Abstract
Acute kidney injury (AKI) poses a severe threat to human health. MicroRNAs (miRNAs/miRs) are known to be involved in the progression of AKI; however, the function of miR-1184 in AKI remains unclear. Thus, the aim of the present study was to examine the role of this miRNA in kidney injury. In order to mimic AKI in vitro, HK-2 cells were treated with cisplatin. Bioinformatics analysis was performed to explore the differentially expressed miRNAs in AKI. A Cell Counting Kit-8 assay and flow cytometry were performed to examine cell viability and apoptosis, respectively. mRNA expression levels were detected via reverse transcription-quantitative PCR, and protein levels were investigated by western blot analysis. ELISA was performed to examine the levels of IL-1β and TNF-α in the cell supernatants. The results revealed that miR-1184 expression was downregulated in AKI. Exosomes derived from miR-1184 agomir-treated mesenchymal stem cells (MSCs) significantly reversed cisplatin-induced cell growth inhibition by inhibiting apoptosis. Moreover, forkhead box O4 (FOXO4) was found to be the direct target of miR-1184, and exosomes expressing miR-1184 notably inhibited cisplatin-induced inflammatory responses in HK-2 cells via the mediation of IL-1β and TNF-α. Furthermore, exosomes derived from miR-1184 agomir-treated MSCs significantly induced G1 phase arrest in HK-2 cells via the regulation of FOXO4, p27 Kip1 and CDK2. In conclusion, the present study demonstrated that exosomal-miR-1184 derived from MSCs alleviates cisplatin-associated AKI. Thus, the findings presented herein may shed new light onto the exploration of novel strategies for the treatment of AKI.
Keywords: microRNA-1184, exosome, acute kidney injury
Introduction
Acute kidney injury (AKI) is known to be associated with a decline in kidney function within 48 h (1). It is often associated with a high mortality rate in patients, specifically in 1% of the affected population (2). Ischemia-reperfusion injury, sepsis and nephrotoxic insults are the major risk factors for AKI (3–5). Meanwhile, renal tubular epithelial cells are the main cell type in kidney tissue; therefore, the dysfunction of these cells is the main pathophysiological process of AKI (6). Studies have shown that the injury of renal tubular epithelial cells plays an important role in the pathogenesis of AKI (7,8). Moreover, the main features of AKI include inflammatory cell infiltration and the massive secretion of inflammatory factors, necrosis, apoptosis and delayed renal resident cell proliferation (9,10). Although significant efforts have been made in the treatment of AKI, patient outcomes remain dismal. Thus, there is an urgent need for the development of novel strategies for the treatment of AKI.
MicroRNAs (miRNAs/miRs) are a novel class of non-coding small ribonucleic acids, which can regulate gene expression by suppression of mRNA translation or degradation of mRNAs (11,12). Moreover, miRNAs participate in cellular process, including the growth of renal tubular epithelial cells (13,14). Meanwhile, miRNAs have been reported to be associated with AKI progression (15–17). For instance, miR-21 has been shown to inhibit the progression of AKI (18); Jiang et al (19) found that miR-500a-3p alleviated kidney injury by targeting mixed lineage kinase domain like pseudokinase. However, other miRNAs associated with the progression of AKI warrant further investigation.
Exosomes are microvesicles ranging from 70 to 120 nm in diameter and are derived from multivesicular bodies (20). In addition, exosomes participate in cell communication by transferring proteins and nucleic acids, and this function can lead to the mediation of intercellular communication (21,22). In recent studies, a number of exosomal proteins, miRNAs and lncRNAs have been reported to promote the progression of AKI. For example, Cao et al (14) found that exosomal miR-125b-5p deriving from mesenchymal stem cells (MSCs) could promote tubular repair by suppression of p53 in ischemic AKI; Zhang et al (23) indicated that endothelial progenitor cells-derived exosomal miR-21-5p could alleviate sepsis-induced AKI by inhibiting runt-related transcription factor 1 expression. However, the function of exosomes in AKI needs to be further explored. On the other hand, among the potential targets of miR-1184, forkhead box O4 (FOXO4) has been found to be associated with cell growth (24). Thus, the present study focused on the relationship between miR-1184 and the FOXO signaling pathway.
On the other hand, MSCs, characterized by the abilities of self-renewal, differentiation, immunomodulation and trophic support, are essential in regenerative medicine owing to the capacity to create a microenvironment conducive to the repair of injured tissues (14). Previous studies indicated that exosomes derived from MSCs have been proposed as an alternative to MSC-based therapy for several diseases (14,25). In addition, MSC-derived exosomes are known to be involved in AKI progression. For instance, Cao et al (26) found that exosomes derived from MSCs (MSC-exos) could significantly attenuate cisplatin-induced murine AKI through inhibiting inflammation; Ji et al (25) demonstrated that platelet-rich plasma could promote MSC-derived exosome paracrine signaling to repair AKI via the AKT/Rab27 pathway. Thus, exosomes derived from MSCs can play a vital role in AKI progression.
Based on this background, the present study aimed to detect the differentially expressed miRNAs closely associated with the progression of AKI. The findings presented herein may provide new insight on the role of miRNAs in AKI and may aid in the development of novel treatment methods for AKI.
Materials and methods
Cell culture and treatment
HK-2 cell lines (American Type Culture Collection) and MSCs (American Type Culture Collection) were maintained in RPMI-1640 medium (Thermo Fisher Scientific, Inc.), supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.), 1% penicillin (Thermo Fisher Scientific, Inc.) and 1% streptomycin (Thermo Fisher Scientific, Inc.) in a condition with 5% CO2 and 37°C. To mimic AKI in vitro, HK-2 cells were treated with 20 µM cisplatin (MedChemExpress) for 48 h according to previous refs. (8,27).
Cell transfection
HK-2 cells or MSCs (5×103 cells/well) were transfected with miR-1184 agomir or agomir-negative control (NC) using Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.) at 37°C for 48 h. miR-1184 agomir (50 nM) and agomir-NC (50 nM) were obtained from Shanghai GenePharma Co., Ltd. The sequences were as follows: miR-1184 agomir, 5′-CCUUCGGUAGUUCAGCGACGUCC-3′ and agomir-NC, 5′-UUCUCCGAACGUGUCACGUUU-3′. After 48 h of transfection, cells were used in subsequent experiments.
For FOXO4 overexpression, MSCs (5×103 cells/well) were transfected with pcDNA3.1 (1 µg/µl; Shanghai GenePharma Co., Ltd.) or pcDNA3.1-FOXO4 (1 µg/µl; Shanghai GenePharma Co., Ltd.) using Lipofectamine 2000 for 48 h at 37°C, according to the manufacturer's instructions. After 48 h of transfection, cells were used in subsequent experiments.
