Abstract
The response to disease outbreaks, such as SARS-CoV-2, can be constrained by a limited ability to measure disease prevalence early at a localized level. Wastewater based epidemiology is a powerful tool identifying disease spread from pooled community sewer networks or at influent to wastewater treatment plants. However, this approach is often not applied at a granular level that permits detection of local hot spots. This study examines the spatial patterns of SARS-CoV-2 in sewage through a spatial sampling strategy across neighborhood-scale sewershed catchments. Sampling was conducted across the Reno-Sparks metropolitan area from November to mid-December of 2020. This research utilized local spatial autocorrelation tests to identify the evolution of statistically significant neighborhood hot spots in sewershed sub-catchments that were identified to lead waves of infection, with adjacent neighborhoods observed to lag with increasing viral RNA concentrations over subsequent dates. The correlations between the sub-catchments over the sampling period were also characterized using principal component analysis. Results identified distinct time series patterns, with sewersheds in the urban center, outlying suburban areas, and outlying urbanized districts generally following unique trends over the sampling period. Several demographic parameters were identified as having important gradients across these areas, namely population density, poverty levels, household income, and age. These results provide a more strategic approach to identify disease outbreaks at the neighborhood level and characterized how sampling site selection could be designed based on the spatial and demographic characteristics of neighborhoods.
Keywords: SARS-CoV-2, Wastewater based epidemiology, Spatial cluster detection, Viral RNA concentration
Graphical abstract
1. Introduction
Public health entities and local governments around the world faced unprecedented challenges in deploying clinical testing to monitor and inform policy in order to slow the spread of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) disease, COVID-19. The accuracy of the clinical testing data collected by public health officials was limited by a multitude of factors, such as strained supply systems, public access, the high occurrence of asymptomatic viral infections, and public acceptance. Respiratory viruses like SARS-CoV-2 are typically shed at lower concentrations in human stool compared to viruses that infect the gastrointestinal tract, which often spread through fecal-oral transmission (Wigginton et al., 2015). Although the transmission of SARS-CoV-2 is primarily through fine droplets and an oral-oral route, there is evidence that SARS-CoV-2 does replicate in the gastrointestinal tract (Ahmadiara, 2020; Kitajima et al., 2020; Yeo et al., 2020; Zhou et al., 2017) resulting in abundant concentrations measured from human stool (Foladori et al., 2020; Lee et al., 2020; Zhang et al., 2020; Zhou et al., 2020). Viral RNA concentrations shed into sewage may range from 102 to 106.5 gene copies/L, when typical dilution rates of sewage are accounted for (Saawarn and Hait, 2021). Wastewater-based epidemiology (WBE) arose as a promising strategy to acquire less biased data regarding the viral prevalence in a community and to provide earlier detection of outbreaks.
As a member of the Coronaviridae family, SARS-CoV-2 are characterized by an enveloped structure with single-stranded, positive-sense RNA genome (Kitajima et al., 2020). Although the survivability and partitioning of SARS-CoV-2 in sewer networks has not yet been reported, several studies have explored the potential fate and transport characteristics of the virus based on its morphology and similarity to other enteric viruses (Kitajima et al., 2020; Mohapatra et al., 2021; Saawarn and Hait, 2021). Viruses with enveloped structures are more readily inactivated in environments like sewer networks (Wigginton et al., 2015). Ye et al. (2016) observed that enveloped viruses had shorter survival times in sewage compared to nonenveloped viruses. Thus, measurement of SARS-CoV-2 at neighborhood scale sewer sub-catchments may provide superior characterization of disease spread due to the shorter retention time compared to monitoring at the influent to water reclamation facilities (WRFs). These characteristics contribute to the value of WBE to monitor COVID-19 disease spread through communities, particularly at more granular community scales.
The analysis of spatial patterns in pathogen levels through WBE may provide several benefits in developing the understanding of social and geographical characteristics that correlate to the distribution of disease while also providing insight into the representativeness of sewage from WRFs versus in neighborhood sub-catchments. Several studies have explored spatial patterns in disease with the goal of identifying social characteristics that correlated to virus infection rates or the probability of infection (McLafferty, 2015; Odoi et al., 2003; Samphutthanon et al., 2014). This spatial approach has recently been extended to understanding the COVID-19 pandemic at the province scale to evaluate the social factors that best correlated to geographic patterns in the reported COVID-19 cases (Mollalo et al., 2020; Ramírez-Aldana et al., 2020; Ren et al., 2020). Few studies have applied spatio-temporal sewershed sampling strategies to characterize the distribution of disease and data biases across a catchment (e.g., Freeman et al., 2017; McCall et al., 2017; Wang et al., 2012). As demonstrated by Hart and Halden (2020a), the heteroskedasticity of socio-economic characteristics across a WRF sewershed is an important spatial parameter to consider when evaluating the potential biasing of WBE data because the biomarkers in sewage are expected to decay according to their residence time within the sewer network. These researchers demonstrated that biomarker decay resulted in the overrepresentation of the populations closest to the WRF when WBE monitoring was performed from WRF influent. An extensive study by McCall et al. (2017) explored uncertainty in WBE monitoring at numerous sewer catchment scales, ranging from 14,000 to 370,000 persons, and evaluated the variability of HRT and loss in the biomarkers of interest across the spatial sampling areas. Notably, this study determined that the biomarkers that were transformed due to biotic in-sewer processes expressed the greatest variability and that these biomarker transformation rates were also the greatest source of uncertainty when modeling the source population. Thus, the size of the sewer catchment captured by a WBE sampling site is likely to influence the magnitude of data biasing as well as the representativeness of the measured biomarker to a community. Spatial sampling methods may address important sources of data bias (Hart and Halden, 2020a; McCall et al., 2017) and also enhance the detection of spatial clustering patterns (Wang et al., 2012).
WBE has been widely proposed as a necessary approach to avoid the data biases associated with clinical testing of COVID-19 (Ahmed et al., 2020; Thompson et al., 2020), however no studies were found that applied WBE across small sewer catchments for spatially resolute early outbreak detection. Additionally, there was a need to examine the spatial biasing of SARS-CoV-2 data collected from WRFs, which may limit the applicability of such data as a means of detecting hot spots or localized trends in SARS-CoV-2 spread. To this end, this study aimed to examine the spatial patterns of SARS-CoV-2 viral shedding through a spatial sampling strategy across neighborhood-scale sewershed catchments. Also, it identified spatially clustered patterns of viral RNA concentrations in sewage according to geography, temporal patterns in viral shedding, and to classify WRF and neighborhood monitoring sites into groups based on spatio-temporal patterns in viral RNA concentrations.
2. Methods and materials
2.1. Study area
The spatial sampling protocol was implemented in the Reno-Sparks metropolitan area (pop. 360,000) as the main population center for Washoe County, Nevada, USA. Approximately 80% of the wastewater generated in the region is conveyed to the main regional WRF. The WRF has two main catchment areas; the northern channel catchment includes sewer lines that largely fall within the boundaries of the City of Sparks (pop. 100,600) while the southern channel catchment is representative of the population from the City of Reno (pop. 246,500), and the rest in the suburban areas belonging to Washoe County, Nevada (USA). The sampling program included both urban and suburban neighborhoods. Some characteristics of the neighborhoods selected for sampling are provided in Table SI-1, with additional descriptions presented in the Supporting Information. The sampling strategy focused on spatial distribution over the WRF service area, with one to two sampling locations in each of the residential postal zip codes. Demographic characteristics of each sampling site were obtained from the American Community Survey according to the location of sewershed sub-catchments in census tracts (U.S. Census Bureau, American Community Survey, 2019).
2.2. Geospatial site selection
Sewershed sampling locations were established using geographic information systems (GIS) (ESRI, Redlands, CA, USA) to analyze spatial locations, such as zip codes, sewer lines, manholes, water reclamation facilities, parcels, and land use (ESRI, 2011). The application of GIS in developing the sewershed sampling locations for epidemiological surveillance was developed from the field of medical geography (Ahmad et al., 2015; Kassié et al., 2017). The GIS analysis encompassed the sewershed of Truckee Meadows Water Reclamation Facility (TMWRF) with population distribution, sanitary sewer network, and land use. This approach allowed for identification of residential neighborhoods (> 90% residential land use for parcels in each neighborhood sub-catchment) (Table SI-1).
