Abstract
Poly(ethylene terephthalate) (PET) is a petroleum-based plastic that is massively produced and used worldwide. A promising PET recycling process to circumvent petroleum feedstock consumption and help to reduce environmental pollution is microbial or enzymatic biodegradation of post-consumer (PC) PET packages to its monomers—terephthalic acid (TPA) and ethylene glycol (EG)—or to key intermediates in PET synthesis—such as mono- and bis-(2-hydroxyethyl) terephthalate (MHET and BHET). Two species of filamentous fungi previously characterized as lipase producers (Penicillium restrictum and P. simplicissimum) were evaluated in submerged fermentation for induction of lipase production by two inducers (BHET and amorphous PET), and for biodegradation of two substrates (BHET and PC-PET). BHET induced lipase production in P. simplicissimum, achieving a peak of 606.4 U/L at 49 h (12.38 U/L.h), representing an almost twofold increase in comparison to the highest peak in the control (without inducers). Microbial biodegradation by P. simplicissimum after 28 days led to a 3.09% mass loss on PC-PET fragments. In contrast, enzymatic PC-PET depolymerization by cell-free filtrates from a P. simplicissimum culture resulted in low concentrations of BHET, MHET and TPA (up to 9.51 µmol/L), suggesting that there are mechanisms at the organism level that enhance biodegradation. Enzymatic BHET hydrolysis revealed that P. simplicissimum extracellular enzymes catalyze the release of MHET as the predominant product. Our results show that P. simplicissimum is a promising biodegrader of PC-PET that can be further explored for monomer recovery in the context of feedstock recycling processes.
Keywords: Biodegradation, Depolymerization, Esterase, Fungi, Lipase, PET
Introduction
Poly(ethylene terephthalate) (PET) is a semi-aromatic and thermoplastic polyester that was patented in 1941, and since then it has become one of the most important industrial polymers due to properties such as high tensile impact strength, chemical resistance, processability, transparency and thermal stability (Lepoittevin and Roger 2011). Virgin PET has an estimated annual worldwide production of more than 70 million tons, and its main applications are in the production of textile fibers and packaging (IHS Markit 2018; Crippa and Morico 2019). However, extensive production and use of plastics (including PET) engendered three pressing problems, one related to feedstock supply, owing to the depletion of readily available and cheap petroleum reservoirs (Hall 2017), another due to greenhouse gas emissions related to plastics production (Zheng and Suh 2019), and a third related to environmental pollution, caused by disposal mismanagement and persistence in the environment due to high recalcitrance to degradation (Geyer et al. 2017; Kumar et al. 2020).
One effort that may contribute to mitigate both problems is to expand and improve reverse logistics processes for PET, especially for post-consumer (PC) PET (Raheem et al. 2019). One aspect of reverse logistics is recycling, and there are three available PET recycling processes: (1) mechanical grinding followed by melting; (2) chemical depolymerization of PET scraps by glycolysis, methanolysis or hydrolysis; and (3) incineration for thermal energy generation (Sinha et al. 2010). Only the first two of these processes enable feedstock recycling to resynthesize PET, but both require high energy input and generate undesirable byproducts and effluents (Crippa and Morico 2019).
A promising alternative to traditional recycling processes is PET bio-depolymerization to recover its monomers (Wei and Zimmermann 2017a; Koshti et al. 2018; Salvador et al. 2019; Zimmermann 2020; Carniel et al. 2021). Biocatalysts employed in this route can be either cultivated microbes or isolated enzymes with the ability to selectively break the ester bonds of PET molecules and enable the recovery of terephthalic acid (TPA) and ethylene glycol (EG), which are the constituent monomers of PET, or the recovery of key intermediates in PET synthesis, such as mono- and bis-(2-hydroxyethyl) terephthalate (MHET and BHET, respectively) (Fig. 1). In comparison to physicochemical processes, biotechnological processes can usually be performed at milder temperature, pressure and pH conditions, and avoid the use and formation of hazardous chemicals (Wei and Zimmermann 2017b).
Fig. 1.
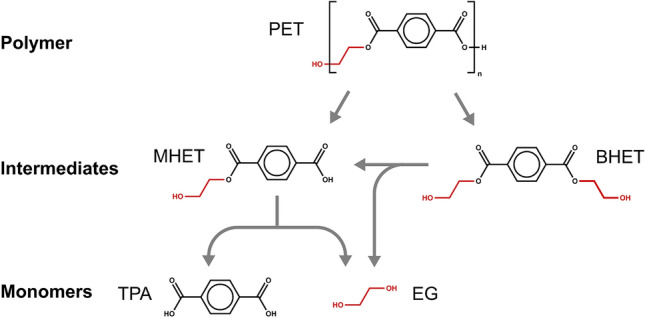
Molecular structures of polyethylene terephthalate (PET), bis-(2-hydroxyethyl) terephthalate (BHET), mono-(2-hydroxyethyl) terephthalate (MHET), terephthalic acid (TPA) and ethylene glycol (EG), and the catalyzed hydrolysis reactions involved in biological PET depolymerization
However, PET molecules did not occur on Earth before humans began producing it, so we would not expect that living organisms have evolved metabolic capabilities for PET biodegradation. On the other side, there are known bacterial, archaeal and fungal species that can metabolize other highly hydrophobic compounds containing ester bonds, such as cutin, suberin, long-chain triglycerides and lignin-carbohydrate complexes, so that these organisms and their enzymes may be able to biodegrade PET due to substrate promiscuity (Copley 2017; Zimmerman 2020; Carniel et al. 2021). Additionally, the increasing presence of PET in different environments during the last eight decades may have resulted in the evolution by natural selection of the adaptive metabolization of PET as an energy and carbon source by organoheterotrophic microorganisms, which seems to be the case of the bacterium Ideonella sakaiensis 201-F6 (Yoshida et al. 2016). The naturally occurring PET-degrading microbial strains and enzymes discovered through bioprospection can serve as starting points to the artificial optimization of biocatalysts for industrial processes, which can be pursued through rational design, directed evolution, or a combination of both (e.g., Tournier et al. 2020).
In addition to the already mentioned bacterium I. sakaiensis (Yoshida et al. 2016), to the best of our knowledge at least three other naturally occurring microbes are known to display in vivo PET biodegradation abilities: two isolates of filamentous fungus Microsphaeropsis arundinis: CBMAI 2109 and CBMAI 2110 (Malafatti-Picca et al. 2019), and the yeast Yarrowia lipolytica IMUFRJ 50,682 (Costa et al. 2020; Sales et al. 2020). In contrast, a larger number of enzymes have been characterized as PET hydrolases, and all of them are esterases (EC 3.1.1.-, carboxylic ester hydrolases) from bacterial and fungal species (Kawai et al. 2019, 2020; Taniguchi et al. 2019). These esterases include carboxylesterases (EC 3.1.1.1, carboxyl ester hydrolases), cutinases (EC 3.1.1.74, cutin hydrolases) and lipases (EC 3.1.1.3, triacylglycerol hydrolases) (Guebitz and Cavaco-Paulo 2007; Zimmermann and Billig 2011; Wierckx et al. 2018; Gan and Zhang 2019).
All the isolated enzymes or enzyme preparations from fungi described until now as capable of PET depolymerization are derived from 11 species of yeasts and filamentous fungi affiliated to the phylum Ascomycota (Gan and Zhang 2019; Sánchez 2020), namely: Aspergillus oryzae (Wang et al. 2008), Diutina rugosa (formerly Candida rugosa) (Carniel et al. 2017), F. oxysporum (Nimchua et al. 2007), F. solani pisi (Silva et al. 2005; Nimchua et al. 2008), Moesziomyces antarcticus (formerly Candida antarctica) (Carniel et al. 2017), Penicillium citrinum (Liebminger et al. 2007), Saccharomonospora viridis (= Micropolyspora internatus) (Hantani et al. 2018), Thermomyces insolens (formerly Humicola insolens) (Ronkvist et al. 2009), T. lanuginosus (Eberl et al. 2009), and Y. lipolytica (Costa et al. 2020).
