ABSTRACT
There is an increasing interest in studying bacterial-fungal interactions (BFIs), also the interactions of Pleurotus ostreatus, a model white-rot fungus and important cultivated mushroom. In Europe, P. ostreatus is produced on a wheat straw-based substrate with a characteristic bacterial community, where P. ostreatus is exposed to the microbiome during substrate colonisation. This study investigated how the bacterial community structure was affected by the introduction of P. ostreatus into the mature substrate. Based on the results obtained, the effect of the presence and absence of this microbiome on P. ostreatus production in an experimental cultivation setup was determined. 16S rRNA gene-based terminal restriction fragment length polymorphism (T-RFLP) and amplicon sequencing revealed a definite succession of the microbiome during substrate colonisation and fruiting body production: a sharp decrease in relative abundance of Thermus spp. and Actinobacteria, and the increasing dominance of Bacillales and Halomonas spp. The introduced experimental cultivation setup proved the protective role of the microbial community against competing fungi without affecting P. ostreatus growth. We could also demonstrate that this effect could be attributed to both living microbes and their secreted metabolites. These findings highlight the importance of bacterial-fungal interactions during mushroom production.
Keywords: Pleurotus ostreatus, T-RFLP, amplicon sequencing, bacterial community structure, bacterial-fungal interactions, succession and spatial heterogeneity
Bacterial community composition has changed significantly during Pleurotus ostreatus production, and this microbiome has a role in inhibiting competing microbes without affecting P. ostreatus growth.
INTRODUCTION
The study of bacterial-fungal interactions (BFIs) has become increasingly important. Of particular importance is that BFIs can be considered as one of the simplest cross-kingdom (prokaryotic-eukaryotic models) interactions whose interplay has a major role in a number of fields, including nutrient cycling, host productivity, biocontrol, food production, and bioremediation (Deveau et al. 2018). One of the more specialised areas is wood colonisation and degradation (Johnston, Boddy and Weightman 2016), where white-rot fungi involved in the process may interact with bacteria in a variety of ways. Bacterial presence can have negative effects on fungi, as bacteria can rapidly consume easily degradable substrates (Messner et al. 2003); however, some bacterial products can serve as fungal growth factors (Clausen 1996). On the other hand, some white-rot fungi, such as Hypholoma fasciculare and Resinicium bicolor, decreased the number of wood-colonising bacteria, while the number of bacteria adhering to exploratory hyphae in the surrounding soil increased (Folman et al. 2008). Hervé et al. (2016) reported that Phanerochaete chrysosporium selected certain bacteria in its mycosphere during wood decay, which are less inhibitory to fungal growth and are involved in cellulose and hemicellulose degradation, and iron mobilisation. It is also possible that fungal species do not select bacteria directly, but rather indirectly through changing conditions during wood decay (Kielak et al. 2016).
Pleurotus ostreatus (oyster mushroom), a model organism for white-rot fungi, is the second most important cultivated edible mushroom in the world (Royse, Baars and Tan 2017) and has a remarkable role in bioremediation (Stella et al. 2017). Several studies have reported on the interactions between bacteria and P. ostreatus. Pleurotus ostreatus was reported to lyse bacterial colonies under low nutrient conditions (Barron 1988), decrease soil bacterial colony counts by several orders of magnitude (Lang et al. 1997), and kill most indigenous soil bacteria expected to take part in polycyclic aromatic hydrocarbon degradation (Gramss, Voigt and Kirsche 1999; Andersson et al. 2003). The antioxidant and antimicrobial (e.g. volatile compound production) properties of P. ostreatus (Beltran-Garcia, Estarron-Espinosa and Ogura 1997; Martínez et al. 2015; Smolskaite, Venskutonis and Talou 2015) may contribute to this impact. However, there are some examples of positive effects. Cho et al. (2003) demonstrated that fluorescent pseudomonads promoted the fruiting body formation of P. ostreatus on agar plates. Additionally, P. ostreatus appeared to stimulate bacterial growth in some cases (Stella et al. 2017), whereas Adamski and Pietr (2019) revealed at least 34 different bacterial genera living in association with eight P. ostreatus strains. Some of them had laccase-like genes, so these microorganisms could support the biodegradative capacity of P. ostreatus.
Notwithstanding the examples above, there is little information on the interactions between bacteria and P. ostreatus during mushroom production. The initial step of mushroom cultivation is the so-called substrate production. In Europe, a wheat straw-based substrate, prepared via partial composting, pasteurisation, and conditioning, is used (Rühl and Kües 2007; Vajna et al. 2012). First, the straw is moistened, and easily degradable substrates are consumed during partial composting. The elimination of easily degradable substrates hinders substrate contamination by competing microbes with no complete lignocellulose-degrading enzyme set during substrate colonisation of P. ostreatus. Also the accessibility of cellulose is increasing (Vajna et al. 2010). Second, pasteurisation eliminates competing and pathogenic microbes of P. ostreatus, and third, during conditioning, the substrate is colonised with the surviving non-competing and non-pathogenic bacteria (Ororbia and Núñez 2001; Oei 2016). We previously described microbial succession during substrate preparation (Vajna et al. 2010, 2012). Fungal CFU (colony-forming unit) values reached their maxima during partial composting with species from genera Thermomyces, Myceliophthora, and Rhizomucor having versatile thermostable cellulolytic enzymes, suggesting their role in the initial straw polysaccharide decomposition (Vajna et al. 2010). Using sequence-aided terminal restriction fragment length polymorphism (T-RFLP), it was revealed that Proteobacteria (Pseudomonas and Sphingomonas spp.) dominated in the first phase of substrate preparation, Firmicutes (Bacillus, Geobacillus, and Ureibacillus spp.), Pseudoxanthomonas, and Thermobispora spp. at the end of partial composting, and several genera of Actinobacteria, Thermus spp., and Firmicutes (Bacillus, Geobacillus, Thermobacillus, and Ureibacillus spp.) in the mature substrate (Vajna et al. 2012).
P. ostreatus is exposed to this microbial community after spawning the mature substrate during substrate colonisation. However, it is not completely understood how microbial community structure changes during this process and what kind of interactions develop among microbial community members and P. ostreatus. Our first aim was to test using molecular fingerprint techniques and amplicon sequencing whether known bacterial community structure of the mature mushroom substrate was transformed during P. ostreatus production. Subsequently, a laboratory-scale experimental cultivation setup was established in which the effect of the presence and absence of microbiome on P. ostreatus cultivation was investigated. With the help of this system we addressed the following questions: (i) if microbes have any effect on P. ostreatus growth (ii) if there is an effect, whether it is attributed to the entire microbiome or to its extracellular metabolites (iii) a single indigenous bacterial strain could have an antagonistic effect on P. ostreatus? To monitor fungal functioning in the model, activity patterns of lignocellulose-degrading enzymes and hyphal growth rate were used as indicators.
