Abstract
Background & Aims
The development of nonalcoholic fatty liver disease (NAFLD) can be modulated by microRNAs (miRNA). Dietary polyphenols modulate the expression of miRNA such as miR-467b-3p in the liver. In addition, 6-gingerol (6-G), the functional polyphenol of ginger, has been reported to ameliorate hepatic steatosis; however, the exact mechanism involved and the role of miRNA remain elusive. In this study, we assessed the role of miR-467b-3p in the pathogenesis of hepatic steatosis and the regulation of miR-467b-3p by 6-G through the hepatocyte nuclear factor 4α (HNF4α).
Methods
miR-467b-3p expression was measured in free fatty acid (FFA)-treated hepatocytes or liver from high-fat diet (HFD)-fed mice. Gain- or loss-of-function of miR-467b-3p was induced using miR-467b-3p–specific miRNA mimic or miRNA inhibitor, respectively. 6-G was exposed to FFA-treated cells and HFD-fed mice. The HNF4α/miR-467b-3p/GPAT1 axis was measured in mouse and human fatty liver tissues.
Results
We found that miR-467b-3p was down-regulated in liver tissues from HFD-fed mice and in FFA-treated Hepa1-6 cells. Overexpression of miR-467b-3p decreased intracellular lipid accumulation in FFA-treated hepatocytes and mitigated hepatic steatosis in HFD-fed mice via negative regulation of glycerol-3-phosphate acyltransferase-1 (GPAT1). In addition, miR-467b-3p up-regulation by 6-G was observed. 6-G inhibited FFA-induced lipid accumulation and mitigated hepatic steatosis. Moreover, it increased the transcriptional activity of HNF4α, resulting in the increase of miR-467b-3p and subsequent decrease of GPAT1. HNF4α/miR-467b-3p/GPAT1 signaling also was observed in human samples with hepatic steatosis.
Conclusions
Our findings establish a novel mechanism by which 6-G improves NAFLD. This suggests that targeting of the HNF4α/miR-467b-3p/GPAT1 cascade may be used as a potential therapeutic strategy to control NAFLD.
Keywords: 6-gingerol, NAFLD, MicroRNA, HNF4α, GPAT1
Abbreviations used in this paper: ACC, acetyl-CoA carboxylase; cDNA, complementary DNA; ChIP, chromatin immunoprecipitation; DARTS, drug-affinity–responsive target stability; DNL, de novo lipogenesis; FFA, free fatty acid; GPAT, glycerol-3-phosphate acyltransferase; HF, high-fat diet–fed group; HFD, high-fat diet; HNF4α, hepatocyte nuclear factor 4α; LysoPA, lysophosphatidic acid; miRNA, microRNA; mRNA, messenger RNA; NAFLD, nonalcoholic fatty liver disease; 6-G, 6-gingerol; TAG, triacylglycerol; TLDA, TaqMan low-density arrays; UTR, untranslated region
Graphical abstract
Summary.
MicroRNA467b-3p is downregulated in fatty liver tissues and its direct target is glycerol-3-phosphate acyltransferase-1. The overexpression of microRNA-467b-3p improves hepatic steatosis. 6-Gingerol, the main active polyphenol of ginger, improves hepatic steatosis via hepatocyte nuclear factor 4 alpha- dependent upregulation of microRNA-467b-3p and subsequent decrease in glycerol-3-phosphate acyltransferase-1 levels.
Nonalcoholic fatty liver disease (NAFLD) comprises a spectrum of stages, from simple steatosis to nonalcoholic steatohepatitis, which can progress to fibrosis, cirrhosis, and, ultimately, to hepatocellular carcinoma. Despite being one of the most common chronic liver diseases, the pathogenesis of NAFLD remains largely unknown. Accumulating evidence suggests that plasma free fatty acids (FFAs) are the primary source for triacylglycerol (TAG) in hepatocytes.1,2 Therefore, one possible method to decrease hepatic steatosis would be to diminish hepatic FFA uptake and TAG accumulation.
MicroRNAs (miRNAs) are short noncoding RNA transcripts that regulate biological processes and diseases by RNA-mediated gene-silencing mechanisms.3 There is a growing body of evidence supporting a significant role for miRNAs in NAFLD pathogenesis,4 and it has been observed that the expression of certain miRNAs is altered in high-fat diet (HFD)-fed mice5 and NAFLD patients.6 Therefore, understanding the role of miRNAs in NAFLD is crucial for the development of new therapies. Previously, we reported that hepatic miR-467b-5p is up-regulated in HFD-fed obese mice and targets lipoprotein lipase in the process of NAFLD.7 Although both miR-467b-5p and miR-467b-3p are processed into mature miRNAs from the same precursor, the mechanism by which miR-467b-3p regulates NAFLD has not yet been elucidated. Interestingly, miR-467b-3p was reported to be one of the commonly regulated miRNAs by polyphenols in the liver.8 In a previous study, we suggested the possibility that 6-gingerol (6-G), a ginger polyphenol, could ameliorate NAFLD.9 However, the involvement of miRNA in improvement of hepatic steatosis induced by 6-G was not examined.
Glycerol-3-phosphate acyltransferase (GPAT) regulates the first commitment step in TAG synthesis via the acylation of glycerol 3-phosphate,10 and 4 GPAT isoforms (GPAT1–4) have been reported in mammals. In particular, GPAT1 is located on the outer mitochondrial membrane and initiates the TAG synthesis pathway by esterifying a long-chain fatty acetyl-CoA to lysophosphatidic acid (LysoPA). GPAT1 comprises 30%–50% of the total activity in the liver and its overexpression causes hepatic steatosis.11 Conversely, its absence confers protection against the development of hepatic steatosis caused by a HFD.12 Therefore, the regulation of GPAT1 is one of the key steps to control TAG synthesis in the liver.
Polyphenols are micronutrients widely present in plants and the regulation of NAFLD by dietary polyphenol has been reviewed extensively.13 Accordingly, 6-G, the main active polyphenol of ginger, has been reported to exert antioxidant,14 anti-inflammatory,15 anticancer,16 neuroprotective,17 anti-obesity,18 and antihepatic steatosis19,20 effects. However, the exact mechanism involved in the prevention of hepatic steatosis and the role of miRNAs in its regulation remain unknown.
In the present study, we aimed to determine the role of miR-467b-3p in the pathogenesis of NAFLD, and we also examined the transcriptional regulation of miR-467b-3p by 6-G.
Results
Down-Regulation of miR-467b-3p Is Associated With Hepatic Steatosis
We measured miR-467b-3p expression in liver tissues from HFD-fed mice and found that miR-467b-3p decreased markedly to 4.75% of its normal level (Figure 1A). We also observed that FFA treatment decreased miR-467b-3p and induced intracellular lipid deposition in Hepa1–6 cells (Figure 1B–D). Next, we examined the effect of increased miR-467b-3p expression on FFA-induced intracellular lipid accumulation and found that intracellular fat accumulation was inhibited effectively by miR-467b-3p overexpression.
Figure 1.
miR-467b-3p decrease in fatty liver and FFA-treated Hepa1–6 cells. (A) Real-time PCR analysis of miR-467b-3p level in hepatic tissues of normal diet (NC) or HFD-fed (HF) mice (n = 5). ∗∗P < .01. (B) Relative miR-467b-3p levels, (C) intracellular lipid accumulation, and (D) Nile red fluorescence intensity in miRDIANTM miRNA mimic control (mimic_CTL) or miRIDIAN miR-467b-3p mimic (mimic-miR-467b-3p) transfected Hepa1–6 cells, after exposure to 50 μmol/L palmitate for 24 hours (n = 4). ∗∗P < .01. (E) Hepatic miR-467b-3p level, (F) histologic examination of H&E (top row) and Oil red O stain (bottom row), (G) hepatic lipid profile, (H) body weight gain, and (I) liver and white adipose tissue (WAT) weights after administration of miRVana miR-467b-3p mimic (n = 4-5). ∗P < .05 vs mimic_CTL. CTL, control; DAPI, 4′,6-diamidino-2-phenylindole; TC, total cholesterol.
