Abstract
The standard circulating biomarker of liver injury in both clinical settings and drug safety testing is alanine aminotransferase (ALT). However, ALT elevations sometimes lack specificity for tissue damage. To identify novel serum biomarkers with greater specificity for injury, we combined unique animal models with untargeted proteomics, followed by confirmation with immunoblotting. Using proteomics, we identified 109 proteins in serum from mice with acetaminophen (APAP)-induced liver injury that were not detectable in serum from mice with benign ALT elevations due to high-dose dexamethasone (Dex). We selected 4 (alcohol dehydrogenase 1A1 [Aldh1a1], aldehyde dehydrogenase 1 [Adh1], argininosuccinate synthetase 1 [Ass1], and adenosylhomocysteinase [Ahcy]) with high levels for further evaluation. Importantly, all 4 were specific for injury when using immunoblots to compare serum from Dex-treated mice and mice with similar lower ALT elevations due to milder models of APAP or bromobenzene-induced liver injury. Immunoblotting for ALDH1A1, ADH1, and ASS1 in serum from APAP overdose patients without liver injury and APAP overdose patients with mild liver injury revealed that these candidate biomarkers can be detected in humans with moderate liver injury as well. Interestingly, further experiments with serum from rats with bile duct ligation-induced liver disease indicated that Aldh1a1 and Adh1 are not detectable in serum in cholestasis and may therefore be specific for hepatocellular injury and possibly even drug-induced liver injury, in particular. Overall, our results strongly indicate that ALDH1A1, ADH1, and ASS1 are promising specific biomarkers for liver injury. Adoption of these biomarkers could improve preapproval drug safety assessment.
Keywords: hepatotoxicity, drug-induced liver injury, drug safety, regulatory science, transaminitis
Alanine aminotransferase (ALT)/serum glutamic-pyruvic transaminase was identified as a serum biomarker of liver injury more than 60 years ago (McGill, 2016). Since then, serum ALT activity has been the standard biomarker for noninvasive diagnosis and monitoring of hepatocellular liver injury and disease, both in the clinical setting and in the drug regulation process. However, it has several major limitations, including less-than-desirable specificity for tissue damage.
The use of serum ALT to assess liver injury is based on the idea that damaged hepatocytes release the enzyme into the extracellular space. There are 2 major mammalian isoforms of ALT (ALT1 and ALT2) encoded by 2 different genes, glutamic-pyruvic transaminase (GPT) 1 and GPT2, respectively. Both are highly expressed in the liver compared with other tissues (LaDue and Wroblewski, 1956). The dominant mechanism of ALT release into serum is thought to be loss of membrane integrity in necrotic hepatocytes. However, there is strong evidence from animal and cell culture models that other mechanisms exist (McGill, 2016). Furthermore, there are numerous reports of serum ALT elevations in the absence of necrosis and liver dysfunction in humans (Harrill et al., 2012; Rautou et al., 2008; Singhal et al., 2014; Watkins et al., 2006). This lack of specificity for significant injury can pose a fundamental challenge in drug regulation, and sometimes even in patient care.
In the current study, our goal was to identify complementary serum biomarkers that are more specific for liver injury than ALT. To do that, we applied a proteomics approach to animal models of injury caused by acetaminophen (APAP) overdose and a model of benign ALT elevations due to acute treatment with a large dose of dexamethasone (Dex). Acetaminophen overdose causes massive liver necrosis in both mice and humans (McGill et al., 2012), whereas Dex has been shown to cause transient elevations of serum ALT in rodents without causing liver injury (Jackson et al., 2008; Reagan et al., 2012). We then tested several of those biomarkers using samples from APAP overdose patients with mild ALT elevations to determine if they can be measured in humans as well. Finally, we tested 2 of the candidate biomarkers in a rat model of cholestatic liver injury to determine if they may be detectable in other forms of liver injury. Overall, we have identified several novel biomarkers that may be useful to distinguish hazardous and benign serum ALT elevations.
MATERIALS AND METHODS
Animals
All animals were kept in a temperature-controlled room with a 12-h light/dark cycle. All were between 8 and 12 weeks of age at the time of our studies. Male C57Bl/6J mice were purchased from Jackson Laboratories (Bar Harbor, Maine). The mice had free access to food prior to initiation of our experiments and had free access to water for the duration of our studies. For the initial proteomics experiment, some mice were fasted overnight, then treated with either 250 mg/kg APAP (Sigma, St Louis, Missouri) dissolved in warm 1× phosphate-buffered saline (PBS) or an equivalent volume of PBS alone. Blood and liver tissue were collected 6 h later. Other C57Bl/6J mice were allowed ad libitum access to food and were treated with either 100 mg/kg Dex (Sigma) dissolved in dimethylsulfoxide (DMSO) or an equivalent volume of DMSO alone. Blood, liver tissue, and muscle tissue were collected 24 h later. To confirm our results from proteomics using animals with ALT elevations of lower magnitude than the traditional APAP overdose model, an additional experiment was performed. Some C57Bl/6J mice were fasted overnight, then treated with 175 mg/kg APAP in PBS. Blood was collected 6 h later. Other C57Bl/6J mice were allowed ad libitum access to food and treated with 400 mg/kg APAP. Blood was collected 24 h later. We also performed a time course experiment in which C57Bl/6J mice were fasted overnight, then treated with 250 mg/kg APAP. Blood and liver tissue were collected 0, 6, 24, and 48 h later. Finally, we performed an experiment in which C57Bl/6J mice were allowed ad libitum access to food and treated with 1.1 g/kg bromobenzene (BB) mixed in corn oil vehicle. Blood and liver were collected 24 h later. Drug doses were selected based on existing literature (Reagan et al., 2012) and results from preliminary experiments. For each mouse, 1 liver section and 1 muscle section were fixed in 10% phosphate-buffered formalin for histology. Other liver pieces were flash frozen in liquid nitrogen for later biochemical analysis. Male Wistar rats were purchased from Charles River (Wilmington, Massachusetts) and allowed free access to food and water. The rats were subjected to either sham surgery or bile duct ligation (BDL) to induce cholestasis, as previously described (Hambuchen et al., 2019). Blood was collected by venipuncture 7 days postsurgery. Liver tissue was collected 9 days postsurgery. All treatments were administered i.p. All animal experiments were reviewed and approved by the Institutional Animal Care and Use Committees of the University of Arkansas for Medical Sciences and of Marshall University.
APAP overdose patients
Patients ≥ 18 years of age with a diagnosis of APAP overdose were recruited. Inclusion criteria were a physician’s diagnosis of APAP-induced liver injury or elevated serum APAP levels based on the Rumack-Matthew nomogram, and abnormal liver function test results. Exclusion criteria were reasonable evidence of another major cause of acute liver injury, such as a recent history of alcohol abuse. Informed consent was obtained from each patient or next of kin, and at least 1 blood sample was collected in an EDTA plasma or serum tube. The study protocol was approved by the Institutional Review Board of the University of Kansas Medical Center and conformed to the ethical guidelines of the 1975 Declaration of Helsinki.
Clinical chemistry
Alanine aminotransferase was measured using the traditional coupled enzyme reaction of Karmen et al. (1955) with either a kit from Pointe Scientific Inc (Canton, Michigan) (all APAP, Dex, and other drug experiments) or an Abaxis VetScan instrument (BDL experiment). Creatine kinase (CK) was measured in serum using a kit from Pointe Scientific Inc.
