Highlights
-
•
Novel imine reductase from yeast Candida parapsilosis purified and characterized.
-
•
CpIM1 belongs to unexplored Ornithine cyclodeaminase/Mu crystallin protein family
-
•
CpIM1 catalyzed stereospecific alkylamination of α-ketoacids/ketoesters.
-
•
CpIM1 also reduced cyclic and aryl imines.
-
•
First report on enzymatic alkylamination of α-ketoesters and reduction of arylimines.
Keywords: Imine reductase, μ-crystallin/Ornithine cyclodeaminase family, Candida parapsilosis, Alkylamination, N-alkyl amino acid/esters, Chiral amines
Abstract
We report a stereospecific imine reductase from Candida parapsilosis ATCC 7330 (CpIM1), a versatile biocatalyst and a rich source of highly stereospecific oxidoreductases. The recombinant gene was overexpressed in Escherichia coli and the protein CpIM1 was purified to homogeneity. This protein belongs to the Ornithine cyclodeaminase/ μ-crystallin (OCD-Mu) family of proteins which has only a few characterized members. CpIM1 catalyzed the alkylamination of α-keto acids/esters producing exclusively (S)-N-alkyl amino acids/esters e.g. N-methyl-l-alanine with > 90% conversion and > 99% enantiomeric excess (ee). The enzyme showed the highest activity for the alkylamination of pyruvate and methylamine leading to N-methyl-l-alanine with an apparent KM of 15.04 ± 2.8 mM and Vmax of 13.75 ± 1.07 μmol/min/mg. CpIM1 also catalyzed (i) the reduction of imines e.g. 2-methyl-1-pyrroline to (S)-2-methylpyrrolidine with ∼30% conversion and 75% ee and (ii) the dehydrogenation of cyclic amino acids e.g. l-Proline (as monitered by reduction of cofactor NADP+ spectrophotometrically).
1. Introduction
Chiral amines are essential functional groups in many bioactive molecules, and ∼40% of active pharmaceutical ingredients and 20% of agrochemicals contain one or more chiral amine moieties [1]. The synthesis of these bioactive molecules in their optically pure form traditionally involves heavy metal catalysts, reagents, solvents that pose environmental problems and the extensive use of protecting groups. Thus, reducing the efficiency of modern manufacturing processes. Therefore, it is necessary to develop improved environmentally benign methods. For these reasons, importance of biocatalysts, which are stereo-, regio-and chemo-selective, requiring mild operating conditions and water as the solvent, is ever-increasing [2], [3], [4].
The last decade saw several reports for the biocatalytic synthesis of chiral amines, including resolution of racemic amines using lipases and monoamine oxidases, asymmetric synthesis using transaminases, imine reductases, and amine dehydrogenases [5], [6], [7], [8], [9]. Enzymatic imine reduction is the most promising approach since the single-step asymmetric reduction of prochiral imines to the corresponding amines can theoretically yield 100% of the required enantiomer. Further, the reduction of imines generated in situ by the condensation of amines and carbonyl compounds can be used to synthesize almost any primary, secondary, or tertiary amine [1]. Enzymes of several structurally unrelated protein families catalyze the reduction of C=N bonds in pathways responsible for the synthesis of folate, alkaloids, siderophores, and antibiotics; however, many of these enzymes have been studied only for their physiological and biomedical relevance [1].
Two enantio-complementary NADPH-dependent imine reductases (IREDs), that natively reduced 2-methyl-1-pyrroline (2-MPN) from Streptomyces spp. by were first reported by Mitsukura et al.,[10], [11], [12]. The application of these enzymes in the asymmetric reduction of a variety of cyclic imines, iminium ions, and biocatalytic cascades has generated interest in IREDs as catalysts for the synthesis of chiral amines. Several research groups worldwide have screened and characterized numerous IRED homologs [13], [14], [15], [16], [17], [18], [19] but successfully isolated only a few IRED homologs that are capable of reductive amination of carbonyl substrates with amines [19], [20], [21]. In addition to IREDs ketimine reductases (KIREDs) and Δ1-piperidine-2-carboxylate/Δ1-pyrroline-2-carboxylate reductases (PipC/PyrC REDs) reportedly reduce several naturally occurring cyclic imino-acids [22]. Some amine dehydrogenases are known to catalyze reductive amination of ketones with ammonia [23]. At the same time, proteins like l-amino acid dehydrogenases and opine dehydrogenases (which natively couple α-amino acids with α-keto acids) have been engineered to broaden the substrate scope to improve their utility in chiral amine synthesis [24], [25], [26].
The present study reports the isolation and characterization of a stereospecific imine reductase CpIM1 from Candida parapsilosis ATCC 7330, a rich source of oxidoreductases [27], [28], [29]. Imine reducing activities of various yeasts are known, and our earlier work on Candida parapsilosis ATCC 7330 reported the reduction of aryl imines using whole cells of this yeast [6,30]. Notably, no imine reductases from yeasts have yet been characterized, and only recently ene-reductases from Saccharomyces spp. have been reported to reduce C=N group of oximes promiscuously [31]. To the best of our knowledge, this is the first report of a purified and characterized imine reductase from yeast.
2. Materials and methods
2.1. Gene cloning
The genomic DNA of Candida parapsilosis ATCC 7330 was isolated from liquid culture using the lithium acetate (LiOAc)-SDS method [32]. Briefly, 1 ml of culture was centrifuged, and the cell pellet was resuspended in 100 mM lithium acetate, 1% sodium dodecyl sulfate (LiOAc-SDS) solution to lyse the cells. Subsequently, DNA was precipitated by adding three parts of ethanol, followed by centrifugation at 18000 g. The obtained pellet was resuspended in 100 µl of ultrapure water and again centrifuged, and the supernatant transferred to a fresh centrifuge tube and stored at -20 °C.
