Abstract
The detrimental effects of increased homozygosity due to inbreeding have prompted the development of methods to reduce inbreeding. The detection of runs of homozygosity (ROH), or contiguous stretches of homozygous marker genotypes, can be used to describe and quantify the level of inbreeding in an individual. The estimation of inbreeding coefficients can be calculated based on pedigree information, ROH, or the genomic relationship matrix. The aim of this study was to detect and describe ROH in the turkey genome and compare estimates of pedigree-based inbreeding coefficients (FPED) with genomic-based inbreeding coefficients estimated from ROH (FROH) and the genomic relationship matrix (FGRM). A total of 2,616,890 pedigree records were available. Of these records, 6,371 genotyped animals from three purebred turkey (Meleagris gallopavo) lines between 2013 and 2019 were available, and these were obtained using a dense single nucleotide polymorphism array (56,452 SNPs). The overall mean length of detected ROH was 2.87 ± 0.29 Mb with a mean number of 84.87 ± 8.79 ROH per animal. Short ROH with lengths of 1 to 2 Mb long were the most abundant throughout the genome. Mean ROH coverage differed greatly between chromosomes and lines. Considering inbreeding coefficient means across all lines, genomic derived inbreeding coefficients (FROH = 0.27; FGRM = 0.32) were higher than coefficients estimated from pedigree records (FPED = 0.14). Correlations between FROH and FPED, FROH and FGRM, and FPED and FGRM ranged between 0.19 to 0.31, 0.68 to 0.73, and 0.17 to 0.30, respectively. Additionally, correlations between FROH from different lengths and FPED substantially increased with ROH length from -0.06 to 0.33. Results of the current research, including the distribution of ROH throughout the genome and ROH-derived inbreeding estimates, can provide a more comprehensive description of inbreeding in the turkey genome. This knowledge can be used to evaluate genetic diversity, a requirement for genetic improvement, and develop methods to minimize inbreeding in turkey breeding programs.
Key words: genomic relationship matrix, inbreeding coefficient, pedigree, runs of homozygosity, turkey
INTRODUCTION
The accumulation of inbreeding resulting from strong directional selection is a concern in livestock production populations. Directional selection in poultry has led to genetic gain in economically important traits over the years, leading for instance to an increase in body weight and egg production (Nestor et al., 2008; Aslam et al., 2011a; Abdalla et al., 2019; Emamgholi Begli et al., 2019). However, inbreeding leads to an increase in homozygosity, which can reduce the performance of production, reproduction, and survival traits (Leroy, 2014; Baes et al., 2019). To effectively monitor inbreeding and its consequences in livestock breeding programs, accurate and reliable estimates of inbreeding are essential.
The inbreeding coefficient (F) is the probability that 2 alleles at any given locus in an individual are identical-by-descent (IBD) and, for example, have descended from a common ancestor (Wright, 1922). Pedigree records have traditionally been used to estimate the level of inbreeding for an individual by assessing parentage relationships (Meuwissen and Luo, 1992). This method has limitations as pedigrees may be incomplete or contain errors, it is unable to accurately capture the Mendelian sampling that occurs during mating, and it assumes that founders are unrelated (Keller et al., 2011). Consequently, inbreeding coefficients based on pedigree records (FPED) have been shown to underestimate the true level of inbreeding in an individual (Forutan et al., 2018).
The availability of SNP arrays have allowed for the estimation of closer-to-true inbreeding levels (Forutan et al., 2018). With genomic data, an inbreeding coefficient based on the genomic relationship matrix (FGRM) can be estimated. This method estimates the inbreeding coefficients from the diagonal of the genomic relationship matrix, assuming that founders are unrelated and that allele frequencies are equal to 0.5 (VanRaden, 2008; Yang et al., 2011). This approach often overestimates true inbreeding coefficients due to the inability to distinguish between alleles that are truly IBD and alleles that are identical-by-state (IBS) (Forutan et al., 2018).
Runs of homozygosity (ROH), defined as contiguous stretches of homozygous segments of DNA in an individual that have been passed down from a common ancestor, can also be used to accurately describe genomic inbreeding levels (Broman and Weber, 1999; Gibson et al., 2006). Using ROH has become a more common approach to estimate inbreeding (FROH; McQuillan et al., 2008) as it allows for the distinction between IBD and IBS alleles (MacLeod et al., 2009; Keller et al., 2011; Bjelland et al., 2013). The characterization of ROH can provide information on the history of a population and also insight into when an inbreeding event may have occurred (Bosse et al., 2012; Purfield et al., 2012). Long ROH are a consequence of more recent inbreeding and short ROH are indicative of more distant ancestral inbreeding where the short ROH may be a result of recombination events breaking long chromosomes into segments (Browning and Browning, 2012; Mastrangelo et al., 2016). Other advantages are the ability to differentiate local versus genome-wide inbreeding, and the ability to reveal selection signatures that potentially harbor genes associated with economically important traits targeted for genetic improvement (Strillacci et al., 2018; Almeida et al., 2019).
The increased availability of medium- and high-density genomic data for many livestock species (cattle, swine, sheep, etc.) has promoted a large number of studies on ROH and genomic inbreeding (Bosse et al., 2012; Marras et al., 2014; Signer-Hasler et al., 2019; Makanjuola et al., 2020). However, limited research is available on this topic in poultry, especially in turkeys (Meleagris gallopavo). The objectives of this study were 1) to detect and characterize the distribution of ROH in the turkey genome; 2) to estimate and compare FPED, FROH, and FGRM; and 3) to determine correlations between alternate methods to estimate inbreeding coefficients.
MATERIALS AND METHODS
Ethics Statement
No Animal Care Committee approval was necessary for the purposes of this study, as all information required was obtained from existing databases.
Turkey Population
Data from 3 purebred lines of turkeys (A, B, and C) with breeding objectives balanced between commercial and reproductive traits were used in this study. A stronger selection emphasis was placed on reproductive traits in female lines A and B compared to the male line C. In total, 6,371 genotyped individuals were available for the 3 lines, collected between 2013 and 2019 (Table 1). Pedigree records consisted of known ancestors of all individuals with genotypes and were available for each line with 916,973 records from line A, 854,999 records from line B, and 844,918 records from line C. The maximum generation depth for each line was 36 for line A, 35 for line B, and 31 for line C, which was calculated in R statistical software (R Core Team, 2016) using the Pedigree package (Coster, 2013).
