Abstract
Rhizospheres are microecological zones at the interface of roots and soils. Interactions between bacteria and roots are critical for maintaining plant and soil health but are difficult to study because of constraints inherent in working with underground systems. We have developed an in-situ rhizosphere imaging system based on transparent soils and molecular probes that can be imaged using confocal microscopy. We observed spatial patterning of polysaccharides along roots and on cells deposited into the rhizosphere and also co-localised fluorescently tagged soil bacteria. These studies provide insight into the complex glycan landscape of rhizospheres and suggest a means by which root / rhizobacteria interactions can be non-disruptively studied.
Keywords: Rhizobacteria, Transparent Soil, Rhizosphere, Polysaccharide, Fluorescence Microscopy
The interface of roots and soil, the rhizosphere, is a microecological zone harbouring a rich diversity of microbes (the rhizobiome) (Daniel, 2005, Wagg et al., 2014). Interactions between the root and rhizobiome are complex, dynamic and important for plant health in relation to nutrient and water uptake, defence against pathogenic and herbivorous organisms, and root colonisation by arbuscular mycorrhizal fungi (Ferreira et al., 2020). Healthy soils are characterised by having diverse rhizobiomes, and diversity is known to be strongly influenced by molecules deposited by roots into the rhizosphere (rhizodeposits) (Richardson, 2001, Mendes et al., 2013, Ofek-Lalzar et al., 2014, Tkacz and Poole, 2015, Jacoby and Kopriva, 2019). Metabolites such as hormones (e.g. strigalactone) and phenols (e.g. coumarins) have long been implicated in promoting favourable root-microbe interactions (Jacoby and Kopriva, 2019). The roles of complex polysaccharides in root-microbe interactions has received less attention, although they are abundant in the rhizobiome both as exuded mucilage, on the surface of epidermal cells and sloughed off border cells and as components of microbial biofilms. Evidence from other terrestrial and marine ecosystems demonstrates that polysaccharide complexity drives cognate microbial diversity via evolution of highly specialised carbohydrate active enzymes (CAZymes) that enable microbes to utilise glycan substrates (Flint et al., 2012, Bennke et al., 2016, Grondin et al., 2017, Lapébie et al., 2019). There is also evidence that qualitative and quantitative variations in polysaccharide-rich root exudates and those that are surface bound have a major influence on soil microbiomes (Kuzyakov and Domanski, 2000, Dennis et al., 2010, Galloway et al., 2018, Jacoby and Kopriva, 2019). In some cases, glycans have been identified that are associated with colonisation by specific microbes, for example, galactans in potato root exudates and Pectobacterium atrosepticum (Koroney et al., 2016). Structurally complex proteoglycans (extensins and arabinogalactan-proteins) are abundant in exudates and appear to have roles in both plant growth promotion and defence (Nguema-Ona et al., 2013, Castilleux et al., 2018). However, a detailed understanding of the roles that glycans either in rhizodeposits or on root surfaces (rhizoglycans) play in maintaining rhizobiome diversity is lacking, partly because of the technical challenges associated with sampling and analysing glycans at the precise locations where these complex root-microbe interactions occur in this underground microecological zone.
Prior work by the authors and others using polysaccharide-directed monoclonal antibodies (mAbs) and carbohydrate binding modules (CBMs) has revealed how epitope distribution varies markedly along the root surface, with characteristic and consistent glycan labelling associated with the main body of the root, the root tip, root hairs and exudates including border cells (Willats et al., 2001, McCartney et al., 2003, Guillemin et al., 2005, Wilson et al., 2015). These in vitro experiments show that root glycan patterning is complex and dynamic. However, they do not provide information on how this patterning changes over time with root growth and how glycan deposition may be influenced by interactions between roots and soil particles, and the effects of microbial degradation.
