Abstract
Leaf senescence can be triggered by multiple abiotic stresses including darkness, nutrient limitation, salinity, and drought. Recently, heatwaves have been occurring more frequently, and they dramatically affect plant growth and development. However, the underlying molecular networks of heat stress-induced leaf senescence remain largely uncharacterized. Here we showed that PHYTOCHROME INTERACTING FACTOR 4 (PIF4) and PIF5 proteins could efficiently promote heat stress-induced leaf senescence in Arabidopsis. Transcriptomic profiling analysis revealed that PIF4 and PIF5 are likely to function through multiple biological processes including hormone signaling pathways. Further, we characterized NAC019, SAG113, and IAA29 as direct transcriptional targets of PIF4 and PIF5. The transcription of NAC019, SAG113, and IAA29 changes significantly in daytime after heat treatment. In addition, we demonstrated that PIF4 and PIF5 proteins were accumulated during the recovery after heat treatment. Moreover, we showed that heat stress-induced leaf senescence is gated by the circadian clock, and plants might be more actively responsive to heat stress-induced senescence during the day. Taken together, our findings proposed important roles for PIF4 and PIF5 in mediating heat stress-induced leaf senescence, which may help to fully illustrate the molecular network of heat stress-induced leaf senescence in higher plants and facilitate the generation of heat stress-tolerant crops.
Keywords: Circadian clock, heat stress, leaf senescence, PHYTOCHROME INTERACTING FACTOR 4 (PIF4), PIF5, transcriptional regulation
PIF4 and PIF5 proteins promote heat stress-induced leaf senescence by regulating the expression of NAC019, SAG113, IAA29, and CBF2 during daytime after heat treatment in Arabidopsis.
Introduction
Leaf senescence is the final stage of plant leaf growth and development, and can be induced by a variety of internal factors, such as aging and phytohormones, and external environmental factors, including darkness, UV-B, nutrient limitation, salinity, drought, and high ambient temperature (Lim et al., 2007; Kim et al., 2020). The onset of senescence is usually accompanied by a number of physiological, biochemical, and molecular changes, including loss of chloroplasts, degradation of chlorophylls, proteins, nucleic acids, and lipids, accumulation of reactive oxygen species, and up-regulation of defense-related genes (Avila-Ospina et al., 2014; Sakuraba et al., 2014b; Kim et al., 2016; Woo et al., 2019).
Age-dependent senescence is closely related to the regulation of hormonal pathways (Schippers, 2015). Ethylene (ET), which accumulates as senescence progresses, is a major promoter of leaf senescence. The mRNA level and activity of ETHYLENE-INSENSITIVE 3 (EIN3), a core component of ET signaling, are significantly increased during leaf senescence. EIN3 also directly activates a series of downstream senescence-associated genes (SAGs), such as ORESARA 1 (ORE1), NAC DOMAIN CONTAINING PROTEIN 83 (NAC083), NAC102, and SENESCENCE-ASSOCIATED GENE 29 (SAG29) (Kim et al., 2009; Chang et al., 2013; Li et al., 2013). Abscisic acid (ABA) is involved in promoting environmental stress-induced senescence. The content of ABA in leaves was increased by drought, high salt, and cold stress (Guo and Gan, 2006; Schippers, 2015). ABA-activated ABA INSENSITIVE 5 (ABI5) promotes the transcription of NAC-LIKE ACTIVATED BY AP3/PI (NAP), while NAP inhibits stomatal closure by activating SAG113 (Zhang and Gan, 2012), and induces the expression of NON-YELLOW COLORING 1 (NYC1) to promote chloroplast degradation (Yang et al., 2014). The key regulators of the brassinosteroid (BR) signaling pathway BRASSINAZOLE-RESISTANT 1 (BZR1) and BRI1-EMS-SUPPRESSOR 1 (BES1) can directly bind to promoters of NAC transcription factors, such as: NAC002, NAC019, NAC055, and NAC072, inhibiting their expression and delaying senescence (Sun et al., 2010; Yu et al., 2011; Chung et al., 2014). Jasmonic acid (JA) also plays critical roles in regulating leaf senescence. TEOSINTE BRANCHED1/CYCLOIDEA/PROLIFERATING CELL FACTOR4 (TCP4) activates the expression of the JA biosynthesis gene LIPOXYGENASE2 (LOX2), while TCP9 and TCP20 bind to the promoter of LOX2 to inhibit JA biosynthesis (Danisman et al., 2012). These proteins coordinately activate JA signaling gradually with the process of senescence. In addition, JA and auxin antagonistically regulate leaf senescence by WRKY57 (Lim et al., 2003; Jiang et al., 2014). WRKY57 can directly represses the transcription of SENESCENCE4 (SEN4) and SAG12. In addition, INDOLE-3-ACETIC ACID INDUCIBLE 29 (IAA29) and JASMONATE ZIM-DOMAIN4/8 (JAZ4/8) competitively interact with WRKY57 to integrate the JA and auxin signaling pathways to regulate leaf senescence (Jiang et al., 2014). Thus, all these hormone signaling pathways integrated to cooperatively regulate leaf senescence.
When plants are under stress conditions, all aspects of their growth and development will be affected, which will induce precocious senescence of older leaves, thereby removing the injured tissues and delivering nutrients to younger tissues to improve adaptability (Schippers et al., 2015; Quint et al., 2016). Many external factors, such as darkness, nutrient limitation, drought, heat or cold, high salinity, and pathogen attacks or wounding, can trigger senescence during leaf development (Balazadeh et al., 2010; Sade et al., 2018). Temperature is one of the most important environmental factors affecting the seasonal growth of plants (Ding et al., 2020). When the ambient temperature rises moderately (~28 °C), it will markedly stimulate the thermomorphogenesis of plants (Quint et al., 2016). When plants survive after a non-lethal high temperature (~37 °C) for a period of time, their tolerance to lethal high temperature (≥42 °C) is enhanced. This process is called acquired heat tolerance (Mittler et al., 2012; Li et al., 2019). When plants grow in an environment above their optimal growth temperature for a prolonged period of time, they will experience heat stress (Bita and Gerats, 2013). Heat stress will inhibit seed germination, reduce fertility, decrease crop yield, and activate programmed cell death in specific cells or tissues, and, in severe cases, cause plant death (Mittler et al., 2012; Quint et al., 2016; Li et al., 2019). However, there are few studies on the induction of plant leaf senescence by heat stress, and the underlying mechanism of heat stress-induced senescence is still largely unclear.
PIF4 was shown to be an integrator in the high temperature signaling pathway (Quint et al., 2016). The core component of the circadian clock, TIMING OF CAB EXPRESSION1 (TOC1), can interact with PIF4 to inactivate it in order to inhibit the thermomorphogenesis of Arabidopsis in the evening (Zhu et al., 2016). In addition, the transcription of TCP5 is rapidly induced and its protein is more stable at high temperature. Further, TCP5 enhances PIF4’s activity at both transcriptional and post-transcriptional levels to positively regulate the plant response to high temperature (Han et al., 2019; Zhou et al., 2019). Previously, PIF4 and PIF5 proteins were shown to be involved in dark-induced and age-triggered leaf senescence by directly activating expression of SAGs, such as EIN3, ABI5, and ENHANCED EM LEVEL (EEL) (Seaton et al., 2015), which are critical components of ET and ABA signaling pathways (Sakuraba et al., 2014a; Song et al., 2014; Liebsch and Keech, 2016). Very recently, it was reported that the increased PIF4 protein binds to the ORE1 promoter at high ambient temperature to accelerate leaf senescence (Kim et al., 2020). However, the network of PIF4 and PIF5 proteins in regulating heat stress-induced leaf senescence still remains elusive.
To investigate whether PIF4 and PIF5 are involved in heat stress-induced leaf senescence, we examined the sensitivity of rosette leaves of both mutants and overexpression lines of PIF4 and PIF5 in response to heat stress. We found that PIF4 and PIF5 could positively regulate heat stress-induced leaf senescence in Arabidopsis thaliana. Further, we found that heat stress-induced leaf senescence could be gated by the circadian clock, and plants were more resistant to heat stress-induced leaf senescence in the daytime. Moreover, the accumulation of PIF4 and PIF5 proteins is accompanied by recovery after heat treatment. Finally, our findings established a molecular framework for fully deciphering the underlying mechanisms of PIF4 and PIF5 in mediating heat stress-induced leaf senescence, and provided a candidate target for generating heat-resistant crops to better cope with climate warming.