Compared to bacteria, fungi are promising microbial biocatalysts to industrial process aiming at depolymerization of solid hydrophobic substrates such as plastics, especially due to their high production and secretion of hydrolytic enzymes (Hyde et al. 2019). Compared to yeasts, filamentous fungi show the ability of adhering and penetrating solid substrates through their hyphae, and some also produce surfactants like hydrophobins that contribute to the attachment to hydrophobic substrates (Kim & Rhee 2003; Sánchez 2020).
Regarding the Penicillium genus, two strains of P. simplicissimum (YK and H11) were shown as capable of degrading solid substrates: polyethylene (PE) and cellulose, respectively (Yamada-Onodera et al. 2001; Bai et al. 2013). An isolated polyesterase from P. citrinum was shown to be capable of hydrolyzing PET fabrics and BHET (Liebminger et al. 2007), while P. funiculosum was evaluated for biodegradation of mixtures between PET films and Bio-nolle®, an aliphatic synthetic polyester (Nowak et al. 2011.) In the present work, we investigated two other strains of Penicillium for PC-PET depolymerization for monomer recovery (P. restrictum and P. simplicissimum 28f2). These strains were characterized as lipase producers in previous works (Freire et al. 1997a; Gutarra et al. 2005). The induction of lipase production in these two species was evaluated under the presence of BHET and amorphous PET as inducers. Subsequently, we carried out a microbial biodegradation assay consisting of submerged fermentation with PC-PET fragments as substrate. Finally, we investigated the enzymatic biodegradation of two different substrates (PC-PET fragments and BHET) by cell-free filtrates of P. simplicissimum cultures. This is the first work to obtain experimental evidence that a Penicillium species is capable of PC-PET in vivo depolymerization and monomer recovery.
Methods
Fungal strains and culture conditions
The two strains of P. restrictum and P. simplicissimum 28f2 (acession number 071113-05 at the International Depository Authority of Canada—IDAC) were previously isolated from wastes of a Brazilian babassu palm (Attalea speciosa) oil industry and identified as lipase producers (Freire 1996; Freire et al. 1997a; Gutarra et al. 2005). For all experiments, P. simplicissimum and P. restrictum were cultivated in duplicates under submerged fermentation at 30 °C and 170 rpm, and the inocula were adjusted to 2 × 10 spores/mL before being transferred to a 500 mL erlenmeyer flask containing 120 mL of culture medium.
Composition of culture media for cultivation were based on previous reports (Freire et al. 1997b; Gutarra et al. 2007). The culture medium for P. simplicissimum was composed of: 1.2 g of glucose; 1.2 g of meat peptone; 0.6 g of yeast extract; 0.36 g of olive oil; 0.06 g of magnesium sulfate heptahydrate (MgSO4 · 7H2O); 0.06 g of potassium chloride (KCl); 0.0012 g of iron(II) sulfate (FeSO4); and 120 mL of sodium phosphate buffer 200 mM pH 7.0 (final pH was adjusted to 7.6). The culture medium for P. restrictum was composed of: 2.4 g of meat peptone; 1.2 g of olive oil; 0.6 g of sodium chloride (NaCl); 0.12 g of yeast extract; and 120 mL of distilled water (final pH was adjusted to 7.6).
Induction of lipase production
P. restrictum and P. simplicissimum were cultivated in duplicates for 4 days under three conditions: (1) standard culture medium (the control); (2) culture medium + 0.2 g/L of BHET; and (3) culture medium + 1.0 g/L of milled amorphous PET. The selection of these concentrations was based on previous works (Wang et al. 2008; Carniel et al. 2017; Sales et al. 2021). BHET (> 94.5% purity, Sigma-Aldrich) was autoclaved in powder form and subsequently added to the culture medium, which was previously sterilized by autoclaving. Milled amorphous PET was obtained by grinding amorphous PET chips kindly supplied by PQS (http://pqspe.com.br/), sieving the ground PET through a 4.0 mm mesh sieve, and then adding it to the culture medium before sterilization of the mixture by autoclaving. Samples of 10 µL were collected at nine different time points between 0 and 100 h to evaluate lipase activity, and samples of 50 µL were colected at four different time points (one at each day) to evaluate the concentrations of TPA, MHET and BHET.
Microbial PC-PET biodegradation
P. restrictum and P. simplicissimum were cultivated in duplicates for 28 days under submerged fermentation with 1.0 g of PC-PET added as substrate (for an initial concentration of 8.33 g/L). PC-PET was added to the culture medium in the form of square fragments of 5.0 mm sides and 0.1 mm thickness, and they were cut from waste bottles of noncarbonated mineral water (brand Crystal©). It was previously determined that this PC-PET has a molar mass of 42,737 ± 288 g/mol and a crystallinity of 36.6 ± 0.5% (Carniel et al. 2017). Samples of 10 µL and 50 µL were collected on four different time points (at 7, 14, 21, and 28 days) and evaluated respectively for lipase activity and concentrations of TPA, MHET and BHET. The mass loss of PC-PET fragments was calculated as the difference between initial and final mass.
Enzymatic PC-PET and BHET hydrolysis
To obtain the preparations for enzymatic depolymerization assays, P. simplicissimum was initially cultivated for 52 h under submerged fermentation in the standard culture medium described above, without the addition of any inducers. The resulting fermentation broth was filtered through membranes with 0.45 µm diameter pores to retain cells, and the cell-free filtrate was evaluated for lipase activity, and for protein concentration by the colorimetric Bradford assay (Bradford 1976). The crude filtrate was lyophilized and stored for subsequent use, being resolubilized in ultrapure water before it was utilized in the assays.
The enzymatic hydrolysis assays were performed in triplicate at 37 °C and 25 rpm, in 15 mL conical centrifuge tubes inside a hybridization incubator (combi-D24, FINEPCR) for a total incubation time of 42 days. Final reaction volume was 10 mL, with sodium phosphate buffer 200 mM pH 7.0 as solvent. Two different substrates were evaluated: 0.5 g/L of BHET and 20 g/L of PC-PET fragments (obtained as described in the previous sections). The cell-free filtrate containing extracellular enzymes was added to final concentrations of 0.04 g of protein per gram of BHET, or 0.01 g of protein per gram of PC-PET. Samples of 50 µL were collected on each time point to determine concentrations of TPA, MHET and BHET.
The observation of MHET build up during enzymatic BHET hydrolysis assays motivated the addition of Lipozyme© CALB (Novozymes) after 42 days of incubation at the concentration of 0.1 g of protein per gram of BHET. An additional 16 h of incubation was carried out, and concentrations of BHET, MHET and TPA were evaluated by collecting a sample of 50 µL at the end of incubation. Lipozyme© CALB was chosen because it was the most efficient one in catalyzing BHET hydrolysis to TPA according to a previous screening of ten commercial enzymes (Carniel et al. 2017).
Lipase activity measurement
Lipase hydrolytic activity was quantified by a spectrophotometric method based on the detection of p-nitrophenol, the chromophore product of the hydrolysis of p-nitrophenyl alkanoate esters by lipases (Hasan et al. 2009; Syedd-León et al. 2020). The substrate utilized was 4-nitrophenyl dodecanoate (≥ 98.0% purity, Sigma-Aldrich), more commonly known as p-nitrophenyl laurate (pNL). Samples of 10 µL were collected from fermentation assays and mixed with 40 µL of phosphate buffer 200 mM pH 7.0 and 50 µL of a 0.5 mM pNL solution inside the wells of microtiter plates. The released p-nitrophenol was detected on each well at the wavelength of 410 nm in 5 min scannings by a Multiskan™ GO Microplate Spectrophotometer (Thermo Scientific). Results were expressed as units of enzyme activity per liter (U/L), with one unit of enzyme activity being defined as the quantity of enzyme that catalyzes the release of 1.0 µmol of p-nitrophenol per minute, at reaction conditions. Productivity (U/L.h) was calculated for each time point by dividing lipase activity (U/L) by the incubation time (h) at that time point.
