Summary
Mantle cell lymphoma (MCL) is an aggressive subtype of non-Hodgkin's lymphoma and one of the most challenging blood cancers to combat due to frequent relapse after treatment. Here, we developed the first-in-class BTK/PI3K/BRD4 axis inhibitor SRX3262, which simultaneously blocks three interrelated MCL driver pathways – BTK, PI3K-AKT-mTOR and MYC. SRX3262 concomitantly binds to BTK, PI3K, and BRD4, exhibits potent in vitro and in vivo activity against MCL, and overcomes the Ibrutinib resistance resulting from the BTK-C481S mutation. Our results reveal that SRX3262 inhibits IgM-induced BTK and AKT phosphorylation and abrogates binding of BRD4 to MYC loci. SRX3262 promotes c-MYC destabilization, induces cell cycle arrest and apoptosis, and shows antitumor activity in in vivo xenograft models. Together, our study provides mechanistic insights and rationale for the use of the triple BTK/PI3K/BRD4 activity inhibitors as a new approach to treat MCL.
Subject area: Medical Biochemistry, Molecular Physiology, Cancer
Graphical abstract

Highlights
-
•
BTK/PI3K/BRD4 axis inhibitor SRX3262 simultaneously blocks three MCL driver pathways
-
•
SRX3262 binds to BTK, PI3K, and BRD4 and exhibits potent anti-MCL activity in vivo
-
•
SRX3262 inhibits BTK and AKT phosphorylation and abrogates binding of BRD4 to MYC
-
•
SRX3262 promotes c-MYC destabilization and induces cell cycle arrest and apoptosis
Medical biochemistry; Molecular physiology; Cancer;
Introduction
Mantle cell lymphoma (MCL) is an aggressive subtype of non-Hodgkin's lymphoma characterized by poor prognosis. MCL represents one of the most challenging B-cell lymphomas to combat due to the disease complexity and frequent relapse after treatment (Jain and Wang, 2019; Maddocks, 2018). Despite recent advances in therapeutic approaches, MCL remains incurable, with the median extended lifespan of patients with MCL being two to five years and decreasing to one to two years in patients with relapsed MCL (Kumar et al., 2019). The complexity of MCL pathogenesis arises from diverse molecular alterations, and no unified treatment is currently available for patients with MCL. A number of genes and signaling pathways, including BTK, PI3K–AKT–mTOR, CDK4/6, and MYC, have become targets for therapeutic intervention.
Bruton's tyrosine kinase (BTK) is constitutively activated in MCL and is recognized as one of the crucial mediators of cancer cell survival. BTK is a member of the TEC family of nonreceptor protein tyrosine kinases. It signals downstream of the B-cell receptor (BCR) and activates the phospholipase PLCγ, an enzyme that produces a phospholipid signaling messenger resulting in calcium mobilization and activation of several signaling cascades, including mammalian PI3K–AKT–mTOR (Merolle et al., 2018). The prosurvival PI3K–AKT–mTOR signaling cascade has been identified as a major pathway which is abnormally activated in cancer cells when BTK is inhibited. In normal B cells, PI3K is required for cell development and functions as a transducer of BCR signaling, regulating proliferation, differentiation, apoptosis, and survival (Fruman et al., 1999). In MCL, the PI3Kδ isoform is predominantly overexpressed and activated, while the PI3Kα isoform supports the survival of B cells with defective BCR signaling (Chiron et al., 2014). Activation of AKT, which is linked to phosphorylation of downstream proteins, is also found in many MCL tumors.
Another key driver of MCL pathogenesis is the MYC oncogene. The epigenetic regulator bromodomain-containing protein 4 (BRD4) mediates MYC transcription and therefore has emerged as an attractive target to downregulate MYC expression (Dang, 2012). BRD4 is recruited to gene promoters through binding of its bromodomains to acetylated lysine residues in histone proteins, and a number of inhibitors have been developed to disrupt this interaction. These include JQ1 and I-BET151 inhibitors which displace BRD4 from acetylated chromatin (Andrews et al., 2017; Filippakopoulos et al., 2010; Guo et al., 2020), leading to transcriptional repression of MYC, cell cycle growth arrest, and apoptosis of cancer cells (Dawson et al., 2011). While the development of inhibitors for the BET protein family and particularly BRD4 has rapidly grown, less progress is seen in the development of selective PI3Kδ inhibitors. Although Idelalisib, a PI3Kδ inhibitor, has begun to show significant efficacy toward relapsed-refractory MCL, more effective PI3Kδ inhibitors with enhanced response rate and safety profile are required.
Ibrutinib was the first BTK inhibitor approved by the Food and Drug Administration (FDA) in 2013 for relapsed-refractory MCL with 68% of objective response rate and 23% complete response (Byrd et al., 2013; Sarkozy and Ribrag, 2020). While Ibrutinib treatment tends to initially be effective for MCL, very few patients achieve complete remission. Furthermore, the selective pressure of Ibrutinib monotherapy has been shown to lead to the development of drug resistance. Ibrutinib promotes mutation of cysteine 481 of BTK – which is required for irreversible binding of Ibrutinib to BTK – and/or mutations in PLCγ. These BTK pathway mutations are culprits in the drug-resistance mechanisms of not only MCL but also chronic lymphocytic leukemia (CLL) (Woyach et al., 2017). The median progression-free survival of patients with relapsed-refractory MCL after Ibrutinib monotherapy is one year (Sarkozy and Ribrag, 2020). The poor prognosis after Ibrutinib treatment underscores the importance of the development of novel therapeutic strategies to improve the outcomes for patients with MCL.
As efficacy of a single inhibitor appears to be limited, many patients are increasingly being treated with multiple regimens. Recent preclinical studies report that cotreatment of primary MCL cells with JQ1 (BRD4 inhibitor) and Ibrutinib (BTK inhibitor) is synergistically lethal (Sun et al., 2015). Phase II clinical studies have shown that a combination of PI3Kδ and BTK inhibitors is beneficial in treatment of relapsed-refractory MCL and CLL. Although the combination of single-agent-targeted therapies allows for adjusting dose individually, this approach is also fraught with challenges. Differences in pharmacokinetics, adsorption, distribution, and metabolism of each drug and additive off-target toxicities can be significant enough to warrant dose reductions that may compromise efficacy (Davids et al., 2019; Matulonis et al., 2017). Furthermore, adjusting dose schedule/regimen in the clinic to elicit optimum therapeutic outcome remains challenging and is further affected by patients' compliance to adhering to multiple drugs. To overcome the limitations of the combination-based therapy and increase efficacy while decreasing drug-induced adverse effects, we have focused on developing single-molecule inhibitors highly specific toward multiple synergistic targets.
In this study, we report the first-in-class BTK/PI3K/BRD4 axis inhibitor SRX3262, which simultaneously blocks three MCL targets, BTK, PI3K, and BRD4. SRX3262 exhibits potent in vitro and in vivo activity against MCL cell lines and MCL tumor models. SRX3262 overcomes Ibrutinib resistance resulting from the treatment-induced C481S mutation in BTK. Our study provides a unique therapeutic strategy for treatment of MCL and Ibrutinib-resistant MCL tumors.
