Abstract
The potent transactivator Tat recognizes the transactivation response RNA element (TAR) of human immunodeficiency virus type 1 and stimulates the processivity of elongation of RNA polymerase (Pol) II complexes. The cellular proteins Tat-SF1 and human SPT5 (hSPT5) are required for Tat activation as shown by immunodepletion with specific sera and complementation with recombinant proteins. In nuclear extracts, small fractions of both hSPT5 and Pol II are associated with Tat-SF1 protein. Surprisingly, the RAP30 protein of the heterodimeric transcription TFIIF factor is associated with Tat-SF1, while the RAP74 subunit of TFIIF is not coimmunoprecipitated with Tat-SF1. Overexpression of Tat-SF1 and hSPT5 specifically stimulates the transcriptional activity of Tat in vivo. These results suggest that Tat-SF1 and hSPT5 are indispensable cellular factors supporting Tat-specific transcription activation and that they may interact with RAP30 in controlling elongation.
The human immunodeficiency virus (HIV) Tat protein is expressed early in the viral life cycle and is required for viral replication and progression to disease (5–7, 16). Unlike other transcriptional activators which bind DNA, Tat binds to RNA in the transactivation response element (TAR), which forms a stem-loop structure in the 5′ terminus of HIV-1 transcripts (8, 27). Tat activates transcription by increasing the processiveness of elongation by the RNA polymerase (Pol) II. Several cellular proteins which are important for Tat activation of elongation have been biochemically characterized and purified. Among these are the Tat specific factor Tat-SF1 (44), the positive transcription elongation factor P-TEFb (19, 22, 23, 45), and TFIIH (reference 16 and references therein; see also references 4, 24, and 40).
P-TEFb is a general elongation factor and was initially identified biochemically in Drosophila melanogaster as a factor which suppressed the activity of an inhibitor of elongation (22, 23). P-TEFb is composed of several protein subunits including the novel kinase, Cdk9/PITALRE, and its cyclin partner, cyclin T (19, 26, 33, 45). P-TEFb will efficiently phosphorylate the carboxy-terminal domain (CTD) of Pol II, and this may be its critical activity in the stimulation of elongation. Several experimental results suggest that P-TEFb plays a critical role in Tat activation. First, the depletion of P-TEFb inactivates Tat stimulation of HIV transcription in vitro (45). Second, the sensitivity of Tat activation to a spectrum of different drugs mirrors those which inhibit Cdk9 kinase activity in vitro (19). Third, a cellular kinase complex termed TAK (Tat-activated kinase) that interacts with the activation domain of Tat and phosphorylates the CTD of Pol II has been identified as P-TEFb (12, 13, 39, 45). Fourth, in the presence of Tat, cyclin T binds to TAR RNA in a loop sequence-dependent manner (26, 33). Recent studies have shown that human cyclin T contains a cysteine residue that is critical for the specific binding to TAR and which is not found in the rodent homolog (1, 9). The rodent cyclin T protein does not recognize the loop sequence of TAR, and this difference in cyclin T sequence likely explains the specificity of Tat activation for human cells compared to rodent cells. Finally, overexpression of a mutant Cdk9 kinase blocks Tat activation of elongation in human cells (19).
The TFIIH kinase complex also has an important role in Tat activation, probably by stimulating the phosphorylation of the CTD of Pol II (10, 24, 40). The TFIIH factor, particularly its Cdk7 kinase of the CAK type, is important for transcription of most promoters in vivo (14). Furthermore, a blockage of the kinase activity of TFIIH reduces Tat-dependent transcription activation (4, 24, 40). Therefore, it has been suggested that both P-TEFb and TFIIH phosphorylate the CTD of Pol II, perhaps in a sequential manner, promoting the processivity of Pol II by Tat (7, 15).
A recent study has suggested that the human homolog of the yeast transcription factor SPT5 is also involved in Tat-activated elongation of transcription (35). Both genetic and biochemical data suggest that the two yeast proteins SPT4 and SPT5 are stably associated in an active transcription complex and that SPT5 interacts with Pol II (11, 34). The human SPT4 and SPT5 proteins also form a complex, denoted as DSIF (DRB sensitive inducing factor [31]), which arrests the elongation of Pol II at sites proximal to the promoter and release from this pause-state is sensitive to DRB (5,6-dichloro-1-β-d-ribofuranosylbenzimidazole) (31, 32, 38). It was subsequently shown that P-TEFb positively regulates Pol II processivity by, at least in part, suppressing the activity of DSIF in a phosphorylation step that is DRB sensitive (32, 38).
Previous studies have demonstrated that a partially purified reconstituted transcription reaction supported Tat-specific and TAR-dependent activation of HIV-1 transcription (43, 44). This reconstituted reaction requires the cellular factor, Tat-SF1 (Tat-stimulatory factor 1) (18, 43, 44). Tat-SF1 is a phosphoprotein which directly binds wild-type Tat protein but not a transcriptionally inactive mutant Tat protein (reference 44 and unpublished results). The amino acid sequence of Tat-SF1, as deduced from the cDNA sequence, contains two RNA recognition motifs and a highly acidic carboxyl terminal motif (44). This study shows that Tat-SF1 efficiently associates with the RAP30 subunit of TFIIF and inefficiently associates with Pol II and human SPT5 (hSPT5). Consistent with previous observations, depletion of either Tat-SF1 or hSPT5 from nuclear extract inactivates Tat stimulation of transcription. In addition, we show for the first time that complementation with either recombinant Tat-SF1 or hSPT5 protein restores the Tat activation of transcription. Finally, overexpression of Tat-SF1 and hSPT5 increases the level of transcription in a Tat-specific manner in vivo.
MATERIALS AND METHODS
In vitro transcription.
Transcription reactions were performed as described previously with minor modifications (43). Briefly, these reactions contained 13 mM HEPES, 60 mM KCl, 10 mM creatine phosphate, 7 mM MgCl2, 7 mM dithiothreitol (DTT), 300 ng of poly(I)-poly(C), 50 ng of poly(dG)-poly(dC), 0.1 mM EDTA, 10% (vol/vol) glycerol, and 100 to 200 ng of template DNA (pHIV-LTR+TAR and pHIV-LTRΔTAR), as well as 25 ng of purified Tat protein when indicated, and 25 to 30 μg of HeLa nuclear extracts or immunodepleted HeLa nuclear extracts. As control template DNAs, 12.5 ng of adenovirus E4 promoter and 112.5 ng of major late promoter (MLP), which produced G400 and G300 transcripts, respectively, were used (2, 25). Immunodepletions of Tat-SF1 and hSPT5 were performed as described previously (31, 32, 44). Reactions were assembled on ice and then preincubated at 30°C for 30 min to allow for preinitiation complex formation, followed by the addition of 10 mCi of [α-32P]CTP (800 Ci/mmol) and 1 μl of an A/G/U/CTP mixture (5 mM of ATP, GTP, and UTP and 0.125 mM CTP). After a further incubation of 30 min, RNase T1 (5 U) was added to the reaction at 37°C for 5 min. In vitro-transcribed RNAs were extracted with phenol-chloroform and precipitated with ethanol. Transcripts were analyzed by electrophoresis on 5% polyacrylamide gels containing 8 M urea, 90 mM Tris base, 89 mM boric acid, and 2 mM EDTA. The gels were dried and exposed to X-ray film. Reaction products were quantitated by using a PhosphorImager and the ImageQuant 3.0 program (Molecular Dynamics).