Proteomics
To avoid problematic masking by abundant albumin and immunoglobulins, the region of each SDS-PAGE gel lane below the albumin band was used. For each sample, the lane was sectioned into 6 segments of equal volume. Each segment was subjected to in-gel trypsin digestion as follows: Gel slices were destained in 50% methanol (Thermo Fisher Scientific, Waltham, Massachusetts), 100 mM ammonium bicarbonate (Sigma), followed by reduction in 10 mM Tris(2-carboxyethyl)phosphine (Pierce, Thermo Fisher Scientific) and alkylation in 50 mM iodoacetamide (Sigma). Gel slices were then dehydrated in acetonitrile (Thermo Fisher Scientific), followed by addition of 100-ng porcine sequencing grade modified trypsin (Promega, Madison, Wisconsin) in 100 mM ammonium bicarbonate (Sigma) and incubation at 37°C for 12–16 h. Peptide products were then acidified in 0.1% formic acid (Pierce, Thermo Fisher Scientific). Tryptic peptides were separated by reverse phase XSelect CSH C18 2.5-μm resin (Waters, Milford, Massachusetts) on an in-line 150 × 0.075-mm column using a nanoAcquity UPLC system (Waters). Peptides were eluted using a 30-min gradient from 97:3 to 67:33 buffer A:B ratio. (Buffer A = 0.1% formic acid, 0.5% acetonitrile; buffer B = 0.1% formic acid, 99.9% acetonitrile.) Eluted peptides were ionized by electrospray (2.15 kV) followed by MS/MS analysis using higher-energy collisional dissociation (HCD) on an Orbitrap Fusion Tribrid mass spectrometer (Thermo Fisher Scientific) in top-speed data-dependent mode. MS data were acquired using the FTMS analyzer in profile mode at a resolution of 240 000 over a range of 375–1500 m/z. Following HCD activation, MS/MS data were acquired using the ion trap analyzer in centroid mode and normal mass range with precursor mass-dependent normalized collision energy between 28.0 and 31.0. Proteins were identified by database search using Mascot (Matrix Science, Boston, Massachusetts) with a parent ion tolerance of 3 ppm and a fragment ion tolerance of 0.5 Da. Scaffold (Proteome Software, Portland, Oregon) was used to verify MS/MS based peptide and protein identifications. Peptide identifications were accepted if they could be established with < 1.0% false discovery by the Scaffold Local FDR algorithm. Protein identifications were accepted if they could be established with < 1.0% false discovery and contained at least 2 identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm (Nesvizhskii et al., 2003).
Quantitative polymerase chain reaction (qPCR)
Liver tissues were homogenized using a bead homogenizer and RNA was extracted using RNA-Bee reagent (Tel-Test Inc, Friendswood, Texas). Chloroform was then added and the samples were shaken, then allowed to incubate at 4°C for 5 min. After centrifugation (12 000 × g, 4°C for 5 min), the aqueous phase was transferred to a new tube and mixed with isopropanol (0.5 ml). The samples were then allowed to sit at room temperature for 10 min, and then RNA was pelleted by centrifugation (12 000 × g, 4°C for 5 min). The RNA pellets were then washed with 75% ethanol and resuspended in RNase-free H2O. RNA concentration and purity were measured using a Nanodrop ND-1000 Spectrophotometer (Thermo Fisher Scientific). Next, 2 μg of RNA was transcribed into cDNA using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Thermo Fisher Scientific) on an Applied Biosystems Veriti thermocycler. For real-time qPCR, cDNA was mixed with PowerUp SYBR Green Master mix (Applied Biosystems, Thermo Fisher Scientific) along with forward and reverse primers (Table 1) from Integrated DNA Technologies and run on an Applied Biosystems ViiA7 real-time qPCR instrument.
Table 1.
Ten Proteins With Highest Spectral Counts in APAP Mouse Serum
| Adenosylhomocysteinase (Ahcy)a |
| Alcohol dehydrogenase 1 (Adh1)a |
| 4-Hydroxyphenylpyruvate dioxygenase (Hpd) |
| Isocitrate dehydrogenase 1 (Idh1) |
| Cluster of retinal dehydrogenase 1 (Aldh1A1)a |
| Fructose-1,6-bisphosphatase 1 (Fbp1) |
| Argininosuccinate synthase (Ass1)a |
| Malate dehydrogenase 1 (Mdh1) |
| Peroxiredoxin 6 (Prdx6) |
| Fumarylacetoacetate hydrolase (Fah) |
Selected for further evaluation based on existing literature and reagent availability.
Western blotting
Serum was diluted 8× in PBS to a final volume of 50 μl, followed by addition of 16 μl of 4× concentrated Laemmli buffer. The samples were then boiled for 1 min and stored at −80°C until use. For electrophoresis and Western blotting, equal volumes of each sample were loaded in the lanes. Liver tissues were homogenized in 25 mM HEPES buffer with 5 mM EDTA (pH 7.4), 0.1% CHAPS, and protease inhibitors, using a bead homogenizer (Thermo Fisher Scientific). Protein concentration in the homogenates was measured using the bicinchoninic acid assay. For electrophoresis and Western blotting, equal amounts of total protein were loaded in the lanes. Tris-glycine gels (4%–20%) were used. After electrophoresis, proteins were transferred to PVDF membranes with 0.45-m pores and blocked with 5% milk in Tris-buffered saline with 0.1% Tween 80. After incubation with the appropriate antibodies, protein bands were visualized using the Odyssey CLX Imaging System (LI-COR Biosciences, Lincoln, NE). ADH1, ALDH1A1, and ASS1 primary antibodies were used at a 1:1000 dilution. AHCY primary antibody was used at a 1:500 dilution. All secondary antibodies were diluted 1:10 000. The primary antibodies were purchased from Cell Signaling Technology (Danvers, Massachusetts) (ALDH1A1 [Cat. No. 12035], ASS1 [Cat. No. 70720], and ADH1 [Cat. No. 5295]) and Proteintech (Rosemont, Illinois) (AHCY [Cat. No. 10757-2-AP]). Secondary antibodies were purchased from LI-COR Biosciences.
Histology and immunohistochemistry
Formalin-fixed tissue was embedded in paraffin wax and 5-μm sections were mounted on glass slides for hematoxylin and eosin (H&E) staining and immunohistochemistry. H&E staining was performed using a standard protocol.
Data analysis and statistics
Proteomics quantitation was based on spectral counts. To maximize specificity, we selected proteins from our proteomics experiment that were detected in serum from the mice with APAP-induced liver injury but either minimally (≤ 12 spectral counts per group on average) or not at all in the other groups. For all experiments, to compare means, data were first tested for normality using the Shapiro-Wilk test. For normally distributed data, 2 groups were compared using a t test, whereas 3 or more groups were compared using one-way analysis of variance (ANOVA) with post hoc Student-Newman-Keul’s test. For nonnormal data, 2 groups were compared using a t test on the ranked data, whereas 3 or more groups were compared using ANOVA on the ranked data with Dunn’s post hoc multiple comparisons. Proteomics data were managed and visualized in R (R Foundation for Statistical Computing, Vienna, Austria). All other statistical analyses were performed in SigmaPlot 12.5 (Systat, San Jose, California).