To evaluate the in vivo consequences of hepatic overexpression of miR-467b-3p, we administered mimic control (mimic_CTL) or miR-467b-3p mimic intraperitoneally. The real-time PCR assay showed that miR-467b-3p hepatic levels increased 4.5-fold (Figure 1E). Importantly, hepatic steatosis was ameliorated, as confirmed by H&E and Oil Red O staining (Figure 1F), and the hepatic lipid profiles and dyslipidemia also were improved by miR-467b-3p overexpression (Figure 1G and Table 1). miR-467b-3p mimic treatment reduced both body and organ weights (Figure 1H and I). Collectively, these findings suggest that modulation of miR-467b-3p ameliorated the development of NAFLD.
Table 1.
Biochemical Analysis of Serum
| Serum | mimic_CTL | mimic_miR-467b-3p |
|---|---|---|
| Glucose, mg/dL | 224.20 ± 9.15 | 205.10 ± 9.66 |
| TAG, mg/dL | 59.41 ± 2.22 | 44.51 ± 3.73a |
| TC, mg/dL | 152.70 ± 3.23 | 128.30 ± 6.54a |
| Insulin, ng/dL | 12.75 ± 1.01 | 6.29 ± 0.58a |
| HOMA-IR | 154.60 ± 13.84 | 110.40 ± 11.17a |
| NEFA, μEq/L | 804.50 ± 102.1 | 1007.00 ± 68.95 |
NOTE. Results are shown as means ± SD.
CTL, control; TAG, triacylglycerol; TC, total cholesterol, HOMA-IR, homeostatic model assessment for insulin resistance; NEFA, non-esterified fatty acid.
P < .05 vs mimic_Ctrl group.
miR-467b-3p Directly Targets GPAT1
To elucidate how overexpression of miR-467b-3p ameliorated NAFLD, we measured the expressions of lipid metabolism-related genes. We observed that these genes were down-regulated in liver tissues (Figure 2A). Among them, Gpat1 down-regulation was the most noticeable because miR-467b-3p mimic treatment markedly decreased Gpat1 messenger RNA (mRNA) levels by 82.4% compared with those of the miRNA mimic control. In accordance, GPAT1 protein expression in liver also was reduced (Figure 2B). In addition, overexpression of miR-467b-3p resulted in a decrease of hepatic LysoPA, which is the byproduct of GPAT1 (Figure 2C).
Figure 2.
GAPT-1 is a direct target of miR-467b-3p. (A) Real-time PCR analysis of lipid metabolism–related genes, (B) Western blot of GPAT1, and (C) relative LysoPA content in liver tissues after administration of miRVana miR-467b-3p mimic (n = 5). ∗P < .05. (D) Complementary sequences to the seed region of miR-467b-3p within the wild and mutated 3’UTR of Gpat1. (E) Luciferase activity in wild and mutant Gpat1-containing cells (n = 4). ∗∗P < .01 vs mimic_CTL-treated wild reporter vector. (F) mRNA expressions of enzymes from the glycerol phosphate pathway for TAG synthesis. (G) Protein expression of enzymes from the glycerol phosphate pathway for TAG synthesis. (H) mRNA and protein expression of GPAT1 in hepatic tissues (n = 5). ∗∗P < .01. (I) Gpat1 mRNA expression in Hepa1–6 cells after 24 hours of exposure to FFA (n = 4). ∗∗P < .01. CTL, control; MGAT, monoacylglycerol acyltransferase; DGAT, diglyceride acyltransferase; AGPAT, acylglycerol phosphate acyltransferase; PAP, phosphatidate phosphatase; NC, normal diet-fed mice; HF, HFD-fed mice.
To determine whether GPAT1 is a potential target of miR-467b-3p, we conducted bioinformatic-based prediction of miRNA targets and found that Gpat1 has a putative miR-467b-3p binding site in its 3’ untranslated region (UTR) (Figure 2D, top). To examine miR-467b-3p and Gpat1 binding, we performed a luciferase reporter assay using wild or mutated 3’UTR of GPAT1 (Figure 2D, bottom). We found that miR-467b-3p mimic treatment significantly decreased luciferase activity in wild-type Gpat1, whereas no change was detected in Gpat1 with the mutated 3’UTR (Figure 2E). These results show that miR-467b-3p directly binds to Gpat1 via seed sequence and down-regulates Gpat1. In addition, other enzymes involved in triglyceride synthesis were measured and we observed that their mRNA and protein levels were decreased by miR-467b-3p (Figure 2F and G).
We measured GPAT1 expression in HFD-fed livers, in which miR-467-3p expression is low, and observed an increase of GPAT1 in mRNA and protein levels (Figure 2H). Moreover, the FFA-induced increase of Gpat1 was abolished when Hepa1–6 cells were transfected with miR-467b-3p mimic (Figure 2I). Taken together, these data suggest that miR-467b-3p down-regulated Gpat1 via direct binding and this led to the improvement of NAFLD by inhibiting intracellular lipid accumulation.
The miR-467b-3p/GPAT1 Axis Is Associated With the Amelioration of NAFLD by 6-G
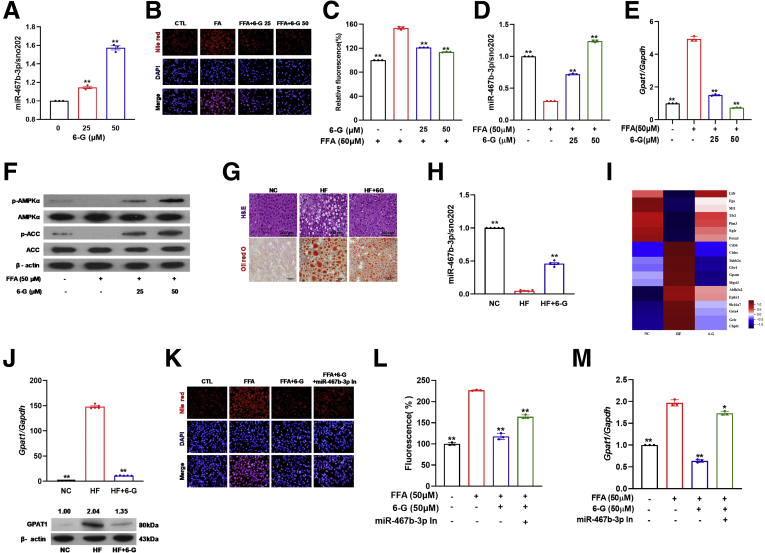
Dietary polyphenol modulates the expression of miRNA in the liver, and miR-467b-3p was identified as one of the miRNAs commonly modulated by polyphenols.8 However, the exact mechanism by which miR-467b-3p regulates the improvement of fatty liver has yet to be identified. In a previous report, we suggested the possibility that 6-G could ameliorate NAFLD9; thus, in this study we selected 6-G to determine the role of miR-467b-3p in the polyphenol-ameliorated NAFLD. We tested the effect of 6-G on miR-467b-3p expression in Hepa1–6 cells and found that 6-G induced a dose-dependent up-regulation of miR-467b-3p (Figure 3A). Moreover, 6-G pretreatment significantly inhibited FFA-induced fat accumulation in Hepa1–6 cells (Figure 3B and C) and allowed the successful recovery of the aberrant expression of miR-467b-3p and Gpat1 observed in Hepa1–6 cells (Figure 3D and E). 6-G also induced phosphorylation of AMP-activated protein kinase (AMPK) and the resultant phosphorylation of acetyl-CoA carboxylase (ACC) (Figure 3F). Subsequently, the effect of 6-G on HFD-induced NAFLD was tested. Histologic examination showed that 8 weeks of 6-G supplementation effectively alleviated HFD-evoked hepatic steatosis (Figure 3G) and improved the hepatic lipid profile (Table 2).
