Abstract
Metastasis accounts for more than 90% of cancer related mortality, thus the most pressing need in the field of oncology today is the ability to accurately predict future onset of metastatic disease, ideally at the time of initial diagnosis. As opposed to current practice, what would be desirable is that prognostic, biomarker-based detection of metastatic propensity and heightened risk of cancer recurrence be performed long before overt metastasis has set in. Without such timely information it will be impossible to formulate a rational therapeutic treatment plan to favorably alter the trajectory of disease progression. In order to help inform rational selection of targeted therapeutics, any recurrence/metastasis risk prediction strategy must occur with the paired identification of novel prognostic biomarkers and their underlying molecular regulatory mechanisms that help drive cancer recurrence/metastasis (i.e. recurrence biomarkers). Traditional clinical factors alone (such as TNM staging criteria) are no longer adequately prognostic for this purpose in the current molecular era. FOXC1 is a pivotal transcription factor that has been functionally implicated to drive cancer metastasis and has been demonstrated to be an independent predictor of heightened metastatic risk, at the time of initial diagnosis. In this review, we present our viewpoints on the master regulatory role that FOXC1 plays in mediating cancer stem cell traits that include cellular plasticity, partial EMT, treatment resistance, cancer invasion and cancer migration during cancer progression and metastasis. We also highlight potential therapeutic strategies to target cancers that are, or have evolved to become, “transcriptionally addicted” to FOXC1. The potential role of FOXC1 expression status in predicting the efficacy of these identified therapeutic approaches merits evaluation in clinical trials.
Keywords: Forkhead Box C1, transcriptional addiction, cancer stem cell, plasticity, epithelial-to-mesenchymal transition, metastasis, targeted therapy
Introduction
Cancer screening strategies have proven to be successful in decreasing cancer-specific mortality in a variety of cancers (1–4). However, the clinical cure rates of patients who are diagnosed with cancer despite such screening measures still largely remain far from ideal (5). Our ability to potentially monitor cancer recurrence and metastasis events in real-time has now become possible with the development of various liquid biopsy modalities (circulating tumor cells, extracellular vesicles, cell-free DNA, etc.) (6). However, if we are to favorably impact current rates of cancer disease progression and exert meaningful decreases in cancer morbidity and mortality, we need to improve upon our ability to predict which newly-diagnosed patients are at the highest risk of suffering recurrence and metastasis events in the future. Biopsy-tissue derived molecular markers that predict heightened future metastasis risk still present the most pragmatic solution to this problem. This is because detection of such biomarkers utilizing standard immunohistochemistry (IHC) has a much higher likelihood of widespread global adoption owing to superior cost effectiveness and greater ease of integration into existing workflow of diagnostic pathology laboratories (7, 8).
Traditional clinical factors alone (such as TNM staging criteria) are no longer prognostically adequate for this purpose and we are in need of new prognostic biomarkers that are superior in their ability to predict cancer recurrence and metastasis risk (9). Absent such markers, our ability to pinpoint and identify which patients stand to derive the greatest clinical benefit of new therapeutic approaches being tested in clinical trials will be lost. As a result of not being able to “enrich” our clinical trial population with those patients who are most likely to derive survival benefit, we will ostensibly dilute the measured therapeutic efficacy in such trials, and may erroneously label a tested approach as being ineffective (10). Such biomarkers, once identified, may or may not play a functional or mechanistic role in driving the underlying aggressive biology contributing to the observed adverse outcomes. For this reason, recurrence/metastasis risk prediction needs to be performed in conjunction with the identification of the pivotal underlying molecular drivers responsible for increasing the probability of suffering a recurrence/metastasis event. This would allow formulation of a rational therapeutic strategy, based on targeting the identified underlying molecular driver mechanism, to ultimately derive clinical benefit.
In recent years, several excellent review articles on the role of the Forkhead box C1 (FOXC1) Transcription Factor (TF) in cancer have been published, highlighting the increasing recognition of its importance as a clinically useful biomarker and potential therapeutic target (11–15). The focus of this review is to summarize and draw inferences from the body of literature that implicates FOXC1 as a transcriptional driver of cancer progression and metastasis (Figure 1). Herein we discuss the current state of evidence that supports the argument that FOXC1 plays this role by virtue of being essential for the emergence, maintenance and proliferation of cancer stem cells (CSCs). We present evidence of how FOXC1 plays a functionally important role in mediating a wide variety of cancer traits all of which are, essentially, cancer stem cell traits. Such traits include cell proliferation, cell plasticity, partial EMT, cell migration, cell invasion, chemoresistance and radio-resistance. In this review we present our viewpoints on the pivotal role that FOXC1 plays in this regard. FOXC1 dependencies develop as a consequence of dysregulated programs in cancer cells and affect clinical progression, therapeutic responsiveness and outcome often emerging to a level of dependence referred to as “transcriptional addiction.” Collectively, the elucidated mechanisms by which FOXC1 modulates aggressive cancer cell traits support our contention that FOXC1 is predominantly a transcriptional driver of cellular plasticity and a cancer stem cell phenotype. In this review, we also highlight several potential targeted therapeutic strategies, based on utilizing already available FDA-approved oral anticancer drugs, that may help achieve improved clinical outcomes for patients diagnosed with FOXC1-overexpressing pro-metastatic cancers. The therapeutic strategies described herein are potentially practice-changing in the field of oncology, and merit being tested in biomarker-driven, adaptive clinical trials.
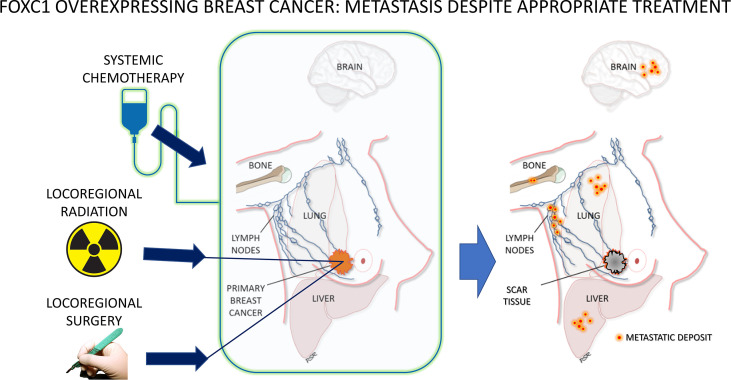
Figure 1.
Clinical hallmarks of FOXC1-overexpressing breast cancer – metastatic recurrence despite adequate surgical resection, lymph node assessment and removal, radiation therapy and chemotherapy. For a biomarker to be associated with such aggressive behavior, resistant to common therapeutic treatment approaches, argues strongly in favor of this being a manifestation of the cancer stem cell phenotype.
FOXC1: A Prognostic Biomarker of Cancer Progression and Metastasis
The earliest reports on the biologic role and function of the FOXC1 TF implicated it in abnormal pathologic conditions like glaucoma (16–23), congenital hydrocephalus (24), congenital renal defects (25, 26), congenital heart defects (21, 27–29) and Axenfeld-Rieger Syndrome (21, 30, 31), a congenital disorder characterized by glaucoma and congenital heart defects. Contemporaneously, several reports detailing the role of FOXC1 in normal physiology were also reported with regard to development of the cornea and the anterior chamber of the eye (32), renal development (33, 34) and cardiovascular development (27, 35). Also elucidated was its role in coordinating the embryonic process of primitive mesoderm cell fate (36) and migration of embryonic tissues (37).
The functional relevance and prognostic significance of FOXC1 in cancer was first reported in breast cancer in 2010 (38). Since that time, we and others have established the pivotal role that FOXC1 plays in coordinating the aggressive biology underlying cancer progression and metastasis in multiple cancers (Tables 1, 2). Studies supporting the role of FOXC1 as a powerful prognostic biomarker are summarized in Table 1 and includes both “solid” as well as “liquid” cancers like acute myelogenous leukemia (AML). Indeed, an excellent and comprehensive systematic review and meta-analysis on the prognostic role of FOXC1 in cancer concluded that FOXC1 expression in cancer is indicative of poor survival outcome (126). In support of FOXC1 playing a role in cancer progression, another meta-analysis reported that FOXC1 is 23.8% more likely to be expressed in late-stage cancers as opposed to early-stage cancers (127). Below, we highlight those studies that demonstrate the role of FOXC1 in predicting an aggressive clinical course, specifically highlighting its role as a predictor of metastatic recurrence.
Table 1.
Clinical evidence supporting the role of FOXC1 in cancer progression and metastasis.
| Cancer Type | Sample Size | FOXC1 in Progression and Metastasis: Clinical Evidence | Outcome Measured | Univariate Analysis | Multivariate Analysis | Reference PMID Number | ||
|---|---|---|---|---|---|---|---|---|
| Hazard ratio (CI) | p-value | Hazard ratio (CI) | p-value | |||||
| AML | 458 | OS | – | – | 1.784(1.29-2.46) | <0.001 | (39) 26373280 | |
| AML | 452 | OS | 1.592(1.263-2.007) | 0.0001 | 1.755(1.355-2.273) | <0.0001 | (40) | |
| 313 | + | OS | 1.539(1.208-1.961) | 0.0002 | 1.678(1.280-2.201) | 0.0001 | ||
| Breast | 295 | OS | – | 0.0001 | 1.25(1.02–1.52) | 0.02 | (38) 20406990 | |
| 286 | + | DMFS | – | <0.0001 | – | – | ||
| 232 | OS | – | 0.0476 | – | – | |||
| 122 | OS | – | 0.0098 | – | – | |||
| 159 | OS | – | 0.0047 | – | – | |||
| Breast | 724 | OS | 3.364(1.758–6.438) | 0.0002 | 3.389(1.928-7.645) | 0.0001 | (41) 21424368 | |
| Breast | 1975 | + | DSS | 1.71(1.31 to 2.23) | <0.001 | 1.55(1.17 to 2.06) | 0.003 | (42) 26041837 |
| Breast | 1986 | + | DSS | 1.973(1.802-2.961) | <0.0001 | (43) 26565916 | ||
| Breast | 120 | + | DFS | 2.62(1.05-6.50) | 0.038 | 2.58(1.04-6.42) | 0.041 | (44) 28493031 |
| Cervical | 219 | + | OS RFS |
– – |
– – |
2.928(0.508-6.585) 2.776(0.207-7.538) |
0.021 0.035 |
(45) 28386355 |
| Cervical | 76 | OS | 0.0094 | (46) 29328284 | ||||
| Colon | 363 |
+ |
OS DMFS |
0.432(0.325−0.573) 0.422(0.319-0.558) |
<0.001 <0.001 |
0.668(0.492−0.907) 0.617(0.457–0.834) |
0.010 0.002 |
(47) 29884889 |
| Colon | 361 185 |
+ |
OS OS DFS |
1.190(1.023-1.389) 3.371(1.745-6.513) 2.557(1.453-4.497) |
0.025 <0.001 0.001 |
(48) 30171256 | ||
| Colon | 361 86 134 |
OS OS OS |
1.20(1.03-1.40) | 0.002 0.004 0.002 |
(49) 31650548 | |||
| Esophageal (SCC) | 82 | OS | – | 0.014 | – | – | (50) 25031703 | |
| Esophageal (SCC) | 147 |
+ |
OS DFS |
– – |
0.023 0.037 |
– – |
– – |
(51) 28861321 |
| Gastric | 120 | OS | 0.273(0.144–0.521) | <0.001 | 0.370(0.184–0.745) | 0.005 | (52) 24329718 | |
| Gastric | 422 | OS | 1.58(1.15-2.15) | 0.0038 | – | – | (53) 33987183 | |
| Hepatic | 406 |
+ |
OS RFS |
0.587(0.453–0.760) 0.566(0.434–0.738) |
<0.0001 <0.0001 |
0.641(0.491–0.837) 0.649(0.495–0.852) |
0.001 0.002 |
(54) 22911555 |
| Lung (NSCLC) | 125 | OS | 1.324(0.657–2.175) | <0.001 | 1.328(0.625–2.021) | <0.001 | (55) 23264086 | |
| Lung (LUAD) Lung (LUSC) |
500 494 |
OS OS |
– – |
0.0484 0.0363 |
– – |
– – |
(56) 30548656 | |
| Lung (NSCLC) | 105 | OS | 2.237(1.220-4.098) | 0.009 | 1.988(1.022-3.860) | 0.043 | (57) 31597217 | |
| Melanoma | Stg III 139 Stg IV 169 |
+ + |
DMFS DMFS |
– – |
<0.05 <0.05 |
– – |
(58) 27533251 | |
| Pancreatic | 85 | OS | 1.432(0.567-2.045) | <0.001 | 1.328(0.586-2.178) | <0.001 | (59) 23242609 | |
| Tongue | 92 | OS | – | 0.006 | – | – | (60) 25130698 | |
“-” indicates that information was not mentioned in the text or supplementary material of the specific publication, AML, acute myelogenous leukemia; CI, confidence interval; DFS, disease free survival; DMFS, distance metastasis free survival; DSS, disease specific survival; FOXC1, Forkhead box C1; LUAD, lung adenocarcinoma; LUS, lung squamous cell; NSCL, non-small cell lung cancer; OS, overall survival; RFS, recurrence free survival.
Table 2.
Functional and mechanistic evidence supporting role of FOXC1 in plasticity, EMT, chemoresistance, cancer progression and metastasis.
| Cancer Type | Transcriptional Addiction | Cell Proliferation | Cellular Plasticity/Stemness | EMT/pEMT/MET | Circulating Tumor Cells (CTCs) | Endocrine Resistance | Chemoresistance | Radioresistance | Cancer Progression/Metastasis | Molecular Pathway | Summary of Findings | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Breast |

|

|
FOXC1 regulates CD44+ normal mammary progenitors | (61) | ||||||||
| Breast |

|

|

|

|
1st Report of FOXC1 as a marker of aggressive traits | (38) | ||||||
| Breast |

|
FOXC1 is associated with chemoresistance | (62) | |||||||||
| Breast |

|
FOXC1 is associated with radioresistance | (63) | |||||||||
| Breast |

|

|

|
ERα | BRCA1 and GATA3 transcriptionally represses FOXC1 | (64) | ||||||
| Breast |

|

|
TGFβ | Elevated FOXC1 in breast CTCs | (65) | |||||||
| Breast |

|
Transient vs Stable overexpression of FOXC1 | (66) | |||||||||
| Breast |

|

|
NFĸB | NFĸB inhibitor blocks FOXC1 mediated M/I | (67) | |||||||
| Breast |

|

|
Raf/MEK/MAPK | FOXC1 downregulation associated with MET | (68) | |||||||
| Breast |

|

|

|
TGFβ | FOXC1 in breast CTCs associated with progression | (69) | ||||||
| Breast |

|
FOXC1 is enriched in mammary luminal progenitors | (70) | |||||||||
| Breast |

|

|
Ras/ERK, PI3K/AKT | EGFR inhibitor blocks FOXC1 mediated M/I | (71) | |||||||
| Breast |

|
FOXC1 is a marker of neoadjuvant chemoresistance | (72) | |||||||||
| Breast |

|

|

|
FOXCUT | FOXC1-FOXCUT form mRNA-lncRNA pair | (73) | ||||||
| Breast |

|

|
C-Wnt, NFĸB | NFĸB blocks stem cell escape following Wnt inhibition | (74) | |||||||
| Breast |

|

|

|

|
FOXC1/FOXA1 transcriptional balance in breast cancer | (74) | ||||||
| Breast |

|

|
NC-Hedgehog | FOXC1 induces Gli2 activation and NC Hedgehog | (43) | |||||||
| Breast |

|
CDK7 | CDK7-dependent SE-mediated FOXC1 addiction | (75) | ||||||||
| Breast |

|
ERα | FOXC1 transcriptionally represses ERα | (76) | ||||||||
| Breast |

|
FOXC1 overexpression induces increased lung metastasis | (77) | |||||||||
| Breast |

|
ERα | FOXC1 transcriptionally represses ERα | (78) | ||||||||
| Breast |

|
FOXC1 is a marker of adjuvant Anthracycline resistance | (44) | |||||||||
| Breast | EGFR, NFĸB | NFĸB mediates EGF-induced FOXC1 expression | (79) | |||||||||
| Breast |

|

|
TGFβ, FGFR1 | TGFβ upregulates FOXC1, triggers FGFR1 isoform switch | (80) | |||||||
| Breast |

|
FOXC1 inhibits ELF5, lobuloalveolar development | (81) | |||||||||
| Breast |

|
NC-Wnt, NFĸB | FOXC1 mediates Wnt5A-NfĸB-MMP7 induced invasion | (82) | ||||||||
| Breast |

|
Alternative splicing switch in FLNB induces FOXC1, EMT | (83) | |||||||||
| Breast |

|

|
FAK-AKT-mTOR | ST8SIA1 regulates FOXC1 | (84) | |||||||
| Breast |

|

|
CXCR4 | CXCR4-inhibitor blocks FOXC1 mediated M/I, metastasis | (85) | |||||||
| Breast |

|
EZH2 | EZH2 epigenetically represses FOXC1 | (86) | ||||||||
| Breast | FOXC1 upregulates LINC01123, ↓miR663a | (49) | ||||||||||
| Breast |

|

|

|

|
CDK7 | FOXC1 contributes to CDK7-inhibitor sensitivity | (87) | |||||
| Breast |

|

|

|
FOXC1 is most significant EMAT/metastasis activator | (88) | |||||||
| Breast |

|

|

|
CDK7 | FOXC1 is most significant invasion/metastasis activator | (89) | ||||||
| Prostate |

|

|
FOXC1 expression associated with AIPC progression | (90) | ||||||||
| Prostate |

|
EGFR | EGFR upregulates FOXC1 | (91) | ||||||||
| Prostate |

|

|

|
MiR-138-5P inhibits FOXC1-mediated M/I | (92) | |||||||
| Prostate |

|

|

|
MiR-138-5P inhibits FOXC1-mediated M/I | (49) | |||||||
| Lung |

|

|

|
FOXC1 drives proliferation, EMT, invasion | (93) | |||||||
| Lung |

|

|

|
HIF1α | HIF1α transcriptionally upregulates FOXC1 | (94) | ||||||
| Lung |

|

|
β-catenin, C-Wnt | FOXC1 transcriptionally upregulates β-catenin | (56) | |||||||
| Lung |

|

|
lncRNA CCAT2 upregulates FOXC1 | (95) | ||||||||
| Lung |

|

|
LOX | FOXC1 transcriptionally upregulates LOX, ↑metastasis | (57) | |||||||
| Lung |

|

|
HIF1α | FOXC1 transcriptionally upregulates LINC00301, ↑HIF1α | (96) | |||||||
| Cervical |

|

|
PI3K/AKT | FOXC1 mediates PI3K/AKT induced EMT, M/I | (45) | |||||||
| Cervical |

|

|
miR-374c-5p inhibits FOXC1-mediated M/I | (45) | ||||||||
| Cervical |

|

|
FOXC1 is associated with radioresistance | (29) | ||||||||
| Cervical |

|
PI3K/AKT | FOXC1 mediates PI3K/AKT induced M/I | (46) | ||||||||
| Endometrial |

|

|
miR-204 inhibits FOXC1-mediated proliferation, M/I | (97) | ||||||||
| Endometrial |

|

|
miR-495 inhibits FOXC1-mediated proliferation, M/I | (98) | ||||||||
| Esophageal |

|
FOXCUT | FOXC1-FOXCUT form mRNA-lncRNA pair | (50) | ||||||||
| Esophageal |

|

|

|
FOXC1 acts as transcriptional coactivator of, PBX1, ↑ZEB2 | (51) | |||||||
| Gastric |

|
β-catenin, C-Wnt | FOXC1 transcriptionally downregulates DKK1, activating Wnt | (53) | ||||||||
| Gastric |

|

|
Wnt | FOXC1 transcriptionally upregulates GPX8, activating Wnt | (99) | |||||||
| Gastric |

|

|

|
LINC00242 inhibits miR-141, upregulates FOXC1 | (100) | |||||||
| Gastric |

|

|

|
MCM3AP-AS1 inhibits miR-148, upregulates FOXC1 | (101) | |||||||
| Gastric | EGFR | EGFR-FOXC1 upregulates histone H3C14 | (102) | |||||||||
| Pancreatic |