Table 1.
Distribution of genotypes available for animals from three purebred turkey lines (A, B, and C) according to sex.
| Line A | Line B | Line C | |
|---|---|---|---|
| Male | 1,270 | 1,890 | 1,763 |
| Female | 508 | 514 | 426 |
| Total | 1,778 | 2,404 | 2,189 |
Genotype Data and Quality Control
Blood samples from the turkey lines were collected for genotyping. DNA was isolated and genotyped using a 65K SNP panel (65,000 SNP, Illumina, Inc., Lincoln, NE) provided by Hybrid Turkeys, Kitchener, Canada. Quality control on the samples was applied to each line separately and was completed using PLINK v1.90b4.1 (Purcell et al., 2007; Chang et al., 2015). SNP markers were retained 1) with a genotype call rate above 90%, 2) with a minor allele frequency greater than 1%, 3) that have Hardy-Weinberg equilibrium exact test P-value above 1 × 10−6 (Wigginton et al., 2005), and 4) that were in autosomal regions. All samples were retained with criteria of including those with a call rate greater than 90% in the analyses. The genotyping rate was 0.999 for all samples and after editing, 53,625, 52,029, and 52,729 SNP markers were analyzed for lines A, B, and C, respectively.
Detection of Runs of Homozygosity
Runs of homozygosity were detected using the R package “detectRUNS” (Biscarini et al., 2019) with the consecutive method which is window-free and scans the genome on a SNP-by-SNP basis (Marras et al., 2014, 2016). To avoid the introduction of artificial ROH that were shorter than a given window size, the sliding window method was not used to detect ROH (Ferenčaković et al., 2013; Marras et al., 2014). ROH were defined in an individual as a stretch of DNA having a minimum number of 50 contiguous SNP with homozygous genotypes and a minimum length of 1 megabase pairs (Mb). This threshold for a minimum length to denote a ROH was used to exclude short and common ROH that occur in individuals, which may arise from strong linkage disequilibrium (McQuillan et al., 2008). In addition, a minimum length of 1 Mb was used as shorter ROH may have been derived from the inheritance of common allozygous haplotypes (Kim et al., 2015). No missing or heterozygous genotypes were allowed within a single ROH and the maximum gap between consecutive SNP was set to 1 Mb. Detected ROH were analyzed as a total length and divided into 5 class lengths: 1 to 2 Mb, 2 to 4 Mb, 4 to 8 Mb, 8 to 16 Mb, and >16 Mb. The mean number of ROH and mean length of ROH, in addition to standard deviations, were calculated per individual.
Measures of Inbreeding
Three measures of inbreeding coefficients (FPED, FROH, and FGRM) were estimated and analyzed. Inbreeding coefficients were estimated from pedigree genealogies (FPED) for each individual in R statistical software (R Core Team, 2016) using the Pedigree package (Coster, 2013). FROH was estimated based on detected ROH for each individual as:
where is the cumulative sum of all ROH lengths in an individual and is the length of the autosomal genome covered by SNP as proposed by McQuillan et al. (2008). FROH values were estimated for each individual on a genome-wide basis where the approximate length of the turkey autosomal genome was 900 Mb. FROH coefficients were also estimated for ROH within the five class lengths (FROH (1 - 2 Mb), FROH (2 - 4 Mb), FROH (4 - 8 Mb), FROH (8 - 16 Mb), and FROH (> 16 Mb)) for the purpose of calculating correlation coefficients. Genomic inbreeding coefficients from the genomic relationship matrix (FGRM) were estimated for each individual following the method proposed by VanRaden (2008). FGRM coefficients were estimated with a fixed allele frequency of 0.5 using the option –ibc from GCTA software (Yang et al., 2011), whereby the genomic relationship matrix was estimated and FGRM values were obtained for each individual from the diagonal of the matrix. FGRM was estimated for each individual j as:
where is the genomic inbreeding of the jth individual and Gjj is the diagonal element of the genomic relationship matrix.
To evaluate similarity among different estimates of inbreeding, Pearson correlation coefficients were calculated between estimates of FPED, FROH, and FGRM. Additionally, correlation coefficients were also calculated between FPED and FROH estimated from ROH within the 5 class lengths. All correlations between inbreeding coefficients were tested to determine whether they were significantly different from zero. Pearson correlations along with corresponding tests of significance values were computed using the cor.test function in R statistical software (R Core Team, 2016).
RESULTS AND DISCUSSION
Genomic Distribution of Runs of Homozygosity
Runs of homozygosity were detected in all individuals within the 3 turkey lines. Mean number of ROH and mean length of ROH (Mb) per animal were calculated in total and for each class length as shown in Table 2. On an individual animal basis, the mean number of ROH ranged from 81.68 to 87.14 for all lines, with values ranging from 20 to 118 for line A, 2 to 110 for line B, and 40 to 108 for line C. Line A marginally showed the highest mean number of ROH per animal. ROH in class length 1 to 2 Mb were the most abundant throughout the genome; 47.60% for line A, 42.18% for line B, and 45.35% for line C of the segments identified accounted for ROH in class length 1 to 2 Mb. Conversely, ROH in class length >16 Mb were the least abundant. The mean ROH length per animal was approximately 3 Mb for all lines. The longest segment was 34.76 Mb in length (1,193 SNP) found on Meleagris gallopavo autosome (MGA) 2 for line A, 48.02 Mb in length (1,568 SNP) found on MGA 2 for line B, and 50.71 Mb in length (1,720 Mb) found on MGA 3 for line C. There is limited research available for ROH analyses in turkey populations. However, with the strong synteny, close ancestral relationship, and similar breeding goals between chickens and turkeys (Griffin et al., 2008), results of this study can be cautiously compared with those of chickens. In an analysis of ROH on a chicken paternal broiler line with similar length restrictions in the definition of ROH, a mean number of ROH per animal of 12.87 (maximum 30 ROH) and a mean ROH length of 5.34 Mb (maximum length of 59.24 Mb) per animal was observed (Marchesi et al., 2018). The values observed by Marchesi et al. (2018) showed a lower mean number of ROH and a higher mean ROH length per animal than the present study. Different results were also observed in a study analyzing three chicken breeds in a conservation program with a higher mean number of ROH per animal (276.90–535.20 ROH per animal) and lower mean length of ROH per animal (0.18–0.19 Mb per animal) in comparison to the present study (Zhang et al., 2018). This difference, however, is likely attributed to the lower length threshold used to detect ROH and the higher density of SNPs used to perform the analyses. Higher numbers of ROH identified in shorter class lengths as seen in the current study corroborates other studies in poultry (Fleming et al., 2016; Marchesi et al., 2018; Almeida et al., 2019), as well as in other livestock species (Bosse et al., 2012; Purfield et al., 2012). ROH size is associated with the degree to which an IBD segment of DNA has been passed down generations (Broman and Weber, 1999). As IBD segments are passed down generations, recombination events may breakdown their length during meiosis (Kirin et al., 2010). As a result, short ROH are likely IBD regions inherited from ancient ancestors indicative of long-term selection and long ROH of more recent selection; short ROH (approximately 1 Mb in length) may be linked to ancestors from up to 50 generations ago and long ROH (approximately 10 Mb in length) may arise due to recent inbreeding from up to 5 generations ago (Howrigan et al., 2011). However, Ferenčaković et al. (2013) suggested to exercise caution when including ROH of less than 4 Mb in analyses as they may not be related to autozygosity. Therefore, it is possible that the majority of regions of homozygosity in these turkey lines are a result of long-term selection while few regions may have arisen due to recent selection. However, this statement must be taken cautiously when extending to turkeys, as the length of the bovine genome is 3 billion base pairs long, which is 3 times the size of the turkey genome. Further investigation of the distribution of these ROH across the genome will provide insights on the demography of these turkey populations.