MATERIALS AND METHODS
Bacterial community composition during large-scale production
Pleurotus ostreatus production and sampling
To study bacterial succession, samples derived from a large-scale P. ostreatus producer (Pilze-Nagy Ltd., Kecskemét, Hungary) were used as described previously (Bánfi et al. 2015). Briefly, mature mushroom substrate (after partial composting, pasteurisation, and conditioning) was inoculated with P. ostreatus HK35 spawn and filled into substrate blocks (22–25 kg; 55 × 35 × 20 cm). Blocks were incubated for substrate colonisation and subsequent fruiting body production in a temperature-, humidity-, and light-controlled production house. The cultivation cycle consisted of three main stages. First, P. ostreatus mycelia colonised the substrate blocks (24–26°C, 75–80% relative humidity [RH]); second, fruiting body production was induced (12–20°C, 85–90% RH); and third, mature fruiting bodies were produced and harvested (10–13°C, 75–80% RH). For further substrate utilisation and mushroom harvesting, a second cultivation cycle was applied (Fig. 1). The sampling period was 11 weeks from spawning (day 1) until the end of the second flush (day 71). Five mushroom substrate blocks (‘A’ to ‘E’) at different locations in a 250 m2 production house were sampled: 30–30 g substrate was collected from surface layers (0–10 cm) of each block at three random regions to avoid the disturbance of the blocks. On day 71, blocks were cut in half, and the inner parts were sampled. Samples were stored at -20°C until further analysis.
Figure 1.

Stages of the P. ostreatus cultivation process and the 11 sampling days. Sampling days are denoted with numbers and arrows on the horizontal axis. ‘NGS’ shows samples processed for amplicon sequencing. Stage 1: 1/1. vegetative growth; 1/2. fruiting body induction; 1/3. first flush and harvest; Stage 2: 2/1. second vegetative growth; 2/2. second fruiting body induction; 2/3. second flush and harvest; columns: biological efficiency/day in the production house; black line: temperature during the investigation period. Biological efficiency was calculated as ratio of weight of fresh fruiting bodies per weight of dried substrate on day 1. Figure modified from Bánfi et al. (2015) with the permission of Fungal Biology.
T-RFLP analysis
DNA was extracted from the samples after grinding in liquid nitrogen as described by Vajna et al. (2010) using a G-spin™ Total DNA Extraction Mini Kit (iNtRON Biotechnology, Seongnam, South Korea). PCR reactions (using HEX-labelled 27F [5’-AGA GTT TGA TCM TGG CTC AG-3’] and 534R [5’-ATT ACC GGG GCT GCT-3’] 16S rDNA primers, Lane 1991), enzymatic digestion, purification of enzymatic digests, and electrophoresis of labelled fragments were carried out as previously described (Székely et al. 2009).
T-RFLP chromatograms were analysed using GeneMapper® Software v3.7 (Applied Biosystems, Foster City, CA, USA). Only T-RFs longer than 50 bp were used. Further data processing was performed according to an updated script of Abdo et al. (2006) (Supplementary Data). The following parameters were applied: noise filtration based on standard deviation (multiplier = 4) of peak area, and T-RFs alignment with a 1 bp clustering threshold. The resulting alignment was compared to raw chromatograms and manually corrected if needed. For normalisation, relative abundance of each detected T-RF within a given T-RFLP profile was calculated. To obtain a more robust result, the data matrix for AluI and Hin6I enzymes was combined.
454 Pyrosequencing
To identify bacterial community members, selected samples (1–1 sample from blocks ‘A’ and ‘B’ at day 1, 30, and 57 corresponding to initial phase, first, and second flush) were subjected to pyrosequencing analysis. 16S rDNA library preparation, using the same primer pair as for T-RFLP, and sequencing were performed as described by Felföldi et al. (2015). Resulting sequence reads were processed as described by Szabó et al. (2017) using the Mothur v1.35 software (Schloss et al. 2009), SINA v1.2.11 aligner tool (Pruesse, Peplies and Glöckner 2012), and the ARB-SILVA SSU NR 99 reference database—SILVA Release 123 (Quast et al. 2013). Raw sequence reads are available in the NCBI Sequence Read Archive under BioProject ID PRJNA630385.
Phylogenetic composition of bacterial communities was visualised with the metacoder package of R software (R Core Team 2020; the used scripts are available as Supplementary Material).
Experimental cultivation setup
Preparation of the cultivation setup
To study the effect of the presence and absence of the microbiome, a P. ostreatus cultivation setup was established. 150 g of unspawned mature P. ostreatus substrate derived from the same production series as used for the large-scale production above were filled into tubes (30 cm, Ø 4 cm) formed from transparent plastic sheets (0.1 mm thick). Substrate density of the model tubes (∼0.6 kg/L) was the same as that in large-scale substrate blocks. Tube ends were covered with plastic foil. For gas exchange and fruiting body development, 15 holes (3 mm) were drilled evenly into the tubes (Fig. S1, Supporting Information).
Applied treatments are summarised in Fig. 2A. About 50 substrate-filled tubes were autoclaved at 121°C for 40 min. (i) Ten autoclaved tubes were used without any treatment, designated as ‘A’ namely ‘autoclaved’. As autoclaving not only kills microbes but may also change the physicochemical structure of substrates (Berns et al. 2008), different inoculation treatments were used. (ii) First, a microbial suspension was prepared by adding 100 g of the same unspawned mature P. ostreatus substrate to 800 mL dechlorinated (0.1% w/v thiosulphate containing) sterilised tap water and shaken at 150 rpm at 28°C for 90 min. Ten autoclaved tubes were inoculated with the resulting microbial suspension, and each tube was injected under aseptic conditions with 10 mL of suspension evenly distributed through 15 holes (∼0.66 mL suspension/hole). The 10 tubes were designated as ‘AM’, namely ‘autoclaved’ substrate, and inoculated with ‘microbial suspension’. (iii) To separate the effect of cells from their metabolites, cell-free extracts were prepared. Adequate volume of microbial suspension was filtered through a 0.2 µm cellulose nitrate membrane filter (Millipore, Billerica, MA, USA) to remove microbial cells. Then 10 mL of cell-free filtrate were used to inoculate 10 autoclaved tubes each in the same way as described above. Tubes were designated as ‘AF’, namely ‘autoclaved’ substrate and inoculated with cell-free ‘filtrate’ of microbial suspension. (iv) A bacterial strain collection from a mature P. ostreatus substrate was established previously. Strains were tested in simple cross-streak antagonism experiments with P. ostreatus on malt extract peptone agar (MEPA, DSMZ Medium 90) plates. Some of them showed inhibitory effects on P. ostreatus hyphal growth. To test the effect of a single antagonistic strain on P. ostreatus in our cultivation model, a suspension from one of the mentioned antagonistic strains (Bacillus sp. Sz55, 16S rRNA gene sequence GenBank accession number MK908096) was prepared in dechlorinated sterilised tap water with 0.04 OD600 density. Further, 10 mL of this suspension was inoculated into another 10 autoclaved tubes each, in the same way as described above. Tubes were designated as ‘AA’, namely ‘autoclaved’ and inoculated with the suspension of an ‘antagonistic’ bacterium. (v) To have an inoculation control, each of the 10 autoclaved tubes were injected with 10 mL of dechlorinated sterilised tap water, in the same way as above. These tubes were designated as ‘AW’, namely ‘autoclaved’ and inoculated with sterile ‘water’. (vi) Finally, 10 tubes designated as ‘N’ were not autoclaved and contained normal, untreated mature P. ostreatus substrate. The fungal inoculum Pleurotus ostreatus strain HK35 (obtained from Sylvan Hungary Ltd.; Dunaharaszti, Hungary) was grown on MEPA plates at 28°C for one week. Thereafter, one end of cultivation model tubes was inoculated with a 2 × 2 cm hypha-covered MEPA culture block (Fig. 2A). Further, additional control tubes not inoculated with P. ostreatus were produced, three tubes for treatment ‘A’, 10 for ‘AM’, 10 for ‘AA’, and two for ‘N’.