Cell cultures and CAT assay.
HeLa cells were cultured in DME containing 10% fetal calf serum and transfected at 60% confluence by use of Superfect (Qiagen). The HIV-LTR chloramphenicol acetyltransferase (CAT) (pU3 CAT) and UAS-CAT reporters were described previously (44). To construct Tat-SF1 expression vector (f-Flag-Tat-SF1), full-length Tat-SF1 cDNA was PCR cloned with EcoRI ends and fused in frame with the Flag moiety of plasmid pFlag-CMV (Sigma). Total cell lysates were prepared after 48 h of transfection. The level of CAT gene expression was determined by measuring CAT enzyme activity. β-Galactosidase (β-Gal) assays were performed by a standard colorimetric procedure with chlorophenol red-β-d-galactopyranoside as the substrate (Boehringer Mannheim). The relative CAT activities were normalized to the β-Gal activity. All transfection experiments were performed in duplicate.
Preparation of recombinant Tat-SF1 protein and recombinant DSIF.
Full-length Tat-SF1 cDNA was PCR cloned to fuse in frame into the pFAST-BacHTa vector (Gibco/BRL). Hi5 cells were infected with baculovirus containing full-length Tat-SF1 cDNA with six-histidine tagging at the amino terminus. The cells were harvested at 48 h postinfection, and recombinant Tat-SF1 proteins were purified by use of a nickel-nitriloacetic acid (Ni-NTA) column as recommended by manufacturer (Gibco/BRL). Purified rTat-SF1 protein was dialyzed against buffer D containing 20 mM HEPES-KOH (pH 7.9), 100 mM KCl, 0.5 mM DTT, 0.5 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride (PMSF), and 20% glycerol. Recombinant DSIF (hSPT5 and hSPT4) proteins were purified as described previously (31). Briefly, bacterial strain BL21(DE3) was transformed with a plasmid encoding histidine-tagged fusion hSPT5 or hSPT4 protein and induced with IPTG (isopropyl-β-d-thiogalactopyranoside) and purified by use of a Ni-NTA column as described above. For further purification, affinity-purified DSIF was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and proteins were recovered from the gel, acetone precipitated, denatured, and renatured as described previously (31).
Immunoprecipitation and Western blot.
Total cell extracts were prepared with lysis buffer containing 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 100 μg/ml PMSF, 10 μg of aprotinin per ml, and 1 mM sodium orthovanadate in phosphate-buffered saline. For immunoprecipitation, each antiserum was incubated with 10 μl of HeLa nuclear extract or 250 μg of total cell extract and incubated for 2 h at 4°C. Then, 30 μl of protein G-Sepharose CL-6B (Pharmacia) was added into mixture for 1 h at 4°C with gentle mixing. Immunoprecipitates were collected by centrifugation and were washed three times with 50 mM Tris (pH 7.9), 150 mM NaCl, 1 mM EDTA, and 1% Nonidet P-40 and then once with phosphate-buffered saline. For Western blotting, isolated immunoprecipitates or nuclear extracts were resolved by SDS-PAGE and then transferred to a polyvinylidene difluoride membrane. The blots were incubated with each of antibodies and visualized by using the ECL kit (Amersham).
Affinity purification of Tat-SF1-associated protein complexes.
HeLa cells were transfected with the f-Tat-SF1 expression vector (see above) as described above. f-Tat-SF1 associated complexes were isolated by mixing ∼4 mg of total cell lysates with 30 μl of M2 anti-Flag beads (Sigma), followed by gentle mixing for 4 h. The beads were pelleted and washed four times in wash buffer containing 25 mM Tris (pH 7.8), 250 mM NaCl, 1 mM DTT, and 0.5% Nonidet P-40. The bound proteins were eluted by adding 20 μl of wash buffer containing 3.75 μg of Flag peptides (Sigma), followed by incubation for 10 min before pelleting the beads and collecting the eluate. This elution step was repeated three or four times. A control experiment was carried out in parallel in which anti-Flag beads were mixed with mock-transfected HeLa cell extracts to examine the level of nonspecific binding of proteins to beads.
RESULTS
Tat-SF1-associated protein complexes.
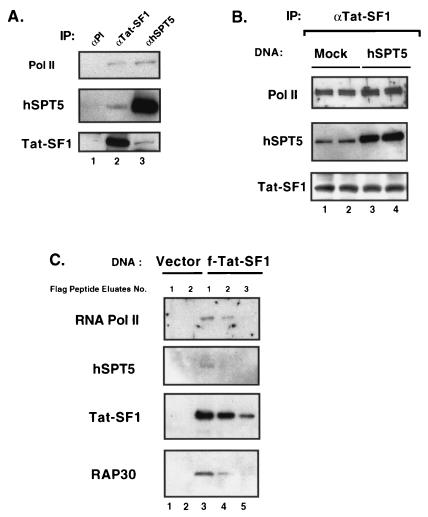
The mechanism by which Tat-SF1 in conjunction with Tat and the general transcription factor stimulates the elongation of transcription was investigated by the identification of proteins associated with Tat-SF1. Surprisingly, coimmunoprecipitation experiments show that Tat-SF1 forms a stable complex with the p30 subunit of the general transcription factor TFIIF (RAP30) (Fig. 1A, lanes 3 and 4). Immunoprecipitation by antiserum to either Tat-SF1 or RAP30 produced similar amounts of the RAP30 and Tat-SF1 polypeptides, respectively. The levels of coimmunoprecipitated Tat-SF1 and RAP30 polypeptides were about 20% of that present in the input nuclear extract (Fig. 1, lane 1). In contrast, coimmunoprecipitates of Tat-SF1 contained negligible amounts of the RAP74 protein, which associates with RAP30 to form the heterodimeric TFIIF (data not shown). Repeated depletion of Tat-SF1, by absorption with specific Tat-SF1 antiserum, reduced the amounts of RAP30 without the depletion of other proteins such as Cdk9 nor RAP74 (Fig. 1B, lanes 2, 3, and 4). The interaction between Tat-SF1 and RAP30 appears to be very specific since a similar association with other elongation factors, such as the elongin A, B, and C complex and TFIIS, was not detected (data not shown). Furthermore, the interaction between Tat-SF1 and RAP30 is probably direct since both polypeptides coimmunoprecipitated with either antiserum when added as purified proteins to a reaction (data not shown).
FIG. 1.