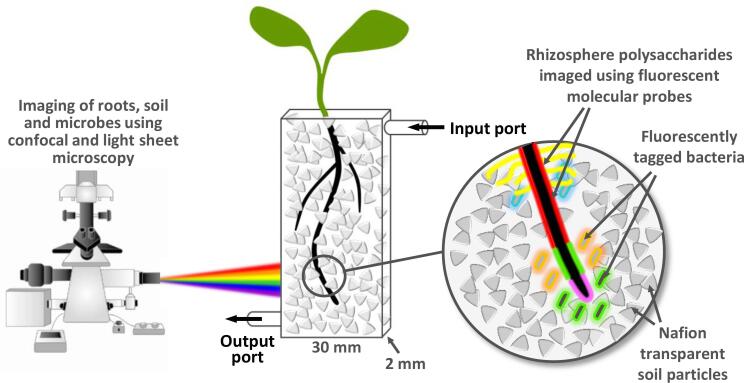
Specialised chambers of various kinds (rhizotrons) have been used for 125 years for observing roots in soil and for sampling material from specific locations (López-Mondéjar et al., 2016; Wilson et al., 2015, McCartney et al., 2003). More recently, X-ray computed tomography and nuclear magnetic resonance have emerged as powerful non-invasive techniques, for studying root bio physics, soil elemental composition and water distribution (Paya et al., 2015, Rogers et al., 2016, Pflugfelder et al., 2017, Yang et al., 2017, Gao et al., 2019). Nevertheless, much of the complex and dynamic biology of the rhizosphere has remained hidden. A breakthrough came with the development of transparent soil systems (TSSs). Several examples have been described and all seek to achieve a balance between reducing opacity whilst replicating nature as closely as possible (Yang et al., 2013, Ma et al., 2019, Sharma et al., 2020). The system employed for this study is based on NafionTM particles, a tetrafluoroethylene based fluoropolymer-copolymer that is physically and chemically adaptable (Downie et al., 2012). NafionTM particle size distribution can be manipulated by freeze milling, and minerals and fluorescent dyes can be adsorbed onto particles for controlling nutrient supply to plants, and fluorescence imaging of soil pore size and particle geometry. Plant development in this TSS is similar to that in natural soils and plants can be maintained for several weeks in this system (Downie et al., 2012, Downie et al., 2014). Here, we report the development of a new method for imaging rhizoglycan distribution and microbial colonisation that combines a NafionTM based TSS with glycan-specific mAbs and Green Fluorescent Protein (GFP) tagged bacteria (Fig. 1). All data presented here was obtained using Leica TCS SP2 or Nikon A1R confocal laser scanning microscopes and objective lenses 10×/0.30, 20×/0.50 or 40×/0.80. Images were generated from 3D dataset using either volume rendering or projection algorithm from ImageJ or Nikon NIS-Elements software.
Fig. 1.
A novel technology platform for an in-situ rhizosphere glycobiology. The transparent soil system is based on a thin glass chamber housing Nafion artificial soil particles in which seedlings grow. Bacteria, molecular probes and other components can be introduced via ports and the system washed with buffers. Using confocal and light sheet microscopy, rhizosphere polysaccharides, fluorescently tagged bacteria and Nafion particles are imaged in-situ.
