Abstract
Emergence of new severe acute respiratory syndrome coronavirus 2 variants has raised concerns related to the effectiveness of vaccines and antibody therapeutics developed against the unmutated wildtype virus. Here, we examined the effect of the 12 most commonly occurring mutations in the receptor-binding domain of the spike protein on its expression, stability, activity, and antibody escape potential. Stability was measured using thermal denaturation, and the activity and antibody escape potential were measured using isothermal titration calorimetry in terms of binding to the human angiotensin-converting enzyme 2 and to neutralizing human antibody CC12.1, respectively. Our results show that mutants differ in their expression levels. Of the eight best-expressed mutants, two (N501Y and K417T/E484K/N501Y) showed stronger affinity to angiotensin-converting enzyme 2 compared with the wildtype, whereas four (Y453F, S477N, T478I, and S494P) had similar affinity and two (K417N and E484K) had weaker affinity than the wildtype. Compared with the wildtype, four mutants (K417N, Y453F, N501Y, and K417T/E484K/N501Y) had weaker affinity for the CC12.1 antibody, whereas two (S477N and S494P) had similar affinity, and two (T478I and E484K) had stronger affinity than the wildtype. Mutants also differ in their thermal stability, with the two least stable mutants showing reduced expression. Taken together, these results indicate that multiple factors contribute toward the natural selection of variants, and all these factors need to be considered to understand the evolution of the virus. In addition, since not all variants can escape a given neutralizing antibody, antibodies to treat new variants can be chosen based on the specific mutations in that variant.
Keywords: protein stability, protein binding, protein function, protein structure, protein expression, COVID-19, SARS-CoV-2, ACE2, antibodies, variants
Abbreviations: ACE2, angiotensin-converting enzyme 2; COVID-19, coronavirus disease 2019; ER, endoplasmic reticulum; HEK, human embryonic kidney; ITC, isothermal titration calorimetry; PDB, Protein Data Bank; RBD, receptor-binding domain; RBM, receptor-binding motif; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; ScFv, single-chain fragment variable; VOC, variant of concern
Coronavirus disease 2019 (COVID-19) pandemic has emerged as a global threat in December 2019. As of September 2021, it has infected over 225 million people and claimed 4.6 million lives (https://coronavirus.jhu.edu/map.html). The causative agent for COVID-19 is severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a single-stranded RNA virus, that belongs to sarbecovirus subgenus of betacoronaviruses (1). It shares close genomic similarity to SARS-CoV (79% identity) and Middle East respiratory syndrome coronavirus (50% identity) that were responsible for SARS outbreak in 2002 and Middle East respiratory syndrome outbreak in 2012, respectively (2, 3, 4, 5, 6). These viruses are thought to have originated in bats and transmitted to other mammals including humans (7).
Both SARS-CoV and SARS-CoV-2 enter the human host through interaction of its spike protein with angiotensin-converting enzyme 2 (ACE2) present on the membrane of host epithelial cells (8, 9, 10). Specifically, the receptor-binding domain (RBD) of the spike protein binds with ACE2 and thus is a major determinant of the viral infectivity and evolution (11, 12, 13). The viral evolution through accumulation of mutations in SARS-CoV-2 is slower than known for other RNA viruses like HIV and influenza (14, 15, 16). Still, SARS-CoV-2 variants pose a major challenge for devising measures to counter the virus threat, as new variants continue to emerge, some of which are believed to be more infectious than the wildtype virus (17, 18, 19). Among all the mutations happening in the viral genome, mutations in RBD are considered to play a significant role in infectivity because of its role in ACE2 binding. Most of the neutralizing antibody responses of the host is generated against RBD (20, 21), because of which RBD is also a major target for most of the therapeutic antibodies developed (22, 23, 24). Mutations in RBD are predicted to dictate the emergence of escape mutants and shape the evolutionary path of the virus through the process of natural selection that would favor the mutants that could evade the antibody response. Apart from this, RBD alone or as part of spike protein is also used as antigen in many prospective vaccines (25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35). The emergence of mutations in RBD is considered to have an impact on the effectiveness of these vaccines. Lower efficacy of some of the vaccines was reported against the emerging variants of concern (VOCs) that include alpha, beta, and gamma variants along with reports of new variants escaping the antibodies approved for emergency use (36, 37, 38, 39).
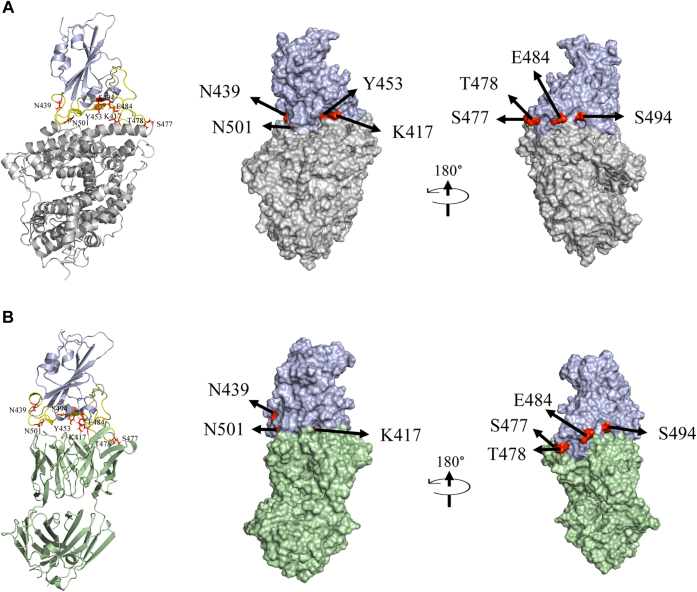
In this work, we examined the effect of variants on RBD protein expression, stability, its binding to ACE2, and antibody escape using a naturally occurring human neutralizing antibody CC12.1. We hypothesize that all these factors act in conjunction and can determine the virus evolution and natural selection of new variants. To test, we selected 12 most frequently occurring mutations in RBD as of January 2021 (https://www.gisaid.org/hcov19-mutation-dashboard). These include nine single-site mutations K417N, N439K, Y453F, S477N, S477I, T478I, E484K, S494P, and N501Y (alpha variant); a double mutant (E484K/N501Y); and two triple mutants (K417N/E484K/N501Y [beta variant] and K417T/E484K/N501Y [gamma variant]). Location of these residues in RBD bound to ACE2 and CC12.1 is shown in Figure 1. All these mutating residues are in the receptor-binding motif (RBM) of RBD, which interacts with ACE2 receptor. Our results presented later indicate that multiple factors contribute toward the natural selection of variants, and all these factors must be considered to understand the natural selection of SARS-CoV-2 variants.
Figure 1.
Structures of SARS-CoV-2 RBD (colored blue) interacting with ACE2 and CC12.1 Fab.A, ACE2 (colored gray; PDB ID: 6M0J). B, CC12.1 Fab (colored green; PDB ID: 6XC2). Both showing the most frequently mutating residues in RBD—K417, N439, Y453, S477, T478, E484, S494, and N501 (colored red). RBM is shown in yellow color. The single mutants of RBD used in this study were K417N, N439K, Y453F, S477N, T478I, E484K, S494P, and N501Y (alpha variant). A double mutant (E484K/N501Y) and triple mutants corresponding to beta variant (K417N/E484K/N501Y) and gamma variant (K417T/E484K/N501Y) were also used. The position of Y453 is not visible in the surface view of RBD interacting with CC12.1 Fab as it is buried at the interface. ACE2, angiotensin-converting enzyme 2; PDB, Protein Data Bank; RBD, receptor-binding domain; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Results
RBD mutations affect protein expression
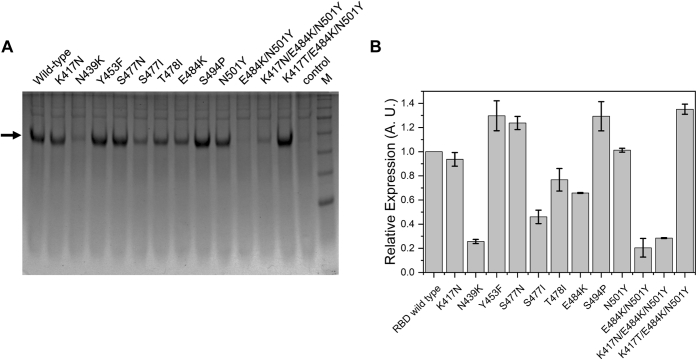
Protein expression was performed in human embryonic kidney (HEK) cells, in which the proteins transport through the secretory pathway, undergo post-translational modifications, and quality control mechanisms ultimately decide the secreted protein levels (40). Expression in HEK cells closely matches the natural infection scenario, where the virus uses the host cell machinery to synthesize its structural proteins. The protein expression levels have a direct bearing on the yield of the viruses, as the virus yield is directly proportional to the amount of the proteins available for virus assembly (41). The infectivity of a particular variant can thus be dependent on the protein expression levels. It should be noted however, that this study only represents the expression level of RBD, and in natural scenario, the expression level of the complete spike protein would ultimately decide the virus yield and thus infectivity. The relative expression of the wildtype RBD along with the mutants was compared using SDS-PAGE after 3 days of expression (Fig. 2). Among nine single-site mutants, two mutants (N439K and S477I) did not express very well, and appreciable amount of protein needed for binding studies could not be obtained for these mutants. The levels of expression were also low for two other single-site mutants T478I and E484K. Expression levels comparable to or higher than the wildtype were obtained for five single-site mutants K417N, Y453F, S477N, S494P, and N501Y. Apart from the single-site mutations, clone carrying double mutations E484K/N501Y did not express. The clone carrying triple mutations (K417N/E484K/N501Y) corresponding to the beta variant also could not be expressed, but the other clone carrying triple mutations (K417T/E484K/N501Y) corresponding to the gamma variant showed high expression. These results suggest that RBD mutations can impact the overall protein expression levels. Similar mutation effects on the expression of the complete spike protein might exist, which can affect the virus yield and infectivity.