Results and discussion
The BTK/PI3K/BRD4 axis inhibitor SRX3262 is cytotoxic to MCL cells
We have previously shown that 5-morpholino-7H-thieno[3,2-b]pyran-7-one (TP) scaffold affords a unique class of dual action inhibitors (Andrews et al., 2017) and, guided by in silico modeling, generated a new series of TP-based inhibitors (Morales et al., 2020) with the ability to bind the catalytic domains of BTK and PI3K and bromodomains of BRD4. Testing these analogues against BTK, PI3K and BRD4 in in vitro binding and kinase assays, we identified a potent compound, SRX3262, which in comparison with previously reported dual BRD4/PI3K activity chemotypes (Andrews et al., 2017), inhibits three protein targets concurrently (Figure 1A). SRX3262 showed high potency against BTK (IC50 = 35 nM), PI3Kδ (IC50 = 16 nM), PI3Kα (IC50 = 64 nM), and both bromodomains of BRD4 (IC50 BD1 = 228 nM, IC50 BD2 = 348 nM) (Figures 1A and 1B). We confirmed the specificity of SRX3262 and lack of off-targeting by screening the inhibitor against more than 460 human kinases in KINOMEscan assays and 40 bromodomains in BROMOscan assays (Figures 1C–1F and Figures S1 and S2).
Figure 1.
SRX3262 is a triple action BTK, PI3K and BRD4 inhibitor
(A) IC50 values (nM) of SRX3262 and other indicated inhibitors of BRD, PI3K and BTK as measured using Alpha Screen assays (BRD4) and in vitro kinase assays (BTK and PI3K). Values for SF2523 are from (Andrews et al., 2017).
(B) Schematic showing that SRX3262 concomitantly binds to and inhibits BTK, PI3K, and BRD4 in mantle cell lymphoma (MCL) cells.
(C) TREEspot analysis of high-throughput BROMOscan. Target effects are indicated by red circles. The BD-containing proteins are divided into eight groups (I-VIII). BET family reside in the group II.
(D) Residual activity (%) of top SRX3262 selected bromodomain targets and controls. See also Figure S1.
(E) Residual activity (%) of top SRX3262 selected kinase targets and controls. See also Figure S2.
(F) TREEspot analysis of high-throughput KINOMEscan.
To examine the inhibition efficacy of SRX3262 in MCL, we performed cytotoxicity assays using three MCL cell lines: JeKo-1, Mino, and Ibrutinib-resistant Granta. Ibrutinib, a BTK covalent inhibitor, the dual PI3K/BRD4 inhibitors SF2523 and SRX3239, and another triple BTK/PI3K/BRD4 inhibitor SRX3240 were tested for comparison. The results of Alamar Blue experiments performed after 48 h of drug exposure revealed that the TP inhibitors show dose-dependent antiproliferative activity in all three MCL cell lines (Figures 2A–2C). The IC50 values of SRX3262 were found to be 0.62 μM, 0.13 μM, and 1.2 μM in JeKo-1, Mino, and Ibrutinib-resistant Granta cells, respectively. In contrast, in the same MCL cells, IC50 values of Ibrutinib were considerably higher, 8 μM (JeKo-1), 6.5 μM (Mino), and 20 μM (Granta). These data demonstrate that SRX3262 has significantly augmented efficacy in MCL and Ibrutinib-resistant MCL cell lines.
Figure 2.
SRX3262 is cytotoxic to MCL cells
(A–C) Graphs showing antiproliferative activity determined by Alamar blue assays of JeKo-1, Mino, and Granta MCL cell lines treated with increasing concentrations of indicated inhibitors. Data are represented as mean of triplicate experiments ±SD.
(D) Cell viability assay of JeKo-1 cells treated with SRX3262 or a combination of Ibrutinib + JQ1+BKM120 at a ratio of 1:1:1 (Ramsey et al., 2018; Sun et al., 2015). Data are represented as mean of triplicate experiments ±SD.
(E and F) Cell toxicity of normal splenic B cells (E) or epithelial cells (F) treated with increasing concentrations of SRX3262 or a cocktail of Ibrutinib, JQ1, and BKM120 (1:1:1) assessed after 48 h post-treatment. Error bars in (E) represent ±SD derived from mean of triplicate experiments, n = 3 (error bars are small). The bar graph in (F) is representative of duplicate experiments. Error bars represent ±SD from the mean value. Grouped analysis with two-way ANOVA shows statistically significant p∗∗≤0.01, p∗∗∗≤0.001.
Treatment of the MCL cell line JeKo-1 with SRX3262 decreased cell proliferation to the same extent as treatment with three individual inhibitors JQ1 (BRD4i), Ibrutinib (BTKi), and BKM120 (PI3Ki) (Figure 2D); however, we observed a substantial difference in toxicity of the combination of the three inhibitors and SRX3262 toward normal cells, such as primary tonsillar epithelial and splenic B-cells (Figures 2E and 2F). Cell viability results demonstrated that SRX3262 is less toxic than similar doses of the combination of three individual inhibitors to both normal cells.
SRX3262 suppresses proliferation by inducing cell cycle arrest and apoptosis in MCL
To explore the inhibitory mechanism, we assessed the apoptotic response in MCL cell lines treated with SRX3262. Treatment of JeKo-1, Mino, and Z-138 cells with increasing concentrations of SRX3262, up to 5 μM for 24 h, dramatically increased apoptosis, as measured by Annexin V and propidium incorporation in flow cytometry (Figure 3A). Unlike SRX3262, Ibrutinib showed no effect in JeKo-1 cells and mild effect in Mino and Z-138 cells at the same concentrations. SRX3262 promoted apoptotic signaling judging by increased levels of cleaved PARP (Figure S3) and induced G2 phase cell cycle arrest in JeKo-1 cells (Figures 3B and 3C).
Figure 3.
SRX3262 induces apoptosis and cell cycle arrest
(A) Flow cytometry analysis of apoptosis induced in MCL cell lines (Jeko-1, Mino and Z-138) treated with DMSO (−) control, Staurosporine (+) control (20 nM), Ibrutinib (0.5–5 μM) ,or SRX3262 (0.5–5 μM) and assessed after 24 h by evaluating the percentage of Annexin V/propidium iodide positively stained cells. The error bars represent ±SD from the mean value of at least two independent cell passage experiment.
(B and C) Graphs showing flow cytometry analysis of cell cycle arrest 24 h after Ibrutinib (1–5 μM) or SRX3262 (1–5 μM) treatment of Jeko-1 and Mino cells. Data represent the mean value of two independent experiments, and error bars represent ± SD from the mean value.