Table 2.
Mean number of runs of homozygosity (ROH) and mean length of ROH in megabase pairs (Mb) per animal for each class length and in total with standard deviations shown in brackets.
| Line A |
Line B |
Line C |
||||
|---|---|---|---|---|---|---|
| ROH Class (Mb) | Mean number of ROH | Mean length in Mb of ROH | Mean number of ROH | Mean length in Mb of ROH | Mean number of ROH | Mean length in Mb of ROH |
| 1 to 2 | 41.48 (6.30) | 1.49 (0.04) | 36.32 (5.90) | 1.49 (0.05) | 37.04 (5.82) | 1.50 (0.05) |
| 2 to 4 | 31.21 (5.69) | 2.75 (0.10) | 32.99 (5.43) | 2.78 (0.10) | 29.67 (5.08) | 2.74 (0.10) |
| 4 to 8 | 12.23 (3.38) | 5.35 (0.32) | 13.58 (3.62) | 5.48 (0.30) | 12.33 (3.29) | 5.36 (0.32) |
| 8 to 16 | 2.43 (1.34) | 10.08 (1.38) | 3.11 (1.60) | 10.38 (1.30) | 2.66 (1.42) | 10.49 (1.46) |
| >16 | 1.15 (0.43) | 19.65 (3.45) | 1.21 (0.50) | 19.93 (3.61) | 1.15 (0.46) | 19.99 (4.10) |
| Total | 87.14 (9.32) | 2.72 (0.25) | 86.10 (8.69) | 2.98 (0.29) | 81.68 (7.45) | 2.86 (0.27) |
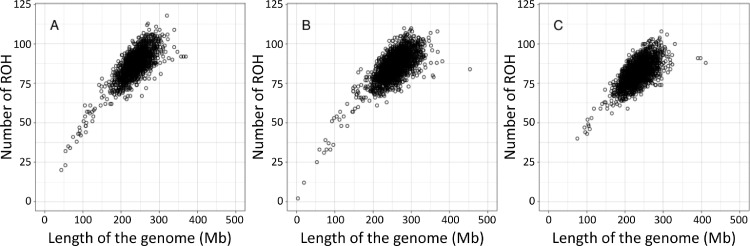
Figure 1 displays number of ROH and length of the genome covered by ROH for individuals in the 3 lines. This provides insight into ROH content of the total length of the genome in an individual. In all 3 lines, animals displaying the same length of the genome covered by ROH had moderate variation in the number of segments composing the total length. This could be a consequence of individuals displaying different distances from common ancestors, a relationship analyzed in cattle by other authors (Mészáros et al., 2015; Peripolli et al., 2018).
Figure 1.
Number of runs of homozygosity (ROH) per individual and the length of the genome covered by ROH in megabase pairs (Mb) for lines A, B, and C.
The mean percent coverage of ROH per chromosome was calculated for each turkey line (Figure 2). ROH were found to display heterogeneity across the genome; ROH did not cluster on specific chromosomes. The highest fraction of a chromosome covered by ROH was found on MGA 8 (42.26% of chromosomal length) for line A, MGA 7 (48.32% of chromosomal length) for line B, and MGA 9 (38.24% of chromosomal length) for line C. No ROH were found on chromosome 18 due to the small size of the chromosome which is only 332,615 base pairs long based on the Turkey_5.1 assembly (Dalloul et al., 2010). Overall, as chromosome length decreased, the mean percent coverage of ROH per chromosome tended to decrease, which is consistent with recent studies in chicken populations (Fleming et al., 2016; Marchesi et al., 2018). Furthermore, this is also supportive of the case that recombination rates are higher on microchromosomes than on intermediate and macrochromosomes (Axelsson et al., 2005) and ROH tend to cluster in regions of the genome where recombination rates are low.
Figure 2.
Mean percent coverage of runs of homozygosity per chromosome along the turkey genome (calculated standard errors for each bar ranged from 0.00 to 0.01).