|

|

|
miR-138-5p inhibits FOXC1,↓ proliferation,↓ tumor growth | (103) | |||||||
| Pancreatic |

|

|

|

|
PI3K/AKT/mTOR | FOXC1 and IGF1R positively regulate each other | (104) | |||||
| Hepatic |

|

|

|

|
VEGF | FOXC1 | (105) | |||||
| Hepatic |

|

|

|
NEDD9 | FOXC1 transcriptionally upregulates NEDD9, ↑metastasis | (54 | ||||||
| Hepatic |

|
IL8,PI3K/AKT,CXCR1 | IL8-dependent PI3K/AKT/HIF1α upregulates FOXC1 | (106) | ||||||||
| Hepatic |

|
HOTAIR | FOXC1 upregulates lncRNA HOTAIR, ↓miRNA-1 | (107) | ||||||||
| Colon |

|
FGFR4 | FOXC1 transcriptionally upregulates ITGA7, FGFR4 | (47) | ||||||||
| Colon |

|

|
FOXC1 transcriptionally upregulates FBP1, ↑Warburg Effect | (48) | ||||||||
| Colon |

|
FOXC1 transcriptionally upregulates miR-31-5p, ↓LATS2 | (108) | |||||||||
| Colon |

|

|
p38MAPK maintain FOXC1 protein stability, ↑metastasis | (49) | ||||||||
| Colon |

|
> | FOXC1 induces resistance to oxaliplatin, irinotecan | (109) | ||||||||
| OSCC |

|

|
FOXCUT | FOXC1-FOXCUT form mRNA-lncRNA pair | (110) | |||||||
| OSCC |

|

|

|
FOXC1 transcriptionally upregulates CCNB1/D1, MMP2/9 | (47) | |||||||
| OSCC |

|

|

|
MCM3AP-AS1 inhibits miR-138, upregulates FOXC1 | (111) | |||||||
| Tongue |

|

|

|
TGFβ | miR-639 inhibits FOXC1-mediated EMT, M/I | (60) | ||||||
| SACC |

|

|

|

|
miR-582-5p inhibits FOXC1, ↓M/I, ↓metastasis | (78) | ||||||
| LSCC |

|
miR-204-5P inhibits FOXC1-mediated M/I | (112) | |||||||||
| NP |

|
FOXC1 associated with low E-cadherin expression | (113) | |||||||||
| NP |

|

|

|
FOXC1 siRNA inhibited EMT, M/I | (114) | |||||||
| NP |

|

|

|
miR-4792 inhibits FOXC1-mediated EMT, M/I | (115) | |||||||
| NP |

|

|
FOXCUT | FOXCUT siRNA inhibited FOXC1-mediated MMP7/9 | (44) | |||||||
| OS |

|

|

|
miR-133b inhibits FOXC1-mediated proliferation, M/I | (116) | |||||||
| OS |

|

|

|
EZH2 | FOXC1 transcriptionally upregulates EZH2 | (117) | ||||||
| OS |

|

|
β-catenin, C-Wnt | Sp1/FOXC1/HOTTIP/LATS2/YAP/β-catenin cascade | (118) | |||||||
| OS |

|

|

|
miR-185-5p inhibits FOXC1-mediated M/I | (119) | |||||||
| Melanoma |

|

|
PI3K/AKT | FOXC1 mediates MST1R mediated colony formation, M/I | (120) | |||||||
| WT |

|

|

|
HOXB2 and FOXC1 synergistically drive progression | (121) | |||||||
| GB |

|

|
miR-133 inhibits FOXC1-mediated M/I | (47) | ||||||||
| GB |

|

|

|
β-catenin, C-Wnt | FOXC1 siRNA inhibited EMT, M/I | (122) | ||||||
| Pituitary |

|
miR-133 inhibits FOXC1, ↓M/I | (120) | |||||||||
| AML |

|

|

|
FOXC1, HOXA9 ↑clonogenic potential, ↓differentiation | (39) | |||||||
| T-ALL |

|
NC-Hedgehog | FOXC1 stabilizes GLI2, drives SMO-inhibitor resistance | (123) | ||||||||
| HL |