RESULTS
Validation of the Dex Model
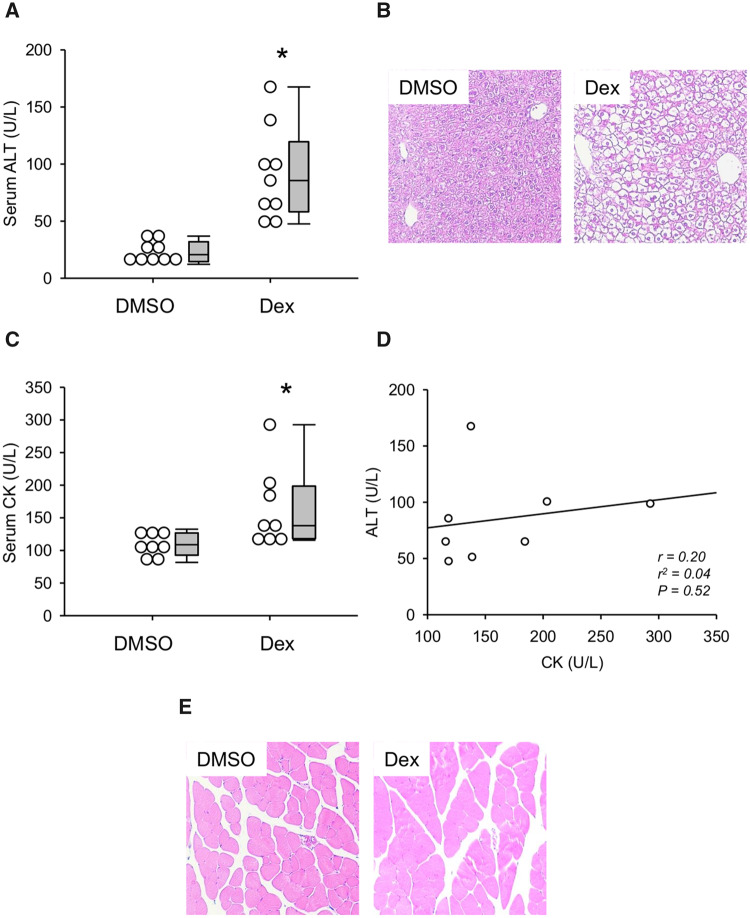
Reagan et al. (2012) previously reported that large doses of Dex can elevate serum ALT in mice without causing liver injury. To confirm their results and to validate high-dose Dex in mice as a model of benign ALT elevations, we first compared serum ALT values between mice treated with Dex or DMSO vehicle. As expected, ALT was significantly increased in the Dex-treated mice (Figure 1A). However, despite evidence of increased glycogen deposition (cytoplasmic clear cell changes in H&E-stained sections), we did not observe any evidence of tissue necrosis or apoptosis by histology (Figure 1B). The lack of injury was confirmed by 2 independent, blinded, fellowship-trained gastrointestinal and hepatobiliary pathologists who agreed that the histological changes were entirely consistent with glycogen deposition, which is expected because Dex stimulates hepatic glycogen synthesis (Margolis and Curnow, 1984). Importantly, it is known that large doses of corticosteroids can cause muscle toxicity, and prior studies did not test whether the ALT elevations caused by high-dose Dex in rodents can be explained in part by muscle damage. To do that, we also measured CK activity in serum from these animals and collected muscle tissue for histology. Although there was a statistically significant difference between the DMSO and Dex groups (Figure 1C), there was no significant correlation between serum ALT and CK values (Figure 1D) and the absolute CK values did not exceed typical reference intervals for CK in C57Bl/6 mice even though ALT values did (Boehm et al., 2007). Finally, there was no obvious evidence of muscle damage by histology (Figure 2E). Thus, if muscle damage does contribute to the increased ALT, its role is likely minor.
Figure 1.
Dexamethasone (Dex) increased serum alanine aminotransferase (ALT). Mice were treated i.p. with 100 mg/kg Dex or DMSO vehicle (DMSO). Blood and tissue were collected 24 h later. A, Serum ALT activity. B, H&E-stained liver sections. C, Serum creatine kinase (CK) activity. D, Correlation between ALT and CK in serum from Dex-treated mice. E, H&E-stained muscle sections. Data expressed as mean ± SE for n = 9–10 mice per group. *p < .05 versus DMSO.
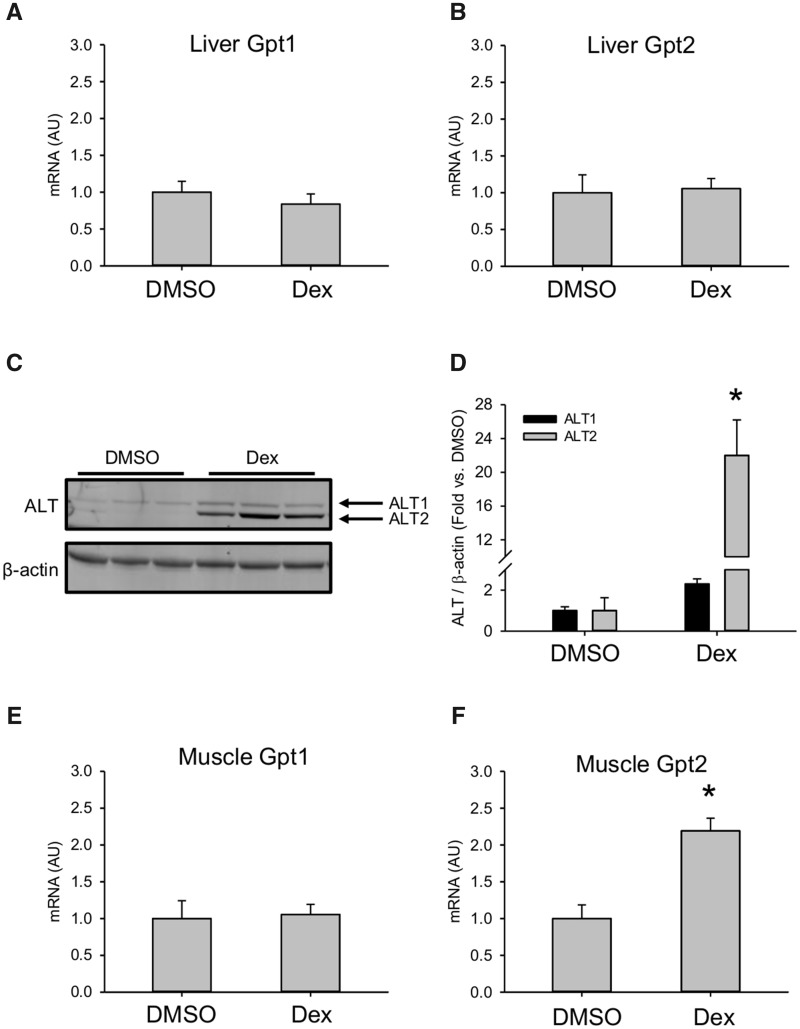
Figure 2.
Dexamethasone (Dex) increased hepatic alanine aminotransferase (ALT). Mice were treated i.p. with 100 mg/kg Dex or DMSO vehicle (DMSO). Liver tissue was collected 24 h later. A, Gpt1 mRNA in liver. B, Gpt2 mRNA in liver. C, Immunoblot for ALT isoforms in liver tissue. D, Densitometry from immunoblot. Data expressed as mean ± SE for n = 5 mice per group for mRNA and n = 3–4 per group mice for immunoblot. *p < .05 versus DMSO.
To determine if increased hepatic expression can explain the serum ALT elevations in our mice, we measured expression of the ALT1 and ALT2 genes, Gpt1 and Gpt2, in the liver. There was no difference in mRNA in the liver between Dex- and vehicle-treated mice (Figs. 2A and 2B). However, there was a clear increase in Alt2 protein encoded by the Gpt2 gene (Figs. 2C and 2D). We also measured mRNA in muscle tissue. Similar to the liver, there was an increase in Gpt2 expression in muscle (Figs. 2E and 2F). These results demonstrate that the mechanism of serum ALT elevation after Dex treatment in mice is increased hepatic and possibly muscle Alt2 protein. Finally, they provide additional evidence that the serum ALT release in the Dex model is not due to liver injury. Altogether, our results confirm previous work demonstrating that acute treatment with large doses of Dex can cause transient serum ALT elevations in mice without liver injury.
