of UHRF1 strongly binds to the physiological substrate for DNMT1.49 UHRF1 recruit DNMT1 to hemimethylated DNA and facilitate DNA methylation.50-52 Further investigations have found that deubiquitylase USP7 and PCNA were also found to form a complex with DNMT1 and influence the E3-ligase activity of UHRF1 and DNA methylation.44,53,54 In normal cells, the expression of hUHRF1 was periodic and highly expressed in the late G1 and G2/M phases.55-57 On the contrary, in cancer cells, the expression of hUHRF1 is always at a high level.58,59
UHRF1 function as an oncogene and were found over-expressed in several cancer types and inversely correlated with survival and prognosis including bladder,60 kidneys,61 breasts,62 cervical,63 ovarian,64 lungs,65 and colorectal cancer.38 The detailed mechanisms may be related to the fact that UHRF1 constitutes a complex that represses tumor suppressor genes expression including MLH1, BRCA1, RB1, and p16INK4A.66-70 UHRF1 mediated tumor suppressor genes such as RASSF1 and CDKN2A inactivation in nonsmall cell lung cancer (NSCLC).71 Knock-down of UHRF1in A549 lung cancer cells results in lower methylation levels of tumor suppressors including CDH13, CYGB, and RASSF1promoters.72 Moreover, knock-down of UHRF was found to inhibit cell proliferation and migration properties.73-75 Down-regulation of ICBP90 and DNMT1 significantly enhanced, p16INK4A and RB1 were observed.76 RNA interference of UHRF1 decreases proliferation and migration, with increased p16INK4A expression in CRC.77 UHRF1 were also regulated by tumor suppressors including TP53 and TP73.78 However, the miRNAs which regulated UHRF1 were little known.
In this study, we constructed miR-9 to 1 over-expressed model and further investigated the biological function and potential target and molecular mechanisms of miR-9 to 1 in NSCLC cell lines.
Materials and Methods
Ethical Approval and Consent to Participate
This study was approved by the Ethics Committee of the School of Medicine, Tongji University (IEC-PAP-15-18). The written informed consents were provided by all patients.
Clinical Tissue Samples
NSCLC and matched noncarcinoma tissue specimens from patients who underwent surgical treatment were obtained between March 2010 to November 2015. Two independent, experienced pathologists confirmed the diagnosis of NSCLC pathologically.
The expression data of miRNAs of 150 paired NSCLC tissues and normal tissues (47 FFPE samples and 103 fresh samples) were procured from GEO datasets GSE36681.
Plasmid Construction and Transfection
The lentivirus vector pLL3.7-miR-9 to 1 was constructed. The sequence was amplified with the primers using primerSTAR®HS (TaKaRa Code DR044A): miR-9 to 1-F-Hpal, 5′-CCCCGTTAACTTTCGGTCTCTGTCGTGT-3′; miR-9 to 1-R-Xhol, 5′-AAAACTCGAGTTGCTCCCCCTCAACTC-3′ by polymerase chain reaction (PCR) and were cloned into the Xho l/Hpa I sites of pLL3.7 vector.
psi-CHECKTM-2-UHRF1 to 3′UTR: UHRF1 3′UTR fragment (NM_001048201.1) containing the full-length 3′UTR of UHRF1 was amplified performed with the same polymerase mentioned above by PCR using the primers 5′-AAAACTCGAGTCTCCAAGCACTTCTCG-3′ and 5′-CCCCGCGGCCGCCTGAGAATCTAAAATCTGGAC-3′ and cloned into Not I/Xho I sites of psi-CHECKTM-2.
psi-CHECKTM-2-UHRF1 to 3′UTR-mut was constructed by using 4 mutated nucleotides of 3′UTR of UHRF1: forward-mut, 5′-TTCTAATTTTAGGTTAGTTTGCAGC-3′and reverse-mut, 5′-AGGCTGCAAACTAACCTAAAATTAG-3′.
Cell Lines
HEK-293T, NCI-H1299, and A549 cells were cultured in DMEM supplemented with 10% FBS (Gibco BRL). A549 cell and NCI-H1299 cells were infected with pLL3.7 (EV) or pLL3.7-miR-9 to 1 for 24 h, and then the medium was refreshed.
Dual-Luciferase Reporter Assay
Dual-luciferase reporter assay was executed in HEK-293T cells. The miR-9 to 1 inhibitor and the control were used to test the inhibitory effect of miR-9 to 1 on UHRF1 3′UTR.
Colony Formation Experiment and Tumor Inoculation Assay
For colony formation assay, lung cancer cells were cultured in the 6 cm diameter culture dish and fixed to stain with 0.5% crystal violet. The colonies numbers in 8 random view fields were counted under an optical microscope.
Tumor inoculation assay was performed in BALB/c nude mice at the age of 6 to 8 weeks. The tumor growth was investigated every 3 days for 4 weeks and the volume was recorded by measuring the length (L) and width (W) with calipers and was calculated with the formula: volume = 1/2(L × W2).25
Total RNA Extraction and Quantitative PCR (qPCR)
Total RNA was extracted from lung cancer cells using Trizol reagent and qPCR was performed with RT Kit in compliance with the recommendations. Specific stem-loop RT primers were used for qPCR as the following: 5′-GCCCGTCTTTGGTTATCTAGC-3′ and 5′-GTGCAGGGTCCGAGGT-3′ for mature miR-9 to 1, 5′-CTCGCTTCGGCAGCACA-3′ and 5′-AACGCTTCACGAATTTGCGT-3′for RNU6; 5′-TGTCAAGGGTGGCAAGAAT-3′ and 5′-GCCAGGCTCATCATCGTC-3′ for UHRF1. RNU6 was used as the internal control.
Western Blot (WB) and Immunohistochemistry
Total protein was extracted using a lysis buffer. The concentration was examined using standard procedures for WB. The blots were incubated with antibodies against UHRF1 (PA5-27969, 1:1000), p14 (MA5-14260, 1:1000), p15 (MA1-12294, 1:1000), p16 (MA5-14260, 1:500) (Invitrogen), and GAPDH (ab9485, Abcam; 1:2000).
Paraffin-embedded tissues were acquired from the sacrificed nude mice and fixed in PBS with 4% formaldehyde for 24 h. Samples were dehydrated and then embedded in paraffin before being sectioned at 8 μM and used for H&E staining.
Proliferation and Interference Experiment
Cell counting kit-8 reagent was used for cellular proliferation assay. The data of optical density (OD) value at 450 nm were recorded through a microplate reader.
Statistical Analysis
Levels of gene expression were recorded as mean ± standard deviation (SD). The independent t-test and the χ2 test were used to calculate the difference between 2 groups or the difference among different groups, respectively. Univariate survival analysis of overall survival (OS) or progression-free survival (PFS) of NSCLC patients was analyzed using the Kaplan-Meier method assay. All statistical analyses were carried out using the SPSS 21.0 software (SPSS Inc.).