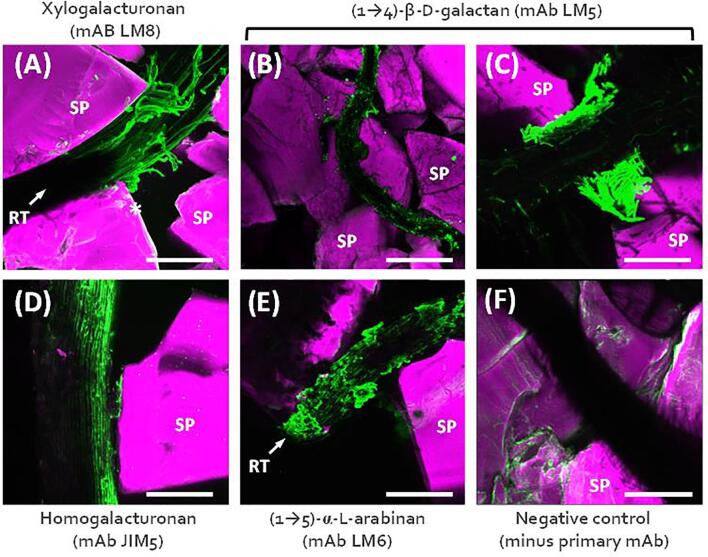
Germinated 5-day old lettuce seedlings were placed at the top of the TSS chamber and roots grew into the Nafion matrix. Roots were blocked using 0.5% Bovine Serum Albumin (BSA) in phosphate buffered saline (PBS) for 30 min, prior to circulation of primary mAbs (diluted to 1:10 with PBS) for 1.5 hr. Both blocking buffer and mAbs were circulated through the chamber via the upper inlet and lower outlet using a peristaltic pump. Primary mAbs were washed out by flushing the system with PBS buffer. Fluorophore tagged secondary antibodies (anti-Rat Alexa Fluor® 488, diluted to 1:100 in PBS) were used to visualise primary mAb binding. The thin and transparent TSS chamber enabled direct non-disruptive in situ imaging of fluorescently labelled entities using a Laser Scanning Confocal Microscopy (LSCM), and Light Sheet Microscopy (Yang et al., 2013). Using this system, it was possible to localise specific epitopes associated with living roots and the rhizosphere in the context of soil particles. We observed localisation of extensin, xylogalacturonan, (1 → 4)-β-D-galactan, homogalacturonan and (1 → 5)-α-L-arabinan on differing tissues around the root tip zone using the primary antibodies LM1 (Smallwood et al., 1995), LM8 (Willats et al., 2004), LM5 (Jones et al., 1997), JIM5 (Knox et al., 1990, Clausen et al., 2003, Verhertbruggen et al., 2009) and LM6 (Willats et al., 1998) respectively. We also observed discrete labelling of specific tissues rich in (1 → 4)-β-D-galactan (LM5 binding to border cells), xylogalacturonan (LM8 biding to sloughed off tissue), and xyloglucan (LM15 particularly abundant on root hairs). Examples of these localisations are shown in Fig. 2. LSCM imaging revealed that soil porosity and particle geometry can significantly impact local deposition of polysaccharides. This was clearly demonstrated for xylogalacturonan and (1 → 4)-β-D-galactan (Fig. 2A and Fig. 2B and 2C respectively), both of which are associated with epidermal and border cells. Where roots grew between closely opposed soil particles, ‘pinch points’ were created that appeared to abrade cells from the root surface, thus creating massive local accumulations of cells and their associated glycans (Fig. 2A-2C). We also observed that some cells, cellular material and associated polysaccharide epitopes appeared to become ‘smeared’ onto soil particles, presumably as roots grew through soil pores. An example of this for homogalacturonan is shown in Fig. 2D. These observations are significant because they provide new insight into how interactions between root growth and soil properties contribute to glycan patterning in the rhizosphere, which is likely to in turn impact the local colonisation of microbes with specialised capacity to utilise those glycans.
Fig. 2.
Imaging root associated polysaccharides using a transparent soil system. Lettuce seedling roots growing through Nafion soil particles (SP) were labelled with mAbs with specificity for xylogalacturonan (A), (1 → 4)- β -D-galactan (B) and (C) homogalacturonan (D) and (1 → 5)- α-L-arabinan (E). RT – root tip. SP – soil particle. Scale bars = 200µm.