Bioinformatics analysis
The differentially expressed miRNAs were presented in a volcano plot and a heatmap using the GSE53771 dataset (28) from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo). In addition, the differentially expressed miRNAs were screened using R analysis (29). Using a t-test, P<0.05 and fold-change >1.5 or <0.667 were considered as differentially expressed miRNAs. On the other hand, the functions of miRNA-targeted mRNAs in terms of ‘Cellular Components’ and ‘Biological Processes’ were investigated by Gene Ontology (GO) analysis (http://www.geneontology.org). Pathway analysis was performed to define biological pathways by using the KEGG orthology-based annotation system (KOBAS; version 3.0; http://kobas.cbi.pku.edu.cn/index.php).
Prediction of miRNA downstream target
The downstream target of miR-1184 was predicted using TargetScan (version 7.2; http://www.targetscan.org/vert_72/) and miRDB (version 2.0; http://www.mirdb.org/).
Exosome extraction and identification
Briefly, MSCs were maintained in RPMI-1640 medium until they reached 80% confluence. The medium was then replaced with the serum-free medium. The supernatants were centrifuged for 1 h (300 × g for 15 min at 4°C, 2,000 × g for 15 min at 4°C and 10,000 × g for 30 min at °C) following 48 h of culture. Subsequently, the supernatants were filtrated and collected to extract exosomes using ultracentrifugation (2,000 × g for 15 min at 4°C). Western blot analysis was used to detect the exosome isolation, and the detailed protocol was in accordance with a previous study (25). The particle sizes were detected by Nanoparticle tracking analysis (NTA).
NTA
A total of ~0.3 ml supernatant was loaded into the sample chamber of an LM10 NanoSight unit (NanoSight, Ltd.) and three videos of either 30 or 60 sec were recorded of each sample. Data analysis was performed using NTA 2.1 software (NanoSight, Ltd.). In NTA, the paths of unlabeled particles acting as point scatterers, undergoing Brownian motion in a 0.25 ml chamber through which a 635-nm laser beam was passed, was determined from a video recording, with the mean squared displacement determined for each possible particle. The diffusion coefficient and sphere-equivalent hydrodynamic radius were subsequently determined using the Stokes-Einstein equation (25).
Fluorescence staining
MSCs (5×104 per well) were seeded overnight. Subsequently, cells were labeled for 24 h at 4°C with phalloidin (1:1,000; Abcam; cat. no. ab176753) or PKH26 red membrane dye (1:1,000; Biolab Co., Ltd.; cat. no. HR9070). The nuclei were stained with 5 µl/ml DAPI (Beyotime Institute of Biotechnology). The results were observed under a fluorescence microscope (magnification, ×200; Olympus Corporation).
Western blot analysis
RIPA lysis buffer (Beyotime Institute of Biotechnology) was used to extract total protein from the HK-2 cells. A BCA protein kit (Thermo Fisher Scientific, Inc.) was used to quantify the total protein. SDS-PAGE (10%) was used to separate the protein (40 µg per lane), and the protein was then transferred to PVDF membranes (Thermo Fisher Scientific, Inc.). After blocking with 5% skimmed milk at room temperature for 1 h, the membranes were incubated overnight at 4°C with anti-CD63 (cat. no. ab134045; 1:1,000), anti-CD81 (cat. no. ab109201; 1:1,000), anti-Bax (cat. no. ab32503; 1:1,000), anti-Bcl-2 (cat. no. ab32124; 1:1,000), anti-cleaved caspase-3 (cat. no. ab32042; 1:1,000), anti-TSG101 (cat. no. ab125011; 1:1,000), anti-FOXO4 (cat. no. ab128908; 1:1,000), anti-p27 Kip1 (cat. no. ab32034; 1:1,000), anti-CDK2 (cat. no. ab32147; 1:1,000) and anti-β-actin (cat. no. ab8227; 1:1,000) primary antibodies (all Abcam). Subsequently, the membranes were incubated with secondary anti-rabbit antibodies (HRP-conjugated; Abcam; cat. no. ab7090; 1:5,000) for 1 h at room temperature. Protein bands were visualized using an ECL kit (Thermo Fisher Scientific, Inc.). β-actin was used as the loading control. All antibodies were purchased from Abcam. ImageJ software (version 6.0; National Institutes of Health) was used for densitometry.
Reverse transcription-quantitative PCR (RT-qPCR)
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) was used to isolate total RNA from HK-2 cells or MSCs. The PrimeScript RT Reagent Kit (Takara Bio, Inc.) was used to reverse transcribe total RNA into cDNA, according to the manufacturer's protocol. Subsequently, qPCR was performed using the SYBR Premix Ex Taq II kit (ELK Biotechnology). RT-qPCR reactions were performed under the following protocol: Initial denaturation for 2 min at 94°C, followed by 35 cycles (30 sec at 94°C and 45 sec at 55°C). The following primer pairs were used for RT-qPCR: miR-1184 forward, 5′-CTGGACTGAGCCGTGCTAC-3′ and reverse, 5′-CTCAACTGGTGTCGTGGAGTC-3′; and U6 forward, 5′-CTCGCTTCGGCAGCACAT-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′. The 2−ΔΔCq method (30) was used to quantify the data. U6 was used as an internal control.
Cell counting kit-8 (CCK-8) assay
HK-2 cells (5×103 cells/well) were seeded and treated with cisplatin (20 µM), MSC-ExomiR-1184 agomir or cisplatin + MSC-ExomiR-1184 agomir for 72 h at 37°C. Subsequently, CCK-8 reagent (10 µl; Beyotime Institute of Biotechnology) was added to the cells for 2 h at 37°C. The absorbance (450 nm) was measured using a microplate reader (Thermo Fisher Scientific, Inc.).
Cell apoptosis analysis
HK-2 cells were trypsinized, washed with PBS, resuspended in Annexin V Binding Buffer, and stained with 5 µl FITC and 5 µl PI for 15 min in the dark. The cells were the analyzed using a flow cytometer (FACSLyric™; BD Biosciences) to assess the incidence of cell apoptosis (early + late apoptosis). FlowJo (version 10.6.2; FlowJo LLC) was used to analyze the data.
Dual-luciferase reporter assay
The FOXO4 3′-untranslated region (UTR) containing putative miR-1184 binding sites were obtained from Sangon Biotech Co., Ltd., and cloned into pmirGLO Dual-Luciferase miRNA Target Expression Vectors (Promega Corporation) to construct FOXO4 wild-type (WT) or mutant (MUT) reporter vectors. The mutated 3′-UTR was generated using a site directed mutagenesis kit (Sangon Biotech Co., Ltd.). FOXO4 (WT) or FOXO4 (MUT) were transfected into HK-2 cells (5×103) together with NC or miR-1184 agomir using Lipofectamine 2000. After 48 h of transfection, relative luciferase activities were then analyzed using a Dual-Glo Luciferase Assay System (Promega Corporation). Renilla luciferase activity was used for normalization.