|

|
FGF2 | OTX2 transcriptionally activates FOXC1 | (124) | |||||||
| DLBCL |

|

|
JUN upregulates FOXC1, drives lymphoma dissemination | (125) |
AIPC, Androgen Insensitive Prostate Cancer; AML, Acute Myelogenous Leukemia; C-, Canonical; CTC, Circulating Tumor Cells; DLBCL, Diffuse Large B Cell Lymphoma; EMT, Epithelial-to-mesenchymal; GB, Glioblastoma; HL, Hodgkins Lymphoma; LSCC, Laryngeal Squamous Cell Carcinoma; MET, Mesenchymal-to-epithelial; NC-, Non-canonical; NP, Nasopharyngeal Carcinoma; OS, Osteosarcoma; OSCC, Oral Squamous Cell Carcinoma; pEMT, Partial Epithelial-to-mesenchymal; SACC, Salivary Squamous Cell Carcinoma; T-ALL, T Cell Acute Lymphocytic Leukemia; WT, Wilms tumor.
Breast Cancer
The clinical evidence with regard to FOXC1 being a powerful prognostic indicator has been most extensively generated in breast cancer. FOXC1 mRNA and/or protein expression status has been demonstrated to be an independent, statistically significant, predictor of metastatic recurrence and poor survival. Multiple studies have now confirmed the clinical utility of FOXC1 expression, both at the mRNA as well as protein level, in predicting poor outcomes. Previous reports by us and others had shown that FOXC1 expression could accurately identify patients with the basal-like breast cancer (BLBC) molecular subtype, the most aggressive subtype of breast cancer (38, 41, 42, 66, 128–131). BLBC has been shown to exist as a “hidden” diagnosis, irrespective of receptor profile status, and is not restricted or confined to only triple negative breast cancers (TNBCs) (42). FOXC1 expression status further correlated with increased incidence of brain and lung metastases and decreased metastasis-free survival in patients without lymph node involvement (38). More recently, in a study involving unbiased bioinformatic screening of transcription factors, FOXC1 was found to display the highest correlation with an epithelial-to-mesenchymal-to-amoeboid (EMAT) cluster having the worst associated disease-specific survival in lymph node negative patients (88). A clinical grade, quantitatively robust, immunohistochemistry-based molecular diagnostic assay has also been developed which measures FOXC1 protein in FFPE tissue and has a high degree of correlation with measurement of FOXC1 mRNA using qRT-PCR from matched FFPE tissue samples (42). The assay has undergone analytical and clinical validation, has achieved regulatory approval for breast cancer with the CE-mark designation and is available for in vitro diagnostic use in the clinic.
Lung Cancer
The prognostic significance of FOXC1 in lung cancer was first reported in 2013 (55) and later confirmed by other independent investigators, both in lung adenocarcinoma as well as lung squamous cell carcinoma (56, 57). Studies have shown that high FOXC1 expression is more frequently associated with adverse clinical parameters and poor overall survival independent of other clinicopathological prognostic factors, including lymph node status. The data are clear on elevated FOXC1 expression being a predictor of lung cancer progression, but are not yet available with regard to predicting spread outside of the primary organ. It is important to note that in the case of lung cancer, twice as many patients ultimately succumb to respiratory failure than as a consequence of distant metastasis to sites outside the lung (132). Thus, lung cancer metastasis to locations or organs outside the lung does not appear to be the predominant cancer-related cause of death as in some other cancers that arise from the breast, colon, or skin (e.g. melanoma).
Gastrointestinal Cancers
FOXC1 expression status has now also been shown to be a poor prognostic indicator in multiple gastrointestinal cancers including esophageal cancer, gastric cancer, liver cancer, pancreatic cancer and colon cancer. Two independent published reports provided initial insight into the potential prognostic significance of FOXC1 expression in esophageal squamous cell carcinoma with regard to overall survival. However, univariate and multivariate hazard ratio values were not available in the published reports (50, 51). One of these studies did however demonstrate that FOXC1 expression displayed a statistically significant association with disease-free survival as well (51). Similarly, two independent studies in gastric cancer thus far have provided preliminary confirmation of the prognostic significance of FOXC1 expression with regard to overall survival (52, 53). Additional independent validation studies performed with greater statistical rigor are therefore required to ascertain the true prognostic value of FOXC1 expression in the case of both esophageal as well as gastric cancer.
A fairly large investigation of patients diagnosed with hepatocellular carcinoma (HCC) demonstrated that FOXC1 expression was a powerful, statistically significant prognostic indicator of worse overall survival as well as recurrence free survival, independent of other clinical variables (54). This result was confirmed on both univariate as well as multivariate analysis. While this is a retrospective, single institution study, the sample size and level of statistical significance suggest that FOXC1 expression status will likely be of clinical value in predicting the prognosis of patients diagnosed with HCC. With regard to pancreatic cancer, there is a single report investigating the potential prognostic relevance of FOXC1 expression in pancreatic cancer (59). While the sample size was not large, the study was successful in demonstrating that FOXC1 is a statistically significant and independent prognostic indicator of adverse outcomes in terms of poor overall survival. This result was strengthened by the fact that it was valid on both univariate as well as multivariate analysis.
Amongst the gastrointestinal cancers, the evidence in support of the prognostic utility of FOXC1 is perhaps most robust in colon cancer. Three independent studies have confirmed the fact that FOXC1 expression status can in fact predict worse overall survival in colon cancer (47–49). One of these studies reports convincing data with regard to both overall survival and disease-free survival (48). Yet another provides further confirmation of FOXC1 being a statistically significant independent predictor of shortened distant-metastasis-free survival on both univariate as well as multivariate analysis (47). Thus, after breast cancer, clinical assessment of FOXC1 expression status for prognostic stratification of patients is most likely to be useful in colon cancer.
Other Cancers
Within head and neck cancers, some functional and mechanistic studies have been published implicating FOXC1 in the biology of these tumors. However, investigations examining the prognostic role of FOXC1 are not yet available. A preliminary report on tongue cancer did demonstrate FOXC1 to be a statistically significant predictor of worse overall survival (60). However, the results of univariate and multivariate analysis were not accessible in the publication for review. Thus, information with regard to the prognostic role of FOXC1 in head and neck cancers is nascent at best, but does hold promise based on the findings of some initial studies on the functional significance of FOXC1. Studies investigating the prognostic significance of FOXC1 in nasopharyngeal carcinomas, laryngeal and oral squamous cell carcinomas are, therefore, needed. Similarly, with regard to melanoma, there is a single publication that reports FOXC1 to have prognostic significance in advanced Stage III or Stage IV melanoma in terms of distant metastasis-free survival. While highly insightful, the univariate and multivariate hazard ratios were not accessible in the publication for review. Thus, additional investigations are needed before we are able to draw any conclusions regarding the prognostic utility of FOXC1 expression in these cancers.
In contrast, data regarding the prognostic impact of FOXC1 expression in cervical cancer is more developed. Two independent studies have reported that FOXC1 possesses prognostic value with regard to predicting shorter overall survival (45, 46). More importantly, one of these confirmed FOXC1 to be a predictor of shorter recurrence-free survival on multivariate analysis (45). FOXC1 is a promising prognostic biomarker in cervical cancer and further studies should be performed to better delineate its clinical utility in this regard. In AML, high FOXC1 expression was found to be associated with adverse prognosis in comparison to low FOXC1 expression (39). Subsequently, high FOXC1 expression was significantly correlated with both refractoriness to induction chemotherapy as well as an increased risk of relapse (40). While further studies are needed in this regard, patients diagnosed with FOXC1+ AML should probably be recommended to enroll in clinical trials examining the efficacy of combination therapy protocols that combine various targeted therapies with standard induction chemotherapy. Such approaches may lead to their achieving minimal residual disease burden having no evidence of leukemic cancer stem cell markers, a known predictor of long-term remission following subsequent allogeneic bone marrow stem cell transplantation.
FOXC1 Expression Associated With Good Prognosis
It is important to note, however, that in two very specific cancer types, elevated FOXC1 expression, in contradistinction to the above evident trend, proved to be a predictor of favorable prognosis. These departures from the norm are unexpected exceptions to the seemingly apparent rule that is supported by the overwhelming majority of research findings in support of FOXC1 expression being an adverse prognostic indicator. Thus, while the preponderance of evidence supports elevated expression of FOXC1 being an accurate predictor of adverse clinical outcomes, the two notable exceptions include ovarian cancer and Luminal B molecular subtype of breast cancer where elevated expression of FOXC1 predicts good prognosis.
In the case of ovarian cancer, a retrospective study (133) demonstrated that positive immunostaining for FOXC1 protein significantly decreased with advancing International Federation of Gynecology and Obstetrics Stage (I-II vs. III-IV) as well as pathologic subtypes from benign to borderline and malignant tumors trending towards good prognosis. However, it should be noted that the sample size was small for cystadenocarcinomas (n=40) and pathologic subtype (n=80). Although nuclear and cytoplasmic FOXC1 staining was observed in cell lines, the above conclusion was based solely upon cytoplasmic FOXC1 as FOXC1 was not detected in the nucleus of the ovarian tumors.
The second report on FOXC1 expression in Luminal B breast cancer (134) looks at FOXC1 as being predictive of favorable outcome and establishes the use of EZH2 methyltransferase inhibitors as a strategy to block metastasis. Specifically higher expression of FOXC1 was associated with increased relapse-free survival in Luminal B patients (HR=0.68 p=0.001), but not in BLBC patients (HR=1.01 p-0.94). This study demonstrated that FOXC1 activates certain anti-metastatic genes in the Luminal B breast cancer subset and that any pro-tumor effects of FOXC1 are overridden by the anti-metastatic functions of FOXC1 leading to an overall pro-survival effect in Luminal B patients expressing higher FOXC1 levels.
These highly exceptional findings merit further investigation into the context-specific subcellular localization of FOXC1, and factors that control and determine its post-translational stability and degradation. While initial insight into this aspect of FOXC1 regulation was provided in two published investigations of FOXC1 protein release, stability and degradation (83, 135), it remains to be established whether these or similar mechanisms may help explain the above observations related to FOXC1 expression in ovarian cancer and Luminal B breast cancer.
FOXC1: A Functional Driver of Cancer Progression and Metastasis
The insight into FOXC1 as a prognostic predictor of cancer progression and metastasis was accompanied by a growing body of work that also demonstrated its functional importance as a molecular driver of these processes utilizing both in vitro and in vivo models Figure 2. The literature supporting this is summarized in Table 2. Several other cancer types than are not featured in Table 2 have also been reported in which significant associations between the malignant phenotype and FOXC1 overexpression have been demonstrated. However, prognostic or functional roles of FOXC1 have yet to be elucidated in these cancers. These include carcinomas of the thyroid (136, 137), gallbladder (138), kidney (139, 140), non-melanoma skin cancer (141) and synovial sarcoma (142). Further investigations are required to explore the potential prognostic and functional significance of these initial reports.
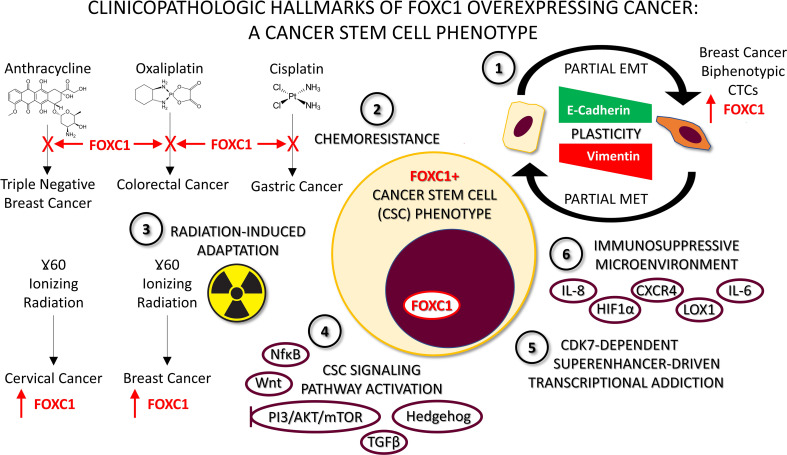
Figure 2.
Characteristics of FOXC1-overexpressing pro-metastatic cancers. 1. Plasticity whereby FOXC1+ cells undergo partial Epithelial-to-Mesenchymal Transition (EMT), and can revert back by undergoing a partial Mesenchymal-to-Epithelial Transition (MET). Detection of FOXC1+ E/M hybrid biphenotypic Circulating Tumor Cells (CTCs) in the peripheral blood of breast cancer patients provides clinical evidence supporting the occurrence of this phenomenon in vivo. 2. Chemoresistance to a variety of chemotherapeutic agents driven by FOXC1 has been described in multiple cancer types with varied mechanisms. 3. Radiation-induced adaptation and subsequent resistance has been described in two different types of cancer cell line models to be characterized by FOXC1 overexpression. 4. Stem cell pathway activation, particularly of the non-canonical variety has been described for NFĸB, Wnt, Hedgehog, PI3K/AKT/mTOR and TGFβ signaling pathways in cancer. Moreover, they all converge on FOXC1. This has led to the suggestion that combination therapy with two or more pathway inhibitor drugs may be necessary to block FOXC1-driven cancer metastasis. 5. Superenhancer-driven transcriptional addiction to FOXC1 mediated by CDK7 has been described in breast cancer and was shown to be effectively thwarted using a CDK7 inhibitor drug. 6. FOXC1 contributes to an immunosuppressive microenvironment by upregulating multiple immunosuppressive factors including HIF1α, CXCR4, CXCR1 and LOX1. This helps explain how the cancer stem cell phenotype helps evade immune detection, and how FOXC1+ cancers are a valid target for immunotherapy approaches like immune checkpoint inhibitors.
By virtue of being a transcription factor, and being a central hub gene controlling a network of hundreds of genes, it is not surprising that upregulation of FOXC1 in cancer does cast wide influence on a number of biologic processes critical for tumor survival and propagation. While this includes proliferation as reported in a number of studies, what has been a hallmark feature of FOXC1+ status is that it appears to predominantly be responsible for a pro-metastatic phenotype. This is evident in Table 2 based on the number of studies across a wide variety of cancers where this has been demonstrated to be the case. In addition to the clinical correlative studies outlined above, clear examples of the critical importance of FOXC1 in metastagenesis includes studies performed in breast cancer (77, 85), lung cancer (57), HCC (54, 143), colon cancer (49) and salivary gland cancer (144). In these studies, overexpression led to the development of an increased number of metastatic lesions in preclinical animal models. Conversely, siRNA-mediated knockdown of FOXC1 practically abolished metastatic propensity (49, 54, 85) in most of these models, confirming the critical dependence of these cancers on FOXC1 to drive the metastatic program.
Several molecular mechanisms have also been elucidated in this regard. These include induction of partial EMT that promotes enhanced cancer cell migration (discussed below), increased production of matrix metalloproteinases that promotes cancer cell invasion (Table 2), and interaction with other cell types in the tumor microenvironment by way of molecular crosstalk that further supplements and enhances these aggressive cancer cell characteristics (discussed below). Crosstalk with other cell types also leads to a state of pervasive immune suppression in the TME that significantly assists with cancer immune evasion. This also aids and abets the cancer cell in being able to continue on its aggressive pro-metastatic course. Cell crosstalk mechanisms pertinent to FOXC1+ cancers are discussed in more detail in a separate section below.
FOXC1: A Driver of Aberrant Cellular Plasticity and Partial EMT
An epithelial-mesenchymal transition (EMT) enables cancer cells to depart from the primary tumor, invade surrounding tissue, and disseminate to distant organs. Several excellent reviews have implicated FOXC1’s role in EMT. After multiple initial reports that linked FOXC1 with EMT in various cancers, some recent reports have further refined our understanding that FOXC1 is in fact associated with a partial EMT phenotype comprising of hybrid E/M cells (Figure 2). Here we briefly summarize the literature in this regard.
Maheswaran and colleagues had found that mesenchymal cells expressing known EMT regulators, including TGF-β pathway components and the FOXC1 transcription factor, were highly enriched in circulating tumor cells (CTCs) and these mesenchymal CTCs were associated with disease progression (69). Similarly, Agelaki and colleagues found that EMT markers (Twist and Vimentin) are expressed in CTCs of patients with metastatic breast cancer (145). Additionally, Maheswaran and colleagues also noticed small populations of CTCs that were positive for both epithelial and mesenchymal markers by RNA-in situ hybridization, and these hybrid E/M CTCs were often enriched in patients with progressive disease after chemotherapy (69). In this same study, an index patient demonstrated dynamic switching between mesenchymal and epithelial CTCs upon each cycle of therapy, suggesting that CTCs maintain dynamic E/M plasticity (69, 146). This data supports the result from a study by Gupta and colleagues which utilized a DNA barcoding approach in the human breast cancer cell line MDA-MB-157 whereby a distinct clonal population of tumor cells was observed to fluctuate between epithelial and mesenchymal states, demonstrating intrinsic E/M plasticity (147). Additionally, they further demonstrated that a single clonal population of tumor cells maintained stable co-expression of both epithelial-to-mesenchymal markers, suggesting the fact, that it is possible for cells to be plastic enough to maintain both epithelial and mesenchymal characteristics.
In an independent study performed by Blanpain and colleagues that utilized a model of squamous cell carcinoma, the clonal population of tumor cells which maintain a hybrid E/M state, has been demonstrated to possess greater metastatic potential than either complete E polarized or complete M polarized cancer cells (148). In an HCC study, elevated FOXC1 expression was associated with enhanced trans-endothelial migration and microvascular invasion (105). However, siRNA-mediated knockdown of FOXC1 was only able to exert an incomplete reversion of EMT, characterized by decreased expression of mesenchymal markers (Vimentin, N-cadherin), but without an accompanying increase in a key epithelial marker (E-cadherin). Epithelial traits were only partially impacted in this condition, and E-cadherin remained unchanged in both expression level and distribution. In summary, this study provides evidence that metastasis may be more dependent on cells maintaining a high FOXC1 expression, which drives a partial EMT (having more of hybrid E/M characteristics) than it is on cells undergoing a complete EMT. This pool of highly plastic cells is more likely to survive in the bloodstream and represents the primary pool of cells from which metastatic lesions are seeded and arise. As such CTC FOXC1+ expression status may in fact help define those CTCs which are Metastasis Initiating Cells (MICs) (149).
FOXC1: Role in Cancer Cell Crosstalk with Other Cells in the Tumor Microenvironment
Tumor cells are surrounded by a heterogeneous and complex tumor microenvironment (TME). The TME consists of a tumor specific extracellular matrix, which recruits an abundance of non-cancer cells including epithelial cells, endothelial cells, mesenchymal cells, immune cells and fibroblasts, all of which interact with the primary tumor cell contributing to tumor progression and metastasis. TME-tumor signaling actively secretes chemokines, cytokines, growth factors, and other metabolites to create a dynamic changing environment (150, 151). A chronic inflammatory, pro-angiogenic and immunosuppressive environment is created through ECM remodeling and through TME-tumor crosstalk, cancer progression and resistance to therapy (152, 153). Infiltrating inflammatory cells can provide a chemotactic escape route for migrating cancer cells from the bulk tumor and modulate cell invasiveness (154, 155). Breast cancer cells and macrophages, through a reciprocal paracrine loop involving EGF, CSF-1, CSF-2 or CCL18, leads to EMT, increased cell motility, invasion and metastasis (156, 157).