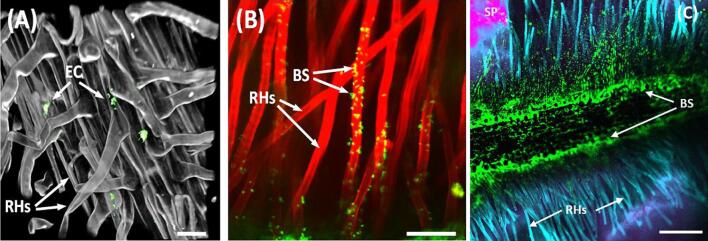
We further explored associations between root polysaccharides and microbial colonisation by combining both GFP-tagged strains of soil bacteria and mAbs. Bacteria were inoculated onto 0.5 cm2 filter paper placed on the top surface of NafionTM particles in the TSS and allowed to migrate through and freely colonise the TSS (Fig. 3). We visualised GFP-tagged E. coli colonies adhering to root hairs (Fig. 3A), and by inoculating with GFP-tagged Bacillus subtilis NRS1473 (Hobley et al., 2013) and simultaneously probing with mAb (LM15) that recognises xyloglucan (Marcus et al., 2008), we were able to co-localise B. subtilis and xyloglucan on root hairs (Fig. 3B). The observation that xyloglucan is abundant on root hairs and at the elongation zone is consistent with previous work on in vitro labelling studies (Freshour et al., 2003, Larson et al., 2014). The co-localisation with bacteria observed in this study may indicate that xyloglucan serves as a nutrient source at this location, thus creating a micro-ecological niche exploitable by microbes with the appropriate CAZymes to utilise xyloglucan.
Fig. 3.
Imaging of microbes and glycans.(A) colonies of GFP-tagged E. coli 0157:H7 (EC, green punctate labelling) observed within the root hair (RH) matrix of a Latuca sativa root; (B) GFP-tagged B. subtilis (BS, green punctate labelling) were inoculated into the TSS and also colonised Latuca sativa root hairs. Roots were simultaneously labelled with the anti-xyloglucan mAb LM15 (red, Alexa594) revealing that this polysaccharide was abundant on the root hair surface; (C) as in (B) showing B. subtilis colonies (BS, green punctate labelling) located on the main root surface with xyloglucan labelling (mAb LM15, Alexa594) displayed in blue to contrast with Naphion soil particles. Scale bars, A = 50µm, B = 100µm, C = 200µm.
Our results clearly show that a significant amount of polysaccharide-rich material is retained on or near the root surface, despite the washing steps employed. It seems reasonable to speculate that the same may also be true to roots growing naturally in soils subject to rainwater. Nevertheless, since it is likely that some polysaccharides are washed through the TSS and therefore were not observed, the system only provides partial insight onto glycan rhizodeposits. However, we envisage that the novel methodology described here can make a significant contribution to our understanding of the roles that glycans play in establishing and maintaining rhizobiome diversity. This in turn may help us identify novel means to fine tune rhizobiomes to enhance plant health and crop performance.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This research was supported by the N8 Agrifood Program Newcastle Pump priming. This work received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (Grant agreement No. 647857-SENSOILS).
References
- Bennke C.M., Krüger K., Kappelmann L., Huang S., Gobet A., Schüler M., Barbe V., Fuchs B.M., Michel G., Teeling H., Amann R.I. Polysaccharide utilisation loci of Bacteroidetes from two contrasting open ocean sites in the North Atlantic. Environ. Microbiol. 2016;18(12):4456–4470. doi: 10.1111/1462-2920.13429. [DOI] [PubMed] [Google Scholar]
- Castilleux R., Plancot B., Ropitaux M., Carreras A., Leprince J., Boulogne I., Follet-Gueye M.L., Popper Z.A., Driouich A., Vicre M. Cell wall extensins in root-microbe interactions and root secretions. J. Exp. Bot. 2018;69(18):4235–4247. doi: 10.1093/jxb/ery238. [DOI] [PubMed] [Google Scholar]