Figure 2.
Comparison of relative expression of RBD and its mutants.A, SDS-PAGE showing relative amounts of expressed RBD and its mutants. M represents molecular weight markers (from top to bottom: 180, 130, 100, 70, 55, 35, and 25 kDa). B, relative expression of mutants quantified from the band intensities in SDS-PAGE in panel A. RBD, receptor-binding domain.
For the mutants that did not express well as secretory proteins, we suspected the endoplasmic reticulum (ER) quality control mechanisms to play a key role. ER quality control determines the protein quality and levels secreted outside the cell. Proteasomal degradation is one of the major pathways of ER quality control in which misfolded/unfolded and aggregation-prone non-native proteins are tagged for degradation in the cytosol and well-folded proteins are passed through the secretory pathway. To test the role of proteasomal degradation in determining the expression levels of less expressed proteins, we used a commonly used proteasome inhibitor MG-132 (42, 43). Use of MG-132 increased the expression levels of beta variant (Fig. S1), indicating that the proteasomal pathway might play a big role in controlling the expression of SARS-CoV-2 variants.
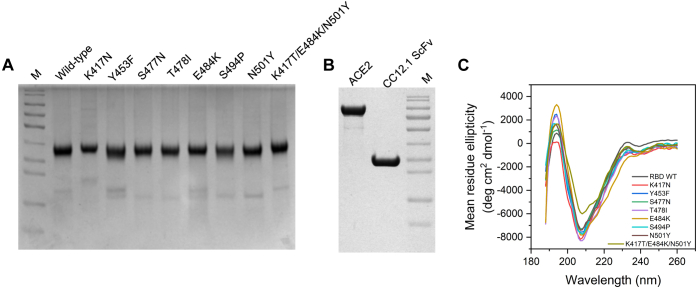
Because of protein expression constraints, further studies on the mutant proteins were carried out on seven single-site mutants (K417N, Y453F, S477N, T478I, E484K, S494P, and N501Y) and the triple mutant K417T/E484K/N501Y, which were purified to homogeneity along with ACE2 and CC12.1 single-chain fragment variable (ScFv) (Fig. 3, A and B).
Figure 3.
Secondary structure characterization of RBD and its mutants.A, purified RBD and its mutants. B, ACE2 and CC12.1 ScFv. M represents molecular weight marker (from top to bottom: 180, 130, 100, 70, 55, 35, 25, 15, and 10 kDa). C, comparison of secondary structures of RBD and its mutants using far-UV CD spectroscopy. Table 1 lists the proportion of various secondary structures when the spectra were deconvoluted using BeStSel software. ACE2, angiotensin-converting enzyme 2; RBD, receptor-binding domain; ScFv, single-chain fragment variable.
Mutations do not significantly affect the global protein structure
Most of the random mutations in proteins do not affect the protein structure and are thus considered neutral. Some mutations though can bring significant structural changes and can have either a beneficial or a deleterious effect on virus fitness. In the absence of other selection pressures, the deleterious mutations are lost, but the beneficial mutations get selected and prevail. Here, we investigated the effect of eight frequently occurring mutations on the RBD structure using far-UV CD spectroscopy (Fig. 3C). The CD spectra for the wildtype protein showed a major negative band at 208 nm and a positive band at 192 nm. The CD spectra were similar to previously reported spectra for wildtype RBD (44). The far-UV CD spectra of all eight mutants were similar to that of the wildtype protein with most significant difference observed for the gamma variant. Deconvolution using BeStSel web software (45) reveals a low percentage of alpha-helix along with high beta-sheet content for all the variants. The proportion of the random coil structure is substantial for RBD accounting for about 50% (Table 1), primarily originating from the RBM (Fig. 1). These CD results suggest that the mutations do not significantly affect the global structure of RBD, and all mutants adopt a similar fold like the wildtype RBD. This is also consistent with the soluble expression of variants (Fig. 3A). Proteins that are expressed as secretory proteins in general adopt well-folded structures. This shows that only those mutants that do not have any deleterious effect on the protein structure are getting naturally selected. Since software used for deconvoluting CD spectra in general accounts only for regular secondary structures, differences in the CD spectra of the triple mutant K417T/E484K/N501Y could be due to changes in short, irregular, and nonrepeating secondary structures most probably in the RBM region.
Table 1.
Proportion of the secondary structural elements calculated by deconvolution of far-UV CD spectra of RBD and its mutants using BeStSel software (45)
| RBD variant | Secondary structure (%) |
||||
|---|---|---|---|---|---|
| Helix | Antiparallel | Parallel | Turn | Others | |
| Wildtype | 13.7 | 25.0 | 0.0 | 13.9 | 47.3 |
| K417N | 12.8 | 17.9 | 4.4 | 14.1 | 50.7 |
| Y453F | 15.3 | 19.2 | 1.5 | 13.9 | 50.2 |
| S477N | 13.5 | 22.0 | 2.0 | 13.2 | 49.3 |
| T478I | 13.8 | 17.8 | 3.2 | 13.3 | 51.9 |
| E484K | 15.7 | 16.3 | 4.0 | 13.4 | 50.6 |
| S494P | 14.1 | 21.4 | 2.1 | 13.3 | 49.1 |
| N501Y | 11.4 | 22.9 | 2.8 | 13.0 | 49.9 |
| K417T/E484K/N501Y | 10.4 | 24.3 | 2.9 | 13.1 | 49.3 |
Most RBD mutants have similar thermal stability as that of wildtype protein
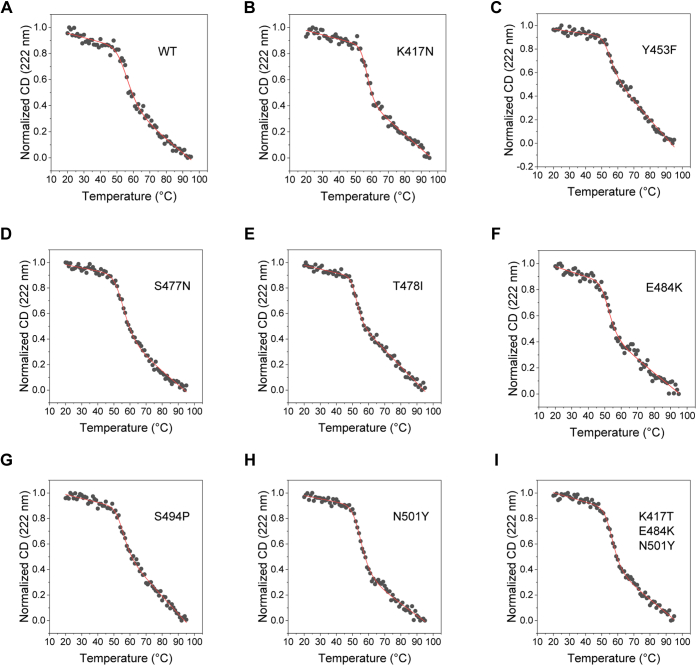
Protein stability can be an important factor in protein evolution (46). More stable proteins can accommodate wide range of mutations and determine the evolvability of the proteins (47). It is thus important to determine the stability of protein to be able to determine its evolutionary path. The stability of the wildtype RBD and its mutants was assessed with thermal denaturation melts using far-UV CD spectroscopy (Fig. 4). Thermal denaturation of the wildtype protein shows a cooperative transition with sloped native and denatured baselines. At the end of the thermal melt, we did not see any protein aggregates. However, the CD spectra of the refolded protein showed slight differences compared with the native protein (Fig. S2). The unfolding transition could be fitted well to a two-state model (Equation 1 in the Experimental procedures section), giving Tm (midpoint temperature of thermal denaturation) value of 56.1 °C ± 0.7 °C (Table 2). The thermal denaturation curves of the mutant proteins were similarly obtained and found to be cooperative, which fitted well to a two-state unfolding model. Although the thermal denaturation is not completely reversible, they can be qualitatively used to compare the relative stabilities of mutants of a given protein (48). All the mutants showed similar Tm values as that of the wildtype protein, indicating that none of the mutations are causing drastic changes in the RBD stability. Compared with other mutants, T478I and E484K showed slightly lesser thermal stability. The same two mutants also showed lesser expression levels (Fig. 2B).