A number of chromosomes in the turkey genome displayed higher levels of homozygosity than others. These chromosomes may be defined based on those harboring the longest ROH detected in each line, which was found on MGA 2 and 3, and the chromosomes displaying higher percent coverages by ROH. MGA 1, 6, 7, 8, 9, 10, 11, 13, and 22 showed percent coverages greater than 30.0% in at least one of the turkey lines, as seen in Figure 2. Selection pressure has printed regions along the genome, known as selection signatures, which have been found to harbor functionally important sequence variants. Selection signatures can be detected by using combinations of statistical measures such as ROH and FST mapping (Elbeltagy et al., 2019) or the presence of exceptionally extended haplotype homozygosity (Liu et al., 2016). Additionally, human studies have shown the existence of QTL in ROH (Lencz et al., 2007; Hildebrandt et al., 2009). Aslam et al. (2011b, 2012, 2014) completed studies in turkey populations to detect areas of the genome under selection. A comparison of these results with the chromosomes displaying increased homozygosity in the present study showed many similarities. Aslam et al. (2011b) found QTL for breast meat yield, meat quality, body weight, and those affecting growth on MGA 3, 7, 8, 11, 13, and 22. Aslam et al. (2012) found regions displaying low nucleotide variation that showed state of fixation towards alleles different than wild alleles on MGA 3, 9, and 22. Aslam et al. (2014) found regions showing significant reduction in genomic variation on MGA 2, 7, 9, and 22, which were enriched with genes known to affect growth. Considering that the populations in the current study have been selected for meat-type traits, these similarities show evidence that the ROH detected in the present study may have arose due to the selection that has been put in place for the development of these turkey lines. However, further investigation into intrachromosomal regions and specific ROH genotypes in the currently studied turkey lines is necessary for a more comprehensive comparison of these studies. Therefore, the detection of ROH can aid in the identification of selection signatures, which can provide valuable insights about genomic regions and genes that have been under selection pressure, and hence develop an understanding of how these regions are affecting traits of interest.
Measures of Inbreeding
Descriptive statistics for FPED, FROH, and FGRM are shown in Table 3. Distributions of FPED, FROH, and FGRM are displayed in Figure 3. FROH, estimated as the percentage of the genome that is autozygous, had an estimated mean value of 26.24%, covering 237.02 Mb of the autosomal genome, for line A, 28.46% (257.04 Mb) for line B, and 25.82% (233.18 Mb) for line C. Overall, inbreeding coefficients estimated in these three turkey lines were relatively high. These high inbreeding coefficients can likely be attributed to the purebred nature of these lines. Purebred lines are developed through selective breeding and maintained over time. These lines are subsequently crossed to produce the commercial populations to attain the benefits of heterozygosity, or hybrid vigor.
Table 3.
Mean inbreeding coefficients based on pedigree information (FPED), runs of homozygosity (FROH), and genomic relationships (FGRM) with standard deviations (SD) in brackets and range of minimum to maximum observed inbreeding coefficients for three turkey lines.
| Line A |
Line B |
Line C |
||||
|---|---|---|---|---|---|---|
| Mean (SD) | Range | Mean (SD) | Range | Mean (SD) | Range | |
| FPED | 0.14 (0.01) | 0.12–0.26 | 0.16 (0.02) | 0.12–0.31 | 0.12 (0.01) | 0.09–0.34 |
| FROH | 0.26 (0.04) | 0.05–0.41 | 0.28 (0.04) | 0.00–0.50 | 0.26 (0.03) | 0.08–0.46 |
| FGRM | 0.32 (0.04) | 0.15–0.48 | 0.32 (0.04) | 0.16–0.48 | 0.32 (0.03) | 0.21–0.52 |
Figure 3.
Distribution of inbreeding coefficients based on pedigree information (FPED), runs of homozygosity (FROH), and the genomic relationship matrix (FGRM) for each turkey line.
Limited literature is available in regard to inbreeding levels in turkey populations. According to Marchesi et al. (2018), average inbreeding coefficients estimated in a paternal chicken broiler line was found to be 0.07 and 0.04 for FROH and FPED, respectively, however these coefficients are very low. In a study by Muir et al. (2008), lower mean inbreeding coefficients, based on excess homozygosity, for commercial chicken pure lines were estimated to be between 0.14 and 0.16. However, the authors explain that these estimates may have been an underestimate of the true level of inbreeding, since their samples were not representative of the true ancestral population. Additionally, it may be reasonable to expect that the estimates of F, in the study by Muir et al. (2008), have increased over the past decade with strong directional selection that occurs the commercial chicken populations. Therefore, the estimates of inbreeding provided by Muir et al. (2008) may only be cautiously compared to the estimated coefficients of the present study. As expected, FGRM estimates were greater than FROH estimates which were both greater than FPED estimates in all 3 lines (Howrigan et al., 2011). This finding is in agreement with studies in poultry and other livestock species where genomic-based inbreeding coefficients generally are found to be greater than pedigree-based inbreeding coefficients (Hammerly et al., 2013; Marras et al., 2014; Marchesi et al., 2018). FPED may not be an accurate estimate of the true inbreeding in a population due to a number of limitations such as there being errors in the pedigree records and the inability for the coefficient to capture the stochastic nature of Mendelian sampling and recombination (Hill and Weir, 2011). This has also been demonstrated in simulation studies by authors Liu et al. (2014) and Forutan et al. (2018). As observed in a simulation study in cattle by Forutan et al. (2018), FGRM may be overestimating the level of inbreeding due to the presence of alleles that are IBS and IBD and the use of 0.5 as the fixed allele frequency (VanRaden, 2008; Pryce et al., 2014). This may explain why FGRM was observed to be greater than FROH in the present study. Therefore, the level of inbreeding estimated from FROH may be closer to the true level of inbreeding since ROH are a direct measure of autozygosity.
Distributions of FGRM and FROH were larger than distributions of FPED, as seen in Figure 3, which corroborates the study by Hammerly et al. (2013). FPED estimates the expected inbreeding, which in a highly structured livestock population, tends to have similar values for a large proportion of the animals. FPED distributions are therefore narrow, with only a few animals displaying extreme values. In contrast, genomic data allows the estimation of more realized inbreeding coefficients. As a consequence, FGRM and FROH distributions are wider with a larger interquartile range and a finer differentiation between animals. It is also noted that the distribution of FROH was larger than the distribution of FGRM for line B; specifically, more animals were displaying lower FROH than FGRM (Figure 3). This observation may be attributed to the criteria used to detect ROH in an individual. A minimum number of 50 contiguous SNP with homozygous genotypes and a minimum length of 1 Mb may have allowed for a low number of ROH detected and therefore, resulted in a low level of inbreeding calculated from ROH. Additionally, FGRM have been shown to provide an overestimate of the true level of inbreeding, compared to those estimated from FROH, due to the inability to distinguish IBD and IBS alleles (Forutan et al., 2018). This may have contributed to the higher FGRM estimates.
