Abstract
Background & Aims
Chronic inflammation in colon section is associated with an increased risk of colorectal cancer (CRC). Proinflammatory cytokines were produced in a tumor microenvironment and correlated with poor clinical outcome. Tumor-infiltrating T cells were reported to be greatly involved in the development of colon cancer. In this study, we demonstrated that kynurenine (Kyn), a metabolite catalyzed by indoleamine 2,3-dioxygenase (IDO), was required for IDO-mediated T cell function, and adaptive immunity indeed played a critical role in CRC.
Methods
Supernatant of colon cancer cells was used to culture activated T cells and mice spleen lymphocytes, and the IDO1-Kyn-aryl hydrocarbon (AhR) receptor axis was determined in vitro. In vivo, an azoxymethane (AOM)/dextran sodium sulfate (DSS)–induced CRC model was established in IDO–/–, Rag1–/–, and wild-type mice, and tumor-associated T lymphocyte infiltration and Kyn/AhR signaling pathway changes were measured in each group.
Results
Kyn promoted AhR nuclear translocation increased the transcription of Foxp3, a marker of regulatory T cells (Tregs), through improving the interaction between AhR and Foxp3 promoter. Additionally, compared WT mice, IDO–/– mice treated with AOM/DSS exhibited fewer and smaller tumor burdens in the colon, with less Treg and more CD8+ T cells infiltration, while Kyn administration abolished this regulation. Rag1–/– mice were more sensitive to AOM/DSS-induced colitis-associated colon cancer (CRC) compared with the wild-type mice, suggesting that T cell–mediated adaptive immunity indeed played a critical role in CRC.
Conclusions
We demonstrated that inhibition of IDO diminished Kyn/AhR-mediated Treg differentiation and could be an effective strategy for the prevention and treatment of inflammation-related colon cancer.
Keywords: IDO, Kyn, AhR, Treg, Colitis-Associated Colon Cancer
Abbreviations used in this paper: 1-MT, 1-methyltryptophan; 1-L-MT, 1-methyl-L-tryptophan; AhR, aryl hydrocarbon receptor; AOM, azoxymethane; BSA, bovine serum albumin; ChIP, chromatin immunoprecipitation; CM_, conditional medium; CRC, colorectal cancer; DSS, dextran sodium sulfate; FBS, fetal bovine serum; GZB, granzyme B; IDO, indoleamine 2,3-dioxygenase; INCB, epacadostat; Kyn, kynurenine; PBS, phosphate-buffered saline; PCR, polymerase chain reaction; PI, propidium iodide; siRNA, small interfering RNA; TIL, tumor-infiltrating lymphocyte; Treg, regulatory T cell; Trp, tryptophan; WT, wild-type
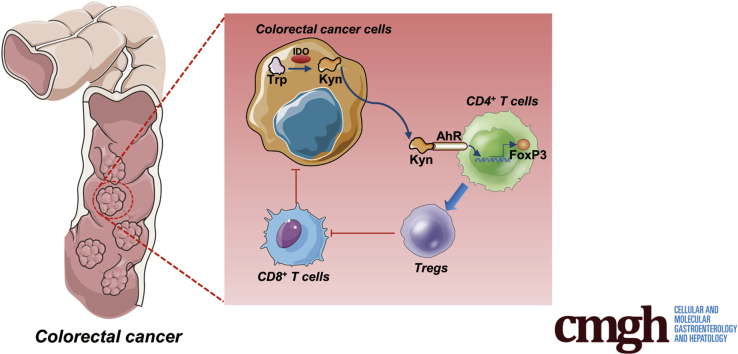
Graphical abstract
Summary.
Proinflammatory cytokines produced in tumor microenvironment resulted in eradication of anti-tumor immunity and enhanced tumor cell survival. Kynurenine (Kyn) was required for IDO-mediated T cells function via aryl hydrocarbon receptor (AhR)/Foxp3. Additionally, T cell–mediated adaptive immunity indeed played a critical role in CRC progression. Inhibition of IDO could be an effective strategy for the prevention and treatment of inflammation-related CRC.
Colorectal cancer (CRC) is one of the most common cancers and remains the second-leading cause of cancer-related mortality in the Western worl.1 It has been associated with gene mutations, unhealthy eating habits, chronic intestinal inflammation, and changes in the gut microbiota.2,3 Indeed, chronic inflammation in the colon increases the risk for CRC4 and correlates with poor clinical outcomes.5 In addition, on the one hand, the milieu of inflammatory microenvironment might enhance the development of T cell resistance by the cancer cells.6 On the other hand, tumor-infiltrating T cells, a vital component of adaptive immunity, have been shown to mediate the development of colon cancer.1,7,8 Therefore, it is feasible to explore the interaction between colon cancer cells and immune response, which could contribute to the development of more effective treatment options for inflammation-associated cancers.
Indoleamine 2,3-dioxygenase (IDO) mediates the development and progression of many cancer types.9, 10, 11, 12 IDO catalyzes the cleavage of tryptophan (Trp) in the rate-limiting step of the kynurenine (Kyn) pathway, resulting in the depletion of Trp and production of immunoregulatory molecules, including Kyn.13, 14, 15, 16 The IDO-induced production of Kyn and other metabolites in the tumor microenvironment could suppress the proliferation of functional T cells and natural killer cells while promoting the differentiation and activation of FoxP3+ (forkhead box P3+) regulatory T cells (Tregs).17,18 Moreover, IDO activation correlates with poor clinical outcomes in patients with endometrial carcinoma, ovarian carcinoma and CRC.19, 20, 21 Furthermore, many IDO inhibitors such as 1-methyltryptophan (1-MT) and epigallocatechin gallate reduced the occurrence of preneoplastic lesions in colon section.22,23 Although studies have reported IDO activation in CRC, the regulatory mechanism of the IDO in the progression of CRC remains unknown.
Tregs, which continuously express the transcription factor Foxp3, have been associated with immunological tolerance.24,25 They inhibit immune responses by releasing many anti-inflammatory cytokines, including transforming growth factor beta and interleukin-10.26 In addition, Tregs confer cytotoxicity against other T cells. In particular, activated CD4+ and CD8+ T cells were demonstrated to be preferentially attacked by Tregs, thereby protecting the tumor against immune surveillance.27, 28, 29 It has been shown that there is almost a 2-fold upregulation of Tregs in colonic tumors compared with the normal colonic mucosa, and the differentiation of Tregs from CD4+ T cells could worsen CRC.30 However, the mechanisms behind the activation of IDO and IDO-mediated T cell activities in CRC are not well established.
In our previous study, we found that the antiproliferation effect of 1-methyl-L-tryptophan (1-L-MT) mediated by IDO-induced cell cycle disaster in colon cancer cells and identified 1-L-MT as a promising candidate for the chemoprevention of CRC.31 We speculate that in addition to accelerates tumor cell proliferation, IDO activation in tumors may also mediate tumoral immune tolerance. Here, we evaluate and report that both the expression and activation of IDO were significantly induced by inflammatory microenvironment in an azoxymethane (AOM)/dextran sodium sulfate (DSS)–induced CRC model. Compared with wild-type (WT) mice, IDO–/– mice were less sensitive to AOM/DSS, with fewer and smaller tumors in the colon section. Both in vivo and in vitro assays attested that Kyn played a crucial role in the regulation of CD4+ T cell functions through the aryl hydrocarbon receptor (AhR)/FoxP3 pathway. Taken together, our data demonstrate the effect of chronic inflammation on IDO and IDO-induced immunosuppression and highlight IDO inhibition as a potential therapeutic option for colon cancer.
Results
IDO Activation Inhibited the Proliferation of Activated T Cells
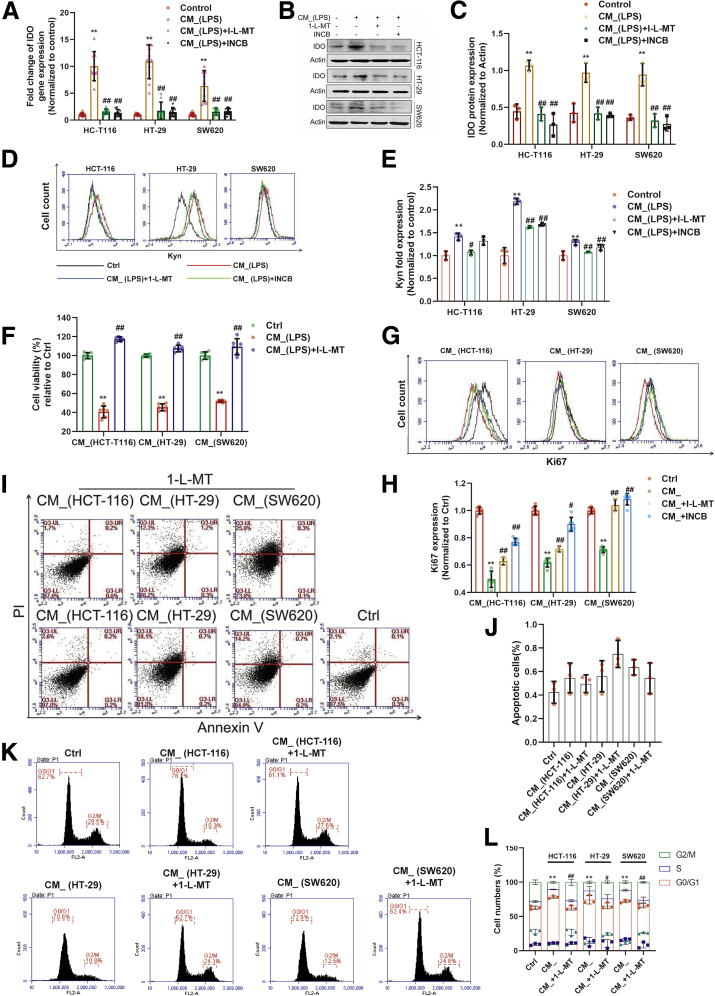
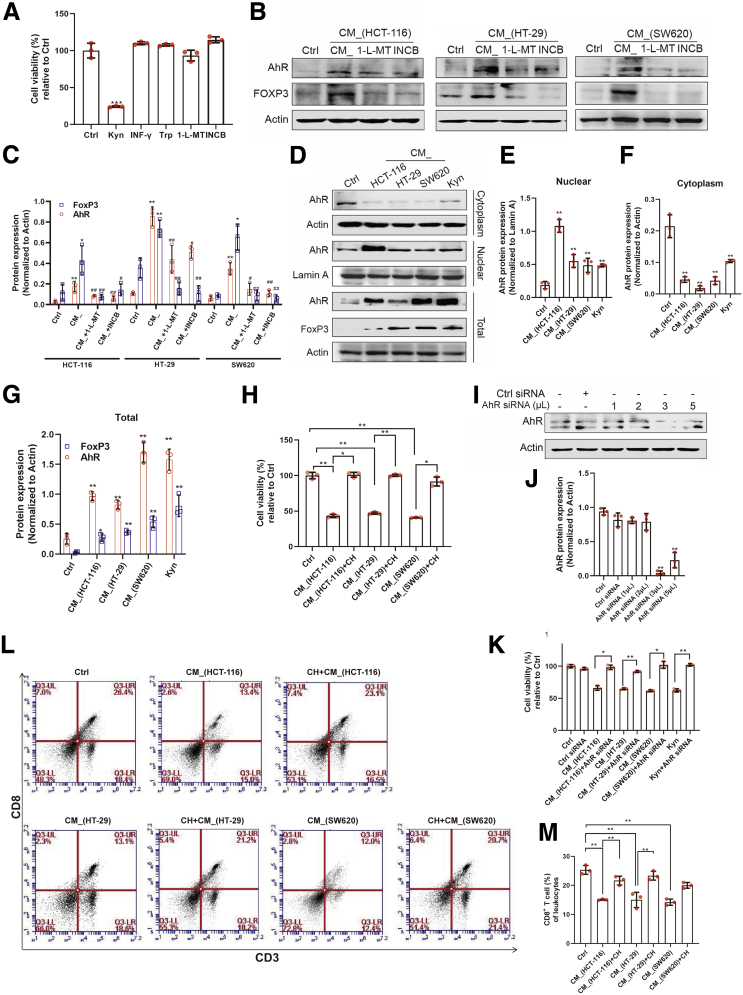
We explored whether the expression of IDO in colon cancer cells is involved in the regulation of T cell functions. Two IDO inhibitors, 1-L-MT (1 mM) and epacadostat (10 μM) (INCB024360) (INCB), were used to inhibit both the expression (Figure 1A–C) and activation (Figure 1D and E) of IDO in HCT-116, HT-29, or SW620 cells. We then cultured activated T cells (Jurkat cell line) with the conditional medium (CM_) from colon cancer cells for 24 hours. The results showed that CM_ stimulation significantly suppressed the proliferation of Jurkat cells, while 1-L-MT abolished this inhibition (Figure 1F–H). Notably, there was no detectable apoptosis of the activated T cells after incubation of CM_ (Figure 1I and J), while cell cycle distribution of the Jurkat cells showed a remarkable CM_-specific G0/G1 phase arrest (Figure 1K and L), which was rescued by 1-L-MT. Taken together, these results demonstrated that IDO activation inhibits T cells proliferation by blocking cell cycle progression.