Materials and methods
Plant material and growth conditions
Arabidopsis thaliana plant material used in this study includes pif3-1 (CS2101452), pif4-2 (CS66043), pif4-2 pif5-3 (CS68096), pifq (CS66049), 35S::PIF3, PIF4pro:PIF4-HA (Li et al., 2020), PIF5pro:PIF5-HA (Li et al., 2020), and wild-type Col-0. Arabidopsis thaliana seeds were sterilized with 75% ethyl alcohol for 3 min and 10% sodium hypochlorite for 10 min, washed with sterile water three or five times, and then placed on Murashige and Skoog (MS; Schaffer et al., 1998) solid medium containing 3% sucrose. After 3 d at 4 °C, they germinated and were grown in a 22 °C light incubator under 12 h light and 12 h dark cycles (light quality, LED light; light intensity, 200 μmol m−2 s−1). After 10 d, the seedlings were transplanted into the soil to grow under 16 h light and 8 h dark cycles (light quality, LED light; light intensity, 50 μmol m−2 s−1).
Measurements of chlorophyll and membrane ion leakage
The A. thaliana plants grown in soil under 16 h light/8 h dark conditions for 3 weeks were used for heat treatment at 42 °C for 5 h at Zeitgeber time (ZT) 2. After 3 d of recovery growth at 22 °C, the third and fourth rosette leaves of the treated and untreated plants were collected. Each sample contains 6–8 leaves and the weight was recorded. A 1 ml aliquot of 80% acetone solution (v/v) was added to immerse the leaves, then they were placed at room temperature in the dark for >24 h. The absorbance at 645 nm and 663 nm was measured by a microplate reader (TECAN Infinite M200 Pro), and the chlorophyll content was calculated by the formula (8.02 A663+20.21 A645)×V/W as previously described (Zhang et al., 2018). For measuring the membrane ion leakage, the leaves were washed three times with sterilized deionized water. Then 6 ml of deionized water was added to each sample and the leaves were immersed and shaken gently overnight. The conductivity C1 (before boiling) and the conductivity C2 (after boiling for 10 min) were measured by a conductivity meter (CD400, Alalis). The ion leakage rate is the ratio of C1 to C2, as previously described (Zhang et al., 2018).
RNA-sequencing analysis
The A. thaliana Col-0 and pif4 pif5 plants grown in soil under 16 h light/8 h dark conditions for 3 weeks were used for heat treatment at 42 °C for 5 h. After 3 d of recovery growth, the third and fourth rosette leaves of the treated and untreated plants were collected for RNA extraction. RNA-sequencing (RNA-seq) and differential gene expression analyses were performed at BerryGenomics (Beijing). Sequencing libraries were generated using the NEB Next Ultra™ RNA Library Prep Kit for Illumina (NEB, USA) following the manufacturer’s recommendations, and index codes were added to attribute sequences to each sample. The library preparations were sequenced on an Illumina Hiseq Xten platform and paired-end reads were generated. Genes with q≤0.05 and fold change ≥1.5 were identified as differentially expressed genes (DEGs). RNA-seq raw data generated in this study have been deposited in the Gene Expression Omnibus under accession number GSE155710.
RNA extraction and quantitative real-time PCR
Seedlings of Col-0 and pif4 pif5 were grown under 12 h light/12 h dark cycle conditions (LED light; light intensity 200 μmol m−2 s−1) at 22 °C for 18 d. Heat treatment was at 42 °C for 2 h at two time points, ZT6 and ZT18. The third and fourth rosette leaves were collected immediately, while the untreated material was collected at the same time (ZT8 and ZT20). The third and fourth rosette leaves of the heat-treated and untreated materials were also collected at the corresponding time points (ZT8 and ZT20) for the next 3 d, and the total RNA was extracted by TRIzol reagent (Life Technologies). The guide DNA (gDNA) eraser (Takara) was used to remove the potential DNA contamination, and reverse transcription into cDNA was performed with the PrimeScript RT kit (Takara) according to the manufacture’s instruction. Quantitative PCR was performed on a Quant Studio 3 instrument (Applied Biosystems, USA) using a SYBR Green Real-Time PCR Master Mix (Toyobo, Japan). The PCR program was: 95 °C for 2 min, followed by 40 cycles of 95 °C for 15 s, 55 °C for 15 s, and 72 °C for 15 s, followed by melting curve analysis. Gene expression was normalized by the geometric mean of TUB4 and ACTIN2 expression. At least three biological and three technical replicates were conducted. Data represent the mean ±SD of three biological replicates. All of the primers used for real-time PCR analysis are listed in Supplementary Table S1.
Purified GST-tagged PIF4 protein and EMSA
The GST-PIF4 plasmid was transformed into the Escherichia coli BL21 strain. An 8 ml overnight culture was seeded to 400 ml of LB for continued growth at 37 °C for 2 h, then induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at 28 °C for 6 h. The cells in the pellet were collected by centrifuging at 4000 rpm for 10 min, then resuspended with 10 ml of extraction buffer [5 mM EDTA, 250 mM NaCl, 50 mM Tris–HCl, pH 8.0, 1 mM PMSF (phenylmethylsulfonyl fluoride), 1 μg ml–1 pepstatin, 1 μg ml–1 aprotinin, and 5 μg ml–1 leupeptin]. Then, 500 μl of lysozyme solution (10 mg of lysozyme in 1 ml of water) was added and incubated on ice for 30 min. Before sonication, 100 μl of 1 M DTT and 1 ml of 10% sarkosyl (w/v) were added and mixed thoroughly. The lysate was sonicated until it become transparent, then 2.3 ml of Triton X-100 was added and mixed thoroughly. After centrifuging at 13 000 rpm for 10 min, the supernatant was poured onto 500 μl of glutathione S-transferase (GST)–resin and incubated at 4 °C for 3 h. After washing the beads with buffer (1 mM EDTA, 150 mM NaCl, 50 mM Tris–HCl, pH 8.0, 3 mM DTT, 1 mM PMSF, 0.5% Triton X-100) five times, the GST–PIF4 protein was eluted with 10 mM reduced glutathione (Aladdin) solution. The Lightshift Chemiluminescent EMSA kit (Thermo Scientific) was used for EMSAs, as previously described (Li et al., 2020).
Immunoblot for detecting PIF proteins
The third and fourth leaves of the transgenic materials of PIF4pro:PIF4-HA and PIF5pro:PIF5-HA which were grown under 12 h light/12 h dark cycle conditions (LED light; light intensity 200 μmol m−2 s−1) at 22 °C for 18 d, treated at 42 °C for 2 h at ZT6 and ZT18, and harvested immediately after treatment. They were also harvested at corresponding time points over the next 2 d. Total protein was extracted as described previously (Li et al., 2020). PIF4-HA and PIF5-HA were detected by western blotting with HA antibody (3F10, Roche).
Statistical analysis
Differences between means were statistically analyzed by one-way ANOVA using Tukey’s honestly significant difference (HSD) mean separation test (IBM SPSS Statistics Software). Statistically significant differences at the P<0.05 level are indicated with different letters in the figures.
Results
PIF4 and PIF5 are critical regulators of heat stress-induced leaf senescence
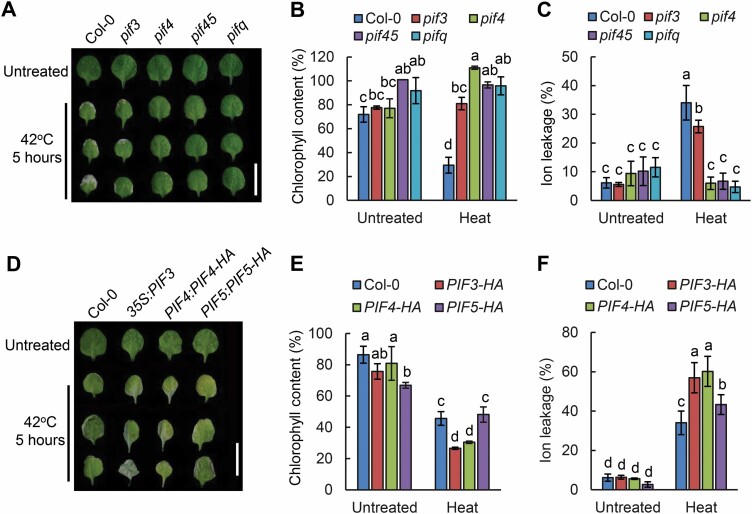
Leaf senescence can be induced by a number of external abiotic factors, including drought, salinity, and high ambient temperature (Kim et al., 2020). Whether leaf senescence can be triggered by heat stress and the underlying mechanisms still remain unclear, knowledge of which is important given the increased frequency of extreme heatwaves (Kim et al., 2020). PIF4 was shown to function as a central hub in the regulation of high temperature-induced architectural adaptations in Arabidopsis (Koini et al., 2009; Quint et al., 2016). In addition, PIF4 and PIF5 proteins were implicated in the regulation of dark-induced and age-triggered leaf senescence (Sakuraba et al., 2014a; Song et al., 2014; Liebsch and Keech, 2016). Thus, we questioned whether PIF proteins are involved in heat stress-induced leaf senescence. To examine this, we treated 3-week-old Col-0 and pif4 pif5 mutant plants grown in soil at 42 °C for 3, 4, or 5 h, and then left to recover at 22 °C for 3 d. We found that heat treatment could trigger leaf senescence in the wild-type Col-0, whereas the pif4 pif5 mutant exhibited a pronounced delayed heat stress-induced leaf senescence phenotype, particularly with 5 h of heat treatment (Supplementary Fig. S1). Further, we comprehensively examined the effect of heat stress-induced leaf senescence in other PIF mutants such as pif3, pif4, pif4 pif5, and pifq (pif1 pif3 pif4 pif5). Consistently, all the tested mutants displayed a significantly delayed heat stress-induced leaf senescence phenotype (Fig. 1A), with higher chlorophyll contents (Fig. 1B) and lower levels of membrane ion leakage (Fig. 1C). Moreover, we found that mutation in PIF4 and PIF5 resulted in more severe phenotypes than mutation in PIF3 (Fig. 1A–C). Intriguingly, the pif4 pif5 double mutants are overall similar to the pifq mutant with regard to chlorophyll content and membrane ion leakage, suggesting that PIF4 and PIF5 play a major role in regulating heat stress-induced leaf senescence. Further, we analyzed the heat stress-induced leaf senescence phenotypes produced by elevated expression levels of PIF genes including 35S:PIF3, PIF4pro:PIF4-HA, and PIF5pro:PIF5-HA. We found the elevated PIF gene levels in those transgenic lines could accelerate heat stress-induced leaf senescence with lower chlorophyll contents and much higher membrane ion leakage (Fig. 1D–F). Taken together, all these results suggested that PIF3, PIF4, and PIF5 acted as positive regulators in mediating heat stress-induced senescence, in which PIF4 and PIF5 play more critical roles.