ELISA
HK-2 cell supernatants were collected by centrifugation (500 × g, 10 min, 4°C). Subsequently, the levels of TNF-α (cat. no. H052-1) and IL-1β (cat. no. H002) were investigated using ELISA kits (Nanjing Jiancheng Bioengineering Institute), according to the manufacturers' protocols.
Cell cycle analysis
In brief, HK-2 cells were harvested, fixed with 75% ethanol on ice for 20 min, permeabilized with 0.25% Triton X-100 and stained with PI/RNase (BD Pharmingen; BD Biosciences). Following incubation at 4°C for 15 min, cells were analyzed using a flow cytometer (BD FACSAria III; BD Biosciences). The data were quantified using FlowJo software (version 3.0; FlowJo LLC).
Statistical analysis
Each group was examined in three independent experiments and all data are expressed as the mean ± SD. Western blot analysis, RT-qPCR, flow cytometry, CCK-8 assay and immunofluorescence staining were repeated three times. An unpaired Student's t-test was used to analyze the differences between two groups, and one-way ANOVA followed by Tukey's test were used to analyze differences among multiple groups (>2 groups, using GraphPad Prism 7; GraphPad Software, Inc.). P<0.05 was considered to indicate a statistically significant difference.
Results
Differentially expressed miRNAs in AKI
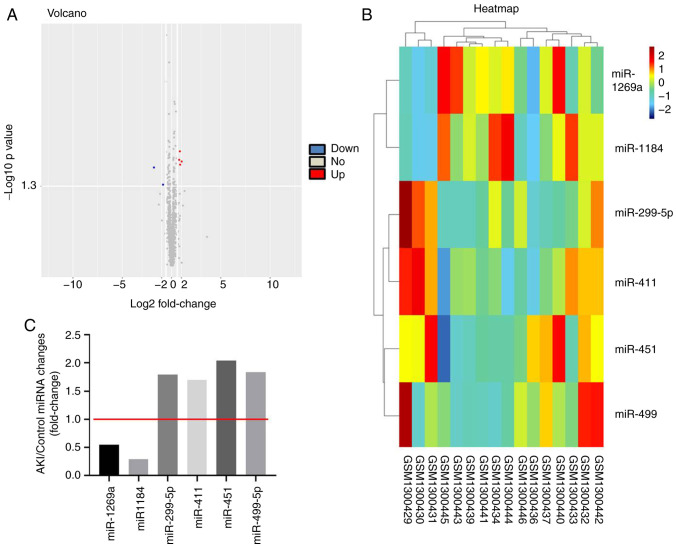
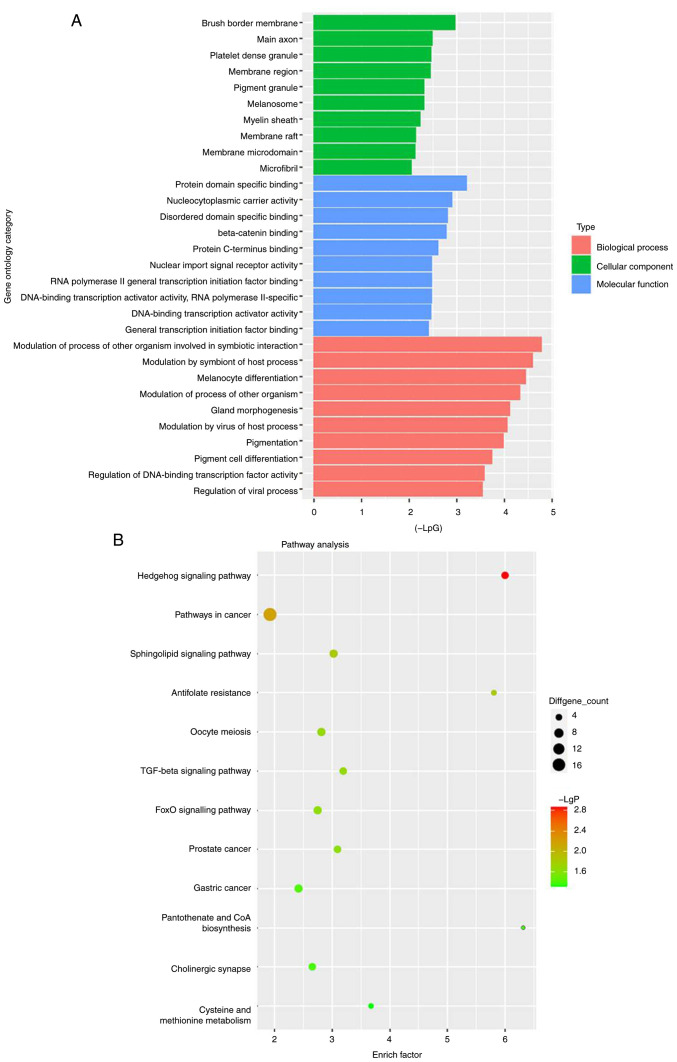
To detect differentially expressed miRNAs in AKI, bioinformatics analysis was performed. As indicated in Fig. 1A and B, differentially expressed miRNAs (miR-1269a, miR-1184, miR-299-5p, miR-411, miR-451 and miR-499) are presented using a volcano plot and heatmap. In addition, the six differentially expressed miRNAs (miR-1269a, miR-1184, miR-299-5p, miR-411, miR-451 and miR-499; miR-1269a and miR-1184 were downregulated, while the other four miRNAs were upregulated) in AKI are presented (Fig. 1C). Moreover, GO and pathway analyses were performed to determine the most common ‘Biological Process’ of miR-1184. The results indicated that miR-1184 was enriched in the following ‘Cellular Component’ terms: ‘Brush border membrane’, ‘main axon’, ‘platelet dense granule’, ‘membrane region’, ‘pigment granule’, ‘melanosome’, ‘myelin sheath’, ‘membrane raft’, ‘membrane microdomain’ and ‘microfibril’ (Fig. 2A). miR-1184 was enriched in the following ‘Molecular Function’ terms: ‘Protein domain specific binding’, ‘nucleocytoplasmic carrier activity’, ‘disordered domain specific binding’, ‘β-catenin binding’, ‘protein C-terminus binding’, ‘nuclear import signal receptor activity’, ‘RNA polymerase II general transcription initiation factor binding’, ‘DNA-binding transcription activator activity’ and ‘RNA polymerase II-specific’ (Fig. 2A). miR-1184 was enriched in the following ‘Biological Process’ terms: ‘Modulation of process of other organism involved in symbiotic interaction’, ‘modulation by symbiont of host process’, ‘melanocyte differentiation’, ‘modulation of process of other organism’, ‘gland morphogenesis’, ‘modulation by virus of host process’, ‘pigmentation’, ‘pigment cell differentiation’, ‘regulation of DNA-binding transcription factor activity’ and ‘regulation of viral process’ (Fig. 2A). Notably, the most enriched ‘Cellular Component’ was ‘brush border membrane’, the most enriched ‘Molecular Function’ was ‘protein domain specific binding’ and the most enriched ‘Biological Process’ was ‘modulation of process of other organism involved in symbiotic interaction’ (Fig. 2A). Pathway analysis revealed that the ‘Hedgehog signaling pathway’, ‘pathways in cancer’, ‘sphingolipid signaling pathway’, ‘antifolate resistance’, ‘oocyte meiosis’, ‘TGF-beta signaling pathway’, ‘FOXO signaling pathway’, ‘prostate cancer’, ‘gastric cancer’, ‘pantothenate and CoA biosynthesis’, ‘cholinergic synapse’ and ‘cysteine and methionine metabolism’ were associated with the development of AKI (Fig. 2B). Based on the aforementioned data, miR-1184 was selected for further analysis.