Correlation coefficients and associated tests of significance for lines A, B, and C are presented in Table 4. Low to moderate correlations were observed between pairwise comparisons of FPED, FROH, and FGRM. Similar correlations of 0.65 and 0.62 for Holstein and Jersey cattle, respectively, were seen between FROH and FGRM in a study by Pryce et al. (2014). Bjelland et al. (2013) reported correlations of 0.81 in Holstein cattle between FROH and FGRM when an allele frequency of 0.5 was used to estimate the genomic relationship matrix. Low correlations observed between FPED and genomic inbreeding coefficients in the current study may be related to the different distributions of inbreeding estimated from pedigree records vs. those estimated with genomic data (as shown in Figure 3). Low correlations between FPED and FROH may also suggest that FPED may not be the most suitable method for capturing ancient inbreeding as FROH can capture more ancient inbreeding than FPED. The low correlations observed in the present study are in agreement with low correlations of 0.06 observed between FPED and genomic-based inbreeding coefficients in a paternal chicken broiler line (Marchesi et al., 2018) and of 0.02 in a Large White pig population (Zanella et al., 2016). Therefore, this may be supportive of FROH and other genomic-based measures providing a more accurate description of inbreeding than traditional methodologies due to it being able to better detect the proportion of the genome that is IBD. A substantial increase in correlations of −0.06 to 0.19 for line A, −0.06 to 0.23 for line B, and −0.06 to 0.33 for line C between FPED and FROH as class length increases is supportive of the argument that FPED is not the most suitable method for capturing ancient inbreeding. As longer ROH are considered to estimate FROH, which tend to be associated with more recent inbreeding (Howrigan et al., 2011), the correlation between FPED and FROH tends to increase. This is in agreement with correlations observed in a study by Peripolli et al. (2018) in dairy cattle as well as by authors displaying increasing correlations between FROH and FPED when pedigrees have an increased number of generations available (Ferenčaković et al., 2012; Purfield et al., 2012; Marras et al., 2014).
Table 4.
Correlations coefficients of genomic inbreeding coefficients (FROH, FROH (1 - 2 Mb), FROH (2 - 4 Mb), FROH (2 - 4 Mb), FROH (4 - 8 Mb), FROH (8 - 16 Mb), FROH (> 16 Mb), and FGRM) and pedigree-based inbreeding coefficients (FPED) for lines A, B, and C.
| Line A | Line B | Line C | |
|---|---|---|---|
| FROH, FPED | 0.19⁎⁎ | 0.24⁎⁎ | 0.31⁎⁎ |
| FROH, FGRM | 0.68⁎⁎ | 0.69⁎⁎ | 0.73⁎⁎ |
| FGRM, FPED | 0.17⁎⁎ | 0.21⁎⁎ | 0.30⁎⁎ |
| FROH (1 - 2 Mbps), FPED | -0.06⁎⁎ | -0.06⁎⁎ | -0.06⁎⁎ |
| FROH (2 - 4 Mbps), FPED | 0.02 | 0.02 | 0.03 |
| FROH (4 - 8 Mbps), FPED | 0.11⁎⁎ | 0.11⁎⁎ | 0.11⁎⁎ |
| FROH (8 - 16 Mbps), FPED | 0.18⁎⁎ | 0.17⁎⁎ | 0.22⁎⁎ |
| FROH (> 16 Mbps), FPED | 0.19⁎⁎ | 0.23⁎⁎ | 0.33⁎⁎ |
P ≤ 0.01.
A limitation to this study, as with all studies analyzing ROH, is the lack of consensus criteria for defining a ROH (Ku et al., 2010). This discrepancy between ROH definitions makes comparison between studies difficult and therefore, caution should be taken when interpreting and comparing results of this nature.
The goal of this study was to lay the groundwork for future inbreeding and selection investigations in turkey populations. Further investigation into the regions of the genome showing increased levels of homozygosity, in combination with other statistical measures to evaluate areas of the genome that have been under intense selection, can lead to the detection of candidate genomic regions and genes related to economically important traits. This will provide a more thorough understanding of genotype-phenotype relationships and how selection has shaped the turkey genome.
CONCLUSIONS
The detection and characterization of ROH and inbreeding levels estimated using different methods in purebred turkey lines were investigated in this study. Long and abundant ROH were detected and ROH did not cluster on specific chromosomes. Genomic-based inbreeding coefficients were higher than pedigree-based inbreeding coefficients. Low to moderate correlations between respective inbreeding coefficients were observed. These results provide first insights of the genomic architecture of inbreeding and inbreeding levels in purebred turkey populations.
Acknowledgments
ACKNOWLEDGMENTS
The authors extend their gratitude to the managers and personnel of the Hybrid Turkey pedigree farm (Kitchener, Canada) for collecting and providing data used in this study. The study was part of the project entitled, “Application of genomic selection in turkeys for health, welfare, efficiency, and production traits”. This project was funded by the Government of Canada through Genome Canada and the Ontario Genomics Institute (OGI-133) as part of their Genomic Application Partnership Program (recipients: B.J. Wood (Industry) and C.F. Baes (Academic)). The authors would also like to acknowledge Hybrid Turkeys and NSERC for their financial support.
Authors’ contributions: All authors (SA, MD, BM, GM, BW, and CB) made substantial intellectual contributions to the conception of the study and interpretation of the data. SA conducted the statistical analyses with assistance from MD, BM, and GM. SA drafted the first version of the manuscript. MD, BM, GM, BW, and CB provided substantive input and contributions to the manuscript revision. All authors (SA, MD, BM, GM, BW, and CB) read and approved the final manuscript.
Ethics approval and consent to participate: No Animal Care Committee approval was necessary for the purposes of this study, as all information required was obtained from existing databases.
Consent for Publication : Not applicable
Availability of data and materials: Data that support the findings of this study are available from Hybrid Turkeys upon reasonable request, but restrictions apply to the availability of these data, which were used under a license of a material transfer agreement for the current study, and thus are not publicly available.
DISCLOSURES
The authors declare that B.J. Wood was an employee at Hybrid Turkeys at the time of the study. All remaining authors declare that they have no known competing financial interests or personal relationships that influence the work reported in this paper.