Figure 1.
IDO activation in colon cancer cells inhibited proliferation of activated T cells. (A–C) Lipopolysaccharide (2 μg/mL) stimulated THP-1 for 24 hours, and the supernatant was collected. (A) HCT-116, HT-29, and SW620 cells were incubated with such supernatant with or without the stimulation of 1-L-MT or INCB for 24 hours, and the gene of IDO expression was determined (n = 9). (B, C) The protein of IDO expression was determined by Western blot (n = 3). (D, E) The synthesis of Kyn was measured by flow cytometry. (F) The supernatant of lipopolysaccharide-stimulated THP-1 was incubated with HCT-116 (n = 3), HT-29 (n = 3), and SW620 (n = 3) cells with or without 1-L-MT or INCB for 24 hours, and then the supernatant (CM_) was collected. Jurkat cell lines (activated T cells) were stimulated with such CM_ for 24 hours. The cell viability was determined by CCK8. (G, H) Ki67 expression of Jurkat cells was detected by flow cytometry. (I, J) Activated T cells exposed to previous stimulation were analyzed for the extent of apoptosis by annexin V–tagged FITC-PI flow cytometry (n = 3). (K, L) Cell cycle distribution was measured using flow cytometry. The percentage of cells in each population are shown as the mean ± SD (n = 3). ∗P < .05, ∗∗P < .01 with control group. #P < .05, ##P < .01 with CM_ or CM_(lipopolysaccharide) group. The results were performed at least in triplicate and expressed as the mean ± SD. n.s, no significant difference.
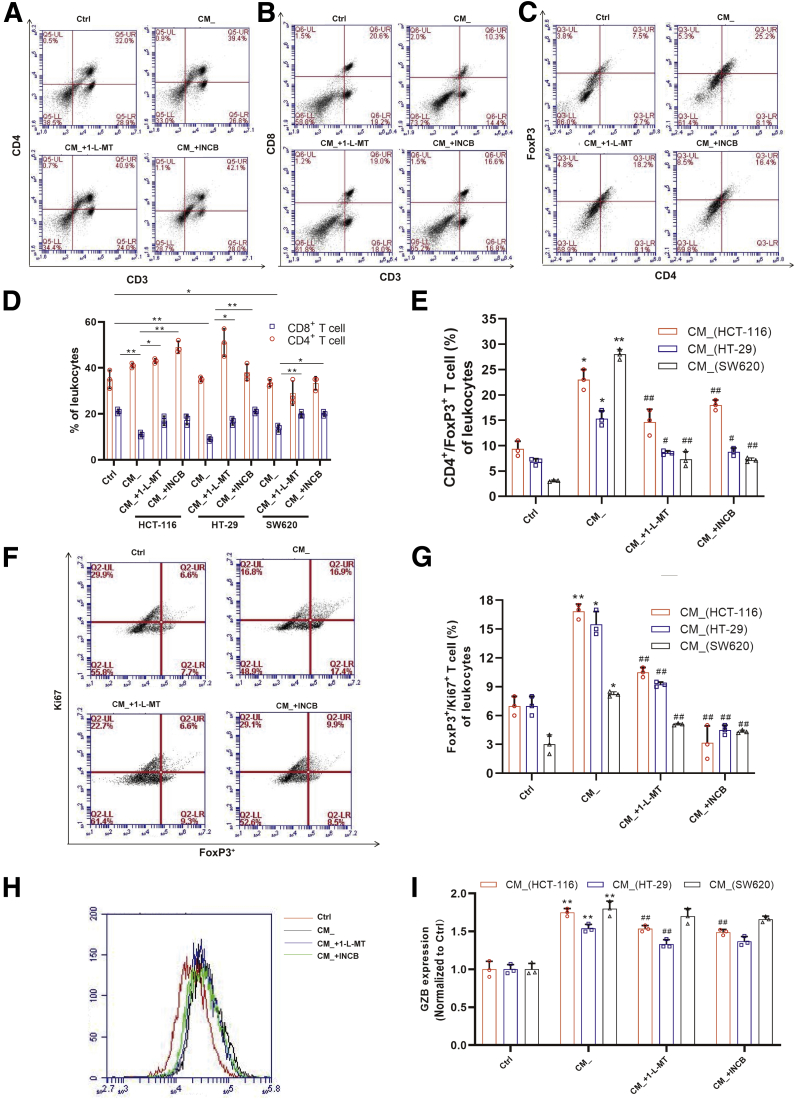
Because Tregs are involved in tumor-induced immune suppression, we hypothesized that tumor cells with higher IDO expression may possess immune regulatory functions, thus conferring self-defense from immune attack and promoting tumor progression. To determine whether the inflammatory microenvironment-induced IDO activation in colon cancer cells could regulate the differentiation of CD4+ T cells, we used the CM_ to culture mice spleen lymphocyte. On the one hand, after incubation for 24 hours, the number of CD4+ T cells was not affected (Figure 2A and D). On the other hand, there was a significant decrease (~10%) in CD8+ T cells, which was restored by both 1-L-MT or INCB (Figure 2A, B, and D). To ascertain whether IDO activation affected Tregs differentiation, spleen lymphocytes were stimulated with CM_ and then stained with FoxP3 or CD4. The data showed upregulation of Tregs following IDO activation, which was reversed by 1-L-MT and INCB (Figure 2C–E). The change of Ki67 expression in Tregs proved that the effect of IDO on T cell transdifferentiation was at least partly via the induction of Treg proliferation (Figure 2F and G). Upon CM_ stimulation, the expression of granzyme B (GZB) in FoxP3+ T cells exhibited a distinct upregulation (Figure 2H and I), which was then blocked by 1-L-MT or INCB treatment. These data demonstrated that IDO activation suppressed CD8+ T cell response via stimulating of the proliferation of Tregs.
Figure 2.
Inflammatory microenvironment–induced IDO activation promoted CD4+T cell transdifferentiation into Tregs. (A) Mice spleen lymphocytes were exposed to the supernatant of colon cancer cells for 24 hours. The percentage of CD3+/CD4+ T cells was measured. (B, C) The percent of (B) CD3+/CD8+ T cells and (C) Tregs was detected (n = 3). (D) The distributions of CD4+ and CD8+ T cells were determined by representative fluorescence-activated cell sorter blots (n = 3). (E) The percentage of Tregs in each population. (F, G) The expression of Ki67 in Tregs was determined by flow cytometry (n = 3). (H, I). The expression of GZB in Tregs was detected (n = 3). The results are representative of 3 independent experiments and are expressed as the mean ± SD. ∗P < .05, ∗∗P < .01 compared with the control, #P < .05, ##P < .01 compared with CM_. n.s, no significant difference.
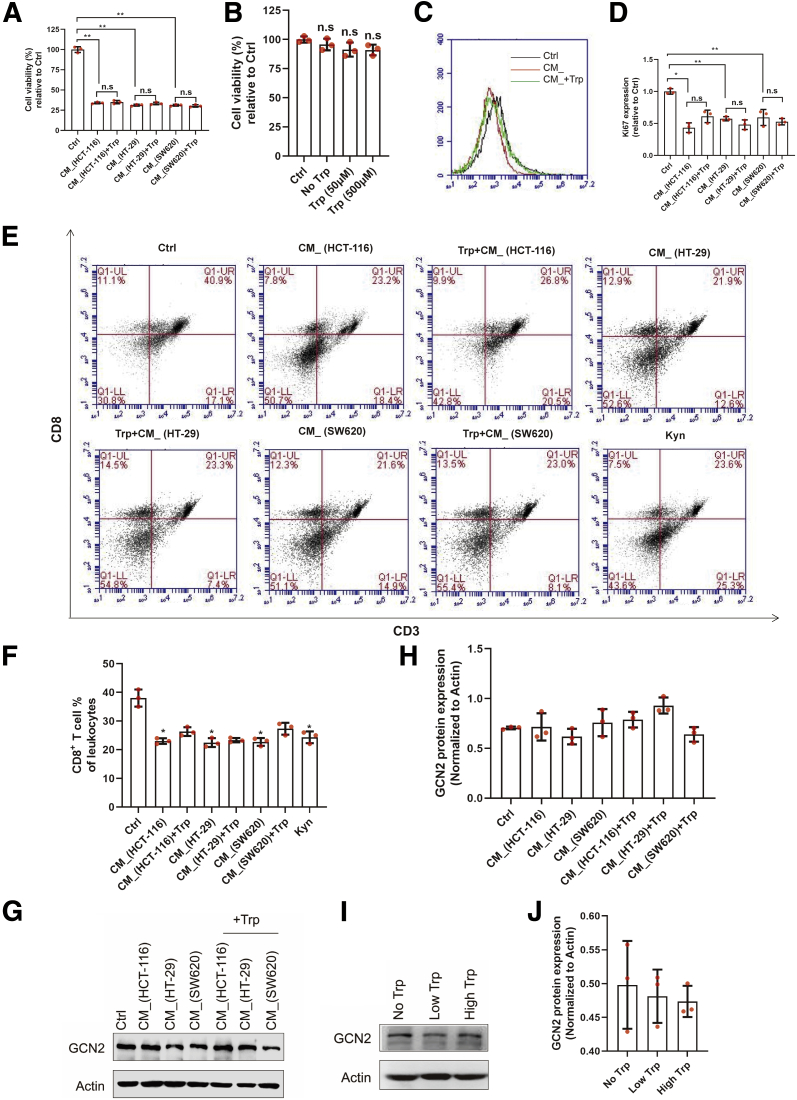
Previous reports have shown that IDO activation not only produces kynurenine-based metabolites, but also enhances localized depletion of tryptophan (Trp). The shortage of Trp was reported to induce activated T cell apoptosis and the differentiation of Tregs. To further interrogate the role of Trp in IDO-mediated immunosuppression, cells exposed to the previously mentioned stimulations were incubated with Trp. To our surprise, compared with CM_ incubation, Trp did not increase the viability of the activated T cells (Figure 3A). Moreover, Trp-deficient media did not affect the proliferation of Jurkat cells, compared with media with low or high concentration of Trp (Figure 3B). Additionally, supplementation with Trp did not rescue the decreased expression of Ki67 in the CM_-induced activated T cells (Figure 3C and D). Similarly, when mice spleen lymphocytes were exposed to CM_, there was significant suppression of CD8+ T cells (Figure 3E and F), which could not be rescued by increased concentrations of Trp, whereas it has been shown that GCN2 (general control nonderepressible 2) kinase is sensitive to Trp deficiency and is essential in the IDO-mediated immunomodulation. Here, we found that neither CM_ nor Trp changed the GCN2 expression (Figure 3G–J). Therefore, our data excluded the role of Trp starvation in IDO-mediated T cell function.