Fig. 1.
Heat stress-induced leaf senescence phenotype of PIF-related mutants. (A) Representative third or fourth rosette leaves from 3-week-old Col-0, pif3-1, pif4-2, pif4-2 pif5-3, and pifq plants grown in soil and left to recover for 3 d after heat treatment at 42 °C for 5 h at ZT2 under 16 h light/8 h dark cycles. Scale bar=1 cm. (B and C) Chlorophyll content (B) and ion leakage (C) of the third and fourth rosette leaves from 3-week-old Col-0, pif3-1, pif4-2, pif4-2 pif5-3, and pifq plants grown in soil and left to recover for 3 d after heat treatment at 42 °C for 5 h. (D) Representative third or fourth rosette leaves from 3-week-old Col-0, 35S::PIF3, PIF4pro:PIF4-HA, and PIF5pro:PIF5-HA transgenic lines grown in soil and left to recover for 3 d after heat treatment at 42 °C for 5 h at ZT2 under 16 h light/8 h dark cycles. Scale bar=1 cm. (E and F) Chlorophyll content (E) and ion leakage (F) of the third or fourth rosette leaves from 3-week-old Col-0, 35S::PIF3, PIF4pro:PIF4-HA, and PIF5pro:PIF5-HA transgenic lines after recovery for 3 d following heat treatment at 42 °C for 5 h. For (A–F), three biological replicates were performed. Error bars represent the SD. Different letters indicate statistically significant differences among means by Tukey’s HSD mean separation test with SPSS statistics software (P<0.05).
Multiple biological processes are involved in PIF4- and PIF5-mediated heat stress-induced leaf senescence
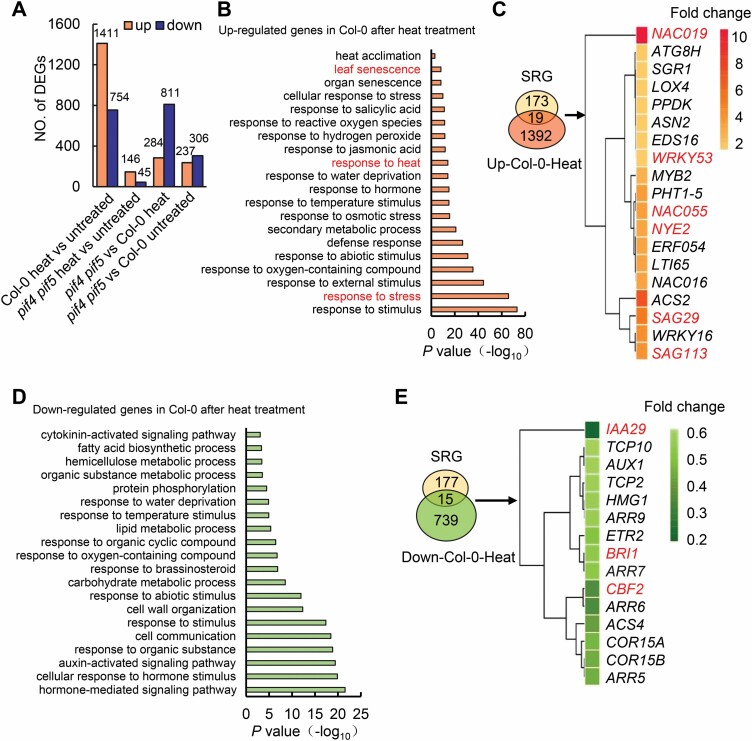
To investigate potential mechanisms underlying PIF4- and PIF5-mediated heat stress-induced leaf senescence, we performed RNA-seq analysis. Three-week-old Col-0 and pif4 pif5 double mutant grown in soil were treated at 42 °C for 5 h, and then left to recover at 22 °C for 3 d. The third and fourth rosette leaves were collected for RNA-seq analysis. In total, we identified 2165 DEGs after heat treatment compared with the untreated group in Col-0, comprising 1411 up-regulated and 754 down-regulated genes. However, only 291 DEGs were found in the pif4 pif5 double mutant, comprising 146 up-regulated and 45 down-regulated genes, which was significantly lower than in Col-0 (Fig. 2A; Supplementary Dataset S1), consistent with PIF4 and PIF5 playing critical roles in mediating heat stress-induced leaf senescence. Functional assignment of the heat stress-induced DEGs in Col-0 by Gene Ontology (GO) enrichment analysis further revealed that the DEGs were involved in a series of biological processes, including response to stress, response to heat, response to hormone stimulus, and leaf senescence (Fig. 2B, D). Further, we compared the up-regulated DEGs activated by heat stress with 192 previously documented senescence-regulatory genes (SRGs) (Li et al., 2014). In total, 19 SRGs were present in the heat stress-up-regulated DEGs (Fig. 2C) and 15 SRGs in the heat stress-down-regulated DEGs (Fig. 2E). GO analysis of DEGs with heat treatment in pif4 pif5 showed that hormone- and heat stress-related biological processes were also highly enriched (Supplementary Fig. S2). Taken together, the transcriptomic profiling analysis suggests that PIF4 and PIF5 regulate heat stress-induced leaf senescence probably through integrating multiple pathways.
Fig. 2.
Transcriptomic analysis of pif4 pif5 and Col-0 with heat stress treatment. (A) The number of differentially expressed genes (DEGs) in the pif4 pif5 mutant and Col-0 with heat treatment. The samples were harvested from the third and fourth rosette leaves of 3-week-old wild-type Col-0 and pif4 pif5 plants grown in soil under 16 h light/8 h dark conditions and left to recover for 3 d after heat treatment at 42 °C for 5 h, and untreated plants. (B) Gene Ontology (GO) analysis of the up-regulated genes between heat-treated and untreated wild-type Col-0. (C) Venn diagram showing the overlap of up-regulated genes in heated Col-0 with leaf senescence-regulatory genes (SRGs). The heatmap shows the up-regulated SRGs in Col-0 after heat treatment. The scale represents fold change. (D) GO analysis of the down-regulated genes between heat-treated and untreated wild-type Col-0. (E) Venn diagram showing the overlap of down-regulated genes in heated Col-0 with leaf SRGs. The heatmap shows the down-regulated SRGs in Col-0 after heat treatment. The scale represents fold change.