Quantification of MHET, BHET, and TPA
Concentrations of TPA, MHET and BHET were determined by high-performance liquid chromatography (HPLC). Samples of 50 µL were collected at each selected time point, mixed with 450 µL of pure methanol, homogenized with a vortex, and filtered through membranes with 0.22 μm diameter pores prior to application into a system composed of a chromatographer Dionex UltiMate 3000 (Thermo Scientific), a pre-column Zorbax SB C18 (4.6 × 12.5 mm, 5 µm), and a column Agilent Eclipse Plus C18 (4.6 × 250 mm, 5 µm). A gradient mixture of acetonitrile (TEMED) and 0.05% formic acid (Sigma-Aldrich) was used as mobile phase at the flow rate of 0.5 mL/min and 30 °C, and detection was performed in a ultraviolet detector at the wavelength of 254 nm. TPA (98% purity, Sigma-Aldrich), MHET [(95% purity, obtained from enzymatic hydrolysis of BHET as described by Carniel et al. (2017)] and BHET (> 94.5% purity, Sigma-Aldrich) powders were used to construct standard curves to correlate to the peak areas of samples to calculate the concentrations of the analytes (expressed as µmol/L).
Molar fractions for the enzymatic BHET biodegradation assay were calculated as percentages by dividing the concentration of each compound by the sum of the concentrations of the three compounds (BHET, MHET and TPA).
Mass loss determination
PC-PET fragments added to each flask were collectively weighed on an analytical balance before and after microbial biodegradation assays to calculate the percentage of mass loss. At the end of the assay, PC-PET fragments were collected, washed with a 2.0% Tween™ 80 solution, and subsequently rinsed with ultrapure water. Then, PC-PET fragments were dried in an oven at 105 °C until constant weight.
Results and discussion
Induction of lipase production
Lipases are one of the three types of esterases that may display PET hydrolase activity, and a previous work showed that the addition of BHET or short PET fibers to the culture medium has induced the expression of PET-degrading lipases by A. oryzae CCUG 33812 (Wang et al. 2008). Based on that result, we evaluated if the addition of 0.2 g/L of BHET or 1.0 g/L of amorphous PET would also induce lipase production in P. restrictum and P. simplicissimum. Amorphous PET was selected as a polymer source for enzyme production trials instead of short PET fibers because it is a more accessible substrate for PET-degrading enzymes, as discussed previously by Castro et al. (2017).
Under all treatments and for all sampled time points, lipase hydrolytic activity was much higher for P. simplicissimum than for P. restrictum (Table 1). Considering P. restrictum, the highest value for lipase hydrolytic activity was observed in the control treatment, without any inducers (8.75 U/L at 51 h, or productivity of 0.17 U/L.h), but the highest productivity was achieved under induction by amorphous PET (1.77 U/L.h at 4 h). In contrast, for P. simplicissimum both the highest peak of lipase hydrolytic activity (606.4 U/L) and productivity (12.38 U/L.h) were observed under BHET induction at 49 h, which represented respectively an almost two- and threefold increase in comparison to the peaks in the control treatment (331.2 U/L and 4.47 U/L.h, both at 74 h).
Table 1.
Peaks of lipase hidrolytic activity and respective incubation time for P. restrictum and P. simplicissimum cultivated in submerged fermentation under control treatment (standard culture medium) and the addition of BHET (0.2 g/L) or amorphous PET (1.0 g/L)
| Species | Peak of lipase hydrolytic activity, U/L (fermentation time) | ||
|---|---|---|---|
| Control | + BHET | + Amorphous PET | |
| P. restrictum | 8.75 (51 h) | 4.63 (22 h) | 7.08 (4 h) |
| P. simplicissimum | 331.2 (74 h) | 606.4 (49 h) | 354.3 (100 h) |
The highest lipase hydrolytic activity was obtained with P. simplicissimum (under induction by 0.2 g/L of BHET at 49 h) and it was two orders of magnitude lower than that reported by Wang et al. (2008) for A. oryzae (under the presence of 0.1 g/L of BHET at 84 h): 606.4 versus 16.884 U/L; but considering productivity, the difference was of one order of magnitude (12.38 versus 201 U/L.h). However, Wang et al. (2008) employed a different method to measure lipase hydrolytic activity, one that offers the triglycerides of olive oil instead of p-nitrophenyl alkanoate esters as substrates, and subsequently measures the quantity of liberated fatty acids by titration with sodium hydroxide (NaOH). Triglycerides are the natural substrates of lipases, while p-nitrophenyl alkanoate esters are structural analogs of triglycerides, so the differences observed can be explained by the higher affinity of lipases for triglycerides than for pNL.
BHET was also the best inducer of lipase production for the yeast Y. lipolytica cultivated in YP medium (Yeast extract, Peptone) under two different orbital agitations: 118.19 U/L (160 rpm) and 385 U/L (250 rpm), both at 24 h (Costa et al. 2020). These values are more comparable to the values we obtained for P. simplicissimum, since Costa et al. (2020) also employed a spectrophotometric method, even though the substrate offered was p-nitrophenyl butyrate (pNB) instead of pNL (i.e., a C4 instead of a C12 alkyl chain). The peak of lipase hydrolytic activity was higher for P. simplicissimum than for Y. lipolytica (606.4 U/L at 49 h versus 385 U/L at 24 h), although the productivity was higher for Y. lipolytica (12.38 U/L.h versus 16.04 U/L.h).
By comparing the peak of lipase hydrolytic activity under the presence of BHET with the respective controls, it is possible to reduce the influence of the different methodologies employed to measure this activity. By doing this comparison, we obtained an increase of 183% in P. simplicissimum, which is similar to the increase obtained for the other 2 species: 134% in A. oryzae (Wang et al. 2008), and 196% (160 rpm) or 150% (250 rpm) in Y. lipolytica (Costa et al. 2020).
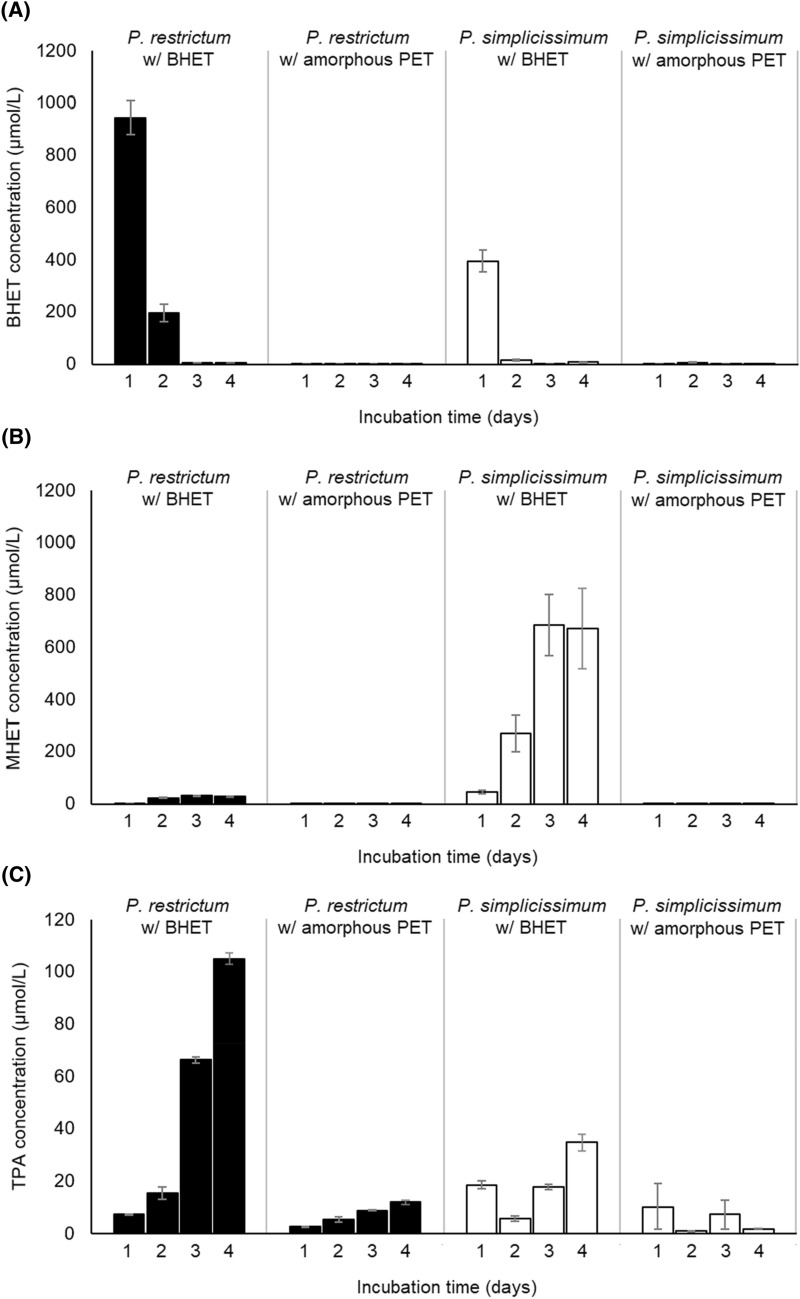
The concentrations of BHET, MHET and TPA were also evaluated during the lipase induction assays to check if amorphous PET and BHET were hydrolyzed by the produced enzymes (Fig. 2). When BHET was added to the culture medium, it showed high initial concentrations, which declined with incubation time, reaching very low values, close to the detection limit of the method, at days 3 and 4 for both fungal strains (Fig. 2A). However, there was a marked difference in relation to which BHET biodegradation products accumulated in the broth of each strain. At the end of the incubation time of 4 days, P. simplicissimum led to a higher concentration of MHET than TPA in the culture medium (673.21 versus 34.89 µmol/L), while P. restrictum resulted in a higher concentration of TPA than MHET (105.11 versus 31.71 µmol/L) (Fig. 2B, C). Considering that the presence of whole organisms involves additional variables in comparison to isolated enzymes, such as cell uptake and assimilation into biomass, it is not possible to infer that MHET acts as an inhibitor of MHET-degrading enzymes in P. simplicissimum.