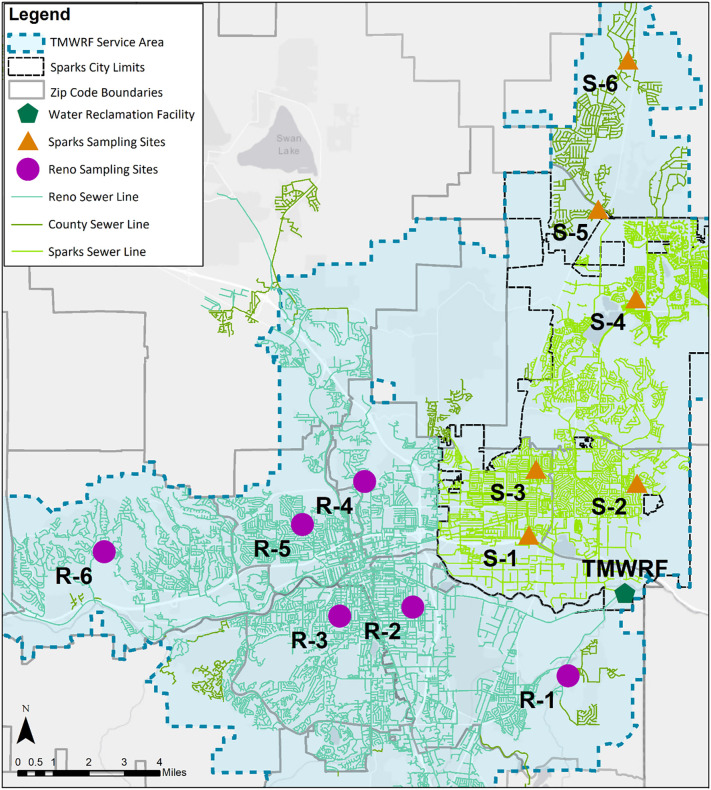
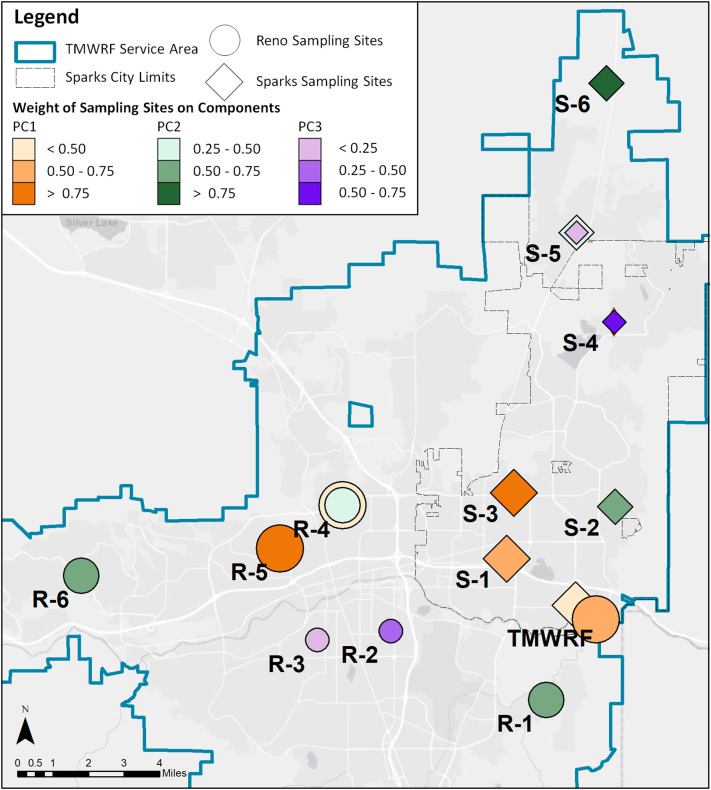
The sampling site selection methodology was developed for geospatial representation of neighborhood scale data (Fig. SI-1). These criteria identified sewershed sub-catchments in which sewage flows from neighborhoods approximating 500 to 1000 people piped to a single sampling point and located within the WRF and single zip code areas. Subareas this size are also large enough to assure individual resident anonymity. The focus was to locate the subareas which were predominantly residential and with minimal commercial area contribution that provided diverse geographic representation across the TMWRF service area (Fig. 1 ). Further, population from residential areas were more likely to correspond accurately to regional land-use/population density estimates and census data. The summary of the number of residential units and estimated residential population, assuming 2.5 individuals per household, was presented in Table SI-1. The site numbering system used in the study identifies sites according to their location within either the Reno or Sparks (R or S) sewersheds, and with numbers (1-6) corresponding to their ranked distance from TMWRF, as measured by the length of sewer pipe.
Fig. 1.
Map of sewershed sampling sites across the study area.
2.3. Sample collection
Sample campaign was held since late October to December 2020 during the peak of the COVID-19 pandemic in the region. A total of 186 samples were collected during the sampling collection phase across twelve sampling locations throughout the WRF service area of the between 7:00 am to 11:00 am to coincide with the approximate peak diurnal flow in residential neighborhoods (Table SI-8), two or three times per week for each location. The collection of 24-hour composite samples was not feasible due to constrained resources and the ability to power autosamplers at manholes across the urban area. Thus, manual composite sampling techniques were used based on an equal time/equal volume approach, with four grab samples measuring 250 mL collected every 15 min over the one-hour sampling period for a 1-liter total volume in sterile Nalgene bottles. Samples were stored in a cooler at 4 °C, transported to the laboratory for further processing and analyses.
2.4. Virus recovery, RNA extraction and reverse transcription and quantitative polymerase chain reaction
To detect SARS-CoV-2 viral RNA in the sewage samples, samples were first inactivated using heat treatment as described by Wu et al. (2020), then step filtration processes were followed to concentrate and recover the viral RNA. The concentration and filtration processes could concentrate the sample up to 70× the original volume and included centrifugation and sequential membrane filtration. The sample was first processed by centrifugation at 3500g for 15 min to remove the big particles. The supernatant was processed through step filtration with 1.5 μm, 0.8 μm, and 0.45 μm MilliporeSigma (Burlington, MA, USA) membrane filters. Following these pretreatment steps, 15 mL filtrate was then transferred into a 100 kDa Amicon® Ultra-15 Centrifugal Filter Cartridge Units (MilliporeSigma, USA). The ultrafiltration cartridges were used to concentrate the sample to a final volume of 500 μL in each cartridge through centrifugation at 3500g for 5 min.
The process to extract RNA from the concentrated samples used the AllPrep PowerViral DNA/RNA kit (QIAGEN, Inc., Germantown, MD, USA), with RT-qPCR analysis conducted on the CFX96 Touch Real-Time PCR Detection System (Bio Rad, Hercules, CA, USA) according to MIQE guidelines (Johnson et al., 2014). The reaction contained 1× Reliance One-Step Multiplex Supermix (Bio Rad, Hercules, CA, USA), 5 μL of the total genomic RNA template, probes (0.2 μM) and primers (0.4 μM each) in a total volume of 20 μL. qPCR standards were prepared in each assay in the range from 200,000 to 2 gc/μL by 10-fold serial dilutions. Non-template control was included in each qPCR assay to confirm no contamination. Field blank and RNA extraction blank were conducted once per month, and no contamination were identified. RT-qPCR was carried out by the following program: Reverse transcription at 50 °C for 10 min, denaturation at 95 °C for 10 min, followed by 45 cycles of 3 s denaturation at 95 °C, 30 s annealing/extension and plate read at 60 °C.
