Topoisomerase I is the target for a number of widely prescribed anticancer drugs that are based on camptothecin. In this issue of Cell Chemical Biology, Flor et al. demonstrate that the cellular response to camptothecin is mediated by lipid-derived electrophiles that are generated as a result of drug-induced oxidative stress.
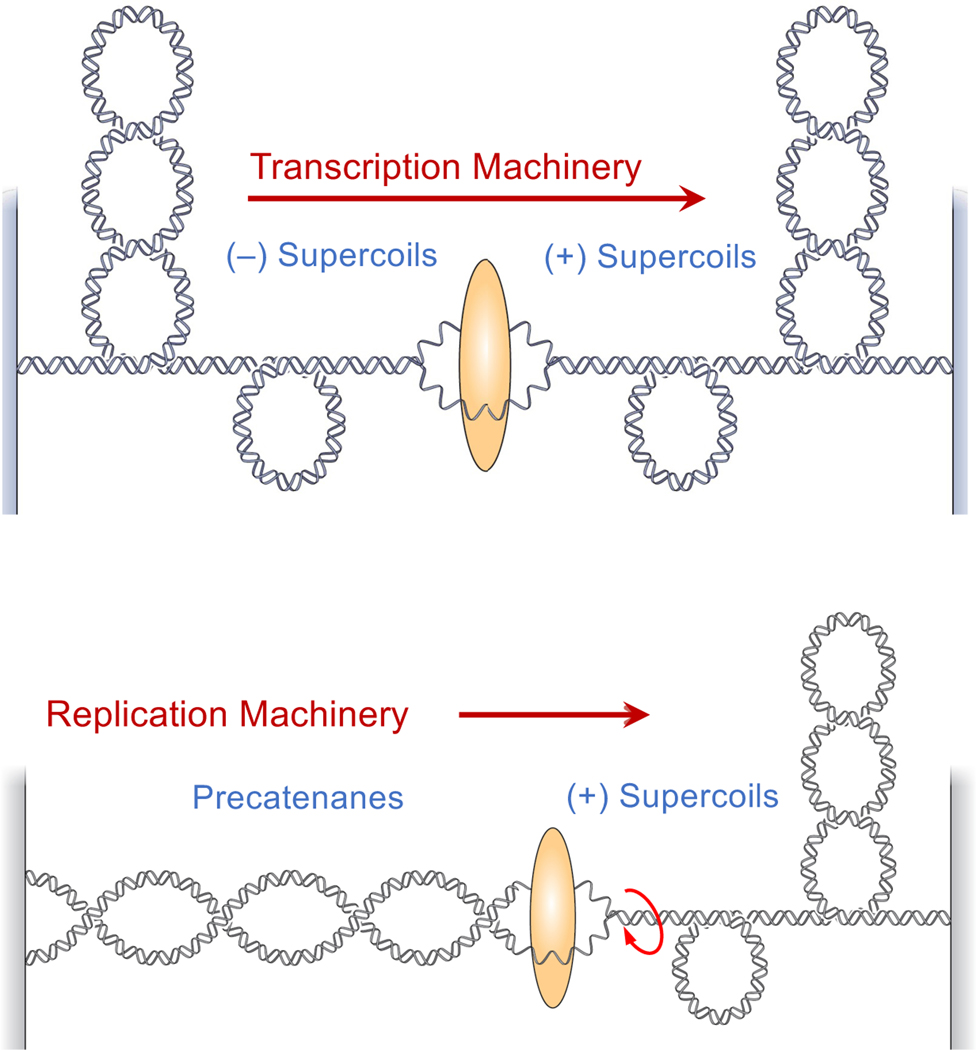
DNA topoisomerases are enzymes that regulate DNA underwinding (i.e., negative supercoiling) and overwinding (i.e., positive supercoiling) (Ashley and Osheroff, 2019; Deweese and Osheroff, 2009; Pommier et al., 2016). These enzymes relieve the torsional strain caused by the accumulation of positive DNA supercoils ahead of replication and transcription complexes and the accumulation of negative supercoils behind transcription complexes (Figure 1) by making transient breaks in the nucleic acid backbone (Ashley and Osheroff, 2019; Deweese and Osheroff, 2009; Pommier, 2013). Topoisomerase I plays an important role in alleviating this strain in human cells. The enzyme removes positive and negative supercoils by generating a single-stranded break in the double helix and allowing controlled rotation about the break (Ashley and Osheroff, 2019; Pommier, 2013). Human topoisomerase IIα and IIβ also play critical roles in alleviating torsional and axial (knotting and tangling) strain in DNA. These latter enzymes act by generating a double-stranded break in the genetic material and passing a separate double helix through the break (Ashley and Osheroff, 2019; Deweese and Osheroff, 2009; Pommier, 2013).