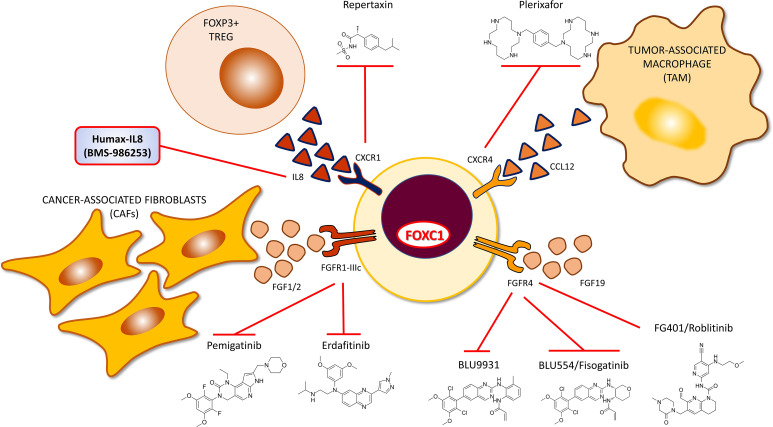
Akin to the above general trend, crosstalk between FOXC1+ cancer cells and other cells of the TME has also been a prominently noted feature. Upregulated FOXC1 in tumor cells induces production and release of cytokines, chemokines and growth factors which mediates recruitment of stromal cells to the TME (Figure 2). Proinflammatory cytokine Interleukin-8 (IL-8) is a member of the CXC chemokine family of angiogenesis/inflammation-related chemokines, secreted by stromal (endothelial cells and fibroblasts) and tumor cells. All biological effects of IL-8 are mediated by 2 receptors designated as CXCR1 (IL-8RA) and CXCR2 (IL-8RB). IL-8 induces FOXC1 upregulation by activation of the PI3K/AKT pathway and Hypoxia Inducible Factor 1α (HIF1α) (106). Consequently, activated FOXC1 transactivates CXC chemokine receptor 1 (CXCR1), a crucial promoter of cancer cell motility through activation of Rho-GTPases (158), that increases invasion and metastasis in HCC (106). CCL2 (monocyte chemoattractant protein-1, MCP-1) is a potent chemokine for monocytes, and a variety of other immune cells, known to activate JAK2/STAT3 signaling (159). CCL2 was also transcriptionally upregulated by FOXC1 in a FOXC1 overexpressed HCC cell line, and appropriately repressed in a FOXC1 knockdown HCC cell line. This transactivation of CCL2 by FOXC1 significantly promoted macrophage infiltration and cancer metastasis in HCC mouse models. These findings were corroborated in human HCC tissues, where FOXC1 expression was found to correlate with levels of IL-8, CXCR1 and CCL2 expression, and infiltration of tumors by macrophages. What is important to note here is that while IL-8 induces FOXC1 transcriptional upregulation via a PI3K/AKT-HIF1α-driven mechanism, FOXC1 in turn transcriptionally upregulates the cognate receptor of IL-8 which is CXCR1, thereby establishing a self-sustaining positive feedback loop. With regard to a different cytokine, FOXC1 overexpression robustly increased NFκB-driven luciferase activity in breast cancer MDA-MB-231 and MCF-7 cells (67). Upregulated NFκB, in turn, induced interleukin-6 (IL-6) generation in MDA-MB-231 cells. The IL-6/STAT3/NFκB positive feedback loop is known to persistently activate breast stromal fibroblasts (160). Pharmacologic targeting of the IL-8/FOXC1/CXCR1 and FOXC1/NFκB/IL-6 positive feedback loops hold promise for deriving clinical therapeutic benefit in FOXC1+ pro-metastatic cancers. The chemokine CXCL12 and its cognate receptor CXCR4, a transcriptional target of FOXC1 was shown to play central roles in cancer proliferation, angiogenesis, invasion, tumor microenvironment, as well as drug resistance induced by chemotherapy (85). CXCL12 affects tumor cell biology via 1) direct stimulation of signaling pathways that promote cancer cell growth, metastasis, and angiogenesis; 2) indirect effects, including the recruitment of CXCR4/CXCR7-positive cancer cells to CXCL12-expressing organs.
FGFR1 is a proven transcriptional target of FOXC1 in breast cancer, following its own transcriptional upregulation by TGFβ pathway activation (80). FGFR1 inhibition is known to promote infiltration of myeloid-derived suppressor cells (MDSCs), that exhibit strong immunosuppressive activity, into the breast cancer TME and promote both cancer progression and metastasis. Inhibition of FGFR1 markedly diminished the level of MDSC infiltration (161) and efficiency of metastatic dissemination (113). Crosstalk between FGFR1 and macrophage derived chemokines CXCL1 and CXCL5 promote tumor formation and progression (162). FGFR1 also promotes the release of inflammatory chemokine CX3CL1 which recruits macrophages to the TME and promotes angiogenesis, both processes being effectively blocked upon treatment with a CX3CL1 inhibitor (163). FOXC1 also transcriptionally upregulates FGFR4 in colon cancer (47). FGFR4 upon ligation with FGF19 is known to promote drug resistance, cancer progression and metastasis. FGFR4 has also been demonstrated to be a poor prognostic indicator in colon cancer (164). The activated FGF19-FGFR4 pathway enhances GSK3β-βcatenin signaling, consequently inducing EMT and resulting in increased HCC metastasis (165, 166) and CRC metastasis (47).
FOXC1: Interplay With Signal Transduction Pathways in Cancer
FOXC1 has been shown to play a critical role in the development and progression of multiple malignancies. Aberrant FOXC1 expression is involved in diverse tumorigenic processes, such as abnormal cell proliferation, cancer stem cell maintenance, cancer migration, and angiogenesis. However, although FOXC1 overexpression often drives aggressive traits in a wide array of human carcinomas, the mechanisms of FOXC1 deregulation that influence the oncogenic and metastagenic processes seem specific to each tumor setting. Here we present an overview of FOXC1 and its correlation with various signal transduction pathways that have been reported in various cancers and highlight how the signal transduction-FOXC1 connection may provide an effective modality to therapeutically intervene and block the aggressive progression of FOXC1+ cancers.
FOXC1: Interconnected with EGFR, NF-κB, Ras/Raf/MEK/ERK, and PI3K/Akt/mTOR Signaling
FOXC1 and EGFR are both critical markers and functional regulators of BLBC. Cui and colleagues reported that EGFR activation regulates FOXC1 expression through ERK and AKT-mediated pathways in BLBC cells (71). In a separate study by the same group, NFκB transcription factor was found to regulate FOXC1 expression in BLBC cells through EGFR signaling (79). EGFR activation also promoted nuclear translocation of NFκB, which binds to the FOXC1 promoter. In the highly aggressive BLBC subtype of TNBC, FOXC1 regulated Pin1/NFκB signaling (67) (Figure 2). Further, FOXC1 overexpression in basal-like MDA-MB-231 breast cancer cells markedly induced phosphorylation of NFκB p65 subunit at Ser-546 and its translocation into the nucleus. Battula and coworkers discovered that ganglioside GD2 expression defined breast cancer stem cells (BCSCs) and ST8SIA1 regulated GD2 expression and breast cancer stem cell (BCSC) function by activation of the FAK-AKT-mTOR signaling pathway. They also showed that in primary TNBC, ST8SIA1 was highly expressed and its expression positively correlated with the expression of FOXC1 (84). In HCC cell lines, IL-8 activated expression of FOXC1 via the phosphoinositide 3-kinase/AKT signaling and HIF 1α. FOXC1 transcriptionally activated CXCR1 and CCL2, which promoted inflammation, invasion and metastasis (106). In cervical cancers and melanoma, it was shown that FOXC1 increased MST1R and activated the PI3K/AKT pathway to drive invasion and migration in melanoma cells (58). Overexpression of FOXC1 in MIA PaCa-2 Pancreatic Cancer cells resulted in increasing the active forms of AKT, PI3K, ERK, and p70s6k (104). Taken together, these studies supported the finding that therapeutic targeting of the EGFR/FOXC1/NFκB pathway and PI3K/AKT/mTOR in BLBC, PI3K/AKT/HIF-1α/FOXC1 axis in HCC and MST1R/PI3K/AKT in cervical cancers, melanoma and pancreatic cancers, may provide effective modalities for treatment.
FOXC1 and Wnt Signaling
Both WNT5A and FOXC1 are up-regulated in TNBC cells and play a significant role in invasion and metastasis. Han and colleagues showed that FOXC1 binds directly to the promoter of WNT5A and up-regulates WNT5A expression in TNBC cells via NFκB signaling (82) (Figure 2). Increased WNT5A expression in TNBC cells is also associated with increase in MMP7 expression. Collectively, FOXC1-WNT5A-MMP7-NFκB signaling axis plays an important role in the migration, invasion, and distant metastasis of TNBC cells. Further, FOXC1 negatively regulates DKK1 (a WNT inhibitor) expression by binding to its promoter region, thereby activating Wnt pathway in gastric cancer cells. FOXC1 also forms a complex with unphosphorylated β-catenin protein in the cytoplasm thereby promoting the entry of β-catenin into the nucleus. Once inside the nucleus, it dissociates from β-catenin, thus regulating transcription of c-MYC, which promotes the proliferation of gastric cancer cells (53).
FOXC1 and Non-Canonical Hedgehog Signaling
FOXC1 induces cancer stem cell (CSC) properties in BLBC cells via activation of Smoothened-independent Hedgehog (Hh) signaling (43) (Figure 2). This non-canonical activation of Hh signaling is specifically mediated by N-terminal domain of FOXC1 binding directly to Gli2, enhancing transcription-activating capacity of Gli2. Together with regulating non-canonical Hh signaling, FOXC1 overexpression also induces resistance to SMO-inhibitors targeting canonical Hedgehog signaling, thus further confirming that actions exerted by FOXC1 are in agreement with it being a marker of CSC function.
FOXC1 and MAPK, IGF1/IGF1R, and LOX Signaling Axis
Aberrant expression of FOXC1 and activation of the FOXC1-p38-MAPK loop promotes tumor metastasis in colorectal cancer (CRC) (49). IGF-1R and FOXC1 regulate each other in pancreatic cancer and FOXC1 is a direct downstream signaling molecule of IGF-1/IGF-1R axis. IGF-1R upregulation in pancreatic cancer cells contributes to cancer progression and metastasis (104). A positive correlation between FOXC1 expression and lysyl oxidase (LOX) expression was established in NSCLC patient samples wherein FOXC1 activated LOX transcription to drive cancer progression through the FOXC1-LOX axis (57). These results suggest that FOXC1 has oncogenic properties that favor metastasis of various cancers.
FOXC1 and TGF-Beta Signaling
Exogenous exposure to TGFβ1 increased FOXC1 mRNA during TGFβ1-induced EMT via Smad2 and Smad3 transcription factors. Hopkins and colleagues demonstrated that FOXC1 expression was activated during TGF-β1-mediated EMT events through the binding of Smad3 proteins to a region in the FOXC1 promoter, ~800 bp upstream of the transcription start site (80). In thus study, while FOXC1 was not essential for TGFβ-induced EMT, it was however critical for effecting an FGFR1 isoform switch that was critical for driving invasion and metastasis, that followed the TGFβ-induced EMT.
FOXC1: Transcriptional Addiction in Triple Negative Breast Cancer
Transcription factors have been implicated in controlling extensive gene expression and regulating various cellular responses. Enhancers are regions of non-coding DNA, which mediate the transcription of adjacent genes, serving as a cis-regulatory element. Superenhancers (SEs) are a hyper active subset of enhancers, that recruit transcription factors, cofactors, and chromatin regulators to drive abundant expression of some significant genes (e.g., oncogenes) in the cancer context. FOXC1 is an SE-associated transcription factor that contributes to invasion, migration and metastasis in TNBC (75, 89). Young and colleagues had found that TNBC cells and ER-negative cells are exceptionally dependent on the expression of at least a subset of the active genes that are transcriptionally regulated by cyclin-dependent kinase 7 (CDK7) (Figure 2). Additionally, in this study it was demonstrated that TNBC cell proliferation is selectively sensitive to THZ1, a newly developed CDK7 inhibitor while ER and or PR-positive cells were largely unaffected by treatment of THZ1 (75). To seek potential biomarkers of THZ1 sensitivity, Tang and coworkers analyzed the mRNAs profile in breast cancer cells treated with THZ1 from the previous study and demonstrated that elevated expression of SOX9 was significantly associated with the sensitivity of THZ1 in TNBC (87). Furthermore, SOX9 and FOXC1 interacted with each other, to co-regulate the MYC signaling pathway in TNBC, while CDK7 inhibitor, THZ1 significantly disrupted the binding of SOX9 to FOXC1 and several enhancer and SE-associated transcription factors, increasing apoptotic cell death. In summary, these findings demonstrate that a collection of SE-associated TNBC genes (EGFR, FOSL1, FOXC1 and MYC) play a significant regulatory role in the proliferation, survival, invasion and metastasis of these CDK7-sensitive and TNBC-enriched cancers. As a logical conclusion, CDK7 inhibitors that block these targetable oncogenes could potentially serve as a rational targeted therapy option for patients diagnosed with TNBC.
FOXC1: Regulator of Cancer Stem Cells, Quiescence and Tumor Escape Mechanism From Conventional Chemotherapy, Radiation Therapy and Targeted Therapy
FOXC1 plays an important role in mediating normal as well as cancer stem cell traits. In normal physiology, Yi and colleagues demonstrated that murine hair follicle stem cells induce FOXC1 to re-establish quiescence (167). In this study, FOXC1 was shown to help preserve quiescent stem cell identity by activating NFATC1 and BMP signaling. In an independent study, hair follicle stem cells were demonstrated to have significantly higher FOXC1 expression, where it helps govern their proliferation and conserves their tissue-regenerating potential compared to downstream, more differentiated hair follicle cells (168). FOXC1 also helps reprogram murine epidermal cells to induced functional sweat gland-like cells, thus proving its potential to determine sweat gland fate in vitro (169). In murine reticular cells, FOXC1 expression is essential for maintenance of the niche where adult hematopoietic stem cells reside (170). These finding were later corroborated in the human system as well (171). These studies collectively highlight the role FOXC1 as a key transcriptional regulator of normal stem cell activity.
Cancer stem cells (CSCs), are a small population of cancer cells that recapitulate most normal stem cell traits, but also play an essential role in tumor initiation, maintenance, progression, metastasis, drug resistance to anti-cancer drugs and metastatic recurrence. Recent studies have indicated that FOXC1, which is associated with a wide variety of cancers, is also strongly associated with mediating CSC activity (Figure 2). Cui and coworkers reported that FOXC1-overexpressed MDA-MB-231 cells, when injected orthotopically into the mammary glands of BALB/c nude mice, resulted in a marked increase in tumor formation efficiency compared to the control group (43). On the contrary, when FOXC1-knockdown BT549 cells were injected into the mouse mammary glands, tumorigenesis was completely inhibited. In vitro, FOXC1 overexpressed SUM159 and MDA-MB-468 cells showed enhanced aldehyde dehydrogenase activity, the increase of which is used for characterizing breast CSC. The above results indicate that FOXC1 positively regulates CSC properties of BLBC cells in vivo and in vitro. In NSCLC, Xu and colleagues demonstrated that FOXC1 knockdown reduced CD133+ cell percentage, suppressed self-renewal ability, decreased expression of stemness-related genes (OCT4, NANOG, SOX2 and ABCG2) and inhibited NSCLC cell tumorigenicity in vivo (56). Battula and coworkers discovered that ganglioside GD2 expression defines breast cancer stem cells (BCSCs) (84). In thus study, ST8SIA1, which regulates GD2 expression was found to be positively correlated with FOXC1 upregulation.
Chemotherapeutic drug resistance is a well-established cancer stem cell property. Mullan and colleagues were the first to show that FOXC1 acts as a mediator of drug resistance to a chemotherapeutic drug regimen comprising 5-fluorouracil/epirubicin/mitomycin C (FEM) as well as to docetaxel (64) (Figure 2). Basal levels of FOXC1 expression were increased in FEM-resistant clones compared to parental MDA-MB-468 cells. Additionally, knockdown of FOXC1 in MDA-MB-231 cells resulted in increased sensitivity to treatment with docetaxel. In NSCLC, FOXC1 knockdown increased cisplatin and docetaxel sensitivity and reduced gefitinib resistance, whereas FOXC1 overexpression enhanced CSC-like properties and resistance to cisplatin and docetaxel (56). Oxaliplatin (OXA) is currently used as first-line chemotherapy to treat stage III and stage IV metastatic colorectal cancer (CRC). Transcription factor FOXC1 binds to the miR-31 promoter to increase the expression of miR31-5p and regulate LATS2 expression in CRC, resulting in cancer cell resistance to OXA (107). LincRNA MCM3AP−AS1 induced upregulation of FOXC1 expression, indicating cisplatin resistance in gastric cancer patients (101). Xu and coworkers investigated the effects of FOXC1 on chemosensitivity in TNBC patients and found that only a minority of TNBC patients have an excellent outcome after receiving standard chemotherapy (44). Despite receiving standard cytotoxic chemotherapy, approximately 30–40% of patients with early-stage TNBC develop metastatic disease. They showed that a significant percentage of TNBC patients who had suboptimal outcomes with anthracycline-based standard chemotherapy were FOXC1 positive.
Therapeutic resistance in cancer also includes resistance to endocrine therapy with anti-estrogen drugs. In breast cancer, Cui and colleagues demonstrated that ectopic FOXC1 expression in ERα-positive MCF7 luminal breast cancer cells greatly diminished the effects of growth-stimulatory B-estradiol and growth-inhibitory antiestrogen treatment with Tamoxifen and Fulvestrant (76). Furthermore, in breast cancer patients with ER-positive primary tumors who received tamoxifen treatment, FOXC1 expression is associated with decreased or undetectable ER expression in recurrent tumors post endocrine treatment. In another independent study by Wang and coworkers, overexpression of FOXC1 decreased expression of ERα protein and reduced cellular responses to estradiol and tamoxifen, while knockdown of FOXC1 induced expression of ERα protein and improved cellular responses to estradiol and tamoxifen (78).
FOXC1: Role in Metabolic Programming in Cancer
Metabolic reprogramming is another essential hallmark of cancer (172). Specific metabolic processes can be directly involved in the transformation process or biological processes that support tumor growth. FOXC1, which has been shown to play an important role in the development and progression of multiple malignancies, also plays a pivotal role in metabolism. Xia and colleagues reported that FOXC1 could inhibit the cysteine metabolism-related genes, cystathionine γ-lyase (CTH) through upregulation of de novo DNA methylase 3B (DNMT3B) expression, which resulted in the decrease of cysteine levels and increase reactive oxygen species (ROS) levels (143). In human HCC cells FOXC1 was in turn upregulated by ROS-ERK1/2-p-ELK1 signaling axis. This positive feedback loop of ROS-FOXC1-cysteine metabolism promotes liver cancer proliferation and metastasis, and this pathway may provide a prospective clinical treatment approach for HCC. Altered glycolysis metabolism is a well characterized signature of invasive cancers. Li and coworkers investigated the role of FOXC1 in regulating glycolysis in CRC cells and found that knockdown of FOXC1 expression in LoVo and RKO cells in vitro markedly reduced glucose uptake and lactate production, while ectopic expression of FOXC1 in HT29 and SW480 cells increased glucose consumption and lactate production. Further, FOXC1 promoted glycolysis and proliferation in CRC cells by inhibiting a key gluconeogenesis regulating enzyme, fructose-1,6-bisphosphatase 1 (FBP1) expression by binding directly to the promoter regions of the FBP1 gene and negatively regulating its transcription.
Therapeutically Targeting FOXC1+ Prometastatic Cancers
It is evident from the above narrative that multiple signaling pathways converge upon and regulate the FOXC1 transcription factor. FOXC1, in turn, influences and coordinates multiple biological processes, again utilizing a variety of downstream signaling pathways, by which FOXC1 pro-metastatic cancers participate in the maturation of the aggressive migratory phenotype leading to metastasis. In some cases, self-sustaining positive-feedback loops are created which make their targeted interruption particularly attractive therapeutic strategies to test in the clinic. While further mechanistic elucidation of the upstream regulators and downstream mediators of FOXC1 activity are required, certain preliminary therapeutic strategies have begun to emerge based on the foundational investigations that have thus far been completed. Below we describe some of these potential strategies which hold promise and merit further consideration during the clinical trial design process.
NFĸB Signal Transduction Pathway