Tat-SF1 associates with RAP30 of TFIIF. (A) Tat-SF1 coimmunoprecipitated with RAP30. HeLa total cell extracts (250 μg) were used for immunoprecipitation with antibodies against preimmune (PI, lane 2), Tat-SF1 (lane 3) (44), and RAP30 of TFIIF (lane 4) (Santa Cruz Biotechnology). The input lane contained 20% of the extract used for the immunoprecipitation (lane 1). Western blots of the immunoprecipitates were probed with anti-Tat-SF1 (top) and anti-RAP30 (bottom) antibodies. (B) Tat-SF1-depleted HeLa nuclear extracts contained decreased levels of RAP30. HeLa nuclear extracts were immunodepleted with anti-Tat-SF1 antibody. After each depletion step, aliquots were taken and Western blotting was performed with antibodies against Tat-SF1 and RAP30. Anti-Cdk9 antibody and anti-RAP74 antibodies (Santa Cruz Biotechnology) were used for the control.
Interestingly, Pol II (largest subunit: Pol II-LS) and human SPT5 (hSPT5) were also found to be associated with Tat-SF1 (Fig. 2A). Immunoprecipitations with antiserum to either Tat-SF1 or hSPT5 produced protein complexes containing Pol II, hSPT5, and Tat-SF1 (Fig. 2A, lanes 2 and 3, respectively). When compared with the interaction between Tat-SF1 and RAP30, the association between Tat-SF1 and hSPT5 is inefficient, since less than 5% of the input SPT5 protein was immunoprecipitated with α-Tat-SF1 sera. The reciprocal experiment gave a similar level (<5%) of Tat-SF1 protein after immunoprecipitation with hSPT5 antibodies (Fig. 2A). The interaction between Tat-SF1 and hSPT5 proteins is probably direct because each protein could be coimmunoprecipitable by antiserum to the other protein from a mixture of purified proteins (data not shown). To ascertain whether the in vivo interaction between Tat-SF1 and hSPT5 is independent of Pol II, HeLa cells were transfected in duplicate with either vector alone (mock) or vector expressing hSPT5. Total cell extracts were prepared from transfected cells and then immunoprecipitated with Tat-SF1 antibody for analysis by Western blotting. As shown in Fig. 2B, the Tat-SF1 immunoprecipitated complex contained the same levels of Pol II-LS and Tat-SF1 (a shorter exposure than that of Fig. 2A) in both the mock (lanes 1 and 2)- and hSPT5 (lanes 3 and 4)-transfected cell lysates. In contrast, the amounts of hSPT5 present in the Tat-SF1 immunocomplex were increased by the overexpression of hSPT5. These results suggest that Tat-SF1 and hSPT5 can associate independently of Pol II binding.
FIG. 2.
Tat-SF1 interacts with hSPT5 and Pol II. (A) Tat-SF1 coimmunoprecipitated with hSPT5 and Pol II. HeLa total cell extracts were immunoprecipitated with antibodies against preimmune (lane 1), Tat-SF1 (lane 2), and hSPT5 (lane 3). Western blots of immunoprecipitates were probed with anti-Pol II CTD (top) (38), anti-hSPT5 (middle) (31), and anti-Tat-SF1 (bottom) antibodies. (B) Expression of hSPT5 in vivo increases the level of association with Tat-SF1. HeLa cells were transfected with vector control (mock) or hSPT5 expression vectors. Aliquots of total cell extracts were immunoprecipitated with anti-Tat-SF1 antibody, and immunoprecipitates were probed with antibodies against Pol II CTD (top), hSPT5 (middle), and Tat-SF1 (bottom) for Western blotting. (C) Isolation of f-Tat-SF1 and its associated protein complexes. An extract of HeLa cells transfected with f-Tat-SF1 was incubated with anti-Flag beads and washed as described in Materials and Methods. As a control, mock-transfected extract was prepared (lanes 1 and 2). To elute bound complex, Flag peptides (∼3.75 μg) were incubated with the beads. After each round of elution, aliquots of eluted fractions were separated by SDS-PAGE, and the Western blots were developed with antibodies against Pol II CTD, hSPT5, Flag (Sigma), and RAP30 of TFIIF.
Cross-reactivity by antiserum is always a concern in coimmunoprecipitation experiments. Therefore, the nature of Tat-SF1-associated protein complexes was examined by using a Flag epitope-tagged Tat-SF1 (f-Tat-SF1) cDNA construct. The Tat-SF1-associated protein complex was purified from total HeLa cell extracts by chromatography with immobilized monoclonal Flag antibody. After an extensive washing, Tat-SF1-associated factors were eluted from the antibody column with synthetic Flag epitope peptides (see Materials and Methods) and analyzed by Western blotting. In accordance with Fig. 1 and 2, these eluted fractions contained Pol II-LS, RAP30, and hSPT5, as well as Tat-SF1 (Fig. 2C, lanes 3, 4, and 5). However, it is not likely that the Tat-SF1-associated complex is a previously defined Pol II holoenzyme complex since Western analysis shows that this complex does not contain polypeptides from TFIIH (Cdk7 and cyclinH), RAP74, and SRB proteins such as Cdk8 and cyclin C (data not shown). These results confirm that Tat-SF1 forms protein complexes with Pol II, RAP30, and hSPT5.
Depletion of either Tat-SF1 or hSPT5 reduces Tat activation of transcription.
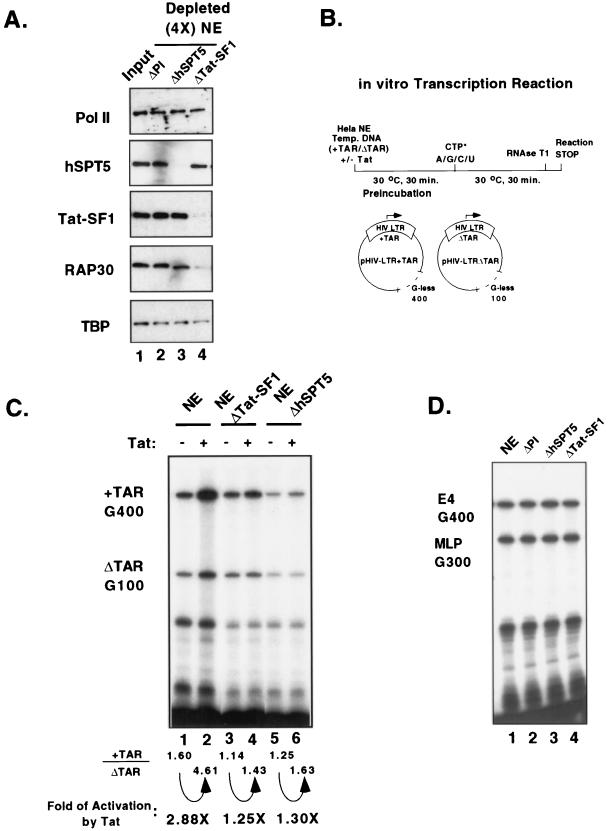
The functional role of Tat-SF1 and hSPT5 in Tat activation of transcription was studied by depletion of nuclear extracts with antiserum to either of these proteins. The levels of depletion were determined by Western blot analysis, and the activities of the depleted extracts were tested for Tat-dependent activation of transcription. Four rounds of immunodepletion produced extracts which contained only 5 to 10% of the Tat-SF1 or hSPT5 protein present in the input HeLa nuclear extract (Fig. 3A). Except for RAP30, no significant changes in the amounts of other general transcription factors, such as Pol II and TBP, were detected (Fig. 3A). Since only a small fraction of Tat-SF1 is associated with hSPT5 and vice versa, the immunodepleted extracts contained nearly input levels of the other protein.