Figure 3.
The change of Trp hardly affected IDO-mediated T cell function. (A) CM_-stimulated activated T cells were exposed to Trp for 24 hours. The cell viability was determined by CCK8 (n = 3). (B) Jurkat cells were cultured in the medium with no, low (50 μM), or high (500 μM) Trp concentration (n = 3). Cell viability was determined by CCK8. (C, D) Ki67 expression of activated T cells, which was stimulated by CM_ and additional Trp, was measured by flow cytometry (n = 3). (E, F) Mice spleen lymphocytes were exposed to the supernatant of colon cancer cells and Trp for 24 hours (n = 3). Percentage of CD3+/CD8+ T cells was measured. (G–J) The GCN2 expression of activated T cells with previous stimulation was analyzed by Western blot (n = 3). The results are representative of 3 independent experiments and are expressed as the mean ± SD. ∗P < .05, ∗∗P < .01 indicate significant difference. n.s, no significant difference.
Kyn/AhR Is Essential for IDO-Mediated Regulation of T Cells and Promotes FoxP3 Transcription
To evaluate whether Kyn modulates the functions of CD4+ T cells, we stimulated Jurkat cells with interferon gamma, Trp, or Kyn and found that only Kyn significantly inhibited the viability of the activated T cells (Figure 4A). On the one hand, the CM_ remarkably enhanced the expression for AhR protein, a major receptor of Kyn (Figure 4B and C). Besides, the expression of FoxP3, one of the AhR target genes, was also upregulated. On the other hand, CM_ containing 1-L-MT or INCB decreased the expression of both AhR and FoxP3 proteins. Furthermore, Kyn at 50 μM promoted the nuclear translocation of AhR, similar to the observations made with the CM_ incubation (Figure 4D–G). The CM_ incubation increased both the expression and activation of AhR. To evaluate whether AhR was required for the regulation of Kyn, activated T cells were incubated with CM_ after 1-hour pretreatment with an AhR inhibitor, CH-223191, and it was showed that the inhibitory effects of CM_ on the proliferation of Jurkat cells were compromised (Figure 4H). The AhR knockdown in the activated T cells rescued the CM_-inhibited proliferation (Figure 4I–K). In addition, CH-223191 pretreatment resulted in an increase in CD8+ T cells, which was suppressed by CM_ incubation (Figure 4L and M). Taken together, the findings demonstrate that the modulatory effect of CM_ on T cells required AhR activation.
Figure 4.
Kyn/AhR was essential for IDO-mediated regulation of T cells. (A) Jurkat cells were exposed to the same dose (500 μM) of Kyn, INF-γ, Trp, 1-L-MT(1 mM), and INCB (10 μM) for 24 hours (n = 3). Cell viability was measured by CCK8. (B, C) The protein levels of AhR and FoxP3 in activated T cells after incubation with different CM_ were detected. (D–G) Jurkat cells were treated with CM_ or Kyn for 24 hours, and the protein level of AhR and its nuclear translocation were determined by Western blot (n = 3). (H) CM_-pretreated activated T cells were stimulated with CH-223191 (10 μM) for 24 hours, and cell viability was measured (n = 3). (I, J) Western blot analysis of AhR expression in Jurkat cells transfected with different concentrations of AhR siRNA for 24 hours. (K) AhR siRNA-transfected activated T cells were stimulated with CM_ for the indicated periods. Cell viability was determined by CCK8 (n = 3). (L, M) Mice spleen lymphocytes were cultured in the supernatant of colon cancer cells and CH-223191 (10 μM) for 24 hours. Percentage of CD3+CD8+ T cells was measured (n = 3). The results are representative of 3 independent experiments and are expressed as the mean ± SD. ∗P < .05, ∗∗P < .01 compared with the control, #P < .05, ##P < .01 compared with CM_ (A, C, E, F, G, and J). ∗P < .05, ∗∗P < .01 indicates significant difference (H, M).
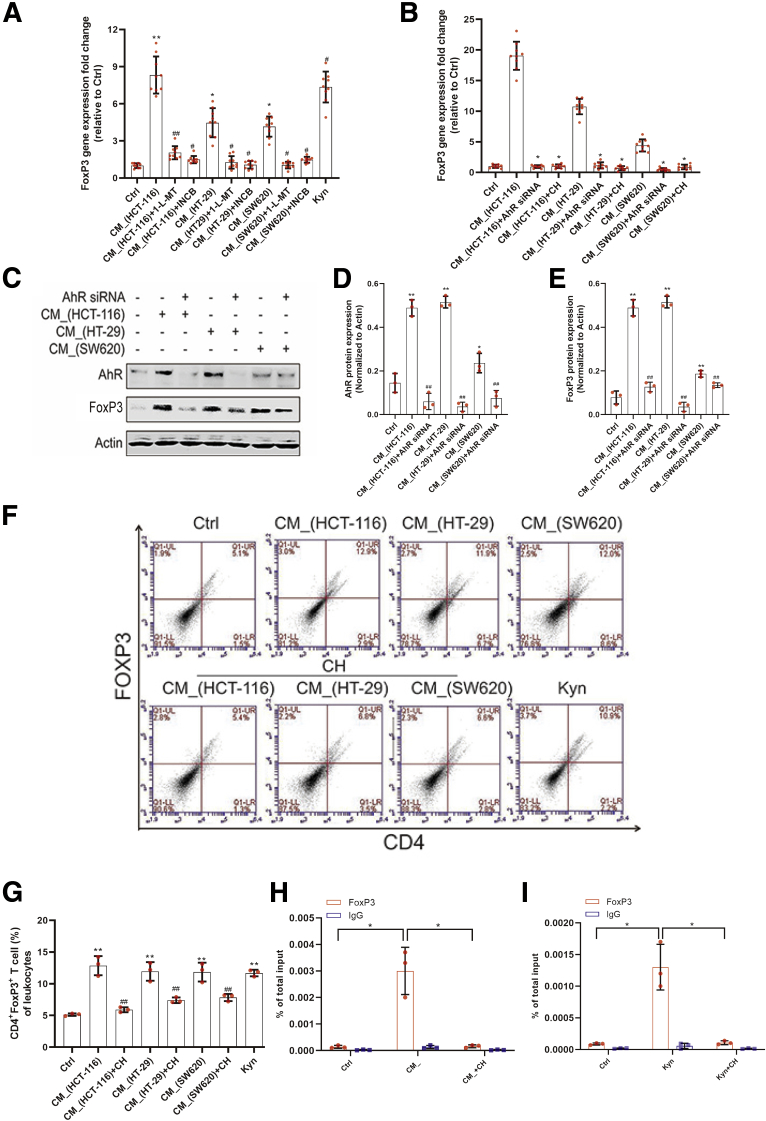
Based on the fact that IDO activation inhibited the number of CD8+ T cells but not the number of CD4+ T cells, we hypothesized that CD4+ T cells, including naive and activated T cells, may be differentiating into Tregs. Previous studies have shown that, upon stimulation, naive T cells could transform into functional FoxP3+ Tregs. Here, we investigated whether colon cancer cell expressed IDO could affect FoxP3 in activated T cells. Transcript analysis at 24 hours of different CM_ incubation revealed that the supernatant of HCT-116, HT-29, or SW620 cells upregulated the expression FoxP3, with CM_ induced FoxP3-encoding genes to a relatively higher extent than Kyn did (Figure 5A), and the increased FoxP3 expression was blocked by the IDO inhibitors. Both pharmacologic and genetic means of AhR inhibition confirmed that CM_-induced FoxP3 expression was dependent on the Kyn/AhR signaling (Figure 5B). Furthermore, FoxP3 protein expression was decreased when activated T cells were transfected with AhR small interfering RNA (siRNA) (Figure 5C–E). The CM_-enhanced levels of Tregs were compromised by the treatment of AhR inhibitor (Figure 5F and G). To determine how Kyn-enriched CM_ contributed to FoxP3 transcription, chromatin immunoprecipitation (ChIP) assay was performed, and it was demonstrated that CM_ and Kyn enhanced the interaction between AhR and FoxP3 promoter (Figure 5H and I). The results suggested that CM_ upregulated the expression of FoxP3 via AhR nuclear translocation–mediated FoxP3 transcription.
Figure 5.
Kyn-mediated AhR activation promoted FoxP3 transcription. (A) Jurkat cells were treated with different CM_ or Kyn (500 μM) for 24 hours, and the messenger RNA level of FoxP3 was detected by real time PCR (n = 9). (B) AhR siRNA-transfected or CH-223191 (10 μM) (CH)–pretreated Jurkat cells were stimulated with different CM_, and the FoxP3 messenger RNA level was determined by real-time PCR (n = 9). (C–E) AhR siRNA-transfected Jurkat cells were stimulated with CM for 24 hours and FoxP3 expression was detected by Western blot (n = 3). (F, G) Mice spleen lymphocytes were cultured in the supernatant of colon cancer cells and CH-223191 (10 μM) for 24 hours (n = 3). The percentage of Tregs was measured. (H, I) ChIP assay was performed to illustrate the interaction between AhR and FoxP3 promoter (n = 3). The results were performed at least in triplicate and expressed as the mean ± SD. ∗P < .05, ∗∗P < .01 compared with the control, #P < .05, ##P < .01 compared with CM_ (A, B, D, E, G). ∗P < .05, ∗∗P < .01 indicates significant difference (H, I).∗P < .05, ∗∗P < .01 indicates significant difference.