- Clausen M.H., Willats W.G.T., Knox J.P. Synthetic methyl hexagalacturonate hapten inhibitors of anti-homogalacturonan monoclonal antibodies LM7, JIM5 and JIM7. Carbohydr. Res. 2003;338(17):1797–1800. doi: 10.1016/s0008-6215(03)00272-6. [DOI] [PubMed] [Google Scholar]
- Daniel R. The metagenomics of soil. Nat. Rev. Microbiol. 2005;3(6):470–478. doi: 10.1038/nrmicro1160. [DOI] [PubMed] [Google Scholar]
- Dennis P.G., Miller A.J., Hirsch P.R. Are root exudates more important than other sources of rhizodeposits in structuring rhizosphere bacterial communities? FEMS Microbiol. Ecol. 2010;72(3):313–327. doi: 10.1111/j.1574-6941.2010.00860.x. [DOI] [PubMed] [Google Scholar]
- Downie H., Holden N., Otten W., Spiers A.J., Valentine T.A., Dupuy L.X. Transparent soil for imaging the rhizosphere. PLoS ONE. 2012;7(9) doi: 10.1371/journal.pone.0044276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downie H.F., Valentine T.A., Otten W., Spiers A.J., Dupuy L.X. Transparent soil microcosms allow 3D spatial quantification of soil microbiological processes in vivo. Plant Signal Behav. 2014;9(10) doi: 10.4161/15592316.2014.970421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira D.A., da Silva T.F., Pylro V.S., Salles J.F., Andreote F.D., Dini-Andreote F. Soil microbial diversity affects the plant-root colonization by Arbuscular Mycorrhizal fungi. Microb. Ecol. 2020 doi: 10.1007/s00248-020-01502-z. [DOI] [PubMed] [Google Scholar]
- Flint H.J., Scott K.P., Duncan S.H., Louis P., Forano E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes. 2012;3(4):289–306. doi: 10.4161/gmic.19897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway A.F., Pedersen M.J., Merry B., Marcus S.E., Blacker J., Benning L.G., Field K.J., Knox J.P. Xyloglucan is released by plants and promotes soil particle aggregation. New Phytol. 2018;217(3):1128–1136. doi: 10.1111/nph.14897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freshour G., Bonin C.P., Reiter W.-D., Albersheim P., Darvill A.G., Hahn M.G. Distribution of Fucose-Containing Xyloglucans in Cell Walls of the mur1 Mutant of Arabidopsis. Plant Physiol. 2003;131:1602–1612. doi: 10.1104/pp.102.016444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W., Schlüter S., Blaser S.R.G.A., Shen J., Vetterlein D. A shape-based method for automatic and rapid segmentation of roots in soil from X-ray computed tomography images: Rootine. Plant Soil. 2019;441(1-2):643–655. [Google Scholar]
- Grondin J.M., Tamura K., Déjean G., Abbott D.W., Brumer H., O'Toole G. Polysaccharide Utilization Loci: Fueling Microbial Communities. J. Bacteriol. 2017;199(15) doi: 10.1128/JB.00860-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemin F., Guillon F., Bonnin E., Devaux M.-F., Chevalier T., Knox J.P., Liners F., Thibault J.-F. Distribution of pectic epitopes in cell walls of the sugar beet root. Planta. 2005;222(2):355–371. doi: 10.1007/s00425-005-1535-3. [DOI] [PubMed] [Google Scholar]
- L. Hobley A. Ostrowski F.V. Rao K.M. Bromley M. Porter A.R. Prescott C.E. MacPhee D.M.F. van Aalten N.R. Stanley-Wall Proceedings of the National Academy of Sciences 2013 13600 13605. [DOI] [PMC free article] [PubMed]
- Jacoby R.P., Kopriva S. Metabolic niches in the rhizosphere microbiome: new tools and approaches to analyse metabolic mechanisms of plant-microbe nutrient exchange. J. Exp. Bot. 2019;70(4):1087–1094. doi: 10.1093/jxb/ery438. [DOI] [PubMed] [Google Scholar]