FIG. 3.
Tat-SF1 and hSPT5 are required for Tat-dependent activation. (A) Analysis of Tat-SF1- or hSPT5-depleted nuclear extracts with Western blotting. HeLa nuclear extracts were immunodepleted four times with preimmune (PI) (lane 2), anti-hSPT5 (lane 3), and anti-Tat-SF1 (lane 4) antibodies. Aliquots of each depleted extract were probed with antibodies against Pol II CTD, hSPT5, Tat-SF1, RAP30, and TBP. An equal volume (2 μl) of input nuclear extract was used for the control (lane 1). (B) Scheme of the Tat-dependent TAR-specific transcription reaction. HeLa nuclear extracts were preincubated with template DNAs including pHIV-LTR+TAR and pHIV-LTRΔTAR at 30°C for 30 min. Radiolabelled CTP and 200 μM of cold nucleotide mixtures (ATP, GTP, CTP, and UTP) were added into the reactions for transcription elongation. RNase T1 digestion was carried out to distinguish the Tat-dependent TAR-specific transcripts (+TAR G400) versus the TAR-independent transcripts (ΔTAR G100). (C) Tat-SF1 depleted- and hSPT5 depleted-nuclear extracts are defective for Tat-dependent activation. Immunodepleted nuclear extracts (ΔTat-SF1, lanes 3 and 4; ΔhSPT5, lanes 5 and 6), as well as control nuclear extracts (lanes 1 and 2), were used for transcription in vitro. Purified Tat protein (∼25 ng) was added during the preincubation step as indicated (lanes 2, 4, and 6). TAR-specific transcription activities were calculated by quantitation of the synthesized transcripts from +TAR/ΔTAR. Fold levels of Tat-specific transcription activities were obtained by normalization of TAR-specific transcription activities in the presence or absence of Tat protein as described previously (20). (D) Depletion of Tat-SF1 and hSPT5 did not change the transcription activities from the adenovirus E4 promoter and the MLP. In vitro transcription activities from each of the depleted nuclear extracts, as well as control nuclear extracts, were determined for E4 and MLP (see Materials and Methods).
Two different template DNAs, pHIV+TAR and pHIV ΔTAR, were used to test Tat-dependent TAR-specific transcription activity (Fig. 3B). RNase T1 digestion of transcription products yields two different transcript lengths, 400 and 100 nucleotides, from the pHIV+TAR and pHIV ΔTAR templates, respectively. Since both of these G-less tracks were inserted into the template at a site ca. 1,000 nucleotides downstream of the initiation site, the RNase T1-resistant transcripts indicate elongation products. The pHIV+TAR template, containing the wild-type HIV-LTR (−329 to +82), is responsive to the Tat protein, while the pHIV ΔTAR template, containing a deletion in the TAR sequence (+35 to +38), is not responsive to the Tat protein (21, 43). In control HeLa nuclear extracts, the addition of the Tat protein stimulated a three- to fivefold increase in specific activation (G400 transcripts) without a significant increase in transcription activity from the ΔTAR template (G100 transcripts; Fig. 3C, lanes 1 and 2). In contrast, Tat-SF1- or hSPT5-depleted nuclear extracts failed to support Tat-dependent transcription activation (Fig. 3C, lanes 3 to 6). However, the reduced Tat activation potential produced by the depletion of either Tat-SF1 or hSPT5 did not extend to Tat-independent transcription as shown in the sustained level of ΔTAR G100 transcripts (Fig. 3C, lanes 3 to 6). To ensure that the effects of depletion of either Tat-SF1 or hSPT5 protein are specific for Tat-activated transcription, the same nuclear extracts were tested with other promoters such as the adenovirus E4 and the MLP. After normalization of the protein concentrations, the depleted nuclear extracts showed similar levels of transcription activity when compared to the normal nuclear extracts or preimmune depleted nuclear extracts (Fig. 3D, compare lanes 2 to 4 with lane 1). These results suggest that both Tat-SF1 and hSPT5 are important for Tat-activated transcription are but not essential for general transcription from the HIV-LTR or other promoters.
Recombinant Tat-SF1 and SPT5 proteins complement Tat activation of transcription.
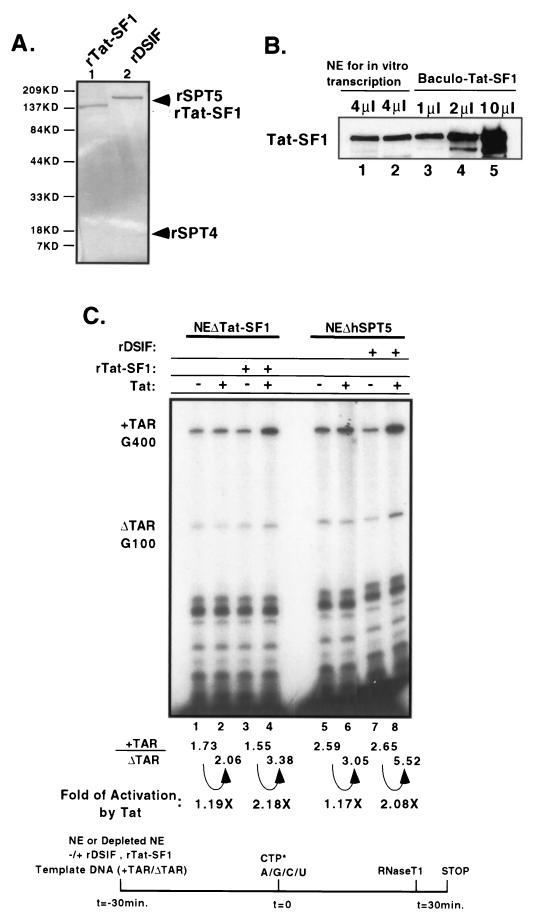
The activity of purified Tat-SF1 or hSPT5 protein in Tat-dependent transcription activation was assayed in vitro by using transcription reactions containing depleted nuclear extracts. Recombinant Tat-SF1 (rTat-SF1) was purified from insect cells infected with baculovirus (Fig. 4A, lane 1), and its concentration in transcription reactions was titrated to levels comparable to that found in HeLa nuclear extracts. Approximately 50 ng of baculovirus-expressed rTat-SF1 protein contained the same level of Tat-SF1 as 30 μg of HeLa nuclear extract (Fig. 4B). The purified DSIF factor contains hSPT4 and hSPT5 and has been shown to affect transcription elongation in vitro (31). It is likely that immunodepletion of hSPT5 also resulted in depletion of hSPT4. To test the activity of this complex, recombinant DSIF (rDSIF), composed of hSPT5 and hSPT4 polypeptides, was purified from Escherichia coli (Fig. 4A), and the activities of these proteins have been normalized in previous studies (31). The addition of rTat-SF1 or rDSIF to Tat-SF1- or hSPT5-depleted extracts, respectively, restored Tat-dependent transcription activity (Fig. 4B, lanes 1 to 4 and lanes 5 to 8, respectively). However, the addition of rTat-SF1 protein into hSPT5-depleted nuclear extracts did not support Tat-dependent transcription nor did the addition of rDSIF into Tat-SF1-depleted extracts (data not shown). The addition of 10-fold-higher amounts of rTat-SF1 than the endogenous level did not confer further Tat-specific transcription activity, indicating that the system was saturated for Tat-SF1 in support of Tat-specific transcription elongation. Furthermore, the addition of recombinant TFIIF (RAP30 and RAP74) into Tat-SF1-depleted nuclear extract increased both Tat-dependent and Tat-independent transcription (data not shown). This finding is in good agreement with a previous report which demonstrated that TFIIF facilitates transcriptional elongation in both types of reactions (17). These results demonstrate that the addition of rTat-SF1 and rDSIF proteins could specifically restore Tat-activated transcriptional elongation to their depleted nuclear extracts without altering the level of general transcription. In addition, it indicates that both factors are required independently to support the Tat-dependent TAR-specific transcription process.