The recovery rate of the viral RNA concentration method was evaluated using human coronavirus OC43 strain (HCoV-OC43) as a surrogate for SARS-CoV-2 due to its similar enveloped structure. Wastewater sample volumes of 180 mL were spiked with 100 μL HCoV-OC43. This was followed by the previously described Amicon ultrafiltration concentration methods and then quantification of HCoV-OC43 following the method described by Uppal et al. (2021). The recovery rate was determined to be 24 ± 2%. qPCR inhibitor was evaluated by ∆Ct values of 10- and 100- folds dilution of extracted RNA of HCoV-OC43 spiked samples according to Li et al. (2020b). Sample limits of detection (SLoD) was determined by the lowest dilution of the SARS-CoV-2 standards (IDT, Skokie, IL, USA) with positive signals, which was 2 gc/μL in of RNA elution. The SLoD of each sewage sample was calculated by the concentrated factors when virus recovered, which is 2 gc/μL divided by the overall concentration factors of the samples with the unit of gc/L in sewage.
2.5. Statistical analysis
Recent studies have applied numerous indicators to evaluate spatial autocorrelation in the SARS-CoV-2 pandemic based on clinical testing to investigate the suitability of spatial modeling due to the epidemiological relevance of community spread and other geospatial characteristics that may be deterministic in community prevalence (Kang et al., 2020; Li et al., 2020a; Ramírez-Aldana et al., 2020). Importantly, these studies have revealed spatial correlations in disease spread based on regional demographic and environmental variables (Kang et al., 2020; Ramírez-Aldana et al., 2020). Analysis was carried out with the N1 viral gene marker concentrations detected in sewage samples from sampling sites over the study period due to the more frequent detection of N1 compared to N2. Over the sampling period, the monitoring across all sewershed sampling sites produced 11 observations out of 186 that were below the detection limit, while no WRF influent observations were observed below the detection limit. Although less robust than distributional methods, Farnham et al. (2002) demonstrated that substitution with zero can produce acceptable results with less bias in PCA analysis than substitution when less than 25% of the data is non-detect; thus, this method was selected to handle non-detects. The heatmap reduced spatial dimensions to a single axis based on the sewer pipe length from the sampling site to TMWRF, with the horizontal axis presenting the sampling dates for time series trends in viral RNA concentrations. Sewer pipe lengths were determined from sewage network maps in ArcGIS software version 10.7.1 ESRI, Redlands, CA, USA. The plots were produced in R studio (version) (R Core Team, 2020) using ggplot2 (Wickham, 2016) and FactoMineR (Lê et al., 2008). The statistical methods required data to conform to the assumption of normality. The Shapiro-Wilk test was used to evaluate the normality of each variable. To adjust for the skewness of the data and achieve a distribution that met the assumption of normality, (Fig. SI-1) all statistical analysis were carried out using logarithmic transformations of the viral RNA concentrations measured in raw sewage.
2.5.1. Local spatial autocorrelation methodology
GIS has techniques and tools that focus on spatial autocorrelation of the data. Parameters associated with spatial locations can be evaluated for patterns with spatial autocorrelation tools to determine if the attribute values are either defined as clusters/outliers or hot/cold spots. This study selected two of the widely used local spatial statistics, Anselin Local Moran's I and Getis-Ord Gi*, following the methodologies of Anselin (1995) and Getis and Ord (1992). These examine localized patterns in autocorrelation in order to identify local pockets of hot and/or cold spots (Gi* analysis) and to assess the influence of individual locations to identify outliers (Anselin I). Identifying autocorrelation at a local scale is powerful for determining spatial characteristics of disease spread that can be impacted by environmental impacts (e.g., Ahmad et al., 2015; Guo et al., 2021) and socioeconomic characteristics (Mollalo et al., 2020; Samphutthanon et al., 2014). The local patterns of spatial autocorrelation were evaluated based on the viral RNA concentrations in sewage for each sampling date. Cluster and outlier analysis (Anselin I) and hot spot analysis (Getis-Ord Gi*) were selected based on their wide use in geospatial epidemiology research. These analyses were conducted in ArcGIS, following the methodologies of Anselin (1995) and Getis and Ord (1992). Reno and Sparks sampling events were evaluated together by pairing the adjacent sampling dates (Monday-Tuesday; Wednesday-Thursday).
Anselin I measured local patterns of spatial clustering and outliers through a decomposition of the global spatial autocorrelation between all sampling sites. This method to identify clusters was performed using the average viral RNA concentrations measured among neighboring sample sites. Hot and cold spot analysis through the Gi* autocorrelation test calculated spatial autocorrelation by comparing individual sampling locations to neighboring sampling locations. Locations at which the local average viral RNA concentration was significantly different from the expected value were identified as a hot or a cold spot. Neighboring sites were identified based on the maximum distance between each site and its two nearest neighbors (determined to be a maximum of 8.3 km). This distance was then applied as a radius at each site to identify neighbors. A p-value <0.05 was considered statistically significant for Anselin I autocorrelation analysis (Li et al., 2020a). The strength of clustering was evaluated based on p-values (p < 0.1 was associated with a confidence interval of at least 90%, p < 0.05 was associated with a confidence interval of at least 95%) was considered statistically significant for Gi* autocorrelation analysis.
2.5.2. Multivariate analysis of site similarity from time series
Principal component analysis (PCA) was adopted as a transformational approach to examine the similarity of spatial sampling sites based on the time series (Benedetti et al., 1994; Lhermitte et al., 2011; Lobo and Maisongrande, 2008). This analysis has a powerful ability to identify the most significant variations in the viral RNA concentration according to spatial and temporal variations. Prior to PCA analysis, data suitability was evaluated based on the Shapiro-Wilk test, which identified the necessity of conducting analysis on the log-transformed viral RNA concentrations to account for the assumption of normality during analysis. The dataset was also assessed using Bartlett's test to ensure that no spatial sampling locations characterized an identity matrix, and that the strength of correlations was sufficient to demonstrate that observed patterns in variance could not be attributed to sampling error and the Kaiser-Meyer-Olkin test to evaluate sampling adequacy (Field et al., 2012).
The PCA technique used the Pearson correlation between sites with similar temporal patterns in viral RNA concentration to project the data onto axes that are linear combinations of the variables. Principal components (PCs) for Reno and Sparks were developed as linear aggregates of the sampling sites that shared similar patterns in data variance over time. Although data at sampling sites had a similar range after log-transformation, the variance observed across sampling sites varied widely, with the smallest sampling sites exhibiting the greatest variability. To ensure that sites with more frequent non-detect observations did not exert undue bias in the PCA, the analysis was conducted on the correlation matrix rather than the covariance matrix. While the components that describe the largest variance in the data are likely to capture the desired phenomena, noise in the data is generally captured in the high order components (Jolliffe, 2003).
3. Results and discussion
3.1. Spatio-temporal distribution of SARS-CoV-2
The dynamics of the viral RNA surveillance across the spatial sampling sites was first evaluated in collapsed space (latitude and longitude collapsed into one dimension) using Hovmöller plots, or heatmaps (Fig. 2a and b). These figures compared the viral RNA concentrations observed at each sampling site over time using a color gradient to indicate the measured viral RNA concentration. The heatmaps illustrated two important patterns in the data: the potential biasing of WRF surveillance (shown as S-0-WRF and R-0-WRF) to certain sites, and the similarity in the time series trends observed between sites (e.g., S-1 and S-3, R-4 and R-5). In both regions the mean viral RNA concentrations exhibited an increasing trend to mid-November, with observed viral RNA concentrations ranging from 2.62 × 104 to 2.16 × 106 gc/L. At both Reno and Sparks sampling sites, the mean viral RNA concentrations began to decline around November 10. In the City of Reno, this trend continued through early December and reached the lowest observed mean concentration on December 8. This was followed by an increasing trend in viral RNA concentration through to the end of the study period. In the City of Sparks, the daily mean viral RNA concentrations across all sampling sites were at their highest, due to several observations at sites S-4 through S-6 that ranged from 7.67 × 105 to 1.73 × 106 gc/L until a decrease in early December.
Fig. 2.