As described above, the NFĸB signaling pathway and FOXC1 can in certain contexts reinforce one another’s actions indefinitely, thereby constituting a self-perpetuating positive feedback loop that contributes to the maintenance of cancer stem cell traits. This suggests that it is a potential therapeutic vulnerability that could potentially be exploited to improve survival outcomes (Figure 3). Significant preliminary data in support of such an approach being therapeutically actionable in the clinic was provided by a preclinical study (173). Utilizing a genome-wide siRNA screen, proteasome addiction was identified as a vulnerability of basal-like TNBC cells. Basal-like TNBC cell lines, known to have elevated FOXC1 expression status, were selectively sensitive to proteasome inhibitor drugs, proportionate to their relative level of FOXC1 expression. Proteasome inhibition effectively blocked tumor-initiating cell function in vitro, and significantly reduced tumor growth and metastatic dissemination to the lungs in vivo. Further evidence to support such an approach was obtained in another preclinical study wherein the Wnt signaling pathway (a CSC-associated pathway) was therapeutically inhibited in an in vitro FOXC1+ cancer model (74). While targeted inhibition of Wnt signaling was initially successful in markedly inhibiting mammosphere formation efficiency (a surrogate marker of cancer stem cell activity), resistant clones did emerge and mammosphere formation ability was regained. However, concurrent treatment with Bortezomib prevented the emergence of such resistant clones and effectively blocked the cancer stem cell escape mechanism.
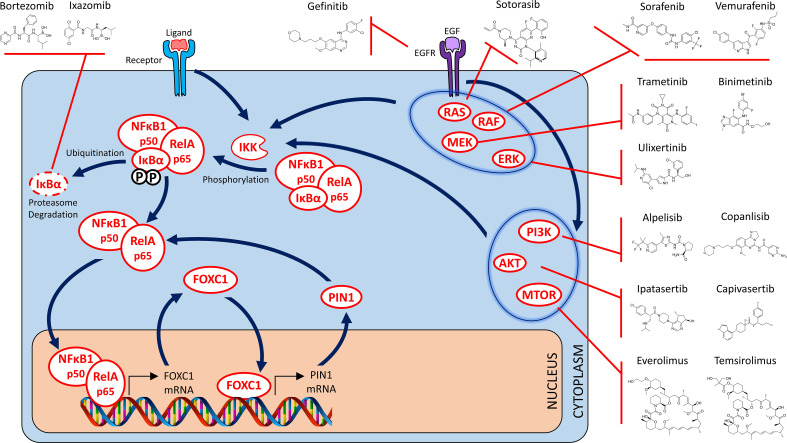
Figure 3.
Targeted NFĸB, RAS/MAPK and PI3K/AKT/MTOR therapy strategies for FOXC1-overexpressing prometastatic cancers – I. Drugs that target the NFkB signaling pathway (Binding of transcriptional coactivator and RNA polymerase to p50/p65 heterodimer not shown). Indirect targeting opportunities are also depicted where targeted inhibition of alternate signaling pathways effectively blocks activation of the NFĸB signaling pathway as well. These include targeting of the EGFR receptor, various members of the RAS/MAPK signaling pathway (including RAS, RAF, MEK and ERK), as well as the individual members of the PI3K/AKT/mTOR signaling pathway.
Bortezomib and Ixazomib are two examples of FDA-approved drugs that are approved for use in the clinic to treat multiple myeloma. While Bortezomib is required to be administered intravenously (IV), Ixazomib is an oral drug with a superior toxicity profile, making it an ideal candidate for evaluating therapeutic efficacy against FOXC1+ pro-metastatic cancers by targeting the NFĸB signaling pathway via proteasome inhibition. Based on this rationale, a Phase I/II clinical trial (AGMT MBC-10) is currently underway to examine the efficacy of Ixazomib in combination with Carboplatin in previously treated advanced TNBC (174). Marizomib, a next generation IV/oral, brain-penetrant, proteasome inhibitor which also displays dual oxidative phosphorylation inhibitory action, has been shown to possess excellent efficacy in an in vivo preclinical study of TNBC utilizing both nude mouse/syngeneic animal models, as well as patient-derived xenografts (175). Marizomib not only caused a marked decline in tumor growth, it significantly reduced the development of lung and brain metastasis as well. Marizomib is currently being evaluated in multiple Phase I/II and III clinical trials in a variety of cancer types. Thus, FOXC1 expression status may serve as a companion/complementary diagnostic for the proteasome inhibitor class of drugs by virtue of their ability to disrupt the NFĸB-FOXC1 positive feedback loop.
RAS/RAF/MEK/ERK/MAPK Signal Transduction Pathway
Utilizing a bioinformatic screening method, the breast cancer molecular subtype that was found to be most susceptible to treatment with small molecular MEK inhibitors was the basal-like subtype (176). The MAPK signal transduction cascade has been demonstrated to regulate and promote CSC traits in an in vivo mouse model of BLBC (177). Treatment with a MEK inhibitor in this model was successful in decreasing tumor growth. Pertinent to this review, RAS/MAPK pathway was demonstrated to regulate FOXC1 expression in breast cancer (68), suggesting that targeted inhibition of this pathway may offer therapeutic benefit in FOXC1+ pro-metastatic cancers (Figure 3). This and other preclinical evidence led to inhibitors of this pathway being tested as a rational treatment strategy for TNBC (178). Subsequently it was demonstrated that RAS/MAPK activation was associated with reduced tumor infiltrating lymphocytes in TNBC and likely contributed towards immune evasion (179). This suggested the possibility that efficacy of immune checkpoint blockade might be further improved if combined with MEK inhibitor therapy. Since activation of this pathway was found to be elevated in patients diagnosed with TNBC who had already been treated with anthracycline-based chemotherapy regimens, the RAS/MAPK pathway may be an attractive therapeutic target in the subset of patients who relapse or have refractory disease (180, 181). Emergence of resistance is a recognized phenomenon when undertaking targeted therapy of any signal transduction pathway in oncology, and the RAS/MAPK pathway proved to be no exception. Several groups reported a seemingly compensatory activation of the PI3K/AKT pathway in response to RAS/MAPK inhibitor therapy, and therapeutic response was obtained upon employing PI3K/AKT inhibitor therapy in animal models of RAS/MAPK inhibitor-resistance (182). This argued in favor of utilizing PI3K/AKT inhibitors to address therapeutic resistance to RAS/MAPK inhibitor therapy, or to undertaking combination therapy with inhibitors of both the RAS/MAPK and PI3K/AKT pathway (183). Another proposed approach to overcome therapeutic resistance encountered with MEK inhibitor therapy, or to achieve greater efficacy than could be achieved with MEK inhibitor therapy alone, was to undertake simultaneous blockade of multiple members of this signaling cascade. Thus, dual inhibition of both MEK and ERK (184) or of MEK and RAF (185), in preclinical models, proved to be successful in blocking the emergence of resistance as well as to overcome acquired resistance to MEK inhibitor therapy.
PI3K/AKT/mTOR Signal Transduction Pathway
PI3K activating mutations and PTEN deactivating mutations are both known to increase and augment signal transduction through the PI3K/AKT/mTOR pathway. And both of these pathway activating mutations are known to occur with greater frequency in TNBC (186). Furthermore, with regard to pathway activation status itself, the PI3K/AKT/mTOR pathway is known to be significantly more upregulated in TNBC as compared to other receptor subtypes of breast cancer (187–189), and is known to contribute to both hormonal therapy resistance as well as chemotherapy resistance. Preclinical data was generated supporting the efficacy of PI3K/AKT/mTOR inhibitors in in vitro and in vivo models of TNBC (190). Based on these promising findings, a concerted effort was undertaken to examine the therapeutic efficacy of mTOR inhibitors in various clinical trials, in both ER+ breast cancer as well as TNBC. Everolimus was found to prolong progression free survival in patients diagnosed with ER+ breast cancer in the advanced/metastatic setting who had developed resistance to hormonal therapy (191). However, two separate Phase II trials failed to show any demonstrable efficacy in TNBC (192, 193). However, an appropriate and suitable companion diagnostic was never established to better guide therapy in either ER+ breast cancer or TNBC. Therefore, at the present time, there is no way to predict which patients diagnosed with ER+ breast cancer, are most likely to respond to treatment with this class of targeted therapeutics. It is also therefore not known whether a specific subset within the TNBC patients tested in trials, did actually derive some clinical benefit, again because no correlation was found between therapeutic efficacy and the candidate companion diagnostic markers tested thus far.
From the clinical trial experience with targeted inhibition of both the RAS/RAF/MEK/ERK/MAPK and the PI3K/AKT/mTOR signal transduction pathways, we have learned that the responses have been variable and not consistent. Furthermore, undertaking a dual inhibitory approach of two different nodes (i.e. MEK and ERK, MEK and RAF, PI3K and mTOR) or dual inhibition of both pathways is often fraught with severely limiting toxicity issues. One potential reason is that the bulk of the preclinical data that provided the initial rationale for these trials was obtained in models of BLBC. However, at the time of patient recruitment for these trials, the TNBC criterion was employed instead. As we have learned time and time again, the BLBC and TNBC definitions are definitely not synonymous and have a widely variable degree of overlap. The assumption that TNBC status is a close enough approximation and adequate surrogate equivalent of BLBC status is known to be incorrect. This might very well be an important factor contributing to potential miscalculation of the therapeutic efficacy in these trials as BLBC or high FOXC1+ status was not employed for patient enrichment, nor to assess potential companion diagnostic utility (Figure 3). A retrospective reassessment of the prospectively accrued patient samples from concluded trials on these lines may help to shed light on how trials evaluating the efficacy of this class of drugs might be better designed in the future.
TGFβ Signal Transduction Pathway
Targeted inhibition of the TGFβ pathway to target tumor growth, progression and metastasis has been controversial. This is primarily because activation of this pathway was demonstrated by two independent studies to variably exert anti-tumorigenic effects on the one hand, but pro-metastagenic effects on the other (194, 195). Interest was however, rekindled in utilizing this approach for therapeutic benefit, when it was demonstrated that TGFβ signaling, in certain subsets of cancer patients like BLBC, contribute to immunosuppression in the tumor microenvironment and immune evasion by the tumor. This was first suggested by a study in which the therapeutic effects of a TGFβR1 inhibitor being evident in a syngeneic, immunocompetent mouse model with an intact immune system, but not demonstrable in an athymic nude mouse model that lacks the ability to mount an immune response (196). Independent validation followed in another report wherein use of an anti-TGFβ antibody successfully suppressed metastasis mainly by inducing a highly significant enhancement of the CD8+ T-cell-mediated antitumor immune response (197). A subsequent independent study highlighted how such an approach held promise in the therapy of BLBC (198). Since then, multiple independent studies have reported that targeting TGFB with a therapeutic antibody (199), with a small molecule inhibitor (200) or with a bifunctional fusion protein (201) was able to overcome therapeutic resistance to immune checkpoint blockade, resulting in complete and durable therapeutic responses in otherwise poorly immunogenic tumors. Recently, another elegant and comprehensive study provided independent confirmation that therapeutic approaches based on TGFβ inhibition in breast cancer were most likely to be successful in patients with high grade, ER-negative disease of the claudin-low and basal subtypes (202). In summary, targeted inhibition of the TGFβ signaling pathway merits investigation in clinical trials to examine whether efficacy thereof can be predicted based on FOXC1 expression status (Figure 4).
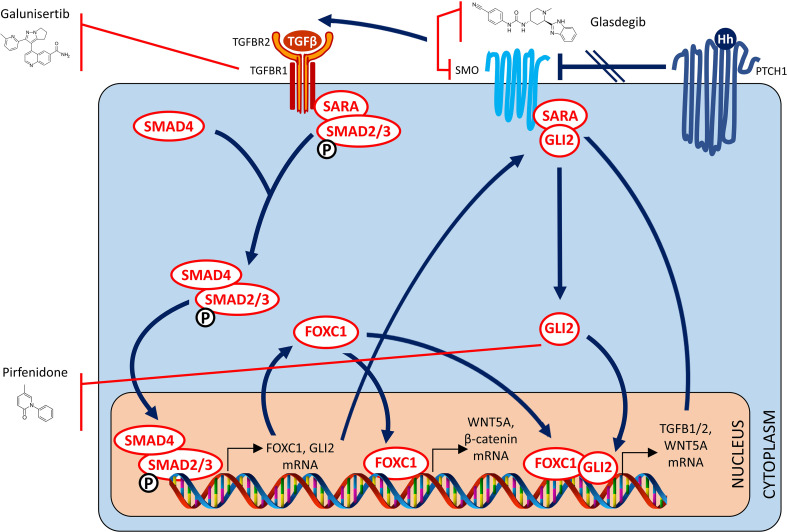
Figure 4.
Targeted TGF-β and hedgehog therapy strategies for FOXC1-overexpressing pro-metastatic cancers – II. Drugs that target the TGF Beta and Hedgehog signaling pathways.
GLI2 Non-Canonical Hegdehog Signal Transduction Pathway
In a study by Cui and colleagues, FOXC1 was demonstrated to regulate CSC maintenance through activation of Smoothened-independent hedgehog signaling and binding to GLI2 in BLBC cells (43). Furthermore, FOXC1 over-expression also induced resistance to SMO-inhibitors targeting canonical Hedgehog signaling Therefore, blockade of the non-canonical pathway described above through targeted inactivation of Gli2 might be a potential strategy to overcome acquired resistance to canonical Hedgehog SMO-inhibitors, and attenuate FOXC1 induced tumorigenicity and metastatic dissemination. Glasdegib is an oral SMO-inhibitor which is FDA-approved for the treatment of newly-diagnosed AML in adults older than 75 years or those who have comorbidities that preclude use of intensive induction chemotherapy (203). In a preclinical study, Glasdegib was demonstrated to successfully target leukemia stem cells (204). Interestingly, although Glasdegib is a SMO-inhibitor, its effects downstream of its SMO-inhibitory action include GLI2 inhibition which abrogates leukemia stem cell dormancy (205). As such, efficacy of Glasdegib in AML and/or other relevant cancers may be predicted by FOXC1 expression status.
Perfenidone is an oral drug approved for treatment of Idiopathic Pulmonary Fibrosis (206, 207). Perfenidone has been proven to decrease fibrosis by decreasing TGFβ1 through a GLI2 inhibitory mechanism (208). However, by virtue of this same mechanism, Pirfenidone was recently demonstrated to be capable of inhibiting fibroblast activation and tumor-fibroblast crosstalk in the TME (209). Another study reported that treatment with Perfenidone was successful in reducing immunosuppressive capacity of cancer-associated fibroblasts in the TME (210). Most importantly, treatment with Pirfenidone was recently reported to augment the observed therapeutic benefit of PD1/PDL1 immune checkpoint blockade in syngeneic, immunocompetent mouse models of lung, liver and colon cancer (211). Collectively, these findings present a unique opportunity for testing in cancer clinical trials where FOXC1 expression status may be found to be useful in predicting efficacy of Perfenidone in ameliorating tumor growth and/or metastatic dissemination (Figure 4).
CXCR1 Driven Cell-Cell Interaction
FOXC1 transcriptionally upregulates CXCR1 in breast cancer (106). CXCR1 is a breast CSC marker (212), and upregulation has been reported to increase recruitment of FOXP3+ T regulatory cells to the TME and contribute to an immunosuppressive state (213). Repartaxin is a small molecule inhibitor of CXCR1 and has been demonstrated to have therapeutic efficacy in preclinical models of breast cancer (214) and gastric cancer (215). In another study utilizing preclinical models of HER2-positive breast cancer, Repartaxin successfully reduced breast CSC activity and demonstrated enhanced therapeutic efficacy when combined with Lapatinib (216). In a window of opportunity neoadjuvant trial, Repartaxin was successful in decreasing the breast CSC count in by more than 20% in preoperative tumors (217). Newer CXCR1 inhibitors have also been reported to have efficacy in preclinical models of RCC and HNSCC (218). Utilizing CXCR1 inhibitors to target CSCs is a promising therapeutic approach with sufficient pre-clinical data to merit examination of their therapeutic efficacy in FOXC1+ pro-metastatic cancers, especially in the setting of advanced/metastatic and/or recurrent cancers which are predicted to be enriched for FOXC1+ CSCs (Figure 5).
Figure 5.
Targeted TME therapy strategies for targeting FOXC1-overexpressing pro-metastatic cancers – III. Drugs that target the IL8, CXCR1, CXCR4, FGFR1 and FGFR4 mediated cell-cell interactions in the tumor microenvironment.
CXCR4 Driven Cell-Cell Interaction
CXCR4 is a direct transcriptional target of FOXC1 in breast cancer that helps mediate increased invasion and metastasis in a preclinical model (85). In this model, treatment with Plerixafor, a CXCR4 antagonist was successful in reversing the CXCR4-mediated invasion and metastasis. Plerixafor is an oral inhibitor of CXCR4 currently FDA-approved as an HPSC mobilizer. The antitumor efficacy of Plerixafor in combination with a standard radio-chemotherapy protocol was evaluated in a murine model of cervical cancer (219). In another study, Plerixafor and another CXCR4 antagonist were observed to successfully reduce the incidence of bone metastasis in an animal model of prostate cancer (220). More recently, Plerixafor was found to mobilize cancer stem cells from their niche in both AML as well as glioblastoma (221). Beyond CXCR4 expression status itself, FOXC1 expression status may provide additional information for more accurately predicting the therapeutic efficacy of CXCR4 inhibitors in various cancers and merits examination in clinical trials (Figure 5).
FGFR1 Signal Transduction Pathway
As discussed above, FGFR1 is a proven transcriptional target of FOXC1 in breast cancer. Erdafitinib is a pan-FGFR oral inhibitor currently FDA-approved for the treatment of advanced or metastatic urothelial carcinoma (222). Pemigatinib is another oral inhibitor of FGFR1/2/3 currently FDA-approved for the treatment of locally advanced or metastatic cholangiocarcinoma (223). FOXC1 expression status may help predict therapeutic efficacy and improve patient selection for treatment with this class of drugs (Figure 5).
FGFR4 Signal Transduction Pathway
As earlier discussed, FOXC1 transcriptionally upregulates FGFR4 in colon cancer (47). Several selective small molecule inhibitors of FGFR4 have been developed (224–228) whose efficacy in HCC and various other cancers is currently being evaluated in early phase clinical trials. FGFR4 activation is closely associated with its specific ligand FGF19. Beyond FGF19 expression status, FOXC1 expression status may provide additional information for more accurately predicting the therapeutic efficacy of FGFR4 inhibitors (Figure 5).
Conclusion
Over the course of the past decade, much has been learned regarding the important role played by the transcription factor FOXC1 in cancer. It is now well accepted that FOXC1 regulates a diverse set of biologically aggressive traits in cancer. Beyond studies of association, FOXC1 has been demonstrated to play a causative role in cancer stem cell biology that contributes not only to an aggressive phenotype but to an aggressive clinical course as well, often culminating in metastatic dissemination and death. From the first report of its central role in aggressive BLBCs more than 10 years ago, we have come to understand that FOXC1 plays a pivotal functional role in more than 16 different cancer types. This list is likely to keep growing. During this interval, our understanding has advanced significantly. From an ever-expanding understanding of the functional and mechanistic role of FOXC1 in cancer, we have also developed an appreciation of its potential application as a powerful prognostic biomarker. FOXC1 expression in cancer tissues, has been demonstrated to be capable of identifying those patients who are at heightened risk of suffering metastatic recurrence. This is of particular relevance in those patient subsets (e.g. lymph node negative patients) where, based on traditional clinical factors, a heightened recurrence risk would have been least suspected. The fact that FOXC1 expression can now be measured quite easily using an inexpensive, clinically validated and commercially available assay (42) means that testing can truly be accomplished on a global scale, even in resource-challenged settings. This is a huge step from a global oncology care perspective. By accurately identifying those patients with FOXC1+ pro-metastatic cancer who are at greatest risk, such a test serves to refine and improve resource utilization, to optimize delivery of life-saving chemotherapy and targeted therapy to the patients who need it the most. Testing to allow identification of metastasis risk early in the disease process is crucial in the fight to reduce glaring inequalities in global cancer control. Even by conservative estimates, a lack of such testing, if not remedied very quickly, will allow cancer to assume global pandemic proportions within the next decade (229).