Fig. 2.
Concentrations of BHET (A), MHET (B) and TPA (C) in the broth of submerged fermentation with P. restrictum and P. simplicissimum under the presence of BHET and milled amorphous PET. Error bars denote the standard error of two independent biological replicates
When amorphous PET was added to the culture medium, concentrations of BHET, MHET and TPA were always lower when compared to the addition of BHET (Fig. 2). The highest concentration for a biodegradation product from amorphous PET was 5.05 µmol/L of BHET for P. simplicissimum at 2 days (Fig. 2A), and 12.09 µmol/L of TPA for P. restrictum at 4 days (Fig. 2C).
Microbial PC-PET biodegradation
Microbial biodegradation assay was conducted with 8.33 g/L of PC-PET square fragments as substrates. Corroborating the results from the lipase induction assays, P. simplicissimum achieved a higher peak of lipase hydrolytic activity than P. restrictum: 32.39 U/L at 7 h versus 15.41 U/L at 21 h, respectively. After the incubation time of 28 days, P. simplicissimum imposed a mass loss of 3.09% on PC-PET fragments, while P. restrictum led to a mass loss of 0.08%, which is almost indistinguishable from the respective abiotic control (Fig. 3). In comparison to other results for PET biodegradation reported in the literature, the performance of P. simplicissimum is one of the best (Table 2), only surpassed by two isolates of M. arundinis (CBMAI 2109 and CBMAI 2110) which were able to achieve a similar mass loss in 14 days (Malafatti-Picca et al. 2019). Therefore, P. simplicissimum is a promising biodegrader of a real, assorted PC-PET sample from residual sources, and it will probably achieve even higher biodegradation if this fungal strain and the reaction conditions are optimized.
Fig. 3.
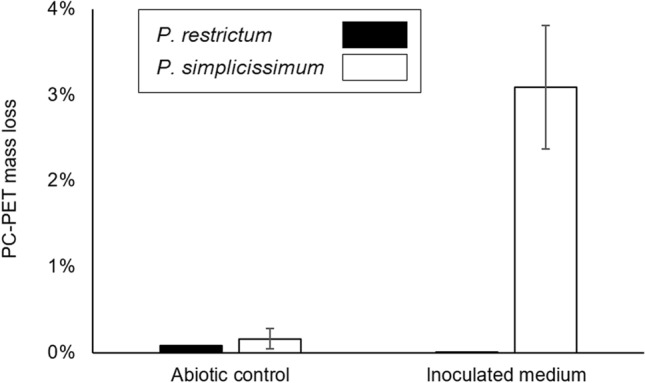
Mass loss of PC-PET fragments after 28 days of microbial biodegradation by P. restrictum and P. simplicissimum under submerged fermentation. Error bars denote the standard error of two independent biological replicates
Table 2.
Summary of PET biodegradation results
| Biocatalyst | Substract | Incubation time | Result | References |
|---|---|---|---|---|
| P. funiculosum | PET films (40 × 10 mm) | 84 days | 0.08% mass loss | Nowak et al. (2011) |
| Solution of extracellular extract of P. funiculosum | PET films (40 × 10 mm) | 84 days | 0.06% mass loss | Nowak et al. (2011) |
| A mixed culture of environmental microbes | PET transparent sheets (10 × 10 mm) | 200 days | ~ 2.0% mass loss | Sharon and Sharon (2012) |
| Four mixed cultures of microbes from activated sludges | PET fibers | 14 days | shallow erosion of the PET surface | Zhang et al. (2004) |
| Two isolates of M. arundinis (CBMAI 2109 and CBMAI 2110) | PET fragments (45 × 25 mm, 500 mg) at 25 g/L | 14 days | 2.0–3.0% mass loss | Malafatti-Picca et al. (2019) |
| P. simplicissimum | PC-PET fragments (5.0 × 5.0 × 0.1 mm) at 8.33 g/L | 28 days | 3.09% mass loss | This work |
| P. restrictum | PC-PET fragments (5.0 × 5.0 × 0.1 mm) at 8.33 g/L | 28 days | 0.08% mass loss | This work |
Besides mass loss of PC-PET fragments, the concentrations of BHET, MHET and TPA in the fermentation broth were also evaluated (Fig. 4). BHET was detected at very low concentrations (> 0.7 µmol/L) with the exception of the time point at 7 days: 23.44 and 9.14 µmol/L for P. restrictum and P. simplicissimum, respectively (Fig. 4A). MHET was also present in low concentrations (> 4.0 µmol/L), with the notable exception of biodegradation by P. restrictum at 14 days (Fig. 4B). In contrast, TPA concentrations were higher during biodegradation by P. restrictum, with a maximum value of 247.91 µmol/L at 7 days, and values between 38.00 and 72.98 µmol/L for the other three time points (Fig. 4C). For P. simplicissimum, the highest TPA concentration was also observed at 7 days (32.94 µmol/L), and did not surpass 2.09 µmol/L at the remaining time points (Fig. 4C).
Fig. 4.