A series of senescence-associated genes were transcriptionally regulated by PIF4 and PIF5 in mediating heat stress-induced leaf senescence
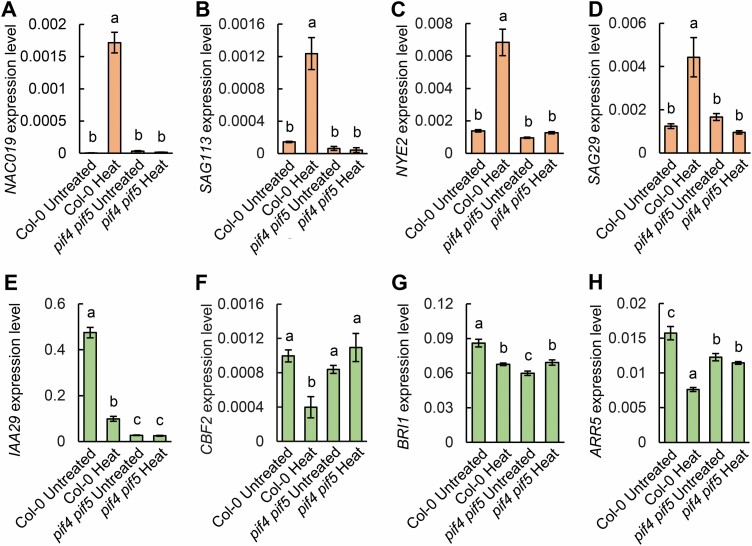
To investigate whether the expression of SAGs was related to heat stress-induced leaf senescence, we detected the expression profiles of these genes in 3-week-old Col-0 and pif4 pif5 mutants grown in soil and left to recover for 3 d after heat treatment or no treatment. The results showed that SAG113, NAC019, Non-yellowing 2 (NYE2), SAG29, NAC055, and WRKY6 were significantly up-regulated in Col-0 after heat treatment, but the expression levels were continuously lower and not significantly changed in pif4 pif5 (Fig. 3A–D; Supplementary Fig. S3B–F). In contrast, the expression levels of IAA29, C-REPEAT/DRE BINDING FACTOR 2 (CBF2), BRASSINOSTEROID INSENSITIVE 1 (BRI1), and ARR5 were significantly down-regulated in Col-0, but did not significantly change in the pif4 pif5 double mutant after heat treatment (Fig. 3E–H; Supplementary Fig. S3A). Therefore, we suggested that PIF4 and PIF5 promote heat stress-induced leaf senescence by transcriptional regulation of a series of SAGs.
Fig. 3.
The transcript levels of senescence-associated genes after heat stress-induced leaf senescence. (A–H) Quantitative real-time PCR analysis of NAC019 (A), SAG113 (B), NYE2 (C), SAG29 (D), IAA29 (E), CBF2 (F), BRI1 (G), and ARR5 (H) in Col-0 and pif4 pif5 of the heat-treated and untreated groups. Three-week-old wild-type Col-0 and pif4 pif5 plants were grown in soil under 16 h light/8 h dark conditions and were treated at 42 °C for 5 h. The third and fourth rosette leaves which began to turn yellow at the edge were collected for RNA extraction after 3 d of recovery. Three biological replicates were performed. Error bars represent the SD. Different letters indicate statistically significant differences among means by Tukey’s HSD mean separation test with SPSS statistics software (P<0.05).
Potential direct target genes of PIF4 and PIF5 in regulating heat stress-induced leaf senescence
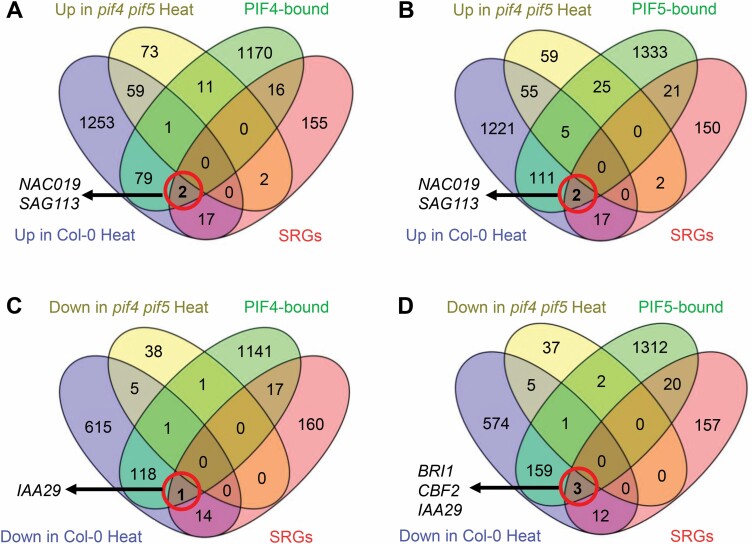
As PIF4 and PIF5 function as transcription factors (Leivar and Quail, 2011; Leivar and Monte, 2014), we screened the potential direct targets of PIF4 and PIF5 to mediate heat stress-induced leaf senescence. Previously reported ChIP-Seq analyses have identified 1279 PIF4-bound genes and 1497 PIF5-bound genes (Hornitschek et al., 2012; Pfeiffer et al., 2014). Thus, we first compared and identified genes that were up-regulated in Col-0 but not in pif4 pif5, then overlapped those genes with PIF4/5-bound genes and leaf SRGs. We indentified NAC019 and SAG113 as potential direct target genes of PIF4 and PIF5 proteins (Fig. 4A, B). Similarly, we compared and identified genes that were down-regulated in Col-0 but not in pif4 pif5, then overlapped these genes with PIF4- and PIF5-bound genes and leaf SRGs. We found that IAA29, BRI1, and CBF2 were potential direct target genes of PIF4 and PIF5 in this process (Fig. 4C, D). Taken together, these results suggested that NAC019, SAG113, IAA29, CBF2, and BRI1 were potential direct targets of PIF4 and PIF5 proteins in mediating heat stress-induced leaf senescence.
Fig. 4.
Screening the potential targets of PIF4 and PIF5 proteins to mediate heat stress-induced leaf senescence. (A and B) Venn diagrams showing the overlap among up-regulated genes in Col-0 or pif4 pif5 after heat treatment, the genes bound to PIF4 (A) or PIF5 (B) proteins, and the leaf senescence-regulatory genes (SRGs). (C and D) Venn diagrams showing the overlap among down-regulated genes in Col-0 or pif4 pif5 after heat treatment, the genes bound to PIF4 (C) or PIF5 (D) proteins, and the leaf SRGs.
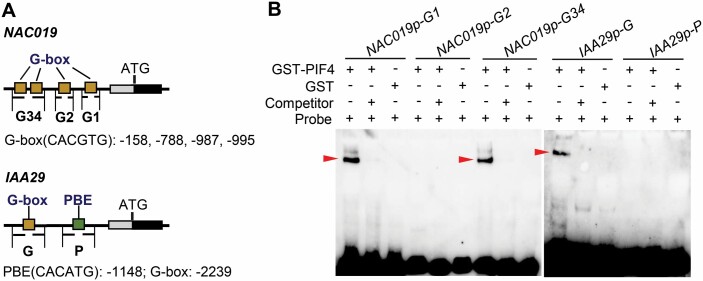
PIF4 directly binds to the promoters of NAC019 and IAA29
To further analyze the direct binding of PIF4 and PIF5 to the promoters of their target genes, we next examined the binding affinity of PIF4 proteins for the promoters of NAC019 and IAA29 by EMSAs. The EMSAs were performed with purified PIF4 protein tagged by GST. Bioinformatic analysis of the potential PIF protein binding sites found that there were four G-boxes (G4, base pairs –1001 to –995; G3, base pairs –993 to –987; G2, base pairs –794 to –788; and G1, base pairs –164 to –158) in the NAC019 promoter. Thus, the 60 bp probes containing the G-box sequence were designated as NAC019pro-G1, NAC019pro-G2, and NAC019pro-G34 (because G3 is very close to G4, we designed a 60 bp probe containing both G-box 3 and G-box 4) (Fig. 5A) There was one G-box (base pairs –2245 to –2239) and one PBE (PIF-binding element, base pairs –1154 to –1148) in the IAA29 promoter and the probes were referred to as IAA29pro-G and IAA29pro-p (Fig. 5A). The results of the EMSAs revealed that GST–PIF4 could bind NAC019pro-G1, NAC019pro-G34, and IAA29pro-G, but not NAC019pro-G2 and IAA29pro-p, while the negative control of GST alone did not bind any probes (Fig. 5B). Moreover, the intensities of the shifted bands were significantly inhibited by non-labeled competitor probes, suggesting that PIF4 could specifically bind the promoter of NAC019 and IAA29. These EMSA results, together with previous publicly accessible ChIP-seq data, suggested that NAC019 and IAA29 could be bound by PIF4 protein. In addition, our analysis found that there were two G-boxes (base pairs –321 to –315 and –1865 to –1859) on the promoter of SAG113, one G-box (base pairs –251 to –245) and one PBE (base pairs –324 to –318) on the promoter of CBF2, and one PBE (base pairs –238 to –232) on the promoter of BRI1. Therefore, the promoters of SAG113, CBF2, and BRI1 contain the binding motifs of PIF4 and PIF5 proteins, and combined with the data of ChIP-Seq, we suggested that SAG113, CBF2, and BRI1 may also be bound by PIF4 and PIF5 proteins.
Fig. 5.
PIF4 protein can directly bind the promoters of NAC019 and IAA29. (A) Schematic diagram of the promoter regions of NAC019 and IAA29. Yellow boxes represent the G-box elements. The green box represents the PBE (PIF-binding element). There are four G-boxes in the NAC019 promoter. NAC019pro-G1, NAC019pro-G2, and NAC019pro-G34 (the third and fourth boxes are adjacent, so this probe includes the third and fourth G-boxes) represent probes containing G-box sequences. The IAA29 promoter sequence has a G-box and a PBE, and the probes are designed as follows: IAA29pro-G and IAA29pro-p. (B) EMSA show that GST–PIF4 can bind the NAC019 and IAA29 promoter. The 100-fold unlabeled competitor (Competitor) and GST alone were used as negative controls. Red arrowheads mark the shifted bands.