SRX3262 overcomes Ibrutinib resistance
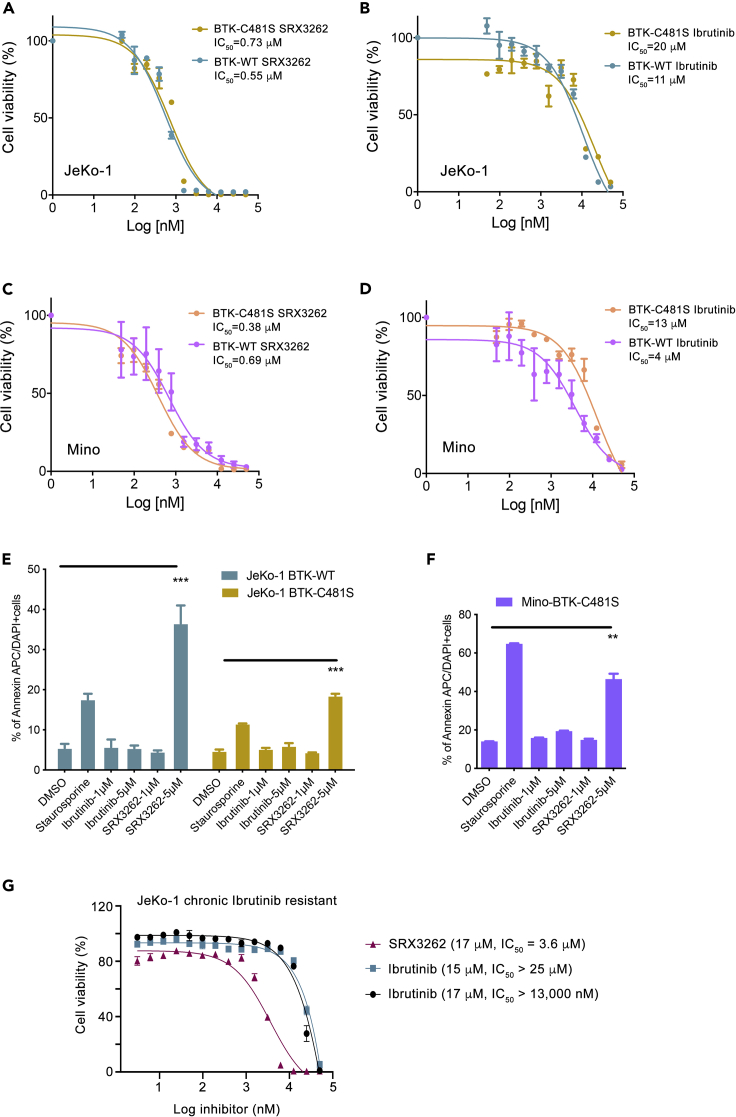
Ibrutinib resistance in MCL was shown to be due to treatment-induced C481S mutation in BTK (BTK- C481S) or due to intrinsic resistance (activation of PI3K/AKT and other pathways), therefore we compared the efficacy of SRX3262 in BTK wildtype (BTK-WT) and engineered Ibrutinib-resistant BTK-C481S JeKo-1 and Mino cell lines using cell viability assays. We found that SRX3262 is ∼20–27 times more effective than Ibrutinib in both JeKo-1 BTK-WT and JeKo-1 BTK-C481S cells. Specifically, SRX3262 exhibited an IC50 of 0.55 μM in JeKo-1 BTK-WT cells and 0.73 μM in BTK-C481S cells, whereas Ibrutinib had IC50 values of 11 μM and 20 μM in the same cells, respectively (Figures 4A and 4B). Similar results were obtained in Mino cells. IC50 values of SRX3262 were 0.69 μM and 0.38 μM in Mino WT and C481S cells, respectively, while the corresponding IC50 values for Ibrutinib were 4 μM and 13 μM (Figures 4C and 4D). These data suggest that SRX3262 is substantially more potent than Ibrutinib and is capable of overcoming the Ibrutinib resistance. In further support, SRX3262 treatment led to an increase in apoptotic cell death in either JeKo-1 BTK-WT or Ibrutinib-resistant cells JeKo-1 BTK-C481S and Mino BTK-C481S (Figures 4E and 4F). In contrast, Ibrutinib failed to induce apoptosis at the same concentrations as SRX3262, up to 5 μM. Furthermore, treatment of the JeKo-1 chronic Ibrutinib-resistant cell line with SRX3262 decreased the cell viability by ∼3–6 times as compared to the treatment of these cells with Ibrutinib (Figure 4G).
Figure 4.
SRX3262 overcomes Ibrutinib resistance
(A–D). Cell proliferation assay of JeKo-1 and Mino cells expressing BTK-WT or Ibrutinib resistant BTK-C481S treated with increasing concentration of SRX3262 and Ibrutinib for 48 h. Data are represented as mean of triplicate experiment ±SD.
(E–G) (E and F) Apoptosis analysis of JeKo-1 WT and C481S mutant cells (E) and Mino C481S mutant cells (F) treated with SRX3262 and Ibrutinib at indicated dosage. Error bars represent ±SD from the mean value of two independent experiments. p∗∗≤0.01, p∗∗∗≤0.001 shows statistically significant by two-way ANOVA and student t-test. (G) Cell viability assessed by Cell titer Glow assay of chronic Ibrutinib resistant (treated with 17 μM or 15 μM Ibrutinib for 5 months) JeKo-1 cells treated with increasing concentrations of Ibrutinib and SRX3262 for 48 hr. Data are represented as mean of triplicate experiments ±SD.
SRX3262 blocks activation of BTK-PI3K signaling in MCL cells
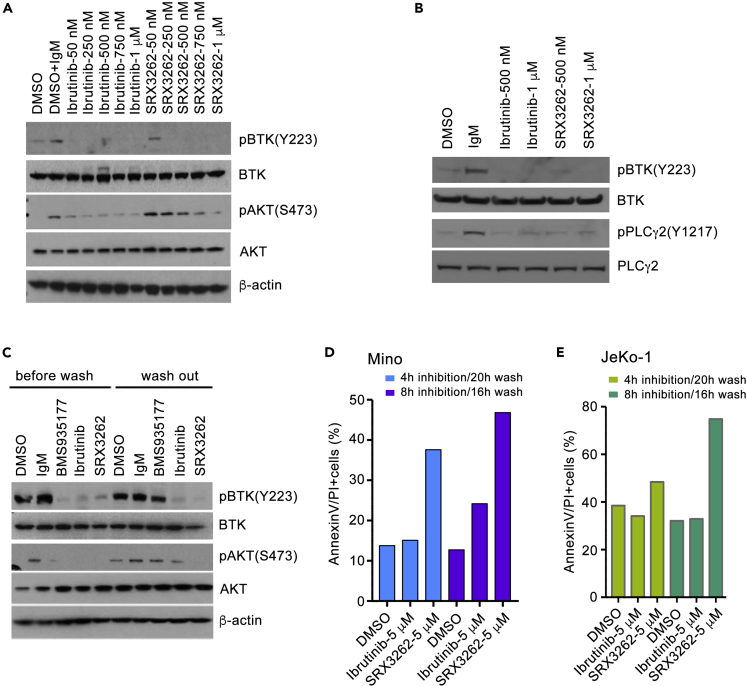
Given the persistent activation of BTK/PI3K signaling cascades in MCL cells, we next investigated the effect of SRX3262 on phosphorylation of BTK and its downstream target PLCγ2 as well as AKT, a downstream target of PI3K, in IgM stimulated MCL cells. As shown in Figure 5A, SRX3262 inhibited phosphorylation of Y223 in BTK and S473 in AKT in a dose-dependent manner in Mino cells, and this inhibition was on par with the inhibitory effect of Ibrutinib. As expected, SRX3262 and Ibrutinib both inhibited phosphorylation of PLCγ2, the downstream target of BTK in JeKo-1 cells (Figure 5B).
Figure 5.
SRX3262 inhibits signaling of BTK and PI3K pathways
(A and B) Western blot analysis of lysates from IgM-stimulated Mino and JeKo-1cells treated with increasing concentrations of Ibrutinib or SRX3262 for 1 hr. BTK and PI3K signaling was assessed by the levels of BTK, phosphorylated at Y223 BTK [pBTK(Y223)], PLCγ2, phosphorylated at Y759 PLCγ2 [pPLCγ2(Y759)], AKT, and phosphorylated at S473 AKT [pAKT(S473)]. See also Figure S4.
(C) Western blots analysis of lysates from anti-IgM-stimulated Mino cells without and with washing out indicated inhibitors (0.5 μM of BMS935177, 1 μM of Ibrutinib and 1 μM of SRX3262) 24 h post washout.
(D and E) Graphs showing flow cytometry analysis of cell apoptosis of Mino and JeKo-1 cells treated for 4 or 8 h followed by drug washout and analyzed 16 or 20 h post washout.