Figure 1. Effects of transcription and replication on DNA topology.
Transcription (top) induces positive (+) DNA supercoils ahead of the advancing transcription machinery and negative (–) DNA supercoils behind the machinery. Replication (bottom) induces positive (+) supercoils ahead of the advancing fork and precatenanes behind the fork. Topoisomerase I is able to remove the accumulated positive and negative supercoils that accumulate in both systems. In contrast, only type II topoisomerases are able to act on the precatenanes that accumulate behind the replication fork.
Beyond their critical cellular functions, human topoisomerases are the targets for some of the most widely prescribed anticancer drugs worldwide (Deweese and Osheroff, 2009; Ketron and Osheroff, 2014; Pommier, 2009, 2013). The article by Flor et al. (Flor et al., 2021) in this issue of Cell Chemical Biology draws an important link between the actions of drugs that target topoisomerase I and oxidative stress in treated cells.
Topoisomerase-targeted anticancer drugs rely on the fact that these enzymes generate DNA strand breaks by the nucleophilic attack of active site tyrosine residues on the sugar-phosphate backbone of the double helix (Ashley and Osheroff, 2019; Deweese and Osheroff, 2009; Pommier, 2013). In order to maintain genomic integrity during DNA cleavage, topoisomerases covalently attach to the newly generated DNA termini through the formation of phosphotyrosine bonds. These hallmark covalent enzyme-cleaved DNA complexes are known as cleavage complexes (Ashley and Osheroff, 2019; Deweese and Osheroff, 2009; Pommier, 2013).
Because the critical nucleic acid functions of topoisomerases require these enzymes to generate DNA strand breaks, topoisomerases are dualistic in nature. While critical to organismal survival, they are also extremely dangerous enzymes (Ashley and Osheroff, 2019; Deweese and Osheroff, 2009; Pommier, 2013) (Figure 2). Topoisomerase-targeted drugs take advantage of this Jekyll/Hyde character and act in an insidious fashion. They bind at the topoisomerase-DNA interface, form specific interactions with the enzyme, and insert themselves between the terminal phosphate and hydroxyl moieties of the cleaved DNA. As a result, these drugs act as “molecular doorstops” and inhibit the enzyme from ligating the DNA break (Ashley and Osheroff, 2019; Deweese and Osheroff, 2009; Pommier, 2013). This action stabilizes cleavage complexes and converts short-lived topoisomerase-generated DNA strand breaks into long-lived proteinaceous roadblocks in the genetic material (Deweese and Osheroff, 2009). When transcription or replication complexes attempt to traverse these stabilized cleavage complexes, critical nucleic process are blocked and the enzymes are often rendered incapable of resealing the DNA breaks (Deweese and Osheroff, 2009; Pommier, 2009, 2013). In this latter case, dedicated DNA damage and recombination pathways must be recruited to repair the resulting DNA lesions. If the DNA strand breaks overwhelm repair systems, cell death pathways may be triggered (Deweese and Osheroff, 2009; Pommier, 2009, 2013). Because topoisomerase-targeted drugs function by converting topoisomerases into potentially lethal agents that fragment the genome (as opposed to robbing the cell of essential enzyme activities), they are referred to as topoisomerase poisons to distinguish them from catalytic inhibitors (Deweese and Osheroff, 2009; Pommier, 2009, 2013).