Figure 1.
Differentially expressed miRNAs in AKI. (A) Volcano plots and (B) heatmap illustrates the miRNAs differentially expressed in AKI. (C) The AKI/Control ratio of miR-1184, miR-1269a, miR-299-5p, miR-411, miR-451 and miR-499-5p was calculated. miR/miRNA, microRNA; AKI, acute kidney injury.
Figure 2.
Profile of miR-1184 in AKI analyzed with GO and pathway analyses. (A) Go analysis exploring the potential functions of differentially expressed miRNAs. (B) Pathway analysis exploring the signaling pathways related to acute kidney injury. miRNA, microRNA; GO, Gene Ontology.
Successful isolation of exosomes from MSCs
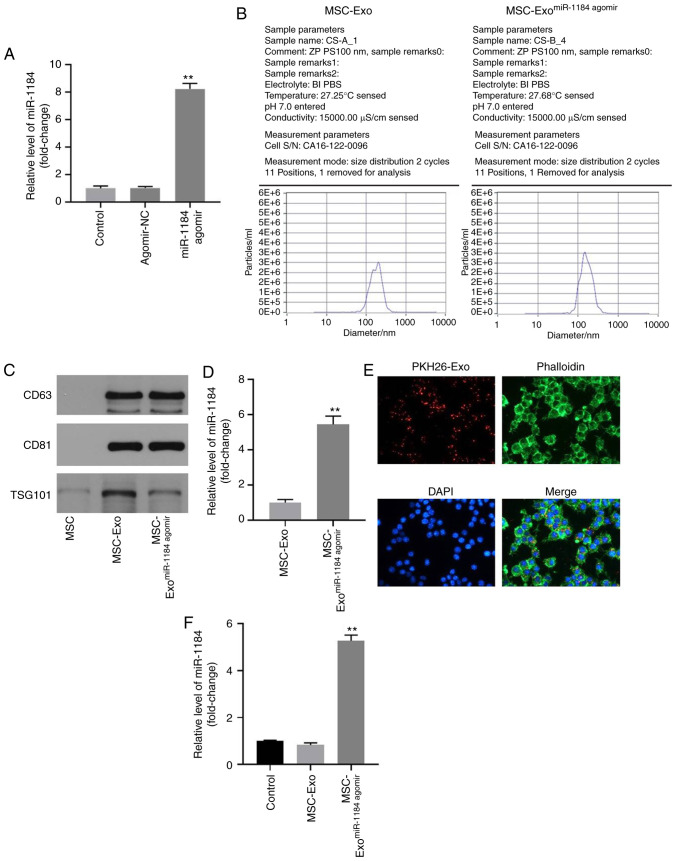
In order to detect the efficiency of cell transfection and exosome isolation, RT-qPCR and NTA was performed. As shown in Fig. 3A, the expression of miR-1184 in MSCs was significantly upregulated by miR-1184 agomir, and NTA revealed a similar size distribution of exosomes (Fig. 3B). Moreover, exosomal proteins (TSG101, CD63 and CD81) were highly expressed in exosomes derived from MSCs, while they were expressed at low levels in MSCs (Fig. 3C). Moreover, the expression of miR-1184 in exosomes derived from MSCs was significantly upregulated by miR-1184 agomir (Fig. 3D), and MSC-derived exosomes labeled with fluorescent PKH26 were internalized by unstained MSCs (Fig. 3E). Furthermore, the expression level of miR-1184 in MSCs was significantly increased by exosomes derived from miR-1184 agomir-treated MSCs (Fig. 3F). Taken together, the data demonstrated that exosomes were successfully isolated from MSCs.
Figure 3.
Exosomes were successfully isolated from MSCs. (A) MSCs were transfected with miR-1184 agomir or agomir-NC. Then, the expression of miR-1184 in MSCs was detected by RT-qPCR. **P<0.01 vs. control. (B) The particle sizes of exosomes were measured by Nanoparticle tracking analysis. (C) The expression levels of CD63, CD81 and TSG101 in MSCs, MSC-Exo or MSC-ExomiR-1184 agomir were detected by western blotting. (D) The expression of miR-1184 in MSC-Exo or MSC-ExomiR-1184 agomir was detected by RT-qPCR. **P<0.01 vs. MSC-Exo. (E) The location of exosomes was observed by immunofluorescence staining. (F) MSCs were co-cultured with MSC-Exo or MSC-ExomiR-1184 agomir. Then, the expression of miR-1184 in MSCs was tested by RT-qPCR. **P<0.01 vs. control. MSCs, mesenchymal stem cells; miR, microRNA; NC, negative control; RT-qPCR, reverse transcription-quantitative PCR; MSC-Exo, exosomes derived from MSCs; MSC-ExomiR-1184 agomir, exosomes derived from miR-1184 agomir-treated MSCs.