Figure 3.
Ginger polyphenol, 6-G, ameliorates hepatic steatosis through the up-regulation of miR-467b-3p. (A) miR-467b-3p expression by real-time PCR after 48 hours of exposure to 6-G (n = 3). ∗∗P < .01. (B and C) Nile red fluorescence intensity, (D) relative miR-467b-3p, (E) Gpat1 levels, and (F) Western blot analysis of total and phosphorylated AMPK and its downstream target, ACC, in Hepa1–6 cells after 24 hours of 6-G pretreatment followed by 24 hours of FFA treatment (n = 3). ∗P < .05 vs FFA-treated. (G) H&E and Oil red O staining of liver sections, (H) relative miR-467b-3p expressions, (I) mRNA sequencing analysis of potential miR-467b-3p targets, and (J) mRNA and protein expression of GPAT1 after HFD with or without 0.05% 6-G for 8 weeks (n = 5). ∗∗P < .01. (K) Nile red staining, (L) Nile red fluorescence intensity, and (M) mRNA expressions of Gpat1 in Hepa1–6 cells transfected with miR-467b-3p inhibitor (In), pretreated with 6-G and subsequent FFA treatment (n = 3). ∗P < .05 vs FFA-treated. CTL, control; DAPI, 4′,6-diamidino-2-phenylindole; FFA, free fatty acid; NC, normal control; AMPK, AMP-activating protein kinase ; p-AMPK, phosphorylated AMPK; ACC, acetyl-CoA carboxylase; p-ACC, phosphorylated ACC.
Table 2.
Effect of 6-G on Liver Weight and Hepatic Lipid Profile
| NC | HF | HF + 6-G | |
|---|---|---|---|
| Liver weight, g | 0.94 ± 0.07a | 1.61 ± 0.20 | 1.28 ± 0.07b |
| Hepatic lipid, mg/g liver | 56.01 ± 3.66a | 145.94 ± 7.82 | 110.89 ± 8.58b |
| Hepatic TAG, mg/g liver | 27.67 ± 0.89a | 60.90 ± 2.66 | 46.56 ± 3.33b |
| Hepatic TC, mg/g liver | 9.15 ± 0.16a | 14.07 ± 0.52 | 12.66 ± 0.52b |
NOTE. Each group consisted of 10 mice. Results shown are means ± SD.
NC, normal control; TC, total cholesterol.
P < .01.
P < .05 vs HF group.
Next, we measured the expression of miR-467b-3p and its target, Gpat1, in liver tissues. The miRNA profiling analysis showed that miR-467b-3p was one of the miRNAs regulated by 6-G (Supplementary Table 1), and 6-G significantly reversed HFD-induced down-regulation of miR-467b-3p (Figure 3G). Next, mRNA sequencing analysis identified 19 differentially expressed mRNAs by 6-G compared with the HFD-fed group (HF) group (Figure 3I). 6-G decreased HFD-induced GPAT1 up-regulation at both the mRNA and protein levels (Figure 3J). These data indicate that up-regulation of miR-467b-3p and the resultant down-regulation of Gpat1 are linked to the inhibitory effect of 6-G against hepatic lipid accumulation.
To confirm the relationship between 6-G and miR-467b-3p for the regulation of lipid accumulation in hepatocytes, we inhibited miR-467b-3p in Hepa1–6 cells and measured the effect of 6-G on intracellular lipid accumulation by FFA. Interestingly, the protective effect of 6-G on FFA-induced lipid accumulation was attenuated when miR-467b-3p was inhibited (Figure 3K and L). Moreover, a diminution in the inhibitory effect of 6-G on FFA-induced Gpat1 up-regulation in the miR-467b-3p–inhibited Hepa1–6 cells was observed (Figure 3M). Overall, these results suggest that the up-regulation of miR-467b-3p by 6-G is related to the improvement of NAFLD via its regulation of Gpat1.
Hepatocyte Nuclear Factor 4α Induction by 6-G Mediates the Up-regulation of miR-467b-3p
Next, we investigated how 6-G induced the up-regulation of miR-467b-3p. To examine the transcriptional regulation of miR-467b-3p, we used the miRGen 2.0 database (Alexander Fleming Biomedical Sciences Research Center, Vari, Greece)21 and found 2 miR-467b-3p hairpin transcripts with a binding site for hepatocyte nuclear factor 4α (HNF4α).
HNF4α is a liver-enriched transcription factor that regulates hepatocyte-specific genes involved in lipid and glucose metabolism,22,23 and the loss of hepatic HNF4α causes fatty liver in mice.23 Thus, we hypothesized that 6-G may regulate HNF4α to induce miR-467b-3p expression. To prove this hypothesis, we examined the effect of 6-G on HNF4 α and observed that 6-G increased Hnf4α expression in a dose-dependent manner (Figure 4A). Furthermore, the chromatin immunoprecipitation (ChIP) assay confirmed that HNF4α recruitment to upstream regions of the miR-467b transcription start site was increased by 6-G (Figure 4B). We also detected 6-G–induced up-regulation of Hnf4α in Hepa1–6 cells and in liver tissues (Figure 4C and D). Measurement of luciferase activity showed that 6-G increased the transcriptional activity of HNF4α (Figure 4E).
Figure 4.
Induction of HNF4α is essential for the up-regulation of miR-467b-3p by 6-G. (A) Hnf4α expression by real-time PCR in Hepa1–6 cells after 48 hours of 6-G treatment (n = 3). ∗P < .05. (B) Schematic representation of putative HNF4α binding site in the proximal pre–miR-467b promoter with the identification of ChIP–DNA fragments with capillary electrophoresis sequencing (left) and ChIP assay for HNF4α in 6-G–treated Hepa1–6 cells (right) (n = 4). ∗∗P < .01 vs IgG. (C) Expression of Hnf4α by real-time PCR in Hepa1-6 cells exposed to FFA for 24 h after 24 h of 6-G pretreatment (n = 3). ∗P < .05 or ∗∗P < .01 vs FFA-treated. (D) Relative Hnf4α hepatic expression in normal control (NC), HF, and HF+6-G mice (n = 5). ∗P < .05 vs HF. (E) Effect of 6-G on HNF4α transcriptional activity (n = 6). ∗∗P < .01 vs HNF4α reporter. (F) Effect of HNF4α-knockdown on the induction of (G) miR-467b-3p, (H) mRNA, and (I) protein expressions of GPAT1, and (J) inhibition of FFA accumulation by 6-G (n = 3-4). ∗P < .05 or ∗∗P < .01 vs small interfering RNA (siRNA)-CTL treated with FFA. #P < .05 vs siRNA-HNF4α treated with FFA. (K) Effect of HNF4α overexpression on (L) miR-467b-3p expression, and (M and N) FFA-induced intracellular fat accumulation in Hepa1–6 cells (n = 4). ∗∗P < .01 vs pGCDN-Cont. (O) Docking of 6-G (green) to HNF4α (orange) with the minimum energy represented by a stick model and ribbon structure, respectively. (P) DARTS test for validation of 6-G and HNF4α binding.