IDO/Kyn Played a Vital Role in Chronic Colitis-Associated Cancer via Regulation of T Cells
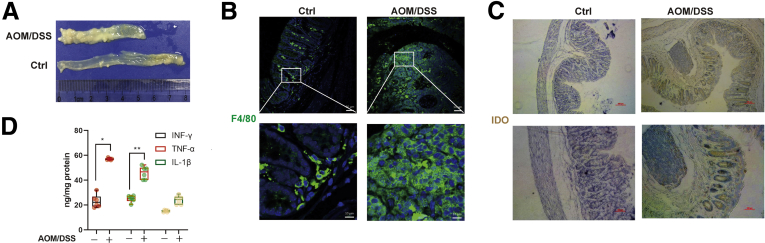
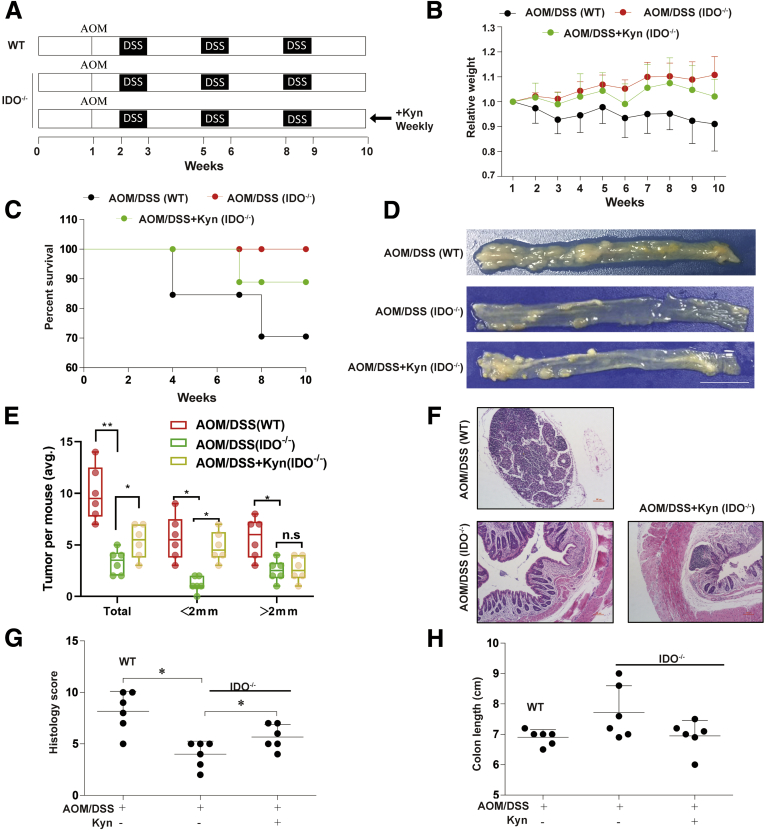
We established the AOM/DSS-induced CRC mouse model to validate our in vitro observations (Figure 6A). The expression of F4/80 was significantly upregulated in the AOM/DSS group (Figure 6B), indicating that there was increased inflammatory cell infiltration including macrophage. Similarly, IDO expression was enhanced after AOM/DSS treatment, together with interferon gamma, interleukin 1β, and tumor necrosis factor alpha (Figure 6C and D) levels, indicating that the inflammatory microenvironment was closely related to CRC progression. An AOM/DSS-induced colitis-associated colon cancer model was also established in WT and IDO–/– mice (Figure 7A). Based on Kaplan-Meier survival curves, IDO–/– mice exhibited increased survival rate, compared with the WT group, while the administration of Kyn (5 mg/wk) led to a higher mortality rate in IDO–/– mice (Figure 7C). In addition, IDO–/– mice had reduced weight loss as compared with the WT and Kyn treatment groups (Figure 7B). After sacrificing the animals, colon length, tumor size, and histology scores were obtained, and we found that IDO–/– mice harbored fewer and smaller tumors compared with the WT group, while the Kyn treatment group had opposite effects (Figure 7D and E). In addition, lower average histology score was seen in IDO–/– mice, in comparison with both WT and Kyn groups (Figure 7F and G). However, there was no difference in colon length in among all the groups (Figure 7H). Taken together, these results indicated that IDO/Kyn played an important role in the AOM/DSS CRC cancer model.
Figure 6.
The AOM/DSS-induced CRC mouse model was established. (A) Number of polyps per mouse in the control and AOM/DSS groups. (B) Serial sections of colon tissues were immunostained with DAPI (blue) and F4/80-FITC (green). They were observed by confocal laser scanning microscope (scale bar = 50 μm, 10 μm). (C) Expression of IDO was analyzed by immunochemical staining of paraffin-embedded colon sections (scale bar = 100 μm). (D) INF-γ, TNF-α, and IL-1β in colonic tissue were determined by enzyme-linked immunosorbent assay (n = 6). The results are representative of at least 6 independent experiments and expressed as mean ± SD. ∗P < .05, ∗∗P < .01 indicates significant difference.
Figure 7.
IDO/Kyn played a vital role in chronic colitis-associated cancer. (A) Induction procedure and groups designed for the AOM/DSS model of chronic colitis-associated cancer (n = 10). (B) Body weight. (C) Survival rate. (D) Number of polyps per mouse. (E) Polyp size (n = 6). (F) Hematoxylin and eosin staining (scale bar = 100 μm). (G) Average Histology Score of colons (n = 6). (H) Colon length (n = 6). The results are representative of at least 6 independent experiments and expressed as mean ± SD. ∗P < .05, ∗∗P < .01 indicates significant difference.
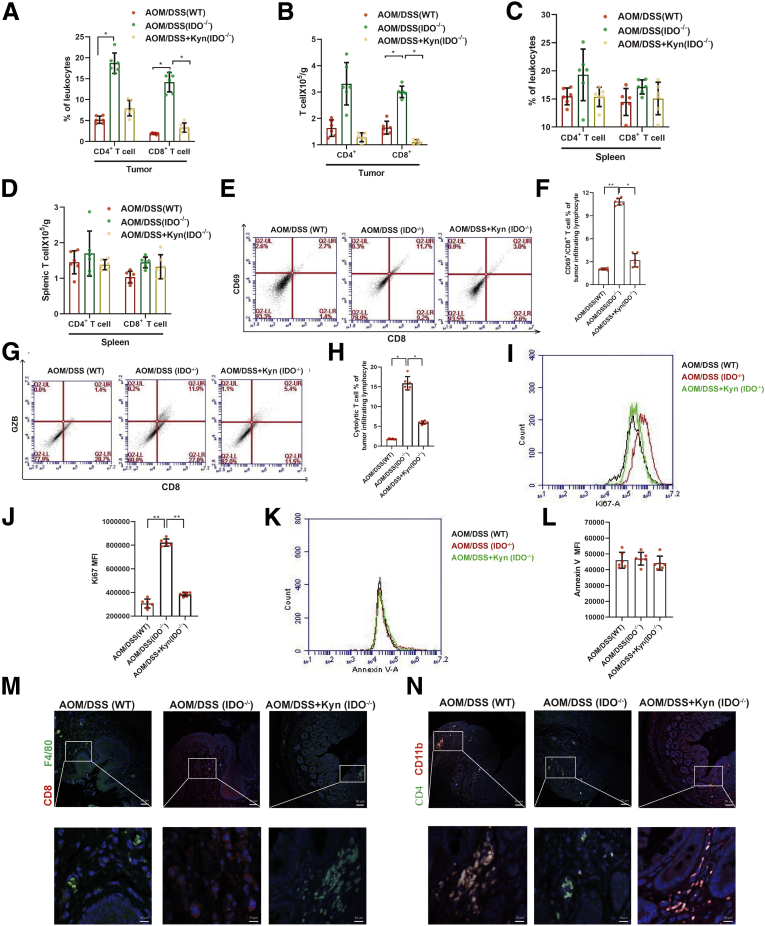
IDO-Deficient Mice Had Increased CD8+ T Cell Infiltration in the Colon
To evaluate the role of IDO/Kyn in adaptive immune response during CRC, we examined the shift on the number of CD8+ T cells in the colon section and spleen. The data showed increased number of CD8+ T cells in the colon sections of the IDO–/– mice but not in the WT mice (Figure 8A and B), while administration of Kyn abolished this enhancement. However, neither IDO nor Kyn affected the number of CD8+ T cells in the spleen (Figure 8C and D). Compared with the WT mice, activation detected by CD69 expression and cytolytic capacity measured by GZB expression of CD8+ T cells were significantly improved in the IDO–/– mice, and Kyn reversed these effects (Figure 8E–H). Besides, the IDO–/– mice showed a dramatic proliferation in the CD8+ T cells, as well as an upregulation of Ki67 in CD8+ T cells, compared with the WT mice (Figure 8I and J), and Kyn reversed this increment. Neither IDO nor Kyn affected apoptosis of the CD8+ T cells (Figure 8K and L). In addition, immunofluorescence analysis revealed that CD8+ and CD4+ T cells diffusely infiltrated the tumor in the IDO–/– mice, but not in the WT or Kyn group (Figure 8M and N). Unlike in the IDO–/– group, inflammatory cells that expressed F4/80 or CD11b were significantly increased in the WT and Kyn mice (Figure 8M and N). These results suggested that the IDO/Kyn axis inhibited CD8+ T cells infiltration in the colon, which contributed to the development of CRC.
Figure 8.
Absence of IDO contributed to increased CD8+T infiltration in colon. (A) Absolute numbers of CD4+ and CD8+ T cells infiltrated in tumor were measured by flow cytometry (n = 6). (B) The percentage of CD4+ and CD8+ T cells in spleen lymphocytes was analyzed by flow cytometry (n = 6). (C, D) Absolute numbers of CD4+ and CD8+ T cells in spleen lymphocytes were detected by flow cytometry (n = 6). (E, F) The frequency of intratumoral CD8+/ CD69+ T cells was determined (n = 6). (G, H) The percentage of CD8+ GZB+ T cells was detected (n = 6). (I, J) The expression of Ki67 and its mean fluorescence intensity (MFI) of CD8+ T were shown (n = 6). (K, L) Annexin V expression on intratumoral CD8+ T cells and its mean fluorescence intensity were determined by flow cytometry (n = 6). (M, N) Serial sections of colon tissues were immunostained with DAPI (blue), anti F4/80 (green), and CD8 (red). They were observed by confocal laser scanning microscope (scale bar = 50 μm, 10 μm). Expressions of CD11b (red) and CD4 (green) were analyzed by immunofluorescence cytochemistry (scale bar = 50 μm, 10 μm). The results are representative of at least 6 independent experiments and expressed as mean ± SD. ∗P < .05, ∗∗P < .01 indicates significant difference.
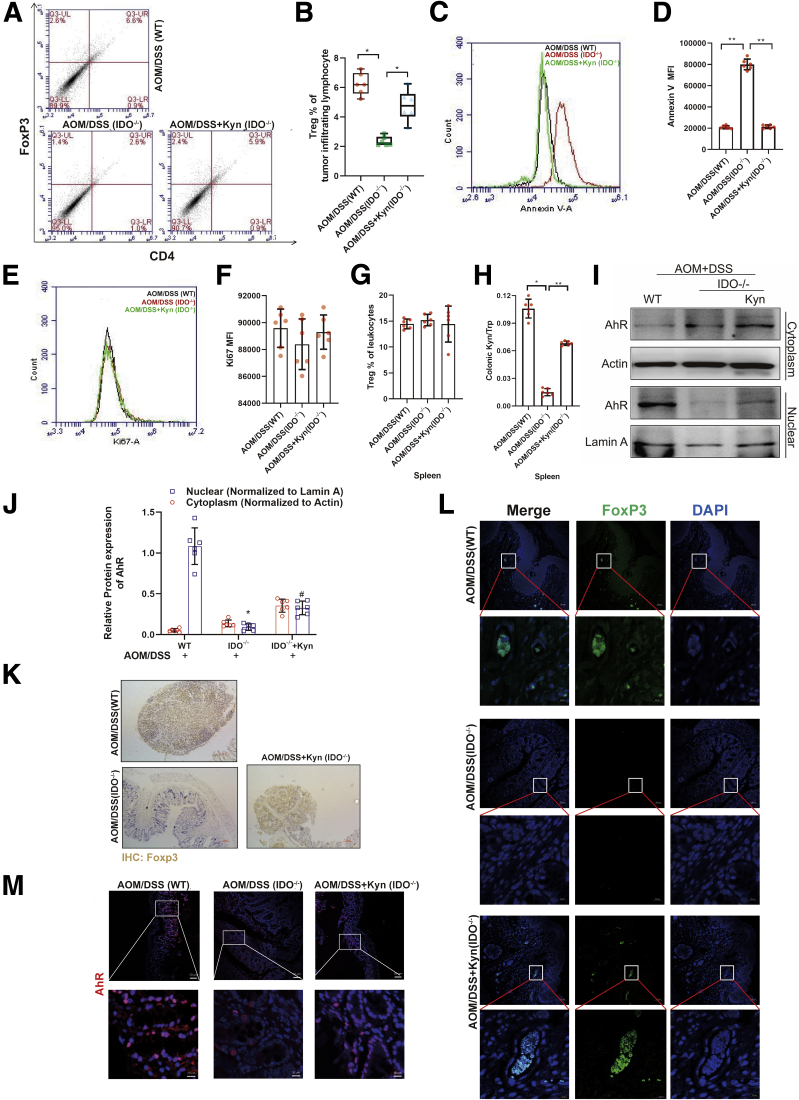
IDO/Kyn/AhR Regulates Treg Differentiation in CRC
We next analyzed whether Tregs were altered by the IDO/Kyn/AhR axis, as they played a key role in regulating the anti-tumor immune responses. The results showed that IDO deficiency suppressed the number of Tregs in the tumors compared with the WT, which was abolished by Kyn administration (Figure 9A and B). There was no significant difference in the number of Tregs in the spleen among 3 groups (Figure 7G). Consistent with these findings, apoptosis of Tregs was found in the tumors in the IDO–/– mice but not in WT- or Kyn-treated ones (Figure 9C and D). However, there was no difference in the expression of Ki67 in Tregs among all the groups, indicating that neither IDO nor Kyn affected the proliferation of Tregs in the colon (Figure 9E and F). In addition, on the one hand, AOM/DSS significantly increased the Kyn-to-Trp ratio in the colon of WT mice, while IDO–/– mice exhibited a lower ratio (Figure 9H). On the other hand, Western blot analysis showed accumulation of AhR in the nucleus of the tumor-infiltrating lymphocyte (TIL) in the WT and Kyn groups (Figure 9I and J). Besides, immunohistochemical and immunofluorescence staining demonstrated that high levels of FoxP3 were induced by AOM/DSS and substantially blocked by IDO deficiency, which was compromised by Kyn treatment (Figure 9K and L). The immunofluorescence assay showed that there was high AhR expression in the WT and Kyn groups (Figure 9M). Thus, our results suggested that IDO/Kyn/AhR regulated Treg function in CRC.