To examine whether the suppression of BTK/PI3K signaling by SRX3262 is sustained, we treated Mino cells with SRX3262 for 1 h and then performed inhibitor-washout experiments (Figure 5C). Ibrutinib, an irreversible inhibitor that covalently binds to BTK, and noncovalent reversible inhibitor BMS-935177 were used as controls. We found that cells treated with SRX3262 and Ibrutinib retained inhibition of BTK phosphorylation even after extensive washes with PBS buffer, whereas cells treated with the reversable inhibitor BMS-935177 regained BTK phosphorylation. While both SRX3262 and Ibrutinib acted as irreversible inhibitors of BTK, their effect on cell apoptosis in washout experiments was different (Figures 5D and 5E). The difference was particularly notable when JeKo-1 and Mino cells were treated with 5 μM of these inhibitors. SRX3262 promoted apoptosis in ∼40–70% cells, whereas only ∼20–30% cells underwent apoptosis upon treatment with Ibrutinib. We note that in contrast to treatment of cells with Ibrutinib, treatment with SRX3262 also led to sustained inhibition of AKT phosphorylation (Figure 5C), indicating that SRX3262 promotes apoptosis by additionally modulating AKT-dependent pathways, one of the major drivers of Ibrutinib resistance in MCL (Zhao et al., 2017). Together, these results suggest that SRX3262 is a potent, irreversible inhibitor that covalently binds to BTK and induces strong apoptotic response even after extensive washes.
SRX3262 inhibits binding of BRD4 to chromatin and destabilizes cMyc
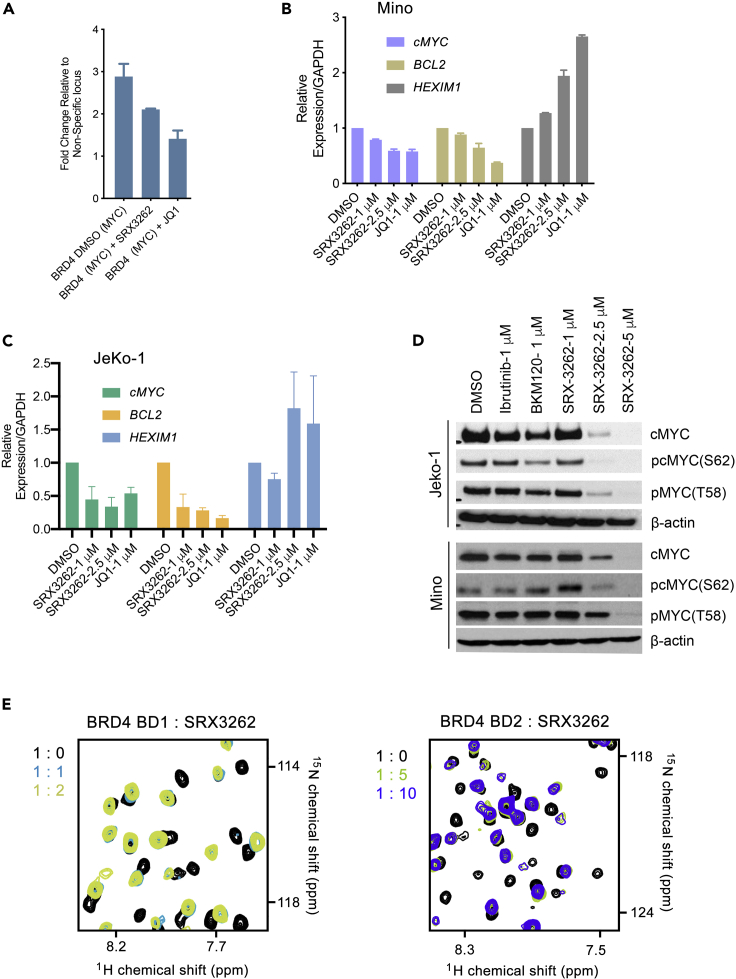
The inhibitory effect of SRX3262 on BRD4 was assessed by NMR titration experiments and chromatin immunoprecipitation (ChIP) assays in JeKo-1 cells. BRD4 has been shown to be involved in regulation of transcription of MYC and other oncogenes through binding to their promoters (Delmore et al., 2011; Mertz et al., 2011). We treated JeKo-1 cells with SRX3262, control DMSO, and JQ1, a commonly used BRD4 inhibitor, and tested occupancy of BRD4 at the cMYC promoter region by ChIP assay (Figure 6A). ChIP analysis revealed that BRD4 localization at the cMYC promoter site was decreased in SRX3262- and JQ1-treated cells, indicating that SRX3262, like JQ1, displaces BRD4 from chromatin. Furthermore, treatment of JeKo-1 and Mino cells with SRX3262 resulted in a decrease of expression of BRD4 target genes, including cMYC and BCL2, and in an increase of expression of HEXIM1 (Figures 6B and 6C). Western blot analysis demonstrated that treatment of Jeko-1 and Mino cells with SRX3262 decreased the cMYC protein level and its phosphorylation at S62, which is necessary for cMYC stability in human cancer cells (Figure 6D). These results suggest that treatment of MCL cell lines with SRX3262 effectively inhibits MYC expression and induces cell death.
Figure 6.
SRX3262 inhibits BRD4 binding
(A) BRD4 ChIP analysis was performed at the cMYC promoter region in JeKo-1 cells treated with SRX3262 (1 μM) or JQ1 (1 μM). Error bars represent GSD from the mean value of two independent experiments.
(B and C) qRT-PCR analysis of cMYC, BCL2 and HEXIM1 expression levels in JeKo-1 and Mino cells treated with DMSO, JQ1 (1μM), or SRX3262 (1 and 2.5 μM). Data represent the mean value of two independent experiments, and error bars represent ± SD from the mean value..
(D) Western blot showing cMYC destabilization in JeKo-1 (top) and Mino (bottom) cell lines in the presence of Ibrutinib, BKM120 (PI3K inhibitor control), or SRX3262.
(E) Superimposed 1H,15N HSQC (heteronuclear single quantum coherence) spectra of uniformly 15N-labeled BRD4 BD1 and BRD4 BD2, recorded while SRX3262 was titrated in. The spectra are color-coded according to the protein:inhibitor molar ratio shown on the left to the spectra.
In agreement with the displacement effect of SRX3262 in cells, titration of SRX3262 into the 15N-labeled BRD4 BD1 and BRD4 BD2 NMR samples led to large chemical shift perturbations (CSPs) in either domain, indicating that SRX3262 binds to both BD1 and BD2 (Figure 6E). CSPs were in the slow exchange regime on the NMR timescale and suggested a tight binding, confirming the IC50 values (Figure 1A). Furthermore, the patterns of CSPs observed in the spectra were similar to the patterns of CSPs induced in these domains by previously reported TP-scaffold compounds (Andrews et al., 2017; Burgoyne et al., 2020; Vann et al., 2020), as well as by acetylated peptides. We concluded that, similar to other TP-scaffold inhibitors, SRX3262 targets the acetyllysine-binding sites of the BRD4 BD1 and BD2.
SRX3262 suppresses tumor growth in a mice xenograft tumor model
Because the BTK/PI3K/BRD4 axis is a major driver of MCL, we next assessed the ability of SRX3262 to block MCL tumor growth of Jeko-1 cells in mouse models. The MCL tumor mouse model was established in 6- to 8-week-old immunodeficient mice using the Jeko-1 cell line. Following tumor growth to ∼100 mm3 during 4–5 weeks post-tumor implantation, mice were randomized into three groups (n = 5) and treated with SRX3262 (60 mg/kg), Ibrutinib (60 mg/kg), or vehicle (solutol with PEG400 in water) daily by oral gavage until tumors were harvested in 3 weeks. Treatment of MCL tumors with SRX3262 resulted in a substantial reduction in tumor volume and tumor mass compared to Ibrutinib-treated and untreated tumors (Figures 7A–7C). We also did not observe any notable changes in body weight for SRX3262 vs Ibrutinib-treated mice (Figure 7D). Collectively, these results demonstrate that SRX3262 is effective in MCL cell tumor growth inhibition and targets oncogenic pathways in vivo and in vitro.
Figure 7.