To confirm the role of HNF4α in the 6-G–induced up-regulation of miR-467b-3p, we knocked down HNF4α in Hepa1–6 cells (Figure 4F). In this context, 6-G failed to induce miR-467b-3p up-regulation and the subsequent decrease of GPAT1 mRNA and protein levels (Figure 4G–I), leading to the loss of the 6-G inhibitory effect on FFA-induced lipid accumulation (Figure 4J). In contrast, overexpression of HNF4α evoked the up-regulation of miR-467b-3p and significantly attenuated FFA-induced lipid accumulation in Hepa1–6 cells (Figure 4K–N). Overall, these data provide experimental evidence that induction of HNF4α is essential for the up-regulation of miR-467b-3p by 6-G.
To further elucidate the regulation mechanisms of HNF4α by 6-G, we tested whether HNF4α is a potential molecular target for 6-G. Molecular docking analysis predicted that HNF4α has the binding pockets necessary for 6-G binding, and that their binding score was -9.0 kcal/mol (Figure 4O). In addition, we conducted a drug-affinity–responsive target stability (DARTS) assay and showed the possibility of 6-G binding to HNF4α (Figure 4P).
Finally, we addressed whether the HNF4α-miR-467b-3p-GPAT1 axis is involved in the development of human NAFLD. We observed a modest decrease of HNF4α and miR-467b-3p in the liver of NAFLD patients (Figure 5A and B, respectively), while the mRNA expression of GPAT1 was increased significantly (P < .05) (Figure 5C). To test whether 6-G regulated the HNF4α-miR-467b-3p-GPAT1 axis could be present in human hepatic tissues we measured this axis in FFA-treated human Huh7 cells (Figure 5D–H). As we observed in murine hepatocytes Hepa1-6 cells, 6-G effectively decreased the intracellular lipid accumulation via regulation of the HNF4α-miR-467b-3p-GPAT1 axis in Huh7 cells. These results suggest that targeting the HNF4α-miR-467b-3p-GPAT1 axis could be a novel therapeutic approach for NAFLD treatment.
Figure 5.
HNF4α/miR-467b-3p/GPAT1 axis in human NAFLD tissues and human hepatocytes. (A) HNF4α, (B) miR-467b-3p, and (C) GPAT1 expression in normal (Nor) and NAFLD human tissues (Nor, n = 5; and NAFLD, n = 6). (D and E) Oil red O staining, (F) relative miR-467b-3p, (G) GPAT1 mRNA expression, and (H) HNF4α and GPAT1 protein expression in Huh7 human hepatocytes after 24 hours of 6-G pretreatment and 24 hours of FFA treatment (n = 3). ∗∗P < .01 vs FFA-treated.
Discussion
In this study, we showed a new mechanism by which miR-467b-3p is implicated in NAFLD (Figure 6). The decrease of miR-467b-3p was observed in FFA-treated hepatocytes and livers from HFD-fed mice. The miR-467b-3p increase generated by miR-467b-3-p mimic or 6-G inhibited FFA-induced intracellular lipid accumulation in hepatocytes and prevented hepatic steatosis in mice. In addition, Gpat1 was a direct target of miR-467b-3p, and 6-G–induced up-regulation of miR-467b-3p was mediated by HNF4α. Therefore, these results suggest that the targeting of the HNF4α/miR-467b-3p/GPAT1 cascade may be used as a novel therapeutic strategy to control NAFLD.
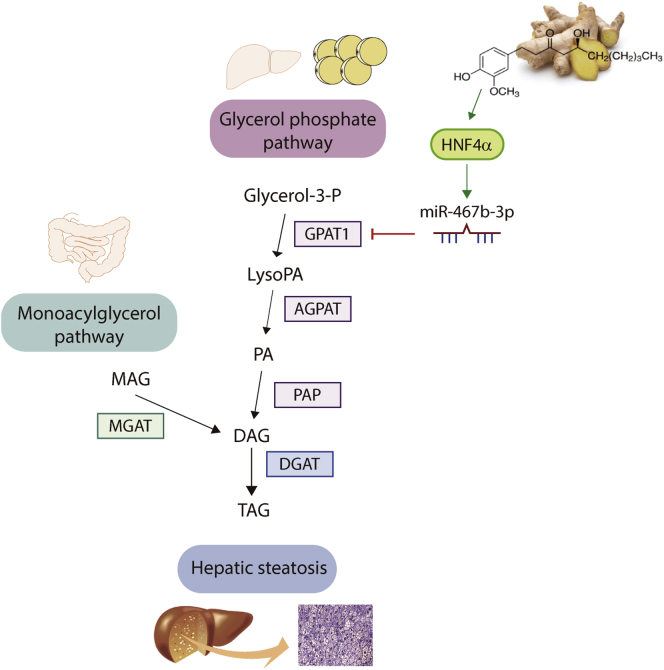
Figure 6.
Schematic representation of the role of the HNF4α/miR-467b-3p/GPAT1 pathway in the improvement of NAFLD. There are 2 major pathways for the synthesis of TAG. In the monoacylglycerol (MAG) pathway, predominantly reported in the intestine, monoacylglycerol acyltransferase (MGAT) produces diacylglycerol (DAG) by covalently joining a fatty acyl moiety to MAG. In the glycerol phosphate pathway, which functions in most cells, glycerol-3-phosphate (glycerol-3-P) is acylated sequentially by GPAT and acylglycerol phosphate acyltransferase (AGPAT) to produce LysoPA and phosphatidic acid (PA), respectively. DAG derived from the dephosphorylation of PA becomes TAG. During the development of hepatic steatosis, miR-467b-3p expression decreases in the liver, which leads to the increase of GPAT1 expression and the consequent increase in TAG synthesis. Conversely, up-regulation of miR-467b-3p inhibits hepatic lipid accumulation and ameliorates NAFLD. HNF4α mediates the induction of miR-467b-3p in hepatocytes. Ginger polyphenol, 6-G, promotes HNF4α induction, the up-regulation of miR-467b-3p, and the improvement of steatosis. Therefore, the regulation of the HNF4α/miR-467b-3p/GPAT1 cascade is a potential therapeutic target for the treatment of NAFLD.
NAFLD is characterized by hepatocyte steatosis and increased serum FFAs.24 Although it is the most common liver disease in the United States, there are no Food and Drug Administration–approved pharmacologic treatments for NAFLD. Because of this, attention has been focused on the use of dietary natural compounds, such as polyphenols from fruits and vegetables, to ameliorate NAFLD. Dietary polyphenols have been shown to modulate hepatic miRNA expression, including that of miR-467b-3p.8 However, the role of miR-467b-3p in hepatic steatosis remains unknown. In this study, we first observed down-regulated miR-467b-3p expression after FFA-induced lipid accumulation in hepatocytes and in HFD-induced hepatic steatosis. Gain- or loss-of-function studies showed that miR-467b-3p plays an important role in reducing lipid accumulation in hepatocytes by directly binding to the 3’UTR of Gpat1 and inhibiting GPAT1 expression at mRNA and protein levels.
GPAT1, which is highly active in liver, catalyzes TAG synthesis by inducing the formation of LysoPA from glycerol-3-phosphate, and its activity is modulated in a manner consistent with the regulation of TAG synthesis.25 It has been reported that in GPAT1 knockout mice, the body weight and liver TAG content decrease.12 Conversely, liver-specific overexpression of GPAT1 induces hepatic steatosis in mice and increases intracellular TAG biosynthesis in rat hepatocytes.26,27 These data suggest that GPAT1 is a crucial target for the prevention and treatment of hepatic steatosis.
Because the importance of miRNA in various human diseases has been shown, an in vivo miRNA delivery strategy for the modulation of miRNA expression may serve as a novel therapeutic modality for such diseases.28 Because the down-regulation of miR-467b-3p and consequent up-regulation of GPAT1 are implicated in hepatic steatosis, we hypothesized that increased expression of miR-467b-3p may ameliorate hepatic steatosis. To evaluate the therapeutic potential of miR-467b-3p for NAFLD in vivo, we administered miR-467b-3p mimic to HFD-fed mice, and, as expected, miR-467b-3p overexpression induced a decrease in GPAT1 and LysoPA, resulting in the improvement of hepatic steatosis.
