Abstract
The purgaty One systems (cap+bottle) are portable stainless-steel water bottles with UV subtype C (UVC) disinfection capability. This study examines the bottle design, verifies disinfection performance against Escherichia coli, Pseudomonas aeruginosa, Vibrio cholerae, and heterotrophic contaminants, and addresses the public health relevance of heterotrophic bacteria. Bottles were inoculated with deliberately contaminated potable water and disinfection efficacy examined using colony forming unit (CFU) assay for each bacterial strain. The heterotrophic plate count (HPC) method was used to determine the disinfection performance against environmental contaminants at day 0 and after 3days of water in stationary condition without prior UVC exposure. All UVC irradiation experiments were performed under stationary conditions to confirm that the preset application cycle of 55s offers the desired disinfection performance under-tested conditions. To determine effectiveness of purgaty One systems (cap+bottle) in disinfection, inactivation efficacy or log reduction value (LRV) was determined using bacteria concentration between UVC ON condition and controls (UVC OFF). The study utilized the 16S ribosomal RNA (rRNA) gene for characterization of isolates by identifying HPC bacteria to confirm if they belong to groups that are of public health concern. Purgaty One systems fitted with Klaran UVC LEDs achieved 99.99% inactivation (LRV4) efficacy against E. coli and 99.9% inactivation (LRV3) against P. aeruginosa, V. cholerae, and heterotrophic contaminants. Based on the 16S rRNA gene analyses, the study determined that the identified HPC isolates from UVC irradiated water are of rare public health concern. The bottles satisfactorily inactivated the target pathogenic bacteria and HPC contaminants even after 3days of water in stationary condition.
Keywords: cholera, disinfection, Escherichia coli, Pseudomonas aeruginosa, public health, UVC LED, Vibrio cholerae, water treatment
Introduction
The low quality of potable water is a major issue in travel medicine, especially when visiting places with poor hygienic conditions due to waterborne diseases, which pose substantial health risk (Ericsson et al., 2002). Waterborne pathogens, predominantly of fecal origin, can be transmitted via contaminated drinking water (Ashbolt, 2004). Even in developed countries, they represent a risk to recreational travelers who have to rely on surface water (Ericsson et al., 2002). For instance, in the United States, it is estimated that each year 560,000 people suffer from severe waterborne diseases due to the consumption of contaminated drinking water, with 7.1million suffering from mild to moderate infections, resulting in estimated 12,000 deaths a year (Medema et al., 2003). Hikers and campers are also exposed to waterborne disease risks if they consume untreated water from rivers and lakes (Schlosser et al., 2001). Diarrheal infections are a major inconvenience in the wilderness during hiking or camping and can easily spread via contaminated water supplies and from person-to-person.
One way to prevent waterborne diseases for healthy travelling in regions with unsafe or underdeveloped water sources is by ensuring adequate supply of potable water. Alternatively, outdoor enthusiasts can use portable and germicidal devices that ensure inactivation of microbial contaminants. UV irradiation in the UVC range (200–280nm) has demonstrated effective inactivation of microbial contaminants in water (Umar et al., 2019). Specifically, as part of the effort to accelerate the sustainable development goals (SDGs) such as clean water and sanitation goal (SDG #6; Fagunwa and Olanbiwoninu, 2020), regulating microbial load is required to control waterborne diseases caused by microorganisms, such as Pseudomonas aeruginosa, Escherichia coli, and Vibrio spp. (Cabral, 2010).
Cholera, caused by Vibrio cholerae remains a serious risk in emerging economies where sanitation is poor, health care limited, and drinking water unsafe (Heidelberg et al., 2000). Additionally, due to global warming, there is association between the spread of pathogenic vibrios and emergence of human diseases toward the temperate world (Vezzulli et al., 2016). Existing water infrastructure including electronic faucets can act as reservoirs and sources of outbreaks once contaminated. Hospital water, for instance, can disseminate opportunistic pathogens such as P. aeruginosa and fecal coliforms, where E. coli is a key species (Kanamori et al., 2016).
Furthermore, heterotrophic bacteria are a concern in drinking water systems if the counts are consistently >500 colony forming units per milliliter (CFU/ml). They can be an indication of general decrease in water quality and potential biofilm formation in municipal water (Lizzadro et al., 2019). Therefore, to eradicate elevated levels of heterotrophic plate count (HPC) and other pathogens, portable bottle devices with disinfection features can act as a form of disinfectant.