SRX3262 suppresses tumor growth
(A–D) 4–5 weeks post-tumor implantation mice were treated with vehicle, Ibrutinib (60 mg/kg), or SRX3262 (60 mg/kg) as described in the Methods section. Tumor volume (mm3), mice weight (g), and tumor mass (g) were assessed post-treatment and/or tumor extraction. Representative images of JeKo-1 MCL tumors extracted from mice treated with vehicle, Ibrutinib (60 mg/kg), or SRX3262 (60 mg/kg) are shown in (B). Values are mean ± SEM (n = 5; ∗∗∗P < 0.001; two-way ANOVA. Graph Pad Prism).
Concluding remarks
MCL is an aggressive, heterogeneous, and incurable white blood cell cancer which is characterized by variable clinical presentations and is associated with poor outcome. There is limited therapy available for treatment of MCL and none of the approaches are curative. Some patients are treated with chemoimmunotherapy, including treatment with Ibrutinib, a first oral-targeted agent approved by the US FDA. Although Ibrutinib may show an initial clinical response, this treatment almost uniformly leads to the development of resistance to the drug. A few other MCL treatment regimens, including a combination of Ibrutinib and rituximab or Ibrutinib and venetoclax, prolong response/remission duration, but are associated with acute and long-term toxicity. Despite the recent notable progress in our understanding of the MCL mechanisms and the identification of therapeutic targets, there is still an urgent need to develop the next generation of anti-MCL agents with improved efficacy and safety.
In this study, we report on the new multitargeted small-molecule compound SRX3262 that disrupts three pathways implicated in MCL pathogenesis – BTK, PI3K-AKT-mTOR, and MYC. SRX3262 simultaneously binds to and inhibits the three oncogenic targets BTK, PI3K, and BRD4 with nanomolar potency. Similar to Ibrutinib, SRX3262 is a selective, irreversible BTK inhibitor, which shows potent in vitro and in vivo activity against BTK in MCL cell lines and MCL tumor models. In contrast to Ibrutinib, SRX3262 overcomes Ibrutinib resistance from the treatment-induced mutation BTK-C481S. SRX3262 is characterized by improved efficacy and toxicity and is less toxic than a combination of an equipotent BTK inhibitor, PI3K inhibitor, and BRD4 inhibitor. Collectively, our findings suggest that the BTK/PI3K/BRD4 axis inhibitor SRX3262 is a highly promising lead for the development of new anti-MCL therapeutics.
Limitations of the study
We acknowledge that in the case of small molecule inhibitors, there could potentially be other targets in the proteome that may or may not have a sequence or functional relationship with BTK, PI3K and BRD4.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| goat F(ab) 2 anti-human IgM antibody | Southern Biotech | Cat. # 2022-01 |
| anti-clPARP antibody | Cell Signaling Tech. | Cat. # 9541S |
| anti-pBTK-Y223 antibody | Cell Signaling Tech. | Cat. # 5082S |
| anti-pAKT-S473 antibody | Cell Signaling Tech. | Cat. # 9271S |
| total anti-BTK antibody | Cell Signaling Tech. | Cat. # 8547S |
| total anti-AKT antibody | Cell Signaling Tech. | Cat. # 4691 |
| anti-cMYC antibody | Cell Signaling Tech. | Cat. # ab32072 |
| anti-pS62MYC antibody | Cell Signaling Tech. | Cat. # 13748S |
| anti-pT58MYC antibody | Abcam | Cat. # ab28842 |
| anti-pPLCγ2-Y759 antibody | Cell Signaling Tech. | Cat. # 3872S |
| total anti-PLCγ2 antibody | Cell Signaling Tech. | Cat. # 3872 |
| anti-β-actin antibody | Cell Signaling Tech. | Cat. # sc69879 |
| BRD4 antibody | Bethyl Laboratories | Cat. # A301-985A50 |
| Rabbit mAb Tri-Methyl-Histone H3 (Lys4) (C42D8) | Bethyl Laboratories | Cat. # 9751S |
| Bacterial and virus strains | ||
| Escherichia coli BL21-CodonPlus (De3) RIL | Agilent Technologies | Cat: # 6366546001 |
| pMSCV-ires-GFP | Gateway (Invitrogen) | N/A |
| pMSCV- ires-tdTomato | Gateway (Invitrogen) | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Ibrutinib | Selleck Chemicals | Cat. # S2680 |
| JQ1 | Selleck Chemicals | Cat. # S7110 |
| BKM120 | Selleck Chemicals | Cat. # S2247 |
| SRX3262 | SignalRx Pharm. | N/A |
| SF2523 | SignalRx Pharm. | N/A |
| SRX3239 | SignalRx Pharm. | N/A |
| SRX3240 | SignalRx Pharm. | N/A |
| Staurosporine | Selleck Chemicals | Cat. # S1421 |
| BMS735177 | Selleck Chemicals | Cat. # S8348 |
| Alamar blue reagent | Thermo Fisher Sci. | Cat. # DAL1100 |
| Cell Titer Glow | Promega | Cat. # G9242 |
| PBS | Thermo Scientific | Cat. # 10010-023 |
| HEPES | Oakwood Chemical | Cat. # 047861 |
| NaCl | Fisher | Cat. # S271-1 |
| TCEP HCL | Goldbio | Cat. # TCEP25 |
| 15NH4Cl | Sigma-Aldrich | Cat. # 299251 |
| Multivitamin Tablet | Centrum Walmart | Cat. # 573963323 |
| ZnCl2 | Sigma-Aldrich | Cat. # 229997 |
| Deuterium oxide (D2O) | Sigma-Aldrich | Cat. # D4501 |
| IPTG | Goldbio | Cat. # I2481 |
| RPMI-1640 | ATCC | Cat. # 30-2001 |
| Fetal bovine serum (FBS) | Thermo Fisher Sci. | Cat. # 26140079 |
| Dulbecco’s modified Eagle’s medium (DMEM) | ATCC | Cat. # 30-2002 |
| RIPA buffer | Thermo Scientific | Cat. # 89900 |
| Penicillin/Streptomycin | Thermo Scientific | Cat. # 15070063 |
| DMSO | Sigma | Cat. # D-8779 |
| iScript cDNA synthesis kit | BIORAD | Cat. # 1708890 |
| Thermo BCA assay Kit | Thermo Fisher | Cat. # PI23227 |
| RNeasy Mini kit | Qiagen | Cat. # 74104 |
| SYBR green supermix | Thermo Fisher | Cat. # 4367659 |
| QIAquick PCR Purification Kit | Qiagen | Cat. # 28104 |
| FITC-Annexin V Apoptosis Kit | BD Pharmingen | Cat. # 556419 |
| Propidium Iodide Staining Solution | Sigma | Cat. # P4170 |
| Annexin APC | Thermo Fisher | Cat. # A35110 |
| RNAse A | Thermo Fisher | Cat. # EN0531 |
| Formaldehyde | Alfa Aesar | Cat. # 43368 |
| PreScission (HRV-3C) protease | Home expressed | N/A |
| Thrombin protease | MP Biomedicals | Cat. #154163 |
| Pierce enhanced chemiluminescence substrate | Thermo Fisher | Cat. # 32106 |
| Matrigel | Thermo Fisher Sci. | Cat. # CB-40234 |
| Critical commercial assays | ||
| Alpha Screen assay | Reaction Biology | N/A |
| Kinase assay | Reaction Biology | N/A |
| Kinase assay | Thermo Fisher | N/A |
| Kinome scan | Eurofins DiscoverX | N/A |
| Bromo scan | Eurofins DiscoverX | N/A |
| Experimental models: Cell lines | ||
| JeKo-1 cell line | Thomas Kipps lab | N/A |
| Mino cell line | Thomas Kipps lab | N/A |
| Granta-519 (Granta) cell line | Thomas Kipps lab | N/A |
| Z-138 cell line | ATCC | Cat. # CRL-2003 |
| BTK C481S mutant JeKo-1 cell line | David Winstock Lab (DFCI) | N/A |
| BTK C481S mutant Mino cell line | David Winstock Lab (DFCI) | N/A |