The expression of miR-467b-3p also is influenced by polyphenol supplementation in hepatic steatosis. We choose to use 6-G because it induced up-regulation of miR-467b-3p in a dose-dependent manner. We showed that 6-G effectively inhibited FFA-induced lipid accumulation in hepatocytes and HFD-induced hepatosteatosis. Interestingly, we observed that FFA and/or HFD-induced down-regulation of miR-467b-3p was inhibited significantly by 6-G treatment. Moreover, 6-G decreased GPAT1 expression at the mRNA and protein levels. Hepatic accumulation of TAG originates from FFA flux (59%), de novo lipogenesis (DNL) (26%), and diet (15%).29 6-G supplementation induced phosphorylation of ACC, a control point that limits DNL rates, leading to inactivation of ACC. This was mediated through phosphorylation of AMPK. Furthermore, 6-G inhibited mRNA expression of DNL-related genes and also normalized dysregulated fatty acid oxidation-related genes. Thus, 6-G improved hepatic steatosis via the up-regulation of miR-467b-3p and the resultant down-regulation of Gpat1.
Transcriptional regulation by nuclear receptors is the primary level of control of miRNA expression.30 miRNA genes also contain a promoter region located near a transcription start site, similar to protein-coding genes.31 Therefore, it is important to provide comprehensive information about the position of miRNA coding transcripts and the binding of functional transcription factors for understanding the regulation of miRNA. HNF4α is a member of the nuclear-receptor superfamily of transcription factors and positively regulates many genes involved in liver-specific functions, and as such is a key regulator of energy metabolism, glucose, and lipid homeostasis.32 Fatty acyl-CoA thioesters and linoleic acid are ligands of HNF4α.33,34 Furthermore, loss of hepatic HNF4α causes fatty liver in mice,23 and nonalcoholic steatohepatitis patients show reduced hepatic HNF4α expression.35 A previous study showed that HNF4α can transactivate the miR-122 promoter and positively regulate miR-122 expression,36 and, recently, HNF4α was reported to be a target of miR-34a, in a pathway that regulates lipoprotein metabolism.35 We found binding sites for HNF4α in the miR-467b promoter region and experimentally confirmed that 6-G increased the transcriptional activity of HNF4α. Moreover, 6-G failed to induce miR-467b-3p up-regulation and to inhibit lipid accumulation when HNF4α was silenced. This is in agreement with a previous study reporting that hepatocyte-specific HNF4 knockout mice show a marked increase in their intracellular hepatocyte lipid levels and disrupted bile acid metabolism,22 which we also observed in our HFD-fed mice. Molecular modeling predicted the possibility of 6-G binding to the pocket of HNF4α, and we confirmed this possibility using the DART assay. The HNF4α-miR-467b-3p-GPAT1 cascade also was confirmed in human NAFLD tissue samples.
In summary, we provide evidence that 6-G–induced up-regulation of miR-467b-3p plays a critical role in the amelioration of hepatic steatosis. Thus, miR-467b-3p could be a promising therapeutic target for the amelioration of NAFLD.
Materials and Methods
Cell Culture and Steatosis Induction
Hepa1–6 and Huh7 cells were purchased from the ATCC (Manassas, VA) and the Korean Cell Line Bank (Seoul, Korea), respectively. These cells were maintained in Dulbecco’s modified Eagle medium (HyClone, Logan, UT) supplemented with 10% fetal bovine serum at 37°C in a 5% CO2 atmosphere. Bovine serum albumin–conjugated FFA was prepared as reported previously.37 Cells were incubated with 50 μmol/L bovine serum albumin–conjugated palmitate for 24 hours. The cells were pretreated for 24 hours with 6-G (G1046; Sigma-Aldrich, St. Louis, MO) and then exposed to palmitate for 24 hours. 6-G was dissolved in dimethyl sulfoxide.
Quantitative Real-Time PCR
Total RNA was isolated with a NucleoSpin RNA II kit (Macherey-Nagel GmbH & Co, Duren, Germany). For miRNA quantification, total RNA was reverse-transcribed with the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Carlsbad, CA) and subjected to real-time PCR with the TaqMan MicroRNA Assay Kit. The miRNA expression was normalized to that of endogenous snoRNA202. For mRNA expression assays, complementary DNA (cDNA) was prepared as previously described,38 and real-time PCR was performed with SYBR Green PCR Master Mix (Toyobo, Osaka, Japan) using a ViiA7 Real-Time PCR System (Applied Biosystems, Foster City, CA). The expression level of each mRNA was normalized to that of Gapdh. The sequences of the primers used in these experiments are listed in Supplementary Table 2.
Nile Red Fluorescence Determination of Neutral Lipid
After fixation with formaldehyde, neutral lipids of hepatocytes were stained with Nile red (Sigma-Aldrich) in acetone and nuclei were stained with 4′,6-diamidino-2-phenylindole (Thermo Fisher Scientific, Waltham, MA) for fluorescence measurement (Infinite M200 PRO; Tecan, Männedorf, Switzerland) and microscopy (IX71; Olympus, Tokyo, Japan). The fluorescence was measured at excitation and emission wavelength settings of 530 nm and 575 nm, respectively.
miR-467b-3p Mimic and Inhibitor Transfection
To modulate miR-467b-3p expression, we used miR467b-3p–specific activator (miRIDIAN miR-467b-3p mimic; Thermo Fisher Scientific) or miR-467b-3p–specific inhibitor (miRIDIAN miR-467b-3p hairpin inhibitor; Thermo Fisher Scientific). For negative control, we used an oligonucleotide sequence based on Caenorhabditis elegans miR-67 (Thermo Fisher Scientific). Each oligonucleotide (1 pmol) was transfected into Hepa1–6 cells using the Lipofectamine RNAiMAX Transfection Reagent (Thermo Fisher Scientific). Overexpression or knockdown of miR-467b-3p was verified by real-time PCR.
Animals and Diets
We performed 2 different in vivo experiments. In the first, NAFLD was induced in 6-week-old male C57BL/6J mice through feeding with a HFD (45% of total calories from fat) for 8 weeks or by miR-467b-3p nanoparticle delivery. In the second, to test the effect of 6-G on NAFLD, 6-week-old male C57BL/6J mice were divided into 3 groups (n = 10/group) and fed with different experimental diets for 8 weeks: (1) normal control group received a normal-fat diet (10% of total calories from fat), (2) HF received a HFD (45% of total calories from fat), and (3) HF+6-G group was fed a HFD containing 0.05% 6-G (Chemfaces, Hubei, China). All of the diets were based on the AIN-76M diet,39 and the composition of each diet is shown in Supplementary Table 3. After 8 weeks, the mice underwent 12 hours of fasting and blood samples were collected just before death. All the animal studies were conducted in accordance with a protocol approved by the Korea Food Research Institute’s Institutional Animal Care and Use Committee.
miR-467b-3p Particle Delivery In Vivo
To induce in vivo overexpression of miR-467b-3p, we used mirVana miR-467b-3p mimic (Thermo Fisher Scientific). The nanoparticles suitable for in vivo application were prepared using mirVana miR-467b-3p mimic and Invivofectamine 2.0 reagent (Thermo Fisher Scientific) according to the manufacturer’s protocol.
After 8 weeks of feeding with a HFD, the mice were injected intraperitoneally with 100 μL (7 mg/kg) of either miR-467b-3p mimic or control. The injections were performed every other day for 10 days. After 2 days of the final injection, the mice were killed and tissues and blood were collected.