Results
The Level of miR-9 to 1 Expression in miRNA Microarray Profiling Data
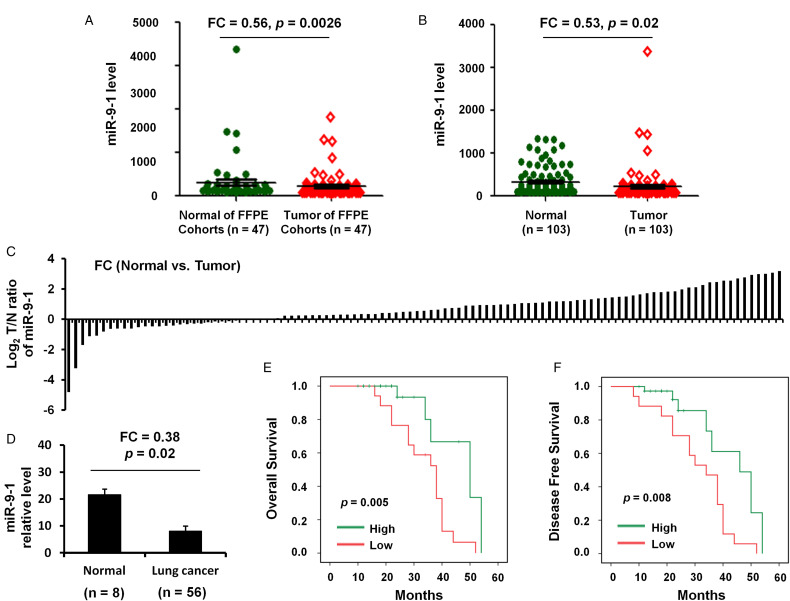
The present study first performed an in silico analysis using GEO database data. Analysis using GSE36681 data showed that the miR-9 to 1 level was both significantly reduced in FFPE (n = 47; FC = 0.56, P = .0026; Figure 1A) and fresh (n = 103; FC = 0.53, P = .02; Figure 1B and C) samples of lung cancer compared with that in normal controls.
Figure 1.
The expression and clinical significance of miR-9 to 1 in NSCLC. (A) miR-9 to 1 expression level in 47 paired FFPC tumor samples and normal lung tissues from NSCLC patients. Raw data was from GSE36681. (B) qRT-PCR was used to quantify miR-9 to 1 expression level in 103 paired frozen tumor samples and normal lung tissues from NSCLC patients. (C) Fold change of miR-9 to 1 expression level in 56 paired frozen tumor samples and normal lung tissues from NSCLC patients. (D) qRT-PCR was used to quantify miR-9 to 1 expression level in 56 frozen tumor samples and 8 normal lung tissues from NSCLC patients. (E, F) Kaplan–Meier survival analysis was used to evaluate the miR-9 to 1 prognostic value for OS (E) and PFS (F) of 56 NSCLC patients.
Abbreviations: NSCLC, nonsmall cell lung cancer; miR-9 to 1, microRNA-9 to 1; FFPC, formalin-fixed paraffin-embedded; qRT-PCR, quantitative real-time polymerase chain reaction; OS, overall survival; PFS, progression-free survival.
Validation of the Reduced miR-9 to 1 Level in NSCLC Tissue Samples
The levels of miR-9 to 1 expression were also found to be lower in all NSCLC tumor biopsies (n = 56) compared with those in normal lung tissues (n = 8) and the differences were showed to be statistically significant (FC = 0.38, P = .02; Figure 1D).
The Association Between miR-9 to 1 Expression and Clinical Characteristics
Next, miR-9 to 1 expression was further analyzed in NSCLC samples based on clinical characteristics (including age, sex, lymph node metastasis, TNM stage, tumor differentiation, smoking history, vascular invasion, invasion of lung membrane, and tumor diameter. Univariate analysis showed that the expression of miR-9-1 was significantly associated with tumor diameter [P = .009], lymph node metastasis [P = .006], and TNM stage [P = .012] in NSCLC patients (Table 1). However, there is no significant association between miR-9-1 expression and age, sex, invasion of the lung membrane, tumor differentiation, vascular invasion, or smoking history (P > .05; Table 1).
Table 1.
The Association Between miR-9 to 1 Expression and Clinical Characteristics.
| Factor | Variable | N | miR-9 to 1 expression (mean ± SD) | P-value |
|---|---|---|---|---|
| Age | ||||
| ≥ 60 | 27 | 0.613 ± 0.029 | .254 | |
| < 60 | 29 | 0.788 ± 0.026 | ||
| Gender | ||||
| Male | 26 | 0.652 ± 0.035 | .238 | |
| Female | 30 | 0.759 ± 0.068 | ||
| Smoking history | ||||
| Ever | 18 | 0.812 ± 0.085 | .065 | |
| Never | 38 | 0.698 ± 0.025 | ||
| Lymph node metastasis | ||||
| Positive | 25 | 0.538 ± 0.021 | .006* | |
| Negative | 31 | 0.833 ± 0.045 | ||
| Tumor differentiation | ||||
| Poorly | 16 | 0.621 ± 0.033 | .079 | |
| Moderately | 11 | 0.756 ± 0.036 | ||
| Well | 29 | 0.812 ± 0.024 | ||
| TNM stage | ||||
| III-IV | 18 | 0.561 ± 0.067 | .012* | |
| I-II | 38 | 0.804 ± 0.055 | ||
| Invasion of lung membrane | ||||
| Positive | 29 | 0.651 ± 0.014 | .068 | |
| Negative | 22 | 0.762 ± 0.036 | ||
| Unknown | 5 | 0.801 ± 0.082 | ||
| Vascular invasion | ||||
| Positive | 25 | 0.621 ± 0.035 | .068 | |
| Negative | 29 | 0.712 ± 0.051 | ||
| Unknown | 2 | 0.798 ± 0.082 | ||
| Diameter | ||||
| ≥ 5 cm | 26 | 0.522 ± 0.031 | .009* | |
| < 5 cm | 30 | 0.813 ± 0.032 |
*P < .05.
Association of miR-9-1 Levels and Clinicopathological Data with Survival of NSCLC Patients
We next studied the correlation of miR-9-1 levels and clinicopathological data with the survival of patients with NSCLC. The results revealed that a reduced miR-9-1 level was associated with a shorter OS (P = .0005; Figure 1E) and DFS (P = .008; Figure 1F) of patients with NSCLC.
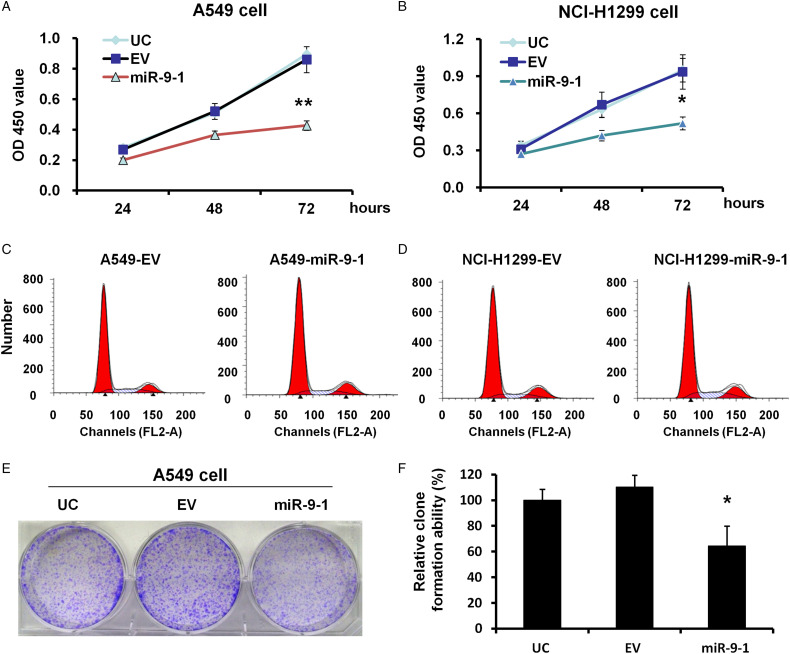
miR-9 to 1 Suppression of Lung Cancer Cell Proliferation In Vitro
To investigate the function of miR-9 to 1 in A549 and NCI-H1299 cells, a CCK-8 assay was performed in stable miR-9 to 1-infected A549 and NCI-H1299 cells. The results showed that the proliferation rate in miR-9 to 1 group was significantly reduced to 49.65% and 55.6% when compared to the EV group in A549 (P < .01) and NCI-H1299 (P < .05) cells, respectively (Figure 2A and B). Moreover, overexpression of miR-9 to 1 significantly increased the cellular apoptosis of A549 and NCI-H1299 cells compared with the control (Figure 2C and D).