| JeKo-1 BTK WT cell line | David Winstock Lab (DFCI) | N/A |
| Mino-BTK WT cell line | David Winstock Lab (DFCI) | N/A |
| Experimental models: Organisms/strains | ||
| Female, 6–8-week- old, athymic nude mice | The Jackson Laboratory | N/A |
| Oligonucleotides | ||
| Primesr: cMYC (RT-PCR) F-GGTGCTCCATGAGGAGACA R- CCTGCCTCTTTTCCACAGAA |
IDT | NA |
| Primers: BCL2 (RT-PCR) F-CTGCACCTGACGCCCTTCACC R- CACATGACCCCACCGAACTCAAAGA |
IDT | NA |
| Primers: HEXIM1 (RT-PCR) F-CCGGGGAGTAGTTCTGTTGT R-AAGGGAGTGGTAGGCAGAAC |
IDT | N/A |
| Primers: GAPDH (RT-PCR) F-GGCTGGGCAAGGTCATCC R- TCCACCACCCTGTTGCTGTA |
IDT | N/A |
| Primers: c-MYC promoter (ChIP) F-GAGCAGCAGAGAAAGGGAGA R- CAGCCGAGCACTCTAGCTCT |
IDT | N/A |
| Primers: BCL2 (RT-PCR) F-CTGCACCTGACGCCCTTCACC R-CACATGACCCCACCGAACTCAAAGA |
IDT | N/A |
| Recombinant DNA | ||
| pGEX-6P-1 BD1 aa 43-180 | Kutateladze lab | N/A |
| pGEX-4T-1 BD2 aa 342–460 | Kutateladze lab | N/A |
| Software and algorithms | ||
| NMRPipe | Delaglio et al., 1995 | https://www.ibbr.umd.edu/nmrpipe/ |
| GraphPad Prism | GraphPad Software, Inc. | https://www.graphpad.com/scientific-software/prism/ |
| TREEspot | Treespot (online) | https://www.discoverx.com/services/drug-discovery-development-services/treespot-data-analysis |
| Other | ||
| HiPrep™ 16/60 Sephacryl® S-100 HR column | GE Healthcare | Cat. # 17-1165-01 |
| Amicon Ultra 15 mL 3K NMWL centrifugal filter unit | Millipore | Cat. # UFC900308 |
| Varian INOVA 600 MHz NMR spectrometer | Agilent Technologiies | N/A |
| Glutathione Sepharose 4B beads | GE Healthcare | Cat. # 17-0756-01 |
| PreScission (HRV-3C) protease | Home expressed | N/A |
| QuantStudio™ 3 Real-Time PCR System | Thermo Scientific | Cat. #4367659 |
| BD Accuri C6 Flow cytometer | BD Biosciences | N/A |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Donald L. Durden (ddurden@ucsd.edu).
Materials availability
All relevant data supporting the key findings of this study are available within the article and its supplemental information files or from the corresponding authors upon reasonable request. There are restrictions to the availability of SRX3239, SRX3240 and SRX3262 compounds, which were developed and synthesized by SignalRx Pharmaceuticals, Inc.
Data and code availability
All data reported in this paper will be shared by the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and subject details
Cell culture
Human Mantle cell lymphoma (MCL) JeKo-1 and Mino cell lines were cultured in RPMI-1640 with 20% fetal bovine serum (FBS), and the Z-138 and Granta cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% FBS. All cells were supplemented with 1% penicillin/streptomycin and incubated at 37°C with 5% CO2. Cells were frequently checked for the presence of mycoplasma contamination by PCR based detection method.
Method details
Cell lines and reagents
Human Mantle cell lymphoma (MCL) cell lines JeKo-1, Mino, and Granta-519 (Granta) and BTK C481S mutant JeKo-1 and Mino cell lines were received from David Weinstock lab and Thomas Kipps lab and Z-138 was obtained from ATCC. BTK WT and BTK C481S mutant cells were cloned into pMSCV-ires-GFP and pMSCV-ires-tdTomato expression vectors, respectively, using the Gateway vector conversion system (Invitrogen), transfected, and selected for homogeneity. Ibrutinib-resistant cell lines were made by gradually increasing the concentration of Ibrutinib (from 20 nM to 17 μM) for a period of five months. Cells were frequently checked for cell viability assay to determine the IC50 value and assess resistance. JeKo-1 and Mino cells were grown in RPMI-1640 with 20% fetal bovine serum (FBS) and Z-138 and Granta cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% FBS. All cells were supplemented with 1% penicillin/streptomycin and incubated at 37°C with 5% CO2. Normal human tonsillar epithelial cells (RRP008) were obtained from the biorepository at Rady Children’s Hospital (San Diego, CA) under an Institutional Review Board approved protocol and were cultured according to the protocol (Burgoyne et al., 2020). Cell lines were tested for mycoplasma and checked for authenticity against the International Cell Line Authentication Committee (ICLAC; http://iclac.org/databases/cross-contaminations/) list. All antibodies were purchased from Cell Signaling Technology. Ibrutinib, JQ1 and BKM120 were purchased from Selleck Chemicals. SF2523, SRX3239, SRX3240, and SRX3262 were in silico designed using crystal structures and fragment-based pharmacophore analysis, synthesized, and provided by SignalRx Pharmaceuticals, Inc. (Cumming, GA).
Inhibitor selectivity screening and displacement assays
The IC50 measurements for inhibition of BRD4 were performed using Alpha Screen assays from Reaction Biology on a set of His-tagged bromodomains against a tetra-acetylated histone H4 peptide (H4K5ac/8ac/12ac/16ac-Biotin) ligand. BTK and PI3K isoform kinase activities were measured using an γ-P32-ATP derived kinase assay from Reaction Biology and ThermoFisher, respectively. KINOMEscan® and BROMOscan® assays were performed at Eurofins DiscoverX and the data were analyzed using the TREEspot online software tool.
Cell viability assay
JeKo-1, Mino and Granta cells or BTK C481S resistant Jeko-1 and Mino cells were plated in 96-well plates at a density of 2x104 in 100 μl media and incubated overnight. Cells were treated with DMSO (0.1% final) or increasing concentrations of Ibrutinib, SRX3262 or indicated inhibitors for 48 h. Then 100 μL of Alamar blue was added to each well and the cells were incubated for 10 min at RT. For Alamar Blue, fluorescence signals were read as emission at 590 nm after excitation at 560 nm. For chronic Ibrutinib resistant cells, 100 μl of cell titer Glow assay reagent (Promega) was added and luminescence was measured. Percentage of cell viability was calculated from the control (DMSO) and analyzed by nonlinear regression then plotted for dose response using GraphPad Prism (GraphPad Software, Inc.).
Mouse splenic B cell isolation and cell proliferation assay
Primary mouse splenic B cells were isolated from C57 black mouse by using the Pan B cell isolation kit (Miltenyi Biotech) according to the manufacturer’s instructions. 5x104 splenic B cells were seeded in 96 well plate. Cells were stimulated with LPS (10 μg/mL) after 30 mins treatment with SRX3262 or JQ1+BKM120+Ibrutinib at a 1:1:1 ratio. Cell proliferation was assessed as described for cell viability assays after treatment with of Alamar blue reagent.