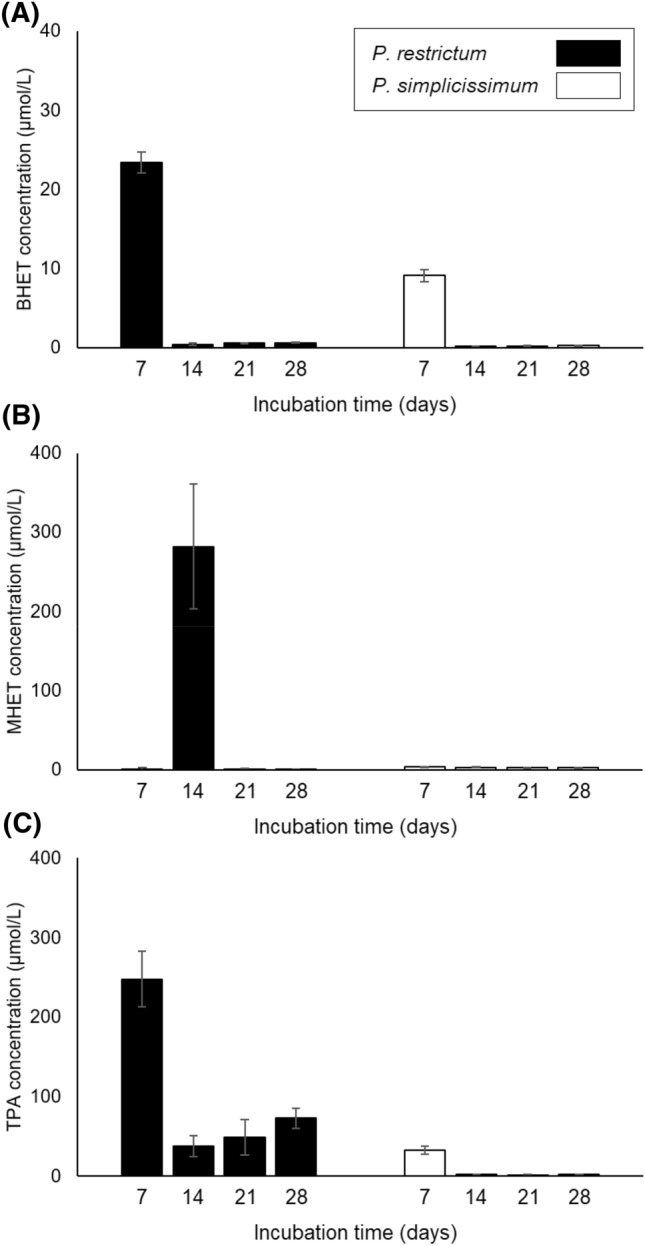
Concentrations of BHET (A), MHET (B) and TPA (C) solubilized in the broth of submerged fermentation for microbial biodegradation of PC-PET fragments by P. restrictum and P. simplicissimum. Error bars denote the standard error of two independent biological replicates
Considering the low concentrations of BHET, MHET and TPA in the fermentation broth, and their projected concentrations according to the mass loss of PC-PET fragments (circa 1300 µmol/L, total), it may be inferred that these compounds were converted into other compounds by P. simplicissimum cells. On the other side, the negligible mass loss imposed by P. restrictum, as well as the low concentrations of BHET, MHET and TPA detected during biodegradation indicate that P. restrictum was not efficient in releasing these monomers from the PC-PET fragments.
Enzymatic PC-PET hydrolysis
Based on the results of the previous assays, in vitro enzymatic biodegradation of PC-PET utilizing cell-free filtrates of P. simplicissimum culture were carried out. In this culture, no lipase inducers were applied and the filtrate achieved a peak of lipase specific activity of 1378.9 U/g of protein at 52 h, with a productivity of 5.05 U/L.h. Gutarra et al. (2009a) reported a higher productivity (27.5 U/L.h after 120 h) but a lower specificity (629 U/g of protein) for P. simplicissimum cultivated in submerged fermentation, while Costa et al. (2020) reported a peak of lipase activity for Y. lipolytica close to 120 U/L at 24 h and a productivity of 5.0 U/L.h when cultivated in YP medium under the presence of BHET.
The hydrolysis here employed were at conditions within the range of the optimal lipolytic activity of crude extracts obtained from solid-state fermentation (SSF) of a castor bean (Ricinus communis) waste by P. simplicissium (temperature from 25–45 °C and pH 6.5–9.0) (Godoy et al. 2011). Considering the products of PC-PET biodegradation, concentrations of BHET and MHET were maintained almost stabilized in the range of 0.46 to 1.06 µmol/L (for BHET) and 2.46 to 3.24 µmol/L (for MHET) (Fig. 5). In contrast, the concentration of TPA began to increase above the detection limit from day 14 onwards and reached a final concentration of 9.51 µmol/L (Fig. 5). For comparison, biodegradation of PET yarns (2 g/L) by enzymatic preparations from F. oxysporum LCH 1 (Nimchua et al. 2007) and F. solani PBURU-B5 (Nimchua et al. 2008) resulted in a final concentration of TPA around 100 µmol/L after 7 days, respectively (enzyme dosage was 80 U of p-nitrophenyl butyrate-hydrolyzing activity). The typical transitions of MHET as a hydrolysis intermediate, observed during PC-PET hydrolysis catalyzed by T. insolens cutinase (Castro et al. 2017) were not noted here, most probably to the low products concentration.
Fig. 5.
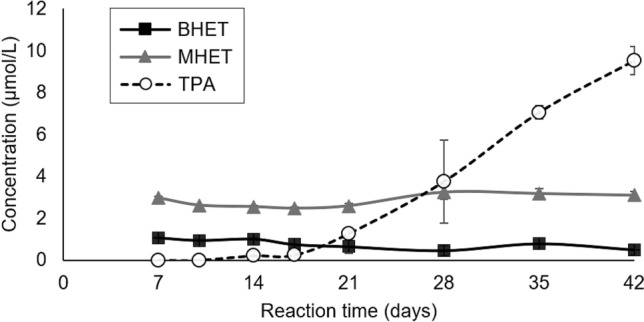
Concentrations of soluble BHET, MHET and TPA in enzymatic biodegradation of PC-PET fragments by cell-free filtrates of P. simplicissimum. Error bars denote the standard error of three independent replicates
The maximum PET conversion of these assays was around 0.15%, well below that found in the microbial biodegradation assay. These low concentrations of BHET, MHET and TPA were expected considering that PC-PET has a high molar mass (42,737 ± 288 g/mol) and high degree of crystallinity (36.6 ± 0.5%), and these properties limit the access of enzymes to the ester bonds, making PC-PET very recalcitrant to biodegradation (Ronkvist et al. 2009; Castro et al. 2017).
In enzymatic assays, the activity of the extracellular enzymes lasts while the proteins are in their active conformation, and no replenishment occurs, in contrast to what happens during microbial biodegradation. Also, the catalytic behavior of extracellular enzymes is dependent on their stability to temperature, pH and other environmental factors. Gutarra et al. (2009b) classified the lipases of the crude extract obtained from SSF of babassu (Attalea speciosa) cake with this same variant of P. simplicissimum as thermostable, displaying > 50% of activity retention at temperatures of up to 40 °C.
In contrast to enzyme preparations and isolated enzymes, whole organisms can resort to mechanisms that facilitate access of enzymes to complex polymers, such as hyphal penetration into the substrate, adsorption onto the hyphal wall and production of natural biosurfactants (Sánchez 2020). As an example of the beneficial effects of biosurfactants, the fusion of a biosurfactant protein (a class II hydrophobin) from the filamentous fungus Trichoderma sp. with the HiC cutinase from T. insolens increased PET hydrolysis by this enzyme (Espino-Rammer et al. 2013). At least one strain from the Penicillium genus is known to produce lipopeptide biosurfactants—P. chrysogenum SNP5 (Gautam et al. 2014)—but P. simplicissimum does not seem to have been investigated for biosurfactant production.
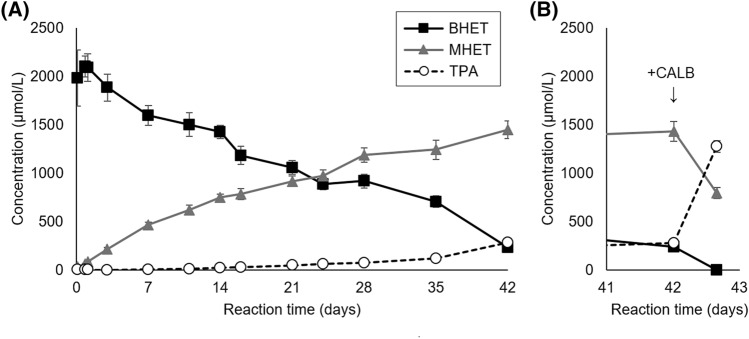
Enzymatic BHET hydrolysis
To better understand the biodegradation mechanism of the extracellular enzymes present in the cell-free filtrate of P. simplicissimum cultures, a second enzymatic biodegradation assay was performed using BHET as substrate. As can be seen in Fig. 6A, BHET concentration gradually decreased during reaction time (from ~ 2000 to 230 µmol/L) while MHET gradually increased (from 30 to 1449 µmol/L), indicating that BHET was predominantly converted into MHET and EG. The concentration of TPA also increased, but at a slower rate, reaching 280 µmol/L at the end of the reaction time of 42 days (Fig. 6A). These higher concentrations of MHET and TPA when comparing with the enzymatic hydrolysis of PC-PET reflect that BHET is a less recalcitrant substrate than PC-PET, which can be attributed to its lower molar mass and higher solubility in the reaction medium, amongst other factors.