Figure 2.
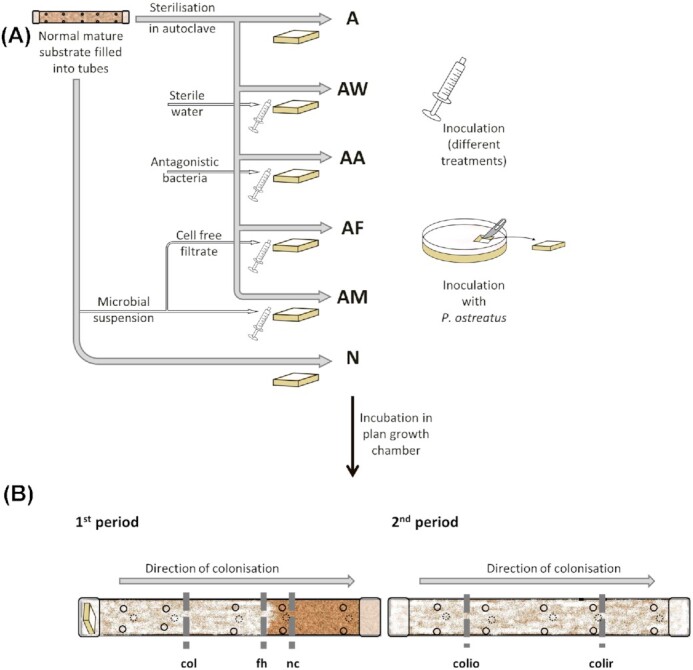
Scheme of applied treatments and sampling regions in the experimental cultivation setup. (A), Scheme of applied treatments (see details in text and Supplementary Fig. 1) A: autoclaved; AW: autoclaved and sterile water injected; AA: autoclaved and inoculated with suspension of an antagonistic bacterium; AF: autoclaved and inoculated with cell-free filtrate of microbial suspension; AM: autoclaved and inoculated with microbial suspension; N: normal, untreated, not autoclaved. (B), Sampling regions at the end of the 1st and 2nd periods of experiment. ‘nc’: noncolonised region, ‘fh’: region of front hyphae, ‘col’: three-week colonised region sampled before induction of the fruiting bodies, ‘colio’: older colonised region sampled after induction of fruiting bodies, ‘colir’: recently colonised region sampled after induction of fruiting bodies.
Incubation and sampling of the tubes
All cultivation model tubes were placed in a plant growth chamber (Sanyo MLR-352H, Japan). In the 1st period, they were incubated for four weeks at 26°C and 90% RH without light, with partial aeration. Hyphal colonisation speed was recorded weekly by measuring the hyphal front distance from the inoculated end of the tubes. The position of the hyphal front was clearly visible by naked eye (Fig. S1, Supporting Information). Despite keeping the tubes in a ‘clean environment’, mould infection was observed during incubation around the tube holes. Contamination of the tubes was recorded at the end of the 1st period (at the end of the 4th week) by counting the number of contaminated and uncontaminated holes. To have colonised and still noncolonised parts in every tube, half of the tubes (i.e. 5 tubes from each of the six treatments: A, AM, AF, AA, AW, and N) were opened at the end of the 1st period, when one-third of every tube was still noncolonised (Fig. 2B). Additionally, half of the control tubes (not inoculated with P. ostreatus) were opened. Samples (10 g of each) were taken from three regions (non-colonised, front hyphae regions, and three-week colonised regions, Fig. 2B) of 30 P. ostreatus inoculated tubes and from two parts of control tubes. Samples were processed immediately for enzyme assays. In the 2nd period, the remaining tubes were incubated under the same conditions for two weeks until complete colonisation of the tubes. The temperature was then decreased by 1.5°C/12 h until 16°C. At this temperature, fruiting body formation was induced by 12-hour light-dark cycle for two weeks. At the end of the 2nd period, the remaining 5 tubes from each of the six treatments and the other half of the control tubes were opened and 10 g of each samples were taken from two regions (recently and longer-colonized regions, Fig. 2B) of the 30 P. ostreatus inoculated tubes, and from two parts of the control tubes. Samples were processed immediately for enzyme assays.
Enzyme assays
Samples from model tubes were subjected to enzyme analysis according to Bánfi et al. (2015). Briefly, lignocellulolytic enzymes were extracted from 3 g of wet samples in 48 mL of 50 mM phosphate buffer (pH 7) with orbital shaking (100 rpm min–1) at 8°C for 2 h. The extract was filtered through a VWR Collection (USA) no. 413 filter paper. The filtrate was used for enzyme activity measurements. Laccase activity was determined using ABTS (2,2’-azino-bis[3-ethylbenzothiazoline-6-sulfonic acid]; Sigma-Aldrich, USA), and manganese peroxidase (MnP) with DMAB-MBTH (3,3-dimethylaminobenzoic acid; Sigma-Aldrich, USA; 3-Methyl-2-benzothiazolinone hydrazone; Sigma-Aldrich, USA) as a substrate. Cellobiohydrolase, β-glucosidase, 1,4-β-xylosidase, and N-acetyl-β-glucosaminidase activities were assayed using p-nitrophenyl substrates (p-nitrophenyl-β-D-cellobioside [pNPC], p-nitrophenyl-β-D-glucoside [pNPG], p-nitrophenyl-β-D-xyloside [pNPX], and p-nitrophenyl-N-acetyl-β-D-glucosaminide [pNPN], respectively; Sigma-Aldrich, USA; Glycosynth, UK). Endo-β-glucanase (cellulase) and endo-1,4-β-xylanase activities were analysed using chromogenic substrates AZO-CM-CELLULOSE and AZO-XYLAN (Megazyme, Ireland), respectively. Spectrophotometric measurements were performed using a microplate reader (Sunrise, Tecan, Switzerland) and a UV-VIS spectrophotometer (Lambda35, PerkinElmer, USA). One unit of enzyme activity was defined as the amount of enzyme releasing 1 μmol of reaction product per minute. Results were calculated as units per gram of dry mass of the P. ostreatus substrate. Dry matter (DM) content was determined by drying the substrate samples at 105°C for 24 h.
Isolation and identification of infectious fungi
From visually different infected parts of the tubes, pure cultures were isolated on MEPA medium. For DNA extraction, a loopful of fungal culture was added to 100 µL 0.1 M Tris buffer (pH 8) containing 100 µL sterile glass beads (0.1 mm). The suspension was shaken in a mixer mill (Mixer Mill MM301, Retsch, Haan, Germany) at 30 Hz for 1 min. Crude lysates were heated at 98°C for 5 min and then centrifuged at 10 000 g for 5 min. The supernatant was used as a template for amplification of the fungal nrDNA ITS region with primers ITS1F forward [5’-TCC GTA GGT GAA CCT GCG G-3’; Gardes and Bruns 1993] and ITS4 reverse [5’-TCC TCC GCT TAT TGATAT GC-3’, White et al. 1990]. Sanger sequencing of the amplicons was carried out by LGC Genomics (Berlin, Germany). Sequences were deposited in GenBank under accession numbers MW365370-MW365376 and were compared with sequences in public databases using BLASTn searches (Altschul et al. 1990).