- Jones, L., Seymour, G.B., Knox, J.P. Localization of Pectic Galactan in Tomato Cell Walls Using a Monoclonal Antibody Specific to (1[->]4)-β-D-Galactan, Plant Physiology, 113, 4, 1405–1412, 1997. [DOI] [PMC free article] [PubMed]
- Knox J.P., Linstead P., King J., Cooper C., Roberts K. Pectin esterification is spatially regulated both within cell walls and between developing tissues of root apices. Planta. 1990;181(4) doi: 10.1007/BF00193004. [DOI] [PubMed] [Google Scholar]
- Koroney A.S., Plasson C., Pawlak B., Sidikou R., Driouich A., Menu-Bouaouiche L., Vicré-Gibouin M. Root exudate of Solanum tuberosum is enriched in galactose-containing molecules and impacts the growth of Pectobacterium atrosepticum. Ann. Bot. 2016;118(4):797–808. doi: 10.1093/aob/mcw128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzyakov Y., Domanski G. Carbon input by plants into the soil. Review. J. Plant Nutr. Soil Sci. 2000;163(4):421–431. [Google Scholar]
- Larson E.R., Tierney M.L., Tinaz B., Domozych D.S. Using monoclonal antibodies to label living root hairs: a novel tool for studying cell wall microarchitecture and dynamics in Arabidopsis. Plant methods. 2014;10(1):30. doi: 10.1186/1746-4811-10-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapébie P., Lombard V., Drula E., Terrapon N., Henrissat B. Bacteroidetes use thousands of enzyme combinations to break down glycans. Nat. Commun. 2019;10(1):2043. doi: 10.1038/s41467-019-10068-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, L., Y. Shi, O. Siemianowski, B. Yuan, T. K. Egner, S. V. Mirnezami, K. R. Lind, B. Ganapathysubramanian, V. Venditti and L. Cademartiri (2019). “Hydrogel-based transparent soils for root phenotyping in vivo.” Proceedings of the National Academy of Sciences 116(22): 11063-11068. [DOI] [PMC free article] [PubMed]
- López-Mondéjar R, Zühlke D, Becher D, Riedel K, Baldrian P. Cellulose and hemicellulose decomposition by forest soil bacteria proceeds by the action of structurally variable enzymatic systems. Sci. Reports. 2016;6 doi: 10.1038/srep25279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus S.E., Verhertbruggen Y., Hervé C., Ordaz-Ortiz J.J., Farkas V., Pedersen H.L., Willats W.GT., Knox J.P. Pectic homogalacturonan masks abundant sets of xyloglucan epitopes in plant cell walls. BMC Plant Biol. 2008;8(1):60. doi: 10.1186/1471-2229-8-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney L., Steele-King C.G., Jordan E., Knox J.P. Cell wall pectic (1–>4)-beta-d-galactan marks the acceleration of cell elongation in the Arabidopsis seedling root meristem. Plant J. 2003;33(3):447–454. doi: 10.1046/j.1365-313x.2003.01640.x. [DOI] [PubMed] [Google Scholar]
- Mendes R., Garbeva P., Raaijmakers J.M. The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 2013;37(5):634–663. doi: 10.1111/1574-6976.12028. [DOI] [PubMed] [Google Scholar]
- Nguema-Ona E., Vicré-Gibouin M., Cannesan M.-A., Driouich A. Arabinogalactan proteins in root-microbe interactions. Trends Plant Sci. 2013;18(8):440–449. doi: 10.1016/j.tplants.2013.03.006. [DOI] [PubMed] [Google Scholar]
- Ofek-Lalzar M., Sela N., Goldman-Voronov M., Green S.J., Hadar Y., Minz D. Niche and host-associated functional signatures of the root surface microbiome. Nat. Commun. 2014;5(1):4950. doi: 10.1038/ncomms5950. [DOI] [PubMed] [Google Scholar]