Figure 2.
miR-9 to 1 suppressed A549 and NCI-H1299 cell growth in vitro. (A, B) Growth curves of miR-9 to 1 and EV-infected A549 (A) and NCI-H1299 (B) cells were analyzed using a CCK-8 assay. The experiments were repeated 3 times independently. (C, D) cell apoptosis in A549 (C) and NCI-H1299 (D) was analyzed by flow cytometry assay before and after miR-9 to 1 overexpression. (E) Colony formation assay in miR-9 to 1 and EV-infected A549 cells. The experiments were repeated 3 times independently. (F) The average number of colonies in EV and miR-9 to 1 group. ** indicates P < .01.
To further evaluate the effect of miR-9 to 1 on cell proliferation, we conducted colony-forming experiments on the A549 cell line. The results showed that the colony-forming ability of the miR-9 to 1 group was significantly inhibited. The colony number of more than 50 cells produced by a single cell in the miR-9 to 1 group was significantly lower when compared to that in the control group (46.37%; P < .01) (Figure 2E and F).
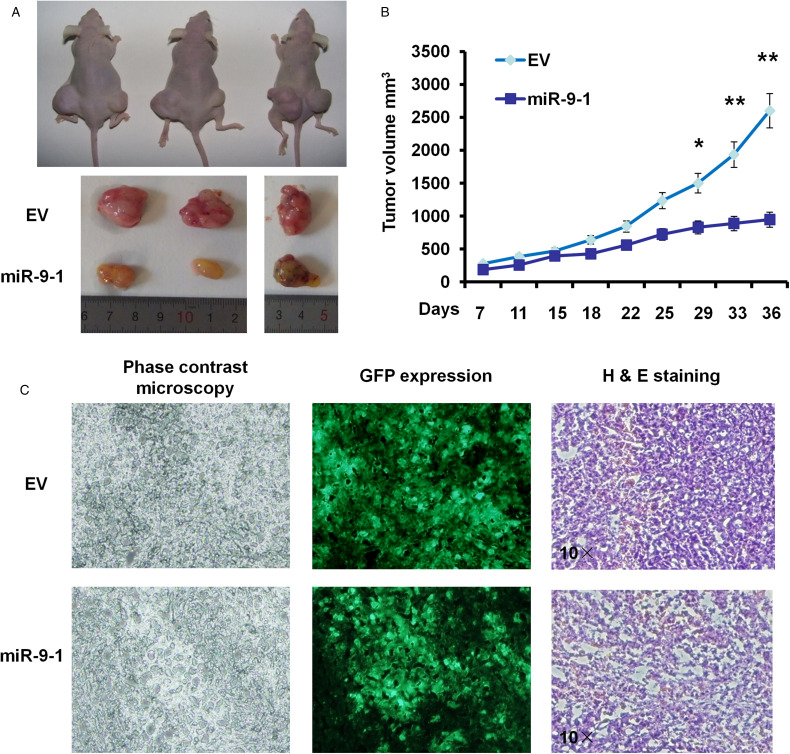
miR-9 to 1 Suppression of Lung Cancer Cell Proliferation In Vivo
In order to further verify the results of in vitro cell experiment, we injected 7 × 106 EV cells or miR-9 to 1 overexpression cells into the skin of the right front leg and the lower part of the left leg, respectively. Tumor mass was observed in all (8 of 8) mice (half male and half female) in the EV group within 7−10 days after inoculation (3 representative mice in Figure 3A and B). All mice were killed at 5 weeks after inoculation, and the tumor weight was weighed. Four weeks after inoculation, the average tumor weight in miR-9 to 1 group was found to be significantly lower than that in the control group (Figure 3A and B). Tumor size was measured every twice a week when a tumor was obvious and our results showed that tumor volumes in miR-9 to 1 group achieved 55.36% and 36.49% of the control (both P < .05) at 4 and 5 weeks after inoculation, respectively (Figure 3B and C), which suggested that miR-9 to 1 suppress cell proliferation of NSCLC cells both in vitro and in vivo.
Figure 3.
miR-9 to 1 suppressed A549 cell growth in vivo. (A) Tumor sizes of 3 representative nude mice.. The tumor volume was calculated every 3 days with calipers when the tumor appeared. * indicates P < .05. (B) Tumor size was measured in nude mice after the inoculation. (C) Hematoxylin and eosin (H&E) examination for tumor tissues in nude mice. Original magnification 10 × . * indicates P < .05 and ** indicates P < .01.
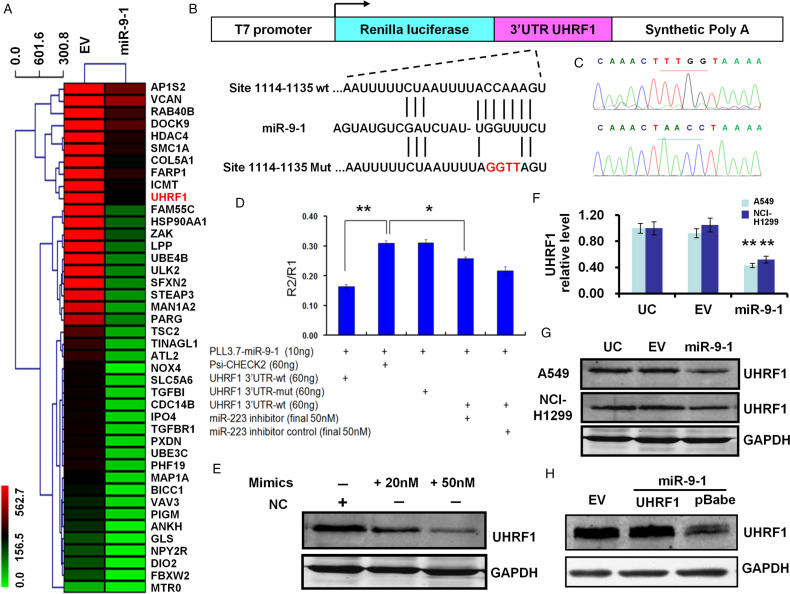
miR-9 to 1 Target UHRF1 in Lung Cancer Cells
To investigate the mechanisms of miR-9 to 1 inhibition effect on proliferation and tumorigenicity of A549 cells, we then searched for potential target genes by using the online prediction algorithm of Targetscan, Targetscan 5.1, Pictar, Mirbase, microiRNA, and RNA22, and found that UHRF1 is found as a putative target of miR-9 to 1.
To establish that miR-9 to 1 targets and regulates the expression of UHRF1, we constructed 3′UTR of UHRF1 downstream of renilla luciferase reporter gene in psi-CHECKTM-2, and then constructed 4 nucleotide mutations in the possible binding region of miR-9 to 1 seed sequence (hereinafter referred to as UHRF1-3′UTR MUT) (Figure 4A and B). Our results indicated that the relative luciferase activity in the reporter that UHRF1 contained wild-type 3′UTR was significantly decreased after miR-9 to 1 overexpression when compared to the EV group (P < .01) (Figure 4C). However, the relative luciferase activity of UHRF1 to 3′UTR-mut was recovered to the same level when compared to the control psi-CHECKTM-2 group (Figure 4C).