Statistical analyses
Statistical analyses were performed using R software (R Core Team, 2020; the used scripts are available as Supplementary Material).
During interpreting bacterial T-RFLP dataset, the following analyses were carried out: (i) For comparing within and among block heterogeneity at selected sampling dates (day 1, 30, and 57), permutational multivariate analysis of variance using Bray-Curtis distances was carried out. Based on this result (see Supplementary Material), only one parallel sample from each block (11 sampling dates, one-one sample from the five mushroom blocks at each date) was used. (ii) Succession of the bacterial community was visualised using non-metric multidimensional scaling (NMDS) using Bray-Curtis distances. (iii) To test whether a priori grouping of samples according to sampling dates were separated significantly from each other, combined cluster and discriminant analysis (CCDA, Kovács et al. 2014; Bánfi et al. 2015) was applied. (iv) To test whether surface and inner samples of the same block at day 71 were different, two groups were compared with permutational multivariate analysis of variance using Bray–Curtis distances. To reveal how groups of inner samples were grouped with other samples, CCDA and NMDS with Bray-Curtis distances were used. Based on this result (see Supplementary Material) samples from the surface layers adequately represent bacterial community changes in the whole substrate block. (v) Shannon diversity indices were calculated. (vi) Spatial and temporal changes in community composition were calculated. Spatial heterogeneity was characterised on the cultivation block level using Bray–Curtis distances among samples within one sampling date. Weekly temporal changes were calculated using Bray-Curtis distances among samples from consecutive sampling dates. Differences among groups were tested using Kruskal-Wallis and Nemenyi post-hoc tests. (vii) To test the effect of P. ostreatus lignocellulose-degrading enzymes on bacterial community composition, enzymatic activities of the same samples were fitted as vectors onto the NMDS ordination, and the significance of fittings was tested with random permutations. Enzyme activity data was derived from Bánfi et al. (2015). (viii) Finally, for comparison, an already published T-RFLP dataset (Vajna et al. 2012) from the first part of the P. ostreatus mushroom production (Supplementary Fig. 2) was involved. That part is the so-called substrate production, from where the mature substrate used in this study was also gained.
For the cultivation model, the following statistical analyses were carried out: (i) Effect of treatments (A, AM, AF, AA, AW, and N) on the speed of hyphal colonisation was tested with a general linear model (LM) using generalised least squares with contamination of the tubes as covariant. The effect of the treatments on substrate contamination was also tested the same way. Based on the results of the later model (see Results and Discussion), a new explanatory variable was introduced as ‘treatment type’ with two categories for treatments with higher (A, AA, and AW) and lower (AM, AF, and N) contamination values. (ii) The effects of different treatments on enzymatic activities were tested separately for the two periods, as in the 1st and the 2nd periods in our cultivation model, different sets of model tubes were used due to destructive sampling. To create new summary variables from enzymatic activities, principal component analysis (PCA) using variance-covariance matrix was carried out on range standardised values separately for samples from 1st and 2nd periods. Only principal components, supported by the broken stick model, were used in further tests. For each principal component, enzyme activities with loadings above 0.53 or below -0.53 were considered for interpretation (Tabachnick and Fidell 2013). Effects of treatment, region, contamination, and treatment type on these new summary variables and with tube number as random effects was tested with general linear mixed-effects models (LMM). Random effect was used to control non-independence of different regions sampled from the same tubes. The best models were found by automated model selection using combinations of fixed effect terms in original models and comparing them based on the Akaike information criteria. For both cases (i and ii), adjusted means were calculated, and post-hoc Tukey's tests were performed.
RESULTS AND DISCUSSION
Bacterial community composition during large-scale production
Bacterial community succession based on T-RFLP
In the first part of the study, samples were collected from a large-scale P. ostreatus producer (11 sampling days, five mushroom blocks with three parallel samples; Fig. 1). 16S rRNA encoding gene based T-RFLP fingerprints of the bacterial community showed a definite succession during 11 weeks of cultivation with the following significantly different groups of samples: day 1, day 8, day 12, day 22–30-36–43, and day 50–57-64–71 (Fig. 3A and Fig. S3, Supporting Information). Bacterial community diversity decreased slightly, based on Shannon indices (Fig. S4, Supporting Information). Spatial heterogeneity of samples within one sampling day was similar during the whole investigated period, whereas weekly shifts (Bray–Curtis distances among samples from consecutive sampling dates) decreased significantly during mushroom production (Fig. S5, Supporting Information), denoting the slowing of changes in the bacterial community. These rates of changes are easier to interpret when compared to the first part of P. ostreatus production, the so called substrate production (Fig. S2, Supporting Information) described by Vajna et al. (2012), where only during 12 days, but under more varied circumstances a much more remarkable change in the bacterial community was observed (Fig. 3C).
Figure 3.
Bacterial community composition changes according to 16S rRNA gene based T-RFLP fingerprints during substrate colonisation and fruiting body production of P. ostreatus. (A) NMDS ordination using Bray-Curtis distances for samples. Different small symbols denote samples from different mushroom blocks (A–E). Filling of symbols is lightened from black to white in parallel with sampling date (day 1 to day 71). Blue filling shows samples subjected to pyrosequencing. Ellipses mark weekly mean ± SD area based on samples’ NMDS coordinates; whereas, arrows point from weekn mean to weekn+1 mean. Filling of ellipses and their respective sampling date label denote significantly different groups of samples according to CCDA method (Supplementary Fig. 3). (B) The same NMDS ordination as in Fig. 3A though only ellipses for each week are plotted. Red arrows show significantly fitted (p < 0.05) P. ostreatus lignocellulose-degrading enzyme activities of the same samples. Enzyme activity data are derived from Bánfi et al. (2015). Enzyme names are abbreviated: β-gluc- β-glucosidase, Cbh- cellobiohydrolase, Chit- N-acetyl-β-glucosaminidase, Lcc- laccase, and MnP- manganese peroxidase. (C) NMDS ordination comparing bacterial community changes in P. ostreatus substrate preparation production series (data derived from Vajna et al. 2012) and samples from the present study. Substrate preparation is the first part of the P. ostreatus mushroom production, where the mature substrate also used in this study was produced. Ellipses correspond to mean ± SD area as in the previous parts. Stages 1, 3 and 7 in darkening green colours correspond to (S1) chopped, wetted wheat straw, (S3) end of partial composting, and (S7) mature substrate (Fig. S2, Supporting Information). For samples from the present study, only weekly ellipses of Fig. 3A are plotted.
Substrate colonisation and fruiting body formation by P. ostreatus had the main influence on the bacterial community. Colonising hyphae produce high amounts of laccases, a known phenomenon when P. ostreatus interacts with other microorganisms (Baldrian 2004; Velázquez-Cedeño et al. 2008; Janusz et al. 2020). This could partially be responsible for changes in bacterial community composition in the first two weeks (Fig. 3B), although it is unclear whether laccases have a direct inhibitory effect on microbial growth (Baldrian 2004). Periodically decreasing temperature, ∼25°C during substrate colonisation and 12–20°C during fruiting body production (Bánfi et al. 2015), and a rapid drop in pH from ∼7 to ∼5 (Viziteu 2004; De Boer et al. 2010) can have a strong effect on the bacterial community. Change of pH is a known process during wood degradation by which fungi control the environment and indirectly bacterial diversity (Johnston et al. 2019). From the third week, high activity of manganese peroxidase and different cellulases were significantly correlated with the transformation of bacterial community structure (Fig. 3B).