The isolated genomic DNA was used as a template for amplifying the Candida parapsilosis imine reductase gene (CpIM1) using the primers:
Im1_F CATATGAAGGTCATTAGAGATAAAGAT (Nde1)
Im1_R GAATTCAAATTTGGTCTAAAAGTCAT (EcoR1)
The amplified product was purified from agarose gel and ligated into pGEMT vector by TA cloning. The resulting construct, pGEMT-IM1, was transformed into Escherichia coli DH5α for bulk production of the circular plasmid. The plasmid pGEMT-IM1 was isolated from respective transformed colonies of E. coli DH5α and subjected to digestion by Nde1 and EcoR1. The insert obtained was ligated into expression vector pET28a by directional cloning.
2.2. Protein expression and purification
After the pET28a-IM1 construct was verified by sequencing, E. coli BL21 (DE3) was transformed by the plasmid pET28a-IM1. The successfully transformed E. coli BL21 (DE3) cells containing the pET28a-IM1 construct were grown in Lysogeny broth to 0.6–0.8 OD600 at 37 °C. Then 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) was added to induce the expression of recombinant CpIM1 at 23 °C incubated for 18 h. The induced cells were harvested and stored at -80 °C until further use.
The purification of overexpressed CpIM1 was carried out by metal affinity chromatography on GE ӒKTA fast protein liquid chromatography system (GE Biosciences), installed inside a cold cabinet. All the purification steps were carried out at 4 °C or on ice. The E. coli BL21 (DE3) cells containing overexpressed CpIM1 were resuspended in a buffer containing 50 mM Tris-Cl (pH 7.5), 300 mM NaCl, and 4 mM benzamidine hydrochloride, and lyzed using ultrasonic cell disruptor (SONICS Vibracell VCX130) set at 60% amplitude for 2 min (1 second on, 2 s off). After ultra-sonication, the lysate was centrifuged at 15000 g to separate cell debris and was loaded onto a HisTrap FF column (5 ml, GE Biosciences). The bound recombinant protein was washed with 50 mM Tris-Cl (pH 7.5) buffer containing 30 mM imidazole and eluted with 50 mM Tris-Cl (pH 7.5) buffer containing 300 mM imidazole. This protein solution was concentrated by ultrafiltration and loaded on to HiTrap Desalting column (5ml, GE Biosciences) to desalt and change the buffer as required. The purified protein samples were routinely quantified by the Bradford method and analyzed by SDS–PAGE, followed by Coomassie Brilliant Blue R250 staining.
2.3. Analytical gel filtration
Analytical gel filtration was performed on a fast protein liquid chromatography (FPLC) system (ӒKTA purifier, GE Biosciences) with a column manually packed with GE Sephacryl G200 resin, the mobile phase with buffer containing 50 mM Tris-Cl (pH 7.5), 300 mM NaCl. Protein standards used were procured from Sigma-Aldrich.
2.4. Comparison of primary and secondary structure
BlastP was used to identify proteins that are similar in sequence to CpIM1 [33]. Multiple sequence alignment was constructed using PSI/TM-Coffee [34]. CpIM1 was modeled with ROBETTA protein structure prediction server, and structural comparisons were made using TM-align [35,36].
2.5. Determination of enzyme activity
The imine reduction/alkylamination activity of the enzyme was determined by monitoring the decrease in the absorbance of the cofactor NAD(P)H at 340 nm using UV visible spectrophotometer (JASCO) at 25 °C. The assay mixture contained an appropriate amount of the protein, 1–4 mM of the imine substrate, and 0.2 mM of NAD(P)H in 50 mM Tris-Cl buffer at pH 7.5. For reductive amination with substrates, the assay mixture consisted of an appropriate amount of the protein, 1–10 mM of the α-ketoacid/ketoester, 1–100 mM amine substrate, and 0.2 mM of NADPH in 50 mM Tris-Cl buffer at pH 7.5. One unit of the enzyme activity is defined as the amount of enzyme that oxidized 1 μmol of NADPH per minute at the specified assay conditions. For measuring the oxidative activity, 10 mM of oxidized cofactor NADP+ was used at pH 10.5, and the increase in absorbance at 340 nm monitored. When necessary, the substrate stocks were prepared in dimethyl sulfoxide (DMSO) and added to the reaction mixture such that the final DMSO concentration was 5% (v/v).
2.6. Determination of pH optima
The pH optima of CpIM1 were determined for using sodium pyruvate and methylamine in case of alkylamination and 2-methyl-1-pyrroline (2-MPN) for imine reduction. Thus, the assay mixture contained an appropriate amount of the protein, 10 mM sodium pyruvate, 50 mM methylamine/4 mM 2-MPN, and 0.2 mM of NAD(P)H in buffers at various pH.
2.7. Kinetics with sodium pyruvate and methylamine as substrates
The kinetic parameters (KM and Vmax) of CpIM1 were determined using sodium pyruvate and methylamine as substrates. Since the enzyme has three substrates, i.e., sodium pyruvate, methylamine, and NADPH, the apparent Vmax and KM were calculated by measuring activities on varying concentrations of one substrate with fixed non-limiting concentrations of the other two substrates. Non-limiting concentrations of 0.2 mM for NADPH, 50 mM for methylamine, and 10 mM for sodium pyruvate were used to invoke pseudo-first-order kinetics. Thus, the assay mixture consisted of 50 mM Tris buffer (pH 7.5), an appropriate amount of the protein, 0.1–50 mM sodium pyruvate, 1–50 mM methylamine, and 0.2 mM of NADPH. All measurements were carried out in triplicates.