Figure 4.
Thermal denaturation melts of RBD and its mutants obtained using far-UV CD spectroscopy.A–I show the data for the wildtype RBD, single amino-acid mutations K417N, Y453F, S477N, T478I, E484K, S494P, N501Y, and for the triple mutant K417T/E483K/N501Y, respectively. The solid lines show the fits to a two-state unfolding equation (Equation 1 in the Experimental procedures section). Table 2 lists the Tm (midpoint melting temperature) and the ΔH (enthalpy change at Tm) values of RBD variants. RBD, receptor-binding domain.
Table 2.
Tm and ΔH values of RBD variants obtained from thermal denaturation curves using far-UV CD
| RBD variant | Tm (°C) | ΔTm (°C) | ΔH (kcal/mol) |
|---|---|---|---|
| Wildtype | 56.1 ± 0.7 | — | −1.7 ± 0.3 |
| K417N | 56.9 ± 0.3 | 0.8 ± 0.8 | −3.0 ± 0.4 |
| Y453F | 54.8 ± 0.4 | −1.3 ± 0.8 | −2.2 ± 0.3 |
| S477N | 55.8 ± 0.3 | −0.3 ± 0.8 | −1.9 ± 0.1 |
| T478I | 52.3 ± 0.2 | −3.8 ± 0.7 | −2.9 ± 0.3 |
| E484K | 52.3 ± 0.5 | −3.8 ± 0.9 | −2.0 ± 0.3 |
| S494P | 55.3 ± 0.4 | −0.8 ± 0.8 | −2.3 ± 0.3 |
| N501Y | 55.1 ± 0.3 | −1 ± 0.8 | −2.1 ± 0.1 |
| K417T/E484K/N501Y | 56.5 ± 0.2 | 0.4 ± 0.7 | −2.4 ± 0.2 |
Errors on ΔTm were calculated using error propagation formulae (74).
Some but not all RBD mutants show increased binding affinity to ACE2
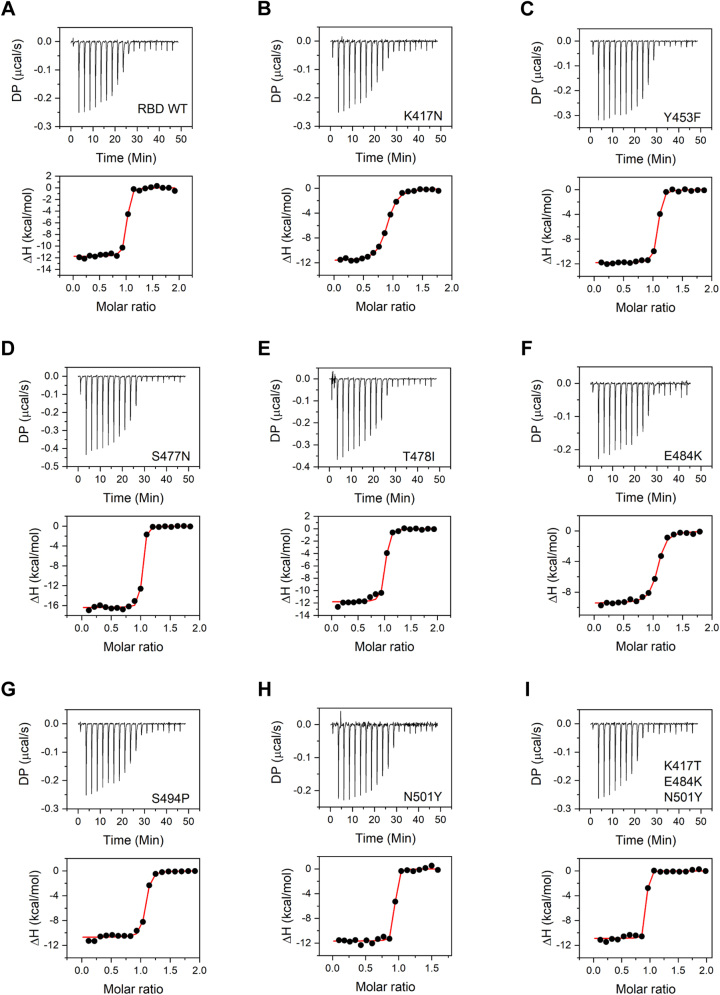
SARS-CoV-2 RBD mediates the interaction of the virus spike protein with the ACE2 receptor on the host cell surface and is thus a major determinant of the viral entry into the host cell. Mutations in the RBD can impact its interaction with ACE2 and can have an important role in determining the infectivity of the virus, with higher affinity interactions contributing to increased infectivity. The binding interactions of the wildtype and mutant RBD proteins with ACE2 were investigated using isothermal titration calorimetry (ITC) (Fig. 5 and Table 3). ITC is particularly advantageous in terms of measuring protein–protein interactions in solution without any covalent modification of proteins. The wildtype RBD interacts with ACE2 in a stoichiometric ratio of 1:1, with a Kd value of 10.0 ± 3.1 nM and enthalpy of interaction (ΔH) of −11.8 ± 0.2 kcal/mol. The measured Kd value is consistent with previously published studies on ACE2–RBD interaction using surface plasmon resonance with immobilized protein (49). All the mutants studied interacted with ACE2 in the same stoichiometric ratio of 1:1. Three of the eight mutants, Y453F, T478I, and S494P, did not show significant difference in their binding interaction with ACE2, with Kd and ΔH values similar to the wildtype protein (Table 3). For S477N mutant, Kd value was similar to that of the wildtype, but an increased ΔH value of −16.4 ± 0.2 kcal/mol was obtained, which may indicate increased interactions between RBD and ACE2 upon mutation. For two other mutants K417N and E484K, the Kd value obtained was higher than the wildtype protein (Table 3), indicating weaker affinity for ACE2. The corresponding ΔH values did not show any significant difference for K417N mutant but showed a decreased value of −9.5 ± 0.2 kcal/mol for E484K mutant. The other single-site mutant N501Y corresponding to the alpha variant and the triple mutant K417T/E484K/N501Y corresponding to the gamma variant showed increased affinity for ACE2 binding with Kd values of 3.0 ± 2.1 and 1.6 ± 1.5 nM, respectively, when compared with the wildtype (Table 3). There was no difference in ΔH values for N501Y mutant and the triple mutant K417T/E484K/N501Y. These ITC results indicate that the ACE2 binding is an important but not the sole parameter determining the natural selection of the variants.
Figure 5.
Binding of RBD and its variants to ACE2 studied using ITC. Panels A–I show the data for the wildtype RBD, single amino-acid mutations K417N, Y453F, S477N, T478I, E484K, S494P, N501Y, and for the triple mutant K417T/E483K/N501Y, respectively. Top panels show the raw thermograms, and the bottom panels show the fit to the integrated heat curve. Table 3 lists the interaction parameters from the data fit. ACE2, angiotensin-converting enzyme 2; ITC, isothermal titration calorimetry; RBD, receptor-binding domain.
Table 3.
Interaction parameters obtained from binding of RBD variants to ACE2 probed by ITC
| RBD variant | Kd (nM) | N | ΔH (kcal/mol) | ΔG (kcal/mol) | −TΔS (kcal/mol) |
|---|---|---|---|---|---|
| Wildtype | 10.0 ± 3.1 | 1.0 ± 0.0 | −11.8 ± 0.2 | −10.7 ± 0.2 | 1.0 ± 0.3 |
| K417N | 94.1 ± 9.5 | 0.9 ± 0.0 | −11.7 ± 0.1 | −9.4 ± 0.1 | 2.3 ± 0.1 |
| Y453F | 11.4 ± 1.7 | 1.0 ± 0.0 | −11.8 ± 0.1 | −10.7 ± 0.1 | 1.2 ± 0.1 |
| S477N | 5.0 ± 1.4 | 1.0 ± 0.0 | −16.4 ± 0.2 | −11.1 ± 0.2 | 5.3 ± 0.3 |
| T478I | 16.7 ± 5.6 | 1.0 ± 0.0 | −11.8 ± 0.2 | −10.4 ± 0.2 | 1.4 ± 0.3 |
| E484K | 51.5 ± 8.2 | 1.0 ± 0.0 | −9.5 ± 0.2 | −9.8 ± 0.1 | −0.3 ± 0.2 |
| S494P | 13.8 ± 3.5 | 1.0 ± 0.0 | −10.7 ± 0.2 | −10.5 ± 0.1 | 0.2 ± 0.2 |
| N501Y | 3.0 ± 2.1 | 0.9 ± 0.0 | −11.7 ± 0.2 | −11.4 ± 0.4 | 0.2 ± 0.5 |
| K417T/E484K/N501Y | 1.6 ± 1.5 | 0.9 ± 0.0 | −10.9 ± 0.2 | −11.8 ± 0.5 | −0.9 ± 0.6 |
Errors on ΔG and −TΔS were calculated using error propagation formulae (74).