With more than 1billion people globally having no access to potable water, and 2.4billion people still living in areas without adequate sanitation systems (World Health Organization, 2000), there is need for portable, durable, appealing, personal, and highly germicidal devices to help curb enteric pathogens. Use of UVC radiation is one of the disinfection methods recognized by WHO (World Health Organization, 2000). Unlike most methods, UV disinfects by striking the target microorganism with sufficient dose of energy, while neither altering the water, nor providing any residue (World Health Organization, 2000). UV is subdivided into three distinct bands: UVA with a wavelength of 315–400nm, UVB with 280–315nm, and UVC with 100–280nm (World Health Organization, 2014). The UVC region has been found to be effective against waterborne microorganisms such as E. coli ATCC 25922 and Staphylococcus aureus ATCC 25923 (Timmermann et al., 2015), where UV systems such as Mountop Water Purifier Bottle and SteriPEN Water Purifier Kit have been used for the disinfection of Mycobacterium abscessus, M. avium, and M. chimaera (Norton et al., 2020). Specifically, the wavelength range between 250 and 270nm is strongly absorbed by the nucleic acids (DNA and RNA) of microbial cells (Dai et al., 2012). The use of UVC for point-of-use (POU) is technically possible at 270nm as demonstrated in a study that utilized E. coli K12 ATCC W3110 and Enterococcus faecalis ATCC 19433 (Lui et al., 2016). This study further revealed that the 310 and >455nm LEDs offer no significant UVC disinfection efficacy. Theoretically, the disinfection performance of a UVC device is a function of the intensity of UVC light (irradiance) and time of exposure resulting in a UVC dose. Greater disinfection efficacy is expected at higher UVC dose (Gora et al., 2019).
The purpose of this study was to investigate the disinfection performance of the recent commercial development of the portable purgaty One system (cap+bottle) by analyzing test bottles against pathogens and heterotrophic contaminants. This study was carried out using US municipal drinking water supplied by Cohoes Water Department in New York State.
Materials and Methods
There are two bottle types of purgaty One on the market, a 650 and a 500ml version. The purgaty brain (cap) can fit either of the bottles. The cap is rechargeable (Figure 1B).
Figure 1.

(A) Test bottles whose ordinary caps can be replaced with UV subtype C (UVC) emitting purgaty brain (B) The UVC emitting purgaty brain threaded onto both bottles (top) and shown individually from a side view and from the bottom (bottom).
Disinfection Cap (Purgaty Brain) Design
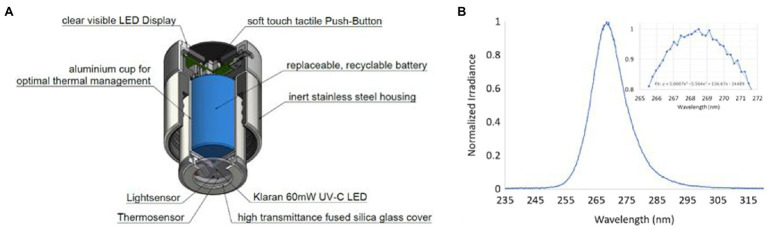
The stainless-steel bottles (Figure 1A) are fitted with a 60mW UVC LED purgaty brain (Figure 1B) with an emission angle of 130° that offered continuous disinfection for 55s and stainless steel that exhibited reflectivity of 25%. The purgaty brain is operated by means of a button on the top of the housing (Figures 1B, 2A). Pressing the button once puts the device in standby mode, while the current charge level of the integrated battery is displayed. A second press of the button for 2s activates the preset disinfection cycle for duration of 55s. The unit is equipped with a safety feature, which allows the disinfection cycle to be started only after the cap has been correctly placed on the bottle (Figure 2A). A light sensor on the Printed Circuit Board (PCB) next to the UVC LED detects ambient light and interrupts the activated UVC LED in case of unthreading the cap or system damage with light entrance during a running disinfection cycle. This mechanism is designed to protect the user from contact with UVC radiation on skin or eyes. During the cycle, the LED and the sensor are periodically monitored to prevent malfunction. At the end of the 55s cycle, the purgaty brain flashes to indicate the end of the treatment process. The cap has one Klaran UVC LED (part number KL265-50U-SM-WD) that emits radiation at 268.5nm peak wavelength (Figure 2B) as confirmed using an Ocean Optics USB4000 photospectrometer.
Figure 2.