Western Blot
JeKo-1 and Mino cells were plated in 10 cm tissue culture dishes at a cell density of 2 x 106 and were incubated overnight. The cells were serum starved for 6 hrs. Cells were treated in the presence of increasing concentration of SRX3262 or Ibrutinib for 1 h followed by stimulated with 10μg/mL of goat F(ab) 2 anti-human IgM for 15 min at 37 °C. Whole cell lysates of the treated and untreated JeKo-1 and Mino cells were then prepared using RIPA buffer supplemented with protease and phosphatase/protease inhibitor cocktails (Thermo Scientific). Protein concentration in cell lysates was determined using a bicinchoninic acid (BCA) assay kit. Equal amount of lysates were resolved on a 4-12% SDS-PAGE, transferred to nitrocellulose membranes and probed with one or more of the following antibodies (Key resource table). Secondary antibodies were chosen according to the species of origin of the primary antibody. Protein bands were detected using the Pierce enhanced chemiluminescence (ECL) substrate.
RNA isolation and qRT-PCR
Jeko-1 and Mino cells were seeded at a density 1x105 cells in six well plate and incubated overnight. The cells were treated for 24 hr with 1 μM SRX3262, 2.5 μM SRX3262, 1 μM JQ1 and DMSO before for RNA isolation. Total RNA was extracted using RNeasy Mini kit (QIAGEN). For cDNA preparation 1 μg total RNA was reverse transcribed using iscript cDNA synthesis kit. Amplification of cDNA was performed with 1× SYBR green supermix on a QuantStudio™ 3 Real-Time PCR System. cDNAs were amplified using specific cMYC, BCL2 and HEXIM1 primers. The data were normalized to GAPDH.
FITC Annexin V apoptosis assay
Jeko-1, Mino and Z-138 cells were seeded at a density of 2x106 cells and incubated overnight. Cells were treated with indicated concentration of Ibrutinib, SRX3262, or control Staurosporine and incubated for 24 hr. Cells were harvested, washed twice with cold PBS and labeled with Annexin V-FITC and PI or Annexin APC according to the manufacturer’s protocol. The labeled cells were analyzed using BD Accuri C6 Flow cytometer.
Cell cycle analysis
JeKo-1 and Mino cells were treated with indicated concentrations of SRX3262, SRX3240, SRX3239, SF2523, Ibrutinib or Staurosporine (control) for 24 h. Cells were harvested, washed once with PBS and then fixed with 70% ethanol and stored at -20°C overnight. The fixed cells were collected by centrifugation, washed twice with PBS, resuspended with assay buffer (PI-100 μL/mL, Triton-X-50 μL/mL and RNase A-100 μg/mL) and incubated in the dark for 30 min at RT. The samples were analyzed using BD Accuri C6 Flow cytometer.
Protein expression and purification
The pGEX6P-BRD4 BD1 (aa 43-180) and BD2 (aa 342-460) constructs were generated as previously described (Vann et al., 2020). Briefly, BRD4 BD1 and BD2 constructs were expressed in E. coli BL21 (DE3) RIL in M19 minimal media supplemented with 15NH4Cl and purified as GST fusion proteins. Once the cells reached log phase, cells were induced with IPTG at a final concentration of 1 mM and grown overnight at 16 °C. The bacteria were harvested by centrifugation, resuspended in 10 mM HEPES pH 7.5, 150 mM NaCl, 1 mM TCEP and lysed by sonication. The proteins were purified from the cleared lysate using glutathione Sepharose 4B beads, and the GST tag was cleaved with PreScission or thrombin protease. The proteins were purified using a S100 column (GE Healthcare) equilibrated in 10 mM HEPES pH 7.5, 150 mM NaCl, 1 mM TCEP. The BRD4 BD1 and BD2 fractions were assessed for purity by SDS-PAGE and concentrated to ∼10-20 mg/mL.
NMR spectroscopy
NMR spectroscopy experiments were carried out on a Varian INOVA 600 MHz spectrometer outfitted with a cryogenic probe. Binding of SRX3262 to uniformly 15N-labeled BRD4 BD1 or BRD4 BD2 was characterized by monitoring CSPs in 1H,15N heteronuclear single quantum coherence (HSQC) spectra of BRD4 BD1 and BRD4 BD2. The spectra were collected in the presence of increasing concentrations of SRX3262 in PBS buffer pH 6.8, 10% D2O and processed with NMRPipe (Delaglio et al., 1995).
Chromatin immunoprecipitation
ChIP assays were performed similar to the method described previously (Andrews et al., 2017). Briefly, JeKo-1 cells (1x108) were treated in the presence or absence of SRX3262 (1 μM) or JQ1 (1 μM) for 24 h, cross-linked in 1% formaldehyde, washed with PBS, and harvested. Cross-linked cells were lysed in buffer supplemented with 1% Triton X-100, washed and sonicated at 4°C. Sonicated samples were cleared and 15 μg of sample was subjected to chromatin immunoprecipitation with antibody-conjugated agarose beads overnight at 4°C. ChIP samples were washed, eluted, reverse cross-linked and digested with RNase A overnight followed by a Proteinase K treatment for 2 hr. Purified ChIP DNA was isolated with a QIAquick PCR Purification Kit (Qiagen) and quantified. Purified ChIP and input DNA were analyzed by real-time PCR analysis using primers against the cMYC promoter. Fold enrichment was analyzed by calculating the immunoprecipitated DNA percentage of input DNA in duplicate for each sample.
BCR pathway stimulation
JeKo-1 and Mino cells (1x 106) were serum starved for 6 h and then incubated with or without SRX3262, Ibrutinib, or controls for 1 hr. Treated cells were then resuspended in 1 ml of RPMI-1640 and stimulated with 10 μg/mL of goat F(ab’)2 antihuman IgM antibody at 37°C for 10 min. Stimulated cells were then harvested and lysed with RIPA buffer followed by Western blot analysis for pBTK and pAKT expression.
Inhibitor washout assay
Mino cells were incubated with SRX3262, Ibrutinib or controls for 1 h, washed three times with PBS, and incubated in RPMI-1640 supplemented with 20% FBS and penicillin/streptomycin for 24 h. The BCR pathways in the washed cells were stimulated with 10 μg/mL of goat F(ab’)2 antihuman IgM and then harvested and lysed. The resulting lysates were further analyzed by Western blot analysis.
Animal studies
All procedures involving animals were performed with the approval of the Institutional Animal Care and Use Committee of The University of California San Diego, Moores Cancer Center, which serves to ensure that all federal guidelines concerning animal experimentation are met. Female, 6- to 8-week-old, athymic nude mice (The Jackson Laboratory) were housed and monitored daily in our animal facility. A matrigel suspension (1:1) of Jeko-1 WT cultured cells (1 × 107) was injected subcutaneously into 6- to 8-week-old mice. Approximately 45 weeks, when tumors reached 100 mm3, mice were randomized into vehicle, SRX3262 and Ibrutinib groups (n = 5 animals per group). Mice were treated once daily with SRX3262 (60 mg/kg) and Ibrutinib (60 mg/kg) by oral gavage until tumors were harvested on day 21. Mice weight was recorded daily, and tumor volumes were measured twice a week by caliper and calculated as tumor volume (length x width2) x 2.
Quantification and statistical analysis
To-way ANOVA and Student t-test was used to evaluate differences observed between experimental groups and to compare differences between SRX3262, Ibrutinib, and vehicle-treated control by GraphPad prism. Differences were considered significant if the p value calculated was p∗∗≤0.01, p∗∗∗≤0.001.
Acknowledgments
We thank Muamera Zulcic for help with experiments. This work was supported in part by grants from NIH CA215651 to D.L.D. and CA252707 and HL151334 to T.G.K.
Author contributions
D.P., K.R.V., N.E.S., and G.A.M. performed experiments and together with S.J., D.E., T.G.K., and D.L.D. analyzed the data. K.R.V., T.G.K., and D.L.D. wrote the manuscript with input from all authors.