Exosomes derived from miR-1184 agomir-treated MSCs significantly reverse cisplatin-induced HK-2 cell growth inhibition
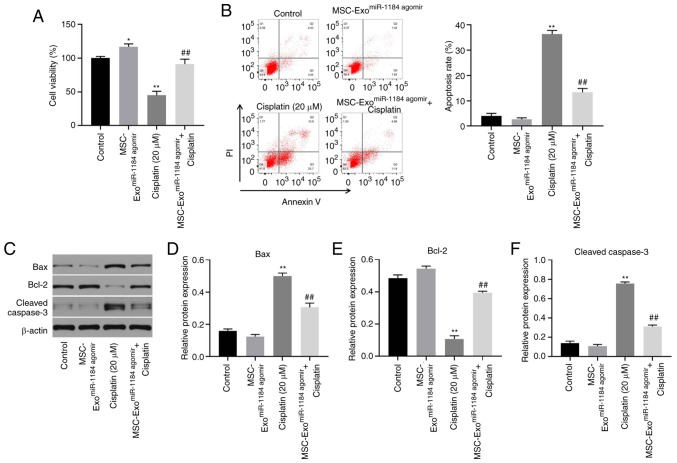
In order to detect the effects of exosomes on AKI in vitro, a CCK-8 assay was used. As illustrated in Fig. 4A, the viability of the HK-2 cells was significantly increased when the cells were co-cultured with exosomes derived from miR-1184 agomir-treated MSCs, and exosomes expressing miR-1184 significantly reversed the cisplatin-induced inhibition of cell viability. In addition, cisplatin significantly induced HK-2 cell apoptosis, while this phenomenon was reversed in the presence of exosomes derived from miR-1184 agomir-treated MSCs (Fig. 4B). Moreover, cisplatin significantly upregulated the expression levels of Bax and cleaved caspase-3 and downregulated the protein expression level of Bcl-2, while this phenomenon was reversed by exosomes derived from miR-1184 agomir-treated MSCs (Fig. 4C-F). Taken together, the results demonstrated that exosomes derived from miR-1184 agomir-treated MSCs significantly reversed cisplatin-induced HK-2 cell growth inhibition.
Figure 4.
MSC-ExomiR-1184 agomir significantly reverses cisplatin-induced HK-2 cell growth inhibition. (A) HK-2 cells were treated with cisplatin, MSC-ExomiR-1184 agomir or cisplatin + MSC-ExomiR-1184 agomir. The viability of HK-2 cells was tested by Cell Counting Kit-8 assay. (B) The apoptosis of HK-2 cells was tested by flow cytometry. (C) The protein levels of Bax, Bcl-2 and cleaved caspase-3 in HK-2 cells were detected by western blotting. (D-F) The relative expression levels were semi-quantified by normalization to β-actin. *P<0.05, **P<0.01 vs. control; ##P<0.01 vs. cisplatin. MSCs, mesenchymal stem cells; miR, microRNA; MSC-ExomiR-1184 agomir, exosomes derived from miR-1184 agomir-treated MSCs.
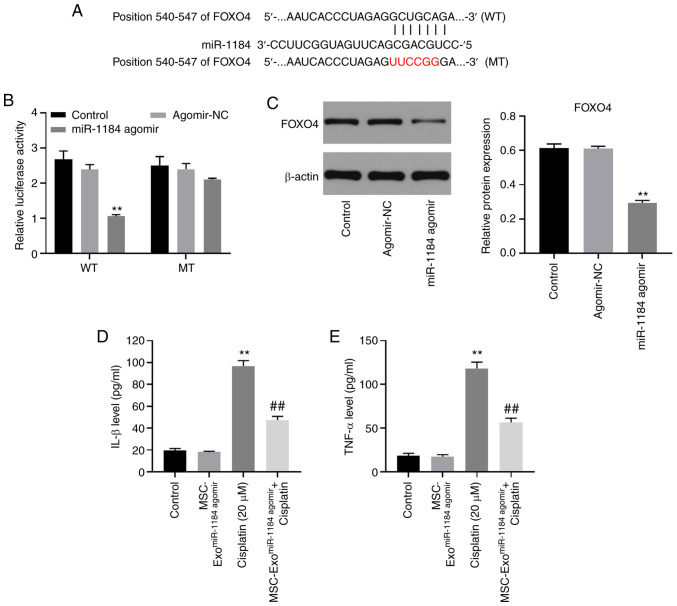
miR-1184 directly targets FOXO4 in HK-2 cells
To explore the potential target of miR-1184, TargetScan and miRDB databases were used. As shown in Fig. 5A, FOXO4 may be the target of miR-1184, and the relative luciferase activity in the WT-FOXO4 group was significantly decreased by miR-1184 agomir (Fig. 5B). Moreover, the expression level of FOXO4 in HK-2 cells was significantly inhibited by the overexpression of miR-1184 (Fig. 5C). Furthermore, the levels of IL-1β and TNF-α in the supernatants of HK-2 cells were significantly upregulated by cisplatin, while this phenomenon was partially reversed by MSC-ExomiR-1184 agomir (Fig. 5D and E). Therefore, miR-1184 directly targeted FOXO4 in HK-2 cells.
Figure 5.
miR-1184 directly targets FOXO4 in HK-2 cells. (A) The target of miR-1184 was predicted by TargetScan and miRDB. (B) The relative luciferase activity was detected by dual-luciferase reporter assay. (C) HK-2 cells were transfected with NC or miR-1184 agomir. The expression of FOXO4 in HK-2 cells was detected by western blotting. The relative expression was semi-quantified by normalization to β-actin. (D) The level of IL-1β in supernatants of HK-2 cells was tested by ELISA. (E) The level of TNF-α in supernatants of HK-2 cells was tested by ELISA. **P<0.01 vs. control; ##P<0.01 vs. cisplatin. miR, microRNA; FOXO4, forkhead box O4; NC, negative control; WT, wild-type; MT, mutant; MSCs, mesenchymal stem cells; MSC-ExomiR-1184 agomir, exosomes derived from miR-1184 agomir-treated MSCs.