PIF4 and PIF5 proteins accumulate with progress of heat stress-induced leaf senescence
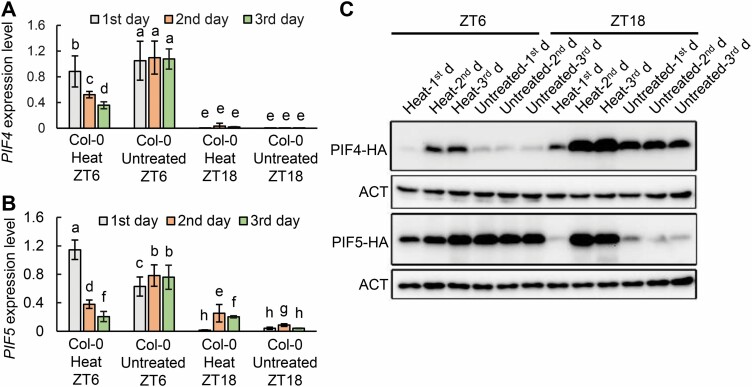
To investigate whether PIF4 and PIF5 were regulated at the transcriptional and post-transcriptional level by the heat treatment, we detected the transcript and protein levels of PIF4 and PIF5 in response to heat treatment. Eighteen-day-old wild-type Col-0 grown in MS agar plates with 3% sucrose under 12 h light/12 h dark cycles were treated at 42 °C for 2 h at ZT6 and ZT18, respectively, and then the third and fourth rosette leaves were collected immediately and also at the same time points over the next 2 d. Interestingly, we found that the transcriptional levels of PIF4 and PIF5 decreased after heat treatment during the recovery stage on the following days (Fig. 6A, B). However, the same treatment and collection method was used to detect the PIF4 and PIF5 proteins in transgenic materials PIF4pro:PIF4-HA and PIF5pro:PIF5-HA, and we found that the proteins levels of PIF4 and PIF5 were decreased on the first day, but were significantly accumulated at the recovery stage (Fig. 6C), suggesting that a post-transcriptional mechanism exists, which could be due to degradation by sequestered phyB (Al-Sady et al., 2006; Shen et al., 2007), as previous reports demonstrated that heat can promote phyB transformation from the Pfr form to the Pr form (Jung et al., 2016; Legris et al., 2016), which may inhibit the degradation of PIF4 and PIF5 proteins. Taken together, these results demonstrated that PIF4 and PIF5 could also be regulated at the transcriptional and post-transcriptional level to respond to heat stress.
Fig. 6.
The expression level of PIF4 and PIF5 during the senescence induced by heat stress. (A and B) The transcript level of PIF4 (A) and PIF5 (B) in heat-treated or untreated wild-type Col-0 determined by qunatitative real-time PCR. The third and fourth leaves from 18-day-old Col-0 grown on MS agar plates with 3% sucrose and left to recover for 3 d after heat treatment at 42 °C for 2 h at ZT6 and ZT18 under 12 h light/12 h dark cycles. The leaves were collected after immediately heat treatment and at the same time of the day over the next 2 d. Different letters indicate statistically significant differences among means by Tukey’s HSD mean separation test with SPSS statistics software (P<0.05). (C) Immunodetection of PIF4-HA and PIF5-HA protein levels in the process of senescence induced by heat treatment. The third and fourth leaves from 18-day-old PIF4pro:PIF4-HA and PIF5pro:PIF5-HA grown on MS agar plates with 3% sucrose and left to recover for 3 d after heat treatment at 42 °C for 2 h at ZT6 and ZT18 under 12 h light/12 h dark cycles. The leaves were collected immediately after heat treatment and at the same time of the day over the next 2 d.
Time of day confers heat stress-induced leaf senescence
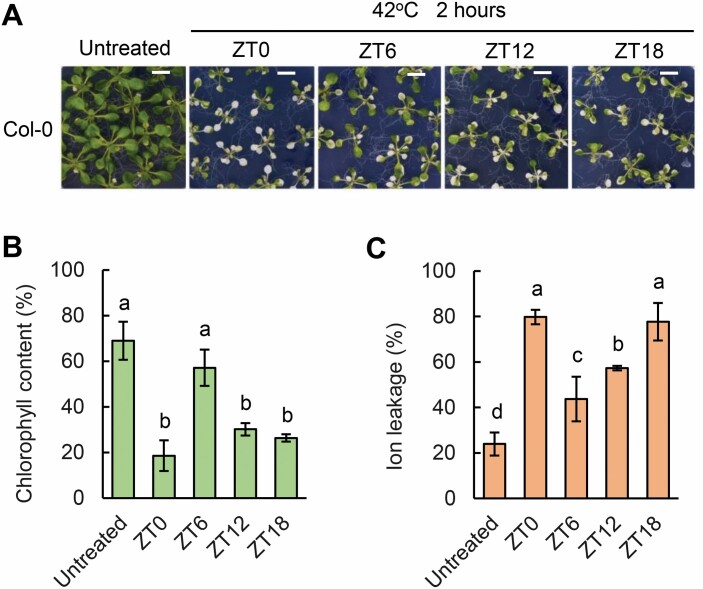
The thermotolerance of plants was shown to be gated by the circadian clock (Li et al., 2019). We wondered whether heat stress-induced leaf senescence is also gated by the circadian clock. To substantiate this notion, we subjected 18-day-old Col-0 seedlings grown in MS agar plates with 3% sucrose under 12 h light/12 h dark to 42 °C for 2 h at four different time points of ZT0, ZT6, ZT12, and ZT18 within a day and night cycle. The seedlings treated with heat at ZT6 displayed a significantly delayed heat stress-induced leaf senescence phenotype compared with those treated at ZT0 and ZT18, showing higher chlorophyll contents and much lower levels of membrane ion leakage (Fig. 7A–C). The seedlings treated with heat at ZT6 showed the lowest rate of leaf senescence. These results indicated that the plants subjected to heat treatment in the dark period display an accelerated leaf senescence rate, while this is relatively delayed during the daytime.
Fig. 7.
The response to heat stress-induced senescence in Col-0 in a time course. (A) The phenotype of 18-day-old Col-0 grown on MS agar plates with 3% sucrose and left to recover for 3 d after heat treatment at 42 °C for 2 h in a time course (ZT0, ZT6, ZT12, and ZT18) under 12 h light/12 h dark cycles. Scale bar=1 cm. (B and C) Chlorophyll content (B) and ion leakage (C) of the third and fourth rosette leaves from 18-day-old Col-0 plants grown on MS plates with 3% sucrose and left to recover for 3 d after heat treatment at 42 °C for 2 h at ZT0, ZT6, ZT12, and ZT18. Three biological replicates were performed. Error bars represent the SD. Different letters indicate statistically significant differences among means by Tukey’s HSD mean separation test with SPSS statistics software (P<0.05).
Heat stress-induced leaf senescence is gated by the circadian clock
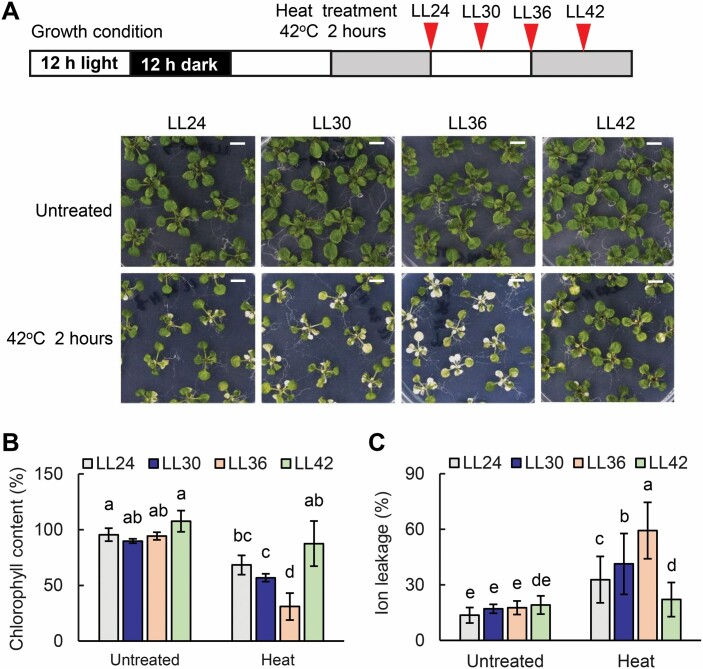
To investigate whether heat stress-induced leaf senescence is gated by the circadian clock, we treated Col-0 seedlings under continuous light (LL) conditions. The Col-0 plants were entrained in 12 h light/12 h dark cycles for 17 d, then transferred to continuous light conditions at dawn, with LL24 as subjective dawn and LL36 as subjective dusk. The plants were treated at 42 °C for 2 h at LL24, LL30, LL36, and LL42. The results showed that Col-0 displayed an accelerated leaf senescence rate during subjective day, and a slower rate of accelerated leaf senescence during subjective night (Fig. 8A–C). This result further confirmed that the heat stress-induced leaf senescence was gated by the circadian clock under free-running condition.