FIG. 4.
Addition of recombinant Tat-SF1 and DSIF proteins rescues Tat-dependent transcription activation. (A) SDS-PAGE analysis of recombinant Tat-SF1 and DSIF proteins. Samples (∼250 ng) of purified recombinant Tat-SF1 (lane 1, open arrowhead) and DSIF, a complex of hSPT4 and hSPT5 (lane 2, closed arrowhead), were resolved by SDS-PAGE on a 4 to 20% gel, and the proteins were visualized by staining gels with Coomassie brilliant blue. (B) Normalization of baculovirus-expressed Tat-SF1. Full-length rTat-SF1 protein was purified from recombinant baculovirus-infected Hi5 cells. Then, 4 μg of HeLa nuclear extracts (lanes 1 and 2) and different amounts of baculovirus-expressed rTat-SF1 proteins (lanes 3 to 5) were analyzed by Western blotting. Approximately 50 ng of purified recombinant Tat-SF1 protein (lane 3) showed the same level of Tat-SF1 as 30 μg of HeLa nuclear extracts (4 μl). (C) Nuclear extracts depleted with Tat-SF1 and hSPT5 can be complemented by recombinant Tat-SF1 and DSIF proteins for Tat-dependent activation. Recombinant Tat-SF1 (lanes 3 and 4) or DSIF, a complex of hSPT4 and hSPT5 (lanes 7 and 8), proteins were added to reactions at the preincubation step. The fold activation by Tat was measured as described previously (20).
Tat-SF1 and hSPT5 specifically stimulate Tat-activated transcription in vivo.
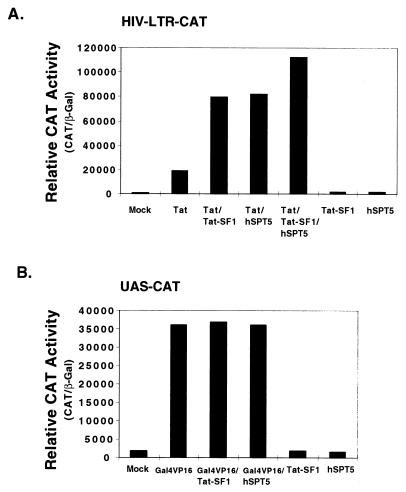
Overexpression of Tat-SF1 in vivo has been previously shown to enhance the level of Tat activation from a TAR-containing vector (44). It is possible that the level of hSPT5 was limiting in these cells. Therefore, the effects of the overexpression of both Tat-SF1 and hSPT5 on Tat activation in vivo were examined. An HIV-LTR CAT reporter construct was cotransfected into HeLa cells with various combinations of plasmids encoding Tat, Tat-SF1, and hSPT5. The transcriptional activity from the HIV-LTR was increased by approximately 50-fold by Tat expression when cotransfected into HeLa cells (Fig. 5A). This Tat-dependent transcriptional activity was further increased (by ca. threefold) by the cotransfection of a plasmid containing Tat-SF1 in combination with the Tat expression plasmid. Overexpression of hSPT5 in the context of cotransfection with a plasmid encoding Tat conferred similar levels of enhanced transcriptional activity as that by Tat-SF1 (Fig. 5A). In the presence of Tat, the overexpression of both Tat-SF1 and hSPT5 conferred a fivefold enforcement beyond that observed by Tat alone. However, the overexpression of either Tat-SF1 or hSPT5 without Tat did not stimulate the transcriptional activity of the HIV-LTR reporter. The stimulatory effects observed with the overexpression of Tat-SF1 and/or hSPT5 appear to be specific for Tat activation. In a previous study (44), overexpression of Tat-SF1 from a plasmid containing a simian virus 40 promoter suppressed the HIV-LTR CAT reporter by threefold in the absence of Tat. A similar suppression was not observed with the current system of plasmids. In both of these studies, the overexpression of Tat-SF1 stimulated Tat-specific activation by three- to sixfold compared to the control of the equivalent transfection in the absence of Tat. A plasmid encoding Gal4-VP16 was cotransfected with an UAS-CAT reporter with or without the cotransfection of Tat-SF1 or hSPT5. Transfection with Gal4-VP16 alone transactivated the UAS-CAT reporter approximately 200-fold (Fig. 5B). In the absence of Gal4-VP16, the transcriptional activities of UAS-CAT were not stimulated above the background (mock) level of transcription by the overexpression of Tat-SF1 or hSPT5. In contrast to Tat-dependent transcription, the overexpression of Tat-SF1 or hSPT5 did not further increase the activation of transcription by Gal4-VP16 in vivo (Fig. 5B). These results strongly suggest that Tat-SF1 and hSPT5 specifically support Tat-dependent transcriptional activation in vivo, as well as in vitro, and that both factors can be the limiting cellular factors for optimal activation in vivo.
FIG. 5.
Ectopic expression of Tat-SF1 and hSPT5 specifically enhances Tat-dependent transcription activity in vivo. (A) Expression of Tat-SF1 and hSPT5 increased Tat-dependent transcription activity. HeLa cells were cotransfected with HIV-LTR CAT reporter DNA (1 μg), 1 μg of Tat-SF1 expression vector (see Materials and Methods), and/or 1 μg of hSPT5 expression vector (38) in the absence or presence of the Tat expression vector (50 ng) as indicated. (B) Expression of Tat-SF1 and hSPT5 did not increase the transcription activities by Gal4-VP16 in vivo. HeLa cells were cotransfected with UAS-CAT reporter (1 μg), 1 μg of Tat-SF1 expression vector, and/or 1 μg of hSPT5 expression vector in the absence or presence of Gal4-VP16 expression vector (200 ng). In both panels A and B, the relative CAT activities were obtained by normalization with β-Gal activities. pCMV–β-Gal expression vector (0.5 μg) was cotransfected in each transfection for the normalization of CAT assay. All transfection experiments were performed in duplicate and independently repeated five times. The results are representative of three separate experiments.