REFERENCES
- Abdalla E.E.A., Schenkel F.S., Emamgholi Begli H., Willems O.W., van As P., Vanderhout R., Wood B.J., Baes C.F. Single-Step methodology for genomic evaluation in Turkeys (Meleagris gallopavo) Front. Genet. 2019;10:1248. doi: 10.3389/fgene.2019.01248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida O.A.C., Moreira G.C.M., Rezende F.M., Boschiero C., de Oliveira Peixoto J., Ibelli A.M.G., Ledur M.C., de Novais F.J., Coutinho L.L. Identification of selection signatures involved in performance traits in a paternal broiler line. BMC Genomics. 2019;20:449. doi: 10.1186/s12864-019-5811-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslam M.L., Bastiaansen J.W.M., Crooijmans R.P.M.A., Ducro B.J., Vereijken A., Groenen M.A.M. Genetic variances, heritabilities and maternal effects on body weight, breast meat yield, meat quality traits and the shape of the growth curve in turkey birds. BMC Genet. 2011;12:14. doi: 10.1186/1471-2156-12-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslam M.L., Bastiaansen J.W.M., Crooijmans R.P.M.A., Vereijken A., Groenen M.A.M. Whole genome QTL mapping for growth, meat quality and breast meat yield traits in turkey. BMC Genet. 2011;12:61. doi: 10.1186/1471-2156-12-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslam M.L., Bastiaansen J.W.M., Elferink M.G., Megens H.J., Crooijmans R.P.M.A., Blomberg L.A., Fleischer R.C., Van Tassell C.P., Sonstegard T.S., Schroeder S.G., Groenen M.A.M., Long J.A. Whole genome SNP discovery and analysis of genetic diversity in Turkey (Meleagris gallopavo) BMC Genomics. 2012;13:391. doi: 10.1186/1471-2164-13-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslam M.L., Bastiaansen J.W.M., Megens H.J., Crooijmans R.P.M.A., Nasreen F., Blomberg L.A., Van Tassell C.P., Sonstegard T.S., Schroeder S.G., Groenen M.A.M., Long J.A. Genome-wide candidate regions for selective sweeps revealed through massive parallel sequencing of DNA across ten turkey populations. BMC Genet. 2014;15:117. doi: 10.1186/s12863-014-0117-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelsson E., Webster M.T., Smith N.G.C., Burt D.W., Ellegren H. Comparison of the chicken and turkey genomes reveals a higher rate of nucleotide divergence on microchromosomes than macrochromosomes. Genome Res. 2005;15:120–125. doi: 10.1101/gr.3021305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baes C.F., Makanjuola B.O., Miglior F., Marras G., Howard J.T., Fleming A., Maltecca C. Symposium review: the genomic architecture of inbreeding: how homozygosity affects health and performance. J. Dairy Sci. 2019;102:2807–2817. doi: 10.3168/jds.2018-15520. [DOI] [PubMed] [Google Scholar]
- Biscarini, F., P. Cozzi, G. Gaspa, and G. Marras. 2019. Detect runs of homozygosity and runs of heterozygosity in diploid genomes. Accessed Apr. 2019. https://cran.r-project.org/web/packages/detectRUNS/detectRUNS.pdf.
- Bjelland D.W., Weigel K.A., Vukasinovic N., Nkrumah J.D. Evaluation of inbreeding depression in Holstein cattle using whole-genome SNP markers and alternative measures of genomic inbreeding. J. Dairy Sci. 2013;96:4697–4706. doi: 10.3168/jds.2012-6435. [DOI] [PubMed] [Google Scholar]
- Bosse M., Megens H.-J., Madsen O., Paudel Y., Frantz L.A.F., Schook L.B., Crooijmans R.P.M.A., Groenen M.A.M. Regions of homozygosity in the porcine genome: consequence of demography and the recombination landscape. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1003100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman K.W., Weber J.L. Long homozygous chromosomal segments in reference families from the Centre d'Etude du Polymorphisme Humain. Am. J. Hum. Genet. 1999;65:1493–1500. doi: 10.1086/302661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning S.R., Browning B.L. Identity by descent between distant relatives: detection and applications. Annu. Rev. Genet. 2012;46:617–633. doi: 10.1146/annurev-genet-110711-155534. [DOI] [PubMed] [Google Scholar]
- Chang C.C., Chow C.C., Tellier L.C.A.M., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coster, A. 2013. Pedigree functions. Accessed Jan. 2019. https://cran.r-project.org/web/packages/pedigree/pedigree.pdf.
- Dalloul R.A., Long J.A., Zimin A.V., Aslam L., Beal K., Blomberg L.A., Bouffard P., Burt D.W., Crasta O., Crooijmans R.P.M.A., Cooper K., Coulombe R.A., De S., Delany M.E., Dodgson J.B., Dong J.J., Evans C., Frederickson K.M., Flicek P., Florea L., Folkerts O., Groenen M.A.M., Harkins T.T., Herrero J., Hoffmann S., Megens H.J., Jiang A., de Jong P., Kaiser P., Kim H., Kim K.W., Kim S., Langenberger D., Lee M.K., Lee T., Mane S., Marcais G., Marz M., McElroy A.P., Modise T., Nefedov M., Notredame C., Paton I.R., Payne W.S., Pertea G., Prickett D., Puiu D., Qioa D., Raineri E., Ruffier M., Salzberg S.L., Schatz M.C., Scheuring C., Schmidt C.J., Schroeder S., Searle S.M.J., Smith E.J., Smith J., Sonstegard T.S., Stadler P.F., Tafer H., Tu Z., van Tassell C.P., Vilella A.J., Williams K.P., Yorke J.A., Zhang L., Bin Zhang H., Zhang X., Zhang Y., Reed K.M. Multi-platform next-generation sequencing of the domestic Turkey (Meleagris gallopavo): genome assembly and analysis. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbeltagy A.R., Bertolini F., Fleming D.S., Van Goor A., Ashwell C.M., Schmidt C.J., Kugonza D.R., Lamont S.J., Rothschild M.F. Natural selection footprints among African chicken breeds and village ecotypes. Front. Genet. 2019;10:376. doi: 10.3389/fgene.2019.00376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emamgholi Begli H., Wood B.J., Abdalla E.A., Balzani A., Willems O., Schenkel F., Harlander-Matauschek A., Baes C.F. Genetic parameters for clutch and broodiness traits in turkeys (Meleagris Gallopavo) and their relationship with body weight and egg production. Poult. Sci. 2019;98:6263–6269. doi: 10.3382/ps/pez446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferenčaković M., Hamzić E., Gredler-Grandl B., Solberg T.R., Klemetsdal G., Curik I., Sölkner J. Estimates of autozygosity derived from runs of homozygosity: empirical evidence from selected cattle populations. J. Anim. Breed. Genet. 2012;130:286–293. doi: 10.1111/jbg.12012. [DOI] [PubMed] [Google Scholar]