Some but not all RBD mutants show CC12.1 antibody escape
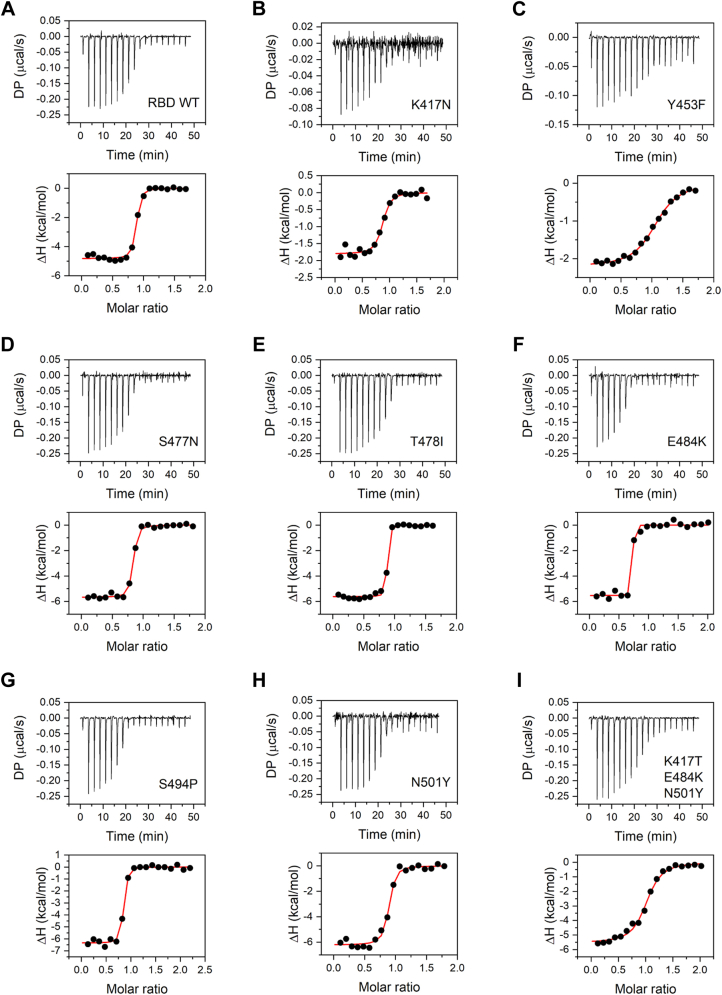
CC12.1 antibody is a monoclonal antibody isolated from the convalescent plasma of a COVID-19 survivor (50, 51). This antibody represents the class of antibodies most elicited by SARS-CoV-2 infection and also in response to current vaccines against the wildtype. The circulating antibodies can play a major role in shaping the evolutionary path of RBD. The mutants that can escape the antibody recognition are naturally selected and represented more in the viral pool. We investigated the binding of the wildtype RBD and its mutants to the ScFv of CC12.1 antibody through ITC (Fig. 6 and Table 4). The CC12.1 ScFv interacts with wildtype RBD at a stoichiometric ratio of 1:1 with a Kd value of 28.0 ± 8.8 nM and ΔH value of −4.8 ± 0.1 kcal/mol. Compared with ACE2 interaction, which is primarily enthalpically driven (Table 3), RBD interaction with CC12.1 has a significant entropic component (Table 4). Of the eight mutants studied, two single-site mutations (S477N and S494P) did not impact the binding affinity of RBD toward CC12.1 ScFv. E484K and T478I mutants however showed increased affinity toward CC12.1 ScFv with a Kd value of 1.7 ± 4.6 and 5.8 ± 3.4 nM, respectively (Table 4), which suggests that CC12.1 may be able to neutralize E484K and T478I mutants. Other four variants, single-site mutants K417N, Y453F, N501Y (alpha variant) and the triple mutant K417T/E484K/N501Y (gamma variant), showed decreased affinity toward CC12.1 binding, with Kd values of 119 ± 50, 827 ± 146, 63 ± 22, and 433 ± 95 nM respectively, representing escape from CC12.1. Another interesting observation is the decrease in ΔH value for K417N and Y453F mutants, indicating decreased strength of interactions between the mutants and CC12.1. These results show that antibody escape can be a very important parameter that shapes virus evolution and natural selection of mutants.
Figure 6.
Binding of RBD and its variants to CC12.1 ScFv studied using ITC. Panels A–I show the data for the wildtype RBD, single amino-acid mutations K417N, Y453F, S477N, T478I, E484K, S494P, N501Y, and for the triple mutant K417T/E483K/N501Y, respectively. Top panels show the raw thermograms, and the bottom panels show the fit to the integrated heat curve. Table 4 lists the interaction parameters from the data fit. ITC, isothermal titration calorimetry; RBD, receptor-binding domain; ScFv, single-chain fragment variable.
Table 4.
Interaction parameters obtained from binding of RBD variants to CC12.1 ScFv probed by ITC
| RBD variant | Kd (nM) | N | ΔH (kcal/mol) | ΔG (kcal/mol) | −TΔS (kcal/mol) |
|---|---|---|---|---|---|
| Wildtype | 28.0 ± 8.8 | 0.9 ± 0.0 | −4.8 ± 0.1 | −10.1 ± 0.2 | −5.3 ± 0.2 |
| K417N | 119 ± 50 | 0.8 ± 0.0 | −1.8 ± 0.1 | −9.3 ± 0.2 | −7.5 ± 0.3 |
| Y453F | 827 ± 146 | 1.0 ± 0.0 | −2.2 ± 0.1 | −8.2 ± 0.1 | −6.0 ± 0.1 |
| S477N | 25.6 ± 6.8 | 0.8 ± 0.0 | −5.7 ± 0.1 | −10.2 ± 0.2 | −4.5 ± 0.2 |
| T478I | 5.8 ± 3.4 | 0.8 ± 0.0 | −5.6 ± 0.1 | −11.0 ± 0.3 | −5.4 ± 0.4 |
| E484K | 1.7 ± 4.6 | 0.7 ± 0.0 | −5.5 ± 0.1 | −11.8 ± 1.6 | −6.2 ± 1.2 |
| S494P | 23.0 ± 7.7 | 0.8 ± 0.0 | −6.4 ± 0.1 | −10.2 ± 0.2 | −3.9 ± 0.2 |
| N501Y | 63 ± 22 | 0.9 ± 0.0 | −6.2 ± 0.2 | −9.7 ± 0.2 | −3.4 ± 0.3 |
| K417T/E484K/N501Y | 433 ± 95 | 1.0 ± 0.0 | −5.6 ± 0.2 | −8.5 ± 0.3 | −3.0 ± 0.2 |
Errors on ΔG and −TΔS were calculated using error propagation formulae (74).
Discussion
The reports of mutations in the spike protein of SARS-CoV-2 started to appear very soon after its emergence in Wuhan, China in December 2019. Most of the mutations in the RBD were noticed during the fall of 2020, and variant B.1.1.7 with mutation N501Y (alpha variant) became the first RBD variant to be labeled as a VOC (https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/). Since the beginning of 2021, there has been an emergence of numerous variants in different parts of the world. The rate of mutations in SARS-CoV-2 genome is lesser than that known for other RNA viruses like influenza and HIV (14, 15, 16). The prime reason for that is the ability of the virus to proofread errors included by RNA-dependent RNA polymerase because of the presence of exonuclease activity (52). Even then, substantial number of variants occurring with high frequency was reported for this virus, with the list continuing to grow. This can be explained by the fact that the appearance of mutations is also dependent on the viral population size. Higher viral population will support greater number of mutants. As this virus has infected human population globally and has emerged as a pandemic, it is not surprising to encounter the different variants. The biggest concern with the emergence of the variants is the ineffectiveness of the currently developed vaccines, which are being used to vaccinate people, and the antibody-based therapeutics, which are approved for emergency use. There are reports of breakthrough infection in already vaccinated individuals (53) and also reports of decreased neutralization ability of vaccinated individuals' serum against variants (36, 37, 38), which substantiate the need to study these variants in more detail.