(A) The purgaty brain (cap emitting UVC) schematic and (B) measured optical spectrum of the UVC LED emission from the cap. Inset: zoom into the peak wavelength range. A trend line has been added to the data to determine the peak wavelength value.
The thermal design is crucial for lifetime management (irreversible power degradation) as well as for ensuring the effective optical output power level (reversible thermal derating) of the UVC LED for achieving the target disinfection performance. In the purgaty brain, an aluminum cup is used to transfer the heat from the PCB to the outer stainless-steel shell, which releases the heat to the ambient air. This design ensures that after a continuous disinfection cycle of 55s, the temperature on the aluminum PCB on which the UVC LED is mounted, does not exceed 45°C at an ambient temperature of 25°C maintaining enough optical output power levels for reliable disinfection performances. A plan-parallel 2mm thick fused silica window covers the UVC LED and seals the electronics section from water and humidity. The LED package is located at 2mm distance from the window.
Bacterial Cultivation and Enumeration of Microorganisms
Three strains of E. coli The American Type Culture Collection (ATCC) 8739, P. aeruginosa ATCC 15442, and V. cholerae ATCC 25872 were obtained from ATCC (Manassas, VA, United States). Stock cultures for E. coli and V. cholerae were propagated in ATCC Medium 3: nutrient agar or nutrient broth. Pseudomonas aeruginosa was propagated in ATCC Medium 18: Trypticase Soy Agar/Broth. All strains were plated and incubated at 37°C for 24h. One isolated colony was picked using sterile inoculation loop and used to inoculate 25ml broth. Flasks with side baffles were used to enhance aeration. Cultures were incubated for 18–20h at 37°C while shaking at 180rpm. Culture storage was done at −80°C (0.7ml of culture with 0.7ml of sterile 40% Glycerol stock). To obtain working cultures, the microorganisms were obtained from −80°C, streaked onto corresponding agar, and incubated under same conditions. Storage of test cultures was done at −4°C.
For UVC disinfection experimental use, each strain was harvested by centrifugation at 4,000rpm for 10min. The pellets were washed using 1× phosphate buffered solution (PBS) three times for 10min. Between each wash, the supernatant was discarded, and the remaining pellet re-suspended by vortexing. After washing thrice, the pellet was resuspended in 1× PBS and used to spike dechlorinated test water to achieve a final UV transmittance (UVT) value of 96% (contaminated drinking water). Dechlorination of test water was verified with a Hach DPD Free Chlorine colorimetric test. All the other test water characteristics were within the NSF/ANSI 55 standard (Supplementary Tables S1–S3). The standard covers UVC disinfection systems within the range of 240 and 300nm for POU and point-of-entry (POE) applications (NSF International Standard, 2019).
Disinfection Experiments
The inactivation efficiency of the purgaty One system was evaluated by inoculating the bottles with 650 or 500ml of contaminated water with a UVT of 96%. A single preset disinfection cycle of 55s was applied, following the manufacturer’s instructions on how to use the tactile push-button of the purgaty brain. Positive control bottles had no UVC activated as there was no use of the push-button, whereas negative control bottles contained uninoculated potable test water. The samples were thoroughly mixed after disinfection, serially diluted, and processed for plating.
Decontamination of Heterotrophic Plate Count Bacteria
The HPC is an analytic method used to measure the variety of bacteria mostly found in water. HPC bacteria have no health effects at lower concentrations, instead at lower concentrations in drinking water, they are an indication of a water system well maintained. Experiments on inactivation of heterotrophic contaminants using purgaty One system were conducted on day 0 (water was not left in stationary condition), and after 3days of water being in stationary condition in bottles without prior UVC exposure. The standard HPC technique was used and incubation at 22°C and 37°C was applied using R2A agar (Gensberger et al., 2015).
HPC Isolation, Identification, and Phylogenetic Analyses
Characteristic colonies (Table 1) were picked from the R2A agar plates and re-streaked for purity, incubated at 22°C prior to being shipped for sequencing. Submitted colony samples underwent a crude Sodium hydroxide lysis and were directly used in PCR amplification. PCR amplification was performed according to Genewiz proprietary protocol. Following amplification, enzymatic cleanup was performed prior to primer extension sequencing (GENEWIZ, Inc., South Plainfield, NJ, United States) using the Applied Biosystems BigDye version 3.1. The reactions were then run on an Applied Biosystem’s 3730xl DNA Analyzer. The primer set used in this study amplifies regions V1–V9 of the 16S gene, which is roughly a 1,400 base pairs amplicon. Internal sequencing primers were utilized in order to allow for the generation of a consensus sequence with the forward and reverse traces. Consensus files were quality trimmed to remove the N’s. The generated 16S ribosomal RNA (rRNA) gene sequences were then compared with those obtained from the NCBI database, using the program BLASTN 2.2.27+.1 For phylogenetic analysis, multiple alignment of Acinetobacter 16S rRNA gene sequences using ClustalW algorithm and tree was constructed using MEGA-X (Kumar et al., 2018). The sequences from type strains used for phylogenetics were retrieved from GenBank (National Centre for Biotechnology Information),2 except for the 16S rRNA gene sequences obtained through this study. Psychrobacter cryohalolentis K5 (Accession no. NR_075055.1) was used for rooting.