Figure 4.
UHRF1 is a direct target of miR-9 to 1. (A) mRNAs microarray analysis to identify key differentially expressed genes after overexpression of miR-9 to 1 in A549 cells. (B) Structure and cloning site of the psi-CHECK-2 vector. (C) Mutations of 4 nucleotides were successfully performed by DNA sequencing. (D) Dual-luciferase assays were performed. Each sample was 6 well repeat. (E) Transient transfection of miR-9 to 1 mimics or its negative control (NC) at indicated concentration in A549 cells and protein were harvested 48 h after transfection. (F) Quantitative PCR results showed the UHRF1 mRNA was reduced at miR-9 to 1stable transfection of A549 and NCI-H1299 cells. (G) UHRF1 protein was decreased in miR-9 to 1 group at A549 and NCI-H1299 cells. (H) Rescue experiments indicate that UHRF1 rescued the inhibition state in the A549 cell line by re-introducing of 3′UTR free UHRF1 coding sequence carried by the pBabe vector. The UHRF1 was detected by its antibody. GAPDH served as a loading control.
Abbreviations: miR-9 to 1, microRNA-9 to 1; PCR, polymerase chain reaction; UHRF1, ubiquitin-like containing PHD and RING finger domains 1; 3′UTR, 3′untranslated region.
UHRF1 expression decrease after miR-9 to 1 overexpression was reversed when adding miR-9 to 1 inhibitor with a final concentration of 50 nM (P < .05) (Figure 4D). The mRNA level of A549 and NCI-H1299 cells were measured by qPCR and the UHRF1 level was reduced to 46.48% and 49.52% at A549 and NCI-H1299 groups, respectively, as compared with the EV control group (both P < .05) (Figure 4E). The protein level of UHRF1 at miR-9 to 1 group was also measured by WB, and the protein reduced to about 1/3 and 1/2 at miR-9 to 1 group as compared with EV group at A549 and NCI-H1299 cell, respectively (Figure 4F). Transfection with UHRF1 coding sequence (CDS) at A549 cell overcome the suppression induced by miR-9 to 1 since the construct contained no 3′UTR. When we used miR-9 to 1 mimics to transfection of A549 cells, the protein of UHRF1 was reduced in a dose-dependent manner (Figure 4G).
MiR-9 to 1 Induces the Expression of Tumor Suppressor Genes Including p15, p16, and p21
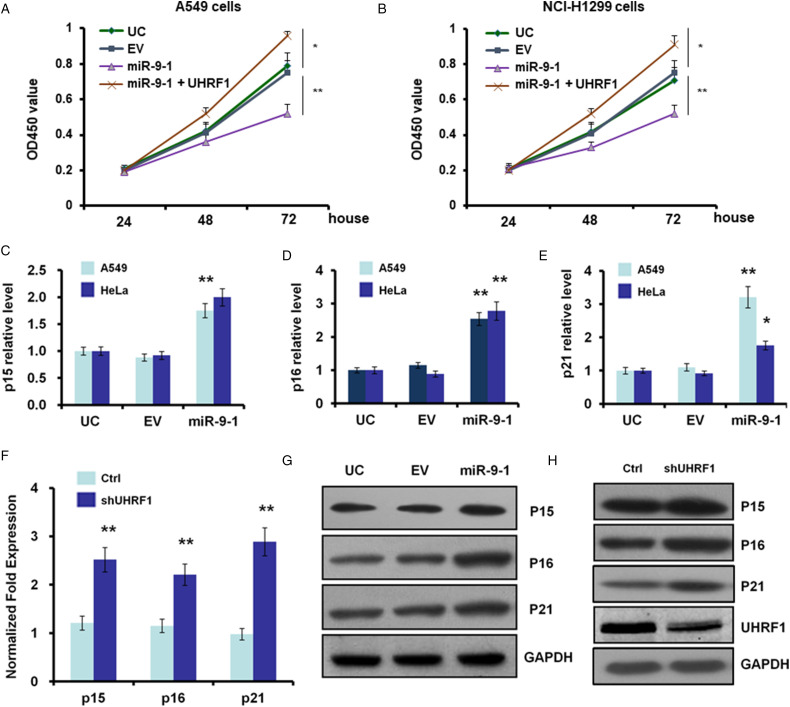
UHRF1 is involved in some tumor suppressor genes inactivation by hypermethylation of its promoter region. And inhibition of URHF1 is also related to the re-expression of tumor suppressors. We next studied whether UHRF1 down-regulation by miR-9 to 1 can re-active the expression of tumor suppressors. Among tumor suppressor genes, we search for some cell cycle inhibitors and found that p15, p16, and p21 are strikingly re-expressed in miR-9 to 1 group in mRNA level (Figure 5A to D) and protein level (Figure 5E). These results are similar to the interference of UHRF1 by specific stem-loop RNA mediated by the pLKO vector (Figure 5F).
Figure 5.
Suppression of UHRF1 by miR-9 to 1 or shRNA causes expression upregulation of p15, p16, p21 in A549 cells. (A, B) Growth curves of miR-9 to 1, UHRF1, and EV-infected A549 (A) and NCI-H1299 (B) cells were conducted by CCK-8 assay. The OD value at 450 nm represented the viable cell numbers. (C-E) Quantitative PCR results showed the mRNA levels of p15 (C), p16 (D), and p21 (E) were induced after miR-9 to 1stable transfection of A549 cells. (F) Quantitative PCR arrays for the mRNA levels of p15 (C), p16 (D), and p21 (E) were induced after UHRF1 RNAi in A549 cells. (G, H) Western blot to quantify protein levels of p15, p16, and p21 after miR-9 to 1 stable transfection (G or UHRF1 RNAi [H] in A549 cells).
Abbreviations: miR-9 to 1, microRNA-9 to 1; PCR, polymerase chain reaction; UHRF1, ubiquitin-like containing PHD and RING finger domains 1.
Expression and Their Correlation of miR-9 to 1 and UHRF1 in NSCLC
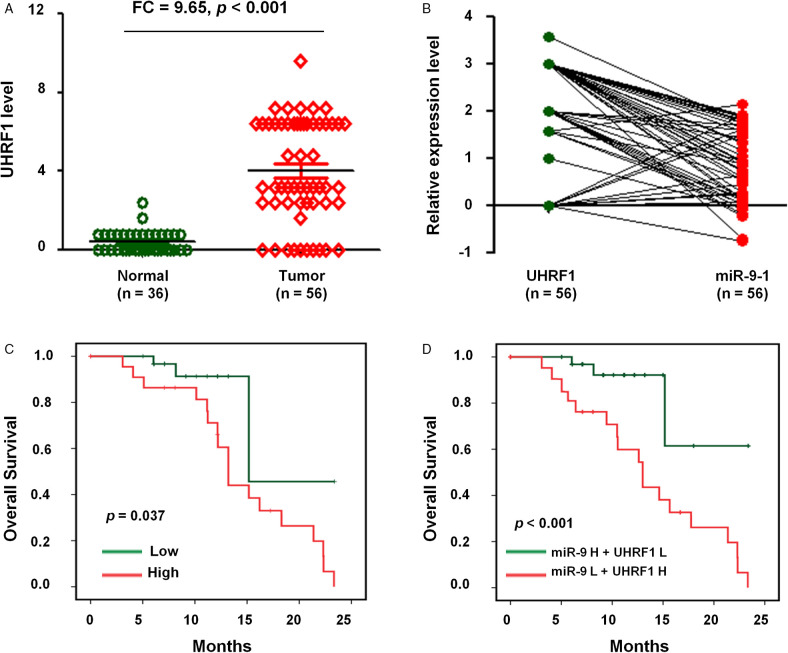
Considering the close association of miR-9 to 1 with UHRF1 in lung cancer cell lines, we next studied the expression and correlation of miR-9 to 1 and UHRF1 in NSCLCs FFPE samples from the human tissue specimen. The results indicated that the expression of UHRF1 was significantly higher in NSCLC samples (n = 56) compared with normal lung tissues (n = 36) (FC = 9.65, P < .001; Figure 6A). Moreover, we observed a negative correlation between miR-9 to 1 and UHRF1 expression in NSCLC samples (n = 56) (Figure 6B).