The occurrence and persistence of a particular mutation in virus pool is dependent on a number of factors. In general, it is considered that mutations that provide a selective advantage to virus fitness are naturally selected. The most important characteristics of proteins that can affect virus fitness are its stability and activity. Mutations in viral proteins can have a neutral, stabilizing, or destabilizing effect on protein stability. Stabilizing mutations offer a fitness advantage to the virus by increasing the proportion of correctly folded protein and increased resistance to protein degradation and aggregation inside the cell (54, 55). Higher stability, similar to higher expression, can thus lead to higher infectivity through increased virus yield (56). Destabilizing mutations on the other hand do not offer fitness advantage and are mostly deleterious (57). They lead to lower virus yield with decreased infectivity. For example, in the case of hemagglutinin protein of influenza virus, it has been shown that the higher stability variants provide fitness advantage and tend to persist longer (56). The mutation rate and population size also affect the stability of the new variants. It has been shown that higher mutation rates and lower population size lead to the emergence of low stability variants, and on the contrary, low mutation rates and high population sizes lead to emergence of variants with higher stability (58). Our results show that the emerging variants are quite resistant to major stability changes (Table 2), despite the nature of the mutation (including many nonconservative mutations with differing physical properties) and multiple mutations accumulating in VOCs such as the gamma variant.
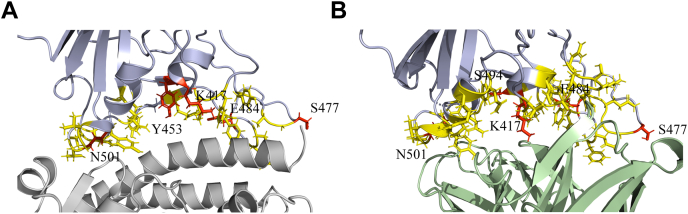
Protein activity is another parameter that affects virus fitness. Unlike protein stability that can be impacted by mutations at many sites on protein, protein activity is controlled by a few key amino acid residues. In case of SARS-CoV-2 RBD, binding to ACE2 with a higher affinity provides a selective advantage toward virus fitness. Higher the affinity, higher the virus infectivity. The key residues of RBD that interact with ACE2 residues are shown in Figure 7A. Most of the frequently occurring mutations in RBD (Fig. 1A) are at the binding interface with ACE2. Our data show that the four mutations Y453F, S477N, T478I, and S494P do not impact ACE2 binding. T478 and S494 are not part of the binding interface with ACE2 (Fig. 7A), and hence, mutations T478I and S494P do not bring any change to the binding affinity. Y453 also does not form any polar contacts with ACE2. Its mutation to phenylalanine is a conservative mutation and thus does not show any difference in binding affinity. Two mutations K417N and E484K show a decrease in ACE2-binding affinity. K417 residue in SARS-CoV-2 RBD has previously been shown to be responsible for increase in binding efficiency of RBD toward ACE2 (49). K417 residue in SARS-CoV-2 RBD helps in bringing a more positive charge on the RBD surface, which better interacts with the negatively charged residues on ACE2. Mutation of lysine to an uncharged amino acid asparagine decreases the positive charge on the surface, thereby decreasing binding affinity toward ACE2. Similarly, E484, a negatively charged amino acid, interacts with a positively charged amino acid, K31 of ACE2. Mutation to lysine, a positively charged amino acid, abolishes this interaction, explaining the decrease in the binding affinity. The other single-site mutation N501Y corresponding to the alpha variant and the triple mutant K417T/E484K/N501Y corresponding to the gamma variant show increased binding affinity to ACE2. From these data, it appears that receptor binding is a big factor in driving SARS-CoV-2 variant evolution, but it is not the only driving force. Some of the RBM mutants are naturally selected even though they do not have increased affinity for ACE2.
Figure 7.
Structures of SARS-CoV-2 RBD colored blue interacting with ACE2 and CC12.1 Fab.A, ACE2 colored gray and B, CC12.1 Fab colored green, showing the interface residues of RBD in yellow along with the side chains. The frequently mutating residues that are part of binding interface are colored red and also shown with their side chains. ACE2, angiotensin-converting enzyme 2; RBD, receptor-binding domain; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Neutral mutations that do not offer any advantage toward virus fitness and even deleterious mutations can get selected and become prevalent in the presence of other selection pressures like escape from human immune responses (59). Neutralizing antibodies are a key component of immune response against natural infection by viruses. Neutralizing antibodies can also be administered as recombinant monoclonal antibody therapeutics and as convalescent plasma to provide passive immunity. The circulating neutralizing antibodies thus provide a selection pressure that can drive the evolution of virus variants (60, 61, 62). Variants that can escape the neutralizing antibodies have a selective advantage and can persist and spread to other individuals. The effectiveness with which a variant can escape the neutralizing antibodies and become prevalent depends on the exposure of that variant to neutralizing antibodies. In the course of normal infection, the levels of antibody during the virus transmission phase are negligible. Thus, the transmitting viruses do not have exposure to neutralizing antibodies and are not considered to contribute to spread of variants with antibody escape potential (63). But reinfection in people with weak immunity increases the chances of exposure of virus to neutralizing antibodies elicited during first infection and contribute to selection of variants with escape potential (64, 65, 66). Similarly, convalescent plasma therapy, monoclonal antibody therapy (especially single antibody therapeutics), and less immunogenic vaccine candidates can increase the chances of variants coming in contact with neutralizing antibodies and drive the emergence of variants that can escape these antibodies (67).
In this study, we tested the potential of the most prevalent variants to escape CC12.1 antibody, a naturally elicited human antibody as a response to SARS-CoV-2 infection (50). It belongs to a class of antibodies that target RBD and is the most abundant class of antibodies naturally produced by humans (20, 51, 68). Antibodies belonging to this class are encoded by the VH3-53 gene segment and represent a set of neutralizing antibodies that bind to the RBD epitope that overlaps with ACE2-binding epitope (Fig. 1) (20). These antibodies are characterized by short H3 CDR and can bind the RBD in “up” conformation. Nevertheless, among the RBD-targeting antibodies, these are the most abundant and commonly found antibodies and represent the general antibody response to SARS-CoV-2 infection in humans (51). Our study examined which of the most prevalent variants have the ability to escape natural immune responses, but it should also be considered that the neutralizing antibody response toward an infection is varied with several neutralizing antibodies generated against different epitopes of spike protein (69, 70). Also, the natural antibody responses can vary from individual to individual. Even then, neutralizing antibodies against RBD represent the most potent and widely used countermeasures against COVID-19 (71). The RBD–CC12.1 binding interface is shown in Figure 7B. Most of the frequently mutating residues we examined are also part of the binding interface, which implies that the mutations are quite likely to impact CC12.1 binding. Our data show that only four of the eight mutants (K417N, Y453F, N501Y and the triple mutant K417T/E484K/N501Y) we examined bind to CC12.1 with weaker affinity (or higher Kd) (Table 4). The single-site mutant Y453F showed the weakest binding or the highest potential to escape CC12.1 ScFv. Interestingly, Y453 is not a part of the binding interface with CC12.1, implying that the residues that do not directly interact with its binding partner can also impact binding after mutation. Other two mutants S477N and S494P did not show any difference in their binding affinity to CC12.1. Interestingly, E484K and T478I bind with a higher affinity (or lower Kd) to CC12.1 (Table 4). These results show that the neutralizing antibody response is one of the driving forces for natural selection of RBD variants and should be considered in closer detail. It should also be considered that the results presented here are only representing the escape toward one class of antibody, and other antibodies that are naturally occurring or administered passively would also have an impact on the emergence of these variants. For example, although E484K and T478I mutations may not escape CC12.1 (Table 4), they may escape other classes of antibodies (72, 73). These results also stress on the need to use viral sequencing to find the variant that has infected a patient and use of those antibodies to treat patients against which the variant does not show escape potential. To achieve this, different therapeutic neutralizing antibodies that target different epitopes on RBD should be developed, and a cocktail antibody drug may work better against new variants.
In summary, we have examined the various factors that might be contributing to the natural selection of most frequent SARS-CoV-2 RBD variants, in particular protein expression, stability, activity in terms binding to ACE2, and antibody escape potential in terms of binding to a human neutralizing antibody. Table 5 summarizes our observations that favor natural selection of variants. These results show that multiple factors contribute to the natural selection of variants and should be considered when evaluating any future variants. For example, the triple mutant K417T/E484K/N501Y (gamma variant) poses a serious threat with many factors, increased protein expression, increased activity, and increased antibody escape potential favoring its emergence and persistence. Followed by the gamma variant, alpha variant N501Y has the most favorable biophysical parameters in terms of increased affinity toward ACE2 and increased escape potential (Table 5). In the case of variants harboring multiple mutations, each mutation might be playing a specific role in virus survival. It can be either stronger binding to ACE2, increased expression, or escape from neutralizing antibodies. For example, when the data in Table 5 are compared between the N501Y (alpha variant), the triple mutant K417T/E484K/N501Y (gamma variant) and the wildtype, increase in ACE2 binding might be significantly determined by the N501Y mutation, since no significant differences were observed in the Kd of ACE2 binding between the N501Y and K417T/E484K/N501Y variants. The effect of the other two mutations K417T and E484K in the gamma variant might be to increase its expression and/or to increase immune escape potential. E484K by itself do not have increased expression or do not escape from CC12.1 compared with the wildtype (Table 5). In fact, E484K binds to CC12.1 with higher affinity, but it is quite likely that this mutation has evolved to show increased immune escape against other neutralizing antibodies (72, 73). Of the eight variants for which complete biophysical data are available in Table 5, only two (N501Y [alpha variant] and the triple mutant K417T/E484K/N501Y [gamma variant]) correspond to the VOCs and the other six are classified as variants of interest. The data suggest that VOCs are evolving by maximizing the biophysical fitness parameters that determine the virus survival. We hope that such fundamental understanding of the physical parameters determining the natural selection of variants will help in designing better countermeasures like new vaccine candidates and antibody therapeutics that work against emerging variants.