2.8. Enzymatic biotransformations and analysis of the reaction product
Enzymatic biotransformations were carried out in 1 ml scale to confirm the formation of the product. For alkylamination reactions, the reaction mixture consisted of 5–10 mM of α-ketoacid, 50–100 mM amine substrate, and 12–15 mM of NADPH in 50 mM Tris-Cl buffer at pH 7.5–8.5. For imine reduction, the reaction mixture consisted of 4 mM of the substrate, 6 mM NAD(P)H in 50 mM Tris-Cl buffer at pH 7.5–8.5. When necessary, the substrate stocks were prepared in DMSO and added to the reaction mixture such that the final DMSO concentration was 5% (v/v). After three-hour incubation at 25 ˚C, the reaction was stopped by adding a one-tenth volume of 10 N sodium hydroxide and further derivatized/extracted (supplementary section) to be analyzed by Gas Chromatography-Mass spectrometry (GC-MS, Shimadzu QP2010).
3. Results and discussion
3.1. Identification of candidate gene
The whole cells of Candida parapsilosis ATCC 7330 catalyze the asymmetric reduction of prochiral imines to chiral amines as reported [6]. This implies the presence of stereospecific imine reductase(s) in this yeast. The preliminary investigation revealed that no gene from Candida parapsilosis showed significant sequence similarity to IRED sequences from Streptomyces spp. or any other IRED homologs which have been reported [10], [11], [12], [13], [14], [15], [16], [17], [18], [19]. The CpIM1 gene was identified using a human Mu crystallin (CRYM) gene sequence as a template, which is reportedly a ketimine reductase that catalyzes the alkylamination of various keto-acids [37]. CpIM1 showed a 24.5% identity with the human CRYM gene and belonged to the ornithine cyclodeaminase (OCD-Mu) family of proteins. The OCD-Mu protein family consists of oxidoreductases acting on carbon-nitrogen bonds, which are widely distributed and contain proteins with diverse functions. Mammalian KIREDs, plant and archaeal PipC/PyrC REDs reduce cyclic imino acids, some of which have been explored for the biocatalytic synthesis of chiral N-alkyl amino acids [22,[37], [38], [39]]. Cyclodeaminases from several gram-negative bacteria catalyze concerted oxidative-deamination cyclization and reduction of ornithine/lysine [40], [41], [42]. The other members of the OCD-Mu protein family include opine dehydrogenases from marine sponges, N-((2S)-2-amino-2-carboxyethyl)-l-glutamate dehydrogenase (ACEGA) from Staphylococcus aureus, alanine dehydrogenase (Adh) from archaeon Archaeoglobus fulgidus and iminosuccinate reductase from Paracoccus denitrificans [43], [44], [45], [46], [47]. It is also true that only a few members of this protein family have been characterized beyond gene annotation, making it an interesting and important area to explore and understand.
3.2. Analysis of the primary structure
As the first step towards the investigation of imine reductases from this yeast, well-characterized homologs of CpIM1 were identified using BlastP [33]. Multiple sequence alignment and a phylogenetic tree were then constructed using PSI/TM-Coffee [34] (Fig. 1, Supplementary Figures S1 and S2). The present study revealed that among the characterized members of the OCD family, CpIM1 was closest to plant OCD homologs (Arabidopsis thaliana and Huperzia serrata), followed by Human CRYM and other mammalian OCD homologs (Supplementary Fig. S2). The conserved motif GXGXXA/G/S (marked by the box in Fig. 1) is present in CpIM1 and the other OCD/ μ-crystallin family members, indicating the presence of the Rossmann fold for cofactor binding [48]. The mechanism and role of residues lining the active pocket in the OCD-Mu family is studied in detail in Streptomyces pristinaespiralis l-Lysine cyclodeaminase (SpLCD) [41] and Pseudomonas putida ornithine cyclodeaminase (PpOCD) [42]. Among these residues Arg48, Met65, Lys76, and Arg118 are conserved in CpIM1 (marked by an arrow in Fig. 1) and most other OCD-Mu homologs. The residues Lys76 and Arg118 interact directly with the carboxyl group of the substrate while Arg48 is thought to form hydrogen bonds with the substrate and the water molecule that transfers a proton during imine reduction. The exact role of Met65 is not understood, although it is conserved in most OCD-Mu proteins. The residues Glu56 (Glu63) and Asp228 (Asp236) from SpLCD (PpOCD), which are considered critical for cyclodeaminase activity are not conserved in CpIM1, plant and mammalian OCD homologs. Instead, the position occupied by Asp228 in SpLCD is occupied by serine in all the mammalian OCD homologs which is also suitably positioned to act as a proton donor [22]. In CpIM1, this position is occupied by bulky tyrosine, marked by the blue arrow in Fig. 1.
Fig. 1.
Multiple sequence alignment showing the key conserved residues among the sequences of CpIM1 and other characterized OCD-Mu proteins.
3.3. Protein overexpression and purification
The CpIM1 gene was amplified using the genomic DNA of Candida parapsilosis ATCC 7330 as the template and was cloned into the pET28a vector system overexpressed in E. coli BL21 (DE3). The His6-tagged CpIM1 was purified by metal affinity chromatography and showed a molecular weight of 42 kDa on SDS-PAGE (Fig. 2a) in agreement with its theoretical value (351 aa). This is slightly higher than other known members of OCD-Mu family proteins (CRYM Homo sapiens 313 aa, Alanine dehydrogenase from Archaeoglobus fulgidus 322 aa, and ornithine cyclodeaminase from Pseudomonas aeruginosa 310 aa) [37,42,46]. Like other members of the OCD-Mu family proteins, CpIM1 was found to exist as a homodimer as confirmed by analytical gel filtration (Fig. 2b).
Fig. 2.
(a) SDS page showing purified CpIM1 (∼42 kDa), protein ladder in lane A; (b) CpIM1 confirmed to be a dimer (∼ 90 kDa) by analytical gel filtration.