Figure 6.
The expression and clinical significance of miR-9 to 1 and UHRF1 in NSCLC. (A) IHC assay to measure UHRF1 level in 56 NSCLC tissues and 36 adjacently normal lung tissues. (B) The correlation between UHRF1 and miR-9 to 1 level in 56 NSCLC tissues. (C) Kaplan–Meier survival analysis was used to evaluate the prognostic value of UHRF1 expression for OS of 56 NSCLC tissues. (D) Kaplan-Meier survival analysis to evaluate the prognostic value of UHRF1 together with miR-9 to 1 expression in NSCLC for OS.
Abbreviations: NSCLC, nonsmall cell lung cancer; miR-9 to 1, microRNA-9 to 1; OS, overall survival; UHRF1, ubiquitin-like containing PHD and RING finger domains 1.
Clinical Significance of UHRF1 and miR-9 to 1 in NSCLC
We then evaluated the prognostic value of UHRF1 and miR-9 to 1 expression in patients with NSCLC. The OS of NSCLC patients with a low expression of UHRF1 was prolonged when compared to NSCLC patients with high expression of UHRF1 (P = .037; Figure 6C).
We next explored the clinical significance of UHRF1 expression together with miR-9 to 1. Our results indicated that NSCLC patients with UHRF1high and miR-9 to 1low levels show significantly decreased OS (P < .001) (Figure 6D), suggesting that UHRF1 and miR-9 to 1 might have potential prognostic value and could be useful as tumor biomarkers for the diagnosis of NSCLC patients.
Discussion
MiRNAs are noncoding RNA molecules, which regulate gene expression through translational repression or deregulating of messenger RNAs.79-82 Accumulating evidence suggests miRNAs can play various roles in lung cancers.83-85 In the present study, we firstly investigated the biological function miR-9 to 1 in lung cancer cell lines and identified that miR-9 to 1 suppresses lung cancer cell growth, colony formation in vitro, and tumorigenesis in vivo, which suggests that miR-9 to 1 function as a tumor suppressor in the lung cancer cell.
MiR-9 to 1 has 3 loci in the genome, including miR-9 to 1, miR-9 to 1-2, and miR-9 to 1-3.86 MiR-9 to 1 is the well-studied one. In this study, we search for the miR-9 to 1 direct target by informatics through a public prediction website and identified that UHRF1 is the putative target of miR-9 to 1. UHRF1 to 3′UTR dual-luciferase experiments have shown a significant decrease in renilla luciferase activity when miR-9 to 1 were co-transfection. Moreover, mutation of 4 nucleotides of the 3′UTR of UHRF1 with the seed sequence sites abolished the response to miR-9 to 1. Knockdown of miR-9 to 1 by synthetic sponge inhibitor-induced inhibitory function and the renilla luciferase activities rescued to the level similar to control group. Besides, both mRNA level and protein level of UHRF1 were down-regulated in miR-9 to 1 group when compared to the EV group. However, transfection with UHRF1 coding sequence without 3′UTR rescued the expression of UHRF1. Furthermore, transfection of miR-9 to 1 mimics inhibited the UHRF1 protein level in a dose-dependent manner.
Cell cycle activity plays a critical role in many physiological activities of organisms, including organogenesis, epithelial-mesenchymal transition, autophagy, proliferation, differentiation, differentiation, differentiation, maturation, and survival.87-93 Proteins involved in cell cycle regulation are strictly regulated in expression and activity to ensure dynamic balance in the process of embryogenesis and organ development.93-96 UHRF1 was reported to promote G1/S and G2/M transition.80 The decrease of UHRF1 is a critical mechanism at G1/S transition.81 In this study, we found that miR-9 to 1 directly targets UHRF1 by repression of UHRF1 expression cause G1 phase arrest. This result is concordant with the literatures report.
DNA methylation is an important form of epigenetic modification. Studies have confirmed that abnormal DNA methylation can affect a variety of human malignant tumors by regulating the expression of cancer-related genes.97-100 UHRF1 serves as an oncogene in some types of tumor and possesses an essential role in DNA methylation maintenance. We therefore further investigate the possible effect of UHRF1 in cell cycle regulation. We then further screen the possible tumor suppressors which may be methylated and suppressed by UHRF1 by quantitative PCR and found that p15, p16, and 21 were re-expression after miR-9 to 1 overexpression. This results indicate that miR-9 to 1 targeting UHRF1 and lead to the re-expression of tumor suppressor genes in lung cancer cells. The relationship between miR-9 to 1 and UHRF1 may be involved in feedback loop regulation. MiR-9 to 1/2/3 were unmethylated in most normal tissues. However, miR-9 to 1/3 methylations were found to be higher in oropharyngeal and oral carcinomas when compared to that in laryngeal carcinoma. MiR-9 to 1 and or miR-9 to 1-3 were also found merely hypermethylated in clear cell renal cell carcinoma (ccRCC) tissues and breast cancer. Therefore, we speculate that miR-9 to 1 suppresses cell proliferation may involve in the re-expression of tumor suppressor p15, p16, and p21, there may be also a feedback loop that exists between miRNA and its targets.
In summary, in this study, we identified that miR-9 to 1 suppressed lung cancer cell proliferation in vitro and in vivo. MiR-9 to 1 suppresses cell proliferation and induces apoptosis through directly targeting UHRF1 to re-activation of tumor suppressor genes p15, p16, and p21.
Acknowledgments
This study was supported partly by grants from the National Natural Science Foundation of China (81472202, 81372175, 81972214, 82071956, 81302065, and 81772932), Hunan Provincial Natural Science Foundation of China (2020JJ4278), Key Program of Hunan Provincial Department of Science and Technology (2020WK2020 and 2019NK2111), Shanghai Natural Science Foundation (20ZR1472400), The Fundamental Research Funds for the Central Universities (22120170212 and 22120170117), Program of Shanghai Subject Chief Scientist, Shanghai Natural Science Foundation (12ZR1436000 and 16ZR1428900), Construction of Clinical Medical Center for Tumor Biological Samples in Nantong (HS2016004).
Abbreviations
- miR-9 to 1
microRNA-9 to 1
- UHRF1
ubiquitin-like containing PHD and RING finger domains 1
- 3′UTR
3′untranslated region.
Footnotes
Author Contribution: CYJ, YSM, and DF designed the study. All authors performed the experimental analyses and interpreted the data. CYJ, YSM, and DF wrote the manuscript. CYJ, WX, JBL, GXJ, and FS contributed equally to this work. All authors contributed to the final version of the manuscript and approved the final manuscript.
Declaration of Conflicting Interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethical Approval and Consent to Participate: This study was approved by the Ethics Committee of Shanghai Tenth People's Hospital, Tongji University School of Medicine (IEC-PAP-15-18). All patients provided their written informed consent.
The authors are accountable for all aspects of the work (if applied, including full data access, the integrity of the data, and the accuracy of the data analysis) in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the National Natural Science Foundation of China (grant nos. 81972214, 81772932, 81472202, 81201535, 81302065).