Table 5.
Comparison of protein expression, stability, binding to ACE2, and binding to CC12.1 of RBD variants
| RBD variant | Expression | Tm (°C) |
Kd (nM) |
Kd (nM) |
|---|---|---|---|---|
| (ACE2 binding) | (CC12.1 binding) | |||
| Wildtype | — | 56.1 ± 0.7 | 10.0 ± 3.1 | 28.0 ± 8.8 |
| K417N | No change | 56.9 ± 0.3 | 94.1 ± 9.5 | 119 ± 50 |
| Y453F | Increase | 54.8 ± 0.4 | 11.4 ± 1.7 | 827 ± 146 |
| S477N | Increase | 55.8 ± 0.3 | 5.0 ± 1.4 | 25.6 ± 6.8 |
| T478I | Decrease | 52.3 ± 0.2 | 16.7 ± 5.6 | 5.8 ± 3.4 |
| E484K | Decrease | 52.3 ± 0.5 | 51.5 ± 8.2 | 1.7 ± 4.6 |
| S494P | Increase | 55.3 ± 0.4 | 13.8 ± 3.5 | 23.0 ± 7.7 |
| N501Y | No change | 55.1 ± 0.3 | 3.0 ± 2.1 | 63 ± 22 |
| K417T/E484K/N501Y | Increase | 56.5 ± 0.2 | 1.6 ± 1.5 | 433 ± 95 |
Bold values indicate parameters favoring the natural selection of variants.
Experimental procedures
Cloning and expression of RBD, RBD mutants, ACE2, and CC12.1 ScFv
Amino acid sequences of SARS-CoV-2 RBD and human-ACE2 protein were obtained from UniProt (UniProt ID—P0DTC2 and Q9BYF1, respectively). The sequence of heavy and light chain variable regions (VH and VL, respectively) of CC12.1-neutralizing antibody was obtained from Research Collaboratory for Structural Bioinformatics Protein Data Bank (PDB) (PDB ID: 6XC2). The ScFv for CC12.1 was designed as VH-(GGGGS)3-VL. The protein sequences were codon optimized for expression in human cells and synthesized by Twist Biosciences. The RBD variants were generated by site-directed mutagenesis using mutagenic primers (Table S1). The synthesized and mutated genes were cloned into pcDNA 3.4 Topo vector, modified by including a signal sequence of human immunoglobulin heavy chain, His tag and SUMOstar protein. The expression vectors were transfected transiently into Expi293 (modified HEK293) cells using polyethylenimine, and the protein was recovered from the culture supernatant after 5 days. The expression levels for the proteins were compared after running the culture supernatants on SDS-PAGE, staining with Coomassie blue R-250 dye, and quantifying the band intensities corresponding to the target protein using the ImageLab software from Bio-Rad.
Protein purification
The supernatant of the culture was filtered through 0.22-μm filter and purified using nickel–nitrilotriacetic acid chromatography. The eluted protein was dialyzed in buffer containing 50 mM of Tris–HCl, 20 mM NaCl, pH 8.0, and digested using SUMOstar protease overnight to cleave the target protein from SUMOstar and His tag. The digested protein was passed again through the nickel–nitrilotriacetic acid column to obtain the untagged target protein in the flow through. All proteins were dialyzed in buffer containing 50 mM sodium phosphate, 20 mM NaCl, and pH 7.0.
Proteasomal inhibition assay
Expi293 cells were transfected with the plasmid coding for the beta variant of SARS-CoV-2 RBD. The cells were treated with different concentrations of MG-132 (0, 1, 2, and 5 μM) for 24 h after transfection. Culture supernatants were recovered at definite time intervals, and 20 μl was loaded onto SDS-PAGE for comparison of protein expression levels. The gel was stained with Coomassie blue R-250 dye, and the band intensities of the target protein were quantified using the ImageLab software from Bio-Rad.
ITC
ITC experiments were performed using MicroCal-PEAQ-ITC instrument from Malvern in buffer containing 50 mM sodium phosphate, 20 mM NaCl, and pH 7.0 at 20 °C. For ACE2–RBD interactions, ACE2 at a concentration of 12 μM was taken in the cell, and RBD variants at concentration of 120 μM were taken in the syringe. For RBD–CC12.1 ScFv interactions, RBD or its variants were taken in the cell at a concentration of 20 μM, and CC12.1 ScFv was taken in the syringe at 200 μM. The syringe contents were titrated into the cell as 18 injections of 2 μl each with a spacing of 150 s between the injections. ITC data analysis was done using MicroCal PEAQ-ITC Analysis Software from Malvern.
CD spectroscopy
CD spectra of RBD variants were recorded on an Applied Photophysics Chirascan Plus spectrometer at a protein concentration of 5 μM in buffer containing 10 mM sodium phosphate, 4 mM NaCl, and pH 7.0 in a 1-mm path length cuvette. The data were collected at an interval of 1 nm and averaged for 2 s at each wavelength.
The thermal melts for RBD variants were recorded at a protein concentration of 20 μM in buffer containing 50 mM sodium phosphate, 20 mM NaCl, and pH 7.0 in 0.5-mm path length cuvette. The temperature scan rate was 1 °C/min. CD signal at 222 nm averaged for 2 s was plotted against the temperature and fitted to a two-state unfolding model using the equation (48),
| (1) |
where ST is the measured signal as a function of temperature T, SN and SU are the signals corresponding to the native and unfolded baselines, mN and mU are the slopes of linear dependence of SN and SU, ΔHm is the enthalpy change at Tm, R is the universal gas constant, and T is the absolute temperature in Kelvin.
Data availability
All the data are contained within the article and in the supporting information.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank David Bain, John Carpenter, Walter Englander, Mario Santiago, and Eugene Shakhnovich for critical reading of the article and helpful comments and Mark Colbenson and Valeria Zai-Rose for helpful discussions.
Author contributions
V. U. and K. M. G. M. conceptualization; V. U., R. M., and K. M. G. M. methodology; V. U., A. L., and S. P. validation; V. U., A. L., S. P., and K. M. G. M. formal analysis; V. U., A. L., and S. P. investigation; R. M. resources; V. U. and K. M. G. M. writing–original draft; V. U., A. L., S. P., and K. M. G. M. writing–review and editing; V. U. visualization; K. M. G. M. supervision; K. M. G. M. project administration; K. M. G. M. funding acquisition.
Edited by Craig E. Cameron
Footnotes
Present address for Ryuki Miyauchi: Modality Research Laboratories, Daiichi-Sankyo Co, Ltd, Tokyo 134-8630, Japan.