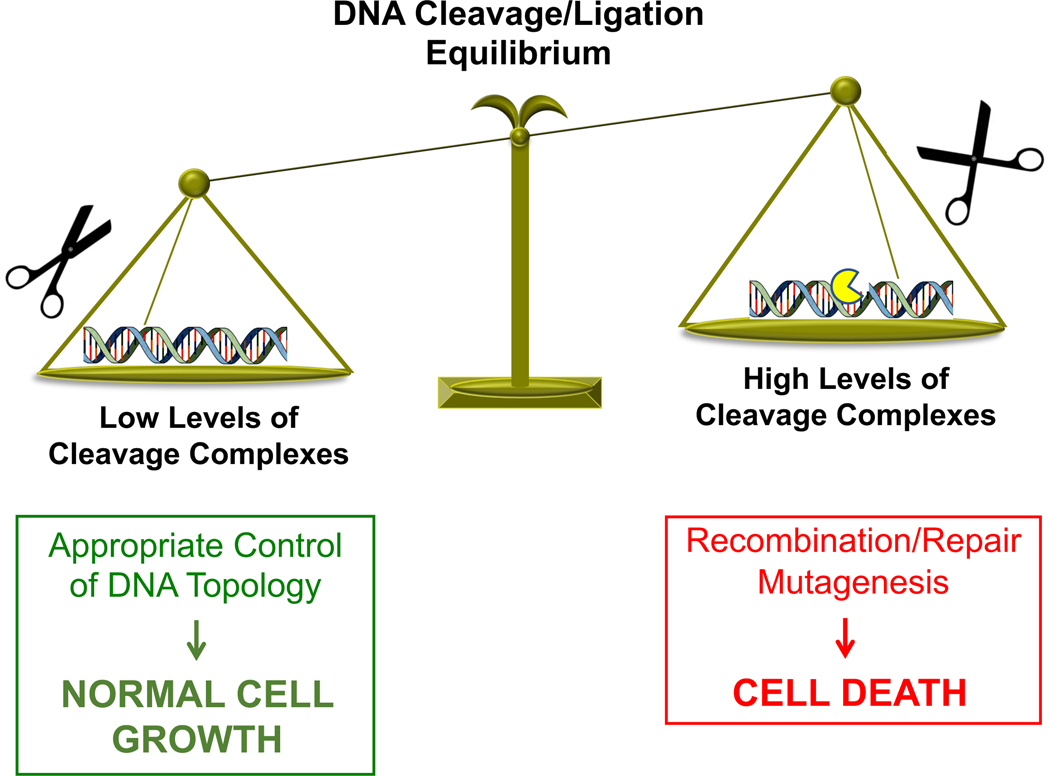
Figure 2. Dualistic nature of topoisomerase I.
Because topoisomerase I (yellow) generates DNA strand breaks as a requisite step in its catalytic cycle, it has a Jekyll/Hyde character. A low level of topoisomerase I-DNA cleavage complexes (left) is essential if the enzyme is to perform its critical cellular functions without harming the cell. Conversely, if the level of topoisomerase I-DNA cleavage complexes becomes too high (right), the actions of DNA tracking systems can convert these transient complexes to DNA strand breaks that no longer can be resealed by the enzyme. The resulting DNA breaks, as well as the inhibition of essential nucleic acid processes, initiate recombination/repair pathways that can generate DNA aberrations and trigger cell death pathways. This is the basis for the actions of the camptothecins and other topoisomerase I-targeted anticancer drugs.
Human topoisomerase I is the target for several poisons that are semisynthetic derivatives of camptothecin, which is derived from the yew tree Camptotheca acuminata (Pommier, 2009, 2013). These derivatives, which include topotecan and the pro-drug, irinotecan, are used to treat a variety of human cancers, including lung, ovarian, and other solid tumors (Pommier, 2009, 2013). Although the actions of camptothecin-based drugs and other topoisomerase I poisons against the human enzyme have been well established, the article by Flor et al. (Flor et al., 2021) adds another layer of complexity to the mechanism by which they induce cell death. These authors report that lipid-derived electrophiles help mediate the cellular effects of topoisomerase I poisons. It has been shown previously that topoisomerase I and II poisons induce redox stress in treated cells, which leads to the generation of reactive oxygen species and lipid peroxidation products (Flor et al., 2016; Flor et al., 2017). Oxidative stress may be caused by drug metabolism or toxicity/cell death pathways. The article by Flor et al. demonstrated that camptothecins induce lipid peroxidation and proteomic signatures of redox stress in tumor cells. Furthermore, treatment of cells with antioxidants prior to camptothecin muted the response to the drug, indicating a role for oxidative stress in drug action.