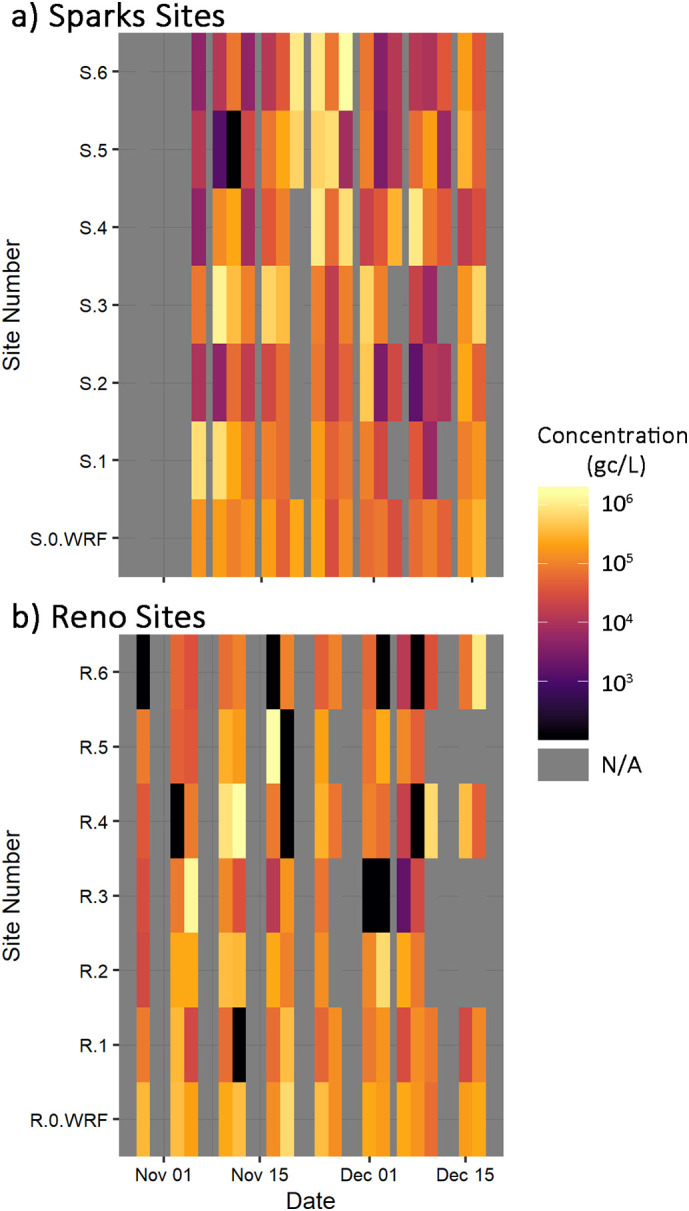
Heatmaps of SARS-CoV-2 genetic marker concentrations in raw sewage at WRF influent and sewershed sampling sites in the Cities of Sparks and Reno, Nevada.
The heatmap of viral RNA concentrations across the WRF sampling sites (S-0-WRF) and six sewershed locations (1-6) further illustrate the similarity between WRF surveillance with the three sites located closest. During the early part of November, the most outlying sites were observed to have the lowest concentrations, while the more central sampling sites (S-1-S-3) generally had moderate to high concentrations. During the following two weeks, viral RNA concentrations increased by three orders of magnitude at the outlying sites (S-4-S-6). This was followed by a period of decreased viral RNA concentrations measured throughout sampling sites in early December and another period of increased viral RNA concentration across city of Sparks sampling sites through mid-December. Importantly, monitoring of concentrations from the WRF (S-0-WRF) did not reflect elevated concentrations on dates when the most distant sewershed sampling sites (S-5 and S-6) were measured to have large increases.
In the Reno sampling areas, the WRF influent showed little variability as did the two sewershed locations closest to the WRF. Conversely, the R-5 and R-4 regions had more notable peaks and valleys in viral RNA concentrations over time (Fig. 2b and Fig. SI-2). Both neighborhoods were located near a university and may have higher incidence of students and university employees compared to other neighborhoods. The R-3, R-4, and R-6 sites had the greatest frequency of observations below the SLoD, but R-3 and R-4 also had the greatest frequency of observations exceeding 1 × 106 gc/L. A statistical summary of the viral RNA concentrations measured at each site during the study period is presented in the Supporting Information. Excluding the skewing from non-detect observations, the R-3 and R-6 sites had the lowest average virus concentrations observed over the sampling period, which may have contributed to the relatively high frequency of non-detects at R-3 and R-5 sites. Additionally, the R-3 site was the smallest population sample size (Table SI-1) as well as the largest observed standard deviation, which indicates that the sample size may not have been large enough to produce consistently representative samples for the neighborhood.
3.2. Spatial clustering through local autocorrelation analysis
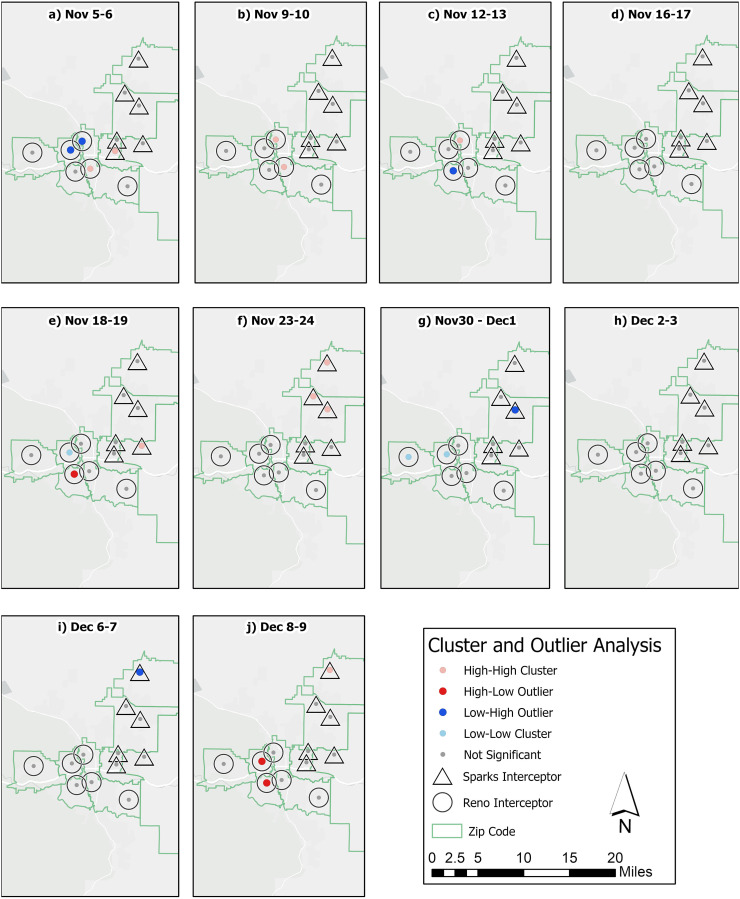
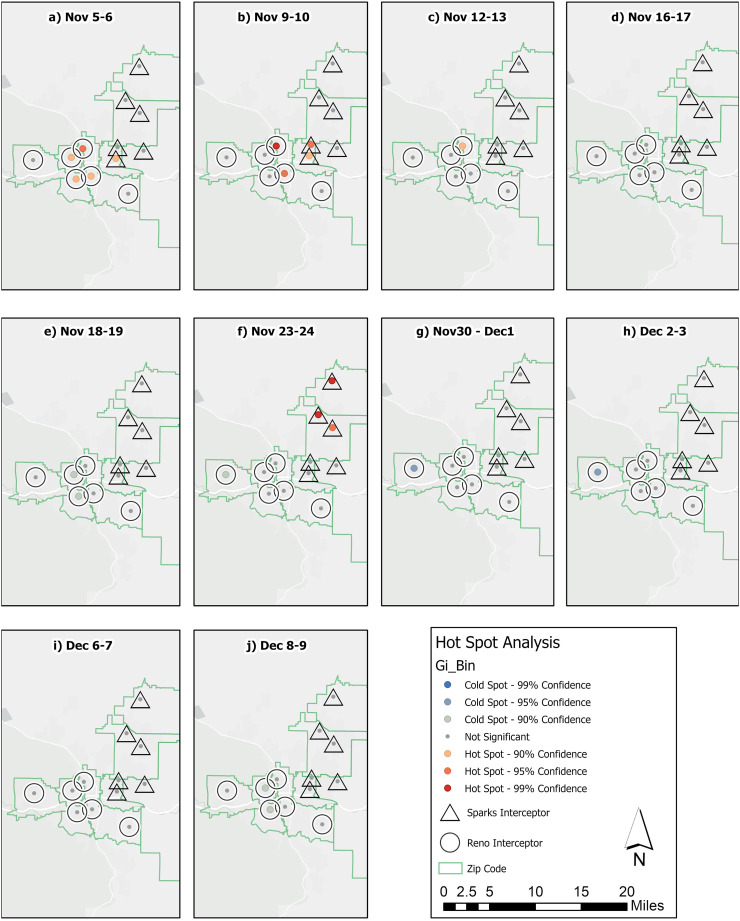
The spread of COVID-19 through respiratory shedding was evaluated to determine whether significant spatial patterns were observable based on the proximity of each sampling location to a hot spot. Local indicators of spatial analysis tests were utilized to evaluate these spatial patterns over the five-week study period (from November 5 to December 10) in the sewer network to identify clusters, outliers, hot spots and cold spots through Anselin I and Getis-Ord Gi* statistical tests. Because not all sampling sites were tested on the same dates, the analysis was accomplished by pairing neighboring date samples across all twelve sampling sites, producing a total of ten sampling events which are discussed in the chronological order below. Monitoring over the first week and a half of November identified high viral RNA concentrations across sites near the urban centers of Sparks and Reno. Both the Anselin I and Gi* statistics classified sampling sites across the urban centers as clusters where viral RNA concentrations were high, referred to as High-High (HH) clusters (Fig. 3 ) and hot spots (Fig. 4 ). The significance of the cluster and hot spot analyses can be found in the supporting information (Tables SI-2 and SI-3).