Table 1.
Identification of the heterotrophic plate count (HPC) bacterial isolated from this study using 16S ribosomal RNA (rRNA) gene.
| Strain name | Colony characteristics | Accession | Organism | Closet relative | % Similarity | Phylum |
|---|---|---|---|---|---|---|
| Poff1 | White | MW167646 | Acinetobacter johnsonii | A. johnsonii ATCC 17909 | 99.18 | Proteobacteria |
| Poff2 | Yellowish | MW167647 | Methylibium petroleiphilum | M. petroleiphilum PM1 | 99.86 | Proteobacteria |
| Poff3 | White | MW167648 | Acinetobacter johnsonii | A. johnsonii ATCC 17909 | 99.39 | Proteobacteria |
| Pon1 | Pink | MW167649 | Methylorubrum populi | M. populi BJ001 | 99.65 | Proteobacteria |
| Pon2 | Yellowish | MW167650 | Sphingomonas ursincola | S. ursincola DSM 9006 | 99.72 | Proteobacteria |
| Pon3 | White | MW167651 | Brevundimonas nasdae | B. nasdae W1-2B | 99.28 | Proteobacteria |
| Pon4 | White | MW167652 | Bradyrhizobium yuanmingense | B. yuanmingense NBRC 100594 | 99.86 | Proteobacteria |
| Pon5 | Pink | MW167653 | Brevibacillus choshinensis | B. choshinensis NBRC 15518 | 99.45 | Firmicutes |
| Pon6 | White | MW167654 | Brevibacillus nitrificans | B. nitrificans DA2 | 99.39 | Firmicutes |
| Pon7 | Pink | MW167655 | Roseomonas mucosa | R. mucosa MDA5527 | 99.72 | Proteobacteria |
| Pon8 | White | MW167656 | Brevibacillus nitrificans | B. nitrificans DA2 | 99.25 | Firmicutes |
| Pon9 | Pink | MW167657 | Methylorubrum rhodesianum | M. rhodesianum NCIMB 13779 | 99.23 | Proteobacteria |
| Pon10 | White | MW167658 | Cupriavidus lacunae | C. lacunae S23 | 98.84 | Proteobacteria |
| Pon11 | White | MW167659 | Caulobacter segnis | C. segnis ATCC 21756 | 99.43 | Proteobacteria |
Poff, purgaty study isolates obtained from UVC off conditions (No UVC applied). These were the most abundant. Pon, purgaty isolates obtained from UVC on conditions (UVC applied).
Results
Disinfection Performances Against Escherichia coli ATCC 8739, Pseudomonas aeruginosa ATCC 15442, and Vibrio cholerae ATCC 25872
Table 2 shows the purgaty One system effectiveness against test strains for both bottle volume types. In all cases, both test bottles obtained a log reduction value (LRV) greater than 3 (equivalent to greater 99.9% reduction) against target microbes. Additionally, the study revealed that E. coli ATCC 29425 was more susceptible to UVC at 268.5nm wavelength compared to other test strains (Table 2; Supplementary Tables S1 and S2). In general, the 500ml bottle obtained slightly better disinfection performances compared to the 650ml bottle.
Table 2.