Supporting information
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim J.M., Chung Y.S., Jo H.J., Lee N.J., Kim M.S., Woo S.H., Park S., Kim J.W., Kim H.M., Han M.G. Identification of coronavirus isolated from a patient in Korea with COVID-19. Osong Public Health Res. Perspect. 2020;11:3–7. doi: 10.24171/j.phrp.2020.11.1.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhong N.S., Zheng B.J., Li Y.M., Poon, Xie Z.H., Chan K.H., Li P.H., Tan S.Y., Chang Q., Xie J.P., Liu X.Q., Xu J., Li D.X., Yuen K.Y., Peiris, Guan Y. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People's Republic of China, in February, 2003. Lancet. 2003;362:1353–1358. doi: 10.1016/S0140-6736(03)14630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 5.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 6.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292.e286. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e278. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perlman S., Netland J. Coronaviruses post-SARS: Update on replication and pathogenesis. Nat. Rev. Microbiol. 2009;7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rausch J.W., Capoferri A.A., Katusiime M.G., Patro S.C., Kearney M.F. Low genetic diversity may be an Achilles heel of SARS-CoV-2. Proc. Natl. Acad. Sci. U. S. A. 2020;117:24614–24616. doi: 10.1073/pnas.2017726117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Dorp L., Acman M., Richard D., Shaw L.P., Ford C.E., Ormond L., Owen C.J., Pang J., Tan C.C.S., Boshier F.A.T., Ortiz A.T., Balloux F. Emergence of genomic diversity and recurrent mutations in SARS-CoV-2. Infect. Genet. Evol. 2020;83:104351. doi: 10.1016/j.meegid.2020.104351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dearlove B., Lewitus E., Bai H., Li Y., Reeves D.B., Joyce M.G., Scott P.T., Amare M.F., Vasan S., Michael N.L., Modjarrad K., Rolland M. A SARS-CoV-2 vaccine candidate would likely match all currently circulating variants. Proc. Natl. Acad. Sci. U. S. A. 2020;117:23652–23662. doi: 10.1073/pnas.2008281117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abdool Karim S.S., de Oliveira T. New SARS-CoV-2 variants - clinical, public health, and vaccine implications. N. Engl. J. Med. 2021;384:1866–1868. doi: 10.1056/NEJMc2100362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Voloch C.M., da Silva Francisco R., Jr., de Almeida L.G.P., Cardoso C.C., Brustolini O.J., Gerber A.L., Guimaraes A.P.C., Mariani D., da Costa R.M., Ferreira O.C., Jr., Covid19-UFRJ Workgroup, LNCC Workgroup, Adriana Cony Cavalcanti. Frauches T.S., de Mello C.M.B., Leitao I.C., Galliez R.M. Genomic characterization of a novel SARS-CoV-2 lineage from Rio de Janeiro, Brazil. J. Virol. 2021;95 doi: 10.1128/JVI.00119-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li M., Lou F., Fan H. SARS-CoV-2 variants: A new challenge to convalescent serum and mRNA vaccine neutralization efficiency. Signal Transduct. Target. Ther. 2021;6:151. doi: 10.1038/s41392-021-00592-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barnes C.O., Jette C.A., Abernathy M.E., Dam K.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E., Robbiani D.F., Nussenzweig M.C., West A.P., Jr., Bjorkman P.J. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang S., Hillyer C., Du L. Neutralizing antibodies against SARS-CoV-2 and other human coronaviruses. Trends Immunol. 2020;41:355–359. doi: 10.1016/j.it.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansen J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E., Fulton B.O., Yan Y., Koon K., Patel K., Chung K.M., Hermann A., Ullman E., Cruz J., Rafique A. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gottlieb R.L., Nirula A., Chen P., Boscia J., Heller B., Morris J., Huhn G., Cardona J., Mocherla B., Stosor V., Shawa I., Kumar P., Adams A.C., Van Naarden J., Custer K.L. Effect of bamlanivimab as monotherapy or in combination with etesevimab on viral load in patients with mild to moderate COVID-19: A randomized clinical trial. JAMA. 2021;325:632–644. doi: 10.1001/jama.2021.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones B.E., Brown-Augsburger P.L., Corbett K.S., Westendorf K., Davies J., Cujec T.P., Wiethoff C.M., Blackbourne J.L., Heinz B.A., Foster D., Higgs R.E., Balasubramaniam D., Wang L., Zhang Y., Yang E.S. The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in non-human primates. Sci. Transl. Med. 2021;13 doi: 10.1126/scitranslmed.abf1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., McGettigan J., Khetan S., Segall N., Solis J., Brosz A. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Perez Marc G., Moreira E.D., Zerbini C., Bailey R., Swanson K.A., Roychoudhury S., Koury K., Li P. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Voysey M., Clemens S.A.C., Madhi S.A., Weckx L.Y., Folegatti P.M., Aley P.K., Angus B., Baillie V.L., Barnabas S.L., Bhorat Q.E., Bibi S., Briner C., Cicconi P., Collins A.M., Colin-Jones R. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: An interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2021;397:99–111. doi: 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sadoff J., Le Gars M., Shukarev G., Heerwegh D., Truyers C., de Groot A.M., Stoop J., Tete S., Van Damme W., Leroux-Roels I., Berghmans P.J., Kimmel M., Van Damme P., de Hoon J., Smith W. Interim results of a phase 1-2a trial of Ad26.COV2.S Covid-19 vaccine. N. Engl. J. Med. 2021;384:1824–1835. doi: 10.1056/NEJMoa2034201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keech C., Albert G., Cho I., Robertson A., Reed P., Neal S., Plested J.S., Zhu M., Cloney-Clark S., Zhou H., Smith G., Patel N., Frieman M.B., Haupt R.E., Logue J. Phase 1-2 trial of a SARS-CoV-2 recombinant spike protein nanoparticle vaccine. N. Engl. J. Med. 2020;383:2320–2332. doi: 10.1056/NEJMoa2026920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu F.C., Guan X.H., Li Y.H., Huang J.Y., Jiang T., Hou L.H., Li J.X., Yang B.F., Wang L., Wang W.J., Wu S.P., Wang Z., Wu X.H., Xu J.J., Zhang Z. Immunogenicity and safety of a recombinant adenovirus type-5-vectored COVID-19 vaccine in healthy adults aged 18 years or older: A randomised, double-blind, placebo-controlled, phase 2 trial. Lancet. 2020;396:479–488. doi: 10.1016/S0140-6736(20)31605-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goepfert P.A., Fu B., Chabanon A.L., Bonaparte M.I., Davis M.G., Essink B.J., Frank I., Haney O., Janosczyk H., Keefer M.C., Koutsoukos M., Kimmel M.A., Masotti R., Savarino S.J., Schuerman L. Safety and immunogenicity of SARS-CoV-2 recombinant protein vaccine formulations in healthy adults: Interim results of a randomised, placebo-controlled, phase 1-2, dose-ranging study. Lancet Infect. Dis. 2021;21:1257–1270. doi: 10.1016/S1473-3099(21)00147-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang J.G., Su D., Song T.Z., Zeng Y., Huang W., Wu J., Xu R., Luo P., Yang X., Zhang X., Luo S., Liang Y., Li X., Huang J., Wang Q. S-Trimer, a COVID-19 subunit vaccine candidate, induces protective immunity in nonhuman primates. Nat. Commun. 2021;12:1346. doi: 10.1038/s41467-021-21634-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chappell K.J., Mordant F.L., Li Z., Wijesundara D.K., Ellenberg P., Lackenby J.A., Cheung S.T.M., Modhiran N., Avumegah M.S., Henderson C.L., Hoger K., Griffin P., Bennet J., Hensen L., Zhang W. Safety and immunogenicity of an MF59-adjuvanted spike glycoprotein-clamp vaccine for SARS-CoV-2: A randomised, double-blind, placebo-controlled, phase 1 trial. Lancet Infect. Dis. 2021;21:1383–1394. doi: 10.1016/S1473-3099(21)00200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang S., Li Y., Dai L., Wang J., He P., Li C., Fang X., Wang C., Zhao X., Huang E., Wu C., Zhong Z., Wang F., Duan X., Tian S. Safety and immunogenicity of a recombinant tandem-repeat dimeric RBD-based protein subunit vaccine (ZF2001) against COVID-19 in adults: Two randomised, double-blind, placebo-controlled, phase 1 and 2 trials. Lancet Infect. Dis. 2021;21:1107–1119. doi: 10.1016/S1473-3099(21)00127-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang J., Wang W., Chen Z., Lu S., Yang F., Bi Z., Bao L., Mo F., Li X., Huang Y., Hong W., Yang Y., Zhao Y., Ye F., Lin S. A vaccine targeting the RBD of the S protein of SARS-CoV-2 induces protective immunity. Nature. 2020;586:572–577. doi: 10.1038/s41586-020-2599-8. [DOI] [PubMed] [Google Scholar]