Figure 9.
IDO/Kyn/AhR regulated Treg differentiation in CRC. (A, B) Representative frequency of intratumoral Tregs was detected by flow cytometry (n = 6). (C, D) Annexin V expression on intratumoral Tregs and its mean fluorescence intensity were determined by flow cytometry (n = 6). (E, F) The expression of Ki67 and its mean fluorescence intensity of Tregs were shown (n = 6). (G) The percentage of Tregs in spleen lymphocytes was analyzed (n = 6). (H) Kyn-to-Trp ratio in the colon section was measured by enzyme-linked immunosorbent assay (n = 6). (I, J) The expressions of cytoplastic protein AhR and nuclear protein AhR in TIL were analyzed by Western blot (n = 6). (K) Expression of FoxP3 was detected by immunochemical staining of paraffin-embedded colon sections (scale bar = 100 μm). (L) Expression of FoxP3 was detected by immunofluorescent staining of paraffin-embedded colon sections (scale bar = 50 μm, 10 μm). (M) Expression of AhR was analyzed by immunofluorescence cytochemistry (scale bar = 50 μm, 10 μm). The results are representative of at least 6 independent experiments and expressed as mean ± SD. ∗P < .05, ∗∗P < .01 indicates significant difference.
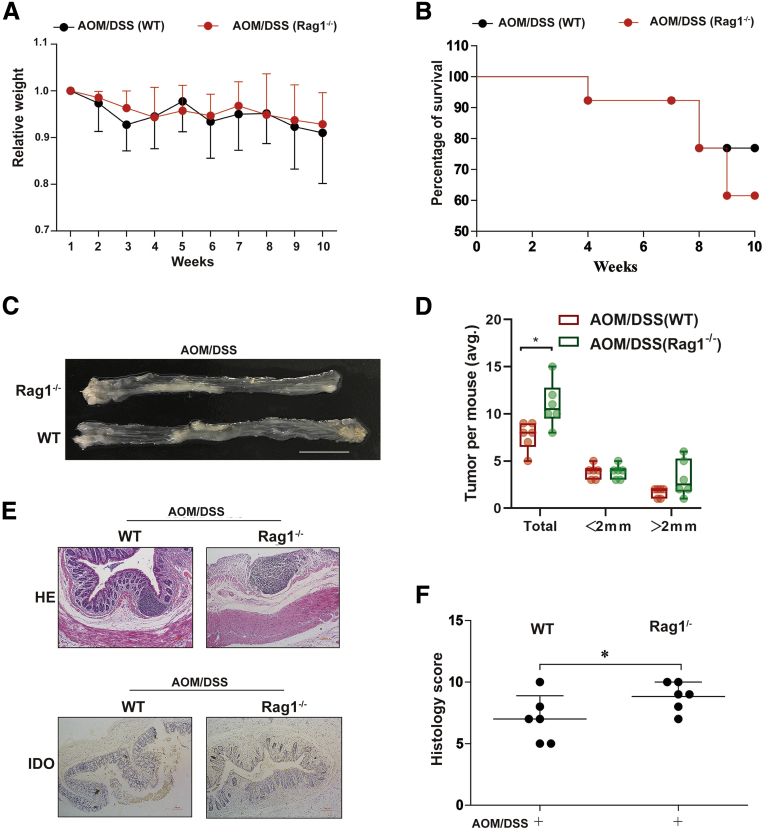
T Cell–Mediated Microenvironment Affects the Development of CRC
To further ascertain the role of the T cells in CRC, we established an AOM/DSS model in Rag1–/– mice, which lacked T cells. The transient weight loss after each cycle of DSS was similar in the WT and Rag1–/– groups (Figure 10A). However, higher mortality rate was observed in the Rag1–/– mice compared with the WT mice (Figure 10B). Rag1–/– mice were found to be more sensitive to AOM/DSS-induced CRC, harboring more tumors in the colon (Figure 10C and D). Microscopy and histology score also revealed more severe injuries in the colon section of Rag1–/– mice than in the WT mice (Figure 10E and F). Taken together, these results demonstrate that the T cell–mediated adaptive immunity was essential in the progression of CRC.
Figure 10.
A T cell–mediated microenvironment was required in the development of CRC. (A) AOM/DSS-induced CRC model was generated (n = 10). The relative weight was measured. (B) Survival rate. (C) Number of polyps per mouse. (D) Polyp size (n = 6). (E) Hematoxylin and eosin staining (scale bar = 100 μm). Expression of IDO was detected by immunochemical staining of paraffin-embedded colon sections (scale bar = 100 μm). (F) Average Histology Score of colons (n = 6). The results are representative of at least 6 independent experiments and expressed as mean ± SD. ∗P < .05, ∗∗P < .01 indicates significant difference.
Discussion
In our previous study, we determined that IDO inhibition suppressed tumor growth via reducing CDC20 transcription and demonstrated the chemopreventive effects of the IDO inhibitor 1-L-MT on CRC.31 However, we did not look at the role of IDO in immune tolerance. In this study, we demonstrated that the presence of multiple cytokines (CM_) enhanced IDO transcription, and the inflammatory microenvironment improved IDO activation. Consistent with our in vitro data, inflammatory cell infiltration and inflammatory cytokine secretion were detected in the colon section of the CRC model, with increased IDO expression.
We showed that the supernatant of colon cancer cells did not inhibit CD4+ T cells but significantly suppressed the levels of CD8+ T cells. Moreover, CM_ stimulation increased both the multiplication and proliferation of Tregs. Previous studies illustrated that IDO and its metabolites exhibited cytotoxicity against both the activated T cells and natural killer cells.32, 33, 34 However, annexin V staining showed no obvious apoptosis in the activated T cells when incubated with a supernatant rich in Kyn and other metabolites induced by IDO activation. Thus, we speculated that inflammation-mediated IDO activation in colon cancer cells might preferentially affect the differentiation rather than apoptosis of CD4+ T cells. It has been shown that naive CD4+ T cells could undergo alternative differentiation fates leading to the production of Tregs.35 Our study showed that whereas activated T cells did not undergo apoptosis, their proliferation was significantly suppressed. We hypothesized that activated T cells might undergo some form of stimulation and become Treg-like cells. As expected, on the one hand, both mRNA and protein levels of FoxP3 in the Jurkat cells were increased following stimulation of different CM_. On the other hand, CM_-induced Tregs or Tregs from other sources could mediate the proliferation of CD8+ T cells, which plays a pivotal role in anti-tumor treatment. However, in our in vivo study, apoptosis was observed in Tregs in the colon sections of IDO–/– mice, which was contrary to the in vitro observations, in which 1-L-MT and INCB did not cause apoptosis in any subtypes of T cells. This might be due to the incubation time of the IDO inhibitors. The stimulation for 24 hours may not be long enough to induce apoptosis. Furthermore, besides tumor cells, dendritic cells, macrophages, or endothelial cells also express IDO,36, 37, 38 which may take part in the regulation of IDO-mediated T cells function in the CRC model. Hence, the activation of IDO may exhibit more complicated biological effects in vivo than in vitro.
Trp metabolism and starvation in the cell microenvironment acted as a survival mechanism in phylogenetically ancient organisms. Yet, Trp consumption could modulate immune system.39 Many immune cells are sensitive to Trp starvation, which activates the stress-response kinase GCN2, whereas the activation of GCN2 in T cells decreases their proliferation and increases Tregs differentiation, which confers immune tolerance.40,41 However, the changes in Trp level did not affect the fate of T cells in our study. In addition, Trp did not inhibit the viability of Jurkat cells or CD8+ T cells. Besides, the addition of Trp did not rescue the inhibitory effects of IDO on T cells. Taken together, the ability of colon-specific IDO expression to locally suppress effector T cells was not dependent on metabolic depletion of Trp. As GCN2 had a direct impact on the phenotype of dendritic cells and macrophages,42,43 we speculated that antigen-presenting cells, rather than activated T cells, may be more sensitive to Trp starvation to change the local milieu from immunogenic to tolerogenic.
Because Trp consumption was not required for the effect of IDO on T cells, we investigated the potential role of Kyn. We did not observe any differences in inflammatory injuries between the Kyn treatment group and the IDO–/– group, which was similar to previous study.43,44 However, Kyn did re-establish and maintain immune tolerance in the colon section of IDO–/– mice. In addition, AhR, a Kyn receptor,45,46 regulates IDO-specific disease development. For example, AhR-deficient mice have been shown to be susceptible to endotoxin shock compared with the WT mice.47 Both pharmacologic and genetic AhR inhibition abolished IDO-induced changes in the differentiation of T cells. Besides, nuclear translocation of AhR increased the levels of FoxP3+ T cells both in vitro and in vivo. A previous study showed that AhR activation is involved in the development of Tregs, which boosted T helper 17 cell differentiation.48 Naive CD4+ T cells usually undergo a perforin/GZB-dependent apoptosis upon activation of AhR. Here, we showed that Kyn mainly enhanced the differentiation of activated T cells, with increased transcription of FoxP3. Besides, after stimulation with CM_, the number and proliferation of Tregs were enhanced. Whereas there was no apoptosis observed in Jurkat cells, there was a remarkable G0/G1 phase arrest induced by CM_ incubation.