Fig. 3.
Clusters and outlier sampling sites for viral RNA concentrations in the Cities of Reno and Sparks over the sampling period.
Fig. 4.
Hot spots and cold spots of viral RNA concentrations measured in sewer sub-areas across the Cities of Reno and Sparks.
At the beginning of the sampling period sampling sites in the urban center of Reno-Sparks were measured as having significant clusters of hot spots, as shown in Fig. 4a–c. The Gi* characterization shown in Fig. 4 was less sensitive to the changes in viral RNA concentrations measured at individual sites compared to the Anselin I results presented in Fig. 3, resulting in the additional detection of outliers within spatially correlated clusters. In Fig. 3a, two sites (R-2 and S-1), were also classified as hot spots with a 90% CI (Fig. 4b). These hot spots also corresponded to HH clusters in Fig. 3a–c. It was notable that several of the hot spot neighborhoods were LH outliers over this same period according to the Anselin I test. Specifically, the R-4 neighborhood was initially a LH outlier (Fig. 3a) before transitioning to part of the HH cluster (p values of 0.030 and 0.044) (Fig. 3b and c). Conversely, several sites (R-2, R-3, and S-1) were observed to have high viral RNA concentrations that were declining over this period, Specifically, R-3 transitions from higher viral RNA concentration as part of the hot spot (Fig. 4a) to an LH outlier, whereas sites R-2 and S-1 transition from hot spots and HH clusters to having no significance in the hot spots by Figs. 3c and 4c. Overall, this period indicates that several neighborhoods in the urban center (R-2, R-3, and S-1) lead the increase in viral RNA concentrations, with other adjacent neighborhoods in the urban center (R-4 and R-5) lagging behind. Significant spatial patterns were also observed in late November; these were largely concentrated in the northern area of Sparks and western Reno. Figs. 3e–f and 4e–f depict a period where sites S-3 through S-6 had elevated viral RNA concentrations (refer to Fig. 2). Similar to the hot spot identified in early November, the late November hot spot was first detected as an HH cluster at the S-3 sampling site (Fig. 4e). This event was then followed by significant hot spot and HH clusters across northern Sparks (Figs. 3f and 4f). This date corresponded to the date that both S-4 and S-6 had viral RNA concentrations of 9.79 × 105 and 9.6 × 105 gc/L, respectively (Fig. 2). Although the sampling sites across northern Sparks had high viral RNA concentrations throughout late November, their concentrations were only significantly different from other neighboring sites on the event where two neighbors had measurements at least an order of magnitude higher than other sampling sites. The concentration of hot spots in outlying sites of northern Sparks strongly contrasted the cold spot (90% CI) in the outlying site of western Reno (R-6) and its neighbor (R-5) over the same period (Figs. 4e–f and 3e–f). These results indicated that while disease spread appeared to flow from population centers outwards, social factors also likely played a role in spread and hot spots development.
As discussed in the previous section, viral RNA concentrations observed across the region over the first week of December were generally moderate, and few spatial patterns were identified. However, several locations were measured to have significant clustering based on their low viral RNA concentrations, resulting in cold spots (Figs. 3g–h and 4g–h). The Anselin I analysis identified two of these sites, R-5 and R-6, as LL clusters due to relatively low viral RNA concentrations (< 1.85 × 104 gc/L). At the same time site S-4, with viral RNA concentration of 2.14 × 104 gc/L, was classified as a LH outlier due its proximity to downtown sampling sites that had higher viral RNA concentrations, such as S-2 and S-5 with 4.48 × 105 and 1.10 × 105 gc/L, respectively (Fig. 3g and 4g). During the following week the viral RNA concentration measured at the R-4 sampling site increased by two orders of magnitude, to 7.05 × 105 gc/L, while neighboring sites had viral RNA concentrations <5 × 104 gc/L. This caused the Anselin I analysis to identify its two neighboring sites, R-5 and R-3 as HL outliers (p-values of 0.016 and 0.026, respectively), while the Gi* characterized the same neighboring sites as cold spots, as illustrated in Figs. 3j and 4j.
Specific trends noted in the study area included repeating pattern of spatial clustering across the study area, with sites R-4 and R-5 (urbanized north Reno neighborhoods near a university) as well as R-2 appearing as either clusters or LH outliers. The results are consistent with recent research from Ramírez-Aldana et al. (2020), who noted that in Iran the sites most likely to be HH clusters were the most urbanized and connected areas. The results also indicated directional patterns in disease spread, with sites that neighbored hot spots frequently becoming hot spots themselves within one to two weeks. As noted by McLafferty (2015), these spatial relationships may have important underlying processes such as commuting to work or recreational activities that are not easily characterized.
3.3. Time series similarity of sampling
The prior section described the significance of spatial correlation patterns at a single point in time, but spatial analysis can fail to identify correlations between sites that were not located in close proximity and only considered observations at a single point in time. Thus, a multivariate approach through PCA was developed to describe how the sites were correlated over time. In both the Reno and Sparks sewersheds, three principal components were able to describe more than 75% in the total data variance. Fig. 5 Illustrates which sampling sites were aggregated into each PC based on their patterns of viral RNA concentrations during the sampling period. Orange represents sampling sites important to PC1; green represents sampling sites important to PC2, and purple represents sampling sites important to PC3. Additionally, PCs were differentiated by the size of site markers: as PC1 described the most important source of data variance the sites were assigned large markers with marker size decreasing for each PC. Because Sparks and Reno sites were analyzed separately, they were represented in Fig. 5 by different shapes, with circles to represent Reno sites and diamonds to represent Sparks. The shade of colors described the weights of each site in characterizing a given PC; these weights represent the scaling coefficient that was assigned to each sampling location based on its importance in describing a given PC.
Fig. 5.
Representation of principal component analysis results across the geographic space of the Reno-Sparks metropolitan area.
Among Sparks sampling sites, the first three components explained 79% of variance across all sampling sites over the sampling period. Among them, a close relationship was observed between the Sparks WRF influent and two of the closest sites to the WRF: S-1 and S-3 (Fig. 5). These three sites had significant positive correlations to PC1, as shown in Table 1 . Although S-2 did have a moderate correlation to S-3 (Pearson correlation of 0.43, p-value = 0.11), it was most correlated to S-6 (correlation of 0.64, p-value = 0.01). Subsequently, sites S-6, S-5, and S-2 were highly correlated to PC2 (Table 1). Finally, S-4 and S-5 were significantly correlated to PC3, but in opposite directions (Table 1). This was the result of their opposing patterns in viral RNA concentrations, with one peaking when the other was at a relatively low observed value.
Table 1.