Bacterial community composition based on 454 pyrosequencing data
The 16S rRNA gene amplicon sequencing of 6 selected samples (1–1 sample from blocks ‘A’ and ‘B’ at day 1, 30 and 57) resulted in a total of 39094 high-quality reads classified within the domain Bacteria. Sequences were randomly subsampled according to the lowest number of sequences in sample block ‘A’ at day 30 (2994 sequences). Sequencing efficiency was high, with Good's coverage values ranging from 0.979 to 0.994. Observed number of OTUs and Shannon diversity indices showed a slight decrease in both blocks (Fig. 4, Table S1, Supporting Information).
Figure 4.
Bacterial diversity based on 16S rRNA gene amplicon sequencing during substrate colonisation and fruiting body production of P. ostreatus(A) Observed number of OTUs and (B) Shannon diversity calculated from the OTU (defined at 97% similarity level) numbers.
Dominant trends in the bacterial community change during substrate colonisation and fruiting body production were the disappearance of Thermus spp., probably due to lower temperatures, a sharp decrease in Actinobacteria relative abundance, and an increase in members of the order Bacillales and Halomonas spp. (Fig. 5; Table S2, Supporting Information). The number of unclassified sequences (order level) decreased from 9%–12% to 1%–2% (Table S2, Supporting Information). Most of the dominant Bacillales genera (Thermobacillus and Ureibacillus) have already been described from the mature substrate of P. ostreatus production (Vajna et al. 2012). Bacillus, Paenibacillus, and Lysinibacillus spp. were also the main isolates identified from the P. ostreatus substrate described by Suarez et al. (2019). Many Bacillus spp. produce antifungal peptides (e.g. fengycin) with selective antagonistic ability as they inhibit green-mould disease causing Trichoderma spp. growth without affecting P. ostreatus (Kim et al. 2008; Nagy et al. 2012; Liu et al. 2015). Paenibacillus polymyxa also supports the competitiveness of P. ostreatus (Velázquez-Cedeño et al. 2008). Moreover, the thick gram-positive cell wall and spore forming potential of these Bacillales may enable their survival. Stella et al. (2017) reported a similar shift due to P. ostreatus toward the dominance of Firmicutes and disappearance of Actinobacteria in a polychlorinated biphenyl degrading soil community. Also in Agaricus bisporus production increasing ratio of gram-positive to gram-negative taxa was observed, suggesting selective suppression of different bacteria (Vos et al. 2017). The genus Halomonas contains moderately halophilic bacteria with broad pH (5–10) and temperature (10–45°C) tolerance (De La Haba et al. 2014), a compact cell wall (Vreeland, Anderson and Murray 1984), and exopolysaccharide production (Mata et al. 2006). Salt concentration of P. ostreatus compost was lower than Agaricus compost (Jasińska 2018), but the lower temperature (5–15°C) after the first fruiting body induction at day 22 might support their growth. Moreover, Halomonas spp. are highly abundant during denitrification and lignocellulose degradation in salty environments (Cortes-Tolalpa et al. 2018; Deng et al. 2020; Zhong et al. 2020), which might contribute to lignocellulose degradation during mushroom production.
Figure 5.
Phylogenetic composition of bacterial communities based on 16S rRNA gene amplicon sequencing during substrate colonisation and fruiting body production of P. ostreatus. Each node represents a taxon from kingdom to family. Tree topology is the same for every sample, whereas colour of the nodes goes from blue (100% relative abundance) through red to yellow (0% relative abundance) according to taxa abundance in each sample. Small numbers on the edges give relative abundance of the taxa, which follow the given edge. Only OTUs having >1% relative abundance at least in one sample were used for calculations. ‘unc’ stands for unclassified bacteria. The original size figure also complemented with genus level is available as Fig. S6 (Supporting Information).
Effect of microbial presence on the cultivation of Pleurotus ostreatus
Monitoring of large-scale production provided an overview of the bacterial community composition, but the role and function of this community was unknown. The P. ostreatus cultivation model was developed to study the effect of the presence and absence of microbes.
Growth of P. ostreatus and contamination of the model tubes
The different applied treatments A, AW, AA, AF, AM and N (Fig. 2A) had a significant influence on colonisation speed (LM: F5,54 = 6.78, P<0.0001). Although P. ostreatus can be produced on a sterilised substrate (Kwon 2004; Oei 2016), P. ostreatus grew significantly slower in autoclaved substrate with no additional treatments (A) and in autoclaved inoculation control (AW), than in normal (N) tubes (Fig. 6A, see Supplementary data). During weekly checking of the model system, mould infection was detected around the tube holes. According to nrDNA ITS sequences, those fungi belonged to the genera Aspergillus and Penicillium. Although the ITS region is not suitable for precise species identification in these groups, we assumed that three Aspergillus spp. and two Penicillium spp. were isolated. Moulds in both genera are common indoor fungi and can be found as competitors during mushroom production (Saǧir and Yildiz 2004; Verdier et al. 2014; Oei 2016) as well. This contamination was significantly treatment-dependent (LM: F5,54 = 34.56, p<0.0001; Fig. 6B, see Supplementary Data). Normal, non-treated tubes (N), and autoclaved tubes inoculated with microbial suspension or its cell-free filtrate (AM and AF) showed significantly lower infections than tubes with no microbiome or microbial metabolites (A, AW, and AA). It was already observed by Gyurkó (1978) that microbes have a protective role in P. ostreatus production, and bacteria may produce antifungal compounds against moulds in Agaricus mushroom production substrate (Liu et al. 2015). Velázquez-Cedeño et al. (2004) showed that on a straw-based cultivation substrate, bacteria reduced the competitive ability of Trichoderma longibrachiatum against P. ostreatus. In a later study (Velázquez-Cedeño et al. 2008), Bacillus spp. and Paenibacillus polymyxa were identified as the taxa responsible for these effects. This finding is consistent with our findings, as large amounts of Firmicutes bacteria were found in the mature substrate (Fig. 5, day 1 samples). Our results also unambiguously showed that this protective role was developed not only by the microbial community, but also by their secreted metabolites. Antifungal activity of microbe cell-free supernatants has been observed in several cases (Shehata et al. 2019; Xu et al. 2019), but this is the first time that it has been described during P. ostreatus production. Another important outcome is that inoculation with a single antagonistic Bacillus sp. (AA) neither reduced hyphal growth nor prevented contamination of the tubes.
Figure 6.
Effects of different treatments (for codes see Fig. 2A) on the (A) speed of P. ostreatus hyphal colonisation in the experimental cultivation setup and (B) contamination of the tubes after the end of the 1st period. Values show estimated marginal means ± standard error calculated from the linear model. Upper letters show significantly different groups based on Tukey's post-hoc test.
Enzymatic activity of P. ostreatus
In the case of the 25 uninoculated control tubes and non-colonized region of the inoculated tubes, higher enzymatic activities (namely 1,4-β-xylosidase and endo-1,4-β-xylanase, Fig. S7, Supporting Information) were measured only in tubes characterised by higher contamination ratios (A, AW, and AA), which can be explained by the hemicellulose degrading capacity of Aspergillus sp. and Penicillium sp. (Andlar et al. 2018). In the non-colonized regions of normal, untreated tubes only activities close to detection limits were measured, showing that the microbial community of mature substrate prior to spawning with P. ostreatus had a negligible effect on measured enzymatic activities. Thus, for further analysis, only the enzymatic activity values of P. ostreatus-colonised regions were used, taking into account the effect of contamination.