Identification of Injury-Specific Biomarkers in Mice Using Proteomics
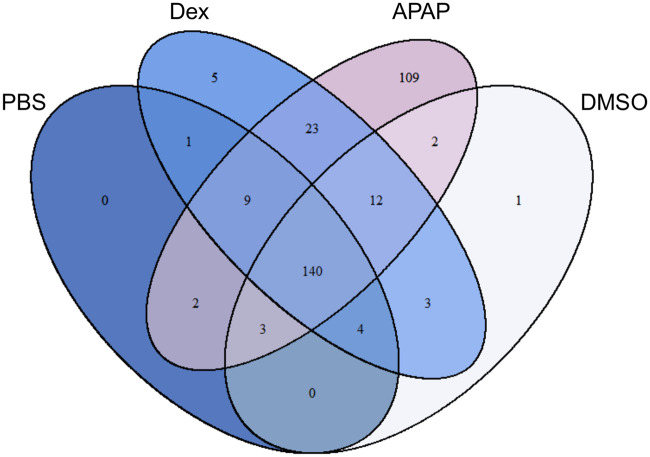
To identify serum proteins that may distinguish hazardous and benign ALT elevations, we compared the serum proteomes of mice in 4 treatment groups: (1) fasted mice treated with either 250 mg/kg APAP or (2) PBS vehicle, and (3) fed mice treated with either 100 mg/kg Dex or (4) DMSO vehicle. Fasting increases liver injury and reduces variation due to hepatic glutathione in the APAP model. A total of 658 proteins were detected. Interestingly, we identified 109 proteins in samples from the APAP overdose group that could not be detected in any samples from the other 3 groups, and others with high average spectral counts in the APAP mice but low counts in the other groups (Figure 3) (Supplementary data). Among the latter, the 10 with the highest spectral counts in the APAP group are listed in descending order in Table 1. Surprisingly, although one might not expect the same proteins to increase in the serum from mice with ALT release due to different mechanisms (necrosis vs increased hepatic expression), 23 proteins were elevated in both the APAP and Dex groups in addition to ALT—more than were shared between any other pairing of groups. It is our contention that this overlap provides even greater legitimacy to our approach using these animal models.
Figure 3.
Proteomics revealed 109 candidate biomarkers that were only in serum from acetaminophen (APAP) overdose mice. Mice were treated i.p. with 100 mg/kg dexamethasone (Dex), 250 mg/kg APAP, DMSO vehicle, or phosphate-buffered saline (PBS) vehicle. Blood was collected 24 h later (Dex, DMSO) or 6 h later (APAP, PBS). Serum aliquots were used for proteomics. n = 5 mice per group.
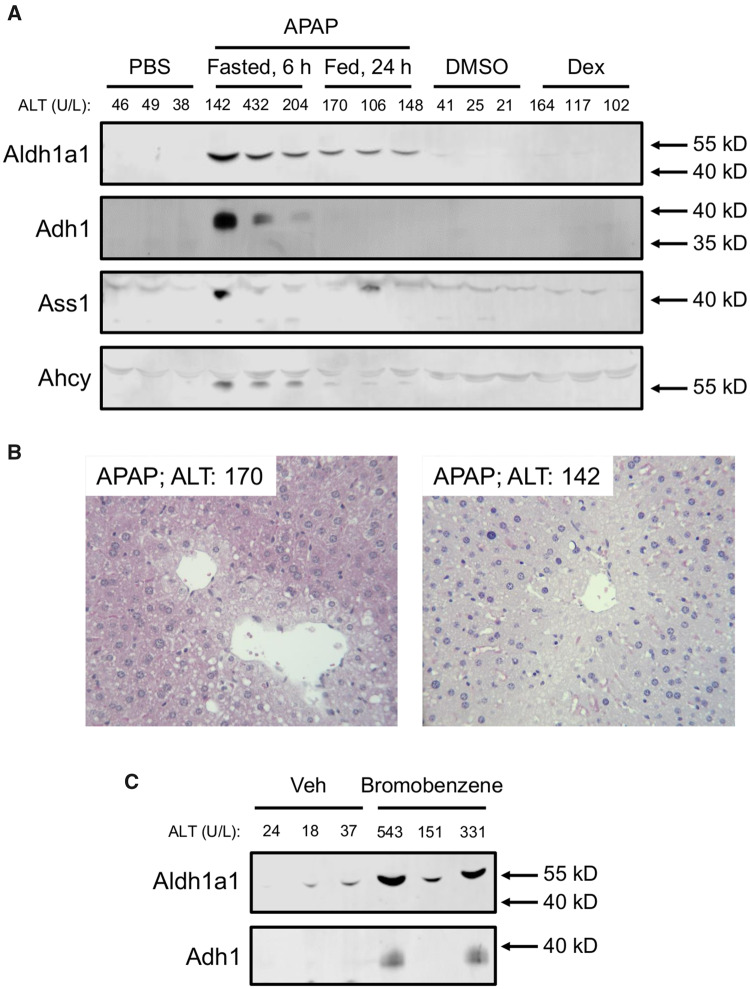
Next, we wanted to verify that the candidate biomarkers identified in the proteomics experiment are detectable after APAP-induced liver injury with lower ALT elevations that more closely resemble both the Dex model and typical benign elevations in humans. The 250 mg/kg dose of APAP following fasting causes severe liver injury with serum ALT values (2768 ± 223 U/l) much greater than those observed in the Dex-treated mice, so it is possible that the proteins we detected solely in the APAP group were merely detectable due to the magnitude of injury and ALT release. To test that, we treated fasted mice with 175 mg/kg and fed mice with 400 mg/kg APAP and collected serum 6 or 24 h later, respectively. Both treatment regimens resulted in ALT values similar to those in the Dex group (Figure 4). We then selected 4 candidate biomarkers from the list in Table 1 for further testing: alcohol dehydrogenase 1a1 (Aldh1a1), aldehyde dehydrogenase 1 (Adh1), adenosylhomocysteinase (Ahcy), and argininosuccinate synthetase 1 (Ass1). Ass1 was chosen because we had previously reported it as a sensitive biomarker of drug-induced liver injury (DILI) (McGill et al., 2014). The remaining 3 candidates were chosen based on the availability of antibodies to detect them. It should be noted that malate dehydrogenase (Mdh1) has also been reported as a biomarker of liver injury in previous studies (Schomaker et al., 2013), though we did not further investigate it here. We then immunoblotted for those 4 proteins in serum from the APAP-, Dex-, and vehicle-treated mice. Importantly, all 4 were much higher in serum samples from the APAP-treated mice than from mice in the other groups (Figure 4A). Necrosis was confirmed in those mice by histology (Figure 4B). Although there were differences in access to food between the Dex group and one of the APAP groups, we do not believe that influenced the results for those biomarkers because both the Dex-treated group and the other APAP-treated group were fed ad libitum throughout the study, yet still exhibited differences.
Figure 4.
Aldh1A1 and Adh1 are specific for liver injury in mice. A, Fed mice were treated i.p. with 100 mg/kg dexamethasone (Dex), 400 mg/kg acetaminophen (APAP), or DMSO vehicle, and blood was collected 24 h later. Fasted mice were treated with 175 mg/kg APAP or phosphate-buffered saline (PBS) vehicle, and blood was collected 6 h later. Serum aliquots were used for immunoblotting for Aldh1a1, Adh1, Ass1, and Ahcy. B, Representative H&E-stained liver sections from the APAP-treated mice showing central veins with characteristic features of necrosis (swelling, karyolysis, karyorrhexis, and eosinophilia) of many surrounding hepatocytes. C, Fed mice were treated i.p. with 1.1 g/kg bromobenzene or vehicle. Blood was collected 24 h later. Serum aliquots were used for immunoblotting for Aldh1a1 and Adh1. Serum ALT values for each animal are displayed above the blots.
Finally, to determine if the candidate biomarkers are elevated in serum after hepatotoxicity caused by other drugs or xenobiotics, we treated mice with 1.1 g/kg BB and collected serum 24 h later. Both Aldh1a1 and Adh1 were higher in serum from the BB-treated mice than the vehicle-treated mice (Figure 4C).