DISCUSSION
Tat activation of transcription in vitro is dependent upon both Tat-SF1 and hSPT5 since Tat stimulation can be specifically restored to immunodepleted extracts by the addition of purified recombinant proteins. Perhaps not surprisingly, small fractions of these two proteins are found associated with one another in transcription reactions, and both proteins are also associated with a fraction of Pol II. Overexpression of either Tat-SF1 or hSPT5 in vivo specifically stimulates Tat activation of transcription, and overexpression of both polypeptides further stimulates Tat activation. Surprisingly, a significant fraction of Tat-SF1 is associated with the RAP30 subunit and not to the other subunit of TFIIF, RAP74. This suggests that RAP30, which is tightly bound to elongating Pol II, may be an important factor in the mechanism of Tat activation.
The general transcription factor TFIIF interacts with Pol II during multiple stages of transcription, including preinitiation complex assembly, initiation, and elongation (29, 30). After initiation, the RAP30 subunit of TFIIF remains tightly bound to the elongating Pol II, while the RAP74 subunit is bound less tightly (41). The RAP30 subunit has several subdomains: a C-terminal region which binds DNA in initiation complexes, a Pol II-binding region which is important for elongation, and a RAP74-binding region in the N terminus. The latter region is important for both initiation and elongation (30). These results are consistent with the proposal that RAP30 which bound to Pol II during elongation mediates the binding of other proteins (41). In particular, RAP30 almost certainly could mediate the binding of RAP74, which suppresses pausing during elongation. RAP30, as part of the elongating complex, probably also associates with Tat-SF1. This association appears to compete with the binding of RAP74 to RAP30 since RAP74 did not efficiently coprecipitate with either Tat-SF1 or RAP30 antiserum. The association of Tat-SF1 with RAP30 might position the former in the elongation complex in close proximity to the Pol II. Since Tat-SF1 also binds the hSPT5 protein which also associates with Pol II, the mutual interaction of these three proteins (RAP30, Tat-SF1, and hSPT5) could greatly stabilize the complex on the Pol II.
Biochemical fractionation of nuclear extracts defined Tat-SF1 as an important factor for Tat activation (43, 44). Immunodepletion with antisera to the pp140 component of Tat-SF1 suggested a critical role for this polypeptide in Tat-activated transcription elongation. Complementation of such depleted extracts with recombinant pp140 indicates that this polypeptide is the only component of Tat-SF1 necessary for the activity. The degrees of specific activation upon addition of Tat to these reactions were about twofold. This suggests that over half of the transcription complexes traversing the G-less cassette 1,000 bp downstream from the promoter were activated by Tat. The fold activation is largely determined by the level of transcription through this region in the absence of Tat activation, which varies with preparations and conditions. Although there is no indication for a cofactor of Tat-SF1 codepleted from the reaction, the limitation of measuring the efficiency of complementation with twofold changes could make detection of such a factor difficult.
Tat-SF1 was specific for Tat activation in these transcription reactions since neither the depletion nor the addition of this protein reduced or stimulated, respectively, general transcription from a number of promoters. These findings agree with previous results (43, 44) but conflict with a recent suggestion that Tat-SF1 is a general elongation factor (18). Several lines of evidence support the idea that Tat-SF1 is not required for general elongation. First, transcription activities from the HIV-ΔTAR promoter and other promoters, including adenovirus E4 and MLP, were not decreased by the depletion of Tat-SF1. Second, complementation of depleted extracts with recombinant Tat-SF1 protein restored stimulation of Tat-activated transcription, while the addition of Tat-SF1 did not change the levels of TAR-independent transcription activities. Third, the overexpression of Tat-SF1 stimulated Tat activation but failed to increase transactivation by Gal4-VP16 in vivo. Thus, these data in vitro and in vivo clearly indicate that Tat activation is more dependent upon Tat-SF1 than general transcription from Tat-independent promoters. However, these results are not conclusive with regard to a potential role of Tat-SF1 in general transcription, and a better mechanistic understanding of Tat-SF1’s role in elongation will be necessary to resolve this issue.
Tat-SF1 tightly associates with the Tat protein in nuclear extracts (18, 43, 44) and has been reported to associate inefficiently with P-TEFb, which is an important factor in the transcription process (42). P-TEFb, containing cyclin T and Cdk9, specifically binds to TAR RNA in the presence of Tat forming a species-specific recognition complex leading to the activation of transcription (1, 33). Although we did not detect an association of Tat-SF1 with Cdk9 (probably because of the high-salt washing conditions), it is likely that Tat-SF1 would associate with Cdk9 in the presence of Tat. The tight association of Tat-SF1 with Tat (18, 44) and P-TEFb with Tat (13, 45) in this complex would place Tat-SF1 in the appropriate location for it to play a role in the activation of elongation.
hSPT5 is also important for Tat activation of transcription both in vitro and probably in vivo. Immunodepletion of hSPT5 reduced the level of Tat activation but not that of general transcription, and the addition of purified recombinant protein complemented this deficiency. Furthermore, the overexpression of hSPT5 in vivo stimulated Tat activation of gene expression but not Gal4-VP16 enhancement of transcription. Previous fractionation studies have suggested that the hSPT5 protein is important for Tat activation, but these experiments did not test complementation by recombinant hSPT5 (35). Studies in yeast have shown that SPT5 is associated with SPT4 and Pol II (11). The DSIF factor (see below) was purified from human extracts as a complex of hSPT4 and hSPT5, a finding consistent with the results found in yeast cells (31). The novel finding that hSPT5 is associated with Tat-SF1 suggests that these two proteins might cooperatively bind to Pol II, perhaps to specifically promote elongation. This proposal is consistent with studies in yeast cells which showed that SPT5 becomes an essential elongation factor when the organism is deficient in the SII elongation factor or when placed under stress by drugs that interfere with transcription elongation by limiting the pool of nucleotide triphosphates (11).