As IDO–/– mice exhibited different levels of T cell subtypes in the colon, we established an AOM/DSS-induced CRC model in Rag1–/– mice. There was high mortality of Rag1–/– mice in the initial cycles of DSS, suggesting that T cells with adaptive immune response may protect against lesions in the colon. A previous study demonstrated that IDO inhibitors suppressed AOM-induced colonic preneoplastic lesions though modulating the immune microenvironment.22 Aberrant crypt foci served as putative precancerous lesions of the colon in experimental models, and it was showed that IDO could be a potential target in this process.49 In our study, depletion of T cells worsened the CRC, and Rag1–/– mice harbored more and larger tumors in the colon. Besides, T cell dysfunction would accelerate the development CRC. However, immune surveillance was disrupted by suppressed CD8+ T cell proliferation and release of anti-inflammatory cytokines.50 A major feature of immune evasion in cancer cells is the expression of multiple inhibitory ligands, notably IDO.51 Thus, an adaptive immune system could improve tumor prognosis. Immunoregulation, including IDO inhibition, may be an effective strategy for the chemoprevention and treatment of colon cancer.
Based on our previous findings and this study, we determined that IDO activation played a critical role in CRC progression, which provided mechanistic evidence for the correlation between high IDO expression and poor prognosis of human CRC. These results confirmed that IDO had multiple effects on CRC progression, including regulation of immune tolerance and suppression of cancer cell proliferation. Nevertheless, how IDO-induced immune response promotes tumorigenesis is still unknown. A previous study indicated that IDO was activated in DSS-induced acute colitis.52 Its expression and activation were elevated in patients with inflammatory bowel disease and positively correlated with inflammation-induced colon tumorigenesis.53 Colitis severity is known to influence the development and progression of colon cancer. Thus, IDO acts as a natural brake to control inflammatory response, and it was initially speculated that IDO might influence CRC via modulating colitis. However, recent studies revealed that IDO deficiency did not promote spontaneous intestinal inflammation.52,54
Conclusion
Taken together, we demonstrate an inflammatory microenvironment–induced immunosuppression in colon cancer through the IDO/Kyn/AhR axis. This regulatory mechanism is critical for colon cancer cells to escape immune surveillance via IDO-mediated Kyn production. Besides, the inhibition of IDO expression and activation in cancer cells suppressed Treg-induced immune tolerance. Thus, targeting IDO in cancer cells presents a potential strategy to prevent and treat inflammation-associated colon cancer.
Materials and Methods
Reagents
1-L-MT (Catalog No. 447439-5G) purchased from Sigma-Aldrich (St. Louis, MO) was dissolved in 0.1M NaOH at 50 mM. INCB (INCB024360; Catalog No. S7910) purchased from Selleck Chemicals (Houston, TX) was dissolved in 50 mM DMSO. Primary antibodies for IDO (Catalog No. BS90685), β-actin (Catalog No. BS6007M), and Lamin-A (Catalog No. BS7013) were purchased from Bioworld Technology, Inc (Nanjing, China). Horseradish peroxidase–conjugated Affinipure Goat Anti-Mouse IgG (Catalog No. SA00001-1) and Goat Anti-Rabbit IgG (Catalog No. SA00001-2) antibodies were purchased from Proteintech Group (Chicago, IL). AhR (Catalog No. A1451), FoxP3 (Catalog No. A12051), GZB (Catalog No. A19592), and GCN2 (Catalog No. A12618) antibody were purchased from Abclonal (Wuhan, Hubei, China). Antibodies for flow cytometry, including CD11b (Catalog No. 50-0112-82), CD3 (Catalog No. 25-0031-81), CD8 (Catalog No. 12-0081-81), CD4 (Catalog No. 17-0049-41), CD69 (Catalog No. 14-0691-82), and F4/80 (Catalog No. 11-4081-82), were purchased from eBioscience (San Diego, CA). AhR siRNA and control siRNA were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Lipopolysaccharide (Catalog No. L2630) was purchased from Sigma-Aldrich. Enzyme-linked immunosorbent assay kits for Trp (Catalog No. SBJ-M0728) and Kyn (Catalog No. SBJ-M0729) were purchased from Senbeijia Biotech (Nanjing, China), and interferon gamma (Catalog No. EK0375) was purchased from Boster Biotech (Wuhan, China).
Cell Culture
Human THP-1 (Catalog No. TCHu57) and Jurkat (Catalog No. TCHU123) cells were obtained from the Cell Bank of Shanghai, Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences and maintained in RPMI 1640 medium (Catalog No. 31800022; Thermo Fisher Scientific, Waltham, MA) supplemented with 10% heat-inactivated fetal bovine serum (Catalog No.10099 and 10091; Thermo Fisher Scientific). Human colon cancer cell lines including HCT-116 (Catalog No. TCHu99), HT-29 (Catalog No. TCHu103), and SW620 (Catalog No. TCHu101) were obtained from Cell Bank of the Chinese Academic of Sciences (Shanghai, China) and cultured in McCoy’s 5a (Catalog No. M4892; Sigma-Aldrich) medium supplemented with 10% fetal bovine serum (FBS), RPMI-1640 medium supplemented with 10% FBS, and Dulbecco’s modified Eagle medium (Catalog No. 12800017; Thermo Fisher Scientific) medium containing 10% FBS. All cell lines were cultured under a humidified 5% (v/v) CO2 atmosphere at 37°C. All cell lines were authenticated with methods of short tandem repeat. All experiments were performed with mycoplasma-free cells.
Western Blot Assay
Total proteins were extracted by adding RIPA lysis buffer (Catalog No. P0013C; Beyotime Biotechnology, Shanghai, China) with 1 mM PMSF (Catalog No. ST506; Beyotime Biotechnology) for 1 hour on the ice and centrifuging at 13,000 rpm for 30 minutes at 4°C. Protein concentration in the supernatants was measured by BCA protein assay (Catalog No.23227; Thermo Fisher Scientific). Then, an equal amount of sample was run on 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis. The proteins were transferred to polyvinylidene difluoride membranes (Catalog No. IPVH00010; Sigma-Aldrich) using a semidry transfer system (Bio-Rad, Hercules, CA). Proteins were detected using specific antibodies of IDO (1:1000), FoxP3 (1:1000), GZB (1:1000), GSCN (1:1000), AhR (1:1000), and β-actin (1:5000) overnight at 4°C. The relative protein expression was calculated by quantifying Western blot band intensities using the ImageJ Gel Analyzer plugin (V1.8.0, National Institutes of Health, Bethesda, MD). Western blots were performed as previously described.55
Nuclear and Cytoplasmic Extraction
Following treatments, cells were harvested by centrifugation and washed twice with phosphate-buffered saline (PBS). Nuclear and cytosol lysates were isolated using a Nuclear/Cytosol Fractionation Kit (Catalog No. K266; BioVision technologies, Milpitas, CA) according to the manufacturer’s instruction. The protein concentration of the nuclear and cytoplasmic extracts was measured with the BCA protein assay reagent (Catalog No. 23227; Thermo Fisher Scientific). Extracts were stored at –80°C until further experimentation.
Analysis of Intracellular Kyn Expression in Jurkat Cells by Flow Cytometry
For intracellular Kyn staining, we used a BD Cytofix/Cytoperm Kit (Catalog No. 554714. BD Pharmingen, San Diego, CA) following the manufacturer's instructions. Intracellular Kyn (Catalog No. sc-69890; Santa Cruz Biotechnology) production was detected with Kyn monoclonal Abs and FITC conjugate-goat anti-mouse IgG (as a secondary antibody; Catalog No. KGAA25; KeyGen Biotech, Nanjing, Jiangsu, China).
RNA Extraction and Real-Time Polymerase Chain Reaction
Total RNA was isolated using the TRIzol reagent (Catalog No. 15596026; Thermo Fisher Scientific) according to the manufacturer’s protocol. RNA samples were reverse transcribed to cDNA and subjected to quantitative polymerase chain reaction (PCR), which was performed with the Light-Cycler 96 Real-Time PCR System (Roche, Indianapolis, IN) using AceQ qPCR SYBR Green Master Mix (Catalog No. Q131; Vazyme Biotech, Nanjing, China). The primer sequences used in this study were as follows:
IDO, 5′-CACTTTGCTAAAGGCGCTGTTGGA-3′ (forward)
5′-GGTTGCCTTTCCAGCCAGACAAAT-3′ (reverse)
GZA, 5′-ACACGGTTGTTCCTCACTCAAGAC-3′ (forward)
5′- TCAATCAAAGCGCCAGCACAGATG-3′ (reverse)
GZB, 5′-TGTGAAGCCAGGAGATGTGTGCTA-3′ (forward)
5′-TCAGCTCAACCTCTTGTAGCGTGT-3′ (reverse)
CCK8 Cell Viability Assay
Cell viabilities were assessed using a Cell Counting Kit-8 (CCK8, Catalog No. CK04; Dojindo Laboratories, Kumamoto, Japan) assay. To this end, Jurkat cells (1.0 × 104/well) were seeded in 96-well plates (3 wells per group) and treated with different stimulations for 24 hours, respectively. Next, 10 μL CCK8was added to the cells, and the viability of the cells was measured with a microplate reader at 490 nm.
Apoptosis Analysis
For fluorescence staining assay, the cells were washed with cold PBS, and incubated with Annexin V-FITC and propidium iodide (PI) (Annexin V-FITC Apoptosis Detection Kit, Catalog No. KGA107; KeyGen Biotech) in turns. After different stimulations, Jurkat cells were resuspended and stained with Annexin V-FITC in dark for15 minutes under 4°C and subsequently treated by PI for 5 minutes under the same conditions. The stained cells were subjected to a flow cytometer for quantitative analysis. Annexin V+/PI– (early apoptosis) together with Annexin V+/PI+ cells (late apoptosis) were deemed as apoptotic portion.
Cell Cycle Assay
After treatment with CM_ with or without 1-L-MT or INCB for 24 hours, Jurkat cells were harvested and spined down and the resulting pellets were fixed in ice-cold 70% ethanol. Fixed cells were centrifuged, washed, and resuspended in PBS-containing RNase A (1 mg/mL), and PI (Cell Cycle Detection Kit, Catalog No. KGA511; KeyGen Biotech) was added (1.0 mg/mL). PI-stained cells were analyzed by a fluorescence-activated cell sorter (Accuri C6; Becton Dickinson, Franklin Lakes, NJ), followed by the determination of the percentage of cells in G0/G1, S, and G2/M.
Preparation of Suspension and Flow Cytometry
Single-cell suspensions of splenocytes were prepared by crushing the spleen with a plunger of a disposable syringe. Cell suspensions were strained through a 70-μm nylon mesh and then suspended in RPMI-1640 medium. The erythrocytes were lysed from splenocytes by using ACK lysis buffer (Catalog No. KGP11100; KeyGen Biotech). The lymphocytes were separated by different density of Percoll layered liquid (Catalog No. P1644; Sigma-Aldrich). The extracted lymphocytes were incubated with FITC-conjugated anti-mouse CD3, antigen-presenting cell–conjugated anti-mouse CD4 and PE-conjugated anti-mouse CD8 antibodies at 4°C for 30 minutes. The suspensions were then washed with sheath fluid. The stained samples were assessed by Accuri C6 (Becton Dickinson).
Transfection of AhR siRNA
AhR siRNA (Catalog No. sc-29654) was purchased from Santa Cruz Biotechnology and transformed according to the manufacturer’s instruction of Exfect Transfection Reagent (Catalog No. T101-01; Vazyme Biotech).