Detection of Candidate Biomarkers in Human Samples
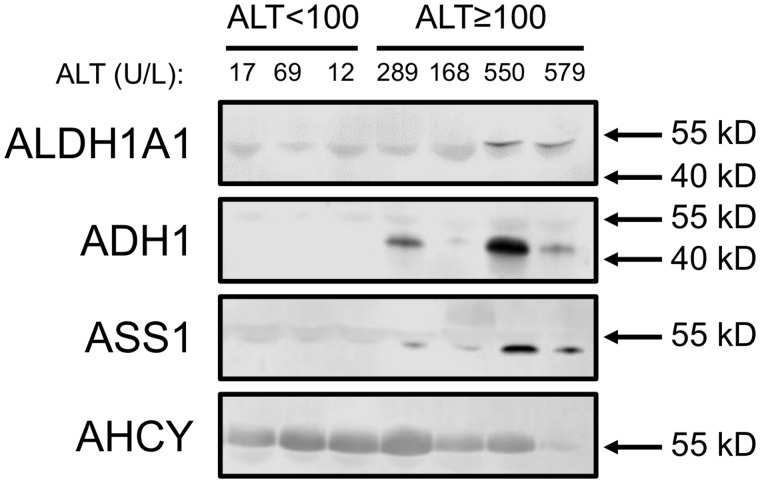
To begin to validate these results in humans, we immunoblotted for all 4 proteins in serum from APAP overdose patients without liver injury and APAP overdose patients with mild liver injury (≥ 100 U/l). Patient demographics and clinical laboratory test results are shown in Table 2. Importantly, all samples in the group with injury were collected at or before the day of peak ALT during hospitalization. It should be noted that a faint albumin band is visible near the bands of interest in all blots (Figure 5), which is common when immunoblotting with serum samples. Importantly, 3 of the 4 biomarkers were detected in 1 or more samples from the APAP overdose patients with liver injury (Figure 5). AHCY was detected in samples from both patients with and without liver injury. These data demonstrate that 3 of the candidate biomarkers (ALDH1A1, ADH1, and ASS1) can detect mild liver injury in humans as well as mice.
Table 2.
Human Subjects
| ALT < 100 U/l | ALT ≥ 100 U/l | |
|---|---|---|
| Age (years) (mean, range) | 49, 28–80 | 35, 21–40 |
| Sex (% female) | 100 | 100 |
| Peak ALT (U/l) (mean, range) | 48, 29–69 | 931, 168–2047 |
| Peak PT (s) (mean, range) | 15, 13–18 | 33, 23–31 |
| Peak INR (mean, range) | 2.7, 1.2–5.3 | 2.3, 1.6–2.8 |
| Peak bilirubin (mean, range) | 1.6, 0.4–3.6 | 11.2, 1.3–20.7 |
| % survival | 100 | 100 |
Figure 5.
ALDH1A1, ADH1, and ASS1 may be specific for liver injury in humans. Immunoblotting for ALDH1A1, ADH1, ASS1, and AHCY was done using aliquots of serum or plasma collected from acetaminophen (APAP) overdose patients without liver injury (peak ALT < 100 U/l) and with modest liver injury (peak ALT ≥ 100 U/l). Circulating ALT values for each subject or patient in the tested specimen are displayed above the blots.
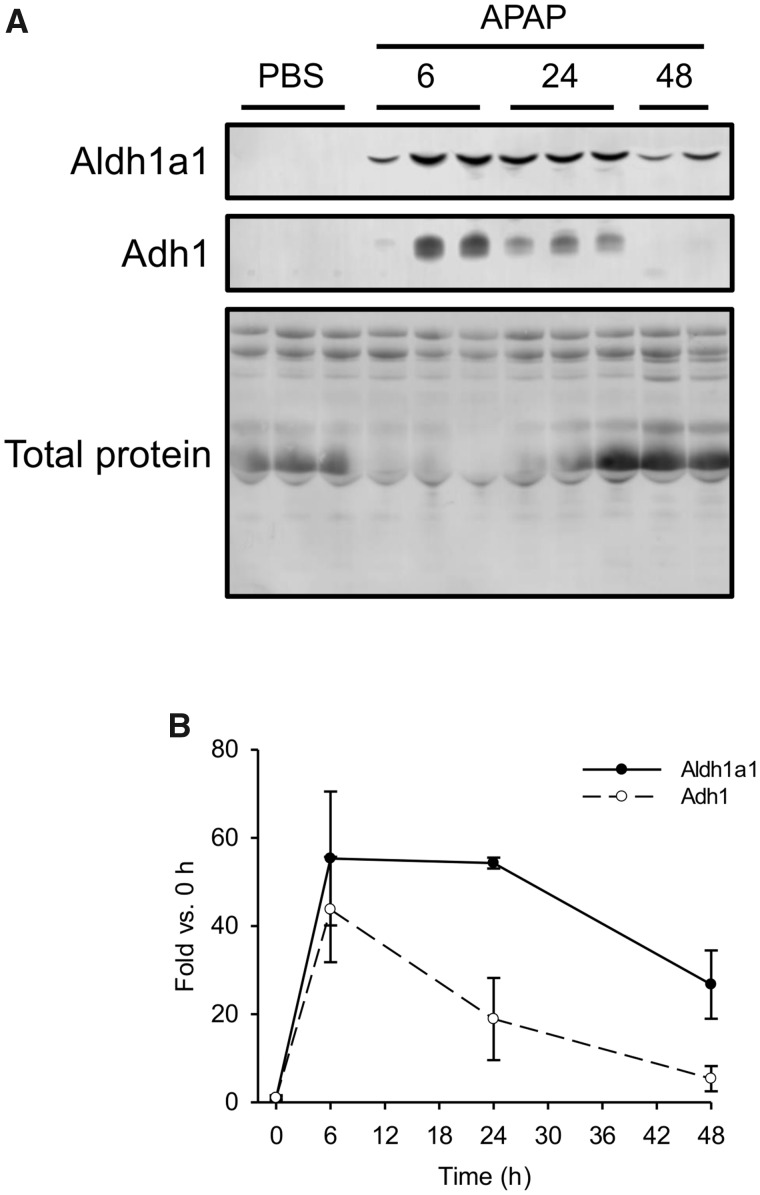
Based on the results from the animal studies, Aldh1A1 and Adh1 appeared to be the most promising biomarkers that we identified (ASS1 was promising in humans, but we could not detect it well in mouse serum), so we next wanted to determine the kinetics of those 2 proteins in serum after liver injury. To do that, we treated fasted mice with 250 mg/kg APAP and collected blood 0, 6, 24, and 48 h later. Both Aldh1a1 and Adh1 were detectable at 6 and 24 h, but only Aldh1a1 could still be detected at 48 h (Figs. 6A and 6B). These data indicate that Adh1 has a shorter serum half-life than Aldh1a1 and may therefore be a better biomarker of active liver injury in early-presenting patients, whereas Aldh1a1 may be more sensitive in late-presenting patients.
Figure 6.
Time course of Aldh1a1 and Adh1 in serum from mice after acute injury. Fasted mice were treated i.p. with 250 mg/kg acetaminophen (APAP) or phosphate-buffered saline (PBS) vehicle. Blood was collected 6, 24, and 48 h later. A, Immunoblotting was done to detect Aldh1a1 and Adh1 in serum. Total serum protein is also displayed. B, Densitometry from immunoblotting. Data expressed as mean ± SE for n = 3 mice per group.