ORCID iD: Da Fu https://orcid.org/0000-0002-0878-2575
Supplemental Material: Supplemental material for this article is available online.
References
- 1.Tipanee J, Di Matteo M, Tulalamba W, et al. Validation of miR-20a as a tumor suppressor gene in liver carcinoma using hepatocyte-specific hyperactive piggyBac transposons. Mol Ther Nucleic Acids. 2020;19:1309-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fu D, Shi Y, Liu JB, et al. Targeting long non-coding RNA to therapeutically regulate gene expression in cancer. Mol Ther Nucleic Acids. 2020;21:712-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cristóbal I, Luque M, Sanz-Alvarez M, Rojo F, García-Foncillas J. Clinical impact and regulation of the circCAMSAP1/ miR-328-5p/E2F1 axis in colorectal cancer. Mol Ther. 2020;28(6):1387-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen S, Gao C, Wu Y, Huang Z. Identification of prognostic miRNA signature and lymph node metastasis-related key genes in cervical cancer. Front Pharmacol. 2020;11:544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jana S, Krishna M, Singhal J, et al. Therapeutic targeting of miRNA-216b in cancer. Cancer Lett. 2020;484:16-28. [DOI] [PubMed] [Google Scholar]
- 6.Takagawa Y, Gen Y, Muramatsu T, et al. miR-1293, a candidate for miRNA-based cancer therapeutics, simultaneously targets BRD4 and the DNA repair pathway. Mol Ther. 2020;28(6):1494-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tan S, Xia L, Yi P, et al. Exosomal miRNAs in tumor microenvironment. J Exp Clin Cancer Res. 2020;39(1):67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao J, Wang G, Wu J, Zuo Y, Zhang J, Jin X. Skp2 expression Is inhibited by arsenic trioxide through the upregulation of miRNA-330-5p in pancreatic cancer cells. Mol Ther Oncolytics. 2019;12:214-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park C, Na KJ, Choi H, et al. Tumor immune profiles noninvasively estimated by FDG PET with deep learning correlate with immunotherapy response in lung adenocarcinoma. Theranostics. 2020;12:204-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuva-Aydemir Y, Simkin A, Gascon E, Gao FB. MicroRNA-9: functional evolution of a conserved small regulatory RNA. RNA Biol. 2011;8(4):557-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abbas M, Ahmed A, Khan GJ, et al. Clinical evaluation of carcinoembryonic and carbohydrate antigens as cancer biomarkers to monitor palliative chemotherapy in advanced stage gastric cancer. Curr Probl Cancer. 2019;43(1):5-17. [DOI] [PubMed] [Google Scholar]
- 12.Wang J, Cheng M, Law IKM, Ortiz C, Sun M, Koon HW. Cathelicidin suppresses colon cancer metastasis via a P2RX7-dependent mechanism. Mol Ther Oncolytics. 2019;12:195-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alwin Prem Anand A, Huber C, Asnet Mary J, et al. Expression and function of microRNA-9 in the mid-hindbrain area of embryonic chick. BMC Dev Biol. 2018;18(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rahmani A, Naderi M, Barati G, Arefian E, Jedari B, Nadri S. The potency of hsa-miR-9-1 overexpression in photoreceptor differentiation of conjunctiva mesenchymal stem cells on a 3D nanofibrous scaffold. Biochem Biophys Res Commun. 2020;529(3):526-532. [DOI] [PubMed] [Google Scholar]
- 15.Chen X, Zhu L, Ma Z, et al. Oncogenic miR-9 is a target of erlotinib in NSCLCs. Sci Rep. 2015;5:17031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu HJ, Hao M, Yeo SK, Guan JL. FAK Signaling in cancer-associated fibroblasts promotes breast cancer cell migration and metastasis by exosomal miRNAs-mediated intercellular communication. Oncogene. 2020;39(12):2539-2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu S, Wu W, Huang H, et al. The p53/miRNAs/Ccna2 pathway serves as a novel regulator of cellular senescence: complement of the canonical p53/p21 pathway. Aging Cell. 2019;18(3):e12918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lavenniah A, Luu TDA, Li YP, et al. Engineered circular RNA sponges act as miRNA inhibitors to attenuate pressure overload-induced cardiac hypertrophy. Mol Ther. 2020;28(6):1506-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu J, Xu R, Mai SJ, et al. LncRNA CSMD1-1 promotes the progression of hepatocellular carcinoma by activating MYC signaling. Theranostics. 2020;12:173-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miroshnichenko S, Patutina O. Enhanced inhibition of tumorigenesis using combinations of miRNA-targeted therapeutics. Front Pharmacol. 2019;10:488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li T, Ju E, Gao SJ. Kaposi sarcoma-associated herpesvirus miRNAs suppress CASTOR1-mediated mTORC1 inhibition to promote tumorigenesis. J Clin Invest. 2019;129(8):3310-3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawasaki H, Takeuchi T, Ricciardiello F, et al. Definition of miRNA signatures of nodal metastasis in LCa:miR-449a targets notch genes and suppresses cell migration and invasion. Mol Ther Nucleic Acids. 2020;20:711-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X, Wang H, Sun Y, et al. Enterovirus A71 oncolysis of malignant gliomas. Mol Ther. 2020;28(6):1533-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Porcelli L, Stolfa D, Stefanachi A, et al. Synthesis and biological evaluation of N-biphenyl-nicotinic based moiety compounds: a new class of antimitotic agents for the treatment of Hodgkin lymphoma. Cancer Lett. 2019;445:1-10. [DOI] [PubMed] [Google Scholar]
- 25.Pavlidis I, Spiller OB, Sammut Demarco G, et al. Cervical epithelial damage promotes Ureaplasma parvum ascending infection, intrauterine inflammation and preterm birth induction in mice. Nat Commun. 2020;11(1):199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morin PJ. Colorectal cancer: the APC-lncRNA link. J Clin Invest. 2019;129(2):503-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mooney R, Majid AA, Batalla-Covello J, et al. Enhanced delivery of oncolytic adenovirus by neural stem cells for treatment of metastatic ovarian cancer. Mol Ther Oncolytics. 2018;12:79-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wan HY, Guo LM, Liu T, Liu M, Li X, Tang H. Regulation of the transcription factor NF-kappaB1 by microRNA-9 in human gastric adenocarcinoma. Mol Cancer. 2010;9:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lehmann U, Hasemeier B, Christgen M, et al. Epigenetic inactivation of microRNA gene hsa-miR-9-1 in human breast cancer. J Pathol, 2008;214(1):17-24. [DOI] [PubMed] [Google Scholar]
- 30.Minor J, Wang X, Zhang F, et al. Methylation of microRNA-9 is a specific and sensitive biomarker for oral and oropharyngeal squamous cell carcinomas. Oral Oncol. 2012;48(1):73-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cabana-Domínguez J, Arenas C, Cormand B, Fernàndez-Castillo N. MiR-9, miR-153 and miR-124 are down-regulated by acute exposure to cocaine in a dopaminergic cell model and may contribute to cocaine dependence. Transl Psychiatry. 2018;8(1):173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amare AT, Schubert KO, Klingler-Hoffmann M, Cohen-Woods S, Baune BT. The genetic overlap between mood disorders and cardiometabolic diseases: a systematic review of genome-wide and candidate gene studies. Transl Psychiatry. 2017;(1):e1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pezuk JA, Salomão KB, Baroni M, Pereira CA, Geron L, Brassesco MS. Aberrantly expressed microRNAs and their implications in childhood central nervous system tumors. Cancer Metastasis Rev. 2019;38(4):813-828. [DOI] [PubMed] [Google Scholar]