- Ferenčaković M., Sölkner J., Curik I. Estimating autozygosity from high-throughput information: effects of SNP density and genotyping errors. Genet. Sel. Evol. 2013;45:42. doi: 10.1186/1297-9686-45-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming D.S., Koltes J.E., Markey A.D., Schmidt C.J., Ashwell C.M., Rothschild M.F., Persia M.E., Reecy J.M., Lamont S.J. Genomic analysis of Ugandan and Rwandan chicken ecotypes using a 600 k genotyping array. BMC Genomics. 2016;17:407. doi: 10.1186/s12864-016-2711-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forutan M., Ansari Mahyari S., Baes C., Melzer N., Schenkel F.S., Sargolzaei M. Inbreeding and runs of homozygosity before and after genomic selection in North American Holstein cattle. BMC Genomics. 2018;19:98. doi: 10.1186/s12864-018-4453-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson J., Morton N.E., Collins A. Extended tracts of homozygosity in outbred human populations. Hum. Mol. Genet. 2006;15:789–795. doi: 10.1093/hmg/ddi493. [DOI] [PubMed] [Google Scholar]
- Griffin D.K., Robertson L.B., Tempest H.G., Vignal A., Fillon V., Crooijmans R.P.M.A., Groenen M.A.M., Deryusheva S., Gaginskaya E., Carré W., Waddington D., Talbot R., Völker M., Masabanda J.S., Burt D.W. Whole genome comparative studies between chicken and turkey and their implications for avian genome evolution. BMC Genomics. 2008;9:168. doi: 10.1186/1471-2164-9-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerly S.C., Morrow M.E., Johnson J.A. A comparison of pedigree- and DNA-based measures for identifying inbreeding depression in the critically endangered Attwater's Prairie-chicken. Mol. Ecol. 2013;22:5313–5328. doi: 10.1111/mec.12482. [DOI] [PubMed] [Google Scholar]
- Hildebrandt F., Heeringa S.F., Rüschendorf F., Attanasio M., Nürnberg G., Becker C., Seelow D., Huebner N., Chernin G., Vlangos C.N., Zhou W., O'Toole J.F., Hoskins B.E., Wolf M.T.F., Hinkes B.G., Chaib H., Ashraf S., Allen S.J., Vega-Warner V., Wise E., Harville H.M., Lyons R.H., Washburn J., MacDonald J., Nürnberg P., Otto E.A. A systematic approach to mapping recessive disease genes in individuals from outbred populations. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill W.G., Weir B.S. Variation in actual relationship as a consequence of Mendelian sampling and linkage. Genet. Res. 2011;93:47–64. doi: 10.1017/S0016672310000480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howrigan D.P., Simonson M.A., Keller M.C. Detecting autozygosity through runs of homozygosity: a comparison of three autozygosity detection algorithms. BMC Genomics. 2011;12:460. doi: 10.1186/1471-2164-12-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller M.C., Visscher P.M., Goddard M.E. Quantification of inbreeding due to distant ancestors and its detection using dense single nucleotide polymorphism data. Genetics. 2011;189:237–249. doi: 10.1534/genetics.111.130922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E.S., Sonstegard T.S., Van Tassell C.P., Wiggans G., Rothschild M.F. The relationship between runs of homozygosity and inbreeding in Jersey cattle under selection. PLoS One. 2015;10 doi: 10.1371/journal.pone.0129967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirin M., McQuillan R., Franklin C.S., Campbell H., Mckeigue P.M., Wilson J.F. Genomic runs of homozygosity record population history and consanguinity. PLoS One. 2010;5:e13996. doi: 10.1371/journal.pone.0013996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku C.S., Naidoo N., Teo S.M., Pawitan Y. Regions of homozygosity and their impact on complex diseases and traits. Hum. Genet. 2010;129:1–15. doi: 10.1007/s00439-010-0920-6. [DOI] [PubMed] [Google Scholar]
- Lencz T., Lambert C., DeRosse P., Burdick K.E., Morgan T.V., Kane J.M., Kucherlapati R., Malhotra A.K. Runs of homozygosity reveal highly penetrant recessive loci in schizophrenia. Proc. Natl. Acad. Sci. U. S. A. 2007;104:19942–19947. doi: 10.1073/pnas.0710021104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy G. Inbreeding depression in livestock species: review and meta-analysis. Anim. Genet. 2014;45:618–628. doi: 10.1111/age.12178. [DOI] [PubMed] [Google Scholar]
- Liu H., Sørensen A.C., Meuwissen T.H.E., Berg P. Allele frequency changes due to hitch-hiking in genomic selection programs. Genet. Sel. Evol. 2014;46:8. doi: 10.1186/1297-9686-46-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Sun C., Qu L., Wang K., Yang N. Genome-wide detection of selective signatures in chicken through high density SNPs. PLoS One. 2016;11 doi: 10.1371/journal.pone.0166146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod I.M., Meuwissen T.H.E., Hayes B.J., Goddard M.E. A novel predictor of multilocus haplotype homozygosity: comparison with existing predictors. Genet. Res. (Camb). 2009;91:413–426. doi: 10.1017/S0016672309990358. [DOI] [PubMed] [Google Scholar]
- Makanjuola B.O., Miglior F., Abdalla E.A., Maltecca C., Schenkel F.S., Baes C.F. Effect of genomic selection on rate of inbreeding and coancestry and effective population size of Holstein and Jersey cattle populations. J. Dairy Sci. 2020;103:5183–5199. doi: 10.3168/jds.2019-18013. [DOI] [PubMed] [Google Scholar]