3.4. Effect of pH on enzyme activity
The enzyme showed maximum activity in the pH range of 6.8–9.0 for alkylamination of sodium pyruvate with methylamine (Fig. 3a). There was a sharp decrease in activity on either side of this pH zone. In a similar study that was carried out using 2-methyl-1-pyrroline (2-MPN), a cyclic imine, which was stable in an aqueous medium around neutral pH, the enzyme showed activity over a broader pH range of 5-9 (Fig. 3b). The human KIRED, which has a pH optimum of 5 for reduction of cyclic imino-acids [49], showed better conversion for alkylaminations of α-ketoacids at pH 8.5 [22]. Since the alkylamination proceeds by the formation of an imine intermediate, the nucleophilic attack by methylamine on the carbonyl carbon of α-ketoacids is favourable at pH near the pKa of the amine (∼10 in case of methylamine). In essence, alkylamination is favoured at basic pH.
Fig. 3.
Activity-pH profile for CpIM1 (a) for methylamination of sodium pyruvate, (b) for reduction of 2-MPN.
The oxidation of proline was preferred towards basic pH with the observed specific activity at pH 10.5 being greater than at pH 9.
3.5. Substrate scope and enantiospecificity
CpIM1 catalyzed alkylamination of various α-ketoacid/ketoesters (Table 1). Among the reactions studied, the highest activity is seen for sodium pyruvate with methylamine (amine) in the presence of NADPH as a cofactor, resulting in the formation of N-methyl alanine (Supplementary Figure S4). This reaction exclusively produced N-methyl-l-alanine, which is the (S) enantiomer (> 99% enantiomeric excess (ee), Supplementary Figure S5) of the product as confirmed by GC-MS after derivatizing with a chiral reagent (R)-menthyl chloroformate (Supplementary scheme S3). The mammalian KIREDs and Thermococcus litoralis OCD are also known for alkyl-amination of various α-ketoacid/ketoesters and produce l-amino acids [22,37,39]. While CpIM1 and mammalian KIREDs use NADPH [22,37], Thermococcus litoralis OCD is known to utilize NADH as cofactor [39]. CpIM1 also catalyzed similar reactions with ethylamine and ammonium chloride producing N-ethyl alanine (Supplementary Fig. S8) and l-alanine (Supplementary Figs. S6 and S7). The specific activities were 20 and 100-fold less, respectively, compared to that of methylamine (Table 1:entries 2 and 3). The human KIRED reportedly shows comparable activities for both, methylamine and ethylamine [37]. The archaeal members of the family Archaeoglobus fulgidus Adh and Thermococcus litoralis OCD show significantly higher activity with ammonia/ammonium chloride as the amine substrate [39,50]. CpIM1 did not show any alkyl-amination activity of sodium pyruvate with dimethylamine, benzylamine, and hydroxylamine, while several mammalian KIREDs are known to show activity with benzylamine [22]. CpIM1 also displayed good activity with ethyl pyruvate and methylamine (Table 1:entry 4). It did not show any significant activity with phenylpyruvic acid and phenylglyoxylic acid (with methylamine) when assayed at pH 7.5; however, on incubation with excess of enzyme at pH 8.5, the formation of corresponding N-methyl α-amino acids was observed as analyzed by GC-MS (Supplementary Figs. S9,10). The human CRYM and other mammalian OCD homologs show moderate activity with phenylpyruvate and methylamine (compared with pyruvate and methylamine) [22,37]. The poor activity of CpIM1 with phenylpyruvic acid and phenylglyoxylic acid may be due to the presence of bulky tyrosine residue instead of serine (in mammalian KIREDs), which projects into the active site [22]. Alkylamination of ethyl 2-oxo-4-phenyl butanoate [a keto-ester] with methylamine was catalyzed by CpIM1 with a considerable activity of 1760 mU (Table 1:entry 7), implying that two carbon length separation between the ketoacid moiety and the phenyl ring allowed proper binding of the substrate. The formation of the product was confirmed by GC-MS (Supplementary Fig. S12,13). CpIM1 also showed formation of product ethyl sarcosinate with ethyl glyoxylate and methylamine (Supplementary Figure S11). The direct alkylamination of α-keto-esters by a member of of OCD-Mu family is reported here for the first time.
Table 1.
General scheme and specific activities for CpIM1 catalyzed alkylaminations
 |
1 mU is defined as the amount of the enzyme that catalyzes the conversion of one nanomole of the substrate (NADPH) per minute under the specified conditions of the assay method.
a Inferred based on comparison with authentic derivatized standard
b Inferred based on expected mass spectra of the product
c Inferred on comparison with the crude chemically synthesized product
d Apparent activity recorded after allowing polymeric ethyl glyoxylate to dissociate in reaction buffer for 1 hr
In addition to alkylamination, CpIM1 catalyzed imine reduction with cyclic imine 2-methyl-1-pyrroline (2MPN) and aromatic imine N-benzylideneaniline (Table 2: entries 1-2), albeit with very low activity when compared to that observed for alkylamination of sodium pyruvate and methylamine. The specific activity observed for the reduction of 2-methyl-1-pyrroline 44 ± 2 mU/mg is comparable to some of the reported IREDs, e.g., 50 ± 20 mU/mg from Streptomyces sp. 3456 [12], 66 mU/mg from Pseudomonas putida KT2440, and 20 mU/mg from Paenibacillus elgii B69 [13]. Interestingly, reduction of 2MPN occurred specifically with NADH producing (S)-2-methylpyrrolidine (75% ee, Supplementary Figure S16). Inhibition was observed at concentrations above 5 mM of 2MPN. The reduction of N-benzylideneaniline occurred with NADH and NADPH. The formation of products with 2MPN and N-benzylidene aniline was confirmed by GC-MS by comparison with authentic product standards (Supplementary Figs. S15 and S17). The switching of cofactor specificity has also been observed in the case of porcine and bovine KIREDs with specific substrates [49]. Although, CpIM1 showed some activity with N-(1-phenylethylidene) benzenamine (52 mU/mg), the conversion was not significant (Supplementary Figs. S18).