While identification of elevated metastasis risk is important, it serves another purpose than the one delineated above. Not only would it allow for potentially life-extending systemic treatment (chemotherapy) to be administered to identified high risk patients, it would also allow for their targeted enrollment in the latest clinical trials. Both of these FOXC1 biomarker status-driven interventions have the potential to exert a significant decrease in overall cancer related morbidity and mortality. Beyond the above outlined points of clinical utility, however, sufficient evidence has now accrued to suggest that FOXC1 expression status might also find use to predict the efficacy of certain classes of targeted therapeutics in oncology practice. We have presented some such potentially therapeutic rationales in this review with the intention of spurring debate and stimulating discussion amongst the global oncology community. This of course will need many years of focused effort to first identify the most promising therapeutic approaches to target FOXC1+ pro-metastatic cancers, followed by carefully conducted clinical trials to test and prove their effectiveness. But if successful, such practice-changing advances hold the promise of diminishing the devastating prognostic impact of metastasis, currently the single largest contributor to cancer-related mortality, and extend the lives of cancer survivors.
Author Contributions
PR and TaR conceptualized the study. PR, TaR and TeR performed critical literature review, wrote the manuscript, critically reviewed and edited the manuscript and were collectively responsible for all manuscript content.
Conflict of Interest
PR, TaR, and TeR are employees of and have equity in Onconostic Technologies, Inc. and 3N Diagnostics, Ltd. PR is a named inventor on patents related to the role of FOXC1 in cancer.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- 1.Tabar L, Fagerberg CJ, Gad A, Baldetorp L, Holmberg LH, Grontoft O, et al. Reduction in Mortality From Breast Cancer After Mass Screening With Mammography. Randomised Trial From the Breast Cancer Screening Working Group of the Swedish National Board of Health and Welfare. Lancet (1985) 1(8433):829–32. 10.1016/S0140-6736(85)92204-4 [DOI] [PubMed] [Google Scholar]
- 2.Mapp TJ, Hardcastle JD, Moss SM, Robinson MH. Survival of Patients With Colorectal Cancer Diagnosed in a Randomized Controlled Trial of Faecal Occult Blood Screening. Br J Surg (1999) 86(10):1286–91. 10.1046/j.1365-2168.1999.01229.x [DOI] [PubMed] [Google Scholar]
- 3.Labrie F, Candas B, Dupont A, Cusan L, Gomez JL, Suburu RE, et al. Screening Decreases Prostate Cancer Death: First Analysis of the 1988 Quebec Prospective Randomized Controlled Trial. Prostate (1999) 38(2):83–91. [DOI] [PubMed] [Google Scholar]
- 4.Andrae B, Andersson TM, Lambert PC, Kemetli L, Silfverdal L, Strander B, et al. Screening and Cervical Cancer Cure: Population Based Cohort Study. BMJ (2012) 344:e900. 10.1136/bmj.e900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin (2021) 71(3):209–49. 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 6.Keup C, Suryaprakash V, Storbeck M, Hoffmann O, Kimmig R, Kasimir-Bauer S. Longitudinal Multi-Parametric Liquid Biopsy Approach Identifies Unique Features of Circulating Tumor Cell, Extracellular Vesicle, and Cell-Free DNA Characterization for Disease Monitoring in Metastatic Breast Cancer Patients. Cells (2021) 10(2):212. 10.3390/cells10020212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sheffield BS. Immunohistochemistry as a Practical Tool in Molecular Pathology. Arch Pathol Lab Med (2016) 140(8):766–9. 10.5858/arpa.2015-0453-RA [DOI] [PubMed] [Google Scholar]
- 8.Swanson PE. Immunohistochemistry as a Surrogate for Molecular Testing: A Review. Appl Immunohistochem Mol Morphol (2015) 23(2):81–96. 10.1097/PAI.0000000000000181 [DOI] [PubMed] [Google Scholar]
- 9.Burke HB. Outcome Prediction and the Future of the TNM Staging System. J Natl Cancer Inst (2004) 96(19):1408–9. 10.1093/jnci/djh293 [DOI] [PubMed] [Google Scholar]
- 10.Wang SJ, Hung HM, O’Neill RT. Adaptive Patient Enrichment Designs in Therapeutic Trials. Biom J (2009) 51(2):358–74. 10.1002/bimj.200900003 [DOI] [PubMed] [Google Scholar]
- 11.Gilding LN, Somervaille TCP. The Diverse Consequences of FOXC1 Deregulation in Cancer. Cancers (Basel) (2019) 11(2). 10.3390/cancers11020184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Z, Jiang S, Cheng Y, Li T, Hu W, Ma Z, et al. FOXC1 in Cancer Development and Therapy: Deciphering Its Emerging and Divergent Roles. Ther Adv Med Oncol (2017) 9(12):797–816. 10.1177/1758834017742576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han B, Bhowmick N, Qu Y, Chung S, Giuliano AE, Cui X. FOXC1: An Emerging Marker and Therapeutic Target for Cancer. Oncogene (2017) 36(28):3957–63. 10.1038/onc.2017.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elian FA, Yan E, Walter MA. FOXC1, the New Player in the Cancer Sandbox. Oncotarget (2018) 9(8):8165–78. 10.18632/oncotarget.22742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J, Li W, Zheng X, Pang X, Du G. Research Progress on the Forkhead Box C1. Oncotarget (2018) 9(15):12471–8. 10.18632/oncotarget.22527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishimura DY, Swiderski RE, Alward WL, Searby CC, Patil SR, Bennet SR, et al. The Forkhead Transcription Factor Gene FKHL7 Is Responsible for Glaucoma Phenotypes Which Map to 6p25. Nat Genet (1998) 19(2):140–7. 10.1038/493 [DOI] [PubMed] [Google Scholar]
- 17.Wang WH, McNatt LG, Shepard AR, Jacobson N, Nishimura DY, Stone EM, et al. Optimal Procedure for Extracting RNA From Human Ocular Tissues and Expression Profiling of the Congenital Glaucoma Gene FOXC1 Using Quantitative RT-PCR. Mol Vis (2001) 7:89–94. [PubMed] [Google Scholar]
- 18.Khan AO, Aldahmesh MA, Al-Amri A. Heterozygous FOXC1 Mutation (M161K) Associated With Congenital Glaucoma and Aniridia in an Infant and a Milder Phenotype in Her Mother. Ophthalmic Genet (2008) 29(2):67–71. 10.1080/13816810801908152 [DOI] [PubMed] [Google Scholar]
- 19.Chakrabarti S, Kaur K, Rao KN, Mandal AK, Kaur I, Parikh RS, et al. The Transcription Factor Gene FOXC1 Exhibits a Limited Role in Primary Congenital Glaucoma. Invest Ophthalmol Vis Sci (2009) 50(1):75–83. 10.1167/iovs.08-2253 [DOI] [PubMed] [Google Scholar]
- 20.Tanwar M, Kumar M, Dada T, Sihota R, Dada R. MYOC and FOXC1 Gene Analysis in Primary Congenital Glaucoma. Mol Vis (2010) 16:1996–2006. 10.1155/2010/212656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du RF, Huang H, Fan LL, Li XP, Xia K, Xiang R. A Novel Mutation of FOXC1 (R127L) in an Axenfeld-Rieger Syndrome Family With Glaucoma and Multiple Congenital Heart Diseases. Ophthalmic Genet (2016) 37(1):111–5. 10.3109/13816810.2014.924016 [DOI] [PubMed] [Google Scholar]
- 22.Medina-Trillo C, Aroca-Aguilar JD, Mendez-Hernandez CD, Morales L, Garcia-Anton M, Garcia-Feijoo J, et al. Rare FOXC1 Variants in Congenital Glaucoma: Identification of Translation Regulatory Sequences. Eur J Hum Genet (2016) 24(5):672–80. 10.1038/ejhg.2015.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Siggs OM, Souzeau E, Pasutto F, Dubowsky A, Smith JEH, Taranath D, et al. Prevalence of FOXC1 Variants in Individuals With a Suspected Diagnosis of Primary Congenital Glaucoma. JAMA Ophthalmol (2019) 137(4):348–55. 10.1001/jamaophthalmol.2018.5646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kume T, Deng KY, Winfrey V, Gould DB, Walter MA, Hogan BL. The Forkhead/Winged Helix Gene Mf1 Is Disrupted in the Pleiotropic Mouse Mutation Congenital Hydrocephalus. Cell (1998) 93(6):985–96. 10.1016/S0092-8674(00)81204-0 [DOI] [PubMed] [Google Scholar]
- 25.Nakano T, Niimura F, Hohenfellner K, Miyakita E, Ichikawa I. Screening for Mutations in BMP4 and FOXC1 Genes in Congenital Anomalies of the Kidney and Urinary Tract in Humans. Tokai J Exp Clin Med (2003) 28(3):121–6. [PubMed] [Google Scholar]
- 26.Wu CW, Mann N, Nakayama M, Connaughton DM, Dai R, Kolvenbach CM, et al. Phenotype Expansion of Heterozygous FOXC1 Pathogenic Variants Toward Involvement of Congenital Anomalies of the Kidneys and Urinary Tract (CAKUT). Genet Med (2020) 22(10):1673–81. 10.1038/s41436-020-0844-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swiderski RE, Reiter RS, Nishimura DY, Alward WL, Kalenak JW, Searby CS, et al. Expression of the Mf1 Gene in Developing Mouse Hearts: Implication in the Development of Human Congenital Heart Defects. Dev Dyn (1999) 216(1):16–27. [DOI] [PubMed] [Google Scholar]
- 28.Sanchez-Castro M, Eldjouzi H, Charpentier E, Busson PF, Hauet Q, Lindenbaum P, et al. Search for Rare Copy-Number Variants in Congenital Heart Defects Identifies Novel Candidate Genes and a Potential Role for FOXC1 in Patients With Coarctation of the Aorta. Circ Cardiovasc Genet (2016) 9(1):86–94. 10.1161/CIRCGENETICS.115.001213 [DOI] [PubMed] [Google Scholar]
- 29.Khalil A, Al-Haddad C, Hariri H, Shibbani K, Bitar F, Kurban M, et al. A Novel Mutation in FOXC1 in a Lebanese Family With Congenital Heart Disease and Anterior Segment Dysgenesis: Potential Roles for NFATC1 and DPT in the Phenotypic Variations. Front Cardiovasc Med (2017) 4:58. 10.3389/fcvm.2017.00058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mears AJ, Jordan T, Mirzayans F, Dubois S, Kume T, Parlee M, et al. Mutations of the Forkhead/Winged-Helix Gene, FKHL7, in Patients With Axenfeld-Rieger Anomaly. Am J Hum Genet (1998) 63(5):1316–28. 10.1086/302109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Micheal S, Siddiqui SN, Zafar SN, Villanueva-Mendoza C, Cortes-Gonzalez V, Khan MI, et al. A Novel Homozygous Mutation in FOXC1 Causes Axenfeld Rieger Syndrome With Congenital Glaucoma. PLoS One (2016) 11(7):e0160016. 10.1371/journal.pone.0160016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kidson SH, Kume T, Deng K, Winfrey V, Hogan BL. The Forkhead/Winged-Helix Gene, Mf1, Is Necessary for the Normal Development of the Cornea and Formation of the Anterior Chamber in the Mouse Eye. Dev Biol (1999) 211(2):306–22. 10.1006/dbio.1999.9314 [DOI] [PubMed] [Google Scholar]
- 33.Kume T, Deng K, Hogan BL. Murine Forkhead/Winged Helix Genes Foxc1 (Mf1) and Foxc2 (Mfh1) are Required for the Early Organogenesis of the Kidney and Urinary Tract. Development (2000) 127(7):1387–95. 10.1242/dev.127.7.1387 [DOI] [PubMed] [Google Scholar]
- 34.Schwab K, Patterson LT, Aronow BJ, Luckas R, Liang HC, Potter SS. A Catalogue of Gene Expression in the Developing Kidney. Kidney Int (2003) 64(5):1588–604. 10.1046/j.1523-1755.2003.00276.x [DOI] [PubMed] [Google Scholar]
- 35.Seo S, Kume T. Forkhead Transcription Factors, Foxc1 and Foxc2, are Required for the Morphogenesis of the Cardiac Outflow Tract. Dev Biol (2006) 296(2):421–36. 10.1016/j.ydbio.2006.06.012 [DOI] [PubMed] [Google Scholar]
- 36.Wilm B, James RG, Schultheiss TM, Hogan BL. The Forkhead Genes, Foxc1 and Foxc2, Regulate Paraxial Versus Intermediate Mesoderm Cell Fate. Dev Biol (2004) 271(1):176–89. 10.1016/j.ydbio.2004.03.034 [DOI] [PubMed] [Google Scholar]
- 37.Mattiske D, Kume T, Hogan BL. The Mouse Forkhead Gene Foxc1 Is Required for Primordial Germ Cell Migration and Antral Follicle Development. Dev Biol (2006) 290(2):447–58. 10.1016/j.ydbio.2005.12.007 [DOI] [PubMed] [Google Scholar]
- 38.Ray PS, Wang J, Qu Y, Sim MS, Shamonki J, Bagaria SP, et al. FOXC1 is a Potential Prognostic Biomarker With Functional Significance in Basal-Like Breast Cancer. Cancer Res (2010) 70(10):3870–6. 10.1158/0008-5472.CAN-09-4120 [DOI] [PubMed] [Google Scholar]
- 39.Somerville TD, Wiseman DH, Spencer GJ, Huang X, Lynch JT, Leong HS, et al. Frequent Derepression of the Mesenchymal Transcription Factor Gene FOXC1 in Acute Myeloid Leukemia. Cancer Cell (2015) 28(3):329–42. 10.1016/j.ccell.2015.07.017 [DOI] [PubMed] [Google Scholar]
- 40.Swaminathan M, W JT, Tania R, D AN, Anit S, Egner JR, et al. ABSTRACT 617: FOXC1 Expression in Acute Myeloid Leukemia: Potential Predictor of Disease Relapse and/or Refractory Disease. Blood (2016) 128(22):5260. 10.1182/blood.V128.22.5260.5260 [DOI] [Google Scholar]
- 41.Ray PS, Bagaria SP, Wang J, Shamonki JM, Ye X, Sim MS, et al. Basal-Like Breast Cancer Defined by FOXC1 Expression Offers Superior Prognostic Value: A Retrospective Immunohistochemical Study. Ann Surg Oncol (2011) 18(13):3839–47. 10.1245/s10434-011-1657-8 [DOI] [PubMed] [Google Scholar]
- 42.Jensen TW, Ray T, Wang J, Li X, Naritoku WY, Han B, et al. Diagnosis of Basal-Like Breast Cancer Using a FOXC1-Based Assay. J Natl Cancer Inst (2015) 107(8). 10.1093/jnci/djv148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Han B, Qu Y, Jin Y, Yu Y, Deng N, Wawrowsky K, et al. FOXC1 Activates Smoothened-Independent Hedgehog Signaling in Basal-Like Breast Cancer. Cell Rep (2015) 13(5):1046–58. 10.1016/j.celrep.2015.09.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu YL, Yao R, Li J, Zhou YD, Mao F, Pan B, et al. FOXC1 Overexpression Is a Marker of Poor Response to Anthracycline-Based Adjuvant Chemotherapy in Sporadic Triple-Negative Breast Cancer. Cancer Chemother Pharmacol (2017) 79(6):1205–13. 10.1007/s00280-017-3319-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang L, Huang Z, Fan Y, He L, Ye M, Shi K, et al. FOXC1 Promotes Proliferation and Epithelial-Mesenchymal Transition in Cervical Carcinoma Through the PI3K-AKT Signal Pathway. Am J Transl Res (2017) 9(3):1297–306. [PMC free article] [PubMed] [Google Scholar]
- 46.Wang L, Chai L, Ji Q, Cheng R, Wang J, Han S. Forkhead Box Protein C1 Promotes Cell Proliferation and Invasion in Human Cervical Cancer. Mol Med Rep (2018) 17(3):4392–8. 10.3892/mmr.2018.8423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu J, Zhang Z, Li X, Chen J, Wang G, Tian Z, et al. Forkhead Box C1 Promotes Colorectal Cancer Metastasis Through Transactivating ITGA7 and FGFR4 Expression. Oncogene (2018) 37(41):5477–91. 10.1038/s41388-018-0355-4 [DOI] [PubMed] [Google Scholar]
- 48.Li Q, Wei P, Wu J, Zhang M, Li G, Li Y, et al. The FOXC1/FBP1 Signaling Axis Promotes Colorectal Cancer Proliferation by Enhancing the Warburg Effect. Oncogene (2019) 38(4):483–96. 10.1038/s41388-018-0469-8 [DOI] [PubMed] [Google Scholar]
- 49.Zhang Y, Liao Y, Chen C, Sun W, Sun X, Liu Y, et al. P38-Regulated FOXC1 Stability Is Required for Colorectal Cancer Metastasis. J Pathol (2020) 250(2):217–30. 10.1002/path.5362 [DOI] [PubMed] [Google Scholar]
- 50.Pan F, Yao J, Chen Y, Zhou C, Geng P, Mao H, et al. A Novel Long non-Coding RNA FOXCUT and mRNA FOXC1 Pair Promote Progression and Predict Poor Prognosis in Esophageal Squamous Cell Carcinoma. Int J Clin Exp Pathol (2014) 7(6):2838–49. [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu X, Wei L, Bai Y, Wu S, Han S. FoxC1 Promotes Epithelial-Mesenchymal Transition Through PBX1 Dependent Transactivation of ZEB2 in Esophageal Cancer. Am J Cancer Res (2017) 7(8):1642–53. [PMC free article] [PubMed] [Google Scholar]
- 52.Xu Y, Shao QS, Yao HB, Jin Y, Ma YY, Jia LH. Overexpression of FOXC1 Correlates With Poor Prognosis in Gastric Cancer Patients. Histopathology (2014) 64(7):963–70. 10.1111/his.12347 [DOI] [PubMed] [Google Scholar]
- 53.Jiang J, Li J, Yao W, Wang W, Shi B, Yuan F, et al. FOXC1 Negatively Regulates DKK1 Expression to Promote Gastric Cancer Cell Proliferation Through Activation of Wnt Signaling Pathway. Front Cell Dev Biol (2021) 9:662624. 10.3389/fcell.2021.662624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xia L, Huang W, Tian D, Zhu H, Qi X, Chen Z, et al. Overexpression of Forkhead Box C1 Promotes Tumor Metastasis and Indicates Poor Prognosis in Hepatocellular Carcinoma. Hepatology (2013) 57(2):610–24. 10.1002/hep.26029 [DOI] [PubMed] [Google Scholar]
- 55.Wei LX, Zhou RS, Xu HF, Wang JY, Yuan MH. High Expression of FOXC1 is Associated With Poor Clinical Outcome in Non-Small Cell Lung Cancer Patients. Tumour Biol (2013) 34(2):941–6. 10.1007/s13277-012-0629-3 [DOI] [PubMed] [Google Scholar]
- 56.Cao S, Wang Z, Gao X, He W, Cai Y, Chen H, et al. FOXC1 Induces Cancer Stem Cell-Like Properties Through Upregulation of Beta-Catenin in NSCLC. J Exp Clin Cancer Res (2018) 37(1):220. 10.1186/s13046-018-0894-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gong R, Lin W, Gao A, Liu Y, Li J, Sun M, et al. Forkhead Box C1 Promotes Metastasis and Invasion of Non-Small Cell Lung Cancer by Binding Directly to the Lysyl Oxidase Promoter. Cancer Sci (2019) 110(12):3663–76. 10.1111/cas.14213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang J, Li L, Liu S, Zhao Y, Wang L, Du G. FOXC1 Promotes Melanoma by Activating MST1R/PI3K/AKT. Oncotarget (2016) 7(51):84375–87. 10.18632/oncotarget.11224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang L, Gu F, Liu CY, Wang RJ, Li J, Xu JY. High Level of FOXC1 Expression Is Associated With Poor Prognosis in Pancreatic Ductal Adenocarcinoma. Tumour Biol (2013) 34(2):853–8. 10.1007/s13277-012-0617-7 [DOI] [PubMed] [Google Scholar]
- 60.Lin Z, Sun L, Chen W, Liu B, Wang Y, Fan S, et al. miR-639 Regulates Transforming Growth Factor Beta-Induced Epithelial-Mesenchymal Transition in Human Tongue Cancer Cells by Targeting FOXC1. Cancer Sci (2014) 105(10):1288–98. 10.1111/cas.12499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bloushtain-Qimron N, Yao J, Snyder EL, Shipitsin M, Campbell LL, Mani SA, et al. Cell Type-Specific DNA Methylation Patterns in the Human Breast. Proc Natl Acad Sci USA (2008) 105(37):14076–81. 10.1073/pnas.0805206105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dejeux E, Ronneberg JA, Solvang H, Bukholm I, Geisler S, Aas T, et al. DNA Methylation Profiling in Doxorubicin Treated Primary Locally Advanced Breast Tumours Identifies Novel Genes Associated With Survival and Treatment Response. Mol Cancer (2010) 9:68. 10.1186/1476-4598-9-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kuhmann C, Weichenhan D, Rehli M, Plass C, Schmezer P, Popanda O. DNA Methylation Changes in Cells Regrowing After Fractioned Ionizing Radiation. Radiother Oncol (2011) 101(1):116–21. 10.1016/j.radonc.2011.05.048 [DOI] [PubMed] [Google Scholar]
- 64.Tkocz D, Crawford NT, Buckley NE, Berry FB, Kennedy RD, Gorski JJ, et al. BRCA1 and GATA3 Corepress FOXC1 to Inhibit the Pathogenesis of Basal-Like Breast Cancers. Oncogene (2012) 31(32):3667–78. 10.1038/onc.2011.531 [DOI] [PubMed] [Google Scholar]
- 65.Powell AA, Talasaz AH, Zhang H, Coram MA, Reddy A, Deng G, et al. Single Cell Profiling of Circulating Tumor Cells: Transcriptional Heterogeneity and Diversity From Breast Cancer Cell Lines. PloS One (2012) 7(5):e33788. 10.1371/journal.pone.0033788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sizemore ST, Keri RA. The Forkhead Box Transcription Factor FOXC1 Promotes Breast Cancer Invasion by Inducing Matrix Metalloprotease 7 (MMP7) Expression. J Biol Chem (2012) 287(29):24631–40. 10.1074/jbc.M112.375865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang J, Ray PS, Sim MS, Zhou XZ, Lu KP, Lee AV, et al. FOXC1 Regulates the Functions of Human Basal-Like Breast Cancer Cells by Activating NF-kappaB Signaling. Oncogene (2012) 31(45):4798–802. 10.1038/onc.2011.635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Leontovich AA, Zhang S, Quatraro C, Iankov I, Veroux PF, Gambino MW, et al. Raf-1 Oncogenic Signaling Is Linked to Activation of Mesenchymal to Epithelial Transition Pathway in Metastatic Breast Cancer Cells. Int J Oncol (2012) 40(6):1858–64. 10.3892/ijo.2012.1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT, et al. Circulating Breast Tumor Cells Exhibit Dynamic Changes in Epithelial and Mesenchymal Composition. Science (2013) 339(6119):580–4. 10.1126/science.1228522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sizemore GM, Sizemore ST, Pal B, Booth CN, Seachrist DD, Abdul-Karim FW, et al. FOXC1 is Enriched in the Mammary Luminal Progenitor Population, But is Not Necessary for Mouse Mammary Ductal Morphogenesis. Biol Reprod (2013) 89(1):10. 10.1095/biolreprod.113.108001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jin Y, Han B, Chen J, Wiedemeyer R, Orsulic S, Bose S, et al. FOXC1 is a Critical Mediator of EGFR Function in Human Basal-Like Breast Cancer. Ann Surg Oncol (2014) 21 Suppl 4:S758–66. 10.1245/s10434-014-3980-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Klajic J, Busato F, Edvardsen H, Touleimat N, Fleischer T, Bukholm I, et al. DNA Methylation Status of Key Cell-Cycle Regulators Such as CDKNA2/p16 and CCNA1 Correlates With Treatment Response to Doxorubicin and 5-Fluorouracil in Locally Advanced Breast Tumors. Clin Cancer Res (2014) 20(24):6357–66. 10.1158/1078-0432.CCR-14-0297 [DOI] [PubMed] [Google Scholar]
- 73.Liu J, Shen L, Yao J, Li Y, Wang Y, Chen H, et al. Forkhead Box C1 Promoter Upstream Transcript, a Novel Long Non-Coding RNA, Regulates Proliferation and Migration in Basal-Like Breast Cancer. Mol Med Rep (2015) 11(4):3155–9. 10.3892/mmr.2014.3089 [DOI] [PubMed] [Google Scholar]
- 74.Ray PS, Tsai CY, Ray T, Kim B, Jensen TW. Abstract P2-06-04: Proteosome Inhibitor Bortezomib Inhibits Nfκb and Effectively Overcomes Cancer Stem Cell Escape Triggered by Wnt Inhibitor Therapy in FOXC1+ Basal-Like/claudin-Low Breast Cancer. Cancer Res (2015) 75(9 Supplement):P2-06-4-P2-4. 10.1158/1538-7445.SABCS14-P2-06-04 [DOI] [Google Scholar]