It is important to note that both P-TEFb and DSIF may be involved in the modulation of gene expression during both the “early” and “late” stages of the elongation process. Recent studies involving the nucleoside analog DRB have shown that P-TEFb and DSIF act as positive and negative factors, respectively, during the early stages of elongation. The DSIF complex (hSPT4 and hSPT5) was originally purified as a cellular factor which conferred sensitivity to DRB on transcription in vitro (31). Subsequent results have provided a mechanism explaining this drug effect. The addition of DSIF and another factor, NELF (negative elongation factor), to a reaction arrests Pol II shortly after initiation, making further elongation dependent upon the activity of the Cdk9 kinase, a component of P-TEFb (32, 36). This kinase can phosphorylate the CTD of Pol II, and it is this modification which is thought to signal the release of the Pol II from the inhibitory effects of DSIF (37). The Cdk9 kinase is preferentially sensitive to the drug DRB compared to other Cdk-type kinases known to be important in transcription (19). Since DRB is a general inhibitor of elongation in mammalian cells and generates short promoter proximal transcripts (28), this suggests that DSIF and P-TEFb are important for the “early” stages of basal transcription from most promoters. Tat activation of transcription is unusually sensitive to inhibition by DRB (21) and affects elongation at sites distal from the promoters, suggesting that DSIF and P-TEFb may have an additional late function in Tat activation.
Recent biochemical studies of human P-TEFb are consistent with it having an additional late role in Tat activation (33, 45). Both D. melanogaster and rodent P-TEFb complement a human transcription reaction for the above-mentioned early role in general transcription. However, neither of these factors complement Tat-dependent transcription activation (3, 9, 45). Only human P-TEFb can function in Tat activation correlated with its specific tight binding to TAR RNA in the presence of Tat (1, 33). Apparently, only the human cyclin T subunit of P-TEFb and not the Drosophila or rodent homolog specifically recognize the loop sequence of TAR. These results suggest that a TAR–Tat–P-TEFb complex is required for Tat activation of elongation compared to the promoter proximal basal effects of P-TEFb. This suggests that the TAR–Tat–P-TEFb complex controls elongation as the Pol II traverses promoter-distal sequences or a late stage of elongation. In mediating this late effect, the TAR complex probably interacts with Tat-SF1 and DSIF (hSPT5 and hSPT4) in the activation of elongation. It is possible that this complex also interacts with RAP30 in the elongating complex, providing a mechanism for the dynamic control of elongation by Pol II.
ACKNOWLEDGMENTS
We thank E. Lees, D. Reinberg, and R. Gaynor for valuable materials and T. P. Cujec and B. M. Peterlin for providing the Tat-HA expression vector. We thank A. Mitsui, S. Gilbert, D. Dykxhoorn, D. Tantin, and H. Tang for valuable advice and helpful comments. We also thank M. Siafaca for secretarial support.
This work was supported by U.S. Public Health Service grants RO1-AI32486 and PO1-CA42063 from the National Institutes of Health, by NCI Cancer Center Support (core) grant P30-CA 14051 to P.A.S., by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports and Culture of Japan, and by a research grant from CREST of JST Corporation to H.H. J.B.K. was supported by an Anna-Fuller postdoctoral fellowship. Y.Y. was a JSPS research fellow.
REFERENCES
- 1.Bieniasz P D, Gardina T A, Bogerd H P, Cullen B R. Recruitment of a protein complex containing Tat and cyclin T1 to TAR governs the species specificity of HIV-1 Tat. EMBO J. 1998;17:7056–7065. doi: 10.1093/emboj/17.23.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buratowski S, Hahn S, Sharp P A, Guarente L. Function of a yeast TATA element-binding protein in a mammalian transcription system. Nature. 1988;334:37–42. doi: 10.1038/334037a0. [DOI] [PubMed] [Google Scholar]
- 3.Chen D, Fong Y, Zhou Q. Specific interaction of Tat with the human but not rodent P-TEFb complex mediates the species-specific Tat activation of HIV-1 transcription. Proc Natl Acad Sci USA. 1999;96:2728–2733. doi: 10.1073/pnas.96.6.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cujec T, Okamoto H, Fujinaga K, Meyer J, Chamberlin H, Morgan D O, Peterlin B M. The HIV transactivator Tat binds to the CDK-activating kinase and activates the phosphorylation of the carboxy-terminal domain of RNA polymerase II. Genes Dev. 1997;11:2645–2657. doi: 10.1101/gad.11.20.2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cullen B R. Does HIV-1 Tat induce a change in viral initiation rights? Cell. 1993;73:417–420. doi: 10.1016/0092-8674(93)90126-b. [DOI] [PubMed] [Google Scholar]
- 6.Cullen B R. HIV-1 auxiliary proteins: making connections in a dying cell. Cell. 1998;93:685–692. doi: 10.1016/s0092-8674(00)81431-2. [DOI] [PubMed] [Google Scholar]
- 7.Emerman M, Malim M H. HIV-1 regulatory/accessory genes: keys to unraveling viral and host cell biology. Science. 1998;280:1880–1884. doi: 10.1126/science.280.5371.1880. [DOI] [PubMed] [Google Scholar]
- 8.Feng S, Holland E C. HIV-1 tat trans-activation requires the loop sequence within tar. Nature. 1988;334:165–167. doi: 10.1038/334165a0. [DOI] [PubMed] [Google Scholar]
- 9.Garber M E, Wei P, KewalRamani V N, Mayall T P, Herrmann C H, Rice A P, Littman D R, Jones K A. The interaction between HIV-1 tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev. 1998;12:3512–3527. doi: 10.1101/gad.12.22.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia-Martinez L F, Mavankal G, Neveu J M, Lane W S, Ivanov D, Gaynor R B. Purification of a Tat-associated kinase reveals a TFIIH complex that modulates HIV-1 transcription. EMBO J. 1997;16:2836–2850. doi: 10.1093/emboj/16.10.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hartzog G A, Wada T, Handa H, Winston F. Evidence that SPT4, SPT5 and SPT6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 1998;12:357–369. doi: 10.1101/gad.12.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herrmann C H, Rice A P. Specific interaction of the human immunodeficiency virus Tat proteins with a cellular protein kinase. Virology. 1993;197:601–608. doi: 10.1006/viro.1993.1634. [DOI] [PubMed] [Google Scholar]
- 13.Herrmann C H, Rice A P. Lentivirus Tat proteins specifically associate with a cellular protein kinase, TAK, that hyperphosphorylates the carboxyl-terminal domain of the large subunit of RNA polymerase II: candidate for a Tat cofactor. J Virol. 1995;69:1612–1620. doi: 10.1128/jvi.69.3.1612-1620.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holstege F C P, Young R A. Transcriptional regulation: contending with complexity. Proc Natl Acad Sci USA. 1999;96:2–4. doi: 10.1073/pnas.96.1.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones K A. Taking a new TAK on tat transactivation. Genes Dev. 1997;11:2593–2599. doi: 10.1101/gad.11.20.2593. [DOI] [PubMed] [Google Scholar]