Table 2.
Specific activities for CpIM1 catalyzed imine reduction and oxidations.
| Substrates (reduction) | Specific activity (mU/mg) | |
|---|---|---|
| 1 | 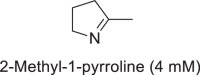 |
44 ± 2 |
| 2 | 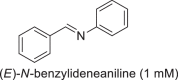 |
28 |
| Substrates (oxidation) | ||
| 3 | 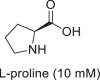 |
420 ± 32 |
1 mU is defined as the amount of the enzyme that catalyzes the conversion of one nanomole of the substrate (NAD(P)H/ NAD(P)+) per minute under the specified conditions of the assay method.
CpIM1 also catalyzed the dehydrogenation of l-proline and 4-hydroxy-l-proline in the basic pH range of 9.0–10.5. With l-proline as substrate, CpIM1 showed the highest activity of 420 mU/mg at pH 10.5 (Table 2:entry 3), while activity with 4-hydroxy-l-proline was about 100-fold less and was barely detectable under the assay conditions. However, this reaction required higher concentrations of cofactor NADP+ (1 mM) for appreciable rates unlike reductions, which required only 0.2 mM of reduced cofactor NADPH. CpIM1 did not show any activity with d-proline when assayed under identical conditions. Interestingly, quite like human KIRED, CpIM1 did not show oxidation of N-methyl-l-alanine (the product from methylamination of sodium pyruvate) under any of the tested conditions.
3.6. Steady-state kinetic parameters
Since the enzyme has the highest specific activity for methylamination of sodium pyruvate, which involves three substrates, i.e., sodium pyruvate, methylamine, and NADPH, two sets of experiments were carried to for determination of kinetic parameters to invoke pseudo-first-order kinetics: (i) concentration of pyruvate was varied with non-limiting concentrations of the other two substrates (0.2 mM for NADPH, 50 mM for methylamine) (ii) concentration of methylamine was varied with fixed concentration of pyruvate (10 mM) and NADPH (0.2 mM). CpIM1 followed Michaelis-Menten kinetics in the first set of experiments with pyruvate as a substrate with an apparent KM of 15.04 ± 2.8 mM and Vmax of 13.75 ± 1.07 μmol/min/mg (Fig. 4a). In the second set of experiments, the determination of KM and Vmax with methylamine was inconclusive since the rate of reaction did not saturate even with 50 mM of methylamine, and further addition of methylamine led to a drastic increase in pH (Fig. 4b). Human KIRED is reported to behave similarly [37]. The apparent KM observed for CpIM1 was higher than that of human CRYM (2.9 ± 0.3 mM) [37] but comparable to that of N-Methylalanine dehydrogenase from Pseudomonas MS ATCC 25262 (KM of 15 mM) [51], which is structurally unrelated but catalyzes the same reaction. However, Vmax of CpIM1 is higher than values reported for both human CRYM (2 ± 0.3 μmol/min/mg) and N-Methylalanine dehydrogenase from Pseudomonas MS ATCC 25262 (0.37 μmol/min/mg) [37,51]. Two other enzymes, which are structurally unrelated to CpIM1, PipC/PyrC reductases from P. putida [52], and P. syringae [53], have higher specific activities (42 μmol/min/mg and 140 μmol/min/mg) for the same reaction.
Fig. 4.
Plot of specific activity vs substrate concentration for CpIM1 at various concentrations of (a) pyruvate (0.05–50 mM) with 50 mM methyl amine and 0.2 mM NADPH (b) methyl amine (0.5–50 mM) with 10 mM pyruvate and 0.2 mM NADPH
Despite the high observed KM, the biotransformations with sodium pyruvate and methylamine proceeded to completion, making it very apparent that imine intermediate is the actual substrate for the enzymatic reduction. Since apparent saturation was observed with pyruvate but not with methylamine, it can be concluded that the enzyme has binding site for pyruvate. This is supported by the crystal structure of mouse KIRED (4BV9), which has pyruvate in the active site [54]. Thus, imine formation can occur within the active site by the action of methylamine on the enzyme-bound pyruvate.
4. Conclusion
A novel imine reductase gene CpIM1 isolated, cloned, and overexpressed from the versatile biocatalyst Candida parapsilosis ATCC 7330 is reported. Its biochemical properties and the substrate scope and enantiospecificity of the purified CpIM1 were explored. CpIM1 catalyzed alkylamination of α-ketoacids and α-ketoesters producing (S)-alkyl amino acids/esters, which are essential building blocks for the pharmaceutical agents, especially the emerging peptide drugs. It also inherently reduced several imines to corresponding amines and catalyzed dehydrogenation of cyclic amino acids. The enzyme seemed to prefer substrates with a carboxylic acid moiety and would make an attractive candidate for directed evolution to accept ketones as substrates. Further mutational and crystallographic studies will not only provide insights into the structure and mechanism of CpIM1 but also aid in understanding the structure-function relationship in the OCD-Mu protein family.
Declaration of Competing Interest
The authors whose names are listed immediately below certify that they have no affiliations with or involvement in any organization or entity with any financial interest (such as honoraria; educational grants; participation in speakers’ bureaus; membership, employment, consultancies, stock ownership, or other equity interest; and expert testimony or patent-licensing arrangements), or non-financial interest (such as personal or professional relationships, affiliations, knowledge or beliefs) in the subject matter or materials discussed in this manuscript.
Acknowledgments
One of the authors (M V N Uma Mahesh) acknowledges the Council of Scientific and Industrial Research (CSIR, India) for fellowship. Facilities at IITM are gratefully acknowledged.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.btre.2021.e00664.