Histologic Analysis and Measurement of Liver Lipid Content
For histologic analysis, liver samples were fixed in 10% buffered formalin and stained with H&E or Oil Red O. Total hepatic lipid content of liver homogenates obtained from chloroform–methanol extraction was measured as described previously40 and expressed as milligrams of lipid per gram of wet liver weight. Tissue TAG and total cholesterol levels were measured with commercially available kits according to the manufacturer’s instructions (Wako Chemicals, Osaka, Japan). The content of lysophosphatidic acid was measured by commercial enzyme-linked immunosorbent assay kit (MyBioSource, San Diego, CA)
Western Blot
Total proteins were isolated using RIPA lysis buffer (Thermo Fisher Scientific), and their concentration was determined with an enhanced BCA protein assay kit (Thermo Fisher Scientific). Protein samples (20 μg) were separated by 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes (Bio-Rad, Hercules, CA). The membranes were blocked with 5% skim milk in Tris buffered saline with 0.1% Tween 20 detergent (TBST) buffer and incubated with primary antibodies overnight at 4°C. After washing several times with TBST buffer, the membranes were incubated with the appropriate horseradish-peroxidase–conjugated secondary antibody. The immunoreactive proteins were visualized with the Clarity ECL Western Blot Substrate (Bio-Rad). The list of antibodies used in this study is shown in Supplementary Table 4.
Luciferase Reporter Assays
The pMIR-REPORT System (Thermo Fisher Scientific) was used to identify miR-467b-3p binding sites in the 3ʹUTR of Gpat1. The β-galactosidase reporter control vector was used for normalization. The putative or mutated miR-467b-3p binding sites of Gpat1 3ʹUTR were cloned into a luciferase miRNA expression reporter vector ,which contains firefly luciferase under the control of a mammalian promoter/terminator system. Hepa1–6 cells were co-transfected with the Gpat1 3ʹUTR luciferase reporter and the control construct, and with the miR-467b-3p mimic or mimic control, using the Lipofectamine 2000 system (Thermo Fisher Scientific). After 48 hours of transfection, luciferase activity was measured using the Dual-Light System (Applied Biosystems) and normalized to the corresponding β-galactosidase activity.
For the measurement of HNF4α signaling, Hepa1–6 cells were transfected with negative control or inducible HNF4α reporter plasmid from the Cignal HNF4α Reporter Assay Kit (SABiosciences, Hilden, Germany), along with a constitutively expressed Renilla construct. After 24 hours of transfection, cells were treated with 6-G for 48 hours. Luciferase activity was measured using the Dual-Light System.
mRNA Sequencing Analysis
Total RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA). RNA quantity was assessed with a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific). For mRNA sequencing analysis, RNA quality was determined on a Bioanalyser 2100 (Agilent, Santa Clara, CA) and only RNA samples with an RNA integrity number between 8 and 10 were used. Libraries were prepared from total RNA using the NEBNext Ultra II Directional RNA Sequencing Kit (New England BioLabs, Inc, Hitchin, UK). The isolation of mRNA was performed using the Poly(A) RNA Selection Kit (Lexogen, Inc, Vienna, Austria), indexing with the Illumina indexes 1–12. High-throughput sequencing was performed as paired-end 100 sequencing with the NovaSeq 6000 (Illumina, Inc., San Diego, CA). Gene expression levels were estimated using fragments per kilobase per million reads values by Cufflinks (University of Washington, Seattle, WA).41 Data mining and graphic visualization were performed using ExDEGA (Ebiogen, Inc, Seoul, Korea).
Measurement of Hepatic miRNAs Profiles Using TaqMan Low-Density Arrays Rodent Cards
Megaplex profiling using rodent TaqMan low-density arrays (TLDA) (Applied Biosystems) was used to assay the expression of 380 miRNAs as described by the manufacturer. Total RNA from liver tissues was examined by TLDA cards. The cDNA was synthesized from total RNA using Megaplex RT primer Rodent A Pool (4399970; Applied Biosystems) and the TaqMan MicroRNA Reverse Transcription Kit (436659; Applied Biosystems), according to the manufacturer’s instructions. The synthesized cDNA was pre-amplified using Megaplex PreAmp Primers Rodent Pool A (4399203; Applied Biosystems) and Megaplex PreAmp Master Mix (PN4384267; Applied Biosystems). Quantitative PCR was performed using the TaqMan Rodent MicroRNA Array (4398967; Applied Biosystems). miRNA expression data from the TLDAs were analyzed with ExpressionSuite Software v1.0.3 (Life Technologies, Carlsbad, CA) by using the ΔΔ threshold cycle (Ct) method after global normalization. Expression changes are shown as log2 fold change after comparing the normalized expression of each experiment. A list of >2-fold differential changes of miRNAs induced by 6-G is shown in Supplementary Table 1.
RNA Interference of HNF4α
Hepa1–6 cells were transfected with 25 nmol/L Hnf4α small interfering RNA (GE Dharmacon, Lafayette, CO), and control nontargeting small interfering RNA using Lipofectamine RNAiMAX (Thermo Fisher Scientific). After 48 hours of transfection, cells were treated with FFA for 24 hours with or without 6-G pretreatment and then lysed. The silencing of HNF4α was confirmed by Western blot as described earlier.
Retrovirus Production and Transduction of Cells
pGCDNsam-Hnf4a plasmid was supplied by Addgene (Watertown, MA). 293T cells were transfected with the pGCDNsam-Hnf4a plasmid along with pUMVC and pCMV-VSV-G using Fugene6 (Roche, Basel, Switzerland). Twenty-four hours after transfection, the culture media containing the retroviruses were harvested. The polybene (×1000)-supplemented retrovirus solution was added to the Hepa1–6 cells for 3 hours, and this was repeated 4 times. After 12 hours, infected cells were changed to fresh media containing puromycin (2 ug/mL).
ChIP Assay
After treatment with 6-G for 24 hours, Hepa1–6 cells were exposed to formaldehyde 1% for 10 minutes, and washed with phosphate-buffered saline. A ChIP assay was performed with the agarose ChIP kit (Thermo Fisher Scientific) according to the manufacturer’s protocol. Digested chromatin was incubated overnight with 4 μg anti-HNF4α. After DNA recovery, both eluted and total input samples were used in real-time PCR for the quantification of protein-bound DNA. Primers were designed to span an approximately 130-bp region of the putative HNF4α binding site. Primer sequences were as follows: forward: 5'-GTGATGTAGAGTTTTAGC-3' and reverse: 5'-CAGGATCTCTAACTA-3'
Molecular Docking
A HNF4α binding site analysis to determine the potential binding pockets was performed using PockDrug (Paris, France)42 and Autodock Vina (La Jolla, CA).43 The structure of 6-G was downloaded from Pubchem (Bethesda, MD) (Pubchem CID: 442793) in structure data file (SDF) format and converted into PDBQT with PyRx (La Jolla, CA). The crystal structure of HNF4α was obtained from the Protein Data Bank (Piscataway, NJ)(ID: 3FS1). The binding interaction of 6-G and HNF4α was simulated by Autodock Vina. Finally, according to the scoring function, the configuration between 6-G and HNF4α with the lowest binding energy was obtained.
DARTS Test
For the validation of binding between 6-G and HNF4α the DARTS test was performed as described previously.44 Briefly, Hepa1–6 cells were incubated with 25 μmol/L 6-G or 0.1% dimethyl sulfoxide for 48 hours and lysed with M-PER lysis buffer (Sigma-Aldrich) containing protease inhibitor. After centrifugation at 18,000g for 10 minutes, the supernatant was diluted with 0.5 M Tris-HCl containing 0.5 M HCl, 0.1 M NaCl, and 0.1 M CaCl2. The protein concentration was determined by BCA assay. Each sample was divided into 2 aliquots, one was subjected to proteolysis with pronase (Roche, Basel, Switzerland) and the other was used in a mock proteolysis. After digestion, Western blot analysis was performed.