Purgaty One bottles disinfection performances against bacterial strains and environmental contaminants.
| Test strain | 500ml bottle’ LRV | 650ml bottle’ LRV |
|---|---|---|
| Escherichia coli ATCC 29425 | 4.36 | 4.17 |
| Pseudomonas aeruginosa ATCC 15442 | 3.56 | 3.24 |
| Vibrio cholerae ATCC 25872 | 3.34 | 3.31 |
| HPC day zero at 22°C | 4.44 | 4.40 |
| HPC day zero at 37°C | 3.96 | 3.36 |
| HPC after 3days of stationary at 22°C incubation | 3.95 | 3.53 |
| HPC after 3days of stationary at 37°C incubation | 3.62 | 3.11 |
Disinfection Performance Against HPC Bacteria
Heterotrophic plate count bacteria were present in all untreated water samples with concentrations ranging from 5.0×105 to 5.67×105CFU/ml (Supplementary Table S3). Disinfection using the purgaty One system reduced the microbial load of heterotrophic bacteria by >LRV3 (99.9% reduction, Table 2; Supplementary Table S3). Additionally, there was HPC bacterial selection by UVC exposure. The isolation of bacterial strains from UVC irradiated water is not new, as some strains obtained in this study have been isolated previously (Oguma et al., 2018).
Identification and Phylogenetic Analysis of HPC Bacteria
Eleven bacterial monocultures with distinct characteristics isolated from UVC on condition were selected for molecular identification. They were identified as: Methylorubrum populi, Sphingomonas ursincola, Brevundimonas nasdae, Bradyrhizobium yuanmingense, Brevibacillus choshinensis, Brevibacillus nitrificans, Roseomonas mucosa, Methylorubrum rhodesianum, Cupriavidus lacunae, and Caulobacter segnis (Table 1). Two most abundant isolates were however obtained under non-UVC treated conditions (UVC not applied). These were identified as Acinetobacter johnsonii and Methylibium petroleiphilum. These two strains were UVC sensitive, thus accounting for the high HPC bacteria decontamination (Table 2; Supplementary Table S3). All isolates belonged to either Phylum Proteobacteria or Firmicutes (Table 1).
Taxonomic classification of HPC isolates using 16S rRNA gene identified A. johnsonii and M. petroleiphilum to be most dominant in untreated water. These two representatives of Phylum Proteobacteria were sensitive to UVC irradiation and thus not selected by UVC (+UVC condition). Strain Poff1, identified as A. johnsonii, which was isolated under UVC off condition (−UVC) belongs to the same Genus as Acinetobacter baumannii, a multidrug resistant nosocomial pathogen of global concern (Antunes et al., 2014). The study sought to confirm if the two species did not cluster together to rule out any concerns regarding the presence of A. johnsonii in drinking water. Phylogenetic analysis revealed that they do not cluster together (Supplementary Figure S1). Further, based on literature from previous studies, A. johnsonii has been confirmed to rarely cause human infections and has been found to be sensitive to virtually all antibiotics (Montaña et al., 2016).
Discussion
Data from this study revealed >LRV3 in test bacteria, further supporting accumulating evidence for high disinfection activities of portable UVC devices (Timmermann et al., 2015). The study demonstrated high disinfection performance against E. coli ATCC 29425 (99.99% reduction). This can be attributed to relatively low %GC content of E. coli (50.68%; Engelbrecht et al., 2017) and its peak UVC sensitivity (Green et al., 2018). Although the Vibrio Genus has low %GC content (~47%), the presence of more elaborate DNA repair/protection processes make them less susceptible against UVC irradiation (Krin et al., 2018), thus obtaining 99.9% disinfection under similar test conditions (Table 2). This is in comparison with P. aeruginosa ATCC 15442, which has higher %GC content 66.17%.3
Even after 3days of water in stationary condition, the use of purgaty One system effectively decontaminated that HPC bacteria, obtaining <500CFU/ml, as recommended (Kohn et al., 2003). All the UVC selected bacteria are of rare to no public health concern. For instance, B. nitrificans is a heterotrophic nitrifying bacterium (Takebe et al., 2012) whose genera, Brevibacillus is one of the most widespread and is found in diverse environmental habitats, including drinking water (Panda et al., 2014). The study revealed that phylum Proteobacteria as most frequent of the identified isolates (Table 2). Although drinking water has complex microbiota, previous characterization studies have confirmed that phylum Proteobacteria is the most frequent in drinking water (Vaz-Moreira et al., 2017).
Results from this study indicate that UVC exposure of static water in devices such as the purgaty One systems (cap+bottle) can reduce elevated initial bacterial loads. These devices could be useful in environments where people are vulnerable to pathogens. Applications include but are not limited to those related to travel medicine, healthcare facilities where patients are vulnerable to opportunistic pathogens, hiking, remote military installations, and regions having water potability challenges.