Fig. 6.
Concentrations of soluble BHET, MHET and TPA in enzymatic biodegradation of BHET by cell-free filtrates of P. simplicissimum before (A) and after (B) addition of Lipozyme© CALB at 42 days of reaction time. Error bars denote the standard error of three independent replicates
The action of a number of other enzymes also yields MHET as the predominant product of BHET or PET biodegradation. This is the case for: (1) the biodegradation of PET films by a cutinase from the fungus F. solani pisi (Vertommen et al. 2005) and by the PETase from the bacterium I. sakaiensis (Yoshida et al. 2016); (2) the biodegradation of a cyclic PET trimer by a cutinase developed by Novozymes (Hooker et al. 2003); (3) the biodegradation of PET nanoparticles by TfCut2 cutinase from the bacterium Thermobifida fusca KW3 (Barth et al. 2015); (4) the biodegradation of BHET by nine out of ten commercial enzymes (Carniel et al. 2017), by the mutant Cut190 cutinase from the fungus S. viridis AHK190 (Hantani et al. 2018), by the esterase EstB from the bacterium Enterobacter sp. HY1 (Qiu et al. 2020), and by four commercial enzymes (Ion et al. 2020).
Considering the build up of MHET, we tested if the addition of Lipozyme© CALB would be able to convert the remaining BHET and MHET to TPA. CALB is a lipase from the yeast M. antarcticus (formerly C. antarctica) that has been characterized as an efficient catalyst of the conversion of BHET to TPA (Carniel et al. 2017). Indeed, after only 16 h of additional reaction time, the action of CALB drastically reduced the concentrations of BHET and MHET, while it significantly increased TPA concentration (Fig. 6B). The average molar fraction at 42 days for BHET was 11.7%, MHET 73.9%, and TPA 14.3%, and after 16 h of CALB catalysis they change respectively to 0.0%, 38.4% and 61.5%.
Conclusions
Under submerged fermentation, P. simplicissimum attained both the highest lipase hydrolytic activity (606.4 U/L) and productivity (12.38 U/L.h) at 49 h under the presence of 0.2 g/L of BHET, and BHET biodegradation by this fungus resulted in the accumulation of the intermediate product MHET in the fermentation broth. In contrast, P. restrictum did not increase lipase production under the presence of either BHET or amorphous PET, and BHET biodegradation resulted in an accumulation of TPA in the fermentation broth. P. simplicissimum also led to a higher mass loss in microbial biodegradation of PC-PET fragments from bottles, achieving 3.08% after 28 days, while no significant degradation was observed with P. restrictum.
The hydrolysis of PC-PET fragments by the enzymes from the cell-free filtrate of P. simplicissimum culture underperformed the microbial biodegradation, suggesting that mechanisms at the cell level contribute to PET depolymerization, which should be further studied in future works. Enzymatic hydrolysis of BHET catalyzed by the extracellular enzymes of P. simplicissimum released MHET as the main product, suggesting that the fungal enzymes may be inhibited by the reaction intermediate, as similarly found in many other single enzymes or enzymatic preparations characterized so far.
The results presented here supports that P. simplicissimum is a promising biodegrader of post-consumer PET, since it achieved one of the highest rates of mass loss when compared to other results from the literature. Therefore, biodegradation by P. simplicissimum should be further investigated and developed in the context of a circular economy of PET wastes.
Acknowledgements
This study was funded by Petrobras (Petróleo Brasileiro S.A.).
Abbreviations
- BHET
Bis-(2-hydroxyethyl) terephthalate
- EG
Ethylene glycol
- MHET
Mono-(2-hydroxyethyl) terephthalate
- PC-PET
Post-consumer PET
- PET
Poly(ethylene terephthalate)
- SSF
Solid-state fermentation
- TPA
Terephthalic acid
Author contributions
DNS and DAT planned and executed the laboratory work; VAW wrote the original draft; DMGF reviewed the original draft; AMC planned and supervised the laboratory work and reviewed the original draft.
Accession numbers
Penicillium simplicissimum 28f2 (acession number 071113-05 at the International Depository Authority of Canada—IDAC).
Declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Contributor Information
Danuza N. Moyses, Email: danuzamoyses@petrobras.com.br
Danielle A. Teixeira, Email: danielle_altomari@yahoo.com.br
Vinicius A. Waldow, Email: vinicius.waldow@petrobras.com.br
Denise M. G. Freire, Email: freire@iq.ufrj.br
Aline M. Castro, Email: alinebio@petrobras.com.br
References
- Bai H, Wang H, Sun J, Irfan M, Han M, Huang Y, Han X, Yang Q. Production, purification and characterization of novel beta glucosidase from newly isolated Penicillium simplicissimum H-11 in submerged fermentation. EXCLI J. 2013;12:528–540. [PMC free article] [PubMed] [Google Scholar]
- Barth M, Oeser T, Wei R, Then J, Schmidt J, Zimmermann W. Effect of hydrolysis products on the enzymatic degradation of polyethylene terephthalate nanoparticles by a polyester hydrolase from Thermobifida fusca. Biochem Eng J. 2015;93:222–228. doi: 10.1016/j.bej.2014.10.012. [DOI] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Carniel A, Valoni E, Junior JN, Gomes AC, Castro AM. Lipase from Candida antarctica (CALB) and cutinase from Humicola insolens act synergistically for PET hydrolysis to terephthalic acid. Process Biochem. 2017;59:84–90. doi: 10.1016/j.procbio.2016.07.023. [DOI] [Google Scholar]
- Carniel A, Waldow VA, Castro AM. A comprehensive and critical review on key elements to implement enzymatic PET depolymerization for recycling purposes. Biotech Adv. 2021;52:107811. doi: 10.1016/j.biotechadv.2021.107811. [DOI] [PubMed] [Google Scholar]
- Castro AM, Carniel A, Junior JN, Gomes AC, Valoni E. Screening of commercial enzymes for poly(ethylene terephthalate) (PET) hydrolysis and synergy studies on different substrate sources. J Ind Microbiol Biotechnol. 2017;44:835–844. doi: 10.1007/s10295-017-1942-z. [DOI] [PubMed] [Google Scholar]
- Copley SD. Shining a light on enzyme promiscuity. Curr Opin Struc Biol. 2017;47:167–175. doi: 10.1016/j.sbi.2017.11.001. [DOI] [PubMed] [Google Scholar]
- Costa AM, Oliveira Lopes VR, Vidal L, Nicaud JM, Castro AM, Coelho MAZ. Poly(ethylene terephthalate) (PET) degradation by Yarrowia lipolytica: investigations on cell growth, enzyme production and monomers consumption. Process Biochem. 2020;95:81–90. doi: 10.1016/j.procbio.2020.04.001. [DOI] [Google Scholar]
- Crippa M, Morico B. PET depolymerization: a novel process for plastic waste chemical recycling. Stud Surf Sci Catal. 2019;179:215–229. doi: 10.1016/B978-0-444-64337-7.00012-4. [DOI] [Google Scholar]
- Eberl A, Heumann S, Brückner T, Araujo R, Cavaco-Paulo A, Kaufmann F, Kroutil W, Guebitz GM. Enzymatic surface hydrolysis of poly(ethylene terephthalate) and bis(benzoyloxyethyl) terephthalate by lipase and cutinase in the presence of surface active molecules. J Biotechnol. 2009;143(3):207–212. doi: 10.1016/j.jbiotec.2009.07.008. [DOI] [PubMed] [Google Scholar]