Fig. 8.
The response to heat stress-induced senescence in Col-0 under constant light. (A) The phenotype of 18-day-old Col-0 grown on MS agar plates with 3% sucrose and left to recover for 3 d after heat treatment at 42 °C for 2 h in a time course (LL24, LL30, LL36, and LL42) under constant light. The schematic diagram shows the light and heat treatment conditions. The boxes indicate that Col-0 plants were entrained in 12 h light/12 h dark cycles for 17 d, then transferred to continuous light conditions at dawn, where LL24 is subjective dawn and LL36 is subjective dusk. The red arrows indicate that plants were treated at 42 °C for 2 h at LL24, LL30, LL36, and LL42. Scale bar=1 cm. (B and C) Chlorophyll content (B) and ion leakage (C) of the third and fourth rosette leaves from 18-day-old Col-0 grown on MS agar plates with 3% sucrose and left to recover for 3 d after heat treatment at 42 °C for 2 h at LL24, LL30, LL36, and LL42. Three biological replicates were performed. Error bars represent the SD. Different letters indicate statistically significant differences among means by Tukey’s HSD mean separation test with SPSS statistics software (P<0.05).
Time of day confers the transcriptional alterations of NAC019, SAG113, and IAA29 under heat stress
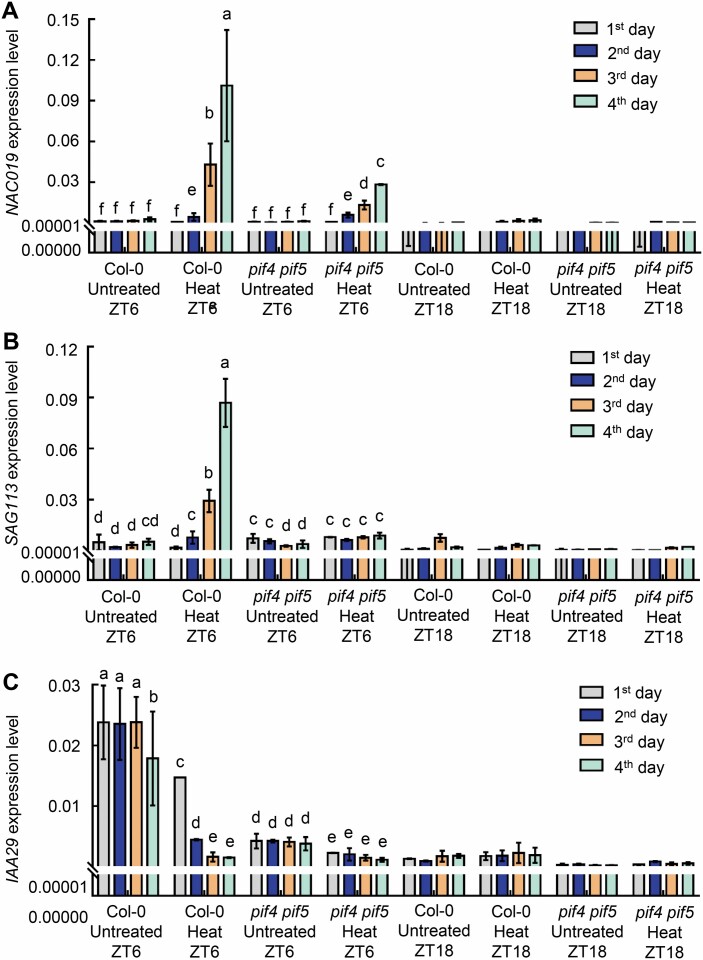
Since heat stress-induced leaf senescence was shown to be gated by the circadian clock, we propose that the heat stress-mediated transcriptional alterations of NAC019, SAG113, IAA29, CBF2, and BRI1 were also gated by the circadian clock. To verify this notion, the expression of these genes in 18-day-old Col-0 and pif4 pif5 seedlings in MS agar plates with 3% sucrose under long-day conditions with or without 42 °C treatment for 2 h at ZT6 and ZT18 was detected by quantitative real-time PCR. The results showed that the mRNA level of NAC019 and SAG113 continuously increased during the recovery period after heat treatment in Col-0 at ZT6, but the rising trend of NAC019 and SAG113 in pif4 pif5 mutants at this time point was slower (Fig. 9A, B). However, the expression level of IAA29 in Col-0 was significantly reduced, but was lower and not significantly changed in pif4 pif5 after heat treatment at ZT6 (Fig. 9C). It is worth noting that the transcriptional expression of the senescence genes changes significantly after heat treatment at ZT6 during the day, but the transcription expression of the senescence genes was lower and there was no significant change at ZT18 at night after heat treatment (Fig. 9; Supplementary Fig. S4). These results suggested that the plants might be more actively responsive to heat stress-induced senescence during the day.
Fig. 9.
The change in transcript levels of NAC019, SAG113, and IAA29 during heat stress-induced senescence. (A–C) Transcript levels of NAC019 (A), SAG113 (B), and IAA29 (C) during heat stress-induced senescence were detected by quantitative real-time PCR. The third and fourth rosette leaves of 18-day-old Col-0 and pif4 pif5 plants grown on MS plates with 3% sucrose were immediately harvested after heat treatment at 42 °C for 2 h at ZT6 and ZT18 under 12 h light/12 dark cycles, and were also collected on the next 3 d at the same time point. Three biological replicates were performed. Error bars represent the SD. Different letters indicate statistically significant differences among means by Tukey’s HSD mean separation test with SPSS statistics software (P<0.05).
Discussion
PIF mutants have a significantly enhanced leaf longevity in age-triggered and dark-induced senescence (Sakuraba et al., 2014a; Song et al., 2014). PIF4 and PIF5 promote leaf senescence through the signaling pathways of two senescence-promoting hormones, ET and ABA, by directly activating expression of EIN3, ABI5, and EEL (Sakuraba et al., 2014a; Song et al., 2014). PIF4 plays a critical role in the regulation of morphogenesis in response to temperature. PIF4 also regulates plant growth and development by integrating multiple signaling pathways to control the expression of downstream growth-related factors. Previous studies have shown that pif4 mutant is more sensitive to high temperatures (Zhu et al., 2016). Further, it was recently reported that high ambient temperature (~28 °C) promotes cotyledon senescence in 7-day-old light-grown seedlings in the dark by increasing both the mRNA and protein levels of PIF4 (Kim et al., 2020). The increased PIF4 protein bound to the promoter of ORE1 at high ambient temperature to accelerate cotyledon senescence, while also activating ABA and ET signaling to accelerate senescence at high ambient temperature (Kim et al., 2020). Besides high ambient temperature, whether PIF proteins are involved in the regulation of senescence of rosette leaves under heat stress (~42 °C) condition is still unclear. Here, we show that heat stress can trigger leaf senescence similar to other abiotic stresses, in which PIF4 and PIF5 positively regulate heat stress-induced leaf senescence in 3-week-old A. thaliana plants. Further we identified that NAC019, IAA29, and SAG113 were direct transcriptional targets of PIF4 and PIF5 in mediating heat stress-induced leaf senescence.