Correlations of sampling sites with principal components, developed separately for Reno and Sparks city data.
| City | PC | Sampling site | Correlation | p-value |
|---|---|---|---|---|
| Reno | PC1 | R-5 | 0.9148 | 3.07E-05 |
| R-4 | 0.6903 | 1.30E-02 | ||
| R-WRF | −0.7466 | 5.27E-03 | ||
| PC2 | R-6 | 0.7928 | 2.10E-03 | |
| R-1 | −0.7272 | 7.36E-03 | ||
| PC3 | R-2 | 0.6509 | 2.19E-02 | |
| Sparks | PC1 | S-3 | 0.8763 | 1.83E-05 |
| S-1 | 0.8443 | 7.54E-05 | ||
| S-WRF | 0.6607 | 7.33E-03 | ||
| PC2 | S-6 | 0.9180 | 1.41E-06 | |
| S-2 | 0.7160 | 2.68E-03 | ||
| S-5 | 0.5364 | 3.93E-02 | ||
| PC3 | S-4 | 0.8560 | 4.67E-05 | |
| S-5 | −0.5320 | 4.12E-02 |
Across the Reno WRF sewershed, two neighborhood sites and the WRF were significant on PC1 (Table 1 and Fig. 5). The central Reno sub-catchments (R-4 and R-5) and WRF monitoring described 42% of the total data variance over the sampling period and comprised PC1. Along this component R-5 had the largest contribution and was R-4 and R-5 were positively correlated to one another (Pearson correlation 0.64, p-value = 0.26), and significantly positively correlated to PC1 (Table 1). Conversely, the WRF had a strong negative correlation to PC1 (−0.75, p-value = 0.005). The second component (PC2) described 32% of the total variance and related the sampling sites R-6, R-1, and R-4 (Fig. 5). This corresponded to a weak positive correlation between R-6 and R-4 (Pearson correlation 0.12, p-value >0.05) and a moderate negative correlation between R-1 and R-4 (Pearson correlation -0.062, p-value = 0.03). Finally, PC3 described 14% of total dataset variance. Despite the proximity of the R-2 and R-3 sampling sites, it was notable that their magnitudes along PC3 were in opposite directions, with R-2 positively correlated to the component and R-3 having a negative correlation that was below the significance level (p > 0.05).
The PCA results did have similarity to the clustering identified through the Getis Ord Gi* spatial autocorrelation tests. As the previous section described, the neighboring S-1, S-3, and R-4, R-5 sites that had the largest weights in PC1 were also among the most likely to have significant clustering and were identified as hot spots, HH clusters, LL clusters, and LH outliers throughout the sampling period. The PCA correlations also highlighted the observation that was depicted in Fig. 4, with the evolution of hot spots first occurring in neighborhoods that were part of the urban center (PC1 sites). Two weeks later, at a time where viral RNA concentrations in sewage decreased across the urban center, a new hot spot emerged across several outlying suburban neighborhoods of Sparks, which were correlated with PC2 and PC3. While analysis with PCA and spatial autocorrelation identified similar patterns, PCA was better able to characterize suburban sites, such as R-6 and S-3, which were often not correlated to the urban sites that they neighbored.
It was notable that in both the Reno and Sparks sewersheds, the trends in WRF influent were grouped with the urban centers, which may represent a relatively large portion of the residential influent flows to the WRF. The most distant sampling sites (S-6, S-5, S-4) were poorly correlated to WRF influent, which has important implications for the inability of WRF monitoring to identify increasing viral RNA trends or the formation of hot spots in outlying communities. Overall, these observations were consistent with the analysis of Hart and Halden (2020b), who identified the importance of considering WRF influent data biasing based on sewer residence time and conditions. However, in the case of Reno and Sparks, WRF monitoring was a good representation of community trends, which may be due to the relatively small sizes of these catchments and the proximity of the WRF to most population centers (McCall et al., 2017). These results also characterize spatial relationships similar to Malaria which was observed to spread differently across urban areas compared to rural areas (Lana et al., 2017).
3.4. SARS-CoV-2 distribution across demographic parameters of principal components
Exploratory data analysis was performed in order to examine differences that could be explain the grouping of sampling sites by PCs. As the previous sections noted, there was a relationship between the north-central sampling sites in Reno and Sparks, which were all grouped into their respective PC1 components and hot spots around the same dates. These were among the most urbanized neighborhoods and indicated that some social-demographic or lifestyle characteristics might be shared by these sites that describes their correlated behavior. Similarly, the more suburban neighborhoods were often clustered together but were not always located in close spatial proximity, additionally these sites were sometimes negatively correlated according to PCA analysis (e.g., R-1 and R-6, S-2 and S-6).
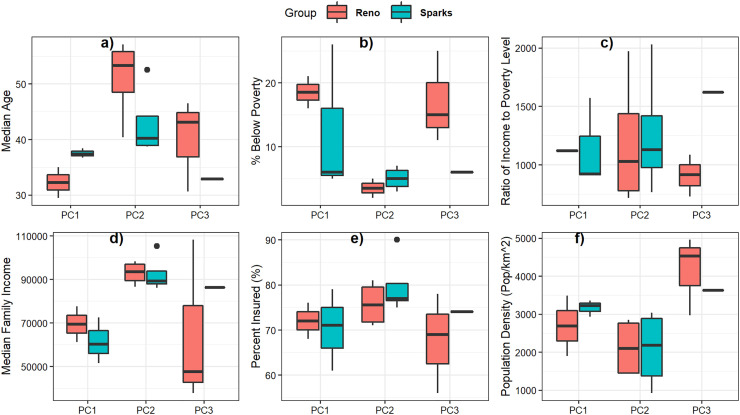
The demographic parameters included differences in economic status (percent of the population below poverty, ratio of income to poverty level, and median family income), median age, percent of population with insurance (including employed and unemployed people), and the population density. The one-way ANOVA test examined the differences in mean values across sampling sites grouped according to PCs in Reno and Sparks. The analysis identified two parameters with a high level of significance (% below poverty and median family income) (refer to Table 2 ). However, neither was significant in both Reno and Sparks sampling areas. Additionally, median age and population density had relatively high levels of significance in Reno sampling sites (p = 0.058 and 0.075, respectively). As illustrated in the boxplots of Fig. 6 , there were generally large differences in the demographic parameters between PC1 and PC2, however the sampling sites grouped in PC3 did not exhibit a clear pattern. Apart from population density the results do not show a consistent geographic structure between the demographic and disease spread patterns in both Reno and Sparks, the results do reveal that certain traits may influence the likelihood of disease. It is important to note that this analysis relied on census data available at the census tract scale, which does not capture all the demographic and economic characteristics that describe how lifestyle and employment factors may impact the timing and level of SARS-CoV-2 outbreaks in a neighborhood.
Table 2.
One-way ANOVA test results of demographic parameters against PCs for Reno and Sparks.
| Parameter | City | Mean Sq | F value | Pr(>F) |
|---|---|---|---|---|
| Median age | Reno | 257.09 | 4.757 | 0.0579 |
| Sparks | 50.84 | 1.975 | 0.233 | |
| % Below poverty | Reno | 222.25 | 10.97 | 0.00989 |
| Sparks | 48.1 | 0.827 | 0.489 | |
| Income to poverty ratio | Reno | 67,778 | 0.385 | 0.696 |
| Sparks | 88,039 | 0.376 | 0.705 | |
| Median family income | Reno | 7.94E+08 | 1.517 | 0.293 |
| Sparks | 8.47E+08 | 9.453 | 0.02 | |
| % Insured | Reno | 56.07 | 0.957 | 0.436 |
| Sparks | 77.29 | 1.265 | 0.359 | |
| Population density (pop/km2) | Reno | 3,618,111 | 4.126 | 0.0746 |
| Sparks | 1,521,926 | 2.364 | 0.189 |
Fig. 6.
Boxplots of demographic parameters across sampling sites, grouped by PCs.