Human Tissue Samples
Human fatty livers (n = 6) and their adjacent normal liver tissues (n = 5) were collected from patients with hepatocellular carcinoma undergoing liver resection at Gachon Medical University (Incheon, Korea). This study was approved by the Ethics Committee of Gachon University Gil Medical Center (GCIRB2016-277), and informed consent was obtained from all individuals.
Statistical Analysis
Data are shown as means ± SD for cell study and means ± SE for animal and human studies. Statistical analyses were performed with GraphPad Prism 6 software (San Diego, CA). Two groups were compared using an unpaired t test with 2-tailed distributions. For 3 or more groups, 1-way analysis of variance was used to compare quantitative data among groups. The Bonferroni post hoc test was used to adjust for multiple comparisons (P < .05).
CRediT Authorship Contributions
Jiyun Ahn, PhD (Conceptualization: Lead; Writing – original draft: Lead; Writing – review & editing: Lead)
Hyunjung Lee (Formal analysis: Supporting)
Chang Hwa Jung (Methodology: Equal)
Seung Yeon Ha (Resources: Supporting)
Hyo-Deok Seo, Ph.D. (Formal analysis: Supporting)
Young In Kim (Formal analysis: Supporting)
Taeyoul Ha (Conceptualization: Lead; Project administration: Lead; Writing – review & editing: Lead)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This work was supported by a grant from the Korea Food Research Institute (E0160500 and E0210101).
Contributor Information
Jiyun Ahn, Email: jyan@kfri.re.kr.
Taeyoul Ha, Email: tyhap@kfri.re.kr.
Supplementary Material
References
- 1.McCullough A.J. Pathophysiology of nonalcoholic steatohepatitis. J Clin Gastroenterol. 2006;40(Suppl 1):S17–S29. doi: 10.1097/01.mcg.0000168645.86658.22. [DOI] [PubMed] [Google Scholar]
- 2.Niklas J., Bonin A., Mangin S., Bucher J., Kopacz S., Matz-Soja M., Thiel C., Gebhardt R., Hofmann U., Mauch K. Central energy metabolism remains robust in acute steatotic hepatocytes challenged by a high free fatty acid load. BMB Rep. 2012;45:396–401. doi: 10.5483/bmbrep.2012.45.7.070. [DOI] [PubMed] [Google Scholar]
- 3.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Vincent R., Sanyal A. Recent advances in understanding of NASH: microRNAs as both biochemical markers and players. Curr Pathobiol Rep. 2014;2:109–115. doi: 10.1007/s40139-014-0049-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park J.H., Ahn J., Kim S., Kwon D.Y., Ha T.Y. Murine hepatic miRNAs expression and regulation of gene expression in diet-induced obese mice. Mol Cells. 2011;31:33–38. doi: 10.1007/s10059-011-0009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheung O., Puri P., Eicken C., Contos M.J., Mirshahi F., Maher J.W., Kellum J.M., Min H., Luketic V.A., Sanyal A.J. Nonalcoholic steatohepatitis is associated with altered hepatic microRNA expression. Hepatology. 2008;48:1810–1820. doi: 10.1002/hep.22569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahn J., Lee H., Chung C.H., Ha T. High fat diet induced downregulation of microRNA-467b increased lipoprotein lipase in hepatic steatosis. Biochem Biophys Res Commun. 2011;414:664–669. doi: 10.1016/j.bbrc.2011.09.120. [DOI] [PubMed] [Google Scholar]
- 8.Milenkovic D., Deval C., Gouranton E., Landrier J.F., Scalbert A., Morand C., Mazur A. Modulation of miRNA expression by dietary polyphenols in apoE deficient mice: a new mechanism of the action of polyphenols. PLoS One. 2012;7 doi: 10.1371/journal.pone.0029837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beattie J.H., Nicol F., Gordon M.J., Reid M.D., Cantlay L., Horgan G.W., Kwun I.S., Ahn J.Y., Ha T.Y. Ginger phytochemicals mitigate the obesogenic effects of a high-fat diet in mice: a proteomic and biomarker network analysis. Mol Nutr Food Res. 2011;55(Suppl 2):S203–S213. doi: 10.1002/mnfr.201100193. [DOI] [PubMed] [Google Scholar]
- 10.Gimeno R.E., Cao J. Thematic review series: glycerolipids. Mammalian glycerol-3-phosphate acyltransferases: new genes for an old activity. J Lipid Res. 2008;49:2079–2088. doi: 10.1194/jlr.R800013-JLR200. [DOI] [PubMed] [Google Scholar]
- 11.Nagle C.A., An J., Shiota M., Torres T.P., Cline G.W., Liu Z.X., Wang S., Catlin R.L., Shulman G.I., Newgard C.B., Coleman R.A. Hepatic overexpression of glycerol-sn-3-phosphate acyltransferase 1 in rats causes insulin resistance. J Biol Chem. 2007;282:14807–14815. doi: 10.1074/jbc.M611550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hammond L.E., Gallagher P.A., Wang S., Hiller S., Kluckman K.D., Posey-Marcos E.L., Maeda N., Coleman R.A. Mitochondrial glycerol-3-phosphate acyltransferase-deficient mice have reduced weight and liver triacylglycerol content and altered glycerolipid fatty acid composition. Mol Cell Biol. 2002;22:8204–8214. doi: 10.1128/MCB.22.23.8204-8214.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Z., Zeng M., Wang Z., Qin F., Chen J., He Z. Dietary polyphenols to combat nonalcoholic fatty liver disease via the gut-brain-liver axis: a review of possible mechanisms. J Agric Food Chem. 2021;69:3585–3600. doi: 10.1021/acs.jafc.1c00751. [DOI] [PubMed] [Google Scholar]
- 14.Dugasani S., Pichika M.R., Nadarajah V.D., Balijepalli M.K., Tandra S., Korlakunta J.N. Comparative antioxidant and anti-inflammatory effects of [6]-gingerol, [8]-gingerol, [10]-gingerol and [6]-shogaol. J Ethnopharmacol. 2010;127:515–520. doi: 10.1016/j.jep.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 15.Abolaji A.O., Ojo M., Afolabi T.T., Arowoogun M.D., Nwawolor D., Farombi E.O. Protective properties of 6-gingerol-rich fraction from Zingiber officinale (Ginger) on chlorpyrifos-induced oxidative damage and inflammation in the brain, ovary and uterus of rats. Chem Biol Interact. 2017;270:15–23. doi: 10.1016/j.cbi.2017.03.017. [DOI] [PubMed] [Google Scholar]
- 16.Poltronieri J., Becceneri A.B., Fuzer A.M., Filho J.C., Martin A.C., Vieira P.C., Pouliot N., Cominetti M.R. [6]-gingerol as a cancer chemopreventive agent: a review of its activity on different steps of the metastatic process. Mini Rev Med Chem. 2014;14:313–321. doi: 10.2174/1389557514666140219095510. [DOI] [PubMed] [Google Scholar]
- 17.Zhang F., Zhang J.G., Yang W., Xu P., Xiao Y.L., Zhang H.T. 6-Gingerol attenuates LPS-induced neuroinflammation and cognitive impairment partially via suppressing astrocyte overactivation. Biomed Pharmacother. 2018;107:1523–1529. doi: 10.1016/j.biopha.2018.08.136. [DOI] [PubMed] [Google Scholar]