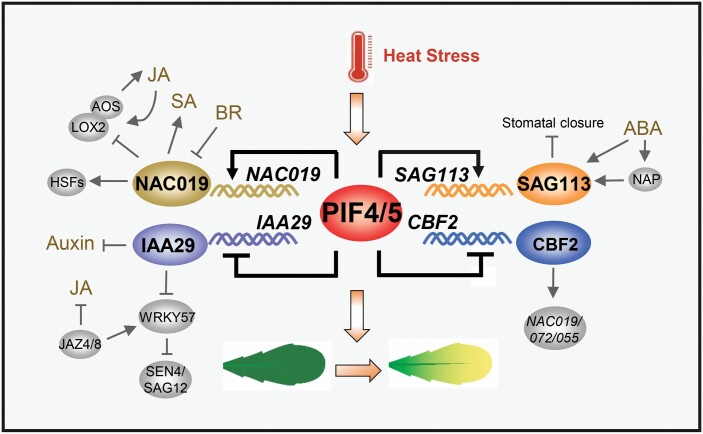
NAC019, NAC055, and NAC072, the three important members of the NAC family, play critical roles in stress responses and leaf senescence (Hickman et al., 2013). While identifying upstream transcriptional regulators of these three genes using yeast one-hybrid assay, CBF1, CBF2, CBF3, and CBF4 were screened out by directly binding the promoter of NAC072 (Hickman et al., 2013). It is worth noting that only CBF1 and CBF2 are involved in the senescence response (Hickman et al., 2013). Therefore, we propose that CBF2 acts as a direct target of PIF4 and PIF5 in the regulation of heat stress-induced leaf senescence by transcriptional regulation of these NAC genes (Fig. 10). It has been reported that NAC019 is also an important regulator in mediating the regulation of thermotolerance and heat stress transcription factors (HSFs) (Guan et al., 2014). Thus, it is reasonable that NAC019 functions downstream of PIF4 and PIF5 to mediate heat stress-induced leaf senescence through integrating multiple hormone signaling pathways, such as JA, salicylic acid (SA), and BR (Fig. 10). SAG113 was activated by NAP to inhibit stomatal closure, hence mediating ABA-induced leaf senescence (Zhang and Gan, 2012). Here, SAG113 acts as a direct target of PIF4 and PIF5 mediating heat stress-induced leaf senescence possibly also through the ABA signaling pathway (Fig. 10). It is worth noting that the transcript abundance of SAG113 does not change significantly in pif4 pif5 mutant grown in soil after heat treatment, and after a 3 d recovery (Fig. 3B). There is a significant heat-induced increase in SAG113 transcript abundance in pif4 pif5 mutant grown in MS agar plates with 3% sucrose (Fig. 9B). We suggest that the differences in the transcript abundance of NAC019 and SAG113 might be due to different growth conditions. IAA29 plays a positive role in regulating JA-induced leaf senescence probably through competitively interacting with WRKY57 to integrate the JA and auxin signaling pathways to regulate leaf senescence (Jiang et al., 2014). In summary, we proposed a downstream molecular regulation network of PIF4 and PIF5 by integrating a series of hormone signaling pathways and leaf senescence to regulate heat stress-induced leaf senescence.
Fig. 10.
A proposed working model for PIF4- and PIF5-mediated heat stress-induced senescence in Arabidopsis. PIF4 and PIF5 proteins act as positive regulators of heat stress-induced senescence via regulation of its direct target genes such as NAC019, IAA29, SAG113, and CBF2, which integrates multiple hormone signaling pathways to inhibit the heat stress-induced leaf senescence process. NAC019 is an important regulator in mediating leaf senescence and stress responses (Hickman et al., 2013; Guan et al., 2014). SAG113 was activated by NAP to inhibit stomatal closure, thus mediating ABA-induced leaf senescence (Zhang and Gan, 2012). We thus suggested that PIF4 and PIF5 can directly activate NAC019 and SAG113 to mediate heat stress-induced leaf senescence through integrating multiple hormone signaling pathways, such as JA, SA, BR, and ABA. IAA29 can integrates the JA and auxin signaling pathways to regulate leaf senescence (Jiang et al., 2014). CBF2 was screened out by directly binding the promoter of NAC019, NAC055, and NAC072, and shown to be involved in the senescence response. We also found that PIF4 and PIF5 can directly repress IAA29 and CBF2 to regulate heat stress-induced leaf senescence through integrating multiple hormone signaling pathways.
It is worth noting that heat treatment appears to significantly increase leaf chlorophyll content in the pif4 mutant. This might be due to the fact that PIF4 directly bound to and regulated the expression of chlorophyll- and chloroplast-related genes, such as NON-YELLOWING 1 (NYE1), GOLDEN 2-LIKE TRANSCRIPTION FACTOR 1 (GLK1), GLK2, and PROTOCHLORO-PHYLLIDE OXIDOREDUCTASE C (PORC), thus negatively regulating chloroplast activity (Song et al., 2014). PIF4 protein accumulates after the heat stress (Fig. 6), thus the chlorophyll degradation genes were activated while the chlorophyll activity maintenance genes were inhibited, and resulted in the yellowing leaf in Col-0 (Fig. 1A, B). However, in the pif4 mutant, these genes lost PIF4 control, resulting in further increased chloroplast activity and chlorophyll content (Fig. 1A, B).
The expression of TCP5 is rapidly induced and its protein stability is also enhanced under high temperature conditions. TCP5 interacts with PIF4 to enhance its transcriptional activation activity and also binds to the promoter of PIF4 to promote the transcription of PIF4 (Han et al., 2019). Moreover, TCP17 interacts with CRY1 at low temperature, and is released at high temperature to up-regulate the expression of PIF4 (Zhou et al., 2019). Therefore, it was proposed that high temperature activates PIF4 by stabilizing TCP to control plant thermomorphogenesis. Here, we found that PIF4 and PIF5 protein levels were temporally decreased with heat treatment, but accumulated at the recovery stage, suggesting that there was a crucial factor involved in the degradation of PIF4 and PIF5 proteins. Whether TCP transcription factors were involved in heat stress-induced leaf senescence, especially at the recovery stage, needs further investigation to decipher the mechanism of PIF4- and PIF5-mediated heat stress-induced senescence.
The thermotolerance of plants is gated by the circadian clock (Li et al., 2019) and exhibits a rhythmic pattern, which is closely related to the diurnal expression patterns of heat-associated genes (Dickinson et al., 2018; Blair et al., 2019). The circadian clock proteins REVEILLE4/8 (RVE4/8) regulate plant heat resistance by directly regulating ERF53/54 around noon, which reveals to some extent how the circadian clock helps plants adapt to high temperatures during the day (Li et al., 2019). CIRCADIAN CLOCK-ASSOCIATED 1 (CCA1), the core component of the circadian clock, directly activates GLK2 and inhibits ORE1 expression to negatively regulate leaf senescence (Song et al., 2018). Here we found that time of day contributes to the heat stress-induced leaf senescence and to whether it is more damaging when it is challenged at night or at dawn. However, plants are more sensitive to heat stress-induced leaf senescence at subjective dusk under free-running conditions. These separate regulations at night or at dawn in a time of day-specific manner and at subjective dusk in free-running conditions indicates that light/dark and the circadian clock are critical for heat stress-induced leaf senescence. The molecular links among the circadian clock, light/dark, and heat stress-induced leaf senescence await further investigation.
Supplementary data
The followng supplementary data are available at JXB online.
Fig. S1. Exploration of experimental conditions for heat treatment.
Fig. S2. Analysis of down-regulated DEGs in Col-0 after heat treatment.
Fig. S3. The transcript levels of senescence-associated genes after heat stress-induced senescence.
Fig. S4. The change in transcript levels of CBF2 and BRI1 during heat stress-induced senescence.
Table S1. Primers used in this study.
Dataset S1. DEGs between pif4 pif5 and Col-0 with heat treatment.
Acknowledgements
We thank Dr Rongcheng Lin (IBCAS) for 35S::PIF3 and pifq seeds.
The work was supported by the National Natural Sciences Foundation of China (31770287 and 31670290), the Strategic Priority Research Program of the Chinese Academy of Sciences, grant no. XDB27030206, Youth Innovation Promotion Association CAS (no. 2017110), and Young Elite Scientists Sponsorship Program CAST (2017QNRC001).
Author contributions
NL, YZ, and CB: performing the experiments; NL, YZ, and LW: data analysis; NL, YZ, and LW: study design; NL, YZ, and LW: writing.
Conflict of interest
The authors declare no competing or financial interests.
Data availability
RNA-seq raw data generated in this study have been deposited in the Gene Expression Omnibus under accession number GSE155710. All data supporting the findings of this study are available within the paper and within its supplementary data published online.