4. Conclusions
The spatial patterns of SARS-CoV-2 in sewage were evaluated using a spatial sampling strategy across twelve neighborhood-scale sewershed sub-catchments. Our results led us to the following conclusions:
-
•
The use of spatial sampling across a sewershed can reveal important relationships as where disease hot spots occur, and the patterns of disease spread. Specifically, both PCA and spatial autocorrelation analysis identified that statistically significant hot spots occurred in outlying suburban neighborhoods approximately two weeks following the emergence of statistically significant hot spots across neighborhoods in the urban center.
-
•
Spatial autocorrelation tests were useful at identifying hot spots when two or more adjacent neighborhoods were impacted, while PCA analysis confirmed these relationships and was also able to detect correlations between sites that were not adjacent but generally had similar characteristics of population density, age, poverty, and income.
-
•
PCA identified that the sites with the highest peaks in viral concentrations (sites located on PC1) had a lower median age, generally had higher rates in poverty and lower family incomes and had higher population densities. These sites were also in the largest hot spot detected, which occurred in early November.
-
•
WRF influent showed evidence of spatial biasing when compared to spatial sampling sites. Specifically, WRF influents were found to be most correlated to sampling sites along PC1, which corresponded to the most urbanized neighborhoods that were generally closest to the WRF.
-
•
The collection of samples from community manholes presented several challenges that may have impacted the variability and representativeness of data collected. Specifically, it was not feasible to deploy autosamplers for collection of 24-h composite samples due to power requirements and safety concerns. Future studies should further explore how this sampling method impacts the representativeness of sewage monitoring data with disease prevalence.
While this research demonstrates how spatial monitoring of SARS-CoV-2 from the sewer network was able to detect hot spots and community patterns of disease spread, the method should be further developed by evaluating the link between sewage monitoring at the neighborhood level and clinical data of disease incidence. This research provides insight into some spatial and socio-economic gradients that correlated to patterns in viral RNA concentrations shed into sewage.
Funding
This study received funding support from the City of Sparks, Nevada, USA utilizing United States Treasury Coronavirus Aid, Relief, and Economic Security (CARES) appropriations.
CRediT authorship contribution statement
Laura Haak: Formal analysis, Methodology, Investigation, Validation, Visualization, Writing – original draft. Blaga Delic: Formal analysis, Methodology, Investigation, Visualization, Writing – original draft. Lin Li: Methodology, Data curation, Validation, Writing – review & editing. Tatiana Guarin: Methodology, Data curation, Validation, Writing – review & editing. Lauren Mazurowski: Methodology, Data curation. Niloufar Gharoon Dastjerdi: Investigation, Data curation, Writing – review & editing. Aimee Dewan: Investigation, Data curation, Methodology. Krishna Pagilla: Conceptualization, Funding acquisition, Supervision, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The team is deeply grateful for the support of the project managers Mr. John Martini, P.E. and Mr. Michael Drinkwater P.E. from the City of Sparks. Rick Warner, of Warner and Associates, provided extensive support coordinating sampling events between the university and agency partners. The researchers also greatly appreciated all the assistance in sampling from university students from multiple research groups, in particular Mingrui Song, Junli Wang, Kenneth Hickenbottom, Nisha Bista, and Yingshi Feng. Staff from the City of Reno, City of Sparks, Washoe County, and contractors provided additional field sampling support and personal safety protective measures and the Truckee Meadows Regional Planning Agency provided technical support.
Editor: Paola Verlicchi
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2021.150390.
Appendix A. Supplementary data
Supplementary material
References
- Ahmad S.S., Aziz N., Butt A., Shabbir R., Erum S. Spatio-temporal surveillance of water based infectious disease (malaria) in Rawalpindi, Pakistan using geostatistical modeling techniques. Environ. Monit. Assess. 2015;187:555. doi: 10.1007/s10661-015-4779-9. [DOI] [PubMed] [Google Scholar]
- Ahmadiara E. Possibility of faecal-oral transmission of novel coronavirus (SARS-CoV-2) via consumption of contaminated foods of animal origin: a hypothesis. J. Food Qual. Hazards Control. 2020 doi: 10.18502/jfqhc.7.1.2445. [DOI] [Google Scholar]
- Ahmed W., Angel N., Bibby K., Bivins A., O’Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anselin L. Local indicators of spatial association—LISA. Geogr. Anal. 1995;27:93–115. doi: 10.1111/j.1538-4632.1995.tb00338.x. [DOI] [Google Scholar]
- Benedetti R., Rossini P., Taddei R. Vegetation classification in the Middle Mediterranean area by satellite data. Int. J. Remote Sens. 1994;15:583–596. doi: 10.1080/01431169408954098. [DOI] [Google Scholar]
- ESRI . Environmental Systems Research Institute; Redlands, CA: 2011. ArcGIS Desktop: Release 10. [Google Scholar]
- Farnham I.M., Singh A.K., Stetzenbach K.J., Johannesson K.H. Chemometrics and Intelligent Laboratory Systems, Fourth International Conference on Environ metrics and Chemometrics Held in Las Vegas, NV, USA, 18-20 September 2000. Vol. 60. 2002. Treatment of nondetects in multivariate analysis of groundwater geochemistry data; pp. 265–281. [DOI] [Google Scholar]
- Field A., Miles J., Field Z. SAGE Publications; 2012. Discovering Statistics Using R. [Google Scholar]
- Foladori P., Cutrupi F., Segata N., Manara S., Pinto F., Malpei F., Bruni L., La Rosa G. SARS-CoV-2 from faeces to wastewater treatment: what do we know? A review. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman M.C., Garn J.V., Sclar G.D., Boisson S., Medlicott K., Alexander K.T., Penakalapati G., Anderson D., Mahtani A.G., Grimes J.E.T., Rehfuess E.A., Clasen T.F. The impact of sanitation on infectious disease and nutritional status: a systematic review and meta-analysis. Int. J. Hyg. Environ. Health. 2017;220:928–949. doi: 10.1016/j.ijheh.2017.05.007. [DOI] [PubMed] [Google Scholar]
- Getis A., Ord J.K. 1992: the analysis of spatial association by use of distance statistics. Geogr. Anal. 1992;24:189–206. [Google Scholar]
- Guo B., Wang Y., Pei L., Yu Y., Liu F., Zhang Donghai, Wang X., Su Y., Zhang Dingming, Zhang B., Guo H. Determining the effects of socioeconomic and environmental determinants on chronic obstructive pulmonary disease (COPD) mortality using geographically and temporally weighted regression model across Xi’an during 2014–2016. Sci. Total Environ. 2021;756 doi: 10.1016/j.scitotenv.2020.143869. [DOI] [PubMed] [Google Scholar]
- Hart O.E., Halden R.U. Computational analysis of SARS-CoV-2/COVID-19 surveillance by wastewater-based epidemiology locally and globally: feasibility, economy, opportunities and challenges. Sci. Total Environ. 2020;730 doi: 10.1016/j.scitotenv.2020.138875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart O.E., Halden R.U. Simulated 2017 nationwide sampling at 13,940 major U.S. sewage treatment plants to assess seasonal population bias in wastewater-based epidemiology. Sci. Total Environ. 2020;727 doi: 10.1016/j.scitotenv.2020.138406. [DOI] [PubMed] [Google Scholar]
- Johnson G., Nour A.A., Nolan T., Huggett J., Bustin S. In: Quantitative Real-time PCR: Methods and Protocols, Methods in Molecular Biology. Biassoni R., Raso A., editors. Springer; New York, NY: 2014. Minimum information necessary for quantitative real-time PCR experiments; pp. 5–17. [DOI] [PubMed] [Google Scholar]
- Jolliffe I.T. Principal component analysis. Technometrics. 2003;45:276. [Google Scholar]