Appendix. Supplementary materials
References
- 1.Schrittwieser J.H., Velikogne S., Kroutil W. Biocatalytic imine reduction and reductive amination of ketones. Adv. Synth. Catal. 2015;357:1655–1685. doi: 10.1002/adsc.201500213. [DOI] [Google Scholar]
- 2.Nakamura K., Yamanaka R., Matsuda T., Harada T. Recent developments in asymmetric reduction of ketones with biocatalysts. Tetrahedron-Asymmetry. 2003;14:2659–2681. doi: 10.1016/s0957-4166(03)00526-3. [DOI] [Google Scholar]
- 3.Hauer B., Lutz S. Editorial overview: beyond native biocatalysts and natural biotransformations. Curr. Opin. Chem. Biol. 2017;37 doi: 10.1016/j.cbpa.2017.04.006. iv–v. [DOI] [PubMed] [Google Scholar]
- 4.Hauer B. Embracing nature's catalysts: a viewpoint on the future of biocatalysis. ACS Catal. 2020;10:8418–8427. doi: 10.1021/acscatal.0c01708. [DOI] [Google Scholar]
- 5.Ghislieri D., Turner N.J. Biocatalytic approaches to the synthesis of enantiomerically pure chiral amines. Top. Catal. 2014;57:284–300. doi: 10.1007/s11244-013-0184-1. [DOI] [Google Scholar]
- 6.Vaijayanthi T., Chadha A. Asymmetric reduction of aryl imines using Candida parapsilosis ATCC 7330. Tetrahedron Asymmetry. 2008;19:93–96. doi: 10.1016/j.tetasy.2007.11.039. [DOI] [Google Scholar]
- 7.Kohls H., Steffen-Munsberg F., Höhne M. Recent achievements in developing the biocatalytic toolbox for chiral amine synthesis. Curr. Opin. Chem. Biol. 2014;19:180–192. doi: 10.1016/j.cbpa.2014.02.021. [DOI] [PubMed] [Google Scholar]
- 8.Grogan G. Synthesis of chiral amines using redox biocatalysis. Curr. Opin. Chem. Biol. 2018;43:15–22. doi: 10.1016/j.cbpa.2017.09.008. [DOI] [PubMed] [Google Scholar]
- 9.Patil M.D., Grogan G., Bommarius A., Yun H. Recent advances in ω-transaminase-mediated biocatalysis for the enantioselective synthesis of chiral amines. Catalysts. 2018;8:254. doi: 10.3390/catal8070254. [DOI] [Google Scholar]
- 10.Mitsukura K., Suzuki M., Tada K., Yoshida T., Nagasawa T. Asymmetric synthesis of chiral cyclic amine from cyclic imine by bacterial whole-cell catalyst of enantioselective imine reductase. Org. Biomol. Chem. 2010;8:4533–4535. doi: 10.1039/c0ob00353k. [DOI] [PubMed] [Google Scholar]
- 11.Kuramoto T., Mitsukura K., Yoshida T., Nagasawa T., Shinoda S., Suzuki M. Purification and characterization of a novel (R)-imine reductase from Streptomyces sp. GF3587. Biosci. Biotechnol. Biochem. 2011;75:1778–1782. doi: 10.1271/bbb.110303. [DOI] [PubMed] [Google Scholar]
- 12.Yamamoto H., Yoshida T., Kuramoto T., Mitsukura K., Kimoto N., Nagasawa T. A NADPH-dependent (S)-imine reductase (SIR) from Streptomyces sp. GF3546 for asymmetric synthesis of optically active amines: purification, characterization, gene cloning, and expression. Appl. Microbiol. Biotechnol. 2012;97:8079–8086. doi: 10.1007/s00253-012-4629-4. [DOI] [PubMed] [Google Scholar]
- 13.Gand M., Müller H., Wardenga R., Höhne M. Characterization of three novel enzymes with imine reductase activity. J. Mol. Catal. B Enzym. 2014;110:126–132. doi: 10.1016/j.molcatb.2014.09.017. [DOI] [Google Scholar]
- 14.Gand M., Thöle C., Müller H., Brundiek H., Bashiri G., Höhne M. A NADH-accepting imine reductase variant: immobilization and cofactor regeneration by oxidative deamination. J. Biotechnol. 2016;230:11–18. doi: 10.1016/j.jbiotec.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 15.Lenz M., Scheller P.N., Richter S.M., Hauer B., Nestl B.M. Cultivation and purification of two stereoselective imine reductases from Streptosporangium roseum and Paenibacillus elgii. Protein Expr. Purif. 2017;133:199–204. doi: 10.1016/j.pep.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 16.Scheller P.N., Nestl B.M. The biochemical characterization of three imine-reducing enzymes from Streptosporangium roseum DSM43021, Streptomyces turgidiscabies and Paenibacillus elgii. Appl. Microbiol. Biotechnol. 2016;100:10509–10520. doi: 10.1007/s00253-016-7740-0. [DOI] [PubMed] [Google Scholar]
- 17.France S.P., Toca-Gonzalez L., Turner N.J., Grogan G., Aleku G.A., Hart S., Leipold F., Turkenburg J.P., Man H., Hussain S., Marchington R. Stereoselectivity and structural characterization of an imine reductase (IRED) from amycolatopsis orientalis. ACS Catal. 2016;6:3880–3889. doi: 10.1021/acscatal.6b00782. [DOI] [Google Scholar]