It must be acknowledged that the current study utilized potable water that was deliberately contaminated with bacteria, and that this water, at 96% UVT, will exhibit lower absorption compared to water of low quality from natural sources like ponds which have abundant phytoplankton population (McKnight et al., 1994), iron rich lakes (Baffico, 2013), and clear alpine lakes, which ordinarily have zooplankton in the upper water layers (Tartarotti et al., 2017), and nutrient rich river estuaries or natural water with high concentrations of organic compounds (Lei et al., 2019). These waters may have UVT which is as low as 50% at 265nm within 1cm. In general, due to the different physical-chemical compositions, these natural waters will shield microorganisms from UVC, leading to lower disinfection efficacies. Additionally, in some cases, the presence of chromophoric dissolved organic matter (CDOM), for instance in arctic lakes, have spectral light attenuation and UVC absorption properties (Markager and Vincent, 2000), which may impact UVC performance. Thus, the results reported in the current study might be different if different water is used by travelers.
Conclusion
This study investigated the efficacy of the purgaty One system (bottle+cap) on bacterial inactivation in a stationary water disinfection setup. The two types of stainless-steel water bottles (650 and 500ml) achieved more than 99.99% inactivation efficacy against E. coli ATCC 29425 after a single treatment cycle of 55s preset by the purgaty brain (cap), including a Klaran UVC LED with peak wavelength of 268.5nm. For P. aeruginosa ATCC 15442 and V. cholerae ATCC 25872, an inactivation efficacy of more than 99.9% was achieved. The bottles were also able to inactivate heterotrophic contaminants with more than 99.9% reduction, even after 3days of water in stationary condition in the bottles without prior exposure to UVC. These results demonstrate the ability of consistent disinfection performances of a mobile, simple-to-use and safe consumer water bottle appliance with a UVC disinfection feature including a single UVC LED only, which has extended application potential during emergency preparedness, such as for flooding situations, outdoor activities like mountain climbing, military use especially during operations in remote areas as well as consumer home use when in doubt of water potability. Lastly, these results offer fundamental evidence on how travel medicine can benefit from the use of personal UVC devices to ensure eradication of enteric pathogens.
Data Availability Statement
The 16S rRNA gene sequences obtained from this study are available through GenBank (https://www.ncbi.nlm.nih.gov/genbank/) under the accession numbers MW167646–MW167659. The Spectral data is available via https://doi.org/10.6084/m9.figshare.13144538.v1.
Author Contributions
RM was a major contributor in study design, coordinated data collection, analysis, and interpretation, and was a substantial contributor in writing the manuscript. SB contributed to study design, data analysis, and interpretation, and was a substantial contributor in writing the manuscript. CB and TG contributed to the understanding of Purgaty One System and ensured provision of study materials. RR contributed to data collection and analysis. All authors contributed to the article and approved the submitted version.
Conflict of Interest
RM, SB, and RR work for Crystal IS Inc., an Asahi Kasei Company that manufactures UVC-LEDs. CB and TG work for purgaty, the innovators of stainless-steel drinking water bottle with cap, which inactivates microorganisms. Purgaty employees did not have any role in the microbial disinfection study design, data collection and analysis, and writing of manuscript, but contributed in the understanding of purgaty One system.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
Authors wish to thank Michelle Lottridge, Chris Scully, Amy Miller, Jeremy Abel, Stephan Gadhof, Paul Bilton, Kevin Kahn, and James Peterson for providing help during research and proofreading the manuscript.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.719578/full#supplementary-material
References
- Antunes L., Visca P., Towner K. J. (2014). Acinetobacter baumannii: evolution of a global pathogen. Pathog. Dis. 71, 292–301. 10.1111/2049-632X.12125, PMID: [DOI] [PubMed] [Google Scholar]
- Ashbolt N. J. (2004). Microbial contamination of drinking water and disease outcomes in developing regions. Toxicology 198, 229–238. 10.1016/j.tox.2004.01.030, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baffico G. D. (2013). Optical properties and light penetration in a deep, naturally acidic, iron rich lake: Lago Caviahue (Patagonia, Argentina). Limnologica 43, 475–481. 10.1016/j.limno.2013.03.003 [DOI] [Google Scholar]