- 34.Hong M, He J, Li D, et al. Runt- related transcription factor 1 promotes apoptosis and inhibits neuroblastoma progression in vitro and in vivo. J Exp Clin Cancer Res. 2020;39(1):52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin CC, Kuo IY, Wu LT, et al. Dysregulated kras/YY1/ZNF322A/Shh transcriptional axis enhances neo-angiogenesis to promote lung cancer progression. Theranostics. 2020;18(3):e12939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xin X, Lu Y, Xie S, et al. miR-155 accelerates the growth of human liver cancer cells by activating CDK2 via targeting H3F3A. Mol Ther Oncolytics. 2020;17:471-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cui C, Yang J, Li X, et al. Functions and mechanisms of circular RNAs in cancer radiotherapy and chemotherapy resistance. Mol Cancer. 2020;19(1):58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ashraf W, Bronner C, Zaayter L, et al. Interaction of the epigenetic integrator UHRF1 with the MYST domain of TIP60 inside the cell. J Exp Clin Cancer Res. 2017;36(1):188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu TM, Liu JB, Liu Y, et al. Power and promise of next-generation sequencing in liquid biopsies and cancer control. Cancer Control. 2020;17:306-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bronner C, Achour M, Arima Y, Chataigneau T, Saya H, Schini-Kerth VB. The UHRF family: oncogenes that are drugable targets for cancer therapy in the near future? Pharmacol Ther. 2007;115(3):419-434. [DOI] [PubMed] [Google Scholar]
- 41.Kim I, Park JW. Hypoxia-driven epigenetic regulation in cancer progression: a focus on histone methylation and its modifying enzymes. Cancer Lett. 2020;17:267-277. [DOI] [PubMed] [Google Scholar]
- 42.Liu L, Wang Y, Wu J, Liu J, Qin Z, Fan H. N6-methyladenosine: a potential breakthrough for human cancer. Mol Ther Nucleic Acids. 2020;19:804-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu C, Peng K, Wu Q, et al. HDAC1 and 2 regulate endothelial VCAM-1 expression and atherogenesis by suppressing methylation of the GATA6 promoter. Theranostics. 2021;11(11):5605-5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nishiyama A, Mulholland CB, Bultmann S, et al. Two distinct modes of DNMT1 recruitment ensure stable maintenance DNA methylation. Nat Commun. 2020;11(1):1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma YS, Liu JB, Wu TM, Fu D. New therapeutic options for advanced hepatocellular carcinoma. Cancer Control. 2020;16:262-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen M, Wong CM. The emerging roles of N6-methyladenosine (m6A) deregulation in liver carcinogenesis. Mol Cancer. 2020;19(1):44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiao J, Liu Y, Wu F, et al. miR-639 expression is silenced by DNMT3A-mediated hypermethylation and functions as a tumor suppressor in liver cancer cells. Mol Ther. 2020;28(2):587-598. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 48.Anvar Z, Acurzio B, Roma J, Cerrato F, Verde G. Origins of DNA methylation defects in Wilms tumors. Cancer Lett. 2019;457:119-128. [DOI] [PubMed] [Google Scholar]
- 49.Giri AK, Aittokallio T. DNMT Inhibitors increase methylation in the cancer genome. Front Pharmacol. 2019;10:385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Macaluso M, Montanari M, Noto PB, Gregorio V, Bronner C, Giordano A. Epigenetic modulation of estrogen receptor-alpha by pRb family proteins: a novel mechanism in breast cancer. Cancer Res. 2007;67(16):7731-7737. [DOI] [PubMed] [Google Scholar]
- 51.Lee HJ, Kim MJ, Kim YS, Choi MY, Cho GJ, Choi WS. UHRF1 Silences gelsolin to inhibit cell death in early stage cervical cancer. Biochem Biophys Res Commun. 2020;526(4):1061-1068. [DOI] [PubMed] [Google Scholar]
- 52.Szabó B, Németh K, Mészáros K, et al. Demethylation status of somatic DNA extracted from pituitary neuroendocrine tumors indicates proliferative behavior. J Clin Endocrinol Metab. 2020;105(6):156. [DOI] [PubMed] [Google Scholar]
- 53.Yin L, Liu Y, Peng Y, et al. PARP Inhibitor veliparib and HDAC inhibitor SAHA synergistically co-target the UHRF1/BRCA1 DNA damage repair complex in prostate cancer cells. J Exp Clin Cancer Res. 2018;37(1):153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schneider M, Trummer C, Stengl A, et al. Systematic analysis of the binding behaviour of UHRF1 towards different methyl- and carboxylcytosine modification patterns at CpG dyads. PLoS One. 2020;15(2):e0229144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vaughan RM, Dickson BM, Whelihan MF, et al. Chromatin structure and its chemical modifications regulate the ubiquitin ligase substrate selectivity of UHRF1. Proc Natl Acad Sci U S A. 2018;115(35):8775-8780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stachecka J, Lemanska W, Noak M, Szczerbal I. Expression of key genes involved in DNA methylation during in vitro differentiation of porcine mesenchymal stem cells (MSCs) into adipocytes. Biochem Biophys Res Commun. 2020;522(3):811-818. [DOI] [PubMed] [Google Scholar]
- 57.Hu Q, Qin Y, Ji S, et al. UHRF1 Promotes aerobic glycolysis and proliferation via suppression of SIRT4 in pancreatic cancer. Cancer Lett. 2019;452:226-236. [DOI] [PubMed] [Google Scholar]
- 58.Oh YM, Mahar M, Ewan EE, Leahy KM, Zhao G, Cavalli V. Epigenetic regulator UHRF1 inactivates REST and growth suppressor gene expression via DNA methylation to promote axon regeneration. Proc Natl Acad Sci U S A. 2018;115(52): E12417-E12426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wei C, Lu N, Wang L, et al. Upregulation of UHRF1 promotes the progression of melanoma by inducing cell proliferation. Oncol Rep. 2018;39(6):2553-2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim KY, Tanaka Y, Su J, et al. Uhrf1 regulates active transcriptional marks at bivalent domains in pluripotent stem cells through Setd1a. Nat Commun. 2018;9(1):2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dong J, Wang X, Cao C, et al. UHRF1 Suppresses retrotransposons and cooperates with PRMT5 and PIWI proteins in male germ cells. Nat Commun. 2019;10(1):4705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Taniue K, Hayashi T, Kamoshida Y, et al. UHRF1-KAT7-mediated Regulation of TUSC3 expression via histone methylation/acetylation is critical for the proliferation of colon cancer cells. Oncogene. 2020;39(5):1018-1030. [DOI] [PubMed] [Google Scholar]
- 63.Hong JH, Jin EH, Kim S, Song KS, Sung JK. LINE-1 hypomethylation is inversely correlated with UHRF1 overexpression in gastric cancer. Oncol Lett. 2018;15(5):6666-6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chow M, Gao L, MacManiman JD, et al. Maintenance and pharmacologic targeting of ROR1 protein levels via UHRF1 in t(1;19) pre-B-ALL. Oncogene. 2018;37(38):5221-5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lin Y, Chen Z, Lin S, et al. MiR-202 inhibits the proliferation and invasion of colorectal cancer by targeting UHRF1. Acta Biochim Biophys Sin (Shanghai ). 2019;51(12):1305-1306. [DOI] [PubMed] [Google Scholar]
- 66.Elia L, Kunderfranco P, Carullo P, et al. UHRF1 epigenetically orchestrate smooth muscle cell plasticity in arterial disease. J Clin Invest. 2018;128(6):2473-2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jiao D, Huan Y, Zheng J, et al. UHRF1 Promotes renal cell carcinoma progression through epigenetic regulation of TXNIP. Oncogene. 2019;38(28):5686-5699. [DOI] [PubMed] [Google Scholar]