Biomarker Testing in Bile Duct–Ligated Rats
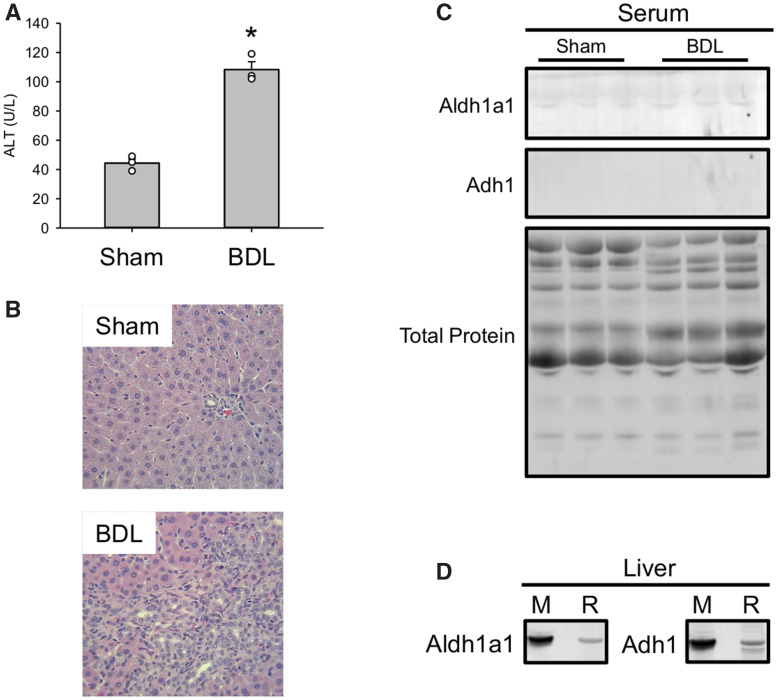
Finally, we wanted to determine if Aldh1a1 and Adh1 can be detected in serum after liver injury of other etiologies. To do that, we subjected rats to sham surgery or BDL to induce cholestasis and collected serum 7 days later. Consistent with earlier studies (Fickert et al., 2013; Hambuchen et al., 2019; Woolbright et al., 2013), BDL caused persistent ALT elevations similar in magnitude to the Dex model (Figure 7A) and clear evidence of bile duct proliferation and liver injury (Figure 7B). Interestingly, there was no obvious increase in Aldh1a1 and Adh1 in serum from the BDL rats compared with the sham group (Figure 7C), even though liver homogenates revealed that the antibody recognized the rat forms in addition to the mouse proteins (Figure 7D). The latter may be due to differences in baseline levels of those proteins in serum between rats and the other 2 species, but it may also indicate that large elevations of those proteins in serum above baseline are not only specific for injury but for hepatocellular injury and possibly even hepatocellular DILI in particular. Although hepatocellular death was also evident in the BDL rats, there may be something qualitatively or mechanistically different about biomarker release in cholestatic or mixed liver injury, and strictly hepatocellular death. That requires much more research to confirm.
Figure 7.
Ald1a1 and Adh1 do not significantly increase in a rat model of cholestasis. Rats were subjected to either bile duct ligation (BDL) or sham surgery (Sham), then allowed to recover. Blood was collected 21 days after surgery. A, Serum ALT activity. B, H&E-stained liver sections. C, Immunoblots for Aldh1a1 and Adh1in serum. D, Detection of both mouse (M) and rat (R) Aldh1a1 and Adh1 using the same antibody. Total protein is also displayed. Data expressed as mean ± SE for n = 3 rats per group. *p < .05 versus Sham.
DISCUSSION
In this study, we have identified biomarkers that may be useful to confirm liver injury in patients or clinical trial subjects with mild ALT elevations. The current approach to assessment of hepatotoxicity during clinical trials is based on monitoring of serum ALT. There are 2 major guiding principles in regulatory assessment of hepatotoxicity: Hy’s Law and Temple’s Corollary. Hy’s Law states that an elevation in serum ALT accompanied by an increase in bilirubin portends liver failure, whereas Temple’s Corollary states that a drug that causes minor ALT elevations in many patients is more likely to cause severe liver injury in at least a few. Based on that, US FDA guidelines (FDA et al., 2009) state that a drug may be considered hepatotoxic when it causes ALT elevations > 3× the upper limit of normal (ULN) in an “excess” number of subjects, > 5× ULN in a “marked” number of subjects, or > 3× ULN with accompanying elevation of serum bilirubin > 2× ULN in any subjects (so-called “Hy’s Law” cases). However, it has been noted, even in the same guidance document, that some drugs more than meet those criteria but have never caused a single confirmed case of severe DILI (FDA et al., 2009). In fact, it is well known that some drugs can cause ALT elevations in a substantial proportion of users without any other evidence of liver injury or dysfunction. Famously, chronic consumption of therapeutic doses of APAP elevates serum ALT activity above baseline in a large proportion of subjects (Heard et al., 2010, 2014; Watkins et al., 2006) with significant elevations (> 3× ULN) reported in approximately 40% in at least 1 study (Watkins et al., 2006). Similar effects have been observed in subjects treated with tacrine (Gracon et al., 1998; Watkins et al., 1994), heparins (Harrill et al., 2012), and several other drugs.
ALT elevations without evidence of liver necrosis or dysfunction have also been reported in some clinical conditions, and even among the general “healthy” population. For example, anorexia nervosa is associated with large ALT elevations in some patients, but liver necrosis is infrequently observed by biopsy despite evidence of other major changes in cell morphology (Rautou et al., 2008). Furthermore, the prevalence of occult serum ALT elevations in the general population may be higher than would be expected based on the typical approach of using the middle 97.5% of a healthy population to establish reference intervals (Clark et al., 2003). Although nonalcoholic fatty liver disease (NAFLD) likely explains some cases of ALT elevation of unknown etiology, it cannot account for all of them. Studies of blood donors selected for elevated ALT at the time of donation have revealed that only half of random ALT elevations are persistent, which would be expected in most NAFLD patients (the rest being sporadic, transient, and unpredictable) (Friedman et al., 1987; Sampliner et al., 1985).
Previous attempts to identify novel biomarkers that are specific for liver injury have been targeted, focusing on already well-characterized mechanistic biomarkers (McGill and Jaeschke, 2018). For example, Harrill et al. (2012) measured sorbitol dehydrogenase, glutamate dehydrogenase activity, microRNA-122 (miR-122), high mobility group box 1 (HMGB1) protein, and full-length and fragmented keratin 18 (K18) in volunteers with asymptomatic ALT elevations secondary to treatment with heparins. They found that all of those biomarkers were elevated in serum along with ALT. Similar results were observed by the same group in patients treated with cholestyramine (Singhal et al., 2014). There are 2 possible conclusions from those results. First, heparins and cholestyramine may cause real liver injury that was not previously recognized. Second, those biomarkers may simply lack specificity for injury. Unfortunately, it is impossible to know for sure which is correct because liver biopsies to assess tissue damage were not available in those human studies due to the unnecessary risk. Thus, although some of those biomarkers are promising for other purposes (Church et al., 2019; Dear et al., 2018; McGill and Jaeschke, 2018), it is not known if they can be used to differentiate between liver injury and benign ALT changes. In the present study, we adopted an untargeted proteomics approach to avoid limiting our work to known or existing biomarkers, and we combined animal models with easily available histopathology and human samples. Although untargeted proteomics has been applied to identification of DILI biomarkers in the past, it has not been used with the specific goal of identifying markers that can differentiate between injury and benign ALT increases.
The dominant mechanism of ALT release from hepatocytes is probably passive release after cell necrosis, but other mechanisms have been proposed (McGill, 2016). The observation of plasma membrane protrusions off of hepatocytes during ischemia-reperfusion liver injury (Lemasters et al., 1981), and the fact that the cytosolic isoform of aspartate aminotransferase (AST) increases in serum before the mitochondrial isoform (Kamiike et al., 1989) led to the idea that hepatocyte membranes can form blebs that burst and release cytosolic contents into the extracellular space without killing the cell (McGill, 2016). Clearly, increased expression in the liver can also lead to increased ALT release. That is demonstrated by the Dex model (Reagan et al., 2012), but also supported by the fact that treatment with cyclohexamide reduces serum aminotransferases but does not affect mortality in mice with CCl4-induced liver injury (Pappas, 1986, 1989), indicating that some of the increase in aminotransferases was due to increased expression and translation. Furthermore, fibrates, which are known to cause ALT elevations in humans, induce expression of ALT in human hepatocyte cell lines (Edgar et al., 1998). The increased hepatic ALT may then be actively released through exosomes, microvesicles, or simply by normal cell turnover. Additionally, a phenomenon known as a macroenzyme, wherein autoantibodies bind to and stabilize an enzyme in serum, may also lead to accumulation of ALT in serum to high concentrations in rare cases. However, macroALT is very rare compared with macroAST and other macroenzymes (Kulecka et al., 2017; Mifflin et al., 1985). Finally, an interesting mechanism of elevated serum enzymes that has been demonstrated in animals is depletion or impairment of Kupffer cells that normally remove enzymes from circulation (Radi et al., 2011).