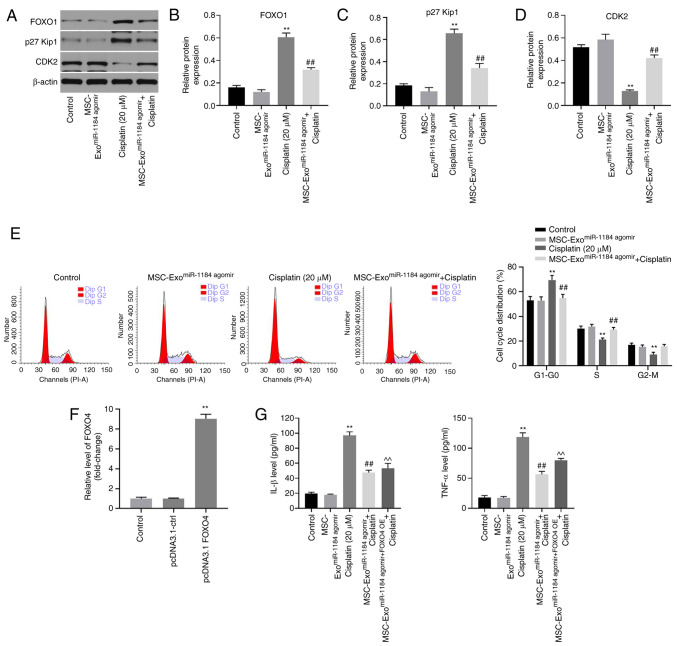
Exosomes derived from miR-1184 agomir-treated MSCs significantly induce G1 arrest in HK-2 cells via the mediation of FOXO1, p27 Kip1 and CDK2
In order to further explore the underlying mechanisms through which exosomes or miR-1184 agomir regulate cisplatin-induced HK-2 cell injury, western blot analysis was performed. As indicated in Figs. 6A-C and S1A-C the protein expression levels of FOXO1 and p27 Kip1 in HK-2 cells were significantly upregulated by cisplatin. By contrast, cisplatin significantly inhibited the expression level of CDK2 (Figs. 6D and S1D). Furthermore, the effect of cisplatin on these three proteins was reversed by MSC-ExomiR-1184 agomir (Figs. 6A-D and S1A-D). In addition, cisplatin-induced G1 arrest was reversed by MSC-ExomiR-1184 agomir (Fig. 6E). The expression of FOXO4 in MSCs was significantly upregulated by transfection with the FOXO4-overexpression vector (Fig. 6F). Meanwhile, the anti-inflammatory effect of exosomes derived from miR-1184 agomir-treated MSCs on cisplatin-treated HK-2 cells was rescued in the presence of exosomes derived from MSCs co-treated with miR-1184 agomir and FOXO4 overexpression (Fig. 6G). Collectively, the results demonstrated that exosomes derived from miR-1184 agomir-treated MSCs significantly induced G1 arrest in HK-2 cells via the mediation of FOXO1, p27 Kip1 and CDK2.
Figure 6.
MSC-ExomiR-1184 agomir significantly induces G1 arrest in HK-2 cells via mediation of FOXO1, p27 Kip1 and CDK2. (A-D) The expression levels of FOXO1, p27 Kip1 and CDK2 in HK-2 cells were detected by western blotting. The relative expression levels were semi-quantified by normalization to β-actin. (E) The cell cycle distribution was tested by flow cytometry. (F) MSCs were transfected with pcDNA3.1-ctrl or pcDNA3.1-FOXO4. The expression of FOXO4 in MSCs was detected via reverse transcription-quantitative PCR. (G) The levels of IL-1β and TNF-α in supernatants of HK-2 cells were tested by ELISA. **P<0.01 vs. control; ##P<0.01 vs. cisplatin; ^^P<0.01 vs. MSC-ExomiR-1184 agomir + cisplatin. MSCs, mesenchymal stem cells; MSC-ExomiR-1184 agomir, exosomes derived from miR-1184 agomir-treated MSCs; FOXO1, forkhead box O1; FOXO4, forkhead box O4; OE, overexpression.
Discussion
It has been reported that miRNAs are involved in the progression of AKI (19,31,32). In the present study, it was found that miR-1184 was downregulated in cisplatin-treated HK-2 cells, and that exosomes derived from miR-1184 agomir-treated HK-2 cells significantly reversed cisplatin-induced HK-2 cell injury. Thus, the present study firstly explored the function of miR-1184 in AKI, suggesting that miR-1184 may function as an inhibitor in AKI. Moreover, miR-1184 is known to be involved in other diseases. For example, Wang et al (33) indicated that hsa_circ_0128846 promoted the tumorigenesis of colorectal cancer by binding to miR-1184 and releasing ajuba LIM protein, and inactivating Hippo/Yes1 associated transcriptional regulator signaling. Chen et al (34) found that miR-1184 mediated the proliferation of colon cancer cells by targeting casein kinase 2 α 1. Thus, other functions of miR-1184 (for example its role in renal cancer) also need to be explored in the future.
It has been reported that exosomes derived from MSCs play important roles in the progression of AKI. For example, Alzahrani (35) found that exosomes derived from MSCs could promote the progression of AKI; Cao et al (14) indicated that exosomal miR-125b-5p derived from MSCs could promote tubular repair by suppression of p53 in ischemic AKI. In the present study, it was found that exosomal miR-1184 derived from MSCs could inhibit cisplatin-induced HK-2 cell injury. Thus, this research first explored the relationship between miR-1184 and exosomes derived from MSCs.
It has been confirmed that miRNAs exert their biological functions due to their target genes (36). In the present study, the results of the dual-luciferase reporter assay indicated that FOXO4 was a downstream target of miR-1184 in AKI. FOXO4 is a member of the FOXO family, and is a crucial mediator of cell proliferation (37). Consistently, the data of the present study revealed that miR-1184 may function as a mediator in AKI by targeting FOXO4. On the other hand, it has been reported that p27 Kip1 is a cell cycle regulator, firstly regarded as a cyclin-dependent kinase antagonist (38). It has been reported that p27 Kip1 expression is upregulated during the progression of AKI (39,40). Taken together with the findings of the present study, it can be concluded that p27 Kip1 is a negative mediator in AKI. Additionally, CDK2 is a mediator of cell cycle distribution and is a downstream protein of p27 Kip1 (41). In the present study, exosomes derived from miR-1184 agomir-treated HK-2 cells reversed cisplatin-induced cell growth inhibition via the mediation of FOXO1, p27 Kip1 and CDK2. Thus, these findings are consistent with those of previous studies. Taken together, the findings presented herein revealed that miR-1184 exerted an inhibitory effect on AKI by targeting FOXO1.
There are some limitations of the present study, which are as follows: i) Other mRNAs targeted by miR-1184 in AKI need to be explored in the future; ii) rescue experiments should be performed in order to further verify the function of exosomes in AKI; iii) electron microscopy analysis is needed to further identify the exosomes; iv) the function of miR-1184 antagomir should be further confirmed; and v) the function of miR-184 in renal cancer needs to be explored. Therefore, further investigations are required in the future.
In conclusion, the present study demonstrated that exosomal-miR-1184 derived from MSCs alleviated cisplatin-associated AKI. Thus, these findings may lead to the development of novel strategies for the treatment of AKI.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
This research was supported by the Construction of Key Projects by Zhejiang Provincial Ministry (grant nos. WKJ-ZJ-1915 and WKJ-ZJ-2017), the Zhejiang Province Chinese Medicine Modernization Program (grant no. 2020ZX001) and the General Project of Zhejiang Education Department (grant no. Y201942823).