ChIP Assay
The Jurkat cells were cross-linked and sonicated according to the protocol published by Nelson et al.56 Briefly, immunoprecipitation was performed in 4 replicates and carried out at 4°C overnight with 1 μg of rabbit anti-human AhR antibody (Catalog No. 83200; Cell Signaling Technology, Danvers, MA) or an irrelevant IgG antibody (Catalog No. A7001. Beyotime Biotechnology) as a negative control. Immunocomplexes were recovered with 50-μL protein A/G beads (Catalog No. P2055. Beyotime Biotechnology). Input DNA and purified immunoprecipitated DNA were analyzed by real-time PCR. Quantification of the ChIP-DNA was performed using the method described by Nelson et al.56
Animal Studies and Colitis-Associated Colon Cancer
Animal welfare and experimental procedures were performed in accordance with the Guide for the Care and Use of Laboratory Animals and the related ethical regulations of our university (No.2020-01-005). Pathogen-free male C57BL/6 and Rag1−/− (C57BL/6-Rag1em1Smoc, Rag1 knockout) mice were purchased from the Model Animal Research Center of Nanjing University (Nanjing, China) at 5 weeks of age, and IDO1 knockout (IDO1−/−) mice, on the C57BL/6J, were originally purchased from the Jackson Laboratory (Bar Harbor, ME). They were exposed to a 12-hour light/dark cycle. At 6weeks of age, mice received intraperitoneal 10-mg/kg AOM (Sigma-Aldrich) followed by 7-day cycles of sterile filtered DSS (TdB Consultancy, Uppsala, Sweden) at 2.5% in their drinking water. A total of 5 mg/kg Kyn injected intraperitoneally every week.
Immunohistochemistry
Immunohistochemical stains against FoxP3 was performed using immunohistochemistry kit (Catalog No. KGSP03; KeyGen Biotech). Briefly, paraffin-embedded slides were deparaffinized, rehydrated, and washed in 1% PBS-Tween. Then they were treated with 3% hydrogen peroxide and blocked with 10% goat serum for 1 hour at 37°C. Slides were incubated with primary antibodies in PBS containing 1% bovine serum albumin (BSA) (1:50) for 1 hour at 37°C. Biotinylated secondary anti-rabbit antibodies were added and incubated at room temperature for 1 hour. Streptavidin-horseradish peroxidase was added, and after 40 minutes the sections were stained with DAB substrate and counterstained with hematoxylin.
Immunofluorescence
Immunofluorescence was performed on paraffin-embedded colonic tissue sections. The sections were deparaffinized, rehydrated, and washed in 1% PBS-Tween. Then, they were treated with 3% hydrogen peroxide, blocked with 10% goat serum, and incubated with CD11b, F4/80, FoxP3, CD4, and CD8 primary antibody in PBS containing 1% BSA (1:100) for 1 hour at 37°C. The slides were stained with DAPI. Images were acquired by confocal laser scanning microscope (Olympus, Lake Success, NY). Settings for image acquisition were identical for control and experimental tissues.
Tumor-Infiltrating Lymphocyte Profile Analysis by CyTOF or Flow Cytometry
Excised tumors were digested in collagenase/hyaluronidase and DNase I (Catalog No. BS137; Biosharp, Hefei, China), and TILs were enriched on a Percoll gradient (Catalog No. P1644; Sigma-Aldrich). TILs were incubated with 5% BSA for 10 minutes, and then TILs were incubated with different antibodies for 30 minutes at room temperature and washed twice. Stained samples were analyzed using Accuri C6 (Becton Dickinson).
Statistical Analyses
All statistical analyses were performed using GraphPad Prism software (V8.0; GraphPad Software, San Diego, CA). Results are presented as the mean ± SD. Statistical analysis was performed using the 2-tailed Student t test for comparison of 2 groups to determine the level of significance. For multiple groups, statistical analyses were performed with 1-way analysis of variance. P values <.05 were considered significant. All experiments were replicated at least 3 times.
Graphical Abstract Preparation
We generated the graphical abstract in PowerPoint, using some of the illustrations available at Servier Medical Art and the Cellular and Molecular Gastroenterology and Hepatology–branded PowerPoint template. Some adapting templates were downloaded from Servier Medical Arts website (https://smart.servier.com/; licensed under a Creative Commons Attribution 3.0 Unported License).
Acknowledgments
The authors thank the Cellular and Molecular Biology Center of China Pharmaceutical University for assistance with immunohistochemistry and immunofluorescence works and Xiao-Nan Ma for her technical help. The authors thank Servier Medical Art for providing access to designed medical elements (https://smart.servier.com/; licensed under a Creative Commons Attribution 3.0 Unported License), supporting the generation of graphical items in this publication.
CRediT Authorship Contributions
Xin Zhang (Data curation: Equal; Investigation: Lead; Writing – original draft: Equal)
Xiuting Liu (Data curation: Equal; Investigation: Equal; Methodology: Equal; Writing –original draft: Equal; Writing – review & editing: Equal)
Wei Zhou (Data curation: Supporting; Investigation: Supporting; Software: Supporting)
Qianming Du (Data curation: Supporting; Investigation: Supporting)
Mengdi Yang (Data curation: Supporting; Investigation: Supporting)
Yang Ding (Investigation: Supporting; Methodology: Supporting)
Rong Hu (Conceptualization: Lead; Funding acquisition: Lead; Investigation: Lead; Writing – review & editing: Lead)
Footnotes
Conflicts of Interest The authors disclose no conflicts.
Funding This work was supported by the National Natural Science Foundation of China (Nos. 81872337 and 81672816).
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Goding Sauer A., Fedewa S.A., Butterly L.F., Anderson J.C., Cercek A., Smith R.A., Jemal A. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020;70:145–164. doi: 10.3322/caac.21601. [DOI] [PubMed] [Google Scholar]
- 3.Pancione M., Remo A., Colantuoni V. Genetic and epigenetic events generate multiple pathways in colorectal cancer progression. Patholog Res Int. 2012;2012:509348. doi: 10.1155/2012/509348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lim S.O., Li C.W., Xia W., Cha J.H., Chan L.C., Wu Y., Chang S.S., Lin W.C., Hsu J.M., Hsu Y.H., Kim T., Chang W.C., Hsu J.L., Yamaguchi H., Ding Q., Wang Y., Yang Y., Chen C.H., Sahin A.A., Yu D., Hortobagyi G.N., Hung M.C. Deubiquitination and Stabilization of PD-L1 by CSN5. Cancer Cell. 2016;30:925–939. doi: 10.1016/j.ccell.2016.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Irrazabal T., Martin A. T Regulatory Cells Gone Bad: An Oncogenic Immune Response against Enterotoxigenic B. fragilis Infection Leads to Colon Cancer. Cancer Discov. 2015;5:1021–1023. doi: 10.1158/2159-8290.CD-15-0987. [DOI] [PubMed] [Google Scholar]
- 6.Geis A.L., Fan H., Wu X., Wu S., Huso D.L., Wolfe J.L., Sears C.L., Pardoll D.M., Housseau F. Regulatory T-cell Response to Enterotoxigenic Bacteroides fragilis Colonization Triggers IL17-Dependent Colon Carcinogenesis. Cancer Discov. 2015;5:1098–1109. doi: 10.1158/2159-8290.CD-15-0447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galon J., Costes A., Sanchez-Cabo F., Kirilovsky A., Mlecnik B., Lagorce-Pages C., Tosolini M., Camus M., Berger A., Wind P., Zinzindohoue F., Bruneval P., Cugnenc P.H., Trajanoski Z., Fridman W.H., Pages F. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–1964. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 8.Saleh R., Sasidharan Nair V., Murshed K., Abu Nada M., Elkord E., Shaheen R. Transcriptome of CD8(+) tumor-infiltrating T cells: a link between diabetes and colorectal cancer. Cancer Immunol Immunother. 2021 Feb 13 doi: 10.1007/s00262-021-02879-7. [E-pub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soliman H., Mediavilla-Varela M., Antonia S. Indoleamine 2,3-dioxygenase: is it an immune suppressor? Cancer J. 2010;16:354–359. doi: 10.1097/PPO.0b013e3181eb3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adams S., Braidy N., Bessede A., Brew B.J., Grant R., Teo C., Guillemin G.J. The kynurenine pathway in brain tumor pathogenesis. Cancer Res. 2012;72:5649–5657. doi: 10.1158/0008-5472.CAN-12-0549. [DOI] [PubMed] [Google Scholar]
- 11.Prendergast G.C., Malachowski W.P., DuHadaway J.B., Muller A.J. Discovery of IDO1 Inhibitors: From Bench to Bedside. Cancer Res. 2017;77:6795–6811. doi: 10.1158/0008-5472.CAN-17-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mellor A.L., Munn D.H. IDO expression by dendritic cells: tolerance and tryptophan catabolism. Nat Rev Immunol. 2004;4:762–774. doi: 10.1038/nri1457. [DOI] [PubMed] [Google Scholar]
- 13.Grohmann U., Fallarino F., Puccetti P. Tolerance, DCs and tryptophan: much ado about IDO. Trends Immunol. 2003;24:242–248. doi: 10.1016/s1471-4906(03)00072-3. [DOI] [PubMed] [Google Scholar]
- 14.Platten M., Friedrich M., Wainwright D.A., Panitz V., Opitz C.A. Tryptophan metabolism in brain tumors - IDO and beyond. Curr Opin Immunol. 2021;70:57–66. doi: 10.1016/j.coi.2021.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Merlo L.M.F., DuHadaway J.B., Montgomery J.D., Peng W.D., Murray P.J., Prendergast G.C., Caton A.J., Muller A.J., Mandik-Nayak L. Differential Roles of IDO1 and IDO2 in T and B Cell Inflammatory Immune Responses. Front Immunol. 2020;11:1861. doi: 10.3389/fimmu.2020.01861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chinnadurai R., Scandolara R., Alese O.B., Arafat D., Ravindranathan D., Farris A.B., El-Rayes B.F., Gibson G. Correlation Patterns Among B7 Family Ligands and Tryptophan Degrading Enzymes in Hepatocellular Carcinoma. Front Oncol. 2020;10:1632. doi: 10.3389/fonc.2020.01632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frumento G., Rotondo R., Tonetti M., Damonte G., Benatti U., Ferrara G.B. Tryptophan-derived catabolites are responsible for inhibition of T and natural killer cell proliferation induced by indoleamine 2,3-dioxygenase. J Exp Med. 2002;196:459–468. doi: 10.1084/jem.20020121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Munn D.H., Mellor A.L. Indoleamine 2,3-dioxygenase and tumor-induced tolerance. J Clin Invest. 2007;117:1147–1154. doi: 10.1172/JCI31178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Craig S.G., Humphries M.P., Alderdice M., Bingham V., Richman S.D., Loughrey M.B., Coleman H.G., Viratham-Pulsawatdi A., McCombe K., Murray G.I., Blake A., Domingo E., Robineau J., Brown L., Fisher D., Seymour M.T., Quirke P., Bankhead P., McQuaid S., Lawler M., McArt D.G., Maughan T.S., James J.A., Salto-Tellez M. Immune status is prognostic for poor survival in colorectal cancer patients and is associated with tumour hypoxia. Br J Cancer. 2020;123:1280–1288. doi: 10.1038/s41416-020-0985-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ino K., Yoshida N., Kajiyama H., Shibata K., Yamamoto E., Kidokoro K., Takahashi N., Terauchi M., Nawa A., Nomura S., Nagasaka T., Takikawa O., Kikkawa F. Indoleamine 2,3-dioxygenase is a novel prognostic indicator for endometrial cancer. Brit J Cancer. 2006;95:1555–1561. doi: 10.1038/sj.bjc.6603477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brandacher G., Perathoner A., Ladurner R., Schneeberger S., Obrist P., Winkler C., Werner E.R., Werner-Felmayer G., Weiss H.G., Gobel G., Margreiter R., Konigsrainer A., Fuchs D., Amberger A. Prognostic value of indoleamine 2,3-dioxygenase expression in colorectal cancer: effect on tumor-infiltrating T cells. Clin Cancer Res. 2006;12:1144–1151. doi: 10.1158/1078-0432.CCR-05-1966. [DOI] [PubMed] [Google Scholar]