It is not clear what mechanism would cause an increase in ALT but not various other liver proteins, such as Adh1 and Aldh1a1, in a model like high-dose Dex. One possibility is that some of the increased hepatic and/or muscle ALT is specifically packaged with cargo targeted for exocytosis and release via exosomes or microvesicles. We are currently investigating that, as well as other possible mechanisms in the animal models used in this study.
The mechanisms of DILI are complicated. There are 2 basic types of DILI, usually referred to as intrinsic and idiosyncratic. Intrinsic DILI is characterized by predictability and a very strong dose-response, whereas idiosyncratic DILI is rare and has a weaker dose-response (though certainly still present) (Uetrecht, 2019). The prevailing view is that idiosyncratic DILI is initiated by an adaptive immune response caused by the interaction of the drug with self-proteins or directly with HLA receptors (Mosedale and Watkins, 2017). The most well-known and widely accepted hypothesis is that the drug or a metabolite covalently modifies self-proteins, leading to identification of self as foreign by the adaptive immune system. However, not all drugs that cause IDILI elicit a clear adaptive immune response, and there are some drugs and metabolites that bind to numerous proteins without causing an adaptive response (eg, APAP), so other hypotheses have been proposed such as mitochondrial dysfunction and bile salt export pump inhibition (Uetrecht, 2019). Unlike idiosyncratic DILI, intrinsic DILI usually occurs before any immune response. The drug or its metabolites directly damage the cells, which then leads to a sterile activation of the innate immune system. The role of sterile inflammation in drug hepatotoxicity is controversial (Jaeschke et al., 2012; Woolbright and Jaeschke, 2018) and probably depends upon the specific toxicant in question.
There are a few weaknesses in the present study. We have used only 1 animal model of benign ALT elevation. However, as far as we know, the Dex model is the only 1 available that definitely causes benign and transient elevations in mice that resemble the low ALT elevations often observed in clinical trials with humans. There is some evidence that serum ALT increases in the methionine choline–deficient mouse model of NAFLD are due to increased hepatic expression (Liu et al., 2009), but that has not yet been clearly demonstrated. Another issue is that we have only been able to obtain a limited number of serum specimens from humans. We are actively engaging in collaborations with clinicians, pharmaceutical companies, and other researchers to obtain samples to address that deficiency in a future study. We hope to expand our results with additional models and additional human samples as we move forward. Finally, we used only semiquantitative (proteomics) and qualitative (immunoblotting) methods in this study. In the future, we plan to validate quantitative methods to measure these biomarkers to facilitate determination of reference intervals and other possible cutoffs for confirmation or rule-out of real liver injury.
CONCLUSIONS
We have identified several biomarkers that are likely specific for liver injury. The most promising markers for further development appear to be ALDH1A1, ADH1, and ASS1. Furthermore, our animal models indicate that those markers may be specific not only for tissue injury but also for hepatocellular injury. Considerable additional work will be needed to determine if they are specific for hepatocellular DILI, in particular. In future studies, we will confirm and expand our results using additional samples from clinical trial subjects, other liver injury and liver disease patients, and other animal models.
DECLARATION OF CONFLICTING INTERESTS
M.R.M. serves as a consultant for Acetaminophen Toxicity Diagnostics, Inc (ATD). ATD had no role in the present study and did not provide funding for it.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank the Department of Laboratory Animal Medicine at UAMS for their excellent animal care and assistance and the Experimental Pathology core laboratory (especially Jennifer D. James, HT[ASCP], HTL, QIHC) for outstanding technical assistance.
FUNDING
This work was supported in part by a Pinnacle Research Award from the American Association for the Study of Liver Diseases (AASLD) Foundation (M.R.M.), by the Marshall University School of Pharmacy Faculty Research Support Program (M.D.H.), by National Institutes of Health (NIH) grants T32 GM106999 (J.H.V. and M.M.C.) and R01 DK102142 (H.J.), and by startup funds provided in part by the UAMS Translational Research Institute supported by NIH grant UL1 TR003107.
The authors certify that all research involving human subjects was done under full compliance with all government policies and the Helsinki Declaration.
Disclaimer: The funding sources had no role in the study design or data interpretation, and this manuscript does not necessarily represent their views.
REFERENCES
- Boehm O., Zur B., Koch A., Tran N., Freyenhagen R., Hartmann M., Zacharowski K. (2007). Clinical chemistry reference database for Wistar rats and C57/BL6 mice. Biol. Chem. 388, 547–554. [DOI] [PubMed] [Google Scholar]
- Church R. J., Kullak-Ublick G. A., Aubrecht J., Bonkovsky H. L., Chalasani N., Fontana R. J., Goepfert J. C., Hackman F., King N. M. P., Kirby S., et al. (2019). Candidate biomarkers for the diagnosis and prognosis of drug-induced liver injury: An international collaborative effort. Hepatology 69, 760–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark J. M., Brancati F. L., Diehl A. M. (2003). The prevalence and etiology of elevated aminotransferase levels in the United States. Am. J. Gastroenterol. 98, 960–967. [DOI] [PubMed] [Google Scholar]
- Dear J. W., Clarke J. I., Francis B., Allen L., Wraight J., Shen J., Dargan P. I., Wood D., Cooper J., Thomas S. H. L., et al. (2018). Risk stratification after paracetamol overdose using mechanistic biomarkers: Results from two prospective cohort studies. Lancet Gastroenterol. Hepatol. 3, 104–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar A. D., Tomkiewicz C., Costet P., Legendre C., Aggerbeck M., Bouguet J., Staels B., Guyomard C., Pineau T., Barouki R. (1998). Fenofibrate modifies transaminase gene expression via a peroxisome proliferator activated receptor alpha-dependent pathway. Toxicol. Lett. 98, 13–23. [DOI] [PubMed] [Google Scholar]
- FDA, CDER, and CBER. (2009). Guidance for Industry Drug-Induced Liver Injury: Premarketing Clinical Evaluation. U.S. Food and Drug Administration https://www.fda.gov/regulatory-information/search-fda-guidance-documents/drug-induced-liver-injury-premarketing-clinical-evaluation. Accessed November 5, 2019.