- 36.Huang B., Dai L., Wang H., Hu Z., Yang X., Tan W., Gao G.F. Serum sample neutralisation of BBIBP-CorV and ZF2001 vaccines to SARS-CoV-2 501Y.V2. Lancet Microbe. 2021;2 doi: 10.1016/S2666-5247(21)00082-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Madhi S.A., Baillie V., Cutland C.L., Voysey M., Koen A.L., Fairlie L., Padayachee S.D., Dheda K., Barnabas S.L., Bhorat Q.E., Briner C., Kwatra G., Ahmed K., Aley P., Bhikha S. Efficacy of the ChAdOx1 nCoV-19 Covid-19 vaccine against the B.1.351 variant. N. Engl. J. Med. 2021;384:1885–1898. doi: 10.1056/NEJMoa2102214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Z., Schmidt F., Weisblum Y., Muecksch F., Barnes C.O., Finkin S., Schaefer-Babajew D., Cipolla M., Gaebler C., Lieberman J.A., Oliveira T.Y., Yang Z., Abernathy M.E., Huey-Tubman K.E., Hurley A. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592:616–622. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Starr T.N., Greaney A.J., Addetia A., Hannon W.W., Choudhary M.C., Dingens A.S., Li J.Z., Bloom J.D. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science. 2021;371:850–854. doi: 10.1126/science.abf9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun Z., Brodsky J.L. Protein quality control in the secretory pathway. J. Cell Biol. 2019;218:3171–3187. doi: 10.1083/jcb.201906047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perlmutter J.D., Hagan M.F. Mechanisms of virus assembly. Annu. Rev. Phys. Chem. 2015;66:217–239. doi: 10.1146/annurev-physchem-040214-121637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khattar V., Lee J.H., Wang H., Bastola S., Ponnazhagan S. Structural determinants and genetic modifications enhance BMP2 stability and extracellular secretion. FASEB Bioadv. 2019;1:180–190. doi: 10.1096/fba.2018-00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gastaldello S., D'Angelo S., Franzoso S., Fanin M., Angelini C., Betto R., Sandona D. Inhibition of proteasome activity promotes the correct localization of disease-causing alpha-sarcoglycan mutants in HEK-293 cells constitutively expressing beta-, gamma-, and delta-sarcoglycan. Am. J. Pathol. 2008;173:170–181. doi: 10.2353/ajpath.2008.071146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Argentinian AntiCovid C. Structural and functional comparison of SARS-CoV-2-spike receptor binding domain produced in Pichia pastoris and mammalian cells. Sci. Rep. 2020;10:21779. doi: 10.1038/s41598-020-78711-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Micsonai A., Wien F., Bulyaki E., Kun J., Moussong E., Lee Y.H., Goto Y., Refregiers M., Kardos J. BeStSel: A web server for accurate protein secondary structure prediction and fold recognition from the circular dichroism spectra. Nucleic Acids Res. 2018;46:W315–W322. doi: 10.1093/nar/gky497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rotem A., Serohijos A.W.R., Chang C.B., Wolfe J.T., Fischer A.E., Mehoke T.S., Zhang H., Tao Y., Lloyd Ung W., Choi J.M., Rodrigues J.V., Kolawole A.O., Koehler S.A., Wu S., Thielen P.M. Evolution on the biophysical fitness landscape of an RNA virus. Mol. Biol. Evol. 2018;35:2390–2400. doi: 10.1093/molbev/msy131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bloom J.D., Labthavikul S.T., Otey C.R., Arnold F.H. Protein stability promotes evolvability. Proc. Natl. Acad. Sci. U. S. A. 2006;103:5869–5874. doi: 10.1073/pnas.0510098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Greenfield N.J. Using circular dichroism collected as a function of temperature to determine the thermodynamics of protein unfolding and binding interactions. Nat. Protoc. 2006;1:2527–2535. doi: 10.1038/nprot.2006.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 50.Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W.T., Limbo O., Smith C., Song G., Woehl J., Yang L., Abbott R.K., Callaghan S., Garcia E., Hurtado J. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yuan M., Liu H., Wu N.C., Lee C.D., Zhu X., Zhao F., Huang D., Yu W., Hua Y., Tien H., Rogers T.F., Landais E., Sok D., Jardine J.G., Burton D.R. Structural basis of a shared antibody response to SARS-CoV-2. Science. 2020;369:1119–1123. doi: 10.1126/science.abd2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Denison M.R., Graham R.L., Donaldson E.F., Eckerle L.D., Baric R.S. Coronaviruses: An RNA proofreading machine regulates replication fidelity and diversity. RNA Biol. 2011;8:270–279. doi: 10.4161/rna.8.2.15013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hacisuleyman E., Hale C., Saito Y., Blachere N.E., Bergh M., Conlon E.G., Schaefer-Babajew D.J., DaSilva J., Muecksch F., Gaebler C., Lifton R., Nussenzweig M.C., Hatziioannou T., Bieniasz P.D., Darnell R.B. Vaccine breakthrough infections with SARS-CoV-2 variants. N. Engl. J. Med. 2021;384:2212–2218. doi: 10.1056/NEJMoa2105000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bershtein S., Mu W., Serohijos A.W., Zhou J., Shakhnovich E.I. Protein quality control acts on folding intermediates to shape the effects of mutations on organismal fitness. Mol. Cell. 2013;49:133–144. doi: 10.1016/j.molcel.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dinner A.R., Abkevich V., Shakhnovich E., Karplus M. Factors that affect the folding ability of proteins. Proteins. 1999;35:34–40. doi: 10.1002/(sici)1097-0134(19990401)35:1<34::aid-prot4>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 56.Klein E.Y., Blumenkrantz D., Serohijos A., Shakhnovich E., Choi J.M., Rodrigues J.V., Smith B.D., Lane A.P., Feldman A., Pekosz A. Stability of the influenza virus hemagglutinin protein correlates with evolutionary dynamics. mSphere. 2018;3 doi: 10.1128/mSphereDirect.00554-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Elena S.F., Carrasco P., Daros J.A., Sanjuan R. Mechanisms of genetic robustness in RNA viruses. EMBO Rep. 2006;7:168–173. doi: 10.1038/sj.embor.7400636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wylie C.S., Shakhnovich E.I. A biophysical protein folding model accounts for most mutational fitness effects in viruses. Proc. Natl. Acad. Sci. U. S. A. 2011;108:9916–9921. doi: 10.1073/pnas.1017572108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weisblum Y., Schmidt F., Zhang F., DaSilva J., Poston D., Lorenzi J.C., Muecksch F., Rutkowska M., Hoffmann H.H., Michailidis E., Gaebler C., Agudelo M., Cho A., Wang Z., Gazumyan A. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. Elife. 2020;9 doi: 10.7554/eLife.61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dowd K.A., Netski D.M., Wang X.H., Cox A.L., Ray S.C. Selection pressure from neutralizing antibodies drives sequence evolution during acute infection with hepatitis C virus. Gastroenterology. 2009;136:2377–2386. doi: 10.1053/j.gastro.2009.02.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Richman D.D., Wrin T., Little S.J., Petropoulos C.J. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. U. S. A. 2003;100:4144–4149. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Petrova V.N., Russell C.A. The evolution of seasonal influenza viruses. Nat. Rev. Microbiol. 2018;16:47–60. doi: 10.1038/nrmicro.2017.118. [DOI] [PubMed] [Google Scholar]
- 63.Wolfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Muller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brunink S., Schneider J., Ehmann R. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 64.Van Elslande J., Vermeersch P., Vandervoort K., Wawina-Bokalanga T., Vanmechelen B., Wollants E., Laenen L., Andre E., Van Ranst M., Lagrou K., Maes P. Symptomatic SARS-CoV-2 reinfection by a phylogenetically distinct strain. Clin. Infect. Dis. 2020;73:354–356. doi: 10.1093/cid/ciaa1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Larson D., Brodniak S.L., Voegtly L.J., Cer R.Z., Glang L.A., Malagon F.J., Long K.A., Potocki R., Smith D.R., Lanteri C., Burgess T., Bishop-Lilly K.A. A case of early re-infection with SARS-CoV-2. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.To K.K., Hung I.F., Ip J.D., Chu A.W., Chan W.M., Tam A.R., Fong C.H., Yuan S., Tsoi H.W., Ng A.C., Lee L.L., Wan P., Tso E., To W.K., Tsang D. COVID-19 re-infection by a phylogenetically distinct SARS-coronavirus-2 strain confirmed by whole genome sequencing. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Baum A., Fulton B.O., Wloga E., Copin R., Pascal K.E., Russo V., Giordano S., Lanza K., Negron N., Ni M., Wei Y., Atwal G.S., Murphy A.J., Stahl N., Yancopoulos G.D. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;369:1014–1018. doi: 10.1126/science.abd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fagiani F., Catanzaro M., Lanni C. Molecular features of IGHV3-53-encoded antibodies elicited by SARS-CoV-2. Signal Transduct. Target. Ther. 2020;5:170. doi: 10.1038/s41392-020-00287-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Luchsinger L.L., Ransegnola B.P., Jin D.K., Muecksch F., Weisblum Y., Bao W., George P.J., Rodriguez M., Tricoche N., Schmidt F., Gao C., Jawahar S., Pal M., Schnall E., Zhang H. Serological assays estimate highly variable SARS-CoV-2 neutralizing antibody activity in recovered COVID-19 patients. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.02005-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., Hagglof T., Oliveira T.Y., Viant C., Hurley A., Hoffmann H.H. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.DeFrancesco L. COVID-19 antibodies on trial. Nat. Biotechnol. 2020;38:1242–1252. doi: 10.1038/s41587-020-0732-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Starr T.N., Greaney A.J., Dingens A.S., Bloom J.D. Complete map of SARS-CoV-2 RBD mutations that escape the monoclonal antibody LY-CoV555 and its cocktail with LY-CoV016. Cell Rep. Med. 2021;2:100255. doi: 10.1016/j.xcrm.2021.100255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu Z., VanBlargan L.A., Bloyet L.M., Rothlauf P.W., Chen R.E., Stumpf S., Zhao H., Errico J.M., Theel E.S., Liebeskind M.J., Alford B., Buchser W.J., Ellebedy A.H., Fremont D.H., Diamond M.S. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe. 2021;29:477–488.e474. doi: 10.1016/j.chom.2021.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bevington P.R., Robinson D.K. 3rd Ed. McGraw-Hill; Newyork, NY: 2003. Data Reduction and Error Analysis for the Physical Sciences. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data are contained within the article and in the supporting information.