- 75.Wang Y, Zhang T, Kwiatkowski N, Abraham BJ, Lee TI, Xie S, et al. CDK7-Dependent Transcriptional Addiction in Triple-Negative Breast Cancer. Cell (2015) 163(1):174–86. 10.1016/j.cell.2015.08.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yu-Rice Y, Jin Y, Han B, Qu Y, Johnson J, Watanabe T, et al. FOXC1 Is Involved in ERalpha Silencing by Counteracting GATA3 Binding and Is Implicated in Endocrine Resistance. Oncogene (2016) 35(41):5400–11. 10.1038/onc.2016.78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zuo HD, Wu Yao W. The Role and the Potential Regulatory Pathways of High Expression of Forkhead Box C1 in Promoting Tumor Growth and Metastasis of Basal-Like Breast Cancer. J BUON (2016) 21(4):818–25. [PubMed] [Google Scholar]
- 78.Wang J, Xu Y, Li L, Wang L, Yao R, Sun Q, et al. FOXC1 is Associated With Estrogen Receptor Alpha and Affects Sensitivity of Tamoxifen Treatment in Breast Cancer. Cancer Med (2017) 6(1):275–87. 10.1002/cam4.990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chung S, Jin Y, Han B, Qu Y, Gao B, Giuliano AE, et al. Identification of EGF-NF-kappaB-FOXC1 Signaling Axis in Basal-Like Breast Cancer. Cell Commun Signal (2017) 15(1):22. 10.1186/s12964-017-0180-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hopkins A, Coatham ML, Berry FB. FOXC1 Regulates FGFR1 Isoform Switching to Promote Invasion Following TGFbeta-Induced EMT. Mol Cancer Res (2017) 15(10):1341–53. 10.1158/1541-7786.MCR-17-0185 [DOI] [PubMed] [Google Scholar]
- 81.Gao B, Qu Y, Han B, Nagaoka Y, Katsumata M, Deng N, et al. Inhibition of Lobuloalveolar Development by FOXC1 Overexpression in the Mouse Mammary Gland. Sci Rep (2017) 7(1):14017. 10.1038/s41598-017-14342-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Han B, Zhou B, Qu Y, Gao B, Xu Y, Chung S, et al. FOXC1-Induced non-Canonical WNT5A-MMP7 Signaling Regulates Invasiveness in Triple-Negative Breast Cancer. Oncogene (2018) 37(10):1399–408. 10.1038/s41388-017-0021-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li J, Choi PS, Chaffer CL, Labella K, Hwang JH, Giacomelli AO, et al. An Alternative Splicing Switch in FLNB Promotes the Mesenchymal Cell State in Human Breast Cancer. Elife (2018) 7. 10.7554/eLife.37184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nguyen K, Yan Y, Yuan B, Dasgupta A, Sun J, Mu H, et al. ST8SIA1 Regulates Tumor Growth and Metastasis in TNBC by Activating the FAK-AKT-mTOR Signaling Pathway. Mol Cancer Ther (2018) 17(12):2689–701. 10.1158/1535-7163.MCT-18-0399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pan H, Peng Z, Lin J, Ren X, Zhang G, Cui Y. Forkhead Box C1 Boosts Triple-Negative Breast Cancer Metastasis Through Activating the Transcription of Chemokine Receptor-4. Cancer Sci (2018) 109(12):3794–804. 10.1111/cas.13823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zheng XJ, Li W, Yi J, Liu JY, Ren LW, Zhu XM, et al. EZH2 Regulates Expression of FOXC1 by Mediating H3K27me3 in Breast Cancers. Acta Pharmacol Sin. 10.1038/s41401-020-00543-x [DOI] [PMC free article] [PubMed]
- 87.Tang L, Jin J, Xu K, Wang X, Tang J, Guan X. SOX9 Interacts With FOXC1 to Activate MYC and Regulate CDK7 Inhibitor Sensitivity in Triple-Negative Breast Cancer. Oncogenesis (2020) 9(5):47. 10.1038/s41389-020-0232-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Emad A, Ray T, Jensen TW, Parat M, Natrajan R, Sinha S, et al. Superior Breast Cancer Metastasis Risk Stratification Using an Epithelial-Mesenchymal-Amoeboid Transition Gene Signature. Breast Cancer Res (2020) 22(1):74. 10.1186/s13058-020-01304-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Huang H, Hu J, Maryam A, Huang Q, Zhang Y, Ramakrishnan S, et al. Defining Super-Enhancer Landscape in Triple-Negative Breast Cancer by Multiomic Profiling. Nat Commun (2021) 12(1):2242. 10.1038/s41467-021-22445-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.van der Heul-Nieuwenhuijsen L, Dits NF, Jenster G. Gene Expression of Forkhead Transcription Factors in the Normal and Diseased Human Prostate. BJU Int (2009) 103(11):1574–80. 10.1111/j.1464-410X.2009.08351.x [DOI] [PubMed] [Google Scholar]
- 91.Peraldo-Neia C, Migliardi G, Mello-Grand M, Montemurro F, Segir R, Pignochino Y, et al. Epidermal Growth Factor Receptor (EGFR) Mutation Analysis, Gene Expression Profiling and EGFR Protein Expression in Primary Prostate Cancer. BMC Cancer (2011) 11:31. 10.1186/1471-2407-11-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Huang H, Xiong Y, Wu Z, He Y, Gao X, Zhou Z, et al. MIR-138-5P Inhibits the Progression of Prostate Cancer by Targeting FOXC1. Mol Genet Genomic Med (2020) 8(4):e1193. 10.1002/mgg3.1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chen S, Jiao S, Jia Y, Li Y. Effects of Targeted Silencing of FOXC1 Gene on Proliferation and In Vitro Migration of Human non-Small-Cell Lung Carcinoma Cells. Am J Transl Res (2016) 8(8):3309–18. [PMC free article] [PubMed] [Google Scholar]
- 94.Lin YJ, Shyu WC, Chang CW, Wang CC, Wu CP, Lee HT, et al. Tumor Hypoxia Regulates Forkhead Box C1 to Promote Lung Cancer Progression. Theranostics (2017) 7(5):1177–91. 10.7150/thno.17895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hu GD, Wang CX, Wang HY, Wang YQ, Hu S, Cao ZW, et al. Long Noncoding RNA CCAT2 Functions as a Competitive Endogenous RNA to Regulate FOXC1 Expression by Sponging miR-23b-5p in Lung Adenocarcinoma. J Cell Biochem (2018). 10.1002/jcb.28077 [DOI] [PubMed]
- 96.Sun CC, Zhu W, Li SJ, Hu W, Zhang J, Zhuo Y, et al. FOXC1-Mediated LINC00301 Facilitates Tumor Progression and Triggers an Immune-Suppressing Microenvironment in Non-Small Cell Lung Cancer by Regulating the HIF1alpha Pathway. Genome Med (2020) 12(1):77. 10.1186/s13073-020-00773-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chung TK, Lau TS, Cheung TH, Yim SF, Lo KW, Siu NS, et al. Dysregulation of microRNA-204 Mediates Migration and Invasion of Endometrial Cancer by Regulating FOXC1. Int J Cancer (2012) 130(5):1036–45. 10.1002/ijc.26060 [DOI] [PubMed] [Google Scholar]
- 98.Xu YY, Tian J, Hao Q, Yin LR. MicroRNA-495 Downregulates FOXC1 Expression to Suppress Cell Growth and Migration in Endometrial Cancer. Tumour Biol (2016) 37(1):239–51. 10.1007/s13277-015-3686-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chen H, Xu L, Shan ZL, Chen S, Hu H. GPX8 is Transcriptionally Regulated by FOXC1 and Promotes the Growth of Gastric Cancer Cells Through Activating the Wnt Signaling Pathway. Cancer Cell Int (2020) 20(1):596. 10.1186/s12935-020-01692-z [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 100.Zhong X, Yu X, Wen X, Chen L, Gu N. Activation of the LINC00242/miR-141/FOXC1 Axis Underpins the Development of Gastric Cancer. Cancer Cell Int (2020) 20:272. 10.1186/s12935-020-01369-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sun H, Wu P, Zhang B, Wu X, Chen W. MCM3AP-AS1 Promotes Cisplatin Resistance in Gastric Cancer Cells via the miR-138/FOXC1 Axis. Oncol Lett (2021) 21(3):211. 10.3892/ol.2021.12472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rashid M, Shah SG, Verma T, Chaudhary N, Rauniyar S, Patel VB, et al. Tumor-Specific Overexpression of Histone Gene, H3C14 in Gastric Cancer Is Mediated Through EGFR-FOXC1 Axis. Biochim Biophys Acta Gene Regul Mech (2021) 1864(4-5):194703. 10.1016/j.bbagrm.2021.194703 [DOI] [PubMed] [Google Scholar]
- 103.Yu C, Wang M, Li Z, Xiao J, Peng F, Guo X, et al. MicroRNA-138-5p Regulates Pancreatic Cancer Cell Growth Through Targeting FOXC1. Cell Oncol (Dordr) (2015) 38(3):173–81. 10.1007/s13402-014-0200-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Subramani R, Camacho FA, Levin CI, Flores K, Clift A, Galvez A, et al. FOXC1 Plays a Crucial Role in the Growth of Pancreatic Cancer. Oncogenesis (2018) 7(7):52. 10.1038/s41389-018-0061-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Xu ZY, Ding SM, Zhou L, Xie HY, Chen KJ, Zhang W, et al. FOXC1 Contributes to Microvascular Invasion in Primary Hepatocellular Carcinoma via Regulating Epithelial-Mesenchymal Transition. Int J Biol Sci (2012) 8(8):1130–41. 10.7150/ijbs.4769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Huang W, Chen Z, Zhang L, Tian D, Wang D, Fan D, et al. Interleukin-8 Induces Expression of FOXC1 to Promote Transactivation of CXCR1 and CCL2 in Hepatocellular Carcinoma Cell Lines and Formation of Metastases in Mice. Gastroenterology (2015) 149(4):1053–67.e14. 10.1053/j.gastro.2015.05.058 [DOI] [PubMed] [Google Scholar]
- 107.Su DN, Wu SP, Chen HT, He JH. HOTAIR, a Long non-Coding RNA Driver of Malignancy Whose Expression Is Activated by FOXC1, Negatively Regulates miRNA-1 in Hepatocellular Carcinoma. Oncol Lett (2016) 12(5):4061–7. 10.3892/ol.2016.5127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hsu HH, Kuo WW, Shih HN, Cheng SF, Yang CK, Chen MC, et al. FOXC1 Regulation of miR-31-5p Confers Oxaliplatin Resistance by Targeting LATS2 in Colorectal Cancer. Cancers (Basel) (2019) 11(10). 10.3390/cancers11101576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Poorebrahim M, Sadeghi S, Ghanbarian M, Kalhor H, Mehrtash A, Teimoori-Toolabi L. Identification of Candidate Genes and miRNAs for Sensitizing Resistant Colorectal Cancer Cells to Oxaliplatin and Irinotecan. Cancer Chemother Pharmacol (2020) 85(1):153–71. 10.1007/s00280-019-03975-3 [DOI] [PubMed] [Google Scholar]
- 110.Kong XP, Yao J, Luo W, Feng FK, Ma JT, Ren YP, et al. The Expression and Functional Role of a FOXC1 Related mRNA-lncRNA Pair in Oral Squamous Cell Carcinoma. Mol Cell Biochem (2014) 394(1-2):177–86. 10.1007/s11010-014-2093-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Li H, Jiang J. LncRNA MCM3AP-AS1 Promotes Proliferation, Migration and Invasion of Oral Squamous Cell Carcinoma Cells via Regulating miR-204-5p/FOXC1. J Investig Med (2020) 68(7):1282–8. 10.1136/jim-2020-001415 [DOI] [PubMed] [Google Scholar]
- 112.Gao W, Wu Y, He X, Zhang C, Zhu M, Chen B, et al. MicroRNA-204-5p Inhibits Invasion and Metastasis of Laryngeal Squamous Cell Carcinoma by Suppressing Forkhead Box C1. J Cancer (2017) 8(12):2356–68. 10.7150/jca.19470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Liu L, Ye TH, Han YP, Song H, Zhang YK, Xia Y, et al. Reductions in Myeloid-Derived Suppressor Cells and Lung Metastases Using AZD4547 Treatment of a Metastatic Murine Breast Tumor Model. Cell Physiol Biochem (2014) 33(3):633–45. 10.1159/000358640 [DOI] [PubMed] [Google Scholar]
- 114.Ou-Yang L, Xiao SJ, Liu P, Yi SJ, Zhang XL, Ou-Yang S, et al. Forkhead Box C1 Induces Epithelialmesenchymal Transition and Is a Potential Therapeutic Target in Nasopharyngeal Carcinoma. Mol Med Rep (2015) 12(6):8003–9. 10.3892/mmr.2015.4427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Li Y, Chen X. miR-4792 Inhibits Epithelial-Mesenchymal Transition and Invasion in Nasopharyngeal Carcinoma by Targeting FOXC1. Biochem Biophys Res Commun (2015) 468(4):863–9. 10.1016/j.bbrc.2015.11.045 [DOI] [PubMed] [Google Scholar]
- 116.Deng L, Liu T, Zhang B, Wu H, Zhao J, Chen J. Forkhead Box C1 is Targeted by microRNA-133b and Promotes Cell Proliferation and Migration in Osteosarcoma. Exp Ther Med (2017) 14(4):2823–30. 10.3892/etm.2017.4870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Qiu ZD, Dan Z, Che X. FoxC1 Promotes Osteosarcoma Cell Proliferation and Metastasis Through the Activation of EZH2. Int J Clin Exp Med (2017) 10(1):376–84. [Google Scholar]
- 118.Liu K, Ni JD, Li WZ, Pan BQ, Yang YT, Xia Q, et al. The Sp1/FOXC1/HOTTIP/LATS2/YAP/beta-Catenin Cascade Promotes Malignant and Metastatic Progression of Osteosarcoma. Mol Oncol (2020). 10.1002/1878-0261.12760 [DOI] [PMC free article] [PubMed] [Retracted]
- 119.Zhao Z, Chen J, Xia D. Knockdown of HCG18 Inhibits Cell Viability, Migration and Invasion in Pediatric Osteosarcoma by Targeting miR-188-5p/FOXC1 Axis. Mol Biotechnol (2021). 10.1007/s12033-021-00343-6 [DOI] [PubMed]
- 120.Wang DS, Zhang HQ, Zhang B, Yuan ZB, Yu ZK, Yang T, et al. miR-133 Inhibits Pituitary Tumor Cell Migration and Invasion via Down-Regulating FOXC1 Expression. Genet Mol Res (2016. a) 15(1). 10.4238/gmr.15017453 [DOI] [PubMed] [Google Scholar]
- 121.Jing P, Zou J, Zhang L, Wang C, Yang Y, Deng L, et al. HOXB2 and FOXC1 Synergistically Drive the Progression of Wilms Tumor. Exp Mol Pathol (2020) 115:104469. 10.1016/j.yexmp.2020.104469 [DOI] [PubMed] [Google Scholar]
- 122.Cao Q, Wang X, Shi Y, Zhang M, Yang J, Dong M, et al. FOXC1 Silencing Inhibits the Epithelialtomesenchymal Transition of Glioma Cells: Involvement of Betacatenin Signaling. Mol Med Rep (2019) 19(1):251–61. 10.3892/mmr.2018.9650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tosello V, Bongiovanni D, Liu J, Pan Q, Yan KK, Saccomani V, et al. Cross-Talk Between GLI Transcription Factors and FOXC1 Promotes T-Cell Acute Lymphoblastic Leukemia Dissemination. Leukemia (2020). 10.1038/s41375-020-0999-2 [DOI] [PubMed]
- 124.Nagel S, Ehrentraut S, Meyer C, Kaufmann M, Drexler HG, MacLeod RA. Aberrantly Expressed OTX Homeobox Genes Deregulate B-Cell Differentiation in Hodgkin Lymphoma. PloS One (2015) 10(9):e0138416. 10.1371/journal.pone.0138416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Blonska M, Zhu Y, Chuang HH, You MJ, Kunkalla K, Vega F, et al. Jun-Regulated Genes Promote Interaction of Diffuse Large B-Cell Lymphoma With the Microenvironment. Blood (2015) 125(6):981–. 10.1182/blood-2014-04-568188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sabapathi N, Sabarimurugan S, Madurantakam Royam M, Kumarasamy C, Xu X, Xu G, et al. Prognostic Significance of FOXC1 in Various Cancers: A Systematic Review and Meta-Analysis. Mol Diagn Ther (2019) 23(6):695–706. 10.1007/s40291-019-00416-y [DOI] [PubMed] [Google Scholar]
- 127.Kume T, Shackour T. Meta-Analysis of the Likelihood of FOXC1 Expression in Early- and Late-Stage Tumors. Oncotarget (2018) 9(93):36625–30. 10.18632/oncotarget.26358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Johnson J, Choi M, Dadmanesh F, Han B, Qu Y, Yu-Rice Y, et al. FOXC1 Identifies Basal-Like Breast Cancer in a Hereditary Breast Cancer Cohort. Oncotarget (2016) 7(46):75729–38. 10.18632/oncotarget.12370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kim S, Moon BI, Lim W, Park S, Cho MS, Sung SH. Feasibility of Classification of Triple Negative Breast Cancer by Immunohistochemical Surrogate Markers. Clin Breast Cancer (2018) 18(5):e1123–e32. 10.1016/j.clbc.2018.03.012 [DOI] [PubMed] [Google Scholar]
- 130.Zhao S, Ma D, Xiao Y, Li XM, Ma JL, Zhang H, et al. Molecular Subtyping of Triple-Negative Breast Cancers by Immunohistochemistry: Molecular Basis and Clinical Relevance. Oncologist (2020) 25(10):e1481–e91. 10.1634/theoncologist.2019-0982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cao PL, Wang K, Wang CB, Li XF, Yang Z, Yang Z, et al. [Expressions of FOXC1 and MMP-7 in Molecular Subtypes of Breast Cancer and Their Association With Clinicopathological Characteristics]. Zhejiang Da Xue Xue Bao Yi Xue Ban (2014) 43(4):406–12. [DOI] [PubMed] [Google Scholar]
- 132.Nichols L, Saunders R, Knollmann FD. Causes of Death of Patients With Lung Cancer. Arch Pathol Lab Med (2012) 136(12):1552–7. 10.5858/arpa.2011-0521-OA [DOI] [PubMed] [Google Scholar]
- 133.Wang LY, Li LS, Yang Z. Correlation of FOXC1 Protein With Clinicopathological Features in Serous Ovarian Tumors. Oncol Lett (2016) 11(2):933–8. 10.3892/ol.2015.3996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hirukawa A, Smith HW, Zuo D, Dufour CR, Savage P, Bertos N, et al. Targeting EZH2 Reactivates a Breast Cancer Subtype-Specific Anti-Metastatic Transcriptional Program. Nat Commun (2018) 9(1):2547. 10.1038/s41467-018-04864-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Elian FA, McMullen TP, Brindley DN, Walter MA. FOXC1 is Over-Expressed and Is More Stable in Triple Negative/Basal-Like Breast Cancer. FASEB J (2018) 32(S1):S648.6. 10.1096/fasebj.2018.32.1_supplement.648.6 [DOI] [Google Scholar]
- 136.Weinberger P, Ponny SR, Xu H, Bai S, Smallridge R, Copland J, et al. Cell Cycle M-Phase Genes Are Highly Upregulated in Anaplastic Thyroid Carcinoma. Thyroid (2017) 27(2):236–52. 10.1089/thy.2016.0285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hossain MA, Asa TA, Rahman MM, Uddin S, Moustafa AA, Quinn JMW, et al. Network-Based Genetic Profiling Reveals Cellular Pathway Differences Between Follicular Thyroid Carcinoma and Follicular Thyroid Adenoma. Int J Environ Res Public Health (2020) 17(4). 10.3390/ijerph17041373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Li C, Shen W, Shen S, Ai Z. Gene Expression Patterns Combined With Bioinformatics Analysis Identify Genes Associated With Cholangiocarcinoma. Comput Biol Chem (2013) 47:192–7. 10.1016/j.compbiolchem.2013.08.010 [DOI] [PubMed] [Google Scholar]
- 139.Yao T, Wang Q, Zhang W, Bian A, Zhang J. Identification of Genes Associated With Renal Cell Carcinoma Using Gene Expression Profiling Analysis. Oncol Lett (2016) 12(1):73–8. 10.3892/ol.2016.4573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wang Y, Guo X, Bray MJ, Ding Z, Zhao Z. An Integrative Genomics Approach for Identifying Novel Functional Consequences of PBRM1 Truncated Mutations in Clear Cell Renal Cell Carcinoma (ccRCC). BMC Genomics (2016) 17(Suppl 7):515. 10.1186/s12864-016-2906-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Singh A, Gupta A, Chowdhary M, Brahmbhatt HD. Integrated Analysis of miRNA-mRNA Networks Reveals a Strong Anti-Skin Cancer Signature in Vitiligo Epidermis. Exp Dermatol (2021). 10.1111/exd.14317 [DOI] [PubMed]
- 142.Fernebro J, Francis P, Eden P, Borg A, Panagopoulos I, Mertens F, et al. Gene Expression Profiles Relate to SS18/SSX Fusion Type in Synovial Sarcoma. Int J Cancer (2006) 118(5):1165–72. 10.1002/ijc.21475 [DOI] [PubMed] [Google Scholar]
- 143.Lin Z, Huang W, He Q, Li D, Wang Z, Feng Y, et al. FOXC1 Promotes HCC Proliferation and Metastasis by Upregulating DNMT3B to Induce DNA Hypermethylation of CTH Promoter. J Exp Clin Cancer Res (2021) 40(1):50. 10.1186/s13046-021-01829-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wang WW, Chen B, Lei CB, Liu GX, Wang YG, Yi C, et al. miR-582-5p Inhibits Invasion and Migration of Salivary Adenoid Cystic Carcinoma Cells by Targeting FOXC1. Jpn J Clin Oncol (2017) 47(8):690–8. 10.1093/jjco/hyx073 [DOI] [PubMed] [Google Scholar]
- 145.Kallergi G, Papadaki MA, Politaki E, Mavroudis D, Georgoulias V, Agelaki S. Epithelial to Mesenchymal Transition Markers Expressed in Circulating Tumour Cells of Early and Metastatic Breast Cancer Patients. Breast Cancer Res (2011) 13(3):R59. 10.1186/bcr2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hinohara K, Polyak K. Intratumoral Heterogeneity: More Than Just Mutations. Trends Cell Biol (2019) 29(7):569–79. 10.1016/j.tcb.2019.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Mathis RA, Sokol ES, Gupta PB. Cancer Cells Exhibit Clonal Diversity in Phenotypic Plasticity. Open Biol (2017) 7(2). 10.1098/rsob.160283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Pastushenko I, Brisebarre A, Sifrim A, Fioramonti M, Revenco T, Boumahdi S, et al. Identification of the Tumour Transition States Occurring During EMT. Nature (2018) 556(7702):463–8. 10.1038/s41586-018-0040-3 [DOI] [PubMed] [Google Scholar]
- 149.Baccelli I, Schneeweiss A, Riethdorf S, Stenzinger A, Schillert A, Vogel V, et al. Identification of a Population of Blood Circulating Tumor Cells From Breast Cancer Patients That Initiates Metastasis in a Xenograft Assay. Nat Biotechnol (2013) 31(6):539–44. 10.1038/nbt.2576 [DOI] [PubMed] [Google Scholar]
- 150.Hu M, Polyak K. Microenvironmental Regulation of Cancer Development. Curr Opin Genet Dev (2008) 18(1):27–34. 10.1016/j.gde.2007.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Balkwill FR, Capasso M, Hagemann T. The Tumor Microenvironment at a Glance. J Cell Sci (2012) 125(Pt 23):5591–6. 10.1242/jcs.116392 [DOI] [PubMed] [Google Scholar]
- 152.Tredan O, Galmarini CM, Patel K, Tannock IF. Drug Resistance and the Solid Tumor Microenvironment. J Natl Cancer Inst (2007) 99(19):1441–54. 10.1093/jnci/djm135 [DOI] [PubMed] [Google Scholar]
- 153.Son B, Lee S, Youn H, Kim E, Kim W, Youn B. The Role of Tumor Microenvironment in Therapeutic Resistance. Oncotarget (2017) 8(3):3933–45. 10.18632/oncotarget.13907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Aras S, Zaidi MR. TAMeless Traitors: Macrophages in Cancer Progression and Metastasis. Br J Cancer (2017) 117(11):1583–91. 10.1038/bjc.2017.356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Qiu SQ, Waaijer SJH, Zwager MC, de Vries EGE, van der Vegt B, Schroder CP. Tumor-Associated Macrophages in Breast Cancer: Innocent Bystander or Important Player? Cancer Treat Rev (2018) 70:178–89. 10.1016/j.ctrv.2018.08.010 [DOI] [PubMed] [Google Scholar]
- 156.Wyckoff J, Wang W, Lin EY, Wang Y, Pixley F, Stanley ER, et al. A Paracrine Loop Between Tumor Cells and Macrophages Is Required for Tumor Cell Migration in Mammary Tumors. Cancer Res (2004) 64(19):7022–9. 10.1158/0008-5472.CAN-04-1449 [DOI] [PubMed] [Google Scholar]
- 157.Su S, Liu Q, Chen J, Chen J, Chen F, He C, et al. A Positive Feedback Loop Between Mesenchymal-Like Cancer Cells and Macrophages Is Essential to Breast Cancer Metastasis. Cancer Cell (2014) 25(5):605–20. 10.1016/j.ccr.2014.03.021 [DOI] [PubMed] [Google Scholar]