Funding
This research was supported by the Construction of Key Projects by Zhejiang Provincial Ministry (grant nos. WKJ-ZJ-1915 and WKJ-ZJ-2017), the Zhejiang Province Chinese Medicine Modernization Program (grant no. 2020ZX001) and the General Project of Zhejiang Education Department (grant no. Y201942823).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
MT and JJ conceived and supervised the present study. JZ and WH designed the experiments and reviewed the results. DZ and QH performed the experiments. All authors have read and approved the final manuscript. MT and JJ confirm the authenticity of all the raw data.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Stephen Inbaraj B, Chen BH. An overview on recent in vivo biological application of cerium oxide nanoparticles. Asian J Pharm Sci. 2020;15:558–575. doi: 10.1016/j.ajps.2019.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang J, Healy HG, Baboolal K, Wang Z, Venuthurupalli SK, Tan KS, Cameron A, Hoy WE, CKD.QLD Collaborative Frequency and consequences of acute kidney injury in patients with CKD: A registry study in queensland australia. Kidney Med. 2019;1:180–190. doi: 10.1016/j.xkme.2019.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guillemet L, Jamme M, Bougouin W, Geri G, Deye N, Vivien B, Varenne O, Pène F, Mira JP, Barat F, et al. Effects of early high-dose erythropoietin on acute kidney injury following cardiac arrest: Exploratory post hoc analyses from an open-label randomized trial. Clin Kidney J. 2020;13:413–420. doi: 10.1093/ckj/sfz068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Güzel C, Yeşiltaş S, Daşkaya H, Uysal H, Sümer I, Türkay M. The effect of gender on acute kidney injury developing in the intensive care unit. Hippokratia. 2019;23:126–130. [PMC free article] [PubMed] [Google Scholar]
- 5.Xiong J, Zhang M, Guo X, Pu L, Xiong H, Xiang P, Liu J, Li A. Acute kidney injury in critically ill cirrhotic patients with spontaneous bacterial peritonitis: A comparison of KDIGO and ICA criteria. Arch Med Sci. 2020;16:569–576. doi: 10.5114/aoms.2019.85148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deng J, Tan W, Luo Q, Lin L, Zheng L, Yang J. Long non-coding RNA MEG3 promotes renal tubular epithelial cell pyroptosis by regulating the miR-18a-3p/GSDMD pathway in lipopolysaccharide-induced acute kidney injury. Front Physiol. 2021;12:663216. doi: 10.3389/fphys.2021.663216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han D, Fang R, Shi R, Jin Y, Wang Q. LncRNA NKILA knockdown promotes cell viability and represses cell apoptosis, autophagy and inflammation in lipopolysaccharide-induced sepsis model by regulating miR-140-5p/CLDN2 axis. Biochem Biophys Res Commun. 2021;559:8–14. doi: 10.1016/j.bbrc.2021.04.074. [DOI] [PubMed] [Google Scholar]
- 8.Yang J, Wu L, Liu S, Hu X, Wang Q, Fang L. Long non-coding RNA NEAT1 promotes lipopolysaccharide-induced injury in human tubule epithelial cells by regulating miR-93-5p/TXNIP axis. Med Microbiol Immunol. 2021;210:121–132. doi: 10.1007/s00430-021-00705-6. [DOI] [PubMed] [Google Scholar]
- 9.Xu Y, Qin S, Niu Y, Gong T, Zhang Z, Fu Y. Effect of fluid shear stress on the internalization of kidney-targeted delivery systems in renal tubular epithelial cells. Acta Pharm Sin B. 2020;10:680–692. doi: 10.1016/j.apsb.2019.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guthrie GD, Bell S. Deprivation and kidney disease-a predictor of poor outcomes. Clin Kidney J. 2019;13:128–132. doi: 10.1093/ckj/sfz151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jia X, Wei L, Zhang Z. NEAT1 Overexpression indicates a poor prognosis and induces chemotherapy resistance via the miR-491-5p/SOX3 signaling pathway in ovarian cancer. Front Genet. 2021;12:616220. doi: 10.3389/fgene.2021.616220. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Zhang F, Sang Y, Chen D, Wu X, Wang X, Yang W, Chen Y. M2 macrophage-derived exosomal long non-coding RNA AGAP2-AS1 enhances radiotherapy immunity in lung cancer by reducing microRNA-296 and elevating NOTCH2. Cell Death Dis. 2021;12:467. doi: 10.1038/s41419-021-03700-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Q, Tao Y, Xie H, Liu C, Liu P. MicroRNA-101 inhibits renal tubular epithelial-to-mesenchymal transition by targeting TGF-β1 type I receptor. Int J Mol Med. 2021;47:119. doi: 10.3892/ijmm.2021.4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao JY, Wang B, Tang TT, Wen Y, Li ZL, Feng ST, Wu M, Liu D, Yin D, Ma KL, et al. Exosomal miR-125b-5p deriving from mesenchymal stem cells promotes tubular repair by suppression of p53 in ischemic acute kidney injury. Theranostics. 2021;11:5248–5266. doi: 10.7150/thno.54550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scullion KM, Vliegenthart ADB, Rivoli L, Oosthuyzen W, Farrah TE, Czopek A, Webb DJ, Hunter RW, Bailey MA, Dhaun N, Dear JW. Circulating argonaute-bound microRNA-126 reports vascular dysfunction and treatment response in acute and chronic kidney disease. iScience. 2020;24:101937. doi: 10.1016/j.isci.2020.101937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun B, Qu Z, Cheng GL, Yang YW, Miao YF, Chen XG, Zhou XB, Li B. Urinary microRNAs miR-15b and miR-30a as novel noninvasive biomarkers for gentamicin-induced acute kidney injury. Toxicol Lett. 2021;338:105–113. doi: 10.1016/j.toxlet.2020.12.006. [DOI] [PubMed] [Google Scholar]
- 17.Jeon BS, Lee SH, Hwang SR, Yi H, Bang JH, Tham NTT, Lee HK, Woo GH, Kang HG, Ku HO. Identification of urinary microRNA biomarkers for in vivo gentamicin-induced nephrotoxicity models. J Vet Sci. 2020;21:e81. doi: 10.4142/jvs.2020.21.e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pan T, Jia P, Chen N, Fang Y, Liang Y, Guo M, Ding X. Delayed remote ischemic preconditioning confersrenoprotection against septic acute kidney injury via exosomal miR-21. Theranostics. 2019;9:405–423. doi: 10.7150/thno.29832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang L, Liu XQ, Ma Q, Yang Q, Gao L, Li HD, Wang JN, Wei B, Wen J, Li J, et al. hsa-miR-500a-3P alleviates kidney injury by targeting MLKL-mediated necroptosis in renal epithelial cells. FASEB J. 2019;33:3523–3535. doi: 10.1096/fj.201801711R. [DOI] [PubMed] [Google Scholar]