- 18.Li H., Zhang G.X., Li L.M., Ou Y.S., Wang M.Y., Li C.X., Zheng G.W., Xu J.H. A novel (R)-imine reductase from paenibacillus lactis for asymmetric reduction of 3H-indoles. ChemCatChem. 2016;8:724–727. doi: 10.1002/cctc.201501170. [DOI] [Google Scholar]
- 19.Roiban G.D., Kern M., Liu Z., Hyslop J., Tey P.L., Levine M.S., Jordan L.S., Brown K.K., Hadi T., Ihnken L.A.F., Brown M.J.B. Efficient biocatalytic reductive aminations by extending the imine reductase toolbox. ChemCatChem. 2017;9:4475–4479. doi: 10.1002/cctc.201701379. [DOI] [Google Scholar]
- 20.Aleku G.A., France S.P., Man H., Mangas-Sanchez J., Montgomery S.L., Sharma M., Leipold F., Hussain S., Grogan G., Turner N.J. A reductive aminase from aspergillus oryzae. Nat. Chem. 2017;9:961–969. doi: 10.1038/nchem.2782. [DOI] [PubMed] [Google Scholar]
- 21.France S.P., Howard R.M., Steflik J., Weise N.J., Mangas-Sanchez J., Montgomery S.L., Crook R., Kumar R., Turner N.J. Identification of novel bacterial members of the imine reductase enzyme family that perform reductive amination. ChemCatChem. 2018;10:510–514. doi: 10.1002/cctc.201701408. [DOI] [Google Scholar]
- 22.Hyslop J.F., Lovelock S.L., Sutton P.W., Brown K.K., Watson A.J.B., Roiban G. Biocatalytic synthesis of chiral N-functionalized amino acids. Angew. Chemie Int. Ed. 2018;57:13821–13824. doi: 10.1002/anie.201806893. [DOI] [PubMed] [Google Scholar]
- 23.Mayol O., Bastard K., Beloti L., Frese A., Turkenburg J.P., Petit J.L., Mariage A., Debard A., Pellouin V., Perret A., de Berardinis V., Zaparucha A., Grogan G., Vergne-Vaxelaire C. A family of native amine dehydrogenases for the asymmetric reductive amination of ketones. Nat. Catal. 2019;2:324–333. doi: 10.1038/s41929-019-0249-z. [DOI] [Google Scholar]
- 24.H. Chen, J. Moore, S.J. Collier, D. Smith, J. Nazor, G. Hughes, J. Janey, G. Huisman, S. Novick, N. Agard, O. Alvizo, G. Cope, W.L. Yeo, J. Sukumaran, S. Ng, Engineered imine reductases and methods for the reductive amination of ketone and amine compounds., 2013.
- 25.Chen F.F., Zheng G.W., Liu L., Li H., Chen Q., Li F.L., Li C.X., Xu J.H. Reshaping the active pocket of amine dehydrogenases for asymmetric synthesis of bulky aliphatic amines. ACS Catal. 2018;8:2622–2628. doi: 10.1021/acscatal.7b04135. [DOI] [Google Scholar]
- 26.Pushpanath A., Siirola E., Bornadel A., Woodlock D., Schell U. Understanding and overcoming the limitations of bacillus badius and caldalkalibacillus thermarum amine dehydrogenases for biocatalytic reductive amination. ACS Catal. 2017;7:3204–3209. doi: 10.1021/acscatal.7b00516. [DOI] [Google Scholar]
- 27.Chadha A., Venkataraman S., Preetha R., Padhi S.K. Candida parapsilosis: a versatile biocatalyst for organic oxidation-reduction reactions. Bioorg. Chem. 2016;68:187–213. doi: 10.1016/j.bioorg.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 28.Sudhakara S., Chadha A. A carbonyl reductase from: Candida parapsilosis ATCC 7330: Substrate selectivity and enantiospecificity. Org. Biomol. Chem. 2017;15:4165–4171. doi: 10.1039/c7ob00340d. [DOI] [PubMed] [Google Scholar]
- 29.Karanam V.K., Chaudhury D., Chadha A. Understanding (R) specific carbonyl reductase from Candida parapsilosis ATCC 7330 [CpCR]: substrate scope, kinetic studies and the role of zinc. Catalysts. 2019;9:702–712. doi: 10.3390/catal9090702. [DOI] [Google Scholar]
- 30.Chimni S.S., Singh R.J. Bio-reduction of a carbon-nitrogen double bond using immobilized baker's yeast - a first report. World J. Microbiol. Biotechnol. 1998;14:247–250. doi: 10.1023/A:1008894416058. [DOI] [Google Scholar]
- 31.Velikogne S., Breukelaar W.B., Hamm F., Glabonjat R.A., Kroutil W. C=C-Ene-Reductases Reduce the C=N Bond of Oximes. ACS Catal. 2020;10:13377–13382. doi: 10.1021/acscatal.0c03755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lõoke M., Kristjuhan K., Kristjuhan A. Extraction of genomic DNA from yeasts for PCR-based applications. Biotechniques. 2011;50:325–328. doi: 10.2144/000113672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.T. Madden, Chapter 16 : The BLAST Sequence Analysis Tool, NCBI Handb. (2002) 1–15.