- Cabral J. P. (2010). Water microbiology. Bacterial pathogens and water. Int. J. Environ. Res. Public Health 7, 3657–3703. 10.3390/ijerph7103657, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai T., Vrahas M. S., Murray C. K., Hamblin M. R. (2012). Ultraviolet C irradiation: an alternative antimicrobial approach to localized infections? Expert Rev. Anti Infect. Ther. 10, 185–195. 10.1586/eri.11.166, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelbrecht K. C., Putonti C., Koenig D. W., Wolfe A. J. (2017). Draft genome sequence of Escherichia coli K-12 (ATCC 29425). Genome Announc. 5:e00574-17. 10.1128/genomeA.00574-17, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericsson C. D., Steffen R., Backer H. (2002). Water disinfection for international and wilderness travelers. Clin. Infect. Dis. 34, 355–364. 10.1086/324747, PMID: [DOI] [PubMed] [Google Scholar]
- Fagunwa O. E., Olanbiwoninu A. A. (2020). Accelerating the sustainable development goals through microbiology: some efforts and opportunities. Access Microbiol. 23:acmi000112. 10.1099/acmi.0.000112, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gensberger E. T., Gössl E.-M., Antonielli L., Sessitsch A., Kostić T. (2015). Effect of different heterotrophic plate count methods on the estimation of the composition of the culturable microbial community. PeerJ 3:e862. 10.7717/peerj.862, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gora S. L., Rauch K. D., Ontiveros C. C., Stoddart A. K., Gagnon G. A. (2019). Inactivation of biofilm-bound Pseudomonas aeruginosa bacteria using UVC light emitting diodes (UVC LEDs). Water Res. 151, 193–202. 10.1016/j.watres.2018.12.021, PMID: [DOI] [PubMed] [Google Scholar]
- Green A., Popović V., Pierscianowski J., Biancaniello M., Warriner K., Koutchma T. (2018). Inactivation of Escherichia coli, Listeria and Salmonella by single and multiple wavelength ultraviolet-light emitting diodes. Innovative Food Sci. Emerg. Technol. 47, 353–361. 10.1016/j.ifset.2018.03.019 [DOI] [Google Scholar]
- Heidelberg J. F., Eisen J. A., Nelson W. C., Clayton R. A., Gwinn M. L., Dodson R. J., et al. (2000). DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406, 477–483. 10.1038/35020000, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamori H., Weber D. J., Rutala W. A. (2016). Healthcare outbreaks associated with a water reservoir and infection prevention strategies. Clin. Infect. Dis. 62, 1423–1435. 10.1093/cid/ciw122, PMID: [DOI] [PubMed] [Google Scholar]
- Kohn W. G., Collins A. S., Cleveland J. L., Harte J. A., Eklund K. J., Malvitz D. M. (2003). Guidelines for infection control in dental health-care settings-2003. MMWR Recomm. Rep. 52, 1–61. PMID: [PubMed] [Google Scholar]
- Krin E., Pierlé S. A., Sismeiro O., Jagla B., Dillies M.-A., Varet H., et al. (2018). Expansion of the SOS regulon of Vibrio cholerae through extensive transcriptome analysis and experimental validation. BMC Genomics 19:373. 10.1186/s12864-018-4716-8, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. 10.1093/molbev/msy096, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei X., Pan J., Devlin A. T. (2019). Characteristics of absorption spectra of chromophoric dissolved organic matter in the pearl river estuary in spring. Remote Sens. 111533. 10.3390/rs11131533 [DOI] [Google Scholar]
- Lizzadro J., Mazzotta M., Girolamini L., Dormi A., Pellati T., Cristino S. (2019). Comparison between two types of dental unit waterlines: how evaluation of microbiological contamination can support risk containment. Int. J. Environ. Res. Public Health 16:328. 10.3390/ijerph16030328, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui G. Y., Roser D., Corkish R., Ashbolt N. J., Stuetz R. (2016). Point-of-use water disinfection using ultraviolet and visible light-emitting diodes. Sci. Total Environ. 553, 626–635. 10.1016/j.scitotenv.2016.02.039, PMID: [DOI] [PubMed] [Google Scholar]
- Markager S., Vincent W. F. (2000). Spectral light attenuation and the absorption of UV and blue light in natural waters. Limnol. Oceanogr. 45, 642–650. 10.4319/lo.2000.45.3.0642 [DOI] [Google Scholar]
- McKnight D. M., Andrews E. D., Spaulding S. A., Aiken G. R. (1994). Aquatic fulvic acids in algal-rich antarctic ponds. Limnol. Oceanogr. 39, 1972–1979. 10.4319/lo.1994.39.8.1972 [DOI] [Google Scholar]
- Medema G., Payment P., Dufour A., Robertson W., Waite M., Hunter P., et al. (2003). “Safe drinking water: an ongoing challenge,” in Assessing Microbial Safety of Drinking Water. Improving Approaches and Method. ed. Fewtrell L. (London: WHO & OECD, IWA Publishing; ). [Google Scholar]
- Montaña S., Schramm S. T., Traglia G. M., Chiem K., Parmeciano Di Noto G., Almuzara M., et al. (2016). The genetic analysis of an Acinetobacter johnsonii clinical strain evidenced the presence of horizontal genetic transfer. PLoS One 11:e0161528. 10.1371/journal.pone.0161528, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton G. J., Williams M., Falkinham J. O., 3rd, Honda J. R. (2020). Physical measures to reduce exposure to tap water-associated nontuberculous mycobacteria. Front. Public Health 8:190. 10.3389/fpubh.2020.00190, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- NSF International Standard (2019). NSF International. NSF Protocol 55-2019: Ultraviolet Microbiological Water Treatment Systems; NSF International: Ann Arbor, MI, USA, 2019.