- Kang D., Choi H., Kim J.-H., Choi J. Spatial epidemic dynamics of the COVID-19 outbreak in China. Int. J. Infect. Dis. 2020;94:96–102. doi: 10.1016/j.ijid.2020.03.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassié D., Roudot A., Dessay N., Piermay J.-L., Salem G., Fournet F. Development of a spatial sampling protocol using GIS to measure health disparities in Bobo-Dioulasso, Burkina Faso, a medium-sized African city. Int. J. Health Geogr. 2017;16:14. doi: 10.1186/s12942-017-0087-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima M., Ahmed W., Bibby K., Carducci A., Gerba C.P., Hamilton K.A., Haramoto E., Rose J.B. SARS-CoV-2 in wastewater: state of the knowledge and research needs. Sci. Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.139076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lana R.M., Riback T.I.S., Lima T.F.M., da Silva-Nunes M., Cruz O.G., Oliveira F.G.S., Moresco G.G., Honório N.A., Codeço C.T. Socioeconomic and demographic characterization of an endemic malaria region in Brazil by multiple correspondence analysis. Malar. J. 2017;16:397. doi: 10.1186/s12936-017-2045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lê S., Josse J., Husson F. FactoMineR: a package for multivariate analysis. J. Stat. Softw. 2008;25:1–18. doi: 10.18637/jss.v025.i01. [DOI] [Google Scholar]
- Lee S., Kim T., Lee E., Lee C., Kim H., Rhee H., Park S.Y., Son H.-J., Yu S., Park J.W., Choo E.J., Park S., Loeb M., Kim T.H. Clinical course and molecular viral shedding among asymptomatic and symptomatic patients with SARS-CoV-2 infection in a community treatment center in the Republic of Korea. JAMA Intern. Med. 2020 doi: 10.1001/jamainternmed.2020.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lhermitte S., Verbesselt J., Verstraeten W.W., Coppin P. A comparison of time series similarity measures for classification and change detection of ecosystem dynamics. Remote Sens. Environ. 2011;115:3129–3152. doi: 10.1016/j.rse.2011.06.020. [DOI] [Google Scholar]
- Li H., Li Hui, Ding Z., Hu Z., Chen F., Wang K., Peng Z., Shen H. Spatial statistical analysis of Coronavirus Disease 2019 (Covid-19) in China. Geospatial Health. 2020;15 doi: 10.4081/gh.2020.867. [DOI] [PubMed] [Google Scholar]
- Li L., You Y., Pagilla K. Density-based separation of microbial functional groups in activated sludge. Int. J. Environ. Res. Pub. Health. 2020;17:376. doi: 10.3390/ijerph17010376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo A., Maisongrande P. Searching for trends of change through exploratory data analysis of time series of remotely sensed images of SW Europe and NW Africa. International Journal of Remote Sensing. 2008;29:5237–5245. doi: 10.1080/01431160802036441. [DOI] [Google Scholar]
- McCall A.-K., Palmitessa R., Blumensaat F., Morgenroth E., Ort C. Modeling in-sewer transformations at catchment scale – implications on drug consumption estimates in wastewater-based epidemiology. Water Res. 2017;122:655–668. doi: 10.1016/j.watres.2017.05.034. [DOI] [PubMed] [Google Scholar]
- McLafferty S. Disease cluster detection methods: recent developments and public health implications. Ann. GIS. 2015;21:127–133. doi: 10.1080/19475683.2015.1008572. [DOI] [Google Scholar]
- Mohapatra S., Menon N.Gayathri, Mohapatra G., Pisharody L., Pattnaik A., Menon N.Gowri, Bhukya P.L., Srivastava M., Singh M., Barman M.K., Gin K.Y.-H., Mukherji S. The novel SARS-CoV-2 pandemic: possible environmental transmission, detection, persistence and fate during wastewater and water treatment. Sci. Total Environ. 2021;765 doi: 10.1016/j.scitotenv.2020.142746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollalo A., Vahedi B., Rivera K.M. GIS-based spatial modeling of COVID-19 incidence rate in the continental United States. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odoi A., Martin S.W., Michel P., Holt J., Middleton D., Wilson J. Geographical and temporal distribution of human giardiasis in Ontario, Canada. Int. J. Health Geogr. 2003;2:5. doi: 10.1186/1476-072X-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . 2020. R: A Language and Environment for Statistical Computing. [Google Scholar]
- Ramírez-Aldana R., Gomez-Verjan J.C., Bello-Chavolla O.Y. Spatial analysis of COVID-19 spread in Iran: insights into geographical and structural transmission determinants at a province level. PLoS Negl. Trop. Dis. 2020;14 doi: 10.1371/journal.pntd.0008875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H., Zhao L., Zhang A., Song L., Liao Y., Lu W., Cui C. Early forecasting of the potential risk zones of COVID-19 in China’s megacities. Sci. Total Environ. 2020;729 doi: 10.1016/j.scitotenv.2020.138995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saawarn B., Hait S. Occurrence, fate and removal of SARS-CoV-2 in wastewater: current knowledge and future perspectives. J. Environ. Chem. Eng. 2021;9 doi: 10.1016/j.jece.2020.104870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samphutthanon R., Tripathi N.K., Ninsawat S., Duboz R. Spatio-temporal distribution and hotspots of hand, foot and mouth disease (HFMD) in Northern Thailand. Int. J. Environ. Res. Public Health. 2014;11:312–336. doi: 10.3390/ijerph110100312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.R., Nancharaiah Y.V., Gu X., Lee W.L., Rajal V.B., Haines M.B., Girones R., Ng L.C., Alm E.J., Wuertz S. Making waves: wastewater surveillance of SARS-CoV-2 for population-based health management. Water Res. 2020;184 doi: 10.1016/j.watres.2020.116181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. Census Bureau, American Community Survey ACS Demographic and Housing Estimates. 2019. https://data.census.gov/cedsci/table?q=ACSDP1Y2019.DP05%20United%20States&g=0400000US32&tid=ACSDP1Y2019.DP05&hidePreview=true
- Uppal T., Khazaieli A., Snijders A.M., Verma S.C. Inactivation of human coronavirus by FATHHOME's dry sanitizer device: rapid and eco-friendly ozone-based disinfection of SARS-CoV-2. Pathogens. 2021;10:339. doi: 10.3390/pathogens10030339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.-F., Stein A., Gao B.-B., Ge Y. A review of spatial sampling. Spat. Stat. 2012;2:1–14. doi: 10.1016/j.spasta.2012.08.001. [DOI] [Google Scholar]
- Wickham H. Springer-Verlag; New York: 2016. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
- Wigginton R., Ye K., Y. M., Ellenberg R. Emerging investigators series: the source and fate of pandemic viruses in the urban water cycle. Environ. Sci. Water Res. Technol. 2015;1:735–746. doi: 10.1039/C5EW00125K. [DOI] [Google Scholar]
- Wu F., Zhang J., Xiao A., Gu X., Lee W.L., Armas F., Kauffman K., Hanage W., Matus M., Ghaeli N., Endo N., Duvallet C., Poyet M., Moniz K., Washburne A.D., Erickson T.B., Chai P.R., Thompson J., Alm E.J. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. mSystems. 2020;5 doi: 10.1128/mSystems.00614-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Ellenberg R.M., Graham K.E., Wigginton K.R. Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environ. Sci. Technol. 2016;50:5077–5085. doi: 10.1021/acs.est.6b00876. [DOI] [PubMed] [Google Scholar]
- Yeo C., Kaushal S., Yeo D. Enteric involvement of coronaviruses: is faecal–oral transmission of SARS-CoV-2 possible? Lancet Gastroenterol. Hepatol. 2020;5:335–337. doi: 10.1016/S2468-1253(20)30048-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N., Gong Y., Meng F., Shi Y., Wang J., Mao P., Chuai X., Bi Y., Yang P., Wang F. Comparative study on virus shedding patterns in nasopharyngeal and fecal specimens of COVID-19 patients. Sci. China Life Sci. 2020 doi: 10.1007/s11427-020-1783-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Li C., Zhao G., Chu H., Wang D., Yan H.H.-N., Poon V.K.-M., Wen L., Wong B.H.-Y., Zhao X., Chiu M.C., Yang D., Wang Y., Au-Yeung R.K.H., Chan I.H.-Y., Sun S., Chan J.F.-W., To K.K.-W., Memish Z.A., Corman V.M., Drosten C., Hung I.F.-N., Zhou Y., Leung S.Y., Yuen K.-Y. Human intestinal tract serves as an alternative infection route for Middle East respiratory syndrome coronavirus. Sci. Adv. 2017;3 doi: 10.1126/sciadv.aao4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., Guan L., Wei Y., Li H., Wu X., Xu J., Tu S., Zhang Y., Chen H., Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material