- Fickert P., Krones E., Pollheimer M. J., Thueringer A., Moustafa T., Silbert D., Halilbasic E., Yang M., Jaeschke H., Stokman G., et al. (2013). Bile acids trigger cholemic nephropathy in common bile-duct-ligated mice. Hepatology 58, 2056–2069. [DOI] [PubMed] [Google Scholar]
- Friedman L. S., Dienstag J. L., Watkins E., Hinkle C. A., Spiers J. A., Rieder S. V., Huggins C. E. (1987). Evaluation of blood donors with elevated serum alanine aminotransferase levels. Ann. Intern. Med. 107, 137–144. [DOI] [PubMed] [Google Scholar]
- Gracon S. I., Knapp M. J., Berghoff W. G., Pierce M., DeJong R., Lobbestael S. J., Symons J., Dombey S. L., Luscombe F. A., Kraemer D. (1998). Safety of tacrine: Clinical trials, treatment IND, and postmarketing experience. Alzheimer Dis. Assoc. Disord. 12, 93–101. [DOI] [PubMed] [Google Scholar]
- Hambuchen M. D., Berquist M. D., Simecka C. M., McGill M. R., Gunnell M. G., Hendrickson H. P., Owens S. M. (2019). Effect of bile duct ligation-induced liver dysfunction on methamphetamine pharmacokinetics and locomotor activity in rats. J. Pharm. Pharm. Sci. 22, 301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrill A. H., Roach J., Fier I., Eaddy J. S., Kurtz C. L., Antoine D. J., Spencer D. M., Kishimoto T. K., Pisetsky D. S., Park B. K., et al. (2012). The effects of heparins on the liver: Application of mechanistic serum biomarkers in a randomized study in healthy volunteers. Clin. Pharmacol. Ther. 92, 214–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard K., Green J. L., Anderson V., Bucher-Bartelson B., Dart R. C. (2014). A randomized, placebo-controlled trial to determine the course of aminotransferase elevation during prolonged acetaminophen administration. BMC Pharmacol. Toxicol. 15, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard K. J., Green J. L., Dart R. C. (2010). Serum alanine aminotransferase elevation during 10 days of acetaminophen use in nondrinkers. Pharmacotherapy 30, 818–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson E. R., Kilroy C., Joslin D. L., Schomaker S. J., Pruimboom-Brees I., Amacher D. E. (2008). The early effects of short-term dexamethasone administration on hepatic and serum alanine aminotransferase in the rat. Drug Chem. Toxicol. 31, 427–445. [DOI] [PubMed] [Google Scholar]
- Jaeschke H., Williams C. D., Ramachandran A., Bajt M. L. (2012). Acetaminophen hepatotoxicity and repair: The role of sterile inflammation and innate immunity. Liver Int. 32, 8–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiike W., Fujikawa M., Koseki M., Sumimura J., Miyata M., Kawashima Y., Wada H., Tagawa K. (1989). Different patterns of leakage of cytosolic and mitochondrial enzymes. Clin. Chim. Acta 185, 265–270. [DOI] [PubMed] [Google Scholar]
- Karmen A., Wróblewski F., LaDue J. S. (1955). Transaminase activity in human blood. J. Clin. Invest. 34, 126–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulecka M., Wierzbicka A., Paziewska A., Mikula M., Habior A., Janczyk W., Dabrowska M., Karczmarski J., Lazniewski M., Ginalski K., et al. (2017). A heterozygous mutation in GOT1 is associated with familial macro-aspartate aminotransferase. J. Hepatol. 67, 1026–1030. [DOI] [PubMed] [Google Scholar]
- LaDue J. S., Wroblewski F. (1956). Serum glutamic pyruvic transaminase SGP-T in hepatic disease: A preliminary report. Ann. Intern. Med. 45, 801–811. [DOI] [PubMed] [Google Scholar]
- Lemasters J. J., Ji S., Thurman R. G. (1981). Centrilobular injury following hypoxia in isolated, perfused rat liver. Science 213, 661–663. [DOI] [PubMed] [Google Scholar]
- Liu R., Pan X., Whitington P. F. (2009). Increased hepatic expression is a major determinant of serum alanine aminotransferase elevation in mice with nonalcoholic steatohepatitis. Liver Int. 29, 337–343. [DOI] [PubMed] [Google Scholar]
- Margolis R. N., Curnow R. T. (1984). Effects of dexamethasone administration on hepatic glycogen synthesis and accumulation in adrenalectomized fasted rats. Endocrinology 115, 625–629. [DOI] [PubMed] [Google Scholar]
- McGill M. R. (2016). The past and present of serum aminotransferases and the future of liver injury biomarkers. EXCLI J. 15, 817–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill M. R., Cao M., Svetlov A., Sharpe M. R., Williams C. D., Curry S. C., Farhood A., Jaeschke H., Svetlov S. I. (2014). Argininosuccinate synthetase as a plasma biomarker of liver injury after acetaminophen overdose in rodents and humans. Biomarkers 19, 222–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill M. R., Jaeschke H. (2018). Biomarkers of drug-induced liver injury: Progress and utility in research, medicine, and regulation. Expert Rev. Mol. Diagn. 18, 797–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill M. R., Sharpe M. R., Williams C. D., Taha M., Curry S. C., Jaeschke H. (2012). The mechanism underlying acetaminophen-induced hepatotoxicity in humans and mice involves mitochondrial damage and nuclear DNA fragmentation. J. Clin. Invest. 122, 1574–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mifflin T. E., Bruns D. E., Wrotnoski U., MacMillan R. H., Stallings R. G., Felder R. A., Herold D. A. (1985). University of Virginia case conference. Macroamylase, macro creatine kinase, and other macroenzymes. Clin. Chem. 31, 1743–1748. [PubMed] [Google Scholar]
- Mosedale, M., Watkins, P.B. (2017) Drug-induced liver injury: Advances in mechanistic understanding that will inform risk management. Clin. Pharmacol. Ther. 101, 469–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. (2003). A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658. [DOI] [PubMed] [Google Scholar]
- Pappas N. J. (1986). Source of increased serum aspartate and alanine aminotransferase: Cycloheximide effect on carbon tetrachloride hepatotoxicity. Clin. Chim. Acta 154, 181–189. [DOI] [PubMed] [Google Scholar]
- Pappas N. J. (1989). Theoretical aspects of enzymes in diagnosis. Why do serum enzymes change in hepatic, myocardial, and other diseases? Clin. Lab. Med. 9, 595–626. [PubMed] [Google Scholar]
- Radi Z. A., Koza-Taylor P. H., Bell R. R., Obert L. A., Runnels H. A., Beebe J. S., Lawton M. P., Sadis S. (2011). Increased serum enzyme levels associated with Kupffer cell reduction with no signs of hepatic or skeletal muscle injury. Am. J. Pathol. 179, 240–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rautou P. E., Cazals-Hatem D., Moreau R., Francoz C., Feldmann G., Lebrec D., Ogier-Denis É., Bedossa P., Valla D., Durand F. (2008). Acute liver cell damage in patients with anorexia nervosa: A possible role of starvation-induced hepatocyte autophagy. Gastroenterology 135, 840–848.e3. [DOI] [PubMed] [Google Scholar]
- Reagan W. J., Yang R. Z., Park S., Goldstein R., Brees D., Gong D. W. (2012). Metabolic adaptive ALT isoenzyme response in livers of C57/BL6 mice treated with dexamethasone. Toxicol. Pathol. 40, 1117–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampliner R. E., Beluk D., Harrow E. J., Rivers S. (1985). The persistence and significance of elevated alanine aminotransferase levels in blood donors. Transfusion 25, 102–104. [DOI] [PubMed] [Google Scholar]
- Schomaker S., Warner R., Bock J., Johnson K., Potter D., Van Winkle J., Aubrecht J. (2013). Assessment of emerging biomarkers of liver injury in human subjects. Toxicol. Sci. 132, 276–283. [DOI] [PubMed] [Google Scholar]
- Singhal R., Harrill A. H., Menguy-Vacheron F., Jayyosi Z., Benzerdjeb H., Watkins P. B. (2014). Benign elevations in serum aminotransferases and biomarkers of hepatotoxicity in healthy volunteers treated with cholestyramine. BMC Pharmacol. Toxicol. 15, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetrecht J. (2019). Mechanistic studies of idiosyncratic DILI: Clinical implications. Front. Pharmacol. 10, 837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins P. B., Kaplowitz N., Slattery J. T., Colonese C. R., Colucci S. V., Stewart P. W., Harris S. C. (2006). Aminotransferase elevations in healthy adults receiving 4 grams of acetaminophen daily: A randomized controlled trial. J. Am. Med. Assoc. 296, 87–93. [DOI] [PubMed] [Google Scholar]
- Watkins P. B., Zimmerman H. J., Knapp M. J., Gracon S. I., Lewis K. W. (1994). Hepatotoxic effects of tacrine administration in patients with Alzheimer’s disease. JAMA 271, 992–998. [PubMed] [Google Scholar]
- Woolbright B. L., Antoine D. J., Jenkins R. E., Bajt M. L., Park B. K., Jaeschke H. (2013). Plasma biomarkers of liver injury and inflammation demonstrate a lack of apoptosis during obstructive cholestasis in mice. Toxicol. Appl. Pharmacol. 273, 524–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolbright B. L., Jaeschke H. (2018). Mechanisms of inflammatory liver injury and drug-induced hepatotoxicity. Curr. Pharmacol. Rep. 4, 346–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.