- Oguma K., Kanazawa K., Kasuga I., Takizawa S. (2018). Effects of UV irradiation by light emitting diodes on heterotrophic bacteria in tap water. Photochem. Photobiol. 94, 570–576. 10.1111/php.12891, PMID: [DOI] [PubMed] [Google Scholar]
- Panda A. K., Bisht S. S., DeMondal S., Kumar N. S., Gurusubramanian G., Panigrahi A. K. (2014). Brevibacillus as a biological tool: a short review. Antonie Van Leeuwenhoek 105, 623–639. 10.1007/s10482-013-0099-7, PMID: [DOI] [PubMed] [Google Scholar]
- Schlosser O., Robert C., Bourderioux C., Rey M., de Roubin M. R. (2001). Bacterial removal from inexpensive portable water treatment systems for travelers. J. Travel Med. 8, 12–18. 10.2310/7060.2001.5143, PMID: [DOI] [PubMed] [Google Scholar]
- Takebe F., Hirota K., Nodasaka Y., Yumoto I. (2012). Brevibacillus nitrificans sp. nov., a nitrifying bacterium isolated from a microbiological agent for enhancing microbial digestion in sewage treatment tanks. Int. J. Syst. Evol. Microbiol. 62, 2121–2126. 10.1099/ijs.0.032342-0, PMID: [DOI] [PubMed] [Google Scholar]
- Tartarotti B., Trattner F., Remias D., Saul N., Steinberg C. E. W., Sommaruga R. (2017). Distribution and UV protection strategies of zooplankton in clear and glacier-fed alpine lakes. Sci. Rep. 7:4487. 10.1038/s41598-017-04836-w, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmermann L. F., Ritter K., Hillebrandt D., Küpper T. (2015). Drinking water treatment with ultraviolet light for travelers – evaluation of a mobile lightweight system. Travel Med. Infect. Dis. 13, 466–474. 10.1016/j.tmaid.2015.10.005, PMID: [DOI] [PubMed] [Google Scholar]
- Umar M., Roddick F., Fan L. (2019). Moving from the traditional paradigm of pathogen inactivation to controlling antibiotic resistance in water – role of ultraviolet irradiation. Sci. Total Environ. 662, 923–939. 10.1016/j.scitotenv.2019.01.289, PMID: [DOI] [PubMed] [Google Scholar]
- Vaz-Moreira I., Nunes O. C., Manaia C. M. (2017). Ubiquitous and persistent Proteobacteria and other gram-negative bacteria in drinking water. Sci. Total Environ. 586, 1141–1149. 10.1016/j.scitotenv.2017.02.104, PMID: [DOI] [PubMed] [Google Scholar]
- Vezzulli L., Grande C., Reid P. C., Hélaouët P., Edwards M., Höfle M. G., et al. (2016). Climate influence on vibrio and associated human diseases during the past half-century in the coastal North Atlantic. Proc. Natl. Acad. Sci. U. S. A. 113, E5062–E5071. 10.1073/pnas.1609157113, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (2000). Global water supply and sanitation assessment 2000 report. World Health Organization.
- World Health Organization (2014). Ultraviolet radiation and health. Available at: https://www.who.int/uv/uv_and_health/en/ (Accessed July 28, 2021).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 16S rRNA gene sequences obtained from this study are available through GenBank (https://www.ncbi.nlm.nih.gov/genbank/) under the accession numbers MW167646–MW167659. The Spectral data is available via https://doi.org/10.6084/m9.figshare.13144538.v1.