To test the effects of different treatments on the enzymatic activities, new independent summary variables were created from original enzymatic activities with PCA for the 1st and 2nd periods separately (Table S3a, Supporting Information). Based on the effect of contamination, a new explanatory variable was introduced as ‘treatment type’ with two categories for treatments having higher (A, AA, and AW) and lower (AM, AF, and N) contamination values. According to the linear mixed-effects model, the combination of regions in model tubes and treatment type were the best explanatory variables (Table 3b, Supporting Information; Fig. 7). The significant effect of the regions was due to enzymatic activities of P. ostreatus in the cultivation model (Fig. S7, Supporting Information) showing a similar temporal trend as in the large-scale cultivation system (Bánfi et al. 2015). In the 1st period, during substrate colonisation, lignin-degrading enzymes were dominant, laccase in the region of front hyphae and MnP in the 3-week-old colonised parts. Treatment type slightly modified the actual values within a region. Unlike large-scale production, xylanases showed higher activity in the region of front hyphae, but only in the case of treatments with higher contamination. Thus, xylanases were produced, or xylanases were induced by the contaminating fungi. After inducing fruiting body production, during the 2nd period, high cellulose, xylan, and chitin-degrading enzyme activity were measured. Activities were similar in the two regions (recently and older colonised), but significantly different according to treatment type (Table S3b, Supporting Information; Fig. 7), so P. ostreatus showed higher activity in case of lower contamination (AM, AF, and N).
Figure 7.
Enzymatic activities in the experimental cultivation setup: Effects of regions and different treatment types on the new PCA-derived summary variables in (A-C) 1st and (D) 2nd period. To understand which enzymes have a large impact on the respective principal components, see loadings in Table 3. ‘Treatment type’ was introduced based on LM for contamination and has two categories: treatments having higher (A, AA, and AW) and lower (AM, AF, and N) contamination ratios (For codes of the original treatments see Fig. 2A). Values show estimated marginal means ± standard error calculated from linear mixed-effects model. Upper letters show significantly different groups based on Tukey's post-hoc test. Lcc stands for laccase and MnP for manganese peroxidase.
Potential role of bacteria during Pleurotus ostreatus production
Our results from large-scale mushroom production showed that the bacterial community composition of the mature mushroom substrate changed significantly during P. ostreatus production. DNA-based methods used did not reflect the activity of the given taxon, that is, whether the detected bacterium has an active role during mushroom production. However, modest community transformation during substrate colonisation and fruiting body production compared to substrate production (Fig. 3C), and the 5–10-fold reduction in relative bacterial 16S rRNA levels observed in a similar cultivation model (Vajna, Frey-Klett and Deveau 2013) support the idea of reduced bacterial activity in general, but does not exclude the active role of some selected lineages.
In order to elucidate the role of microbes, the cultivation model was used to determine how the presence and absence of the known bacterial community affects P. ostreatus growth. Our main result was that the presence of microbes or their cell-free filtrate significantly decreased mushroom substrate contamination, and increased carbohydrate utilisation of P. ostreatus during fruiting body production. It is an important information from P. ostreatus producers, that proper substrate production (partial composting, pasteurisation, and conditioning) and consequently adequate microbial community is needed to avoid substrate contamination and because of this potential yield loss. It is worth mentioning that role of microbes is dependent on the mushroom species. In case of Agaricus bisporus primordia formation requires the presence of Pseudomonas putida or a related pseudomonad (Kertesz and Thai 2018).
Microbes play an important role prior to P. ostreatus colonisation, that is, they produce antimicrobial compounds (Nagy et al. 2012) during substrate production, which decrease the potential of later infection by competitive microbes. They also increase the accessibility of celluloses (Vajna et al. 2010). To test whether some microbe exert further activities during mushroom colonisation and fruiting body formation, or just survive in an inactive form (Mcgee et al. 2017), metaproteomic studies (Alessi et al. 2017) and analysis of secreted metabolites (Pinu et al. 2018; Senges et al. 2018) would be required.
CONCLUSIONS
A definite succession of bacterial community composition during colonisation and fruiting body production of P. ostreatus with increasing dominance of the order Bacillales and Halomonas spp., and decreasing relative abundance of Thermus spp. and Actinobacteria was described. In a cultivation model, it was proven that the presence of these microbes or their cell-free filtrate significantly decreased substrate contamination and increased cellulase and hemicellulase activity of P. ostreatus during fruiting body production. These findings support the hypothesis that the bacterial microbiome plays an important role prior to P. ostreatus spawning (e.g. by production of antimicrobial compounds, increase the accessibility of celluloses), and later P. ostreatus can use microbes partially to build their own biomass, but the microbes or their cell-free filtrate have a role in inhibiting competing microbes without hindering P. ostreatus growth.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are thankful to all employees of Pilze-Nagy Ltd. for their help during fieldwork and Szabina Luzics for her help in some parts of the laboratory work.
Contributor Information
Renáta Bánfi, Department of Microbiology, Eötvös Loránd University, Pázmány Péter sétány 1/C, 1117 Budapest, Hungary.
Zsuzsanna Pohner, Department of Microbiology, Eötvös Loránd University, Pázmány Péter sétány 1/C, 1117 Budapest, Hungary; Institute for Soil Sciences and Agricultural Chemistry, Centre for Agricultural Research, Herman Ottó út 15, H-1022 Budapest, Hungary.
Attila Szabó, Department of Microbiology, Eötvös Loránd University, Pázmány Péter sétány 1/C, 1117 Budapest, Hungary.
Gábor Herczeg, Department of Systematic Zoology and Ecology, Eötvös Loránd University, Pázmány Péter sétány 1/C, 1117 Budapest, Hungary.
Gábor M Kovács, Department of Plant Anatomy, Eötvös Loránd University, Pázmány Péter sétány 1/C, 1117 Budapest, Hungary.
Adrienn Nagy, Pilze-Nagy Ltd., Talfája 50., H-6000 Kecskemét, Hungary.
Károly Márialigeti, Department of Microbiology, Eötvös Loránd University, Pázmány Péter sétány 1/C, 1117 Budapest, Hungary.
Balázs Vajna, Department of Microbiology, Eötvös Loránd University, Pázmány Péter sétány 1/C, 1117 Budapest, Hungary.
FUNDING
This research was supported by: the Hungarian Scientific Research Fund (OTKA K 83764), the European Union and the State of Hungary, co-financed by the European Social Fund in the framework of TÁMOP 4.2.4. A/1–11-1–2012-0001 ‘National Excellence Program’ (A1-MZPD-12–0166), the National Competitiveness and Excellence Program (NVKP_16–1-2016–0035), and partly by the ELTE Thematic Excellence Program 2020 by the National Research, Development and Innovation Office (TKP2020-IKA-05).
Balázs Vajna was supported by the János Bolyai Research Scholarship of the Hungarian Academy of Sciences (Grant No. BO/00 156/21/8).
Conflicts of interest
None declared.