- Marchesi J.A.P., Buzanskas M.E., Cantão M.E., Ibelli A.M.G., Peixoto J.O., Joaquim L.B., Moreira G.C.M., Godoy T.F., Sbardella A.P., Figueiredo E.A.P., Coutinho L.L., Munari D.P., Ledur M.C. Relationship of runs of homozygosity with adaptive and production traits in a paternal broiler line. Animal. 2018;12:1126–1134. doi: 10.1017/S1751731117002671. [DOI] [PubMed] [Google Scholar]
- Marras G., Gaspa G., Sorbolini S., Dimauro C., Ajmone-Marsan P., Valentini A., Williams J.L., MacCiotta N.P.P. Analysis of runs of homozygosity and their relationship with inbreeding in five cattle breeds farmed in Italy. Anim. Genet. 2014;46:110–121. doi: 10.1111/age.12259. [DOI] [PubMed] [Google Scholar]
- Marras G., Rossoni A., Schwarzenbacher H., Biffani S., Biscarini F., Nicolazzi E.L. Zanardi: an open-source pipeline for multiple-species genomic analysis of SNP array data. Anim. Genet. 2016;48:121. doi: 10.1111/age.12485. [DOI] [PubMed] [Google Scholar]
- Mastrangelo S., Tolone M., Di Gerlando R., Fontanesi L., Sardina M.T., Portolano B. Genomic inbreeding estimation in small populations: evaluation of runs of homozygosity in three local dairy cattle breeds. Animal. 2016;10:746–754. doi: 10.1017/S1751731115002943. [DOI] [PubMed] [Google Scholar]
- McQuillan R., Leutenegger A.L., Abdel-Rahman R., Franklin C.S., Pericic M., Barac-Lauc L., Smolej-Narancic N., Janicijevic B., Polasek O., Tenesa A., MacLeod A.K., Farrington S.M., Rudan P., Hayward C., Vitart V., Rudan I., Wild S.H., Dunlop M.G., Wright A.F., Campbell H., Wilson J.F. Runs of homozygosity in European populations. Am. J. Hum. Genet. 2008;83:359–372. doi: 10.1016/j.ajhg.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mészáros G., Boison S.A., Pérez O'Brien A.M., Ferenčaković M., Curik I., Da Silva M.V.B., Utsunomiya Y.T., Garcia J.F., Sölkner J. Genomic analysis for managing small and endangered populations: a case study in Tyrol Grey cattle. Front. Genet. 2015;6:173. doi: 10.3389/fgene.2015.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen T.H.E., Luo Z. Computing inbreeding coefficients in large populations. Genet. Sel. Evol. 1992;24:305–313. [Google Scholar]
- Muir W.M., Wong G.K.S., Zhang Y., Wang J., Groenen M.A.M., Crooijmans R.P.M.A., Megens H.J., Zhang H., Okimoto R., Vereijken A., Jungerius A., Albers G.A.A., Lawley C.T., Delany M.E., MacEachern S., Cheng H.H. Genome-wide assessment of worldwide chicken SNP genetic diversity indicates significant absence of rare alleles in commercial breeds. Proc. Natl. Acad. Sci. U. S. A. 2008;105:17312–17317. doi: 10.1073/pnas.0806569105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestor K.E., Anderson J.W., Patterson R.A., Velleman S.G. Genetics of growth and reproduction in the Turkey. 17. Changes in genetic parameters over forty generations of selection for increased sixteen-week body weight. Poult. Sci. 2008;87:1971–1979. doi: 10.3382/ps.2008-00137. [DOI] [PubMed] [Google Scholar]
- Peripolli E., Stafuzza N.B., Munari D.P., Lima A.L.F., Irgang R., Machado M.A., do C. Panetto J.C., Ventura R.V., Baldi F., da Silva M.V.G.B. Assessment of runs of homozygosity islands and estimates of genomic inbreeding in Gyr (Bos indicus) dairy cattle. BMC Genomics. 2018;19:34. doi: 10.1186/s12864-017-4365-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryce J.E., Haile-Mariam M., Goddard M.E., Hayes B.J. Identification of genomic regions associated with inbreeding depression in Holstein and Jersey dairy cattle. Genet. Sel. Evol. 2014;46:71. doi: 10.1186/s12711-014-0071-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., De Bakker P.I.W., Daly M.J., Sham P.C. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purfield D.C., Berry D.P., McParland S., Bradley D.G. Runs of homozygosity and population history in cattle. BMC Genet. 2012;13:70. doi: 10.1186/1471-2156-13-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2016. R: A Language and Environment for Statistical Computing.https://www.r-project.org/ Accessed Jan. 2019. [Google Scholar]
- Signer-Hasler H., Burren A., Ammann P., Drögemüller C., Flury C. Runs of homozygosity and signatures of selection: a comparison among eight local Swiss sheep breeds. Anim. Genet. 2019;50:512–525. doi: 10.1111/age.12828. [DOI] [PubMed] [Google Scholar]
- Strillacci M.G., Vega-Murillo V.E., Román-Ponce S.I., López F.J.R., Cozzi M.C., Gorla E., Cerolini S., Bertolini F., Fontanesi L., Bagnato A. Looking at genetic structure and selection signatures of the Mexican chicken population using single nucleotide polymorphism markers. Poult. Sci. 2018;97:791–802. doi: 10.3382/ps/pex374. [DOI] [PubMed] [Google Scholar]
- VanRaden P.M. Efficient methods to compute genomic predictions. J. Dairy Sci. 2008;91:4414–4423. doi: 10.3168/jds.2007-0980. [DOI] [PubMed] [Google Scholar]
- Wigginton J.E., Cutler D.J., Abecasis G.R. A note on exact tests of Hardy-Weinberg equilibrium. Am. J. Hum. Genet. 2005;76:887–893. doi: 10.1086/429864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. Coefficients of inbreeding and relationship. Am. Nat. 1922;56:330–338. [Google Scholar]
- Yang J., Lee S.H., Goddard M.E., Visscher P.M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanella R., Peixoto J.O., Cardoso F.F., Cardoso L.L., Biegelmeyer P., Cantão M.E., Otaviano A., Freitas M.S., Caetano A.R., Ledur M.C. Genetic diversity analysis of two commercial breeds of pigs using genomic and pedigree data. Genet. Sel. Evol. 2016;48:24. doi: 10.1186/s12711-016-0203-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Han W., Tang H., Li G., Zhang M., Xu R., Liu Y., Yang T., Li W., Zou J., Wu K. Genomic diversity dynamics in conserved chicken populations are revealed by genome-wide SNPs. BMC Genomics. 2018;19:598. doi: 10.1186/s12864-018-4973-6. [DOI] [PMC free article] [PubMed] [Google Scholar]