- 18.Liu L., Yao L., Wang S., Chen Z., Han T., Ma P., Jiang L., Yuan C., Li J., Ke D., Li C., Yamahara J., Li Y., Wang J. 6-Gingerol improves ectopic lipid accumulation, mitochondrial dysfunction, and insulin resistance in skeletal muscle of ageing rats: dual stimulation of the AMPK/PGC-1alpha signaling pathway via plasma adiponectin and muscular AdipoR1. Mol Nutr Food Res. 2019;63 doi: 10.1002/mnfr.201800649. [DOI] [PubMed] [Google Scholar]
- 19.Li J., Wang S., Yao L., Ma P., Chen Z., Han T.L., Yuan C., Zhang J., Jiang L., Liu L., Ke D., Li C., Yamahara J., Li Y., Wang J. 6-gingerol ameliorates age-related hepatic steatosis: association with regulating lipogenesis, fatty acid oxidation, oxidative stress and mitochondrial dysfunction. Toxicol Appl Pharmacol. 2019;362:125–135. doi: 10.1016/j.taap.2018.11.001. [DOI] [PubMed] [Google Scholar]
- 20.Tzeng T.F., Liou S.S., Chang C.J., Liu I.M. [6]-gingerol dampens hepatic steatosis and inflammation in experimental nonalcoholic steatohepatitis. Phytomedicine. 2015;22:452–461. doi: 10.1016/j.phymed.2015.01.015. [DOI] [PubMed] [Google Scholar]
- 21.Alexiou P., Vergoulis T., Gleditzsch M., Prekas G., Dalamagas T., Megraw M., Grosse I., Sellis T., Hatzigeorgiou A.G. miRGen 2.0: a database of microRNA genomic information and regulation. Nucleic Acids Res. 2010;38(database issue):D137–D141. doi: 10.1093/nar/gkp888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayhurst G.P., Lee Y.H., Lambert G., Ward J.M., Gonzalez F.J. Hepatocyte nuclear factor 4alpha (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol Cell Biol. 2001;21:1393–1403. doi: 10.1128/MCB.21.4.1393-1403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yin L., Ma H., Ge X., Edwards P.A., Zhang Y. Hepatic hepatocyte nuclear factor 4alpha is essential for maintaining triglyceride and cholesterol homeostasis. Arterioscler Thromb Vasc Biol. 2011;31:328–336. doi: 10.1161/ATVBAHA.110.217828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feldstein A.E., Canbay A., Guicciardi M.E., Higuchi H., Bronk S.F., Gores G.J. Diet associated hepatic steatosis sensitizes to Fas mediated liver injury in mice. J Hepatol. 2003;39:978–983. doi: 10.1016/s0168-8278(03)00460-4. [DOI] [PubMed] [Google Scholar]
- 25.Dircks L.K., Sul H.S. Mammalian mitochondrial glycerol-3-phosphate acyltransferase. Biochim Biophys Acta. 1997;1348:17–26. doi: 10.1016/s0005-2760(97)00106-9. [DOI] [PubMed] [Google Scholar]
- 26.Linden D., William-Olsson L., Ahnmark A., Ekroos K., Hallberg C., Sjogren H.P., Becker B., Svensson L., Clapham J.C., Oscarsson J., Schreyer S. Liver-directed overexpression of mitochondrial glycerol-3-phosphate acyltransferase results in hepatic steatosis, increased triacylglycerol secretion and reduced fatty acid oxidation. FASEB J. 2006;20:434–443. doi: 10.1096/fj.05-4568com. [DOI] [PubMed] [Google Scholar]
- 27.Linden D., William-Olsson L., Rhedin M., Asztely A.K., Clapham J.C., Schreyer S. Overexpression of mitochondrial GPAT in rat hepatocytes leads to decreased fatty acid oxidation and increased glycerolipid biosynthesis. J Lipid Res. 2004;45:1279–1288. doi: 10.1194/jlr.M400010-JLR200. [DOI] [PubMed] [Google Scholar]
- 28.Li Z., Rana T.M. Therapeutic targeting of microRNAs: current status and future challenges. Nat Rev Drug Discov. 2014;13:622–638. doi: 10.1038/nrd4359. [DOI] [PubMed] [Google Scholar]
- 29.Donnelly K.L., Smith C.I., Schwarzenberg S.J., Jessurun J., Boldt M.D., Parks E.J. Sources of fatty acids stored in liver and secreted via lipoproteins in patients with nonalcoholic fatty liver disease. J Clin Invest. 2005;115:1343–1351. doi: 10.1172/JCI23621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Z., Wang L. Regulation of microRNA expression and function by nuclear receptor signaling. Cell Biosci. 2011;1:31. doi: 10.1186/2045-3701-1-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Megraw M., Baev V., Rusinov V., Jensen S.T., Kalantidis K., Hatzigeorgiou A.G. MicroRNA promoter element discovery in Arabidopsis. RNA. 2006;12:1612–1619. doi: 10.1261/rna.130506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watt A.J., Garrison W.D., Duncan S.A. HNF4: a central regulator of hepatocyte differentiation and function. Hepatology. 2003;37:1249–1253. doi: 10.1053/jhep.2003.50273. [DOI] [PubMed] [Google Scholar]
- 33.Courage M.L., McCloy U.R., Herzberg G.R., Andrews W.L., Simmons B.S., McDonald A.C., Mercer C.N., Friel J.K. Visual acuity development and fatty acid composition of erythrocytes in full-term infants fed breast milk, commercial formula, or evaporated milk. J Dev Behav Pediatr. 1998;19:9–17. doi: 10.1097/00004703-199802000-00002. [DOI] [PubMed] [Google Scholar]
- 34.Yuan X., Ta T.C., Lin M., Evans J.R., Dong Y., Bolotin E., Sherman M.A., Forman B.M., Sladek F.M. Identification of an endogenous ligand bound to a native orphan nuclear receptor. PLoS One. 2009;4 doi: 10.1371/journal.pone.0005609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu Y., Zalzala M., Xu J., Li Y., Yin L., Zhang Y. A metabolic stress-inducible miR-34a-HNF4alpha pathway regulates lipid and lipoprotein metabolism. Nat Commun. 2015;6:7466. doi: 10.1038/ncomms8466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Z.Y., Xi Y., Zhu W.N., Zeng C., Zhang Z.Q., Guo Z.C., Hao D.L., Liu G., Feng L., Chen H.Z., Chen F., Lv X., Liu D.P., Liang C.C. Positive regulation of hepatic miR-122 expression by HNF4alpha. J Hepatol. 2011;55:602–611. doi: 10.1016/j.jhep.2010.12.023. [DOI] [PubMed] [Google Scholar]
- 37.Huynh F.K., Green M.F., Koves T.R., Hirschey M.D. Measurement of fatty acid oxidation rates in animal tissues and cell lines. Methods Enzymol. 2014;542:391–405. doi: 10.1016/B978-0-12-416618-9.00020-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 39.Bieri J.G. AIN-76 diet. J Nutr. 1979;109:925–926. doi: 10.1093/jn/109.5.925. [DOI] [PubMed] [Google Scholar]
- 40.Bligh E.G., Dyer W.J. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 41.Roberts A., Trapnell C., Donaghey J., Rinn J.L., Pachter L. Improving RNA-seq expression estimates by correcting for fragment bias. Genome Biol. 2011;12:R22. doi: 10.1186/gb-2011-12-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hussein H.A., Borrel A., Geneix C., Petitjean M., Regad L., Camproux A.C. PockDrug-Server: a new web server for predicting pocket druggability on holo and apo proteins. Nucleic Acids Res. 2015;43:W436–W442. doi: 10.1093/nar/gkv462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pai M.Y., Lomenick B., Hwang H., Schiestl R., McBride W., Loo J.A., Huang J. Drug affinity responsive target stability (DARTS) for small-molecule target identification. Methods Mol Biol. 2015;1263:287–298. doi: 10.1007/978-1-4939-2269-7_22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.