- Paya A.M., Silverberg J.L., Padgett J., Bauerle T.L. X-ray computed tomography uncovers root-root interactions: quantifying spatial relationships between interacting root systems in three dimensions. Front. Plant Sci. 2015;6:274. doi: 10.3389/fpls.2015.00274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflugfelder D., Metzner R., van Dusschoten D., Reichel R., Jahnke S., Koller R. Non-invasive imaging of plant roots in different soils using magnetic resonance imaging (MRI) Plant Methods. 2017;13:102. doi: 10.1186/s13007-017-0252-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson A.E. Prospects for using soil microorganisms to improve the acquisition of phosphorus by plants. Funct. Plant Biol. 2001;28(9):897–906. [Google Scholar]
- Rogers E.D., Monaenkova D., Mijar M., Nori A., Goldman D.I., Benfey P.N. X-ray computed tomography reveals the response of root system architecture to soil texture. Plant Physiol. 2016;171(3):2028–2040. doi: 10.1104/pp.16.00397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, K., M. Palatinszky, G. Nikolov, D. Berry and E. A. Shank (2020). “Transparent soil microcosms for live-cell imaging and non-destructive stable isotope probing of soil microorganisms.” eLife 9: e56275. [DOI] [PMC free article] [PubMed]
- Smallwood M., Martin H., Knox J. An epitope of rice threonine- and hydroxyproline-rich glycoprotein is common to cell wall and hydrophobic plasma-membrane glycoproteins. Planta. 1995;196(3):510–522. doi: 10.1007/BF00203651. [DOI] [PubMed] [Google Scholar]
- Tkacz A., Poole P. Role of root microbiota in plant productivity. J. Exp. Bot. 2015;66(8):2167–2175. doi: 10.1093/jxb/erv157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhertbruggen Y., Marcus S.E., Haeger A., Ordaz-Ortiz J.J., Knox J.P. An extended set of monoclonal antibodies to pectic homogalacturonan. Carbohydr. Res. 2009;344(14):1858–1862. doi: 10.1016/j.carres.2008.11.010. [DOI] [PubMed] [Google Scholar]
- Wagg, C., S. F. Bender, F. Widmer and M. G. A. van der Heijden (2014). “Soil biodiversity and soil community composition determine ecosystem multifunctionality.” Proceedings of the National Academy of Sciences 111(14): 5266-5270. [DOI] [PMC free article] [PubMed]
- Willats W.G.T., Marcus S.E., Knox J.P. Generation of a monoclonal antibody specific to (1→5)-α-L-arabinan. Carbohydr. Res. 1998;308(1-2):149–152. doi: 10.1016/s0008-6215(98)00070-6. [DOI] [PubMed] [Google Scholar]
- Willats W.G.T., McCartney L., Steele-King C.G., Marcus S.E., Mort A., Huisman M., van Alebeek G.-J., Schols H.A., Voragen A.G.J., Le Goff A., Bonnin E., Thibault J.-F., Knox J.P. A xylogalacturonan epitope is specifically associated with plant cell detachment. Planta. 2004;218(4):673–681. doi: 10.1007/s00425-003-1147-8. [DOI] [PubMed] [Google Scholar]
- Willats W.G.T., McCartney L., Knox J.P. In-situ analysis of pectic polysaccharides in seed mucilage and at the root surface of Arabidopsis thaliana. Planta. 2001;213(1):37–44. doi: 10.1007/s004250000481. [DOI] [PubMed] [Google Scholar]
- Wilson M.H., Holman T.J., Sorensen I., Cancho-Sanchez E., Wells D.M., Swarup R., Knox J.P., Willats W.G., Ubeda-Tomas S., Holdsworth M., Bennett M.J., Vissenberg K., Hodgman T.C. Multi-omics analysis identifies genes mediating the extension of cell walls in the Arabidopsis thaliana root elongation zone. Front. Cell Dev. Biol. 2015;3:10. doi: 10.3389/fcell.2015.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Varga T., Liu C., Scheibe T.D. What can we learn from in-soil imaging of a live plant: X-ray Computed Tomography and 3D numerical simulation of root-soil system. Rhizosphere. 2017;3:259–262. [Google Scholar]
- Yang Z., Downie H., Rozbicki E., Dupuy L.X., MacDonald M.P. Light sheet tomography (LST) for in situ imaging of plant roots. Opt. Express. 2013;21(14):16239. doi: 10.1364/OE.21.016239. [DOI] [PubMed] [Google Scholar]