- 22.Ogawa K., Hara T., Shimizu M., Ninomiya S., Nagano J., Sakai H., Hoshi M., Ito H., Tsurumi H., Saito K., Seishima M., Tanaka T., Moriwaki H. Suppression of azoxymethane-induced colonic preneoplastic lesions in rats by 1-methyltryptophan, an inhibitor of indoleamine 2,3-dioxygenase. Cancer Sci. 2012;103:951–958. doi: 10.1111/j.1349-7006.2012.02237.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogawa K., Hara T., Shimizu M., Nagano J., Ohno T., Hoshi M., Ito H., Tsurumi H., Saito K., Seishima M., Moriwaki H. (-)-Epigallocatechin gallate inhibits the expression of indoleamine 2,3-dioxygenase in human colorectal cancer cells. Oncol Lett. 2012;4:546–550. doi: 10.3892/ol.2012.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Massoud A.H., Kaufman G.N., Xue D., Beland M., Dembele M., Piccirillo C.A., Mourad W., Mazer B.D. Peripherally Generated Foxp3(+) Regulatory T Cells Mediate the Immunomodulatory Effects of IVIg in Allergic Airways Disease. J Immunol. 2017;198:2760–2771. doi: 10.4049/jimmunol.1502361. [DOI] [PubMed] [Google Scholar]
- 25.Burchill M.A., Yang J., Vogtenhuber C., Blazar B.R., Farrar M.A. IL-2 receptor beta-dependent STAT5 activation is required for the development of Foxp3+ regulatory T cells. J Immunol. 2007;178:280–290. doi: 10.4049/jimmunol.178.1.280. [DOI] [PubMed] [Google Scholar]
- 26.Grossman W.J., Verbsky J.W., Barchet W., Colonna M., Atkinson J.P., Ley T.J. Human T regulatory cells can use the perforin pathway to cause autologous target cell death. Immunity. 2004;21:589–601. doi: 10.1016/j.immuni.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 27.Li F., Sun Y., Huang J., Xu W., Liu J., Yuan Z. CD4/CD8 + T cells, DC subsets, Foxp3, and IDO expression are predictive indictors of gastric cancer prognosis. Cancer Med. 2019;8:7330–7344. doi: 10.1002/cam4.2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng Y., Chen Z., Han Y., Han L., Zou X., Zhou B., Hu R., Hao J., Bai S., Xiao H., Li W.V., Bueker A., Ma Y., Xie G., Yang J., Chen S., Li H., Cao J., Shen L. Immune suppressive landscape in the human esophageal squamous cell carcinoma microenvironment. Nat Commun. 2020;11:6268. doi: 10.1038/s41467-020-20019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y., He M., Zhang G., Cao K., Yang M., Zhang H., Liu H. The immune landscape during the tumorigenesis of cervical cancer. Cancer Med. 2021;10:2380–2395. doi: 10.1002/cam4.3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saito T., Nishikawa H., Wada H. Two FOXP3(+)CD4(+) T cell subpopulations distinctly control the prognosis of colorectal cancers. Nat Med. 2016;22:679–684. doi: 10.1038/nm.4086. [DOI] [PubMed] [Google Scholar]
- 31.Liu X., Zhou W., Zhang X., Ding Y., Du Q., Hu R. 1-L-MT, an IDO inhibitor, prevented colitis-associated cancer by inducing CDC20 inhibition-mediated mitotic death of colon cancer cells. Int J Cancer. 2018;143:1516–1529. doi: 10.1002/ijc.31417. [DOI] [PubMed] [Google Scholar]
- 32.Ravishankar B., Liu H., Shinde R., Chaudhary K., Xiao W., Bradley J., Koritzinsky M., Madaio M.P., McGaha T.L. The amino acid sensor GCN2 inhibits inflammatory responses to apoptotic cells promoting tolerance and suppressing systemic autoimmunity. Proc Natl Acad Sci U S A. 2015;112:10774–10779. doi: 10.1073/pnas.1504276112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu X.H., Zhai X.Y. Role of tryptophan metabolism in cancers and therapeutic implications. Biochimie. 2021;182:131–139. doi: 10.1016/j.biochi.2021.01.005. [DOI] [PubMed] [Google Scholar]
- 34.Campesato L.F., Budhu S., Tchaicha J., Weng C.H., Gigoux M., Cohen I.J., Redmond D., Mangarin L., Pourpe S., Liu C., Zappasodi R., Zamarin D., Cavanaugh J., Castro A.C., Manfredi M.G., McGovern K., Merghoub T., Wolchok J.D. Blockade of the AHR restricts a Treg-macrophage suppressive axis induced by L-Kynurenine. Nat Commun. 2020;11:4011. doi: 10.1038/s41467-020-17750-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wong L.Y., Hatfield J.K., Brown M.A. Ikaros Sets the Potential for Th17 Lineage Gene Expression through Effects on Chromatin State in Early T Cell Development. J Biol Chem. 2013;288:35170–35179. doi: 10.1074/jbc.M113.481440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Banchereau J., Steinman R.M. Dendritic cells and the control of immunity. Nature. 1998;392:245–252. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- 37.Muller A.J., DuHadaway J.B., Chang M.Y., Ramalingam A., Sutanto-Ward E., Boulden J., Soler A.P., Mandik-Nayak L., Gilmour S.K., Prendergast G.C. Non-hematopoietic expression of IDO is integrally required for inflammatory tumor promotion. Cancer Immunol Immunother. 2010;59:1655–1663. doi: 10.1007/s00262-010-0891-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim A.K., Gani F., Layman A.J., Besharati S., Zhu Q., Succaria F., Engle E.L., Bhaijee F., Goggins M.B., Llosa N.J., Pawlik T.M., Yarchoan M., Jaffee E.M., Simons H.C., Taube J.M., Anders R.A. Multiple Immune-Suppressive Mechanisms in Fibrolamellar Carcinoma. Cancer Immunol Res. 2019;7:805–812. doi: 10.1158/2326-6066.CIR-18-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murray P.J. Amino acid auxotrophy as a system of immunological control nodes. Nat Immunol. 2016;17:132–139. doi: 10.1038/ni.3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Munn D.H., Sharma M.D., Baban B., Harding H.P., Zhang Y., Ron D., Mellor A.L. GCN2 kinase in T cells mediates proliferative arrest and anergy induction in response to indoleamine 2,3-dioxygenase. Immunity. 2005;22:633–642. doi: 10.1016/j.immuni.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 41.Fallarino F., Grohmann U., You S., McGrath B.C., Cavener D.R., Vacca C., Orabona C., Bianchi R., Belladonna M.L., Volpi C., Santamaria P., Fioretti M.C., Puccetti P. The combined effects of tryptophan starvation and tryptophan catabolites down-regulate T cell receptor zeta-chain and induce a regulatory phenotype in naive T cells. J Immunol. 2006;176:6752–6761. doi: 10.4049/jimmunol.176.11.6752. [DOI] [PubMed] [Google Scholar]
- 42.Manlapat A.K., Kahler D.J., Chandler P.R., Munn D.H., Mellor A.L. Cell-autonomous control of interferon type I expression by indoleamine 2,3-dioxygenase in regulatory CD19+ dendritic cells. Eur J Immunol. 2007;37:1064–1071. doi: 10.1002/eji.200636690. [DOI] [PubMed] [Google Scholar]
- 43.Liu H., Huang L., Bradley J., Liu K., Bardhan K., Ron D., Mellor A.L., Munn D.H., McGaha T.L. GCN2-dependent metabolic stress is essential for endotoxemic cytokine induction and pathology. Mol Cell Biol. 2014;34:428–438. doi: 10.1128/MCB.00946-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rivollier A., He J., Kole A., Valatas V., Kelsall B.L. Inflammation switches the differentiation program of Ly6Chi monocytes from antiinflammatory macrophages to inflammatory dendritic cells in the colon. J Exp Med. 2012;209:139–155. doi: 10.1084/jem.20101387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stejskalova L., Dvorak Z., Pavek P. Endogenous and exogenous ligands of aryl hydrocarbon receptor: current state of art. Curr Drug Metab. 2011;12:198–212. doi: 10.2174/138920011795016818. [DOI] [PubMed] [Google Scholar]
- 46.Quintana F.J. The aryl hydrocarbon receptor: a molecular pathway for the environmental control of the immune response. Immunology. 2013;138:183–189. doi: 10.1111/imm.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kimura A., Naka T., Nakahama T., Chinen I., Masuda K., Nohara K., Fujii-Kuriyama Y., Kishimoto T. Aryl hydrocarbon receptor in combination with Stat1 regulates LPS-induced inflammatory responses. J Exp Med. 2009;206:2027–2035. doi: 10.1084/jem.20090560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Araujo E.F., Feriotti C., Galdino N.A.L., Preite N.W., Calich V.L.G., Loures F.V. The IDO-AhR Axis Controls Th17/Treg Immunity in a Pulmonary Model of Fungal Infection. Front Immunol. 2017;8:880. doi: 10.3389/fimmu.2017.00880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gupta A.K., Pretlow T.P., Schoen R.E. Aberrant crypt foci: what we know and what we need to know. Clin Gastroenterol Hepatol. 2007;5:526–533. doi: 10.1016/j.cgh.2007.02.014. [DOI] [PubMed] [Google Scholar]
- 50.Krummel M.F., Allison J.P. CD28 and CTLA-4 have opposing effects on the response of T cells to stimulation. J Exp Med. 1995;182:459–465. doi: 10.1084/jem.182.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dong H., Strome S.E., Salomao D.R., Tamura H., Hirano F., Flies D.B., Roche P.C., Lu J., Zhu G., Tamada K., Lennon V.A., Celis E., Chen L. Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat Med. 2002;8:793–800. doi: 10.1038/nm730. [DOI] [PubMed] [Google Scholar]
- 52.Metghalchi S., Ponnuswamy P., Simon T., Haddad Y., Laurans L., Clement M., Dalloz M., Romain M., Esposito B., Koropoulis V., Lamas B., Paul J.L., Cottin Y., Kotti S., Bruneval P., Callebert J., den Ruijter H., Launay J.M., Danchin N., Sokol H., Tedgui A., Taleb S., Mallat Z. Indoleamine 2,3-Dioxygenase Fine-Tunes Immune Homeostasis in Atherosclerosis and Colitis through Repression of Interleukin-10 Production. Cell Metab. 2015;22:460–471. doi: 10.1016/j.cmet.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 53.Ciorba M.A. Indoleamine 2,3 dioxygenase in intestinal disease. Curr Opin Gastroenterol. 2013;29:146–152. doi: 10.1097/MOG.0b013e32835c9cb3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thaker A.I., Rao M.S., Bishnupuri K.S., Kerr T.A., Foster L., Marinshaw J.M., Newberry R.D., Stenson W.F., Ciorba M.A. IDO1 metabolites activate beta-catenin signaling to promote cancer cell proliferation and colon tumorigenesis in mice. Gastroenterology. 2013;145:416–425.e1–4. doi: 10.1053/j.gastro.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu X., Zhang X., Ding Y., Zhou W., Tao L., Lu P., Wang Y., Hu R. Nuclear Factor E2-Related Factor-2 Negatively Regulates NLRP3 Inflammasome Activity by Inhibiting Reactive Oxygen Species-Induced NLRP3 Priming. Antioxid Redox Signal. 2017;26:28–43. doi: 10.1089/ars.2015.6615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nelson J.D., Denisenko O., Bomsztyk K. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat Protoc. 2006;1:179–185. doi: 10.1038/nprot.2006.27. [DOI] [PubMed] [Google Scholar]