- Espino-Rammer L, Ribitsch D, Przylucka A, Marold A, Greimel KJ, Acero EH, Guebitz GM, Kubicek CP, Druzhinina IS. Two novel class II hydrophobins from Trichoderma spp. stimulate enzymatic hydrolysis of poly(ethylene terephthalate) when expressed as fusion proteins. Appl Environ Microbiol. 2013;79(14):4230–4238. doi: 10.1128/AEM.01132-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freire DM, Gomes PM, Bon EPS, Sant’Anna GL., Jr Lipase production by a new promising strain of Penicillium restrictum. Rev Microbiol. 1997;28(Suppl. 1):6–12. [Google Scholar]
- Freire DM, Teles EMF, BonSant’Anna EPSGL., Jr Lipase production by Penicillium restrictum in a bench-scale fermenter: effect of carbon and niltrogen nutrition, agitation, and aeration. Appl Biochem Biotechnol. 1997;63:09–421. doi: 10.1007/BF02920442. [DOI] [PubMed] [Google Scholar]
- Freire DMG (1996) Seleção de microorganismos lipolíticos e estudo da produção de lipase por Penicillium restrictum. Dissertation, Universidade Federal do Rio de Janeiro
- Gan Z, Zhang H. PMBD: a comprehensive plastics microbial biodegradation database. Database. 2019;2019:1–11. doi: 10.1093/database/baz119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautam G, Mishra V, Verma P, Pandey AK, Negi S. A cost effective strategy for production of bio-surfactant from locally isolated Penicillium chrysogenum SNP5 and its applications. J Bioproces Biotech. 2014;4:6. doi: 10.4172/2155-9821.1000177. [DOI] [Google Scholar]
- Geyer R, Jambeck JR, Law KL. Production, use, and fate of all plastics ever made. Sci Adv. 2017;3:e1700782. doi: 10.1126/sciadv.1700782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godoy MG, Gutarra MLE, Castro AM, Machado OLT, Freire DMG. Adding value to a toxic residue from the biodiesel industry: production of two distinct pool of lipases from Penicillium simplicissimum in castor bean waste. J Ind Microbiol Biotechnol. 2011;38(8):945–953. doi: 10.1007/s10295-010-0865-8. [DOI] [PubMed] [Google Scholar]
- Guebitz GM, Cavaco-Paulo A. Enzymes go big: surface hydrolysis and functionalisation of synthetic polymers. Trends Biotechnol. 2007;26(1):32–38. doi: 10.1016/j.tibtech.2007.10.003. [DOI] [PubMed] [Google Scholar]
- Gutarra MLE, Cavalcanti EDC, Castilho LR, Freire DMG, Sant'Anna GL., Jr Lipase production by solid-state fermentation: cultivation conditions and operation of tray and packed-bed bioreactors. Appl Biochem Biotechnol. 2005;121(1–3):105–116. doi: 10.1385/ABAB:121:1-3:0105. [DOI] [PubMed] [Google Scholar]
- Gutarra MLE, Godoy MG, Castilho LR, Freire DMG. Inoculum strategies for Penicillium simplicissimum lipase production by solid-state fermentation using a residue from the babassu oil industry. J Chem Technol Biotechnol. 2007;82(3):313–318. doi: 10.1002/jctb.1674. [DOI] [Google Scholar]
- Gutarra MLE, Godoy MG, Silva JN, Guedes IA, Lins U, Castilho LR, Freire DMG. Lipase production and Penicillium simplicissimum morphology in solid-state and submerged fermentations. Biotechnol J. 2009;4:1450–1459. doi: 10.1002/biot.200800298. [DOI] [PubMed] [Google Scholar]
- Gutarra MLE, Godoy MG, Maugeri F, Rodrigues MI, Freire DMG, Castilho LR. Production of an acidic and thermostable lipase of the mesophilic fungus Penicillium simplicissimum by solid-state fermentation. Bioresour Technol. 2009;100(21):5249–5254. doi: 10.1016/j.biortech.2008.08.050. [DOI] [PubMed] [Google Scholar]
- Hall CAS. Will EROI be the primary determinant of our economic future? The view of the natural scientist versus the economist. Joule. 2017;1(4):635–638. doi: 10.1016/j.joule.2017.09.010. [DOI] [Google Scholar]
- Hantani Y, Imamura H, Yamamoto SA, Yamagami Y, Kato M, Kawai F, Oda M. Functional characterizations of polyethylene terephthalate-degrading cutinase-like enzyme Cut190 mutants using bis(2-hydroxyethyl) terephthalate as the model substrate. AIMS Biophys. 2018;5(4):290–302. doi: 10.3934/biophy.2018.4.290. [DOI] [Google Scholar]
- Hasan F, Shah AA, Hameed A. Methods for detection and characterization of lipases: a comprehensive review. Biotech Adv. 2009;27:782–798. doi: 10.1016/j.biotechadv.2009.06.001. [DOI] [PubMed] [Google Scholar]
- Hooker J, Hinks D, Montero G, Icherenska M. Enzyme-catalyzed hydrolysis of poly(ethylene terephthalate) cyclic trimer. J Appl Polym Sci. 2003;89:2545–2552. doi: 10.1002/app.11963. [DOI] [Google Scholar]
- Hyde KD, et al. The amazing potential of fungi: 50 ways we can exploit fungi industrially. Fungal Divers. 2019;97:1–136. doi: 10.1007/s13225-019-00430-9. [DOI] [Google Scholar]
- Ion S, Voicea S, Sora C, Gheorghita G, Tudorache M, Parvulescu VI. Sequential biocatalytic decomposition of BHET as valuable intermediator of PET recycling strategy. Catal Today. 2020;366:177–184. doi: 10.1016/j.cattod.2020.08.008. [DOI] [Google Scholar]
- Kawai F, Kawabata T, Oda M. Current knowledge on enzymatic PET degradation and its possible application to waste stream management and other fields. Appl Microbiol Biotechnol. 2019;103:4253–4268. doi: 10.1007/s00253-019-09717-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai F, Kawabata T, Oda M. Current state and perspectives related to the PET hydrolases available for biorecycling. ACS Sustain Chem Eng. 2020;8(24):8894–8908. doi: 10.1021/acssuschemeng.0c01638. [DOI] [Google Scholar]
- Kim DY, Rhee YH. Biodegradation of microbial and synthetic polyesters by fungi. Appl Microbiol Biotechnol. 2003;61:300–308. doi: 10.1007/s00253-002-1205-3. [DOI] [PubMed] [Google Scholar]
- Koshti R, Mehta L, Samarth N. Biological recycling of polyethylene terephthalate: a mini-review. J Polym Environ. 2018;26:3520–3529. doi: 10.1007/s10924-018-1214-7. [DOI] [Google Scholar]
- Kumar AG, Anjana K, Hinduja M, Sujitha K, Dharani G. Review on plastic wastes in marine environment—biodegradation and biotechnological solutions. Mar Pollut Bull. 2020;150:110733. doi: 10.1016/j.marpolbul.2019.110733. [DOI] [PubMed] [Google Scholar]
- Lepoittevin B, Roger P. Poly(ethylene terephthalate) In: Thomas S, Visakh PM, editors. Handbook of engineering and speciality thermoplastics: polyethers and polyesters. Beverly: Scrivener Publishing; 2011. pp. 97–126. [Google Scholar]
- Liebminger S, Eberl A, Sousa F, Heumann S, Fischer-Colbrie G, Cavaco-Paulo A, Guebitz GM. Hydrolysis of PET and 3PET with a new polyesterase from Penicillium citrinum. Biocatal Biotransform. 2007;25(2–4):171–177. doi: 10.1080/10242420701379734. [DOI] [Google Scholar]
- Malafatti-Picca L, Chaves MRB, Castro AM, Valoni E, Oliveira VM, Marsaioli AJ, Angelis DF, Attili-Angelis D. Hydrocarbon-associated substrates reveal promising fungi for poly(ethylene terephthalate) (PET) depolymerization. Braz J Microbiol. 2019;50:633–648. doi: 10.1007/s42770-019-00093-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markit IHS (2018) Chemical economics handbook—PET polymer. https://ihsmarkit.com/products/pet-polymer-chemical-economics-handbook.html. Accessed 10 Sept 2020
- Nimchua T, Punnapayak H, Zimmermann W. Comparison of the hydrolysis of polyethylene terephthalate fibers by a hydrolase from Fusarium oxysporum LHC I and Fusarium solani f. sp. pisi. Biotechnol J. 2007;2:361–364. doi: 10.1002/biot.200600095. [DOI] [PubMed] [Google Scholar]