Declaration of interests
D.L.D. and G.A.M. are consultants for and/or have equity interests in SignalRx Pharmaceuticals, Inc., have financial conflicts of interest regarding the SRX3239, SRX3240 and SRX3262 compounds and have a patent related to this work, patent number WO2020023340A1. T.G.K. is a member of the iScience Editorial Board. Other authors declare no competing interests.
Published: September 24, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102931.
Contributor Information
Tatiana G. Kutateladze, Email: tatiana.kutateladze@cuanschutz.edu.
Donald L. Durden, Email: ddurden@ucsd.edu.
Supplemental information
References
- Andrews F.H., Singh A.R., Joshi S., Smith C.A., Morales G.A., Garlich J.R., Durden D.L., Kutateladze T.G. Dual-activity PI3K-BRD4 inhibitor for the orthogonal inhibition of MYC to block tumor growth and metastasis. Proc. Natl. Acad. Sci. U S A. 2017;114:E1072–E1080. doi: 10.1073/pnas.1613091114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgoyne A.M., Vann K.R., Joshi S., Morales G.A., Vega F.M., Singh A., Pal D., Merati A.B., Kutateladze T.G., Durden D.L. A triple action CDK4/6-PI3K-BET inhibitor with augmented cancer cell cytotoxicity. Cell Discov. 2020;6:49. doi: 10.1038/s41421-020-0181-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd J.C., Furman R.R., Coutre S.E., Flinn I.W., Burger J.A., Blum K.A., Grant B., Sharman J.P., Coleman M., Wierda W.G. Targeting BTK with ibrutinib in relapsed chronic lymphocytic leukemia. N. Engl. J. Med. 2013;369:32–42. doi: 10.1056/NEJMoa1215637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiron D., Di Liberto M., Martin P., Huang X., Sharman J., Blecua P., Mathew S., Vijay P., Eng K., Ali S. Cell-cycle reprogramming for PI3K inhibition overrides a relapse-specific C481S BTK mutation revealed by longitudinal functional genomics in mantle cell lymphoma. Cancer Discov. 2014;4:1022–1035. doi: 10.1158/2159-8290.CD-14-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang C.V. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davids M.S., Kim H.T., Nicotra A., Savell A., Francoeur K., Hellman J.M., Bazemore J., Miskin H.P., Sportelli P., Stampleman L. Umbralisib in combination with ibrutinib in patients with relapsed or refractory chronic lymphocytic leukaemia or mantle cell lymphoma: a multicentre phase 1-1b study. Lancet Haematol. 2019;6:e38–e47. doi: 10.1016/S2352-3026(18)30196-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson M.A., Prinjha R.K., Dittmann A., Giotopoulos G., Bantscheff M., Chan W.I., Robson S.C., Chung C.W., Hopf C., Savitski M.M. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–533. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Delmore J.E., Issa G.C., Lemieux M.E., Rahl P.B., Shi J., Jacobs H.M., Kastritis E., Gilpatrick T., Paranal R.M., Qi J. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P., Qi J., Picaud S., Shen Y., Smith W.B., Fedorov O., Morse E.M., Keates T., Hickman T.T., Felletar I. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fruman D.A., Snapper S.B., Yballe C.M., Davidson L., Yu J.Y., Alt F.W., Cantley L.C. Impaired B cell development and proliferation in absence of phosphoinositide 3-kinase p85alpha. Science. 1999;283:393–397. doi: 10.1126/science.283.5400.393. [DOI] [PubMed] [Google Scholar]
- Guo L., Li J., Zeng H., Guzman A.G., Li T., Lee M., Zhou Y., Goodell M.A., Stephan C., Davies P.J.A. A combination strategy targeting enhancer plasticity exerts synergistic lethality against BETi-resistant leukemia cells. Nat. Commun. 2020;11:740. doi: 10.1038/s41467-020-14604-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain P., Wang M. Mantle cell lymphoma: 2019 update on the diagnosis, pathogenesis, prognostication, and management. Am. J. Hematol. 2019;94:710–725. doi: 10.1002/ajh.25487. [DOI] [PubMed] [Google Scholar]
- Kumar A., Sha F., Toure A., Dogan A., Ni A., Batlevi C.L., Palomba M.L.M., Portlock C., Straus D.J., Noy A. Patterns of survival in patients with recurrent mantle cell lymphoma in the modern era: progressive shortening in response duration and survival after each relapse. Blood Cancer J. 2019;9:50. doi: 10.1038/s41408-019-0209-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddocks K. Update on mantle cell lymphoma. Blood. 2018;132:1647–1656. doi: 10.1182/blood-2018-03-791392. [DOI] [PubMed] [Google Scholar]
- Matulonis U.A., Wulf G.M., Barry W.T., Birrer M., Westin S.N., Farooq S., Bell-McGuinn K.M., Obermayer E., Whalen C., Spagnoletti T. Phase I dose escalation study of the PI3kinase pathway inhibitor BKM120 and the oral poly (ADP ribose) polymerase (PARP) inhibitor olaparib for the treatment of high grade serous ovarian and breast cancer. Ann. Oncol. 2017;28:512–518. doi: 10.1093/annonc/mdw672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merolle M.I., Ahmed M., Nomie K., Wang M.L. The B cell receptor signaling pathway in mantle cell lymphoma. Oncotarget. 2018;9:25332–25341. doi: 10.18632/oncotarget.25011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertz J.A., Conery A.R., Bryant B.M., Sandy P., Balasubramanian S., Mele D.A., Bergeron L., Sims R.J., 3rd Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc. Natl. Acad. Sci. U S A. 2011;108:16669–16674. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales, G.A., Garlich, J.R., and Durden, D.L. (2020). Single molecule compounds providing multi-target inhibition of btk and other proteins and methods of use thereof. Patent number WO2020023340A1.
- Ramsey H.E., Fischer M.A., Lee T., Gorska A.E., Arrate M.P., Fuller L., Boyd K.L., Strickland S.A., Sensintaffar J., Hogdal L.J. A novel MCL1 inhibitor combined with venetoclax rescues venetoclax-resistant acute myelogenous leukemia. Cancer Discov. 2018;8:1566–1581. doi: 10.1158/2159-8290.CD-18-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkozy C., Ribrag V. Novel agents for mantle cell lymphoma: molecular rational and clinical data. Expert Opin. Investig. Drugs. 2020;29:555–566. doi: 10.1080/13543784.2020.1760245. [DOI] [PubMed] [Google Scholar]
- Sun B., Shah B., Fiskus W., Qi J., Rajapakshe K., Coarfa C., Li L., Devaraj S.G., Sharma S., Zhang L. Synergistic activity of BET protein antagonist-based combinations in mantle cell lymphoma cells sensitive or resistant to ibrutinib. Blood. 2015;126:1565–1574. doi: 10.1182/blood-2015-04-639542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vann K.R., Pal D., Morales G.A., Burgoyne A.M., Durden D.L., Kutateladze T.G. Design of thienopyranone-based BET inhibitors that bind multiple synthetic lethality targets. Sci. Rep. 2020;10:12027. doi: 10.1038/s41598-020-68964-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woyach J.A., Ruppert A.S., Guinn D., Lehman A., Blachly J.S., Lozanski A., Heerema N.A., Zhao W., Coleman J., Jones D. BTK(C481S)-Mediated resistance to ibrutinib in chronic lymphocytic leukemia. J. Clin. Oncol. 2017;35:1437–1443. doi: 10.1200/JCO.2016.70.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Lwin T., Silva A., Shah B., Tao J., Fang B., Zhang L., Fu K., Bi C., Li J. Unification of de novo and acquired ibrutinib resistance in mantle cell lymphoma. Nat. Commun. 2017;8:14920. doi: 10.1038/ncomms14920. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.