REFERENCES
- Abdo Z, Schüette UME, Bent SJet al. Statistical methods for characterizing diversity of microbial communities by analysis of terminal restriction fragment length polymorphisms of 16S rRNA genes. Environ Microbiol. 2006;8:929–38. [DOI] [PubMed] [Google Scholar]
- Adamski M, Pietr SJ. Biodiversity of bacteria associated with eight Pleurotus ostreatus (Fr.) P. Kumm. strains from Poland, Japan and the USA. Pol J Microbiol. 2019;68:71–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alessi AM, Bird SM, Bennett JPet al. Revealing the insoluble metasecretome of lignocellulose-degrading microbial communities. Sci Rep. 2017;7:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller Wet al. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. [DOI] [PubMed] [Google Scholar]
- Andersson BE, Lundstedt S, Tornberg Ket al. Incomplete degradation of polycyclic aromatic hydrocarbons in soil inoculated with wood-rotting fungi and their effect on the indigenous soil bacteria. Environ Toxicol Chem. 2003;22:1238–43. [PubMed] [Google Scholar]
- Andlar M, Rezić T, Marđetko Net al. Lignocellulose degradation: an overview of fungi and fungal enzymes involved in lignocellulose degradation. Eng Life Sci. 2018;18:768–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldrian P. Increase of laccase activity during interspecific interactions of white-rot fungi. FEMS Microbiol Ecol. 2004;50:245–53. [DOI] [PubMed] [Google Scholar]
- Bánfi R, Pohner Z, Kovács Jet al. Characterisation of the large-scale production process of oyster mushroom (Pleurotus ostreatus) with the analysis of succession and spatial heterogeneity of lignocellulolytic enzyme activities. Fungal Biology. 2015;119:1354–63. [DOI] [PubMed] [Google Scholar]
- Barron GL. Microcolonies of bacteria as a nutrient source for lignicolous and other fungi. Can J Bot. 1988;66:2505–10. [Google Scholar]
- Beltran-Garcia MJ, Estarron-Espinosa M, Ogura T. Volatile compounds secreted by the oyster mushroom (Pleurotus ostreatus) and their antibacterial activities. J Agric Food Chem. 1997;45:4049–52. [Google Scholar]
- Berns AE, Philipp H, Narres HDet al. Effect of gamma-sterilization and autoclaving on soil organic matter structure as studied by solid state NMR, UV and fluorescence spectroscopy. Eur J Soil Sci. 2008;59:540–50. [Google Scholar]
- Cho YS, Kim JS, Crowley DEet al. Growth promotion of the edible fungus Pleurotus ostreatus by fluorescent pseudomonads. FEMS Microbiol Lett. 2003;218:271–6. [DOI] [PubMed] [Google Scholar]
- Clausen CA. Bacterial associations with decaying wood: a review. Int Biodeterior Biodegrad. 1996;37:101–7. [Google Scholar]
- Cortes-Tolalpa L, Norder J, van Elsas JDet al. Halotolerant microbial consortia able to degrade highly recalcitrant plant biomass substrate. Appl Microbiol Biotechnol. 2018;102:2913–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boer W, Folman LB, Klein Gunnewiek PJAet al. Mechanism of antibacterial activity of the white-rot fungus Hypholoma fasciculare colonizing wood. Can J Microbiol. 2010;56:380–8. [DOI] [PubMed] [Google Scholar]
- De La Haba RR, Arahal DR, Sánchez-Porro Cet al. The family Halomonadaceae. The Prokaryotes: Gammaproteobacteria. Vol 9783642389221. Springer-Verlag, Berlin Heidelberg, 2014, 325–60. [Google Scholar]
- Deng Y, Wang R, Wang Yet al. Diversity and succession of microbial communities and chemical analysis in dried Lutianus erythropterus during storage. Int J Food Microbiol. 2020;314:108416. [DOI] [PubMed] [Google Scholar]
- Deveau A, Bonito G, Uehling Jet al. Bacterial-fungal interactions: ecology, mechanisms and challenges. FEMS Microbiol Rev. 2018;42:335–52. [DOI] [PubMed] [Google Scholar]
- Felföldi T, Jurecska L, Vajna Bet al. Texture and type of polymer fiber carrier determine bacterial colonization and biofilm properties in wastewater treatment. Chem Eng J. 2015;264:824–34. [Google Scholar]
- Folman LB, Klein Gunnewiek PJA, Boddy Let al. Impact of white-rot fungi on numbers and community composition of bacteria colonizing beech wood from forest soil. FEMS Microbiol Ecol. 2008;63:181–91. [DOI] [PubMed] [Google Scholar]
- Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes - application to the identification of mycorrhizae and rusts. Mol Ecol. 1993;2:113–8. [DOI] [PubMed] [Google Scholar]
- Gramss G, Voigt KD, Kirsche B. Degradation of polycyclic aromatic hydrocarbons with three to seven aromatic rings by higher fungi in sterile and unsterile soils. Biodegradation. 1999;10:51–62. [DOI] [PubMed] [Google Scholar]
- Gyurkó P. Die Rolle der Bakterien bei der Vorbereitung des Substrates für Pleurotus-Anbau. Mushroom Sci. 1978;10:31–6. [Google Scholar]
- Hervé V, Ketter E, Pierrat JCet al. Impact of Phanerochaete chrysosporium on the functional diversity of bacterial communities associated with decaying wood. PLoS One. 2016;11:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janusz G, Pawlik A, Świderska-Burek Uet al. Laccase properties, physiological functions, and evolution. Int J Mol Sci. 2020;21:966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasińska A. Spent mushroom compost (SMC) – retrieved added value product closing loop in agricultural production. Acta Agraria Debreceniensis. 2018;150:185–202. [Google Scholar]
- Johnston SR, Boddy L, Weightman AJ. Bacteria in decomposing wood and their interactions with wood-decay fungi. FEMS Microbiol Ecol. 2016;92:1–12. [DOI] [PubMed] [Google Scholar]
- Johnston SR, Hiscox J, Savoury Met al. Highly competitive fungi manipulate bacterial communities in decomposing beech wood (Fagus sylvatica). FEMS Microbiol Ecol. 2019;95:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz MA, Thai M. Compost bacteria and fungi that influence growth and development of Agaricus bisporus and other commercial mushrooms. Appl Microbiol Biotechnol. 2018;102:1639–50. [DOI] [PubMed] [Google Scholar]
- Kielak AM, Scheublin TR, Mendes LWet al. Bacterial community succession in pine-wood decomposition. Frontiers in Microbiology. 2016;7:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WG, Weon HY, Seok SJet al. In vitro antagonistic characteristics of Bacilli isolates against Trichoderma spp. and three species of mushrooms. Mycobiology. 2008;36:266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovács J, Kovács S, Magyar Net al. Classification into homogeneous groups using combined cluster and discriminant analysis. Environmental Modelling & Software. 2014;57:52–9. [Google Scholar]
- Kwon H. Bottle cultivation. Mushroom Grow Handb. 2004;1:166–71. [Google Scholar]
- Lane DJ. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds). ‘Nucleic acid techniques in bacterial systematics'. 1991, 15–751. [Google Scholar]
- Lang, Lang E, Kleeberg Iet al. Competition of Pleurotus sp. and Dichomitus squalens with soil microorganisms during lignocellulose decomposition. Bioresour Technol. 1997;60:95–9. [Google Scholar]