- Nimchua T, Eveleigh DE, Sangwatanaroj U, Punnapayak H. Screening of tropical fungi producing polyethylene terephthalate-hydrolyzing enzyme for fabric modification. J Ind Microbiol Biotechnol. 2008;35:843–850. doi: 10.1007/s10295-008-0356-3. [DOI] [PubMed] [Google Scholar]
- Nowak B, Pajak J, Labuzek S, Rymarz G, Talik E. Biodegradation of poly(ethylene terephthalate) modified with polyester "Bionolle®" by Penicillium funiculosum. Polimery. 2011;56(1):35–44. doi: 10.14314/polimery.2011.035. [DOI] [Google Scholar]
- Qiu L, Yin X, Liu T, Zhang H, Chen G, Wu S. Biodegradation of bis(2-hydroxyethyl) terephthalate by a newly isolated Enterobacter sp. HY1 and characterization of its esterase properties. J Basic Microbiol. 2020;60:699–711. doi: 10.1002/jobm.202000053. [DOI] [PubMed] [Google Scholar]
- Raheem AB, Noor ZZ, Hassan A, Hamid MKA, Samsudin SA, Sabeen AH. Current developments in chemical recycling of post-consumer polyethylene terephthalate wastes for new materials production: a review. J Clean Prod. 2019;225:1052–1064. doi: 10.1016/j.jclepro.2019.04.019. [DOI] [Google Scholar]
- Ronkvist AM, Xie W, Lu W, Gross RA. Cutinase-catalyzed hydrolysis of poly(ethylene terephthalate) Macromolecules. 2009;42:5128–5138. doi: 10.1021/ma9005318. [DOI] [Google Scholar]
- Sales JCS, Castro AM, Ribeiro BD, Coelho MAZ. Supplementation of watermelon peels as an enhancer of lipase and esterase production by Yarrowia lipolytica in solid-state fermentation and their potential use as biocatalysts in poly(ethylene terephthalate) (PET) depolymerization reactions. Biocatal Biotransform. 2020;38:457–468. doi: 10.1080/10242422.2020.1782387. [DOI] [Google Scholar]
- Sales JCS, Castro AM, Ribeiro BD, Coelho MAZ. Improved production of biocatalysts by Yarrowia lipolytica using natural sources of the biopolyesters cutin and suberin, and their application in hydrolysis of poly (ethylene terephthalate) (PET) Bioprocess Biosyst Eng. 2021 doi: 10.1007/s00449-021-02603-w. [DOI] [PubMed] [Google Scholar]
- Salvador M, Abdulmutalib U, Gonzalez J, Kim J, Smith AA, Faulon JP, Wei R, Zimmermann W, Jimenez JI. Microbial genes for a circular and sustainable bio-PET economy. Genes. 2019;10:373. doi: 10.3390/genes10050373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez C. Fungal potential for the degradation of petroleum-based polymers: an overview of macro- and microplastics biodegradation. Biotechnol Adv. 2020;40:107501. doi: 10.1016/j.biotechadv.2019.107501. [DOI] [PubMed] [Google Scholar]
- Sharon C, Sharon M. Studies on biodegradation of polyethylene terephthalate: a synthetic polymer. J Microbiol Biotech Res. 2012;2(2):248–257. [Google Scholar]
- Silva CM, Carneiro F, O’Neill A, Fonseca LP, Cabral JSM, Guebitz G, Cavaco-Paulo A. Cutinase—a new tool for biomodification of synthetic fibres. J Polym Sci, Part A: Polym Chem. 2005;43:2448–2450. doi: 10.1002/pola.20684. [DOI] [Google Scholar]
- Sinha V, Patel MR, Patel JV. PET waste management by chemical recycling: a review. J Polym Environ. 2010;18:8–25. doi: 10.1007/s10924-008-0106-7. [DOI] [Google Scholar]
- Syedd-León R, Sandoval-Barrantes M, Trimiño-Vásquez H, Villegas-Peñaranda LR, Rodríguez-Rodríguez G. Revisiting the fundamentals of p-nitrophenol analysis for its application in the quantification of lipases activity. A graphical update. Uniciencia. 2020;34(2):31–43. doi: 10.15359/ru.34-2.2. [DOI] [Google Scholar]
- Taniguchi I, Yoshida S, Hiraga K, Miyamoto K, Kimura Y, Oda K. Biodegradation of PET: current status and application aspects. ACS Catal. 2019;9(5):4089–4105. doi: 10.1021/acscatal.8b05171. [DOI] [Google Scholar]
- Tournier V, Topham CM, Gilles A, David B, Folgoas C, Moya-Leclair E, Kamionka E, Desrousseaux ML, Texier H, Gavalda S, Cot M, Guémard E, Dalibey M, Nomme J, Cioci G, Barbe S, Chateau M, André I, Duquesne S, Marty A. An engineered PET depolymerase to break down and recycle plastic bottles. Nature. 2020;580(7802):216–219. doi: 10.1038/s41586-020-2149-4. [DOI] [PubMed] [Google Scholar]
- Vertommen MAME, Nierstrasz VA, Veer MVD, Warmoeskerken MMCG. Enzymatic surface modification of poly(ethylene terephthalate) J Biotechnol. 2005;120:376–386. doi: 10.1016/j.jbiotec.2005.06.015. [DOI] [PubMed] [Google Scholar]
- Wang X, Lu D, Jönsson LJ, Hong F. Preparation of a PET-hydrolyzing lipase from Aspergillus oryzae by the addition of bis(2-hydroxyethyl) terephthalate to the culture medium and enyzmatic modification of PET fabrics. Eng Life Sci. 2008;8(3):268–276. doi: 10.1002/elsc.200700058. [DOI] [Google Scholar]
- Wei R, Zimmermann W. Biocatalysis as a green route for recycling the recalcitrant plastic polyethylene terephthalate. Microb Biotechnol. 2017;10(6):1302–1307. doi: 10.1111/1751-7915.12714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei R, Zimmermann W. Microbial enzymes for the recycling of recalcitrant petroleum-based plastics: how far are we? Microb Biotechnol. 2017;10(6):1308–1322. doi: 10.1111/1751-7915.12710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierckx N, Narancic T, Eberlein C, Wei R, Drzyzga O, Magnin A, Ballerstedt H, Kenny ST, Pollet E, Avérous L, O’Connor KE, Zimmermann W, Heipieper HJ, Prieto A, Jiménez J, Blank LM. Plastic biodegradation: callenges and opportunities. In: Steffan R, editor. Consequences of microbial interactions with hydrocarbons, oils, and lipids: biodegradation and bioremediation, handbook of hydrocarbon and lipid microbiology. Basel: Springer International Publishing AG; 2018. pp. 1–29. [Google Scholar]
- Yamada-Onodera K, Mukomoto H, Katsuyaya Y, Saiganji A, Tani Y. Degradation of polyethylene by a fungus, Penicillium simplicissimum YK. Polym Degrad Stab. 2001;72:323–327. doi: 10.1016/S0141-3910(01)00027-1. [DOI] [Google Scholar]
- Yoshida S, Hiraga K, Takehana T, Taniguchi I, Yamaji H, Maeda Y, Toyohara K, Miyamoto K, Kimura Y, Oda K. A bacterium that degrades and assimilates poly(ethylene terephthalate) Science. 2016;351(6278):1196–1199. doi: 10.1126/science.aad6359. [DOI] [PubMed] [Google Scholar]
- Zhang J, Wang X, Gong J, Gu Z. A study on the biodegradability of polyethylene terephthalate fiber and diethylene glycol terephthalate. J Appl Polym Sci. 2004;93(3):1089–1096. doi: 10.1002/app.20556. [DOI] [Google Scholar]
- Zheng J, Suh S. Strategies to reduce the global carbon footprint of plastics. Nat Clim Chang. 2019;9:374–378. doi: 10.1038/s41558-019-0459-z. [DOI] [Google Scholar]
- Zimmermann W. Biocatalytic recycling of polyethylene terephthalate plastic. Phil Trans R Soc A. 2020;378:20190273. doi: 10.1098/rsta.2019.0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann W, Billig S. Enzymes for the biofunctionalization of poly(ethylene terephthalate) Adv Biochem Eng/biotechnol. 2011;125:97–120. doi: 10.1007/10_2010_87. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Penicillium simplicissimum 28f2 (acession number 071113-05 at the International Depository Authority of Canada—IDAC).