- 16.Jones K A, Peterlin B M. Control of RNA initiation and elongation at the HIV-1 promoter. Annu Rev Biochem. 1994;63:717–743. doi: 10.1146/annurev.bi.63.070194.003441. [DOI] [PubMed] [Google Scholar]
- 17.Kato H, Sumimoto H, Pognonec P, Chen C H, Rosen C A, Roeder R G. HIV-1 Tat acts as a processivity factor in vitro in conjunction with cellular elongation factors. Genes Dev. 1992;6:655–666. doi: 10.1101/gad.6.4.655. [DOI] [PubMed] [Google Scholar]
- 18.Li X Y, Green M R. The HIV-1 Tat cellular coactivator Tat-SF1 is a general transcription elongation factor. Genes Dev. 1998;12:2992–2996. doi: 10.1101/gad.12.19.2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mancebo H S, Lee G, Flygare J, Tomassini J, Luu P, Zhu Y, Peng J, Blau C, Hazuda D, Price D, Flores O. P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev. 1997;11:2633–2644. doi: 10.1101/gad.11.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marciniak R A, Calnan B J, Frankel A D, Sharp P A. HIV-1 Tat protein trans-activates transcription in vitro. Cell. 1990;63:791–802. doi: 10.1016/0092-8674(90)90145-5. [DOI] [PubMed] [Google Scholar]
- 21.Marciniak R A, Sharp P A. HIV-1 Tat protein promotes formation of more-processive elongation complexes. EMBO J. 1991;10:4189–4196. doi: 10.1002/j.1460-2075.1991.tb04997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marshall N F, Peng J, Xie Z, Price D H. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal kinase. J Biol Chem. 1996;271:27176–27183. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- 23.Marshall N F, Price D H. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J Biol Chem. 1995;270:12335–12338. doi: 10.1074/jbc.270.21.12335. [DOI] [PubMed] [Google Scholar]
- 24.Parada C A, Roeder R G. Enhanced processivity of RNA polymerase II triggered by Tat-induced phosphorylation of its carboxy-terminal domain. Nature. 1996;384:375–378. doi: 10.1038/384375a0. [DOI] [PubMed] [Google Scholar]
- 25.Parvin J D, Shykind B M, Meyers R E, Kim J, Sharp P A. Multiple sets of basal factors initiate transcription by RNA polymerase II. J Biol Chem. 1994;269:18414–18421. [PubMed] [Google Scholar]
- 26.Peng J, Zhu Y, Milton J T, Price D H. Identification of multiple cyclin subunits of human P-TEFb. Genes Dev. 1998;12:755–762. doi: 10.1101/gad.12.5.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ratnasabapathy R, Sheldon M, Johal L, Hernandez N. The HIV-1 long terminal repeat contains an unusual element that induces the synthesis of short RNAs from various mRNA and snRNA promoters. Genes Dev. 1990;4:2061–2074. doi: 10.1101/gad.4.12a.2061. [DOI] [PubMed] [Google Scholar]
- 28.Sehgal P B, Darnell J E, Tamm I. The inhibition by DRB(5,6-dichloro-1-β-d-ribofuranosylbenzimidazole) of hnRNA and mRNA production in HeLa cells. Cell. 1976;9:473–480. doi: 10.1016/0092-8674(76)90092-1. [DOI] [PubMed] [Google Scholar]
- 29.Tan S, Aso T, Conaway R C, Conaway J W. Roles for both the RAP30 and RAP74 subunits of transcription factor IIF in transcription initiation and elongation by RNA polymerase II. J Biol Chem. 1994;269:25684–25691. [PubMed] [Google Scholar]
- 30.Tan S, Conaway R C, Conaway J W. Dissection of transcription factor TFIIF functional domains required for initiation and elongation. Proc Natl Acad Sci USA. 1995;92:6042–6046. doi: 10.1073/pnas.92.13.6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yank K, Hartzog G A, Winston F, Buratowski S, Handa H. DSIF, a novel transcription elongation factor that regulate RNA polymerase II processivity, is composed of human SPT4 and SPT5 homologs. Genes Dev. 1998;12:343–353. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wada T, Takagi T, Yamaguchi Y, Watanabe D, Handa H. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 1998;17:7395–7403. doi: 10.1093/emboj/17.24.7395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wei P, Garber M E, Fang S M, Fischer W H, Jones K A. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- 34.Winston F, Carlson M. Yeast Snf/Swi transcriptional activators and Spt/Sin chromosome connection. Trends Genet. 1992;8:387–391. doi: 10.1016/0168-9525(92)90300-s. [DOI] [PubMed] [Google Scholar]
- 35.Wu-Baer F, Lane W S, Gaynor R B. Role of the human homolog of the yeast transcription factor SPT5 in HIV-1 Tat-activation. J Mol Biol. 1998;277:179–197. doi: 10.1006/jmbi.1997.1601. [DOI] [PubMed] [Google Scholar]
- 36.Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa T, Handa H. NELF, amultisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- 37.Yamaguchi Y, Wada T, Handa H. Interplay between positive and negative elongation factors: drawing a new view of DRB. Genes Cells. 1998;3:9–15. doi: 10.1046/j.1365-2443.1998.00162.x. [DOI] [PubMed] [Google Scholar]
- 38.Yamaguchi Y, Wada T, Watanabe D, Takagi T, Hasegawa J, Handa H. Structure and function of the human transcription elongation factor DSIF. J Biol Chem. 1999;274:8085–8092. doi: 10.1074/jbc.274.12.8085. [DOI] [PubMed] [Google Scholar]
- 39.Yang X, Herrmann C H, Rice A P. The human immunodeficiency virus Tat proteins specifically associate with TAK in vivo and require the carboxyl-terminal domain of RNA polymerase II for function. J Virol. 1996;70:4576–4584. doi: 10.1128/jvi.70.7.4576-4584.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yankulov K, Yamashita K, Roy R, Egly J M, Bently D L. The transcriptional elongation inhibitor 5,6-dichloro-1-β-d-ribofuransyl benzimidazole inhibits transcription factor TFIIH associated protein kinase. J Biol Chem. 1995;270:23922–23925. doi: 10.1074/jbc.270.41.23922. [DOI] [PubMed] [Google Scholar]
- 41.Zawel L, Kumar K P, Reinberg D. Recycling of the general transcription factors during RNA polymerase II transcription. Genes Dev. 1995;9:1479–1490. doi: 10.1101/gad.9.12.1479. [DOI] [PubMed] [Google Scholar]
- 42.Zhou Q, Chen D, Pierstorff E, Luo K. Transcription elongation factor P-TEFb mediates Tat activation of HIV-1 transcription at multiple stages. EMBO J. 1998;17:3681–3691. doi: 10.1093/emboj/17.13.3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou Q, Sharp P A. Novel mechanism and factor for regulation by HIV-1 Tat. EMBO J. 1995;14:321–328. doi: 10.1002/j.1460-2075.1995.tb07006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou Q, Sharp P A. Tat-SF1: cofactor for stimulation of transcriptional elongation by HIV-1 Tat. Science. 1996;274:605–610. doi: 10.1126/science.274.5287.605. [DOI] [PubMed] [Google Scholar]
- 45.Zhu Y, Pe’ery T, Peng J, Ramanathan Y, Marshall N, Marshall T, Amendt B, Mathews M B, Price D H. Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev. 1997;11:2622–2632. doi: 10.1101/gad.11.20.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]