The authors also showed that 4-hydroxy-2-nonenal, a reactive lipid peroxidation product, was sufficient to increase the concentration of topoisomerase I-DNA cleavage complexes in cells and alter enzyme activity in vitro, even in the absence of camptothecin. Moreover, the toxicity of 4-hydroxy-2-nonenal was increased in cells that lacked tyrosyl-DNA phosphodiesterase 1, an enzyme that removes covalently attached topoisomerase I from the ends of cleaved DNA (Pommier, 2013). Taken together, these findings provide strong evidence that the induction of topoisomerase I-mediated DNA cleavage by the lipid peroxidation product contributes to the cellular sensitivity to camptothecin.
A unique aspect of the Flor et al. article is the mechanism by which topoisomerase I-mediated DNA cleavage is induced. Because the presence of DNA lesions generated by reactive oxygen species and lipid peroxidation products are known to increase topoisomerase I- and II-mediated DNA strand breaks, it has been proposed that cleavage enhancement induced by oxidative stress is triggered by genomic damage (Pommier and Osheroff, 2011). However, Flor et al. provides evidence for an alternative mechanism. The authors demonstrate that 4-hydroxy-2-nonenal forms a covalent adduct with topoisomerase I, modifying cysteine 630 by a Michael addition. Thus, this is the first report of a covalent topoisomerase I poison. A variety of reactive quinones, polyphenols, and isothiocyanates have been identified as covalent topoisomerase II poisons (Ketron and Osheroff, 2014). These compounds adduct cysteine residues in the type II enzyme that are outside of the DNA cleavage-ligation active site and are believed to enhance DNA cleavage by closing the N-terminal protein clamp (Ketron and Osheroff, 2014). Although cysteine 630 in human topoisomerase I is proximal to the active site tyrosine 723, the mechanism by which adduction increases the formation of DNA cleavage complexes by the type I enzyme is currently unknown.
The actions of camptothecin as a topoisomerase I poison have been known since the 1980s (Pommier, 2009). The work of Flor et al. demonstrates the complexity of translating enzymology to cellular pathways and emphasizes that we still have much to learn about the actions of camptothecins and the effects of stress on chemotherapy. It also suggests that it may be possible to modulate oxidative stress in order to exacerbate the actions of topoisomerase I-targeted drugs in cancer cells or mute them in non-cancerous tissues.
ACKNOWLEDGEMENTS
The author acknowledges support from grant R01 GM126363 from the National Institutes of Health and Merit Review award I01 Bx002198 from the United States Veterans Administration.
Footnotes
DECLARATION OF INTERESTS
The author declares no competing interests.
REFERENCES
- Ashley RE, and Osheroff N. (2019). Recognition of DNA topology by topoisomerases: mathematics at the molecular level. In Knots, Low-Dimensional Topology and Applications, Adams CC, Gordon CM, Jones VFR, Kauffman LH, Lambropoulou S, Millet K, Przytycki JH, Ricca R, and Sazdanovic R, eds. (New York: Springer; ), pp. 411–433. [Google Scholar]
- Deweese JE, and Osheroff N. (2009). The DNA cleavage reaction of topoisomerase II: wolf in sheep’s clothing. Nucleic Acids Res 37, 738–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flor A, Wolfgeher D, Jing L, Hanakahi LA, and Kron SJ (2021). Lipid-derived electrophiles mediate the effects of chemotherapeutic topoisomerase I poisons. Cell Chem Biol 28, 1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flor AC, Doshi AP, and Kron SJ (2016). Modulation of therapy-induced senescence by reactive lipid aldehydes. Cell Death Discov 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flor AC, Wolfgeher D, Wu D, and Kron SJ (2017). A signature of enhanced lipid metabolism, lipid peroxidation and aldehyde stress in therapy-induced senescence. Cell Death Discov 3, 17075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketron AC, and Osheroff N. (2014). Phytochemicals as anticancer and chemopreventive topoisomerase II poisons. Phytochem Rev 13, 19–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y. (2009). DNA topoisomerase I inhibitors: chemistry, biology, and interfacial inhibition. Chem Rev 109, 2894–2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y. (2013). Drugging topoisomerases: lessons and challenges. ACS Chem Biol 8, 82–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y, and Osheroff N. (2011). Topoisomerase-induced DNA damage. In DNA Topoisomerases and Cancer, Pommier Y, ed. (New York: Springer; ), pp. 145–154. [Google Scholar]
- Pommier Y, Sun Y, Huang SN, and Nitiss JL (2016). Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat Rev Mol Cell Biol 17, 703–721. [DOI] [PMC free article] [PubMed] [Google Scholar]