- 158.Schraufstatter IU, Chung J, Burger M. IL-8 Activates Endothelial Cell CXCR1 and CXCR2 Through Rho and Rac Signaling Pathways. Am J Physiol Lung Cell Mol Physiol (2001) 280(6):L1094–103. 10.1152/ajplung.2001.280.6.L1094 [DOI] [PubMed] [Google Scholar]
- 159.Mellado M, Rodriguez-Frade JM, Aragay A, del Real G, Martin AM, Vila-Coro AJ, et al. The Chemokine Monocyte Chemotactic Protein 1 Triggers Janus Kinase 2 Activation and Tyrosine Phosphorylation of the CCR2B Receptor. J Immunol (1998) 161(2):805–13. [PubMed] [Google Scholar]
- 160.Hendrayani SF, Al-Harbi B, Al-Ansari MM, Silva G, Aboussekhra A. The Inflammatory/Cancer-Related IL-6/STAT3/NF-kappaB Positive Feedback Loop Includes AUF1 and Maintains the Active State of Breast Myofibroblasts. Oncotarget (2016) 7(27):41974–85. 10.18632/oncotarget.9633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Holdman XB, Welte T, Rajapakshe K, Pond A, Coarfa C, Mo Q, et al. Upregulation of EGFR Signaling Is Correlated With Tumor Stroma Remodeling and Tumor Recurrence in FGFR1-Driven Breast Cancer. Breast Cancer Res (2015) 17:141. 10.1186/s13058-015-0649-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Bohrer LR, Schwertfeger KL. Macrophages Promote Fibroblast Growth Factor Receptor-Driven Tumor Cell Migration and Invasion in a CXCR2-Dependent Manner. Mol Cancer Res (2012) 10(10):1294–305. 10.1158/1541-7786.MCR-12-0275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Reed JR, Stone MD, Beadnell TC, Ryu Y, Griffin TJ, Schwertfeger KL. Fibroblast Growth Factor Receptor 1 Activation in Mammary Tumor Cells Promotes Macrophage Recruitment in a CX3CL1-Dependent Manner. PLoS One (2012) 7(9):e45877. 10.1371/journal.pone.0045877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Li CS, Zhang SX, Liu HJ, Shi YL, Li LP, Guo XB, et al. Fibroblast Growth Factor Receptor 4 as a Potential Prognostic and Therapeutic Marker in Colorectal Cancer. Biomarkers (2014) 19(1):81–5. 10.3109/1354750X.2013.876555 [DOI] [PubMed] [Google Scholar]
- 165.Zhao H, Lv F, Liang G, Huang X, Wu G, Zhang W, et al. FGF19 Promotes Epithelial-Mesenchymal Transition in Hepatocellular Carcinoma Cells by Modulating the GSK3beta/beta- Catenin Signaling Cascade via FGFR4 Activation. Oncotarget (2016) 7(12):13575–86. 10.18632/oncotarget.6185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Goetz R, Mohammadi M. Exploring Mechanisms of FGF Signalling Through the Lens of Structural Biology. Nat Rev Mol Cell Biol (2013) 14(3):166–80. 10.1038/nrm3528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wang L, Siegenthaler JA, Dowell RD, Yi R. Foxc1 Reinforces Quiescence in Self-Renewing Hair Follicle Stem Cells. Science (2016) 351(6273):613–7. 10.1126/science.aad5440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Lay K, Kume T, Fuchs E. FOXC1 Maintains the Hair Follicle Stem Cell Niche and Governs Stem Cell Quiescence to Preserve Long-Term Tissue-Regenerating Potential. Proc Natl Acad Sci U S A (2016) 113(11):E1506–15. 10.1073/pnas.1601569113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Yao B, Xie J, Liu N, Hu T, Song W, Huang S, et al. Direct Reprogramming of Epidermal Cells Toward Sweat Gland-Like Cells by Defined Factors. Cell Death Dis (2019) 10(4):272. 10.1038/s41419-019-1503-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Omatsu Y, Seike M, Sugiyama T, Kume T, Nagasawa T. Foxc1 Is a Critical Regulator of Haematopoietic Stem/Progenitor Cell Niche Formation. Nature (2014) 508(7497):536–40. 10.1038/nature13071 [DOI] [PubMed] [Google Scholar]
- 171.Aoki K, Kurashige M, Ichii M, Higaki K, Sugiyama T, Kaito T, et al. Identification of CXCL12-Abundant Reticular Cells in Human Adult Bone Marrow. Br J Haematol (2021) 193(3):659–68. 10.1111/bjh.17396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Hanahan D, Weinberg RA. Hallmarks of Cancer: The Next Generation. Cell (2011) 144(5):646–74. 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 173.Petrocca F, Altschuler G, Tan SM, Mendillo ML, Yan H, Jerry DJ, et al. A Genome-Wide siRNA Screen Identifies Proteasome Addiction as a Vulnerability of Basal-Like Triple-Negative Breast Cancer Cells. Cancer Cell (2013) 24(2):182–96. 10.1016/j.ccr.2013.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Rinnerthaler G, Gampenrieder SP, Petzer A, Burgstaller S, Fuchs D, Rossmann D, et al. Ixazomib in Combination With Carboplatin in Pretreated Women With Advanced Triple-Negative Breast Cancer, a Phase I/II Trial of the AGMT (AGMT MBC-10 Trial). BMC Cancer (2018) 18(1):1074. 10.1186/s12885-018-4979-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Raninga PV, Lee A, Sinha D, Dong LF, Datta KK, Lu X, et al. Marizomib Suppresses Triple-Negative Breast Cancer via Proteasome and Oxidative Phosphorylation Inhibition. Theranostics (2020) 10(12):5259–75. 10.7150/thno.42705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Mirzoeva OK, Das D, Heiser LM, Bhattacharya S, Siwak D, Gendelman R, et al. Basal Subtype and MAPK/ERK Kinase (MEK)-Phosphoinositide 3-Kinase Feedback Signaling Determine Susceptibility of Breast Cancer Cells to MEK Inhibition. Cancer Res (2009) 69(2):565–72. 10.1158/0008-5472.CAN-08-3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Balko JM, Schwarz LJ, Bhola NE, Kurupi R, Owens P, Miller TW, et al. Activation of MAPK Pathways Due to DUSP4 Loss Promotes Cancer Stem Cell-Like Phenotypes in Basal-Like Breast Cancer. Cancer Res (2013) 73(20):6346–58. 10.1158/0008-5472.CAN-13-1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Giltnane JM, Balko JM. Rationale for Targeting the Ras/MAPK Pathway in Triple-Negative Breast Cancer. Discov Med (2014) 17(95):275–83. [PubMed] [Google Scholar]
- 179.Loi S, Dushyanthen S, Beavis PA, Salgado R, Denkert C, Savas P, et al. RAS/MAPK Activation Is Associated With Reduced Tumor-Infiltrating Lymphocytes in Triple-Negative Breast Cancer: Therapeutic Cooperation Between MEK and PD-1/PD-L1 Immune Checkpoint Inhibitors. Clin Cancer Res (2016) 22(6):1499–509. 10.1158/1078-0432.CCR-15-1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Huang J, Luo Q, Xiao Y, Li H, Kong L, Ren G. The Implication From RAS/RAF/ERK Signaling Pathway Increased Activation in Epirubicin Treated Triple Negative Breast Cancer. Oncotarget (2017) 8(64):108249–60. 10.18632/oncotarget.22604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Eralp Y, Derin D, Ozluk Y, Yavuz E, Guney N, Saip P, et al. MAPK Overexpression Is Associated With Anthracycline Resistance and Increased Risk for Recurrence in Patients With Triple-Negative Breast Cancer. Ann Oncol (2008) 19(4):669–74. 10.1093/annonc/mdm522 [DOI] [PubMed] [Google Scholar]
- 182.Hoeflich KP, O’Brien C, Boyd Z, Cavet G, Guerrero S, Jung K, et al. In Vivo Antitumor Activity of MEK and Phosphatidylinositol 3-Kinase Inhibitors in Basal-Like Breast Cancer Models. Clin Cancer Res (2009) 15(14):4649–64. 10.1158/1078-0432.CCR-09-0317 [DOI] [PubMed] [Google Scholar]
- 183.Saini KS, Loi S, de Azambuja E, Metzger-Filho O, Saini ML, Ignatiadis M, et al. Targeting the PI3K/AKT/mTOR and Raf/MEK/ERK Pathways in the Treatment of Breast Cancer. Cancer Treat Rev (2013) 39(8):935–46. 10.1016/j.ctrv.2013.03.009 [DOI] [PubMed] [Google Scholar]
- 184.Hatzivassiliou G, Liu B, O’Brien C, Spoerke JM, Hoeflich KP, Haverty PM, et al. ERK Inhibition Overcomes Acquired Resistance to MEK Inhibitors. Mol Cancer Ther (2012) 11(5):1143–54. 10.1158/1535-7163.MCT-11-1010 [DOI] [PubMed] [Google Scholar]
- 185.Nagaria TS, Shi C, Leduc C, Hoskin V, Sikdar S, Sangrar W, et al. Combined Targeting of Raf and Mek Synergistically Inhibits Tumorigenesis in Triple Negative Breast Cancer Model Systems. Oncotarget (2017) 8(46):80804–19. 10.18632/oncotarget.20534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Cancer Genome Atlas N. Comprehensive Molecular Portraits of Human Breast Tumours. Nature (2012) 490(7418):61–70. 10.1038/nature11412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Ueng SH, Chen SC, Chang YS, Hsueh S, Lin YC, Chien HP, et al. Phosphorylated mTOR Expression Correlates With Poor Outcome in Early-Stage Triple Negative Breast Carcinomas. Int J Clin Exp Pathol (2012) 5(8):806–13. [PMC free article] [PubMed] [Google Scholar]
- 188.Walsh S, Flanagan L, Quinn C, Evoy D, McDermott EW, Pierce A, et al. mTOR in Breast Cancer: Differential Expression in Triple-Negative and Non-Triple-Negative Tumors. Breast (2012) 21(2):178–82. 10.1016/j.breast.2011.09.008 [DOI] [PubMed] [Google Scholar]
- 189.Pelicano H, Zhang W, Liu J, Hammoudi N, Dai J, Xu RH, et al. Mitochondrial Dysfunction in Some Triple-Negative Breast Cancer Cell Lines: Role of mTOR Pathway and Therapeutic Potential. Breast Cancer Res (2014) 16(5):434. 10.1186/s13058-014-0434-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Yunokawa M, Koizumi F, Kitamura Y, Katanasaka Y, Okamoto N, Kodaira M, et al. Efficacy of Everolimus, a Novel mTOR Inhibitor, Against Basal-Like Triple-Negative Breast Cancer Cells. Cancer Sci (2012) 103(9):1665–71. 10.1111/j.1349-7006.2012.02359.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Kornblum N, Zhao F, Manola J, Klein P, Ramaswamy B, Brufsky A, et al. Randomized Phase II Trial of Fulvestrant Plus Everolimus or Placebo in Postmenopausal Women With Hormone Receptor-Positive, Human Epidermal Growth Factor Receptor 2-Negative Metastatic Breast Cancer Resistant to Aromatase Inhibitor Therapy: Results of Pre0102. J Clin Oncol (2018) 36(16):1556–63. 10.1200/JCO.2017.76.9331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Jovanovic B, Mayer IA, Mayer EL, Abramson VG, Bardia A, Sanders ME, et al. A Randomized Phase II Neoadjuvant Study of Cisplatin, Paclitaxel With or Without Everolimus in Patients With Stage II/III Triple-Negative Breast Cancer (TNBC): Responses and Long-Term Outcome Correlated With Increased Frequency of DNA Damage Response Gene Mutations, TNBC Subtype, AR Status, and Ki67. Clin Cancer Res (2017) 23(15):4035–45. 10.1158/1078-0432.CCR-16-3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Park IH, Kong SY, Kwon Y, Kim MK, Sim SH, Joo J, et al. Phase I/II Clinical Trial of Everolimus Combined With Gemcitabine/Cisplatin for Metastatic Triple-Negative Breast Cancer. J Cancer (2018) 9(7):1145–51. 10.7150/jca.24035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Siegel PM, Shu W, Cardiff RD, Muller WJ, Massague J. Transforming Growth Factor Beta Signaling Impairs Neu-Induced Mammary Tumorigenesis While Promoting Pulmonary Metastasis. Proc Natl Acad Sci USA (2003) 100(14):8430–5. 10.1073/pnas.0932636100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Tang B, Vu M, Booker T, Santner SJ, Miller FR, Anver MR, et al. TGF-Beta Switches From Tumor Suppressor to Prometastatic Factor in a Model of Breast Cancer Progression. J Clin Invest (2003) 112(7):1116–24. 10.1172/JCI200318899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Ge R, Rajeev V, Ray P, Lattime E, Rittling S, Medicherla S, et al. Inhibition of Growth and Metastasis of Mouse Mammary Carcinoma by Selective Inhibitor of Transforming Growth Factor-Beta Type I Receptor Kinase In Vivo. Clin Cancer Res (2006) 12(14 Pt 1):4315–30. 10.1158/1078-0432.CCR-06-0162 [DOI] [PubMed] [Google Scholar]
- 197.Nam JS, Terabe M, Mamura M, Kang MJ, Chae H, Stuelten C, et al. An Anti-Transforming Growth Factor Beta Antibody Suppresses Metastasis via Cooperative Effects on Multiple Cell Compartments. Cancer Res (2008) 68(10):3835–43. 10.1158/0008-5472.CAN-08-0215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Ganapathy V, Ge R, Grazioli A, Xie W, Banach-Petrosky W, Kang Y, et al. Targeting the Transforming Growth Factor-Beta Pathway Inhibits Human Basal-Like Breast Cancer Metastasis. Mol Cancer (2010) 9:122. 10.1186/1476-4598-9-122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Mariathasan S, Turley SJ, Nickles D, Castiglioni A, Yuen K, Wang Y, et al. TGFbeta Attenuates Tumour Response to PD-L1 Blockade by Contributing to Exclusion of T Cells. Nature (2018) 554(7693):544–8. 10.1038/nature25501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Holmgaard RB, Schaer DA, Li Y, Castaneda SP, Murphy MY, Xu X, et al. Targeting the TGFbeta Pathway With Galunisertib, a TGFbetaRI Small Molecule Inhibitor, Promotes Anti-Tumor Immunity Leading to Durable, Complete Responses, as Monotherapy and in Combination With Checkpoint Blockade. J Immunother Cancer (2018) 6(1):47. 10.1186/s40425-018-0356-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Lan Y, Zhang D, Xu C, Hance KW, Marelli B, Qi J, et al. Enhanced Preclinical Antitumor Activity of M7824, a Bifunctional Fusion Protein Simultaneously Targeting PD-L1 and TGF-Beta. Sci Transl Med (2018) 10(424). 10.1126/scitranslmed.aan5488 [DOI] [PubMed] [Google Scholar]
- 202.Yang Y, Yang HH, Tang B, Wu AML, Flanders KC, Moshkovich N, et al. The Outcome of TGFbeta Antagonism in Metastatic Breast Cancer Models In Vivo Reflects a Complex Balance Between Tumor-Suppressive and Proprogression Activities of TGFbeta. Clin Cancer Res (2020) 26(3):643–56. 10.1158/1078-0432.CCR-19-2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Cortes JE, Heidel FH, Hellmann A, Fiedler W, Smith BD, Robak T, et al. Randomized Comparison of Low Dose Cytarabine With or Without Glasdegib in Patients With Newly Diagnosed Acute Myeloid Leukemia or High-Risk Myelodysplastic Syndrome. Leukemia (2019) 33(2):379–89. 10.1038/s41375-018-0312-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Fukushima N, Minami Y, Kakiuchi S, Kuwatsuka Y, Hayakawa F, Jamieson C, et al. Small-Molecule Hedgehog Inhibitor Attenuates the Leukemia-Initiation Potential of Acute Myeloid Leukemia Cells. Cancer Sci (2016) 107(10):1422–9. 10.1111/cas.13019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Sadarangani A, Pineda G, Lennon KM, Chun HJ, Shih A, Schairer AE, et al. GLI2 Inhibition Abrogates Human Leukemia Stem Cell Dormancy. J Transl Med (2015) 13:98. 10.1186/s12967-015-0453-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Noble PW, Albera C, Bradford WZ, Costabel U, Glassberg MK, Kardatzke D, et al. Pirfenidone in Patients With Idiopathic Pulmonary Fibrosis (CAPACITY): Two Randomised Trials. Lancet (2011) 377(9779):1760–9. 10.1016/S0140-6736(11)60405-4 [DOI] [PubMed] [Google Scholar]
- 207.King TE, Jr., Bradford WZ, Castro-Bernardini S, Fagan EA, Glaspole I, Glassberg MK, et al. A Phase 3 Trial of Pirfenidone in Patients With Idiopathic Pulmonary Fibrosis. N Engl J Med (2014) 370(22):2083–92. 10.1056/NEJMoa1402582 [DOI] [PubMed] [Google Scholar]
- 208.Didiasova M, Singh R, Wilhelm J, Kwapiszewska G, Wujak L, Zakrzewicz D, et al. Pirfenidone Exerts Antifibrotic Effects Through Inhibition of GLI Transcription Factors. FASEB J (2017) 31(5):1916–28. 10.1096/fj.201600892RR [DOI] [PubMed] [Google Scholar]
- 209.Fujiwara A, Funaki S, Fukui E, Kimura K, Kanou T, Ose N, et al. Effects of Pirfenidone Targeting the Tumor Microenvironment and Tumor-Stroma Interaction as a Novel Treatment for Non-Small Cell Lung Cancer. Sci Rep (2020) 10(1):10900. 10.1038/s41598-020-67904-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Aboulkheyr Es H, Zhand S, Thiery JP, Warkiani ME. Pirfenidone Reduces Immune-Suppressive Capacity of Cancer-Associated Fibroblasts Through Targeting CCL17 and TNF-Beta. Integr Biol (Camb) (2020) 12(7):188–97. 10.1093/intbio/zyaa014 [DOI] [PubMed] [Google Scholar]
- 211.Qin W, Zou J, Huang Y, Liu C, Kang Y, Han H, et al. Pirfenidone Facilitates Immune Infiltration and Enhances the Antitumor Efficacy of PD-L1 Blockade in Mice. Oncoimmunology (2020) 9(1):1824631. 10.1080/2162402X.2020.1824631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Charafe-Jauffret E, Ginestier C, Iovino F, Wicinski J, Cervera N, Finetti P, et al. Breast Cancer Cell Lines Contain Functional Cancer Stem Cells With Metastatic Capacity and a Distinct Molecular Signature. Cancer Res (2009) 69(4):1302–13. 10.1158/0008-5472.CAN-08-2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Eikawa S, Ohue Y, Kitaoka K, Aji T, Uenaka A, Oka M, et al. Enrichment of Foxp3+ CD4 Regulatory T Cells in Migrated T Cells to IL-6- and IL-8-Expressing Tumors Through Predominant Induction of CXCR1 by IL-6. J Immunol (2010) 185(11):6734–40. 10.4049/jimmunol.1000225 [DOI] [PubMed] [Google Scholar]
- 214.Ginestier C, Liu S, Diebel ME, Korkaya H, Luo M, Brown M, et al. CXCR1 Blockade Selectively Targets Human Breast Cancer Stem Cells In Vitro and in Xenografts. J Clin Invest (2010) 120(2):485–97. 10.1172/JCI39397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Wang J, Hu W, Wang K, Yu J, Luo B, Luo G, et al. Repertaxin, an Inhibitor of the Chemokine Receptors CXCR1 and CXCR2, Inhibits Malignant Behavior of Human Gastric Cancer MKN45 Cells In Vitro and In Vivo and Enhances Efficacy of 5-Fluorouracil. Int J Oncol (2016) 48(4):1341–52. 10.3892/ijo.2016.3371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Singh JK, Farnie G, Bundred NJ, Simoes BM, Shergill A, Landberg G, et al. Targeting CXCR1/2 Significantly Reduces Breast Cancer Stem Cell Activity and Increases the Efficacy of Inhibiting HER2 via HER2-Dependent and -Independent Mechanisms. Clin Cancer Res (2013) 19(3):643–56. 10.1158/1078-0432.CCR-12-1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Goldstein LJ, Perez RP, Yardley D, Han LK, Reuben JM, Gao H, et al. A Window-of-Opportunity Trial of the CXCR1/2 Inhibitor Reparixin in Operable HER-2-Negative Breast Cancer. Breast Cancer Res (2020) 22(1):4. 10.1186/s13058-019-1243-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Dufies M, Grytsai O, Ronco C, Camara O, Ambrosetti D, Hagege A, et al. New CXCR1/CXCR2 Inhibitors Represent an Effective Treatment for Kidney or Head and Neck Cancers Sensitive or Refractory to Reference Treatments. Theranostics (2019) 9(18):5332–46. 10.7150/thno.34681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Chaudary N, Pintilie M, Jelveh S, Lindsay P, Hill RP, Milosevic M. Plerixafor Improves Primary Tumor Response and Reduces Metastases in Cervical Cancer Treated With Radio-Chemotherapy. Clin Cancer Res (2017) 23(5):1242–9. 10.1158/1078-0432.CCR-16-1730 [DOI] [PubMed] [Google Scholar]
- 220.Gravina GL, Mancini A, Muzi P, Ventura L, Biordi L, Ricevuto E, et al. CXCR4 Pharmacogical Inhibition Reduces Bone and Soft Tissue Metastatic Burden by Affecting Tumor Growth and Tumorigenic Potential in Prostate Cancer Preclinical Models. Prostate (2015) 75(12):1227–46. 10.1002/pros.23007 [DOI] [PubMed] [Google Scholar]
- 221.Hira VVV, Van Noorden CJF, Molenaar RJ. CXCR4 Antagonists as Stem Cell Mobilizers and Therapy Sensitizers for Acute Myeloid Leukemia and Glioblastoma? Biol (Basel) (2020) 9(2). 10.3390/biology9020031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Loriot Y, Necchi A, Park SH, Garcia-Donas J, Huddart R, Burgess E, et al. Erdafitinib in Locally Advanced or Metastatic Urothelial Carcinoma. N Engl J Med (2019) 381(4):338–48. 10.1056/NEJMoa1817323 [DOI] [PubMed] [Google Scholar]
- 223.Abou-Alfa GK, Sahai V, Hollebecque A, Vaccaro G, Melisi D, Al-Rajabi R, et al. Pemigatinib for Previously Treated, Locally Advanced or Metastatic Cholangiocarcinoma: A Multicentre, Open-Label, Phase 2 Study. Lancet Oncol (2020) 21(5):671–84. 10.1016/S1470-2045(20)30109-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Hatlen MA, Schmidt-Kittler O, Sherwin CA, Rozsahegyi E, Rubin N, Sheets MP, et al. Acquired On-Target Clinical Resistance Validates FGFR4 as a Driver of Hepatocellular Carcinoma. Cancer Discov (2019) 9(12):1686–95. 10.1158/2159-8290.CD-19-0367 [DOI] [PubMed] [Google Scholar]
- 225.Kim RD, Sarker D, Meyer T, Yau T, Macarulla T, Park JW, et al. First-In-Human Phase I Study of Fisogatinib (BLU-554) Validates Aberrant FGF19 Signaling as a Driver Event in Hepatocellular Carcinoma. Cancer Discov (2019) 9(12):1696–707. 10.1158/2159-8290.CD-19-0555 [DOI] [PubMed] [Google Scholar]
- 226.Rezende Miranda R, Fu Y, Chen X, Perino J, Cao P, Carpten J, et al. Development of a Potent and Specific FGFR4 Inhibitor for the Treatment of Hepatocellular Carcinoma. J Med Chem (2020) 63(20):11484–97. 10.1021/acs.jmedchem.0c00044 [DOI] [PubMed] [Google Scholar]
- 227.Weiss A, Adler F, Buhles A, Stamm C, Fairhurst RA, Kiffe M, et al. FGF401, A First-In-Class Highly Selective and Potent FGFR4 Inhibitor for the Treatment of FGF19-Driven Hepatocellular Cancer. Mol Cancer Ther (2019) 18(12):2194–206. 10.1158/1535-7163.MCT-18-1291 [DOI] [PubMed] [Google Scholar]
- 228.Hagel M, Miduturu C, Sheets M, Rubin N, Weng W, Stransky N, et al. First Selective Small Molecule Inhibitor of FGFR4 for the Treatment of Hepatocellular Carcinomas With an Activated FGFR4 Signaling Pathway. Cancer Discov (2015) 5(4):424–37. 10.1158/2159-8290.CD-14-1029 [DOI] [PubMed] [Google Scholar]
- 229.Lopez-Gomez M, Malmierca E, de Gorgolas M, Casado E. Cancer in Developing Countries: The Next Most Preventable Pandemic. The Global Problem of Cancer. Crit Rev Oncol Hematol (2013) 88(1):117–22. 10.1016/j.critrevonc.2013.03.011 [DOI] [PubMed] [Google Scholar]