- Liu C, Sheng J, Chen Let al. Biocontrol activity of Bacillus subtilis isolated from Agaricus bisporus mushroom compost against pathogenic fungi. J Agric Food Chem. 2015;63:6009–18. [DOI] [PubMed] [Google Scholar]
- Martínez MA, Ramírez DO, Simental SSet al. Antibacterial activity of spent substrate of mushroom Pleurotus ostreatus enriched with herbs. J Agric Sci. 2015;7:225–31. [Google Scholar]
- Mata JA, Béjar V, Llamas Iet al. Exopolysaccharides produced by the recently described halophilic bacteria Halomonas ventosae and Halomonas anticariensis. Res Microbiol. 2006;157:827–35. [DOI] [PubMed] [Google Scholar]
- Mcgee CF, Byrne H, Irvine Aet al. Diversity and dynamics of the DNA and cDNA-derived bacterial compost communities throughout the Agaricus bisporus mushroom cropping process. Annals of Microbiology. 2017;67:751–61. [Google Scholar]
- Messner K, Fackler K, Lamaipis Pet al. Overview of white-rot research: where we are today. ACS Symposium Series. 2003;845:73–96. [Google Scholar]
- Nagy A, Manczinger L, Tombácz Det al. Biological control of oyster mushroom green mould disease by antagonistic Bacillus species. Biol Control Fungal Bact Plant Pathog. 2012;78:289–93. [Google Scholar]
- Oei P. Mushroom Cultivation IV. 4th ed.In:Oei P (ed). Amsterdam: ECO Consult Foundation, 2016. [Google Scholar]
- Ororbia MÁM, Núñez JP. La preparación del substrato. In: Sánchez JE, Royse DJ (eds.). La Biología y El Cultivo de Pleurotus Spp. Chiapas: El Colegio de la Frontera Sur, 2001, 157–86. [Google Scholar]
- Pinu FR, Granucci N, Daniell Jet al. Metabolite secretion in microorganisms: the theory of metabolic overflow put to the test. Metabolomics. 2018;14. [DOI] [PubMed] [Google Scholar]
- Pruesse E, Peplies J, Glöckner FO. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics. 2012;28:1823–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz Pet al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . R: A Language and Environment for Statistical Computing. 2020. [Google Scholar]
- Royse DJ, Baars J, Tan Q. Current overview of mushroom production in the world. Edible and Medicinal Mushrooms. John Wiley & Sons, Ltd, 2017, 5–13. [Google Scholar]
- Rühl M, Kües U. Mushroom production. In: Kües U. Wood production, wood technology, and biotechnological impacts. Göttingen: Universitätsverlag Göttingen, 2007, 555–86. [Google Scholar]
- Saǧir A, Yildiz A. Growth of mycelium of Pleurotus spp. on different grains and determination of their competition with some contaminant fungi. Acta Alimentaria. 2004;33:249–57. [Google Scholar]
- Schloss PD, Westcott SL, Ryabin Tet al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senges CHR, Al-Dilaimi A, Marchbank DHet al. The secreted metabolome of Streptomyces chartreusis and implications for bacterial chemistry. Proc Natl Acad Sci. 2018;115:2490–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shehata MG, Badr AN, El Sohaimy SAet al. Characterization of antifungal metabolites produced by novel lactic acid bacterium and their potential application as food biopreservatives. Annals of Agricultural Sciences. 2019;64:71–8. [Google Scholar]
- Smolskaite L, Venskutonis PR, Talou T. Comprehensive evaluation of antioxidant and antimicrobial properties of different mushroom species. LWT - Food Science and Technology. 2015;60:462–71. [Google Scholar]
- Stella T, Covino S, Čvančarová Met al. Bioremediation of long-term PCB-contaminated soil by white-rot fungi. J Hazard Mater. 2017;324:701–10. [DOI] [PubMed] [Google Scholar]
- Suarez C, Ratering S, Weigel Vet al. Isolation of bacteria at different points of Pleurotus ostreatus cultivation and their influence in mycelial growth. Microbiol Res. 2019;234:126393. [DOI] [PubMed] [Google Scholar]
- Szabó A, Korponai K, Kerepesi Cet al. Soda pans of the Pannonian steppe harbor unique bacterial communities adapted to multiple extreme conditions. Extremophiles. 2017;21:639–49. [DOI] [PubMed] [Google Scholar]
- Székely AJAJ, Sipos R, Berta Bet al. DGGE and T-RFLP analysis of bacterial succession during mushroom compost production and sequence-aided T-RFLP profile of mature compost. Microb Ecol. 2009;57:522–33. [DOI] [PubMed] [Google Scholar]
- Tabachnick BG, Fidell LS. Using Multivariate Statistics. 6th ed. Boston: Pearson Education, Inc, 2013. [Google Scholar]
- Vajna B, Frey-Klett P, Deveau A. Transcriptional and enzymatic profiling of Pleurotus ostreatus laccases and manganese peroxidases during mushroom cultivation. Bacterial-Fungal Interactions: A Federative Field for Fundamental and Applied Microbiology. 2013;9. [Google Scholar]
- Vajna B, Nagy A, Sajben Eet al. Microbial community structure changes during oyster mushroom substrate preparation. Appl Microbiol Biotechnol. 2010;86:367–75. [DOI] [PubMed] [Google Scholar]
- Vajna B, Szili D, Nagy Aet al. An improved sequence-aided T-RFLP analysis of bacterial succession during oyster mushroom substrate preparation. Microb Ecol. 2012;64:702–13. [DOI] [PubMed] [Google Scholar]
- Velázquez-Cedeño M, Farnet AM, Mata Get al. Role of Bacillus spp. in antagonism between Pleurotus ostreatus and Trichoderma harzianum in heat-treated wheat-straw substrates. Bioresour Technol. 2008;99:6966–73. [DOI] [PubMed] [Google Scholar]
- Velázquez-Cedeño MA, Farnet AM, Ferré Eet al. Variations of lignocellulosic activities in dual cultures of Pleurotus ostreatus and Trichoderma longibrachiatum on unsterilized wheat straw. Mycologia. 2004;96:712–9. [DOI] [PubMed] [Google Scholar]
- Verdier T, Coutand M, Bertron Aet al. A review of indoor microbial growth across building materials and sampling and analysis methods. Build Environ. 2014;80:136–49. [Google Scholar]
- Viziteu G. Cereal straw and corncobs. Mushroom Grow Handb. 2004;1:86–90. [Google Scholar]
- Vos AM, Heijboer A, Boschker HTSet al. Microbial biomass in compost during colonization of Agaricus bisporus. AMB Express. 2017;7:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vreeland RH, Anderson R, Murray RGE. Cell wall and phospholipid composition and their contribution to the salt tolerance of Halomonas elongata. J Bacteriol. 1984;160:879–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White T, Bruns T, Lee Set al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols a Guide to Methods and Applications. San Diego: Academic Press, 1990, 315–22. [Google Scholar]
- Xu W, Wang H, Lv Zet al. Antifungal activity and functional components of cell-free supernatant from Bacillus amyloliquefaciens LZN01 inhibit Fusarium oxysporum f. sp. niveum growth. Biotechnology & Biotechnological Equipment. 2019;33:1042–52. [Google Scholar]
- Zhong XZ, Zeng Y, Wang SPet al. Insight into the microbiology of nitrogen cycle in the dairy manure composting process revealed by combining high-throughput sequencing and quantitative PCR. Bioresour Technol. 2020;301:122760. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.