- 20.Zhang X, Wang N, Huang Y, Li Y, Li G, Lin Y, Atala AJ, Hou J, Zhao W. Extracellular vesicles from three dimensional culture of human placental mesenchymal stem cells ameliorated renal ischemia/reperfusion injury. Int J Artif Organs. 2021 Jan 19; doi: 10.1177/0391398820986809. (Epub ahead of print). doi: 10.1177/0391398820986809. [DOI] [PubMed] [Google Scholar]
- 21.Jiang Z, Hou Z, Li L, Liu W, Yu Z, Chen S. Exosomal circEPB41L2 serves as a sponge for miR-21-5p and miR-942-5p to suppress colorectal cancer progression by regulating the PTEN/AKT signalling pathway. Eur J Clin Invest. 2021;51:e13581. doi: 10.1111/eci.13581. [DOI] [PubMed] [Google Scholar]
- 22.Pineles B, Mani A, Sura L, Rossignol C, Albayram M, Weiss MD, Goetzl L. Neuronal exosome proteins: Novel biomarkers for predicting neonatal response to therapeutic hypothermia. Arch Dis Child Fetal Neonatal Ed. 2021 May 21; doi: 10.1136/archdischild-2020-321096. (Epub ahead of print). doi: 10.1136/archdischild-2020-321096. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y, Huang H, Liu W, Liu S, Wang XY, Diao ZL, Zhang AH, Guo W, Han X, Dong X, Katilov O. Endothelial progenitor cells-derived exosomal microRNA-21-5p alleviates sepsis-induced acute kidney injury by inhibiting RUNX1 expression. Cell Death Dis. 2021;12:335. doi: 10.1038/s41419-021-03578-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang Y, He Y, Makarcyzk MJ, Lin H. Senolytic peptide FOXO4-DRI selectively removes senescent cells from in vitro expanded human chondrocytes. Front Bioeng Biotechnol. 2021;9:677576. doi: 10.3389/fbioe.2021.677576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ji C, Zhang J, Zhou Z, Shi H, Liu W, Sun F, Zhang C, Zhang L, Sun Z, Qian H. Platelet-rich plasma promotes MSCs exosomes paracrine to repair acute kidney injury via AKT/Rab27 pathway. Am J Transl Res. 2021;13:1445–1457. [PMC free article] [PubMed] [Google Scholar]
- 26.Cao J, Wang B, Tang T, Lv L, Ding Z, Li Z, Hu R, Wei Q, Shen A, Fu Y, Liu B. Three-dimensional culture of MSCs produces exosomes with improved yield and enhanced therapeutic efficacy for cisplatin-induced acute kidney injury. Stem Cell Res Ther. 2020;11:206. doi: 10.1186/s13287-020-01719-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McSweeney KR, Gadanec LK, Qaradakhi T, Ali BA, Zulli A, Apostolopoulos V. Mechanisms of cisplatin-induced acute kidney injury: Pathological mechanisms, pharmacological interventions, and genetic mitigations. Cancers (Basel) 2021;13:1572. doi: 10.3390/cancers13071572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng Q, Wang L. LncRNA XIST serves as a ceRNA to regulate the expression of ASF1A, BRWD1M, and PFKFB2 in kidney transplant acute kidney injury via sponging hsa-miR-212-3p and hsa-miR-122-5p. Cell Cycle. 2020;19:290–299. doi: 10.1080/15384101.2019.1707454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Varet H, Brillet-Guéguen L, Coppée JY, Dillies MA. SARTools: A DESeq2- and EdgeR-based R pipeline for comprehensive differential analysis of RNA-Seq data. PLoS One. 2016;11:e0157022. doi: 10.1371/journal.pone.0157022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 31.Ren GL, Zhu J, Li J, Meng XM. Noncoding RNAs in acute kidney injury. J Cell Physiol. 2019;234:2266–2276. doi: 10.1002/jcp.27203. [DOI] [PubMed] [Google Scholar]
- 32.Zhang W, Zhou X, Zhang H, Yao Q, Liu Y, Dong Z. Extracellular vesicles in diagnosis and therapy of kidney diseases. Am J Physiol Renal Physiol. 2016;311:F844–F851. doi: 10.1152/ajprenal.00429.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X, Chen Y, Liu W, Liu T, Sun D. Hsa_circ_0128846 promotes tumorigenesis of colorectal cancer by sponging hsa-miR-1184 and releasing AJUBA and inactivating Hippo/YAP signalling. J Cell Mol Med. 2020;24:9908–9924. doi: 10.1111/jcmm.15590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen S, Wang Y, Xu M, Zhang L, Su Y, Wang B, Zhang X. miR-1184 regulates the proliferation and apoptosis of colon cancer cells via targeting CSNK2A1. Mol Cell Probes. 2020;53:101625. doi: 10.1016/j.mcp.2020.101625. [DOI] [PubMed] [Google Scholar]
- 35.Alzahrani FA. Melatonin improves therapeutic potential of mesenchymal stem cells-derived exosomes against renal ischemia-reperfusion injury in rats. Am J Transl Res. 2019;11:2887–2907. [PMC free article] [PubMed] [Google Scholar]
- 36.Shin JH, Shin DH, Kim JS. Let-7 miRNA and CDK4 siRNA co-encapsulated in Herceptin-conjugated liposome for breast cancer stem cells. Asian J Pharm Sci. 2020;15:472–481. doi: 10.1016/j.ajps.2019.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang D, Liu K, Hu W, Lu X, Li L, Zhang Q, Huang H, Wang H. Prenatal dexamethasone exposure caused fetal rats liver dysplasia by inhibiting autophagy-mediated cell proliferation. Toxicology. 2021;449:152664. doi: 10.1016/j.tox.2020.152664. [DOI] [PubMed] [Google Scholar]
- 38.Henriques J, Lindorff-Larsen K. Protein dynamics enables phosphorylation of buried residues in Cdk2/cyclin-A-bound p27. Biophys J. 2020;119:2010–2018. doi: 10.1016/j.bpj.2020.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park JW, Cho JW, Joo SY, Kim CS, Choi JS, Bae EH, Ma SK, Kim SH, Lee J, Kim SW. Paricalcitol prevents cisplatin-induced renal injury by suppressing apoptosis and proliferation. Eur J Pharmacol. 2012;683:301–309. doi: 10.1016/j.ejphar.2012.03.019. [DOI] [PubMed] [Google Scholar]
- 40.Coombes JD, Mreich E, Liddle C, Rangan GK. Rapamycin worsens renal function and intratubular cast formation in protein overload nephropathy. Kidney Int. 2005;68:2599–2607. doi: 10.1111/j.1523-1755.2005.00732.x. [DOI] [PubMed] [Google Scholar]
- 41.Guerra B, Dembic M, Siddiqui MA, Dominguez I, Ceppi P, Andresen BS. Down-regulation of CK2α leads to up-regulation of the cyclin-dependent kinase inhibitor p27KIP1 in conditions unfavorable for the growth of myoblast cells. Cell Physiol Biochem. 2020;54:1177–1198. doi: 10.33594/000000308. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article.