- 34.Floden E.W., Tommaso P.D., Chatzou M., Magis C., Notredame C., Chang J.M. PSI/TM-Coffee: a web server for fast and accurate multiple sequence alignments of regular and transmembrane proteins using homology extension on reduced databases. Nucleic Acids Res. 2016;44:W339–W343. doi: 10.1093/nar/gkw300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim D.E., Chivian D., Baker D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004;32 doi: 10.1093/nar/gkh468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y., Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hallen A., Cooper A.J.L., Smith J.R., Jamie J.F., Karuso P. Ketimine reductase/CRYM catalyzes reductive alkylamination of α-keto acids, confirming its function as an imine reductase. Amino Acids. 2015;47:2457–2461. doi: 10.1007/s00726-015-2044-8. [DOI] [PubMed] [Google Scholar]
- 38.Xu B., Fan Z., Lei Y., Ping Y., Jaisi A., Xiao Y. Insights into Pipecolic Acid Biosynthesis in Huperzia serrata. Org. Lett. 2018;20:2195–2198. doi: 10.1021/acs.orglett.8b00523. [DOI] [PubMed] [Google Scholar]
- 39.Watanabe S., Tozawa Y., Watanabe Y. Ornithine cyclodeaminase/μ-crystallin homolog from the hyperthermophilic archaeon Thermococcus litoralis functions as a novel δ1-pyrroline-2-carboxylate reductase involved in putative trans-3-hydroxy-l-proline metabolism. FEBS Open Bio. 2014;4:617–626. doi: 10.1016/j.fob.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muth W.L., Costilow R.N. Ornithine cyclase (deaminating). III. Mechanism of the conversion of ornithine to proline. J. Biol. Chem. 1974;249:7463–7467. doi: 10.1016/S0021-9258(19)81261-9. [DOI] [PubMed] [Google Scholar]
- 41.Min K., Yoon H.-J., Matsuura A., Kim Y.H., Lee H.H. Structural basis for recognition of L-lysine, L-ornithine, and L-2,4-diamino butyric acid by lysine cyclodeaminase. Mol. Cells. 2018;41:331–341. doi: 10.14348/molcells.2018.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alam S., Wang S.C., Ruzicka F.J., Frey P.A., Wedekind J.E. Crystallization and X-ray diffraction analysis of ornithine cyclodeaminase from Pseudomonas putida. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004;60:941–944. doi: 10.1107/S0907444904005256. [DOI] [PubMed] [Google Scholar]
- 43.Kan-No N., Matsu-Ura H., Jikihara S., Yamamoto T., Endo N., Moriyama S., Nagahisa E., Sato M. Tauropine dehydrogenase from the marine sponge Halichondria japonica is a homolog of ornithine cyclodeaminase/mu-crystallin. Comp. Biochem. Physiol. - B Biochem. Mol. Biol. 2005;141:331–339. doi: 10.1016/j.cbpc.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 44.Plese B., Schröder H.C., Grebenjuk V.A., Wegener G., Brandt D., Natalio F., Müller W.E.G. Strombine dehydrogenase in the demosponge Suberites domuncula: Characterization and kinetic properties of the enzyme crucial for anaerobic metabolism. Comp. Biochem. Physiol. - B Biochem. Mol. Biol. 2009;154:102–107. doi: 10.1016/j.cbpb.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 45.Kobylarz M.J., Grigg J.C., Takayama S.I.J., Rai D.K., Heinrichs D.E., Murphy M.E.P. Synthesis of L-2,3-diaminopropionic acid, a siderophore and antibiotic precursor. Chem. Biol. 2014;21:379–388. doi: 10.1016/j.chembiol.2013.12.011. [DOI] [PubMed] [Google Scholar]
- 46.Vadas A., Monbouquette H.G., Lim S., Schroder I., Johnson E. A novel archaeal alanine dehydrogenase homologous to ornithine cyclodeaminase and -crystallin. J. Bacteriol. 2004;186:7680–7689. doi: 10.1128/jb.186.22.7680-7689.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schada von Borzyskowski L., Severi F., Krüger K., Hermann L., Gilardet A., Sippel F., Pommerenke B., Claus P., Cortina N.S., Glatter T., Zauner S., Zarzycki J., Fuchs B.M., Bremer E., Maier U.G., Amann R.I., Erb T.J. Marine Proteobacteria metabolize glycolate via the β-hydroxyaspartate cycle. Nature. 2019;575:500–504. doi: 10.1038/s41586-019-1748-4. [DOI] [PubMed] [Google Scholar]
- 48.Kleiger G., Eisenberg D. GXXXG and GXXXA motifs stabilize FAD and NAD(P)-binding rossmann folds through Cα-H•••O hydrogen bonds and van der Waals interactions. J. Mol. Biol. 2002;323:69–76. doi: 10.1016/S0022-2836(02)00885-9. [DOI] [PubMed] [Google Scholar]
- 49.Hallen A., Cooper A.J.L., Jamie J.F., Haynes P.A., Willows R.D. Mammalian forebrain ketimine reductase identified as μ-crystallin; Potential regulation by thyroid hormones. J. Neurochem. 2011;118:379–387. doi: 10.1111/j.1471-4159.2011.07220.x. [DOI] [PubMed] [Google Scholar]
- 50.Schröder I., Vadas A., Johnson E., Lim S., Monbouquette H.G. A novel archaeal alanine dehydrogenase homologous to ornithine cyclodeaminase and μ-crystallin. J. Bacteriol. 2004;186:7680–7689. doi: 10.1128/JB.186.22.7680-7689.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin M.C., Wagner C. Purification and characterization of N-methylalanine dehydrogenase. J. Biol. Chem. 1975;250:3746–3751. http://www.ncbi.nlm.nih.gov/pubmed/236301 accessed December 31, 2019. [PubMed] [Google Scholar]
- 52.Mihara H., Muramatsu H., Kakutani R., Yasuda M., Ueda M., Kurihara T., Esaki N. N-methyl-L-amino acid dehydrogenase from Pseudomonas putida: a novel member of an unusual NAD(P)-dependent oxidoreductase superfamily. FEBS J. 2005;272:1117–1123. doi: 10.1111/j.1742-4658.2004.04541.x. [DOI] [PubMed] [Google Scholar]
- 53.Goto M., Muramatsu H., Mihara H., Kurihara T., Esaki N., Omi R., Miyahara I., Hirotsu K. Crystal structures of Δ1-piperideine-2-carboxylate/Δ1-pyrroline-2-carboxylate reductase belonging to a new family of NAD(P)H-dependent oxidoreductases: conformational change, substrate recognition, and stereochemistry of the reaction. J. Biol. Chem. 2005;280:40875–40884. doi: 10.1074/jbc.M507399200. [DOI] [PubMed] [Google Scholar]
- 54.Borel F., Hachi I., Palencia A., Gaillard M.-C., Ferrer J.-L. Crystal structure of mouse mu-crystallin complexed with NADPH and the T3 thyroid hormone. FEBS J. 2014;281:1598–1612. doi: 10.1111/febs.12726. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.