References
- Al-Sady B, Ni W, Kircher S, Schäfer E, Quail PH. 2006. Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Molecular Cell 23, 439–446. [DOI] [PubMed] [Google Scholar]
- Avila-Ospina L, Moison M, Yoshimoto K, Masclaux-Daubresse C. 2014. Autophagy, plant senescence, and nutrient recycling. Journal of Experimental Botany 65, 3799–3811. [DOI] [PubMed] [Google Scholar]
- Balazadeh S, Siddiqui H, Allu AD, Matallana-Ramirez LP, Caldana C, Mehrnia M, Zanor MI, Köhler B, Mueller-Roeber B. 2010. A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. The Plant Journal 62, 250–264. [DOI] [PubMed] [Google Scholar]
- Bita CE, Gerats T. 2013. Plant tolerance to high temperature in a changing environment: scientific fundamentals and production of heat stress-tolerant crops. Frontiers in Plant Science 4, 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair EJ, Bonnot T, Hummel M, Hay E, Marzolino JM, Quijada IA, Nagel DH. 2019. Contribution of time of day and the circadian clock to the heat stress responsive transcriptome in Arabidopsis. Scientific Reports 9, 4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang KN, Zhong S, Weirauch MT, et al. 2013. Temporal transcriptional response to ethylene gas drives growth hormone cross-regulation in Arabidopsis. eLife 2, e00675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung Y, Kwon SI, Choe S. 2014. Antagonistic regulation of Arabidopsis growth by brassinosteroids and abiotic stresses. Molecules and Cells 37, 795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danisman S, van der Wal F, Dhondt S, et al. 2012. Arabidopsis class I and class II TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant Physiology 159, 1511–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson PJ, Kumar M, Martinho C, et al. 2018. Chloroplast signaling gates thermotolerance in Arabidopsis. Cell Reports 22, 1657–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Shi Y, Yang S. 2020. Molecular regulation of plant responses to environmental temperatures. Molecular Plant 13, 544–564. [DOI] [PubMed] [Google Scholar]
- Guan Q, Yue X, Zeng H, Zhu J. 2014. The protein phosphatase RCF2 and its interacting partner NAC019 are critical for heat stress-responsive gene regulation and thermotolerance in Arabidopsis. The Plant Cell 26, 438–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Gan S. 2006. AtNAP, a NAC family transcription factor, has an important role in leaf senescence. The Plant Journal 46, 601–612. [DOI] [PubMed] [Google Scholar]
- Han X, Yu H, Yuan R, Yang Y, An F, Qin G. 2019. Arabidopsis transcription factor TCP5 controls plant thermomorphogenesis by positively regulating PIF4 activity. iScience 15, 611–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman R, Hill C, Penfold CA, et al. 2013. A local regulatory network around three NAC transcription factors in stress responses and senescence in Arabidopsis leaves. The Plant Journal 75, 26–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P, Kohnen MV, Lorrain S, et al. 2012. Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. The Plant Journal 71, 699–711. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Liang G, Yang S, Yu D. 2014. Arabidopsis WRKY57 functions as a node of convergence for jasmonic acid- and auxin-mediated signaling in jasmonic acid-induced leaf senescence. The Plant Cell 26, 230–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JH, Domijan M, Klose C, et al. 2016. Phytochromes function as thermosensors in Arabidopsis. Science 354, 886–889. [DOI] [PubMed] [Google Scholar]
- Kim C, Kim SJ, Jeong J, Park E, Oh E, Park YI, Lim PO, Choi G. 2020. High ambient temperature accelerates leaf senescence via PHYTOCHROME-INTERACTING FACTOR 4 and 5 in Arabidopsis. Molecules and Cells 43, 645–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Nam HG, Lim PO. 2016. Regulatory network of NAC transcription factors in leaf senescence. Current Opinion in Plant Biology 33, 48–56. [DOI] [PubMed] [Google Scholar]
- Kim JH, Woo HR, Kim J, Lim PO, Lee IC, Choi SH, Hwang D, Nam HG. 2009. Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science 323, 1053–1057. [DOI] [PubMed] [Google Scholar]
- Koini MA, Alvey L, Allen T, Tilley CA, Harberd NP, Whitelam GC, Franklin KA. 2009. High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Current Biology 19, 408–413. [DOI] [PubMed] [Google Scholar]
- Legris M, Klose C, Burgie ES, Rojas CC, Neme M, Hiltbrunner A, Wigge PA, Schäfer E, Vierstra RD, Casal JJ. 2016. Phytochrome B integrates light and temperature signals in Arabidopsis. Science 354, 897–900. [DOI] [PubMed] [Google Scholar]
- Leivar P, Monte E. 2014. PIFs: systems integrators in plant development. The Plant Cell 26, 56–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Quail PH. 2011. PIFs: pivotal components in a cellular signaling hub. Trends in Plant Science 16, 19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Gao Z, Liu X, Sun D, Tang W. 2019. Transcriptional profiling reveals a time-of-day-specific role of REVEILLE 4/8 in regulating the first wave of heat shock-induced gene expression in Arabidopsis. The Plant Cell 31, 2353–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Zhang Y, He Y, Wang Y, Wang L. 2020. Pseudo response regulators regulate photoperiodic hypocotyl growth by repressing PIF4/5 transcription. Plant Physiology 183, 686–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Peng J, Wen X, Guo H. 2013. Ethylene-insensitive3 is a senescence-associated gene that accelerates age-dependent leaf senescence by directly repressing miR164 transcription in Arabidopsis. The Plant Cell 25, 3311–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Zhao Y, Liu X, Peng J, Guo H, Luo J. 2014. LSD 2.0: an update of the leaf senescence database. Nucleic Acids Research 42, D1200–D1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebsch D, Keech O. 2016. Dark-induced leaf senescence: new insights into a complex light-dependent regulatory pathway. New Phytologist 212, 563–570. [DOI] [PubMed] [Google Scholar]
- Lim PO, Kim HJ, Nam HG. 2007. Leaf senescence. Annual Review of Plant Biology 58, 115–136. [DOI] [PubMed] [Google Scholar]
- Lim PO, Woo HR, Nam HG. 2003. Molecular genetics of leaf senescence in Arabidopsis. Trends in Plant Science 8, 272–278. [DOI] [PubMed] [Google Scholar]
- Mittler R, Finka A, Goloubinoff P. 2012. How do plants feel the heat? Trends in Biochemical Sciences 37, 118–125. [DOI] [PubMed] [Google Scholar]
- Pfeiffer A, Shi H, Tepperman JM, Zhang Y, Quail PH. 2014. Combinatorial complexity in a transcriptionally centered signaling hub in Arabidopsis. Molecular Plant 7, 1598–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quint M, Delker C, Franklin KA, Wigge PA, Halliday KJ, van Zanten M. 2016. Molecular and genetic control of plant thermomorphogenesis. Nature Plants 2, 15190. [DOI] [PubMed] [Google Scholar]
- Sade N, Del Mar Rubio-Wilhelmi M, Umnajkitikorn K, Blumwald E. 2018. Stress-induced senescence and plant tolerance to abiotic stress. Journal of Experimental Botany 69, 845–853. [DOI] [PubMed] [Google Scholar]
- Sakuraba Y, Jeong J, Kang MY, Kim J, Paek NC, Choi G. 2014a. Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nature Communications 5, 4636. [DOI] [PubMed] [Google Scholar]
- Sakuraba Y, Park SY, Kim YS, Wang SH, Yoo SC, Hörtensteiner S, Paek NC. 2014b. Arabidopsis STAY-GREEN2 is a negative regulator of chlorophyll degradation during leaf senescence. Molecular Plant 7, 1288–1302. [DOI] [PubMed] [Google Scholar]
- Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carré IA, Coupland G. 1998. The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93, 1219–1229. [DOI] [PubMed] [Google Scholar]
- Schippers JH. 2015. Transcriptional networks in leaf senescence. Current Opinion in Plant Biology 27, 77–83. [DOI] [PubMed] [Google Scholar]
- Schippers JH, Schmidt R, Wagstaff C, Jing HC. 2015. Living to die and dying to live: the survival strategy behind leaf senescence. Plant Physiology 169, 914–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaton DD, Smith RW, Song YH, et al. 2015. Linked circadian outputs control elongation growth and flowering in response to photoperiod and temperature. Molecular Systems Biology 11, 776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Khanna R, Carle CM, Quail PH. 2007. Phytochrome induces rapid PIF5 phosphorylation and degradation in response to red-light activation. Plant Physiology 145, 1043–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Jiang Y, Kuai B, Li L. 2018. CIRCADIAN CLOCK-ASSOCIATED 1 inhibits leaf senescence in Arabidopsis. Frontiers in Plant Science 9, 280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Yang C, Gao S, Zhang W, Li L, Kuai B. 2014. Age-triggered and dark-induced leaf senescence require the bHLH transcription factors PIF3, 4, and 5. Molecular Plant 7, 1776–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Fan XY, Cao DM, et al. 2010. Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Developmental Cell 19, 765–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo HR, Kim HJ, Lim PO, Nam HG. 2019. Leaf senescence: systems and dynamics aspects. Annual Review of Plant Biology 70, 347–376. [DOI] [PubMed] [Google Scholar]
- Yang J, Worley E, Udvardi M. 2014. A NAP–AAO3 regulatory module promotes chlorophyll degradation via ABA biosynthesis in Arabidopsis leaves. The Plant Cell 26, 4862–4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Li L, Zola J, et al. 2011. A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. The Plant Journal 65, 634–646. [DOI] [PubMed] [Google Scholar]
- Zhang K, Gan SS. 2012. An abscisic acid-AtNAP transcription factor-SAG113 protein phosphatase 2C regulatory chain for controlling dehydration in senescing Arabidopsis leaves. Plant Physiology 158, 961–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang Y, Wei H, Li N, Tian W, Chong K, Wang L. 2018. Circadian evening complex represses jasmonate-induced leaf senescence in Arabidopsis. Molecular Plant 11, 326–337. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Xun Q, Zhang D, Lv M, Ou Y, Li J. 2019. TCP transcription factors associate with PHYTOCHROME INTERACTING FACTOR 4 and CRYPTOCHROME 1 to regulate thermomorphogenesis in Arabidopsis thaliana. iScience 15, 600–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JY, Oh E, Wang T, Wang ZY. 2016. TOC1–PIF4 interaction mediates the circadian gating of thermoresponsive growth in Arabidopsis. Nature Communications 7, 13692. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq raw data generated in this study have been deposited in the Gene Expression Omnibus under accession number GSE155710. All data supporting the findings of this study are available within the paper and within its supplementary data published online.