- 68.He J, Fu X, Zhang M, et al. Transposable elements are regulated by context-specific patterns of chromatin marks in mouse embryonic stem cells. Nat Commun. 2019;10(1):34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang H, Cao D, Wu F. Long noncoding RNA UPAT promoted cell proliferation via increasing UHRF1 expression in non-small cell lung cancer. Oncol Lett. 2018;16(2):1491-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen J, Sheng X, Ma H, et al. WDR79 Mediates the proliferation of non-small cell lung cancer cells by regulating the stability of UHRF1. J Cell Mol Med. 2018;22(5):2856-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang Q, Dai L, Wang Y, et al. Targeted demethylation of the SARI promotor impairs colon tumour growth. Cancer Lett. 2019;448:132-143. [DOI] [PubMed] [Google Scholar]
- 72.Xu W, Xu M, Wang L, et al. Integrative analysis of DNA methylation and gene expression identified cervical cancer-specific diagnostic biomarkers. Signal Transduct Target Ther. 2019;4:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Raeisi F, Mahmoudi E, Dehghani-Samani M, et al. Differential expression profile of miR-27b, miR-29a, and miR-155 in chronic lymphocytic leukemia and breast cancer patients. Mol Ther Oncolytics. 2020;16:230-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang CJ, Zhu CC, Xu J, et al. Correction to the lncRNA UCA1 promotes proliferation, migration, immune escape and inhibits apoptosis in gastric cancer by sponging anti-tumor miRNAs. Mol Cancer. 2019;18(1):129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Luo Q, Song H, Deng X, et al. A triple-regulated oncolytic adenovirus carrying microRNA-143 exhibits potent antitumor efficacy in colorectal cancer. Mol Ther Oncolytics. 2020;16:219-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhou FC, Zhang YH, Liu HT, Song J, Shao J. LncRNA LINC00588 suppresses the progression of osteosarcoma by acting as a ceRNA for miRNA-1972. Front Pharmacol. 2020;11:255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Greally M, Kelly CM, Cercek A. HER2: an emerging target in colorectal cancer. Curr Probl Cancer. 2018;42(6):560-571. [DOI] [PubMed] [Google Scholar]
- 78.Chellini L, Frezza V, Paronetto MP. Dissecting the transcriptional regulatory networks of promoter-associated noncoding RNAs in development and cancer. J Exp Clin Cancer Res. 2020;39(1):51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Burton C, Bartee E. Syncytia formation in oncolytic virotherapy. Mol Ther Oncolytics. 2019;15:131-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cai H, Yang X, Gao Y, et al. Exosomal MicroRNA-9-3p secreted from BMSCs downregulates ESM1 to suppress the development of bladder cancer. Mol Ther Nucleic Acids. 2019;18:787-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Athari SS. Targeting cell signaling in allergic asthma. Signal Transduct Target Ther. 2019;4:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Taverner WK, Jacobus EJ, Christianson J, et al. Calcium influx caused by ER stress inducers enhances oncolytic adenovirus enadenotucirev replication and killing through PKCα activation. Mol Ther Oncolytics. 2019;15:117-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Masoumi E, Jafarzadeh L, Mirzaei HR, et al. Genetic and pharmacological targeting of A2a receptor improves function of anti-mesothelin CAR T cells. J Exp Clin Cancer Res. 2020;39(1):49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Butterfield JSS, Hege KM, Herzog RW, Kaczmarek R. A molecular revolution in the treatment of hemophilia. Mol Ther. 2020;28(4):997-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Taschauer A, Polzer W, Alioglu F, et al. Peptide-targeted polyplexes for aerosol-mediated gene delivery to CD49f-overexpressing tumor lesions in lung. Mol Ther Nucleic Acids. 2019;18:774-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chitapanarux I, Traisathit P, Chitapanarux T, et al. Arginine, glutamine, and fish oil supplementation in cancer patients treated with concurrent chemoradiotherapy: a randomized control study. Curr Probl Cancer. 2020;44(1):100482. [DOI] [PubMed] [Google Scholar]
- 87.Schintu N, Zhang X, Stroth N, Mathé AA, Andrén PE, Svenningsson P. Non- dopaminergic alterations in depression-like FSL rats in experimental parkinsonism and L-DOPA responses. Front Pharmacol. 2020;11:304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lévêque R, Corbet C, Aubert L, et al. ProNGF increases breast tumor aggressiveness through functional association of TrkA with EphA2. Cancer Lett. 2019;449:196-206. [DOI] [PubMed] [Google Scholar]
- 89.Gumlaw N, Sevigny LM, Zhao H, et al. Biab mediated restoration of the linkage between dystroglycan and laminin-211 as a therapeutic approach for α-dystroglycanopathies. Mol Ther. 2020;28(2):664-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hanauer JRH, Koch V, Lauer UM, Mühlebach MD. High-affinity DARPin allows targeting of MeV to glioblastoma multiform in combination with protease targeting without loss of potency. Mol Ther Oncolytics. 2019;15:186-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Krotova K, Day A, Aslanidi G. An engineered AAV6-based vaccine induces high cytolytic anti-tumor activity by directly targeting DCs and improves Ag presentation. Mol Ther Oncolytics. 2019;15:166-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chen X, Xu M, Xu X, et al. METTL14 suppresses CRC progression via regulating N6-methyladenosine- dependent primary miR-375 processing. Mol Ther. 2020;28(2):599-612. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 93.Fang T, Zhang Z, Sun R, et al. RNAm5CPred: prediction of RNA 5-methylcytosine sites based on three different kinds of nucleotide composition. Mol Ther Nucleic Acids. 2019;18:739-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen XY, Zhang J, Zhu JS. The role of m6A RNA methylation in human cancer. Mol Cancer. 2019;18(1):103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Niu Y, Lin Z, Wan A, et al. RNA N6-methyladenosine demethylase FTO promotes breast tumor progression through inhibiting BNIP3. Mol Cancer. 2019;18(1):46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhu B, Xi X, Liu Q, Cheng Y, Yang H. MiR-9 functions as a tumor suppressor in acute myeloid leukemia by targeting CX chemokine receptor 4. Am J Transl Res. 2019;11(6):3384-3397. [PMC free article] [PubMed] [Google Scholar]
- 97.Li Q, Chang Y, Mu L, Song Y. MicroRNA-9 enhances chemotherapy sensitivity of glioma to TMZ by suppressing TOPO II via the NF-κB signaling pathway. Oncol Lett. 2019;17(6):4819-4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wei W, Dong Z, Gao H, et al. MicroRNA-9 enhanced radiosensitivity and its mechanism of DNA methylation in non-small cell lung cancer. Gene. 2019;710:178-185. [DOI] [PubMed] [Google Scholar]
- 99.Sang Z, Jiang X, Guo L, Yin G. MicroRNA-9 suppresses human prostate cancer cell viability, invasion and migration via modulation of mitogen-activated protein kinase 3 expression. Mol Med Rep. 2019;19(5):4407-4418. [DOI] [PubMed] [Google Scholar]
- 100.Chen X, Yang F, Zhang T, et al. MiR-9 promotes tumorigenesis and angiogenesis and is activated by MYC and OCT4 in human glioma. J Exp Clin Cancer Res. 2019;38(1):99. [DOI] [PMC free article] [PubMed] [Google Scholar]